CH25H has received significant attention due to its broad antiviral activity, which it mediates by catalyzing the production of 25HC. Most studies have focused on the antiviral mechanisms of CH25H; however, whether viruses also actively regulate CH25H expression has not yet been reported. Previous studies demonstrated that pCH25H inhibits PRRSV replication not only via production of 25HC but also by ubiquitination and degradation of viral nonstructural protein 1α. In this study, we expanded on previous work and found that PRRSV actively degrades pCH25H through the ubiquitin-proteasome pathway. PRRSV E protein, a viral structural protein, is involved in this process. This study reveals a novel mechanism of interaction between virus and host during PRRSV infection.
KEYWORDS: E protein, porcine reproductive and respiratory syndrome virus, PRRSV, cholesterol 25-hydroxylase, CH25H, ubiquitin-proteasome pathway
ABSTRACT
Porcine reproductive and respiratory syndrome is one of the most important infectious diseases affecting the global pig industry. Previous studies from our group and other groups showed that cholesterol 25-hydroxylase (CH25H), a multitransmembrane endoplasmic reticulum-associated enzyme, catalyzes the production of 25-hydroxycholesterol (25HC) and inhibits porcine reproductive and respiratory syndrome virus (PRRSV) replication. However, PRRSV infection also actively decreases porcine CH25H (pCH25H) expression, through unidentified mechanisms. In this study, we found that the ubiquitin-proteasome pathway plays a major role in pCH25H degradation during PRRSV infection and that the PRRSV-encoded envelope (E) protein interacts with pCH25H. PRRSV E protein degraded pCH25H via ubiquitination, and the ubiquitination site was at pCH25H Lys28. Interestingly, PRRSV E protein appeared to specifically degrade pCH25H but not human CH25H, likely because of a Lys28Arg substitution in the human orthologue. As expected, ubiquitin-mediated degradation by E protein attenuated the antiviral effect of pCH25H by downregulating 25HC production. In addition, we found that knockdown of pCH25H decreased E protein-induced inflammatory cytokine expression and that pCH25H overexpression had the opposite effect. These findings suggested that regulation of pCH25H expression was associated with E protein-induced inflammatory responses. Taken together, our results and those of previous studies of the anti-PRRSV effects of CH25H highlight the complex interplay between PRRSV and pCH25H.
IMPORTANCE CH25H has received significant attention due to its broad antiviral activity, which it mediates by catalyzing the production of 25HC. Most studies have focused on the antiviral mechanisms of CH25H; however, whether viruses also actively regulate CH25H expression has not yet been reported. Previous studies demonstrated that pCH25H inhibits PRRSV replication not only via production of 25HC but also by ubiquitination and degradation of viral nonstructural protein 1α. In this study, we expanded on previous work and found that PRRSV actively degrades pCH25H through the ubiquitin-proteasome pathway. PRRSV E protein, a viral structural protein, is involved in this process. This study reveals a novel mechanism of interaction between virus and host during PRRSV infection.
INTRODUCTION
Since it was first described in the 1980s, porcine reproductive and respiratory syndrome virus (PRRSV) has continuously threatened the global pig industry with serious economic losses (1–3). PRRSV has a single-stranded, positive-sense RNA genome approximately 15 kb in length and is a member of the family Arteriviridae in the order Nidovirales (4, 5). During PRRSV infection, the virus produces two polyproteins, polyprotein 1a (pp1a) and pp1ab, which are subsequently cleaved by its papain-like protease (nonstructural protein 2 [nsp2]) and 3C-like protease (nsp4) into 15 nonstructural proteins (6–8). Eight structural proteins are encoded by the other eight open reading frames (ORFs) (9–12), including glycoprotein 2 (GP2), envelope (E) protein, GP3, GP4, GP5, ORF5a protein, matrix protein, and nucleocapsid (N) protein.
As an economically important disease, porcine reproductive and respiratory syndrome has received significant attention, and many commercial vaccines against PRRSV have been tested and marketed. Unfortunately, none of the commercially available vaccines provides durable control of porcine reproductive and respiratory syndrome (13–19). Thus, mining the host restriction factors for PRRSV has attracted significant interest among PRRSV researchers. Several host restriction factors, such as sterile α-motif histidine-aspartate-domain-containing protein 1 (SAMHD1) (20), virus inhibitory protein, endoplasmic reticulum (ER)-associated, interferon-inducible (viperin) (21), DEAD box helicase 3X (DDX3X) (22), ubiquitin-like protease 18 (USP18) (23), and E74-like factor 4 (ELF4) (24), have been reported to inhibit PRRSV proliferation. More recently, three groups have independently confirmed that cholesterol 25-hydroxylase (CH25H) and its metabolic product, 25-hydroxycholesterol (25HC), have significant anti-PRRSV activity (25–27). CH25H is an ∼31.6-kDa ER-resident hydroxylase whose main function is to catalyze production of 25HC from cholesterol (28). Accumulating evidence suggests that CH25H and 25HC have broad-spectrum antiviral activity and inhibit the proliferation of many viruses, including human immunodeficiency virus (HIV) (29), hepatitis C virus (HCV) (30, 31), and Zika virus (ZIKV) (32). 25HC exerts its antiviral effects through multiple mechanisms. For example, 25HC modifies the cellular membrane to impede HIV entry (33), and treatment with 25HC alters reovirus particle trafficking to late endosomes, delaying viral uncoating (34). In addition, CH25H and 25HC directly interact with some virus-encoded proteins to inhibit viral replication. For example, CH25H interacts with HCV NS5A protein, inhibiting dimer formation (35), and decreases Lassa virus (LASV) G1 glycoprotein N-glycan maturation, reducing the production of infectious LASV virions (36). In the case of PRRSV, 25HC suppresses PRRSV infection by inhibiting viral penetration (25–27). Furthermore, CH25H can degrade the PRRSV nsp1α through the ubiquitin-proteasome pathway. Intriguingly, PRRSV infection downregulates CH25H expression. More recently, Dong et al. reported that two PRRSV-encoded nonstructural proteins, nsp1β and nsp11, could degrade porcine CH25H (pCH25H) via the lysosomal pathway (37). However, no direct interactions between nsp1β or nsp11 and pCH25H were identified, and whether pCH25H can be degraded through the lysosomal pathway during PRRSV infection has not been fully investigated. Thus, further studies are required to elucidate the mechanisms employed by PRRSV to degrade pCH25H.
In this study, we show that the ubiquitin-proteasome pathway plays a major role in the degradation of pCH25H during PRRSV infection. We also demonstrate that PRRSV-encoded E protein directly interacts with pCH25H, resulting in pCH25H degradation through the ubiquitin-proteasome pathway. Our results reveal a previously unrecognized mechanism utilized by PRRSV to counteract the antiviral action of pCH25H.
RESULTS
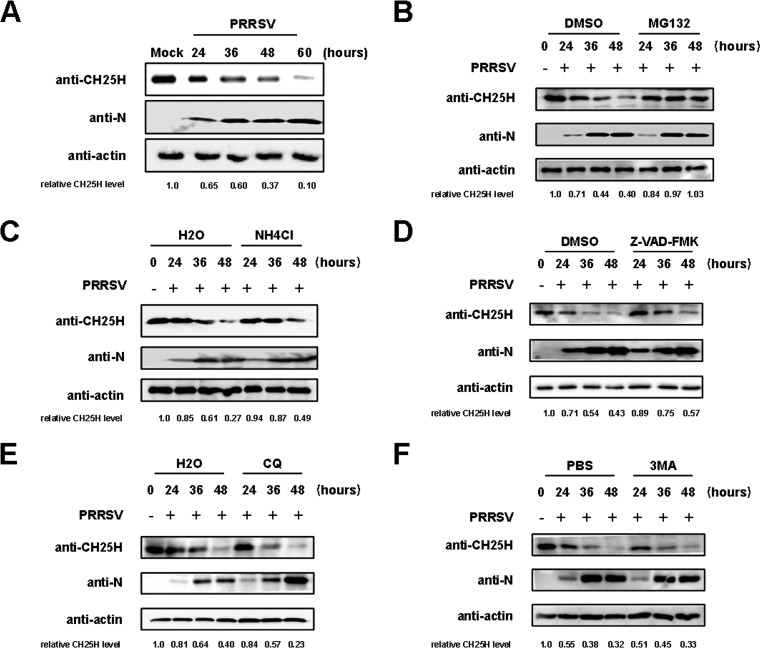
pCH25H is degraded mainly through the ubiquitin-proteasome pathway during PRRSV infection.
In our previous study, we analyzed pCH25H expression after infection with different doses of PRRSV and demonstrated that PRRSV infection resulted in downregulation of pCH25H expression in a dose-dependent manner (27). To further investigate the expression dynamics of pCH25H after PRRSV infection, primary porcine alveolar macrophages (PAMs), the target cells of PRRSV in vivo, were infected with PRRSV strain WUH3 at a multiplicity of infection (MOI) of 1, and the expression of pCH25H was analyzed by Western blotting at different time points (0, 24, 36, 48, and 60 h) postinfection. As shown in Fig. 1A, PRRSV infection resulted in pCH25H downregulation, and this downregulation became more obvious with the progress of virus infection.
FIG 1.
The ubiquitin-proteasome pathway plays a major role in the degradation of pCH25H during PRRSV infection. (A) PAMs were infected with PRRSV (MOI of 1.0). At various time points (24, 36, 48, and 60 h) postinfection, cells were harvested and pCH25H expression was analyzed by Western blotting. (B to F) PAMs were infected with PRRSV (MOI of 1.0) and treated with various inhibitors, including MG132 (10 μM) (B), NH4Cl (10 mM) (C), Z-VAD-FMK (10 μM) (D), CQ (20 μM) (E), or 3-MA (5 mM) (F). At 24, 36, and 48 h postinfection, cells were harvested and pCH25H expression was analyzed by Western blotting. ImageJ software was used to analyze the relative levels of pCH25H in comparison with mock-infected cells, and the ratios are displayed as fold changes below the images. DMSO, dimethyl sulfoxide.
Although a recent study suggested that PRRSV nsp1β and nsp11 can degrade pCH25H via the lysosomal pathway (37), whether pCH25H is degraded via the lysosomal pathway during PRRSV infection remains unknown. The ubiquitin-proteasome system, the autophagy-lysosomal pathway, and apoptosis are three major intracellular protein degradation pathways in eukaryotic cells. In order to explore which pathway is responsible for the degradation of pCH25H during PRRSV infection, the proteasome inhibitor MG132 (10 μM), the autophagy inhibitor 3-methyladenine (3-MA) (5 mM), the lysosomal inhibitors NH4Cl (10 mM) and chloroquine (CQ) (20 μM), and the apoptosis inhibitor Z-VAD-FMK [carbobenzoxy-valyl-alanyl-aspartyl-(O-methyl)-fluoromethylketone] (10 μM) were added to cells infected with PRRSV, and pCH25H expression was analyzed by Western blotting. As shown in Fig. 1B to F, MG132 treatment almost completely restored the expression of pCH25H during PRRSV infection, while treatment with 3-MA or CQ had no effect on expression. Although treatment with NH4Cl or Z-VAD-FMK partially restored its expression, degradation of pCH25H was readily apparent as the infection progressed. These results suggested that the ubiquitin-proteasome pathway plays a major role in the degradation of pCH25H during PRRSV infection, and the lysosomal pathway and the apoptotic pathway may be involved in the pCH25H degradation.
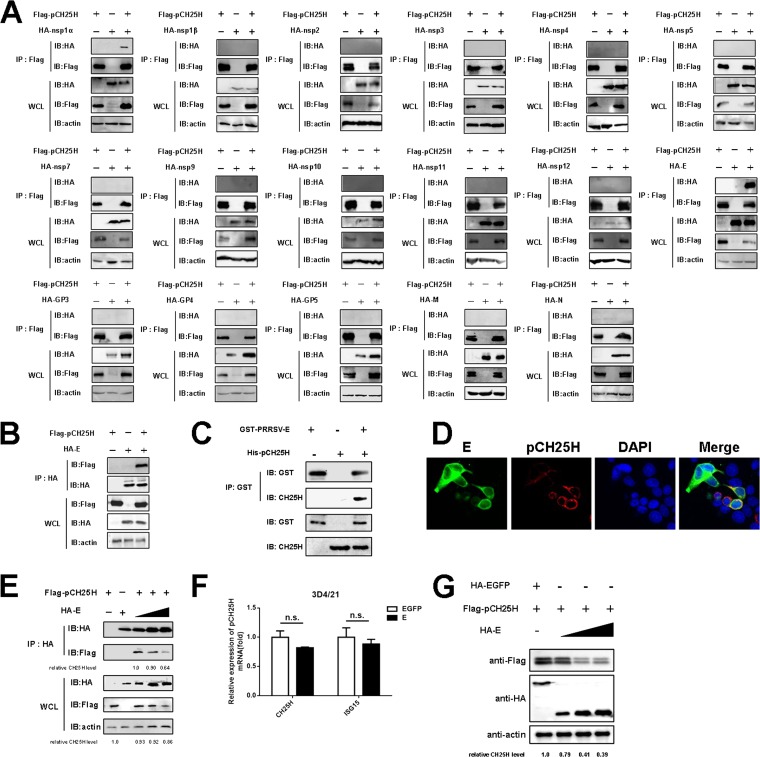
PRRSV E protein interacts with pCH25H and downregulates its expression.
Previously, we identified an interaction between PRRSV nsp1α and pCH25H through yeast two-hybrid screening and a coimmunoprecipitation (Co-IP) assay, but we did not find pCH25H degradation by nsp1α (27). In light of the low expression levels of PRRSV structural proteins in yeast, we screened for interactions between viral nonstructural and structural proteins and pCH25H through Co-IP assays in HEK-293T cells, by cotransfecting the cells with pCAGGS-Flag-pCH25H and expression vectors encoding hemagglutinin (HA)-tagged PRRSV proteins (except for nsp6 and nsp8, because they are very short peptides). We found that, in addition to nsp1α, the PRRSV minor structural E protein interacted with pCH25H (Fig. 2A). In a reverse Co-IP experiment, pCH25H was efficiently coimmunoprecipitated with E protein by an anti-HA antibody (Fig. 2B). We also investigated whether the direct interaction between PRRSV E protein and pCH25H could occur in vitro. To this end, His-pCH25H and glutathione S-transferase (GST)-E proteins were expressed in Escherichia coli and the recombinant proteins were purified for GST pulldown experiments. As shown in Fig. 2C, PRRSV E protein and pCH25H interacted directly. Furthermore, we assessed whether pCH25H and E protein colocalized inside cells in an indirect immunofluorescence assay. The results showed that pCH25H and E protein colocalized in the cytoplasm in cotransfected cells (Fig. 2D). From the results of Co-IP and indirect immunofluorescence assays, we found that the expression levels of pCH25H appeared to be significantly attenuated in cells expressing PRRSV E protein. To confirm these observations, a pCH25H expression construct was cotransfected with different doses of an expression vector encoding PRRSV E protein. At 48 h after cotransfection, cells were collected for Co-IP experiments. As shown in Fig. 2E, with increased PRRSV E protein expression, less pCH25H could be immunoprecipitated with E protein by an anti-HA antibody, indicating that the interaction between pCH25H and PRRSV E protein matches the degradation of pCH25H.
FIG 2.
PRRSV E protein interacts with pCH25H and downregulates its expression. (A) HEK-293T cells were cotransfected with expression vectors encoding FLAG-tagged pCH25H and HA-tagged PRRSV proteins. The cells were lysed at 48 h after transfection and immunoprecipitated with anti-FLAG antibody. Whole-cell lysate (WCL) and immunoprecipitation (IP) complexes were analyzed by immunoblotting (IB) with anti-FLAG, anti-HA, or anti-β-actin antibodies. (B) HEK-293T cells were cotransfected with expression vectors encoding HA-tagged E protein and FLAG-tagged pCH25H. The cells were lysed at 48 h after transfection and immunoprecipitated with an anti-HA antibody. Whole-cell lysate and immunoprecipitation complexes were analyzed by immunoblotting with anti-FLAG, anti-HA, or anti-β-actin antibodies. (C) The purified GST-E protein was incubated with glutathione-Sepharose 4B beads, and then the recombinant protein His-pCH25H was added. After being washed three times, the beads were collected for Western blotting with antibodies against GST or CH25H. (D) HEK-293T cells were cotransfected with FLAG-tagged pCH25H and HA-tagged E protein expression vectors. At 48 h posttransfection, cells were fixed for immunofluorescence assays for pCH25H and E proteins using primary antibodies (rabbit anti-HA and mouse anti-FLAG) followed by secondary antibodies (AF594-conjugated donkey anti-mouse IgG and AF488-conjugated donkey anti-rabbit IgG). Nuclei were counterstained with DAPI. Fluorescence images were acquired with a confocal laser scanning microscope (Olympus FluoView v3.1). (E) HEK-293T cells were cotransfected with expression vectors encoding FLAG-tagged pCH25H and different doses of HA-tagged PRRSV E protein. The cells were lysed at 48 h after transfection and immunoprecipitated with an anti-HA antibody. (F) 3D4/21 cells were cotransfected with expression vectors encoding FLAG-tagged pCH25H and HA-tagged PRRSV E protein. At 48 h posttransfection, cells were harvested for quantitation of pCH25H mRNA by qRT-PCR. Experiments were performed in triplicate, and data are representative of three independent experiments. The standard deviations are indicated by error bars. (G) 3D4/21 cells were cotransfected with an expression vector encoding FLAG-tagged pCH25H and increasing amounts of an expression vector encoding HA-tagged E protein. At 48 h posttransfection, expression of FLAG-pCH25H and HA-E protein was analyzed by Western blotting. The relative levels of pCH25H in comparison with HA-EGFP-transfected cells were analyzed by ImageJ software, and the ratios are displayed as fold changes below the images. n.s., not significant.
To further investigate whether PRRSV E protein mediated downregulation of pCH25H in 3D4/21 cells, a PAM-derived cell line, we cotransfected pCAGGS-FLAG-pCH25H and pCAGGS-HA-E protein or pCAGGS-HA-enhanced green fluorescent protein (EGFP), and changes in pCH25H mRNA abundance were monitored by quantitative reverse transcription-PCR (qRT-PCR). Interferon-stimulated gene 15 (ISG15) was included as a control. As shown in Fig. 2F, E protein did not significantly affect the abundance of mRNA transcripts encoding pCH25H. In addition, we cotransfected 3D4/21 cells with expression vectors encoding pCH25H and different doses of pCAGGS-HA-E protein and then assessed the protein expression of pCH25H. E protein downregulated pCH25H expression at the protein level in a dose-dependent manner (Fig. 2G). These results suggested that PRRSV E protein mediated downregulation of pCH25H expression not at the transcript level but through a protein degradation pathway.
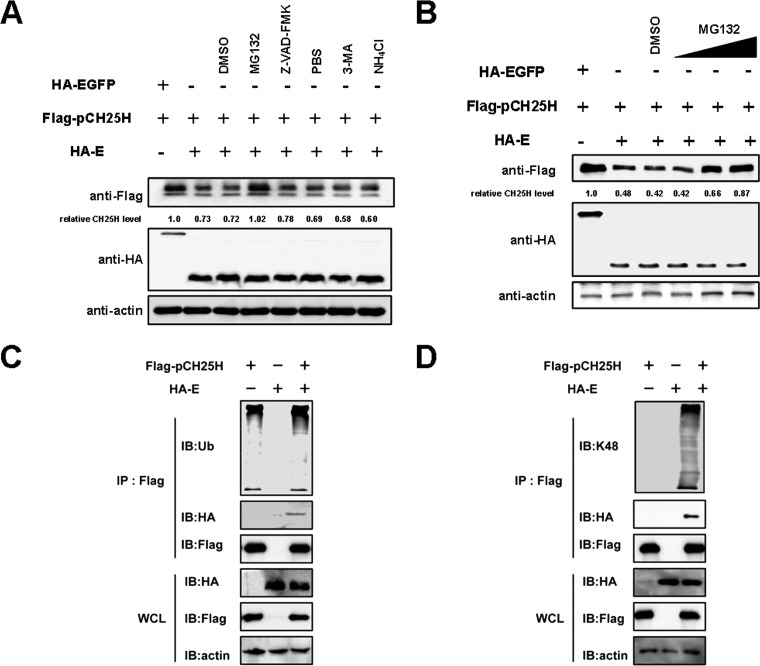
PRRSV E protein degrades pCH25H via the ubiquitin-proteasome pathway.
To identify the pathway through which E protein degrades pCH25H, cells coexpressing E protein and pCH25H were treated with the ubiquitin proteasome inhibitor MG132 (10 μM), the autophagy inhibitor 3-MA (5 mM), the lysosomal inhibitor NH4Cl (10 mM), and the apoptosis inhibitor Z-VAD-FMK (10 μM). We found that, similar to the results observed during PRRSV infection, treatment with Z-VAD-FMK and NH4Cl partially restored expression of pCH25H, and MG132 treatment almost completely restored expression (Fig. 3A). To extend these results, a titration experiment was performed to test the effects of MG132. As shown in Fig. 3B, degradation of pCH25H was inhibited to a greater extend with increasing doses of MG132.
FIG 3.
PRRSV E protein degrades pCH25H via the ubiquitin-proteasome pathway. (A) 3D4/21 cells were cotransfected with expression vectors encoding E protein and pCH25H. At 24 h posttransfection, cells were treated for 24 h with MG132 (10 μM), Z-VAD-FMK (10 μM), 3-MA (5 mM), or NH4Cl (10 mM). Cell lysates were harvested and analyzed by Western blotting for analysis of expression of PRRSV E protein and pCH25H. (B) 3D4/21 cells were cotransfected with expression vectors encoding E protein and pCH25H. Cells were treated with different concentrations of MG132 at 24 h posttransfection. Cell lysates were harvested and analyzed by Western blotting for analysis of pCH25H expression. (C and D) HEK-293T cells were cotransfected with expression vectors encoding HA-tagged E protein and FLAG-tagged pCH25H. Cells were lysed at 48 h posttransfection and immunoprecipitated with anti-FLAG antibody. The whole-cell lysate (WCL) and immunoprecipitation (IP) complexes were analyzed by immunoblotting (IB) with anti-FLAG, anti-HA, anti-β-actin, and anti-ubiquitin (Ub) (C) or anti-K48 (D) antibodies. DMSO, dimethyl sulfoxide.
Because the PRRSV E protein degrades pCH25H via the ubiquitin-proteasome pathway, pCH25H ubiquitination should be elevated in cells cotransfected with the pCAGGS-FLAG-pCH25H and pCAGGS-HA-E expression vectors. As shown in Fig. 3C, the ubiquitination level of pCH25H was enhanced when it was coexpressed with E protein. It is well known that the K48-linked polyubiquitin chain is the primary mediator of degradation. Therefore, we further analyzed the level of pCH25H K48 ubiquitination in cells coexpressing E protein and pCH25H. As expected, levels of pCH25H K48 ubiquitination were higher in cells cotransfected with expression vectors encoding E protein and pCH25H than in cells expressing pCH25H alone (Fig. 3D). Taken together, these results demonstrated that PRRSV E protein degrades pCH25H via the ubiquitin-proteasome pathway.
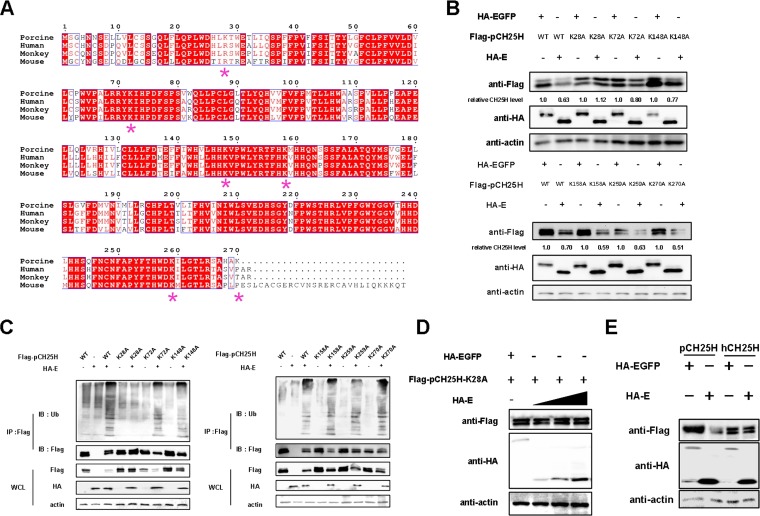
The Lys28 site of pCH25H is ubiquitinated by PRRSV E protein.
By comparing the amino acid sequences of mouse CH25H, human CH25H (hCH25H), monkey CH25H, and pCH25H, we observed four conserved Lys residues in all four proteins, while pCH25H has two additional Lys residues, at positions 28 and 270 (Fig. 4A). To identify possible ubiquitination sites in pCH25H, we cotransfected 3D4/21 cells with expression vectors encoding HA-tagged E protein and either wild-type FLAG-tagged pCH25H or mutant FLAG-tagged pCH25H (K28A, K72A, K148A, K158A, K259A, or K270A). As shown in Fig. 4B, the K28A mutant almost completely abolished the ability of E protein to degrade pCH25H, suggesting that pCH25H K28 is ubiquitinated in the presence of PRRSV E protein. To confirm this result, we cotransfected HEK-293T cells with expression vectors encoding HA-tagged E protein and either wild-type FLAG-tagged pCH25H or the pCH25H mutants. We found that only the K28A mutant of pCH25H was not ubiquitinated by PRRSV E protein (Fig. 4C). Moreover, when an expression vector encoding pCH25H-K28A and different doses of an expression vector encoding E protein were cotransfected, we found that pCH25H-K28A could not be degraded by E protein at any dose (Fig. 4D). These results suggested that K28 of pCH25H is the sole site ubiquitinated by PRRSV E protein. Because of a Lys28Arg substitution in hCH25H (Fig. 4A), it would be expected that this protein would not be degraded by PRRSV E protein. To verify this hypothesis, we coexpressed either hCH25H or pCH25H with E protein in 3D4/21 cells. As expected, PRRSV E protein did not degrade hCH25H (Fig. 4E).
FIG 4.
The Lys28 site of pCH25H is ubiquitinated by PRRSV E protein. (A) CH25H sequences from different species were aligned (*, lysine). (B) 3D4/21 cells were cotransfected with expression vectors encoding FLAG-tagged wild-type (WT) pCH25H or pCH25H mutants (K28A, K72A, K148A, K158A, K259A, or K270A) and HA-tagged PRRSV E protein. At 48 h posttransfection, cells were harvested for analysis of pCH25H expression by Western blotting. The numbers below the images represent the relative levels of pCH25H, compared to that of the corresponding control group, as determined via ImageJ analysis. (C) HEK-293T cells were cotransfected with expression vectors encoding HA-tagged PRRSV E protein and FLAG-tagged wild-type pCH25H or pCH25H mutants (K28A, K72A, K148A, K158A, K259A, or K270A). The cells were lysed at 48 h after transfection and immunoprecipitated with an anti-FLAG antibody. Whole-cell lysate (WCL) and immunoprecipitation (IP) complexes were analyzed by immunoblotting (IB) with anti-FLAG, anti-HA, anti-ubiquitin (Ub), or anti-β-actin antibodies. (D) 3D4/21 cells were cotransfected with expression vectors encoding FLAG-tagged K28A and different amounts of expression vectors encoding HA-tagged E protein. At 48 h posttransfection, cells were harvested for analysis of pCH25H expression by Western blotting. (E) 3D4/21 cells were cotransfected with expression vectors encoding FLAG-tagged pCH25H or hCH25H and HA-tagged E protein. At 48 h posttransfection, cells were harvested for analysis of pCH25H expression by Western blotting.
PRRSV E protein counteracts the anti-PRRSV effect of pCH25H.
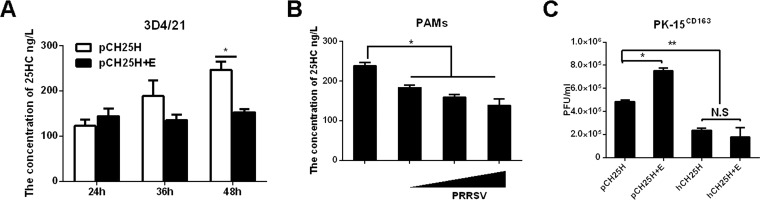
CH25H is a multitransmembrane ER-resident enzyme whose main physiological function is to convert cholesterol into 25HC, reducing cholesterol accumulation (38, 39). Because PRRSV E protein degrades pCH25H, we investigated whether E protein affected the ability of pCH25H to produce 25HC. As shown in Fig. 5A, the 25HC content in the supernatants of 3D4/21 cells coexpressing pCH25H and E protein were decreased at 36 h and 48 h posttransfection, compared with cells coexpressing pCH25H and EGFP, suggesting that PRRSV E protein reduces the ability of pCH25H to produce 25HC. We also analyzed 25HC levels in the supernatants of PAMs infected with PRRSV at different MOIs, and we found that PRRSV infection reduced 25HC production in a dose-dependent manner (Fig. 5B).
FIG 5.
PRRSV protein counteracts the anti-PRRSV effects of pCH25H. (A) 3D4/21 cells were cotransfected with expression vectors encoding FLAG-tagged pCH25H and HA-tagged PRRSV E protein. The cells were harvested for quantitation of 25HC by ELISA at 48 h posttransfection. (B) PAMs were infected with different doses of PRRSV. The cells were harvested for quantitation of 25HC by ELISA at 48 h postinfection. (C) PK-15CD163 cells were cotransfected with expression vectors encoding FLAG-tagged pCH25H or FLAG-tagged hCH25H and HA-tagged PRRSV E protein. At 48 h posttransfection, cells were infected with PRRSV (MOI of 1.0); 24 h later, they were harvested for a plaque assay. All experiments were performed in triplicate, and data shown are representative of three independent experiments. Standard errors of the mean are indicated by error bars. *, P < 0.05; **, P < 0.01. N.S., not significant.
Previous studies have suggested that the antiviral effects of pCH25H could be predominantly attributed to its enzymatic activity and production of 25HC (25–27). Given that PRRSV E protein degrades pCH25H and decreases 25HC production, the antiviral effects of pCH25H should be impaired by PRRSV E protein. To test this hypothesis, we transfected PK-15CD163 cells (a pig kidney cell line stably expressing the PRRSV receptor CD163; a gift from En-ming Zhou at Northwest A&F University, China) with expression vectors encoding pCH25H alone or pCH25H and E protein and then infected the cells with PRRSV (MOI of 1). At 48 h after PRRSV infection, cells were collected and plaque assays were performed to determine viral titers. Because PRRSV E protein could not degrade hCH25H, we used hCH25H as a control. As shown in Fig. 5C, PRRSV E protein did not affect the antiviral effects of hCH25H on PRRSV. However, coexpression of E protein significantly decreased the anti-PRRSV activity of pCH25H. Furthermore, we also found that hCH25H exhibited stronger anti-PRRSV effects than pCH25H (Fig. 5C). Taken together, these results indicated that PRRSV E protein counteracted the antiviral activity of pCH25H by reducing its ability to produce 25HC.
Knockdown of pCH25H enhances E protein-mediated inflammatory responses.
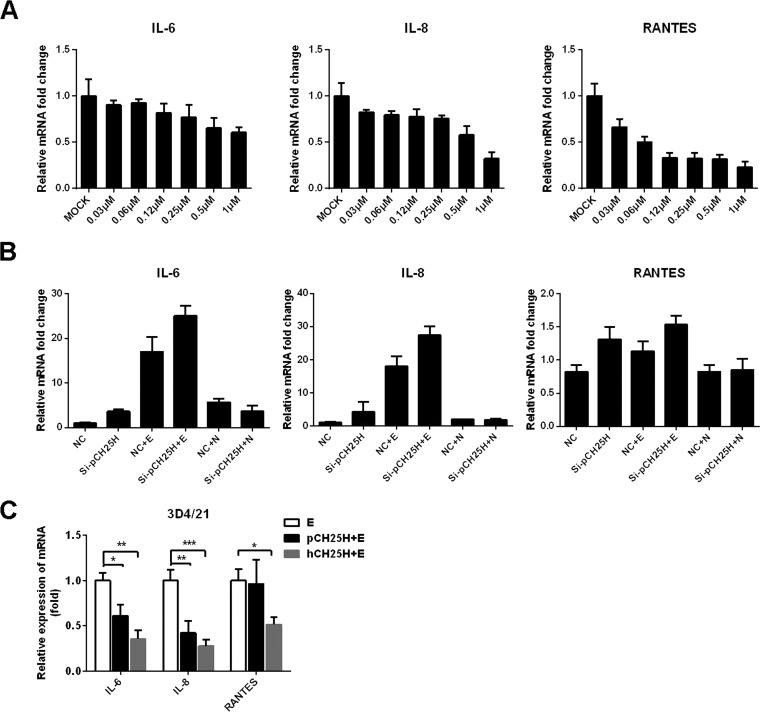
In addition to its antiviral activity, recent studies have suggested that 25HC, the metabolic product of CH25H, also regulates inflammatory cytokine production in vitro and in vivo (40, 41). Coincidentally, PRRSV E protein has been demonstrated to induce inflammatory cytokine production (42, 43), which is potentially associated with the PRRSV-induced inflammatory cytokine storm that occurs in late stages of virus infection. Thus, we further investigated the potential relationship between pCH25H and PRRSV E protein-mediated inflammatory responses. Considering that conflicting results have been reported for the effects of 25HC on inflammatory responses (40, 41, 44, 45), possible reasons for these discrepancies may be that different cell types and/or different concentrations of 25HC were used in the studies. Thus, we first evaluated the inflammatory cytokine expression of 3D4/21 cells after treatment with different concentrations of 25HC. The qRT-PCR results showed that 25HC treatment inhibited production of interleukin 6 (IL-6), IL-8, and RANTES in a dose-dependent manner in 3D4/21 cells (Fig. 6A), suggesting that 25HC suppresses inflammatory cytokine expression in a cell line derived from PAMs, the target cells of PRRSV in vivo. To further test whether pCH25H is associated with PRRSV E protein-induced inflammatory responses, a pCH25H-specific small interfering (siRNA) (si-pCH25H, whose knockdown efficiency was demonstrated in our previous study [27]) was used to deplete endogenous pCH25H in 3D4/21 cells. Cells were then transfected with an expression vector encoding PRRSV E protein, and qRT-PCR was used to assess the expression of IL-6, IL-8, and RANTES. As shown in Fig. 6B, silencing of endogenous pCH25H enhanced E protein-induced inflammatory cytokine expression. We also overexpressed pCH25H or hCH25H and PRRSV E proteins in 3D4/21 cells and collected cell samples at 48 posttransfection to assess inflammatory cytokine expression. As shown in Fig. 6C, overexpression of pCH25H significantly inhibited the proinflammatory effect of PRRSV E protein. Furthermore, the inhibitory effect of hCH25H was stronger than that of pCH25H, which is consistent with the fact that PRRSV E protein does not degrade hCH25H (Fig. 4E).
FIG 6.
Knockdown of pCH25H enhances PRRSV E protein-mediated inflammatory responses. (A) 3D4/21 cells were incubated with various concentrations of 25HC or ethanol (as a control) for 8 h, and then cells were collected for quantitation of IL-6, IL-8, and RANTES mRNA by qRT-PCR. (B) 3D4/21 cells were transfected with pCH25H-specific siRNAs or control siRNA for 24 h and then cells were further transfected with expression vectors encoding PRRSV E protein or N protein. The cells were harvested at 24 h posttransfection for quantitation of IL-6, IL-8, and RANTES mRNA by qRT-PCR. (C) 3D4/21 cells were transfected with expression vectors encoding FLAG-tagged pCH25H or hCH25H and HA-tagged E protein. At 48 h posttransfection, cells were harvested for quantitation of expression of IL-6, IL-8 and RANTES by qRT-PCR. All experiments were performed in triplicate, and data shown are representative of three independent experiments. Standard deviations are indicated by error bars. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
Although CH25H has been demonstrated to have broad antiviral activity, different CH25H expression patterns have been observed after infection with different viruses. For example, CH25H expression was induced in cells infected with some viruses, such as ZIKV (32), HCV (35), vesicular stomatitis virus (33), pseudorabies virus (46), reovirus (34), and mouse cytomegalovirus (47). In contrast, CH25H expression was downregulated during herpes simplex virus 1 infection (48). In this study, we demonstrated that pCH25H expression was also downregulated during PRRSV infection and that PRRSV E protein degraded pCH25H via the ubiquitin-proteasome pathway.
The PRRSV E protein is a small structural protein encoded by PRRSV ORF2b, which mainly localizes to the ER and Golgi apparatus during PRRSV infection (49–51). Previous studies demonstrated that E protein is essential for viral infection, and PRRSV clones lacking ORF2b are not infectious (52). PRRSV E protein is generally considered to have the potential for pore-forming activity and can serve as an ion channel for viral uncoating (49). Considering its shared subcellular localization with CH25H and its ion channel protein-like properties, it is perhaps not surprising that PRRSV E protein interacts with pCH25H and regulates its expression via the ubiquitin-proteasome pathway. It is also possible that the PRRSV E protein degrades pCH25H through the ER-associated degradation (ERAD) response, since both pCH25H and PRRSV E protein are located in the ER. ERAD plays a key role in maintaining ER proteome integrity by continuously monitoring peptide folding under normal and stress conditions (53). ER-resident proteins are largely cleared by ERAD for proteasomal degradation in the ER, while protein aggregates in the ER may be cleared by autophagy (54, 55). We examined the mRNA expression of glucose regulatory protein 78 (GRP78) and GRP94, two key proteins in the ERAD response, in cells overexpressing PRRSV E protein. The results showed that the PRRSV E protein did not significantly change the mRNA expression of GRP78 and GRP94 (data not shown), indicating that PRRSV E protein does not induce ER stress and subsequent degradation through the ERAD response. In addition, a previous study showed that PRRSV infection downregulated host protein expression by inducing host translation shutoff but UV-inactivated PRRSV did not (56). To test whether PRRSV E protein induces host translation shutoff, resulting in the degradation of pCH25H, we determined the levels of puromycin in 3D4/21 cells transfected with PRRSV E protein or cotransfected with PRRSV E protein and pCH25H. The results showed that PRRSV E protein did not induce host translation shutoff (data not shown), indicating that PRRSV E protein-mediated degradation of pCH25H is not associated with host translation shutoff. PRRSV E protein has also been reported to induce apoptosis by interacting with mitochondrial proteins and/or inhibiting ATP production (57). In this study, we found that Z-VAD-FMK and NH4Cl treatment partially restored expression of pCH25H (Fig. 3A). Thus, we could not rule out the possibility that the apoptotic and lysosomal pathways may also be involved in pCH25H degradation by the PRRSV E protein. Although we have demonstrated that PRRSV E protein promotes ubiquitin-proteasome-mediated degradation of pCH25H through direct interaction, the way that PRRSV E protein degrades pCH25H and whether PRRSV E protein is an E3 ubiquitin ligase remain to be further studied.
In addition to its antiviral activity, 25HC can regulate inflammatory responses. The role of 25HC in regulating inflammatory responses remains controversial. Some studies have suggested that 25HC is a negative regulator of inflammatory responses. For example, Reboldi et al. showed that activated macrophages lacking CH25H overproduce inflammatory IL-1-family cytokines and have dysregulated caspase 1-activating inflammasome activity (44). Consistent with these in vitro results, CH25H-deficient mice overproduced IL-1α, IL-1β, and IL-8 following lipopolysaccharide (LPS) injection and died more rapidly from septic shock (58). Ouyang et al. showed that mice pretreated intraperitoneally with 25HC had attenuated LPS-induced inflammatory cytokine release and acute lung injury; these effects were mediated by interaction of 25HC with myeloid differentiation protein 2 (MD-2) and suppression of the Akt/NF‐κB signaling pathway (41). These studies support the repressive action of CH25H/25HC on inflammatory responses. In contrast, some studies have suggested that 25HC acts as an amplifier of inflammatory signaling. For example, Gold et al. found that 25HC inhibited in vitro infection of airway epithelial cells by influenza virus and CH25H-deficient mice were protected from the effects of influenza infection as a result of decreased inflammation-induced pathology (40). Jang et al. found that 25HC was a potent mediator and inducer of cerebral inflammation in X-linked adrenoleukodystrophy through its activation of the NLRP3 inflammasome, supporting the proinflammatory role of 25HC (45). These discrepancies regarding the contribution of 25HC to inflammation across different studies have not been fully explained. One possible explanation lies in the differing concentrations of 25HC used in previous studies; generally, micromolar concentrations of 25HC were used in studies proposing its proinflammatory effects, whereas nanomolar concentrations were used in the studies that observed anti-inflammatory effects (59). In addition, different cell types used in different studies may account for some of the variability across reports. Keeping these factors in mind, we determined the content of 25HC in PAMs, the target cells of PRRSV in vivo, and found that the physiological concentration of 25HC in isolated PAMs was ∼238 ng/liter (Fig. 5B). We also carefully examined the role of 25HC in PAMs using different concentrations of 25HC. We found that consistent decreases in the expression of inflammatory cytokines (IL-6, IL-8, and RANTES) were observed in 3D4/21 cells treated with a range of concentrations (30 nM to 1 μM) of 25HC, providing definitive evidence to support the anti-inflammatory action of 25HC in 3D4/21 cells.
An interesting finding of our study was that PRRSV E protein degraded pCH25H but not hCH25H. The ubiquitination site identified at Lys28 of pCH25H is substituted with Arg in hCH25H. Furthermore, antiviral assays demonstrated that E protein does not degrade hCH25H and does not affect the antiviral effect of hCH25H on PRRSV. However, hCH25H showed greater antiviral activity against PRRSV, compared with pCH25H. Recently, our group showed that the PRRSV 3C-like proteinase nsp4 cleaves porcine mRNA-decapping enzyme 1a (pDCP1a), an ISG (60). Human and monkey DCP1a were not be cleaved by PRRSV nsp4, however, because the cleavage site at Glu237 in pDCP1a is substituted with Asp in the human and monkey orthologues. Moreover, human and monkey DCP1a exhibited greater ability to resist PRRSV infection. Why PRRSV specifically cleaves or degrades porcine ISGs and whether specific cleavage and degradation are related to the fact that pigs are the sole host for PRRSV infection require further investigation.
In summary, we showed that PRRSV infection results in downregulation of pCH25H expression and that PRRSV-encoded E protein specifically degrades pCH25H via the ubiquitin-proteasome pathway. Although pCH25H exhibits antiviral activity, PRRSV has also developed an elaborate strategy to antagonize the antiviral activity of CH25H, which highlights the complex interactions between PRRSV and host CH25H.
MATERIALS AND METHODS
Cell culture and viruses.
HEK-293T and PK-15CD163 cells were cultured at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS), in a humidified atmosphere containing 5% CO2. PAMs and 3D4/21 cells were cultured at 37°C in RPMI 1640 medium (Sigma) supplemented with 10% heat-inactivated FBS, in a humidified atmosphere containing 5% CO2. PRRSV strain WUH3 is a highly pathogenic type 2 (North American) virus isolated in 2006 from the brains of pigs suffering from “high-fever syndrome” in China (61). PRRSV was amplified and titers were determined in PK-15CD163 cells.
Plasmids.
An expression vector containing genes encoding FLAG-tagged wild-type pCH25H (residues 1 to 271) were constructed by PCR amplification of cDNAs from PK-15CD163 cells followed by cloning into the pCAGGS-FLAG vector, yielding the pCAGGS-FLAG-pCH25H plasmid. Lysine-encoding codons at positions 28, 72, 148, 158, 259, and 270 of pCH25H were substituted with alanine by site-directed mutagenesis. Expression vectors containing genes encoding PRRSV proteins were constructed by cloning PRRSV genes into the pCAGGS-HA vector, as described previously (62).
Reagents and antibodies.
Mouse or rabbit monoclonal antibodies (MAbs) against FLAG, HA, or β-actin were purchased from MBL. Mouse anti-CH25H MAb (product no. H00009023-M01) was purchased from Huajing Meida (Beijing, China). Rabbit polyclonal antibodies against ubiquitin and K48-ubiquitin were purchased from Abclonal (Wuhan, China). The proteasome inhibitor MG132, the autophagy inhibitor 3-MA, and the lysosomal inhibitor CQ were purchased from Sigma-Aldrich (St. Louis, MO) and were used at concentrations of 10 μM, 5 mM, and 20 μM, respectively. The pan-caspase inhibitor Z-VAD-FMK and the lysosomal inhibitor NH4Cl were purchased from Beyotime (Jiangsu, China) and were used at concentrations of 10 μM and 10 mM, respectively.
Immunofluorescence staining.
Cells were fixed with 4% paraformaldehyde and then permeabilized with methanol for 15 min prior to the addition of primary antibody (rabbit anti-HA or mouse anti-FLAG) for 1 h at room temperature. The cells were washed with phosphate-buffered saline (PBS) and then incubated with Alexa Fluor 594 (AF594)-conjugated donkey anti-mouse IgG or AF488-conjugated donkey anti-rabbit IgG secondary antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescence images were acquired with a confocal laser scanning microscope (Olympus FluoView v3.1; Olympus, Tokyo, Japan).
RNA extraction and qRT-PCR.
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen) and was reverse transcribed into cDNA using reverse transcriptase (TaKaRa, Japan). qRT-PCR experiments were performed in triplicate. Relative mRNA expression levels were normalized to the expression of β-actin. Absolute mRNA levels were calculated using standard curves. All qRT-PCR experiments were performed using Power SYBR green PCR master mix (Applied Biosystems) and an ABI 7500 real-time PCR system (Applied Biosystems).
Coimmunoprecipitation.
Cells were collected 48 h after transfection with expression vectors and were lysed by resuspension in 50 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and 1% Triton X-100. For immunoprecipitation, lysates were incubated with the appropriate antibodies for 4 h on ice, followed by precipitation with protein A/G-agarose beads (Beyotime). Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After blocking in PBS containing 0.1% Tween 20 and 5% (wt/vol) skim milk, the blots were probed with the indicated antibodies. Western blots were visualized with enhanced chemiluminescence methods.
GST pulldown.
The cDNA encoding pCH25H was cloned into prokaryotic expression vector pET-42b and fused with a His tag. The cDNA encoding PRRSV E protein was also cloned into pET-42b and fused with a GST tag. The fusion proteins His-pCH25H and GST-E were expressed in Escherichia coli BL21 Rosetta strain. The purified recombinant protein GST-E was incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at 4°C and washed three times with PBS. The purified His-pCH25H protein was then added and incubated for 1 h at 4°C. The beads were washed three times with PBS for Western blotting with antibodies against GST and CH25H, respectively.
Western blotting.
Cells were cultured in 60-mm dishes and harvested using lysis buffer (Beyotime). The lysed samples were resolved by SDS-PAGE and transferred to PVDF membranes for analysis of protein expression. Cells were treated with a broad caspase inhibitor (Z-VAD-FMK; Beyotime) or a proteasome inhibitor (MG132; Sigma) at final concentrations of 10 μM, an autophagy inhibitor (3-MA; Sigma) at a final concentration of 5 mM, or lysosomal inhibitors (NH4Cl [Beyotime] and CQ [Sigma]) at final concentrations of 10 mM and 20 μM, respectively.
Plaque assay.
PK-15CD163 cells were cultured in six-well plates and chilled at 4°C for 1 h, and then the culture medium was replaced with PRRSV-containing medium. After incubation at 4°C for another 2 h, the cells were washed with prechilled PBS, covered with overlay medium (DMEM containing 1.8% [wt/vol] Bacto agar and 0.05 mg/ml neutral red), incubated at 37°C for another 72 h, and examined by plaque assay.
Measurement of 25HC levels.
Concentrations of 25HC in the culture medium were determined using a commercial enzyme-linked immunosorbent assay (ELISA) kit (product no. SY-010532; Win-win Biotechnology, China).
Statistical analyses.
GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA) was used for data analyses. Differences between groups were evaluated using two-tailed unpaired t tests and were considered statistically significant when the P value was less than 0.05.
ACKNOWLEDGMENTS
We thank En-ming Zhou for providing cells and expression constructs.
This work was supported by the National Natural Science Foundation of China (grants 31490602, 31372467, and 31225027), the National Basic Research Program of China (grant 2014CB542700), and the Key Technology R&D Program of China (grant 2015BAD12B02).
REFERENCES
- 1.Salguero FJ, Frossard JP, Rebel JM, Stadejek T, Morgan SB, Graham SP, Steinbach F. 2015. Host-pathogen interactions during porcine reproductive and respiratory syndrome virus 1 infection of piglets. Virus Res 202:135–143. doi: 10.1016/j.virusres.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, Renukaradhya GJ. 2016. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- 3.Nelsen CJ, Murtaugh MP, Faaberg KS. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol 73:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol 142:629–633. [PubMed] [Google Scholar]
- 5.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 6.Fang Y, Snijder EJ. 2010. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res 154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Tas A, Snijder EJ, Fang Y. 2012. Identification of porcine reproductive and respiratory syndrome virus ORF1a-encoded non-structural proteins in virus-infected cells. J Gen Virol 93:829–839. doi: 10.1099/vir.0.039289-0. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Tas A, Sun Z, Snijder EJ, Fang Y. 2015. Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase. Virus Res 202:48–59. doi: 10.1016/j.virusres.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 9.Brar MS, Shi M, Hui RK, Leung FC. 2014. Genomic evolution of porcine reproductive and respiratory syndrome virus (PRRSV) isolates revealed by deep sequencing. PLoS One 9:e88807. doi: 10.1371/journal.pone.0088807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan S, Murtaugh MP, Schumann FA, Mickelson D, Faaberg KS. 2004. Characterization of heteroclite subgenomic RNAs associated with PRRSV infection. Virus Res 105:75–87. doi: 10.1016/j.virusres.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. 2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol 92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UB, Atkins JF, Snijder EJ, Posthuma CC. 2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lager KM, Mengeling WL, Brockmeier SL. 1999. Evaluation of protective immunity in gilts inoculated with the NADC-8 isolate of porcine reproductive and respiratory syndrome virus (PRRSV) and challenge-exposed with an antigenically distinct PRRSV isolate. Am J Vet Res 60:1022–1027. [PubMed] [Google Scholar]
- 14.Meng XJ. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol 74:309–329. doi: 10.1016/S0378-1135(00)00196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuckermann FA, Garcia EA, Luque ID, Christopher-Hennings J, Doster A, Brito M, Osorio F. 2007. Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet Microbiol 123:69–85. doi: 10.1016/j.vetmic.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Kimman TG, Cornelissen LA, Moormann RJ, Rebel JM, Stockhofe-Zurwieden N. 2009. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 27:3704–3718. doi: 10.1016/j.vaccine.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 17.Murtaugh MP, Genzow M. 2011. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS). Vaccine 29:8192–8204. doi: 10.1016/j.vaccine.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Vu HLX, Pattnaik AK, Osorio FA. 2017. Strategies to broaden the cross-protective efficacy of vaccines against porcine reproductive and respiratory syndrome virus. Vet Microbiol 206:29–34. doi: 10.1016/j.vetmic.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Mateu E, Diaz I. 2008. The challenge of PRRS immunology. Vet J 177:345–351. doi: 10.1016/j.tvjl.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S, Shan T, Zhou Y, Jiang Y, Tong W, Liu F, Wen F, Zhang Q, Tong G. 2014. Molecular cloning and characterizations of porcine SAMHD1 and its roles in replication of highly pathogenic porcine reproductive and respiratory syndrome virus. Dev Comp Immunol 47:234–246. doi: 10.1016/j.dci.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Fang J, Wang H, Bai J, Zhang Q, Li Y, Liu F, Jiang P. 2016. Monkey viperin restricts porcine reproductive and respiratory syndrome virus replication. PLoS One 11:e0156513. doi: 10.1371/journal.pone.0156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q, Liu Q, Liu D, Wang D, Chen H, Xiao S, Fang L. 2014. Molecular cloning, functional characterization and antiviral activity of porcine DDX3X. Biochem Biophys Res Commun 443:1169–1175. doi: 10.1016/j.bbrc.2013.12.098. [DOI] [PubMed] [Google Scholar]
- 23.Xu D, Lillico SG, Barnett MW, Whitelaw CB, Archibald AL, Ait-Ali T. 2012. USP18 restricts PRRSV growth through alteration of nuclear translocation of NF-κB p65 and p50 in MARC-145 cells. Virus Res 169:264–267. doi: 10.1016/j.virusres.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y, Wang D, Zhu X, Wu Q, Chen H, Xiao S, Fang L. 2016. Molecular cloning and functional characterization of porcine E74-like factor 4 (ELF4). Dev Comp Immunol 65:149–158. doi: 10.1016/j.dci.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Song Z, Zhang Q, Liu X, Bai J, Zhao Y, Wang X, Jiang P. 2017. Cholesterol 25-hydroxylase is an interferon-inducible factor that protects against porcine reproductive and respiratory syndrome virus infection. Vet Microbiol 210:153–161. doi: 10.1016/j.vetmic.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Dong H, Zhou L, Ge X, Guo X, Han J, Yang H. 2018. Antiviral effect of 25-hydroxycholesterol against porcine reproductive and respiratory syndrome virus in vitro. Antivir Ther 23:395–404. doi: 10.3851/IMP3232. [DOI] [PubMed] [Google Scholar]
- 27.Ke W, Fang L, Jing H, Tao R, Wang T, Li Y, Long S, Wang D, Xiao S. 2017. Cholesterol 25-hydroxylase inhibits porcine reproductive and respiratory syndrome virus replication through enzyme activity-dependent and -independent mechanisms. J Virol 91:e00827-17. doi: 10.1128/JVI.00827-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lund EG, Kerr TA, Sakai J, Li WP, Russell DW. 1998. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem 273:34316–34327. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- 29.Moog C, Aubertin AM, Kirn A, Luu B. 1998. Oxysterols, but not cholesterol, inhibit human immunodeficiency virus replication in vitro. Antivir Chem Chemother 9:491–496. doi: 10.1177/095632029800900605. [DOI] [PubMed] [Google Scholar]
- 30.Anggakusuma, Romero-Brey I, Berger C, Colpitts CC, Boldanova T, Engelmann M, Todt D, Perin PM, Behrendt P, Vondran FW, Xu S, Goffinet C, Schang LM, Heim MH, Bartenschlager R, Pietschmann T, Steinmann E. 2015. Interferon-inducible cholesterol-25-hydroxylase restricts hepatitis C virus replication through blockage of membranous web formation. Hepatology 62:702–714. doi: 10.1002/hep.27913. [DOI] [PubMed] [Google Scholar]
- 31.Xiang Y, Tang JJ, Tao W, Cao X, Song BL, Zhong J. 2015. Identification of cholesterol 25-hydroxylase as a novel host restriction factor and a part of the primary innate immune responses against hepatitis C virus infection. J Virol 89:6805–6816. doi: 10.1128/JVI.00587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Deng YQ, Wang S, Ma F, Aliyari R, Huang XY, Zhang NN, Watanabe M, Dong HL, Liu P, Li XF, Ye Q, Tian M, Hong S, Fan J, Zhao H, Li L, Vishlaghi N, Buth JE, Au C, Liu Y, Lu N, Du P, Qin FX, Zhang B, Gong D, Dai X, Sun R, Novitch BG, Xu Z, Qin CF, Cheng G. 2017. 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity 46:446–456. doi: 10.1016/j.immuni.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doms A, Sanabria T, Hansen JN, Altan-Bonnet N, Holm GH. 2018. 25-Hydroxycholesterol production by the cholesterol-25-hydroxylase interferon-stimulated gene restricts mammalian reovirus infection. J Virol 92:e01047-18. doi: 10.1128/JVI.01047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Wang S, Yi Z, Tian H, Aliyari R, Li Y, Chen G, Liu P, Zhong J, Chen X, Du P, Su L, Qin FX, Deng H, Cheng G. 2014. Interferon-inducible cholesterol-25-hydroxylase inhibits hepatitis C virus replication via distinct mechanisms. Sci Rep 4:7242. doi: 10.1038/srep07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrivastava-Ranjan P, Bergeron E, Chakrabarti AK, Albarino CG, Flint M, Nichol ST, Spiropoulou CF. 2016. 25-Hydroxycholesterol inhibition of Lassa virus infection through aberrant GP1 glycosylation. mBio 7:e01808-16. doi: 10.1128/mBio.01808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong H, Zhou L, Ge X, Guo X, Han J, Yang H. 2018. Porcine reproductive and respiratory syndrome virus nsp1β and nsp11 antagonize the antiviral activity of cholesterol-25-hydroxylase via lysosomal degradation. Vet Microbiol 223:134–143. doi: 10.1016/j.vetmic.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Kandutsch AA, Chen HW, Heiniger HJ. 1978. Biological activity of some oxygenated sterols. Science 201:498–501. doi: 10.1126/science.663671. [DOI] [PubMed] [Google Scholar]
- 39.Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. 1999. Structural requirements of ligands for the oxysterol liver X receptors LXRα and LXRβ. Proc Natl Acad Sci U S A 96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gold ES, Diercks AH, Podolsky I, Podyminogin RL, Askovich PS, Treuting PM, Aderem A. 2014. 25-Hydroxycholesterol acts as an amplifier of inflammatory signaling. Proc Natl Acad Sci U S A 111:10666–10671. doi: 10.1073/pnas.1404271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ouyang W, Zhou H, Liu C, Wang S, Han Y, Xia J, Xu F. 2018. 25-Hydroxycholesterol protects against acute lung injury via targeting MD-2. J Cell Mol Med 22:5494–5503. doi: 10.1111/jcmm.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K, Hou Q, Zhong Z, Li X, Chen H, Li W, Wen J, Wang L, Liu W, Zhong F. 2013. Porcine reproductive and respiratory syndrome virus activates inflammasomes of porcine alveolar macrophages via its small envelope protein E. Virology 442:156–162. doi: 10.1016/j.virol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 43.Wang R, Yang L, Zhang Y, Li J, Xu L, Xiao Y, Zhang Q, Bai L, Zhao S, Liu E, Zhang YJ. 2018. Porcine reproductive and respiratory syndrome virus induces HMGB1 secretion via activating PKC-delta to trigger inflammatory response. Virology 518:172–183. doi: 10.1016/j.virol.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 44.Reboldi A, Dang EV, McDonald JG, Liang G, Russell DW, Cyster JG. 2014. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science 345:679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang J, Park S, Jin Hur H, Cho HJ, Hwang I, Pyo Kang Y, Im I, Lee H, Lee E, Yang W, Kang HC, Won Kwon S, Yu JW, Kim DW. 2016. 25-Hydroxycholesterol contributes to cerebral inflammation of X-linked adrenoleukodystrophy through activation of the NLRP3 inflammasome. Nat Commun 7:13129. doi: 10.1038/ncomms13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Zeng L, Zhang L, Guo ZZ, Lu SF, Ming SL, Li GL, Wan B, Tian KG, Yang GY, Chu BB. 2017. Cholesterol 25-hydroxylase acts as a host restriction factor on pseudorabies virus replication. J Gen Virol 98:1467–1476. doi: 10.1099/jgv.0.000797. [DOI] [PubMed] [Google Scholar]
- 47.Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, Lacaze P, Watterson S, Griffiths SJ, Spann NJ, Meljon A, Talbot S, Krishnan K, Covey DF, Wenk MR, Craigon M, Ruzsics Z, Haas J, Angulo A, Griffiths WJ, Glass CK, Wang Y, Ghazal P. 2013. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 38:106–118. doi: 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You H, Yuan H, Fu W, Su C, Wang W, Cheng T, Zheng C. 2017. Herpes simplex virus type 1 abrogates the antiviral activity of Ch25h via its virion host shutoff protein. Antiviral Res 143:69–73. doi: 10.1016/j.antiviral.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Lee C, Yoo D. 2006. The small envelope protein of porcine reproductive and respiratory syndrome virus possesses ion channel protein-like properties. Virology 355:30–43. doi: 10.1016/j.virol.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snijder EJ, van Tol H, Pedersen KW, Raamsman MJ, de Vries AA. 1999. Identification of a novel structural protein of arteriviruses. J Virol 73:6335–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veit M, Matczuk AK, Sinhadri BC, Krause E, Thaa B. 2014. Membrane proteins of arterivirus particles: structure, topology, processing and function. Virus Res 194:16–36. doi: 10.1016/j.virusres.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wissink EH, Kroese MV, van Wijk HA, Rijsewijk FA, Meulenberg JJ, Rottier PJ. 2005. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J Virol 79:12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braakman I, Hebert DN. 2013. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol 5:a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olzmann JA, Kopito RR, Christianson JC. 2013. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol 5:a013185. doi: 10.1101/cshperspect.a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christianson JC, Ye Y. 2014. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol 21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Fang L, Zhou Y, Tao R, Wang D, Xiao S. 2018. Porcine reproductive and respiratory syndrome virus infection induces both eIF2α phosphorylation-dependent and -independent host translation shutoff. J Virol 92:e00600-18. doi: 10.1128/JVI.00600-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pujhari S, Zakhartchouk AN. 2016. Porcine reproductive and respiratory syndrome virus envelope (E) protein interacts with mitochondrial proteins and induces apoptosis. Arch Virol 161:1821–1830. doi: 10.1007/s00705-016-2845-4. [DOI] [PubMed] [Google Scholar]
- 58.Noebauer B, Jais A, Todoric J, Gossens K, Sutterluty-Fall H, Einwallner E. 2017. Hepatic cholesterol-25-hydroxylase overexpression improves systemic insulin sensitivity in mice. J Diabetes Res 2017:4108768. doi: 10.1155/2017/4108768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cyster JG, Dang EV, Reboldi A, Yi T. 2014. 25-Hydroxycholesterols in innate and adaptive immunity. Nat Rev Immunol 14:731–743. doi: 10.1038/nri3755. [DOI] [PubMed] [Google Scholar]
- 60.Tao R, Fang L, Bai D, Ke W, Zhou Y, Wang D, Xiao S. 2018. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 cleaves porcine DCP1a to attenuate its antiviral activity. J Immunol 201:2345–2353. doi: 10.4049/jimmunol.1701773. [DOI] [PubMed] [Google Scholar]
- 61.Li B, Fang L, Liu S, Zhao F, Jiang Y, He K, Chen H, Xiao S. 2010. The genomic diversity of Chinese porcine reproductive and respiratory syndrome virus isolates from 1996 to 2009. Vet Microbiol 146:226–237. doi: 10.1016/j.vetmic.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 62.Jing H, Fang L, Ding Z, Wang D, Hao W, Gao L, Ke W, Chen H, Xiao S. 2017. Porcine reproductive and respiratory syndrome virus nsp1α inhibits NF-κB activation by targeting the linear ubiquitin chain assembly complex. J Virol 91:e01911-16. doi: 10.1128/JVI.01911-16. [DOI] [PMC free article] [PubMed] [Google Scholar]