Superinfection exclusion (SIE) is a widespread phenomenon in which an established virus infection prevents reinfection by closely related viruses. Understanding the mechanisms governing SIE will not only advance our basic knowledge of virus infection cycles but may also lead to improved design of antiviral measures. Despite the significance of SIE, our knowledge about viral SIE determinants and their modes of actions remain limited. In this study, we show that sonchus yellow net virus (SYNV) SIE is mediated by the viral matrix (M) protein. During primary infections, accumulation of M protein in infected nuclei results in coiling of genomic nucleocapsids and suppression of viral transcription. Consequently, nucleocapsids released by potential superinfectors are sequestered and are unable to initiate new infections. Our data suggest that SYNV SIE is caused by M protein-mediated transition from replication to virion assembly and that this process prevents secondary infections.
KEYWORDS: cross-protection, matrix protein, nucleocapsid, plant rhabdovirus, sonchus yellow net virus, superinfection exclusion
ABSTRACT
Superinfection exclusion (SIE) or cross-protection phenomena have been documented for plant viruses for nearly a century and are widespread among taxonomically diverse viruses, but little information is available about SIE of plant negative-strand RNA viruses. Here, we demonstrate that SIE by sonchus yellow net nucleorhabdovirus virus (SYNV) is mediated by the viral matrix (M) protein, a multifunctional protein involved in transcription regulation, virion assembly, and virus budding. We show that fluorescent protein-tagged SYNV variants display mutual exclusion/cross-protection in Nicotiana benthamiana plants. Transient expression of the SYNV M protein, but not other viral proteins, interfered with SYNV local infections. In addition, SYNV M deletion mutants failed to exclude superinfection by wild-type SYNV. An SYNV minireplicon reporter gene expression assay showed that the M protein inhibited viral transcription. However, M protein mutants with weakened nuclear localization signals (NLS) and deficient nuclear interactions with the SYNV nucleocapsid protein were unable to suppress transcription. Moreover, SYNV with M NLS mutations exhibited compromised SIE against wild-type SYNV. From these data, we propose that M protein accumulating in nuclei with primary SYNV infections either coils or prevents uncoiling of nucleocapsids released by the superinfecting SYNV virions and suppresses transcription of superinfecting genomes, thereby preventing superinfection. Our model suggests that the rhabdovirus M protein regulates the transition from replication to virion assembly and renders the infected cells nonpermissive for secondary infections.
IMPORTANCE Superinfection exclusion (SIE) is a widespread phenomenon in which an established virus infection prevents reinfection by closely related viruses. Understanding the mechanisms governing SIE will not only advance our basic knowledge of virus infection cycles but may also lead to improved design of antiviral measures. Despite the significance of SIE, our knowledge about viral SIE determinants and their modes of actions remain limited. In this study, we show that sonchus yellow net virus (SYNV) SIE is mediated by the viral matrix (M) protein. During primary infections, accumulation of M protein in infected nuclei results in coiling of genomic nucleocapsids and suppression of viral transcription. Consequently, nucleocapsids released by potential superinfectors are sequestered and are unable to initiate new infections. Our data suggest that SYNV SIE is caused by M protein-mediated transition from replication to virion assembly and that this process prevents secondary infections.
INTRODUCTION
Superinfection exclusion (SIE) is an extensively described virus-virus interaction phenomenon in which a primary virus infection (primary invader) prevents the infected cells from subsequent infection by the same or a related virus (secondary invader). SIE has been widely documented in viruses ranging from bacteriophages (1, 2) to mammalian viral pathogens (3–6) and plant viruses (reviewed in references 7 and 8). With human and animal viruses, SIE experiments have often been conducted in vitro with cultured cells, and the exclusion effects examined primarily at the cellular level. Entry processes of the challenge viruses have been shown to affect SIE in Rous sarcoma virus, bovine viral diarrhea virus, vaccinia virus, human influenza virus, and human immunodeficiency virus (9–13). Inhibition of virus replication and/or transcription has also been observed during postentry steps with Sindbis virus, hepatitis C virus, and West Nile virus (14–18). In addition, the primary invader may interfere with protein translation or morphogenesis of the secondary invader virus (16, 19).
A phenomenon mechanistically related to SIE, but manifested at the whole-plant level, is cross-protection in virus-infected plants (7, 8). During cross-protection, plants inoculated with mild virus isolates or strains elicit resistance to secondary infections by more severe isolates existing in nature (8, 20, 21). Cross-protection was initially described with tobacco mosaic virus (TMV) in the late 1920s (22) and has since been widely documented for many other plant virus species (reviewed in reference 21). In agricultural practice, cross-protection provided by mild virus strains has been employed in disease management, with prominent sucesses attained for control of viruses infecting perennial crop plants. These include cocoa swollen shoot virus on cocoa (23), papaya ringspot virus on papaya (24), and citrus tristeza virus (CTV) on citrus (7, 25). However, despite extensive studies, the underlying mechanisms leading to cross-protection have yet to be adequately addressed and may involve interference with virus interactions as well as host defense mechanisms that constrain infection either at the cellular or tissue-specific level (revirewed in references 8 and 21).
Early studies of cross-protection involved sequential inoculation of a mild strain of the primary virus, followed by challenge inoculation with a more severe strain and subsequent assessment of protection by symptom inspection and/or molecular detection of the secondary invader. With the availability of infectious virus clones, engineered recombinant plant viruses expressing visual discrimination markers, such as green and red fluorescent proteins (GFP and RFP, respectively), have proved more elegant tools to study temporal-spatial virus interactions at both the cellular and tissue levels. For example, when GFP- and RFP-labeled plum pox virus derivatives were coinoculated onto plants, the fluorescent markers demonstrated that the two viruses coexisted in the same plant leaves but showed that they occupied largely nonoverlapping cell clusters (26). Similar spatial separation or mutual exclusion between variants of the same virus have been observed for apple latent spherical virus and bean yellow mosaic virus (27, 28), TMV (29, 30), soilborne wheat mosaic virus (31), wheat streak mosaic virus (WSMV) and Triticum mosaic virus (TriMV) (32), and turnip crinkle virus (TCV) (33). However, when these differently tagged viruses were inoculated sequentially, the primary virus infection completely blocked subsequent infection by the secondary invader at the whole-plant level (34–41). These observations reinforce the idea that mutual exclusion or SIE occurs at the cellular level and suggest that cross-protection is a manifestation of SIE at the organismal level (8).
Several studies have shown that plant virus-encoded proteins can sufficiently, if not solely, account for cross-protection/SIE. Examples of viral determinants include coat protein (CP) of TMV (42), helper component proteinase (HC-Pro) and CP in potato virus A (43), both CP and Nia protease (NIa-Pro) in WSMV and TriMV (32), p33 and leader proteases L1/LII in CTV (38, 44), and p28 in TCV (33). In agricultural production, an awareness of the importance of virus-encoded factors in cross-protection/SIE has resulted in development of transgenic antiviral crops that have potential to protect plants in the field (45, 46). Although detailed molecular mechanisms underlying the SIE have not been determined in most studies, the phenomenon is known to be closely related to the viral life cycle. However, a recent study by Zhang and colleagues (33) provided an elegant model to explain TCV p28-mediated SIE. In this model, TCV p28 proteins produced from primary infections form mobile, multemeric complexes in a concentration-dependent manner that function to sequester newly synthesized p28 proteins from the superinfector, thus preventing the latter from replicating in the same cells. A follow-up study by the same group has shown that the TCV p28 readthrough product p88, a viral RNA-depedent RNA polymearse, likewise exhibits concentration-dependent repression of virus replication (47).
Although the SIE phenomenon has been studied for several plant positive-strand RNA viruses, no defined information is available for plant negative-strand RNA viruses. Sonchus yellow net virus (SYNV) is an extensively studied nonsegmented, negative-strand RNA virus, belonging to the genus Nucleorhabdovirus, family Rhabdoviridae. The genome of SYNV encodes five structural proteins, designated nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), the large RNA polymerase (L) protein, and a movement protein, sc4, arranged in the order N-P-sc4-M-G-L (48–51). Unlike many other rhabdoviruses infecting vertebrates and plants, nucleorhabdoviruses, including SYNV, undergo replication and virion morphogenesis in the nuclei of infected cells. Upon entry into the cell, nucleocapsids (NCs) released from virus particles are targeted to the nuclei where primary rounds of viral mRNA transcription are initiated. Viral proteins translated from these mRNAs are required for initiation of genome replication to produce antigenomic RNAs (agRNAs,) which are subsequently used as template to direct synthesis of genomic RNAs (gRNAs). Both gRNAs and agRNAs are encapsidated during replication by the N, P, and L core proteins to form progeny gNCs and agNCs. As additional rounds of secondary transcription and genome replication occur, accumulation of NCs results in formation of large viroplasms in the nuclei (reviewed in references 49 and 52). During the later stages of replication, accumulating M proteins presumably function to coil gNCs and assemble into viral cores. Through interactions between the M and G protein carboxyl-terminal tail, the condensed NC cores are directed to inner nuclear membrane surfaces from which they bud to form enveloped bullet-shaped or bacilliform particles that accumulate in the perinuclear spaces (53, 54).
In this paper, we have used engineered GFP- and RFP-labeled recombinant SYNV (rSYNV) variants to show that SYNV infections elicit robust SIE in the experimental host Nicotiana benthamiana. We also used ectopic expression of the M protein and mutant derivatives to show that the M protein is the primary viral SIE determinant and function to inhibit NC transcription. These results provide a model whereby the M protein accumulating in nuclei infected with the primary invading SYNV derivative interacts with incoming secondary invading SYNV NCs to exclude superinfection of the cells.
RESULTS
SYNV variants labeled by different fluorescent proteins exhibit reciprocal SIE.
To investigate possible SIE by SYNV, we used rSYNV-GFP and rSYNV-RFP clones for expression of GFP and RFP markers (Fig. 1A) to visualize infection and movement. The rSYNV derivatives were separately rescued in N. benthamiana plants after agroinfection as described previously (51). Equal volumes of infectious saps from rSYNV-GFP- and rSYNV-RFP-infected leaves were mixed and mechanically coinoculated onto lower leaves of healthy N. benthamiana plants. At 12 days postinoculation (dpi), both GFP and RFP fluorescence were detected by fluorescence microscopy in the upper noninoculated leaves, indicating that both viruses had infected the plants systemically. However, rSYNV-GFP and rSYNV-RFP on these leaves occupied separate tissue “islands” that exhibited clear boundaries, with less than 2% of adjacent cells in the border regions exhibiting both GFP and RFP fluorescence (Fig. 1B). To monitor the interactions of the rSYNV variants in locally infected leaves, we carried out a separate set of coinfection experiments in which Agrobacterium tumefaciens strains carrying plasmids required for recovery of rSYNV-GFP and rSYNV-RFP were coinfiltrated into N. benthamiana leaves. Both rSYNV variants were recovered in the same leaves, as judged from GFP and RFP expression at 8 dpi, and the isolated single-cell foci began to expand to adjacent cells (see reference 51 and data not shown). At 15 dpi when extensive cell-to-cell movement had occurred, some of the expanding GFP and RFP foci converged, and individual viruses at the margins occupied contiguous cells, but the cells rarely exhibited fluorescence of both reporter proteins (Fig. 1C). These observations reveal that the differentially tagged SYNV variants display mutual exclusion at the cellular level.
FIG 1.
Mutual exclusion of SYNV variants tagged with GFP and RFP in N. benthamiana. (A) Schematic diagrams showing rSYNV antigenomes engineered to express GFP (rSYNV-GFP) and RFP (rSYNV-RFP). (B) Confocal microscopy z-stack images showing spatial separation of GFP and RFP fluorescent foci in upper systemically infected leaves of N. benthamiana plants taken at 12 days postinoculation (dpi) with leaf extract containing mixtures of rSYNV-GFP and rSYNV-RFP. The bottom panels show higher magnifications of the boxed areas. Bar, 100 μm. (C) Confocal microscopy z-stack images of contiguous GFP and RFP foci in N. benthamiana leaf tissue at 15 dpi with agrobacterial mixtures harboring rSYNV-RFP and rSYNV-GFP derivatives. Right panels show magnified views of the boxed sectors on the left to highlight the exclusion boundaries of rSYNV-GFP and rSYNV-RFP in adjacent cells of two foci. Bar, 50 μm. (D) GFP and RFP fluorescence in upper leaves of N. benthamiana plants after an initial sap inoculation with one SYNV variant or mock inoculation, followed 12 days later with a challenge inoculation with the other SYNV variant indicated on the left of the panels. Images were captured with a stereo fluorescence microscope with GFP or RFP channels at 20 days after primary inoculation and again 20 days after challenge inoculation. 1st, primary inoculum; 2nd, challenge inoculum. Bar, 2 mm. (E) Confocal microscopy of N. benthamiana leaves after initial sap inoculation with rSYNV-RFP, followed by a PVX-GFP challenge inoculation 6 days later. The images show overlapping GFP and RFP fluorescence in upper infected leaves taken at 6 days after PVX infection. Bar, 100 μm.
We next conducted a second set of challenge inoculation experiments in which lower N. benthamiana leaves were mechanically inoculated to elicit rSYNV-RFP infections, followed by a second inoculation carried out 12 days later in the upper leaves to initiate rSYNV-GFP infections. By 20 days after the second inoculation, the newly expanded upper leaves exhibited only RFP fluorescence (Fig. 1D, top row). In contrast, primary infection with rSYNV-GFP completely blocked secondary infection by rSYNV-RFP (Fig. 1D, third row). In control experiments in which N. benthamiana plants were first rubbed with sap from healthy leaves, secondary inoculations 12 days later with either rSYNV-GFP or rSYNV-RFP resulted in productive infections (Fig. 1D, second and bottom rows). In another control experiment, we first inoculated rSYNV-RFP infectious sap on lower leaves of N. benthamiana plants and challenged upper leaves 6 days later with potato virus X (PVX)-GFP. By 12 dpi, numerous cells of the upper leaves were coinfected with rSYNV-RFP and PVX-GFP as indicated by yellow superimposed of GFP and RFP images (Fig. 1E). These results clearly suggest that primary infection with an rSYNV variant protected the plants from subsequent secondary infection by the other SYNV variant but did not prevent infection by the unrelated PVX.
Overexpression of the M protein inhibits SYNV local infection.
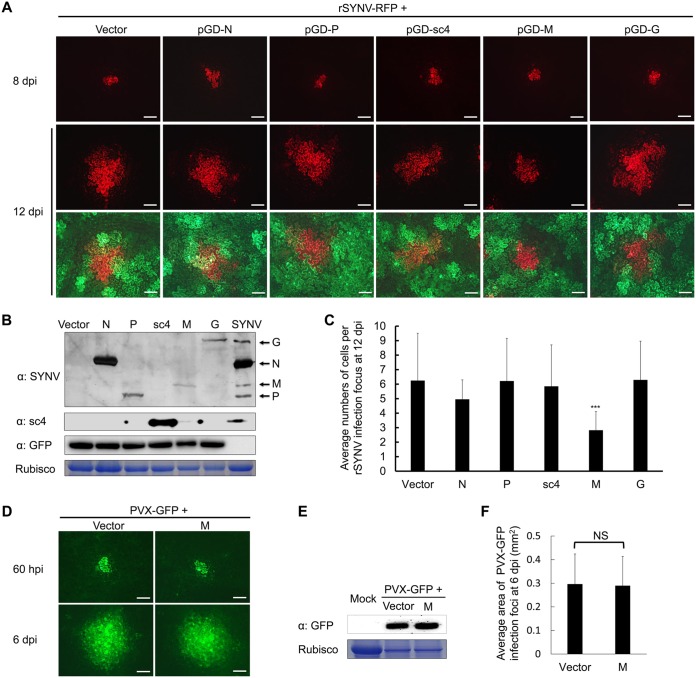
If SYNV SIE is mediated by one or more viral proteins, we posited that ectopic expression of the SYNV proteins would interfere with rSYNV infections. To test this possibility, the SYNV N, P, sc4, M, and G protein genes were inserted into a dual expression vector, pGD-X/GFP, which also contains a GFP expression cassette incorporated into the transfer DNA (T-DNA) region to permit coexpression of each SYNV protein and GFP after agroinfiltration. The SYNV L protein was omitted in this study because we were unable to express it to detectable levels due to its large size (242 kDa). To determine whether localized spread of SYNV was affected by coexpression of the SYNV proteins, we followed the movement of rSYNV-RFP foci in agroinfiltrated leaves by fluorescence microscopy. As observed previously (51), single-cell RFP foci appeared at 8 dpi and then spread to adjacent cells by 12 dpi (Fig. 2A). Foci expressing GFP fluorescence at 12 dpi were considered to have expressed the viral and GFP proteins simultaneously, which was further confirmed by Western blotting of tissue extracts using antibodies against viral proteins and GFP (Fig. 2B).
FIG 2.
Inhibition of rSYNV-RFP localized infections by ectopic expression of SYNV proteins. (A) Fluorescence microscopy observation of N. benthamiana leaves after agroinfiltration to initiate rSYNV local infections and to express each of the SYNV N, P, sc4, M, and G proteins or an empty vector control. Note that each of the viral proteins was expressed from a dual binary plasmid that also contains a GFP expression cassette to visualize the infiltrated leaf regions. Infiltrated leaves were observed under a stereo fluorescence microscope at 8 dpi and again at 12 dpi to estimate local infections during the 4-day movement period. The top two panels show RFP expression, and the bottom panel shows merged images of RFP and GFP expression in leaf cells surrounding the rSYNV RFP foci. Bar, 200 μm. (B) Western blotting to detect GFP expression levels and the presence of viral proteins using polyclonal antibodies against SYNV virions (α:SYNV), sc4 (α:sc4), and GFP (α:GFP). The stained RuBisCO large subunit was used as a loading control. (C) Calculation of the average numbers of RFP-expressing cells per rSYNV-RFP infection focus at 12 dpi. Bars represent standard deviations. Asterisks denote statistical significance of the M protein inhibition by a Student's t test (P < 0.001; n = 24). (D) Observation of the local movement of PVX-GFP coinfiltrated with SYNV M or empty vector at 60 hpi and 6 dpi with a fluorescence microscope. Bar, 200 μm. (E) Western blot analysis of GFP expression levels in leaf tissues infiltrated with PVX-GFP with or without SYNV M protein at 6 dpi. The stained RuBisCO large subunit was used as a loading control. (F) Calculation of the average areas of PVX-GFP infection foci at 6 dpi. NS: not statistically significant (P > 0.05; n = 20) as analyzed by Student's t test.
Coexpression of the M protein resulted in significant decreases in rSYNV-RFP localized spread, whereas coexpression of the N, P, sc4, or G protein had negligible effects on the spread of rSYNV-GFP (Fig. 2A). Statistical analyses of increases in the numbers of cells expressing RFP between 8 and 12 days showed that ectopic expression of the M protein inhibited SYNV cell-to-cell transport by about 2.0- to 2.5-fold compared with movement in tissue containing ectopically expressed SYNV N, P, sc4, or G protein or the negative control (Fig. 2C). To exclude the possibility that the ectopically expressed M protein inhibits SYNV movement by impacting host cells nonspecifically, e.g., by inhibiting protein synthesis, we tested the effect of M protein on local movement of PVX-GFP, a virus that does not display SIE with SYNV (Fig. 1E). Under similar transient expression conditions, SYNV M protein exhibited negligible effects on PVX-GFP movement (Fig. 2D). This conclusion was further supported by protein gel blot analysis of GFP levels (Fig. 2E) and quantitative analysis of the average area of PVX-GFP infection foci (Fig. 2F) at 6 dpi. Thus, these results suggest that ectopic expression of the M protein specifically inhibits SYNV local infections.
SYNV M deletion mutants display compromised SIE.
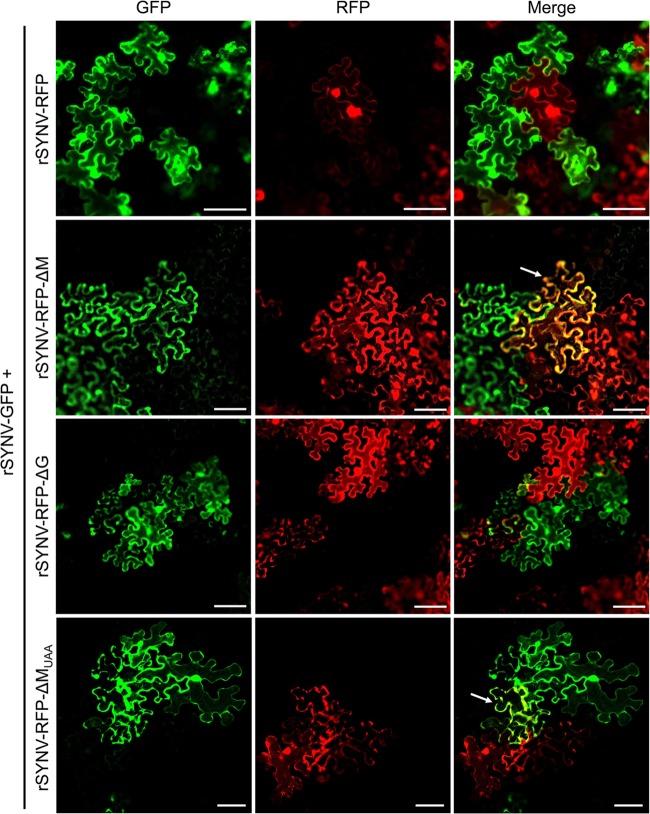
To further investigate involvement of the M protein in SYNV SIE, we tested whether an rSYNV-RFP mutant with a deletion in the M gene (rSYNV-RFP-ΔM) affects SIE by analyzing the ability of the mutant virus to prevent superinfection by rSYNV-GFP. As a control, the SYNV G deletion mutant (rSYNV-RFP-ΔG) was included in this experiment because the G and M proteins have a collaborative role in virion budding and maturation (54). Therefore, N. benthamiana leaves were infiltrated with mixtures of agrobacteria harboring plasmids required for simultaneous rescue of rSYNV-GFP and either rSYNV-RFP, rSYNV-RFP-ΔM, or rSYNV-RFP-ΔG. Cells expressing GFP and/or RFP at the boundaries of merging foci were visualized by fluorescence microscopy at 15 dpi to evaluate SIE interactions between the SYNV derivatives. As shown in Fig. 3, the red and green fluorescent signals in leaves coinfected with rSYNV-GFP and rSYNV-RFP were always spatially separated in adjacent foci of the leaf epidermal cells. Similar separate fluorescent distribution patterns were observed in the neighboring foci of leaves coinfected with rSYNV-GFP and rSYNV-RFP-ΔG. In contrast, a substantial proportion of the cells located at the boundaries of rSYNV-GFP and rSYNV-RFP-ΔM foci exhibited both GFP and RFP fluorescence (Fig. 3, white arrows), suggesting that SIE was less prevalent in the rSYNV-RFP-ΔM infections.
FIG 3.
Identification of the SYNV M protein as a viral superinfection exclusion (SIE) determinant. N. benthamiana leaves were coinfected via agroinoculation with the pair rSYNV-GFP and rSYNV-RFP, rSYNV-GFP and rSYNV-RFP-ΔM, rSYNV-GFP and rSYNV-RFP-ΔG, or rSYNV-GFP and rSYNV-RFP-MUAA. Contiguous GFP and RFP foci in inoculated leaf tissues at 15 dpi were imaged by confocal microscopy to show fluorescence exclusion between foci. The white arrows indicate boundary cells expressing both GFP and RFP. Bar, 100 μm.
To provide more analytical information, the numbers of cells coinfected with rSYNV-GFP/rSYNV-RFP and those infected with rSYNV-GFP/rSYNV-RFP-ΔM or rSYNV-GFP/rSYNV-RFP-ΔG were compared in three independent experiments. Table 1 shows that the average percentages of boundary cells exhibiting both GFP and RFP fluorescence increased to ∼25% for the rSYNV-GFP/rSYNV-RFP-ΔM infections in contrast to ∼1.5% for the rSYNV-GFP/rSYNV-RFP infections and 1.9% for the rSYNV-GFP/rSYNV-RFP-ΔG. These data indicate that deletion of the M protein gene results in a remarkable reduction in SYNV SIE activities.
TABLE 1.
Percentage of boundary cells coinfected with rSYNV-GFP and rSYNV-RFP derivatives in inoculated leaves of Nicotiana benthamiana
| Virus coinoculated with rSYNV-GFPa | Expt 1 |
Expt 2 |
Expt 3 |
Coinfection rate (%)b | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of cells (foci) | No. of cells expressing: |
Total no. of cells (foci) | No. of cells expressing: |
Total no. of cells (foci) | No. of cells expressing: |
||||||||
| GFP and RFP | RFP only | GFP only | GFP and RFP | RFP only | GFP only | GFP and RFP | RFP only | GFP only | |||||
| rSYNV-RFP | 42 (6) | 0 | 19 | 23 | 45 (6) | 1 | 25 | 19 | 45 (6) | 1 | 26 | 18 | 1.5 ± 1.3 |
| rSYNV-RFP-ΔM | 25 (5) | 4 | 12 | 9 | 24 (5) | 5 | 10 | 9 | 23 (5) | 8 | 6 | 9 | 23.8 ± 9.7* |
| rSYNV-RFP-ΔG | 30 (5) | 1 | 17 | 12 | 29 (5) | 0 | 13 | 16 | 47 (6) | 1 | 24 | 22 | 1.8 ± 1.7§ |
| rSYNV-RFP-MUAA | 20 (5) | 3 | 8 | 9 | 23 (5) | 6 | 11 | 6 | 21 (5) | 5 | 8 | 8 | 21.6 ± 5.9* |
| rSYNV-RFP-MRARA | 25 (5) | 3 | 10 | 12 | 28 (5) | 3 | 15 | 10 | 26 (5) | 5 | 9 | 12 | 14.0 ± 4.6* |
| rSYNV-RFP-MAAAA | 20 (5) | 4 | 9 | 7 | 23 (5) | 3 | 11 | 9 | 27 (5) | 5 | 14 | 8 | 17.2 ± 3.7* |
Leaves of N. benthamiana plants were infiltrated with agrobacterium mixtures to initiate mixed infections by rSYNV-GFP and rSYNV-RFP derivatives.
At 15 dpi when the expanding GFP and RFP foci merged, coinfected cells that exhibited both GFP and RFP, and cells expressing GFP only and RFP only, were counted from 5 to 6 infection foci. Coinfection rates were calculated by dividing the number of coinfected cells by the total number of cells bordering the GFP and RFP foci. *, P < 0.01; §, not statistically significant (Student’s t test).
To determine whether the RNA sequence or the protein encoded by the M gene is required in SIE, we created rSYNV-RFP-MUAA, in which the AUG initiation codon of the M cistron was changed to a UAA stop codon to eliminate protein translation. Upon coinfection with rSYNV-GFP in N. benthamiana leaves, the rSYNV-RFP-MUAA mutant also exhibited a compromised SIE ability as both RFP and GFP fluorescence were evident in the same cells (Fig. 3, bottom row). Quantitative analysis showed that ∼21% of the boundary cells exhibited both GFP and RFP fluorescence in tissues coinfected with rSYNV-GFP/rSYNV-RFP-MUAA (Table 1). Taken together, these data indicate that the SYNV M protein, rather than the RNA sequence, is responsible for the SIE activity.
SYNV M protein suppresses transcription of viral mRNAs.
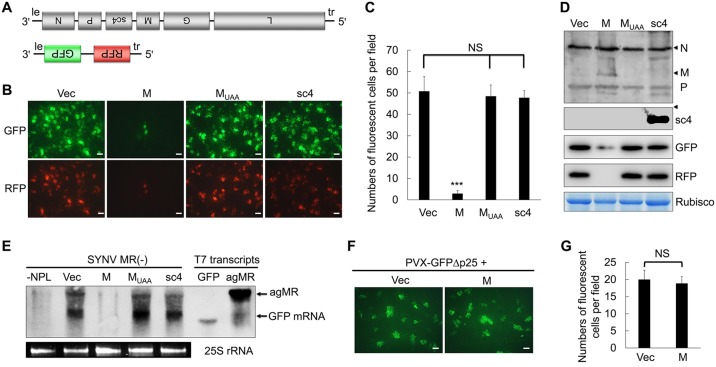
Studies with animal rhabdoviruses have shown that the M protein of these viruses inhibits viral transcription both in vitro and in vivo (55–58). To assess whether transcriptional repression could account for M-mediated SIE in SYNV infections, we next tested whether or not the SYNV M protein suppresses transcription from SYNV genomic RNA using a minireplicon (MR) system shown previously to recapitulate authentic viral RNA synthesis (59). In the SYNV MR system, viral RNA-like MR transcripts encoding GFP and RFP reporter genes substituted for the N and P cistrons (Fig. 4A). N. benthamiana leaves were agroinfiltrated with mixtures of bacteria harboring plasmids to express genomic sense MR [negative-sense MR, (−)MR] transcripts and the N, P, and L core proteins, as well as three viral suppressors of RNA silencing (VSRs), as described previously (59). Upon delivery of constructs expressing the viral components, single-cell foci expressing both the GFP and RFP reporter proteins were observed throughout infiltrated regions of the leaves at 8 dpi (Fig. 4B). When the SYNV MR was coexpressed in combination with the M protein gene, the number of foci exhibiting both reporter proteins had a 50-fold reduction compared to the level of the empty vector control (Fig. 4B and C). As additional controls, coexpression of the SYNV MR with either the MUAA mutant or the sc4 protein had negligible effects on reporter gene expression from the SYNV MR. Protein gel blot analysis showed that the N and P core proteins that are required of MR functions were expressed to similar levels in infiltrated leaf tissues with or without ectopic M protein expression. The pGD-mediated expression of the M and sc4 proteins and the absence of the M protein translation by the MUAA mutant were also verified (Fig. 4D). Protein blots also confirmed that the GFP and RFP protein levels in leaf tissues coexpressing the M protein were greatly reduced compared to coexpression of sc4, MUAA, or the empty vector (Fig. 4D). Because gNCs assembled from the (−)MR transcripts and core proteins in infiltrated leaf tissues can function in viral RNA synthesis, total RNA samples extracted from infiltrated tissues were hybridized with a GFP probe to detect agRNAs and GFP mRNAs (Fig. 4E). The RNA gel blot revealed a band with similar size to the 1,791-nucleotide (nt) MR agRNA size marker in leaf tissues expressing the N, P, and L protein and the (−)MR transcripts. In addition, another band migrating slightly more slowly than the 720-nt GFP marker RNA was also detected. This band is consistent with the size of the MR GFP mRNA that is anticipated to contain the 5′ untranslated region and 3′ poly(A) sequences. Expression of the M protein, but not the MUAA or the sc4 protein, drastically decreased the MR agRNAs and GPF mRNAs to levels similar to those in the negative-control sample in which the three core proteins were not expressed (−NPL). As an additional control, we showed that the SYNV M protein did not appear to affect GFP reporter gene expression from an unrelated PVX-GFPΔp25 replicon in which the p25 movement protein gene was replaced by a GFP gene (Fig. 4F and G), suggesting that the inhibitory effect of the M protein is virus specific. Together, these results suggest that ectopic expression of the M protein significantly suppresses viral RNA synthesis from gNC templates.
FIG 4.
Suppression of viral transcription by the M protein in an SYNV minireplicon (MR) system. (A) Schematic diagrams of the SYNV genome and a negative-sense MR cassette. The (−)MR contains 3′ leader (le) and 5′ trailer (tr) sequences flanking GFP and RFP reporter genes substituted for the SYNV N and P genes. (B) Visualization of GFP and RFP reporter proteins in N. benthamiana leaves agroinfiltrated with Agrobacterium cultures harboring plasmids for expression of the SYNV MR and the N, P, and L core proteins. Additional bacterial cultures containing the empty vector (Vec), pGD-M, pGD-MUAA, and pGD-sc4 plasmids, as indicated on the top of each panel, were also included in the mixtures to test their effects on reporter expression. Infiltrated leaves were photographed at 8 dpi with a fluorescence microscope. Bar, 200 μm. (C) Average numbers of cells per field expressing GFP and RFP at 8 dpi in each SYNV MR treatment. Bars represent standard deviations. Asterisk denotes Student's t test significance of transcription inhibition by the M protein (P < 0.001; NS, not significant; n = 12). (D) Western blots confirming GFP and RFP expression in each group. Expression of the N and P core proteins or of the M or sc4 protein, was detected with polyclonal antibodies against SYNV virion or against the sc4 protein, respectively. The Coomassie blue-stained RuBisCO large subunit provides a loading control. (E) Northern blot hybridization showing inhibitory effects of the M protein on SYNV MR RNA synthesis. Total RNA extracted from N. benthamiana leaf tissues shown in panel B was blotted with a negative-sense GFP probe to detect the MR agRNAs and GFP mRNAs. RNA from leaf tissues infiltrated with (−)MR in the absence of the N, P, and L proteins was used as a negative control. In vitro T7 transcripts corresponding in size to the GFP and MR agRNA (agMR) were used as size markers. The 25S rRNA was stained with ethidium bromide to show equal RNA loading. (F and G) Lack of inhibitory effects of the SYNV M protein on GFP reporter gene expression from a PVX replicon. N. benthamiana leaves were agroinfiltrated with mixtures of Agrobacterium strains harboring the PVX-GFPΔp25 plasmid together with pGD-M or an empty vector. Single-cell GFP foci were imaged with a fluorescence microscope at 3 dpi, and average numbers of cells per field were calculated and statistical significance was analyzed by Student's t test. NS, not statistically significant (P > 0.05; n = 12).
SYNV M mutants defective in nuclear localization fail to inhibit viral transcription.
SYNV replication and virion maturation take place in infected nuclei in which the M protein is also localized (53, 54). An M protein sequence (226GKVVRKRKSRK236) was postulated by Jackson et al. (49) to serve as a nuclear localization signal (NLS). To extend this hypothesis, we used a computer algorithm PSORT (https://wolfpsort.hgc.jp/) to identify four consecutive basic amino acids, 230RKRK233, as key NLS residues. To verify the NLS function of these residues, we mutated the 230RKRK233 residues to 230RARA233 and 230AAAA233 to generate the MRARA and MAAAA mutants. When fused to the cyan fluorescent protein (CFP) and transiently expressed in N. benthamiana epidermal cells, the wild-type M protein localized exclusively to the nuclei, whereas the MRARA and MAAAA mutants exhibited both cytoplasmic and nuclear localizations (Fig. 5A). These data demonstrate that the RKRK motif has an important role in nuclear import of the M protein. We next used bi-molecular fluorescence complementation (BiFC) to determine whether the wild-type and mutated M proteins interact with the N protein and where the interactions are located. As expected, N-M interactions were detected only in the nuclei, where they were associated with large subnuclear inclusions. In contrast, the N-MRARA and N-MAAAA interaction signals were present in both the cytoplasm and the nuclei, and the nuclear yellow fluorescent protein (YFP) foci were more numerous and much smaller than the N-M interaction foci (Fig. 5B), suggesting that they were unable to coalesce to form large foci. Moreover, since the N protein also has nuclear localization signals (60), the appearance of cytoplasmic YFP foci suggests that the interactions between N and M protein mutants occur primarily in the cytoplasm and that cytoplasmic N-M complexes were unable to be imported into the nuclei. These data indicate that the M NLS mutants are deficient in nuclear localization and interactions with N protein in the nuclei.
FIG 5.
Subcellular localization, interactions with the N protein, and the ability to suppress MR transcription by the SYNV M nuclear localization mutants. (A) Confocal micrographs showing RFP-histone 2B reporter protein (RFP-H2B) transgenic N. benthamiana leaf epidermal cells expressing the CFP-M, CFP-MRARA, and CFP-MAAAA fusions. Bar, 20 μm. (B) Bimolecular fluorescence complementation (BiFC) assays to determine interactions of the N protein with the wild-type M or the MRARA or MAAAA mutant protein. Leaf epidermal cells of the N. benthamiana RFP-H2B transgenic plants were agroinfiltrated to express the YFP N-terminal fusion to N (YN-N) and the YFP C-terminal fusions to M derivatives (YC-M). Confocal micrographs of YFP (BiFC), RFP-H2B (nucleus), and the merged channel are shown. Bar, 20 μm. (C) Fluorescence microscopy visualization of SYNV (−)MR reporter gene expression at 8 dpi in N. benthamiana leaves coinfiltrated to express either an empty vector control (Vec) or the M, MRARA, or MAAAA protein. Bar, 200 μm. (D) Average numbers of cells per field expressing GFP and RFP at 8 dpi in each SYNV MR treatment shown in panel C. Bars represent standard deviations. ***, P < 0.001; NS, not significant (by Student's t test; n = 20). (E) Western blots showing expression of the M, MRARA, or MAAAA protein with the N and P proteins in each group. The Coomassie blue-stained RuBisCO large subunit provides a loading control.
We next evaluated the abilities of the M NLS mutants to suppress MR reporter mRNA transcription. The M NLS mutants were transiently expressed in N. benthamiana leaves, along with components required for (−)MR activity, and the GFP and RFP expression levels were examined at 8 dpi by fluorescence microscopy. Despite having retained partial nuclear localization and interaction with the N protein, neither the MRARA nor the MAAAA protein had discernible inhibitory effects on MR reporter expression, as judged by fluorescence imaging (Fig. 5C) and quantitative analyses of the numbers of fluorescent foci (Fig. 5D). As a positive control, the wild-type M protein markedly decreased reporter expression (Fig. 5C and D). Western blot analysis of the protein samples extracted from the infiltrated leaf tissues showed that the two M mutants, especially the MAAAA mutant, accumulated to higher levels than the wild-type M protein (Fig. 5D). Therefore, the lack of inhibitory functions of the M NLS mutants was not a consequence of reduced protein expression but likely was due to compromised M protein interactions with the N protein in the nuclei that affect NC coiling.
rSYNV derivatives carrying the M NLS mutations are defective in SIE activities.
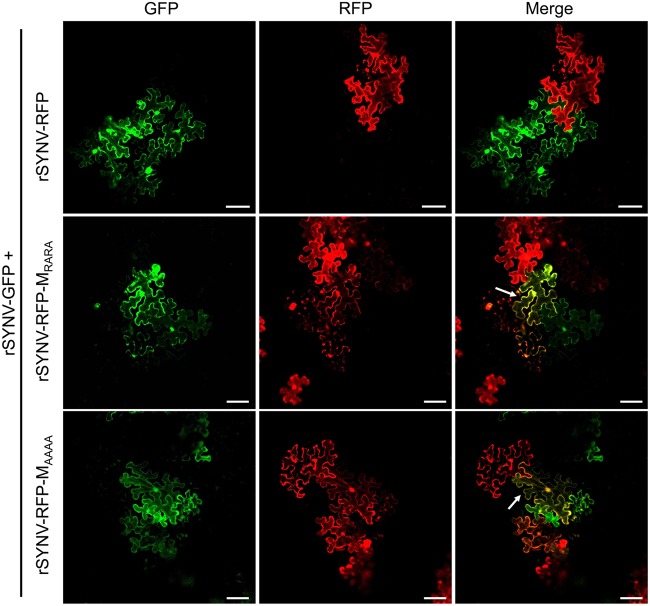
Having demonstrated that the M NLS mutants are unable to suppress viral transcription, we next introduced these mutations into the rSYNV-RFP genome and tested the abilities of the mutant viruses to inhibit superinfection by rSYNV-GFP. In these tests, N. benthamiana leaves were agroinfiltrated to initiate coinfection of rSYNV-GFP and either rSYNV-RFP, rSYNV-RFP-MRARA, or rSYNV-RFP MAAAA. With all combinations, discrete single-cell GFP and RFP foci were observed in infiltrated leaf tissues at 8 dpi, and the foci spread to surrounding cells by 15 dpi. As expected, cells coexpressing GFP and RFP were not observed at the margins of contiguous foci of the parental rSYNV-GFP/rSYNV-RFP coinfections, indicating SIE activities. In contrast, superinfection was evident in substantial numbers of the adjoining cells of merging foci of the rSYNV-GFP/rSYNV-RFP-MRARA and rSYNV-GFP/rSYNV-RFP-MAAAA combinations, as evidenced by yellow fluorescence in these cells (white arrows in Fig. 6). In three independent experiments, ∼14% and ∼17% of the boundary cells of adjacent foci were coinfected with the rSYNV-GFP/rSYNV-RFP-MRARA and rSYNV-GFP/rSYNV-RFP-MAAAA combinations, respectively. The numbers of superinfected cells in these combinations were only moderately lower than the 25% found in the rSYNV-GFP/rSYNV-RFP-ΔM infections but were much greater than the 1.5% coinfection rate in rSYNV-GFP/rSYNV-RFP infections (Table 1). From these data, we conclude that the ability of the M protein to suppress viral transcription is essential for exclusion of superinfection by related SYNV derivatives.
FIG 6.
SIE activities of rSYNV and rSYNV M nuclear localization mutants. N. benthamiana leaves were coinfected via agroinoculation with the virus pairs rSYNV-GFP and rSYNV-RFP, rSYNV-GFP and rSYNV-RFP- MRARA, or rSYNV-GFP and rSYNV-RFP- MAAAA. Contiguous GFP and RFP foci in inoculated leaf tissues at 15 dpi were imaged by confocal microscopy to show fluorescence exclusion between the foci. The white arrows indicate boundary cells expressing both GFP and RFP. Bar, 100 μm.
DISCUSSION
Although the SIE phenomena have been extensively studied with many positive-strand RNA viruses using a variety of methods and are known to be closely related to the viral life cycle, the underlying mechanisms are just beginning to emerge (reviewed in references 61 and 62). However, our knowledge about SIE elicited by plant negative-strand RNA viruses is particularly limited, and this is due in part to technical difficulties associated with genetic engineering of this group of viruses (52). In the current study, we used different fluorophore-tagged rSYNV variants to investigate their interactions in the N. benthamiana model host. Our coinfection and sequential infection experiments provide convincing evidence demonstrating that SIE/cross-protection occurs during SYNV infections at both the cellular and organismal levels. To our knowledge, this study represents the first demonstration of SIE for a negative-strand RNA plant virus.
SIE was documented for the animal rhabdovirus model, vesicular stomatitis virus (VSV), 3 decades ago, and the results demonstrated that the VSV transmembrane G protein is required for SIE and affects entry level processes (63). Further study has shown that G protein expressed from preexisting VSV infections blocks receptor-mediated endocytosis of secondary invasions by VSV strains (64). However, unlike human/animal rhabdoviruses, plant viruses are not known to penetrate host cells through a specific membrane surface receptor-mediated process, and their entry steps are presumably facilitated by insect feeding or physical damage to the cell wall. In addition, the SYNV G protein appears to be dispensable for cell-to-cell movement and entry into distal cells (51). Therefore, the proposed VSV mechanism is unlikely relevant to SYNV-triggered SIE, which probably occurs at postentry levels.
To narrow down possible viral SIE determinants, each of the SYNV proteins was ectopically expressed in SYNV-infected leaves, and these experiments led to the identification of the M protein as a specific inhibitor of SYNV local infections. A loss-of-function approach employing analysis of the rSYNV-RFP-ΔM and rSYNV-RFP-MUAA mutants provided direct evidence for the involvement of the M protein rather than M mRNA in eliciting SIE. Using a similar approach, CTV-encoded p33 has been previously found to be a viral determinant of SIE because a CTV mutant with a deletion in the p33 gene (CTVΔp33) failed to protect infected plants against the same strain at the whole-plant level (38, 39). Unlike the CTV p33 protein, which is dispensable for systemic infection and movement (65), the rSYNV-RFP-ΔM mutant is defective in systemic infections but is capable of localized infections (51). Therefore, we analyzed SIE between rSYNV-RFP-ΔM and rSYNV-GFP in locally infected cells, and the results showed that rSYNV-RFP-ΔM was deficient in SIE at the cellular level. Parallel experiments with the rSYNV-RFP-ΔG mutant failed to demonstrate a role for the G protein in SIE; hence, the SYNV SIE mechanism differs from the virus entry step proposed for VSV SIE.
In rhabdovirus particles, the matrix protein forms a bridge between the helical nucleocapsid core and the transmembrane G protein (66–69). Studies with VSV and other animal rhabdoviruses have shown that late in replication, the M protein coils gNC into a compact structure similar to the bullet-shaped core observed in rhabdovirus virion (70, 71). The condensed gNCs appear to be unable to function in mRNA transcription because transcriptase activity is suppressed under in vitro and in vivo conditions (55–58). Similar functions are likely shared by the M proteins encoded by plant rhabdoviruses, considering their conserved genome structure and replication strategy (49). Indeed, using a (−)MR system that can assemble with coexpressed core proteins into gNC in vivo, we showed that ectopic expression of the SYNV M protein inhibits reporter gene expression significantly.
We found that the SYNV M NLS mutants with weakened nuclear localizations were unable to suppress MR reporter gene expression. These data suggest that NC condensation and inhibition of transcription require high levels of the M protein in the nuclei in which SYNV replication occurs. Consequently, rSYNV-RFP-MRARA and rSYNV-RFP-MAAAA were defective in exerting SIE, most likely because the cell nuclei infected by these virus mutants lack sufficient amounts of M protein to initiate or maintain NC coiling of the challenging secondary SYNV invaders. This hypothesis is supported by in vitro VSV assembly/disassembly kinetics study, which has revealed that high M protein concentrations favor NC-M complex formation, with a dissociation constant in the order of 1 μM (72). An earlier VSV infection dynamics study indicated that virion assembly begins ∼2 h postinfection and peaks at ∼4 to 6 h postinfection (73). During this period of time, the concentration of intracellular M protein rises from about 1 to 30 μM (73), a level sufficient to drive assembly of NC-M complexes (72). After the initial NC-M complex formation, virion core assembly is further facilitated by driving forces provided by cooperative M self-interactions. VSV M protein multimerizes at physiological NaCl concentrations (74) and forms large aggregates assisted by nucleation sites near the N-terminal region (75). Previous BiFC assays have also revealed that SYNV M protein undergoes self-associations that appear as large punctuate subnuclear foci (76).
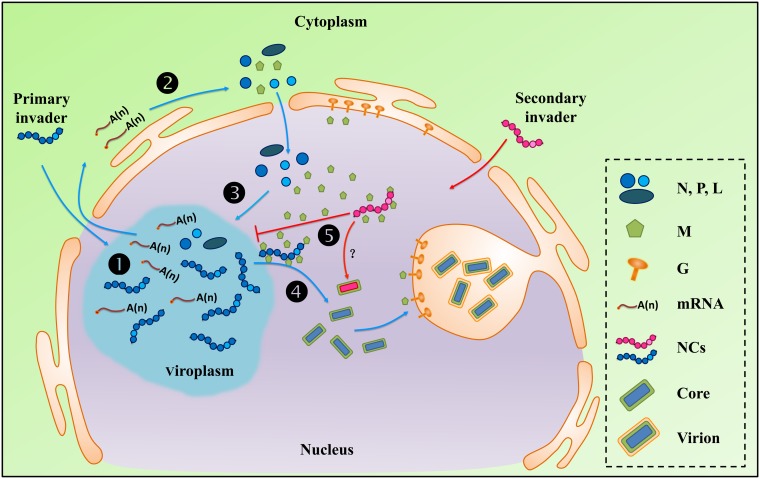
Combining knowledge about rhabdovirus replication cycles (49, 77) with the data presented in this study, we propose an infection phase-associated regulatory mechanism to explain SYNV SIE (Fig. 7). At the early to intermediate stages of replication following primary infection, the M protein accumulates to relatively low levels, owing to the progressive transcriptional attenuation signals present in each of the gene junction sequences (49, 77). This results in lower M mRNA transcription levels that than those of the 3′ N, P, and sc4 genes. At these stages, the M protein may associate with the newly formed gNCs but likely not in sufficient levels for complete NC condensation, and infected cells permit additional cycles of replication and transcription and NC accumulation. However, at later stages of SYNV replication when the M protein accumulates in the nuclei to levels sufficient to trigger NC condensation, a transition from replication to virion assembly occurs. At this stage, nuclei infected with a primary infecting SYNV strain will be actively engaged in nucleocapsid coiling, and incoming gNCs released from secondary invading SYNV will not uncoil sufficiently to initiate new infections. Consequently, cells supporting later stages of infection of the primary infecting virus strain will no longer be permissive for productive replication by the secondary invading virus (Fig. 7).
FIG 7.
Working model to illustrate SYNV M protein-mediated SIE. Upon entry into the cells, genomic nucleocapsids (gNCs) liberated from virions of the primary invader (shown in blue) are imported into the nuclei and initiate primary transcription of viral mRNAs (step 1). The primary mRNAs transcripts are exported to the cytoplasm for synthesis of viral proteins (step 2). The newly synthesized N, P, and L core proteins are subsequently imported into the nuclei where they form viroplasms and participate in secondary rounds of mRNA transcription and progeny antigenomic and genomic RNA replication and encapsidation to form progeny NCs (step 3). The M protein is also imported into the nuclei but accumulates to relatively low levels during the early to intermediate stages of replication owing to polar viral mRNA transcription. However, as cyclic waves of transcription and replication proceed (repeated steps 1 to 3), the accumulating M protein begins to initiate gNC coiling and quenching of viral mRNA transcription. Subsequently, coiled gNC cores interact with the carboxyl-terminal region of the transmembrane G protein and bud through the inner nuclear envelope to form mature virions (step 4). Incoming gNCs released from secondary invaders (shown in magenta) entering these cells are coiled or prevented from uncoiling by the abundant M proteins that have accumulated in the nuclei during the later stages of primary infection (step 5). This NC sequestration process abrogates mRNA transcription by the secondary invaders and results in SIE.
The SYNV cellular infection dynamics have not been characterized in detail, but assuming that they are similar to those of VSV (73), the time window for superinfection permission phase to superinfection exclusion phase can be in the range of a few hours. Given the nonsynchronous nature of virus infections, two SYNV variants that have entered the same cells are unlikely to initiate infection simultaneously or even within a very short interval. Hence, cells with established infections by an earlier SYNV variant would not permit reinfections by later arriving SYNV variants, and coinfections would be manifested by spatial separation of two variants in adjacent foci. However, if two variants do happen to initiate infections in the same cells within the permissive window, superinfections are possible, as reflected by the small percentage (∼1.5%) of cells expressing both GFP and RFP fluorescence after coinoculations of rSYNV-GFP and rSYNV-RFP (Table 1).
Our model resembles the TCV p28 model in that both models posit that a viral protein concentration-dependent infection phase transition accounts for superinfection versus SIE. In the TCV model, accumulating p28 replication protein produced by abundant progeny genomes transforms the infection cycle from a replication-active state to a replication-repressive state (postreplication phase). This transition is initiated by concentration-dependent p28 polymerization resulting in formation of large aggregates that sequester newly synthesized p28 monomers, including those translated from the secondary invading virus genomes (33). Another well-studied example of SIE in plant viruses is the CP-mediated cross-protection described for TMV and several other plant positive-strand RNA viruses (32, 42, 43, 78). It has been proposed that TMV CP-mediated SIE involves multiple modes of interference, including preventing uncoating and/or blocking replicase assembly of the secondary invading virus genomes (42, 79, 80). With TMV, these activities require the ability of CP to assemble into large aggregates by self-association and to bind viral RNA (81–83). Both the TMV SIE model and our model postulate that that an abundant viral assembly factor (TMV CP and SYNV M) produced during the late stages of primary infections is responsible for coating/sequestering of viral infection units (i.e., TMV gRNAs and SYNV gNCs) of secondary invaders to prevent superinfection. Therefore, it appears that SIE by different viruses may involve different functions of specific viral proteins, but they all are manifested as interference with the early infection stages of the secondary invading virus by the more advanced stages involved in synthesis of the primary invading virus. Productive virus infections require coordination of a chain of transient and sequential events including uncoating, translation, replication, cell-to-cell movement, and virion assembly. (Note that in the case of negative-strand RNA viruses, a transcription step precedes translation.) Transition from one phase to another often involves competition for viral proteins or RNAs and may even result in termination of early events (84). Consequently, secondary invaders entering previously infected cells face the challenge of coordinating with existing infection stages to establish superinfections. In this sense, SIE can be seen as a side effect caused by infection phase transitions.
MATERIALS AND METHODS
Plasmids and recombinant viruses.
Plasmids used to recover rSYNV-GFP, rSYNV-RFP, rSYNV-RFPΔM, and rSYNV-RFPΔG have been described in previous studies (51, 54). The PVX-GFP plasmid was constructed as described by van Wezel et al. (85). To engineer rSYNV-RFP variants with mutations in the M gene, the AUG initiation codon of the M coding region was changed to a UAA stop codon by site-directed mutagenesis to generate the MUAA mutant. Similarly, the M 230RKRK233 nuclear import residues were mutated to RARA or AAAA to generate the MRARA and MAAAA mutants. To introduce these M mutations into the rSYNV-RFP genome, an F1 fragment containing a portion of the P gene was amplified from the pSYNV-RFP plasmid by using the primer pair SYNV/P/NheI/F (5′-AAGAGAAGGGGCTAGCATGTC-3′) and SYNV/MUAA/R (5′- GTATATACCTGCTTATCTGAAATACAATAGAGATAACCTTG-3′), and an F2 fragment containing a portion of the M gene was amplified by using the primer pair SYNV/MUAA/F (5′-TAAGCAGGTATATACGCAGTTTCAA-3′) and SYNV/M/PmlI/R (5′-CATCACGACTGACACGTGACCT-3′). The two fragments were infused with NheI and PmlI doubly digested pSYNV-RFP to generate pSYNV-RFP-MUAA. A similar strategy was used to generate the pSYNV-RFP-MRARA and MAAAA mutants with the primers SYNV/sc4/BstZ17I/F (5′-GCAAGTACTTTGGTATACAAGAAAGG-3′) and SYNV/G/BstZ17I/R (5′-TCTTGAATACTGGTATACTTATTCCCACA-3′).
The genomic-sense SYNV MR plasmid [pSYNV-MRGFP-RFP (−)] was generated from the antigenomic-sense minireplicon [MRGFP-RFP (+)] described by Ganesan et al. (59). Briefly, the MR sequence was amplified by PCR with the MR/trailer/F (5′-CCGGTATCCCGGGTCAGAGACAAAAGCTCAGAACAATCC-3′) and MR/leader/R (5′-ATGCCATGCCGACCCAGAGACAGAAACTCAGAAAATACAATC-3′) primers, and the region comprising the vector backbone, the hammerhead ribozyme, and hepatitis D virus ribozyme sequences was amplified with the MR/backbone/F (5′-GGGTCGGCATGGCATCTCCACC-3′) and MR/backbone/R (5′-GACCCGGGATACCGGGTTTCGG-3′) primers. The two fragments were circularized in the presence of the In-Fusion enzyme mixture to generate the MR in the negative-strand orientation. This was facilitated by 15-nt homologous sequences incorporated into the 5′ ends of the primers (underlined in the sequences).
To construct plasmids for transient expression of the M protein, the MUAA, MRARA, and the MAAAA mutant proteins, as well as the sc4 protein, the cDNA sequences of these genes were amplified by PCR using specific primers (these primer sequences will be made available upon request). The recovered PCR products were then inserted into the pGD binary vector (86) by In-Fusion cloning (Clontech). The pGD-CFP-M plasmid used for transient expression of the CFP-M fusion has been described previously (54) and was used to generate the expression plasmids pGD-CFP-MRARA and pGD-CFP-MAAAA.
To permit transient coexpression of the viral proteins and GFP, we first constructed a pGD-X/GFP dual-expression vector. For this purpose, we linearized the pGD-GFP vector (86) by MluI restriction digestion. Then the region spanning the 35S promoter, the multiple cloning sites, and the Nos terminator sequence in the pGD vector was amplified with the pGD/MluI 15 nt/F (5′-CGAATTAATTACGCGTCATGGTGGAGCACGACACTCTCGT-3′) and pGD/MluI 15 nt/R (5′-CTCCACCATGACGCGTCCCGATCTAGTAACATAGATGACA-3′) primers. The PCR product was designed to share a 15-nt homologous sequence (underlined) with the ends of the linearized vector. An In-Fusion cloning mixture (Clontech) was used to assemble the PCR product and the linearized vector in vitro. The coding sequences of the SYNV N, P, sc4, M, and G genes were amplified by PCR and individually inserted into the XbaI restriction site of pGD-X/GFP to generate the respective dual-expression plasmids.
To construct the BiFC vectors, the coding sequences of the N protein, the wild-type M protein, and the M mutants were fused to the N-terminal and C-terminal fragments of the enhanced YFP gene in the p2YN and p2YC vectors (87) by a procedure similar to that of Sun et al. (54).
Agroinfiltration and agroinfection assays.
SYNV minireplicon assays were carried out by infiltration of N. benthamiana leaf tissues with Agrobacterium tumefaciens strains to deliver the pSYNV MRGFP-RFP plasmid, the pGD-NPL plasmid, and the p19, γb, and P1/HC-Pro viral suppressors of RNA silencing (VSR) expression plasmid, as described previously (51, 88). In addition, agrobacterial strains containing individual plasmids for expression of wild-type or mutated M proteins or of the sc4 protein were mixted with the above-mentioned mixtures at a ratio of 1:6 (vol/vol) with a total optical density (OD) A600 of 0.7. Infiltrated leaves were observed under a fluorescence microscope at 8 days postinfiltration (dpi) to test their effects on reporter gene expression. PVX-GFPΔp25 replicon assays were conducted similarly, with the exception that A. tumefaciens strains harboring PVX-GFPΔp25 were diluted 250-fold and mixed in a 1:1:1 ratio with Agrobacterium cultures containing the three viral VSR plasmids (p19, γb, and P1/HC-Pro) (OD A600 = 0.6) and the SYNV M protein (OD A600 = 0.3).
SYNV agroinfection assays were conducted similarly to the minireplicon agroinfiltration assays (59), except that agrobacterial strains harboring the pSYNV-RFP or pSYNV-GFP derivatives were substituted for strains carrying the pSYNV MRGFP-RFP plasmid. To test SIE in mixed agroinfection experiments, agrobacterial strains harboring the pSYNV-RFP and pSYNV-GFP derivatives were mixed in equal volumes prior to infiltration. Infiltrated leaves were observed with a fluorescence microscope at 15 dpi, and the numbers of cells at the boundaries of the foci showing both GFP and RFP were counted. To assess the effects of overexpressed SYNV proteins on the localized spread of rSYNV-RFP, A. tumefaciens strains individually harboring pGD plasmids for expression of the SYNV N, P, sc4, M, or G protein were added to the agroinfection mixtures at ratios of 1:9 (vol/vol) with a total OD A600 of 0.7. Infiltrated leaves were observed with a fluorescence microscope at 8 dpi and subsequently at 12 dpi, and the numbers of cells in each RFP focus were counted.
To assess the effects of the SYNV M protein on the localized movement of PVX-GFP, A. tumefaciens EHA 105 strains harboring PVX-GFP (OD A600 = 1.0) were diluted 1,000-fold and mixed in a 1:1:1 ratio with Agrobacterium cultures containing the three viral VSR plasmids (p19, γb, and P1/HC-Pro) (OD A600 = 0.6) and the SYNV M protein (OD A600 = 0.3). The areas of PVX-GFP infection foci at 6 dpi were measured by LSM software Zen 2012 (Carl Zeiss, Germany).
Mechanical transmission.
SYNV mechanical transmission was carried out as previously described (89). Briefly, young emerging N. benthamiana leaves (∼2 g) systemically infected with the SYNV derivatives were ground with 5-ml of cold (∼4°C) inoculation buffer containing 5% sodium sulfite (grams/liter) and 2% Celite (grams/liter), and the resulting brei was gently rubbed by hand onto young expanded leaves of 6- to 8-week-old N. benthamiana plants. The inoculated plants were placed in growth chambers under the environmental conditions described above.
Protein analysis.
Total protein samples separated by 12% SDS-PAGE were stained with Coomassie blue or transferred to nitrocellulose membranes and detected with polyclonal antibodies specific to disrupted SYNV virions (90) or the sc4 protein (50). In some cases, the membranes were stripped and reprobed with GFP and RFP monoclonal antibodies (Abcam, Cambridge, UK).
BiFC.
BiFC plasmids were introduced individually into A. tumefaciens EHA105 strains by electroporation. Equal volumes of bacterial cultures (A600 = 0.5) harboring the p2YN- and p2YC-based plasmids were mixed and infiltrated into leaves of transgenic N. benthamiana plants expressing the RFP-histone 2B reporter protein (RFP-H2B) (91). YFP and RFP signals were visualized at 36 to 48 h after infiltration into expanded leaves and captured by Zeiss 780 confocal microscopy (Carl Zeiss, Germany).
Fluorescence microscopy and confocal laser scanning microscopy.
An epifluorescence microscope (Zeiss SteREO Lumar V12) was used to visualize SYNV MR fluorescent foci and to determine the movement of rSYNV derivatives and PVX-GFP in infiltrated leaf tissues and systemically infected leaves. Fluorescence in N. benthamiana leaves was captured with a Lumar 31 filter set for RFP detection (excitation, 565/30 nm; emission, 620/60 nm) and a Lumar 38 filter set for GFP detection (excitation, 470/40; emission, 525/50). High-resolution visualization of the fluorescent proteins was performed with a Zeiss 780 confocal microscope. GFP, RFP, YFP, and CFP fluorescence was excited with the 470-, 545-, 488-, and 405-nm laser lines, respectively. For coinfection experiments using recombinant virus derivatives expressing GFP or RFP, a series of 7 to 11 z-stacks were acquired using a 20× objective (NeoFluor air objective; numerical aperture [NA], 0.50[ Zeiss]), spanning approximately 18 to 30 μm in the z dimension, with a 3-μm step size. All images were processed with LSM software Zen 2012 (Carl Zeiss, Germany).
Northern hybridization.
Total RNA was isolated from infiltrated regions of N. benthamiana leaves using TRIzol reagent (TaKaRa, Dalian, China), resolved in 1.5% agarose-formaldehyde gels and probed with a digoxin-labeled complementary GFP oligonucleotide probe (5′-CCTCGAACTTCACCTCGGCGCGGGTCTTGTAGTTGCCGTCGTCCTTGAAGAAGATGGTG-3′) (Invitrogen, Shanghai, China). The blots were developed with horseradish peroxidase-conjugated anti-digoxin antibody and chemiluminescent substrates available in a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics, Basel, Switzerland) and imaged using an ImageQuant LAS 4000 Mini biomolecular imager instrument (GE Healthcare, Little Chalfont, United Kingdom). To generate RNA size markers for GFP and MR, we amplified the cDNAs using the primer pair T7GFPF (5′-TAATACGACTCACTATAGGGATGGTGAGCAAGGGCGAGGA-3′) and GFPR (5′-TTACTTGTACAGCTCGTCCATGC-3′) and the pair T7MRF (5′-TAATACGACTCACTATAGAGAGACAGAAACTCAGAAAATACAATC-3′) and MRR (5′-AGAGACAAAAGCTCAGAACAATCC-3′), respectively. The forward primers contained the T7 promoter positioned at the 5′terminus (underlined), and the PCR products were transcribed in vitro by T7 RNA polymerase as described previously (88).
ACKNOWLEDGMENTS
We thank Michael Goodin (University of Kentucky, USA) for sharing RFP-H2B transgenic N. benthamiana seeds and Fei Yan (Ningbo University, China) for providing the PVX-GFPΔp25 construct.
This work was supported by National Natural Science Foundation of China (grants 31671996 and 31870142).
We declare that we have no conflicts of interest.
REFERENCES
- 1.Sauri CJ, Earhart CF. 1971. Superinfection with bacteriophage T4: inverse relationship between genetic exclusion and membrane association of deoxyribonucleic acid of secondary bacteriophage. J Virol 8:856–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson CW, Eigner J. 1971. Breakdown and exclusion of superinfecting T-even bacteriophage in Escherichia coli. J Virol 8:869–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steck FT, Rubin H. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology 29:628–641. doi: 10.1016/0042-6822(66)90287-x. [DOI] [PubMed] [Google Scholar]
- 4.Bratt MA, Rubin H. 1968. Specific interference among strains of Newcastle disease virus. III. Mechanisms of interference. Virology 35:395–407. doi: 10.1016/0042-6822(68)90218-3. [DOI] [PubMed] [Google Scholar]
- 5.Delwart EL, Panganiban AT. 1989. Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J Virol 63:273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biryukov J, Meyers C. 2018. Superinfection exclusion between two high-risk human papillomavirus (HPV) types during a co-infection. J Virol 92:e01993-17. doi: 10.1128/JVI.01993-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Folimonova S. 2013. Developing an understanding of cross-protection by citrus tristeza virus. Front Microbiol 4:76. doi: 10.3389/fmicb.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XF, Qu F. 2016. Cross protection of plant viruses: recent developments and mechanistic implications, p 241–250. In Wang A, Zhou X (ed), Current research topics in plant virology. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 9.Steck FT, Rubin H. 1966. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology 29:642–653. doi: 10.1016/0042-6822(66)90288-1. [DOI] [PubMed] [Google Scholar]
- 10.Lee YM, Tscherne DM, Yun SI, Frolov I, Rice CM. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J Virol 79:3231–3242. doi: 10.1128/JVI.79.6.3231-3242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laliberte J, Moss B. 2014. A novel mode of poxvirus superinfection exclusion that prevents fusion of the lipid bilayers of viral and cellular membranes. J Virol 88:9751–9768. doi: 10.1128/JVI.00816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marschall M, Meier-Ewert H, Herrler G, Zimmer G, Maassab HF. 1997. The cell receptor level is reduced during persistent infection with influenza C virus. Arch Virol 142:1155–1164. doi: 10.1007/s007050050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2:52. doi: 10.1186/1742-4690-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams RH, Brown DT. 1985. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J Virol 54:351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karpf AR, Lenches E, Strauss EG, Strauss JH, Brown DT. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J Virol 71:7119–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaller T, Appel N, Koutsoudakis G, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J Virol 81:4591–4603. doi: 10.1128/JVI.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tscherne DM, Evans MJ, von Hahn T, Jones CT, Stamataki Z, McKeating JA, Lindenbach BD, Rice CM. 2007. Superinfection exclusion in cells infected with hepatitis C virus. J Virol 81:3693–3703. doi: 10.1128/JVI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou G, Zhang B, Lim PY, Yuan Z, Bernard KA, Shi PY. 2009. Exclusion of West Nile virus superinfection through RNA replication. J Virol 83:11765–11776. doi: 10.1128/JVI.01205-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellenberg P, Linero FN, Scolaro LA. 2007. Superinfection exclusion in BHK-21 cells persistently infected with Junin virus. J Gen Virol 88:2730–2739. doi: 10.1099/vir.0.83041-0. [DOI] [PubMed] [Google Scholar]
- 20.Gal-On A, Shiboleth YM. 2006. Cross-protection, p 261–288. In Loebenstein G, Carr JP (ed), Natural resistance mechanisms of plants to viruses. Springer, Dordrecht, Netherlands. [Google Scholar]
- 21.Ziebell H, Carr JP. 2010. Cross-protection: a century of mystery. Adv Virus Res 76:211–264. doi: 10.1016/S0065-3527(10)76006-1. [DOI] [PubMed] [Google Scholar]
- 22.Mckinney HH. 1929. Mosaic diseases in the Canary Islands, West Africa and Gibraltar. J Agri Res 39:577–578. [Google Scholar]
- 23.Hughes JDA, Ollennu L. 1994. Mild strain protection of cocoa in Ghana against cocoa swollen shoot virus—a review. Plant Pathology 43:442–457. doi: 10.1111/j.1365-3059.1994.tb01578.x. [DOI] [Google Scholar]
- 24.Gonsalves D. 1998. Control of papaya ringspot virus in papaya: a case study. Annu Rev Phytopathol 36:415–437. doi: 10.1146/annurev.phyto.36.1.415. [DOI] [PubMed] [Google Scholar]
- 25.Lee RF, Keremane ML. 2013. Mild strain cross protection of tristeza: a review of research to protect against decline on sour orange in Florida. Front Microbiol 4:259. doi: 10.3389/fmicb.2013.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietrich C, Maiss E. 2003. Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J Gen Virol 84:2871–2876. doi: 10.1099/vir.0.19245-0. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi T, Sugawara T, Yamatsuta T, Isogai M, Natsuaki T, Yoshikawa N. 2007. Analysis of the spatial distribution of identical and two distinct virus populations differently labeled with cyan and yellow fluorescent proteins in coinfected plants. Phytopathology 97:1200–1206. doi: 10.1094/PHYTO-97-10-1200. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi T, Yoshikawa N. 2008. Analysis of cell-to-cell and long-distance movement of apple latent spherical virus in infected plants using green, cyan, and yellow fluorescent proteins. Methods Mol Biol 451:545–554. doi: 10.1007/978-1-59745-102-4_37. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Jara P, Fraile A, Canto T, Garcia-Arenal F. 2009. The multiplicity of infection of a plant virus varies during colonization of its eukaryotic host. J Virol 83:7487–7494. doi: 10.1128/JVI.00636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Julve JM, Gandia A, Fernandez-Del-Carmen A, Sarrion-Perdigones A, Castelijns B, Granell A, Orzaez D. 2013. A coat-independent superinfection exclusion rapidly imposed in Nicotiana benthamiana cells by tobacco mosaic virus is not prevented by depletion of the movement protein. Plant Mol Biol 81:553–564. doi: 10.1007/s11103-013-0028-1. [DOI] [PubMed] [Google Scholar]
- 31.Miyashita S, Kishino H. 2010. Estimation of the size of genetic bottlenecks in cell-to-cell movement of soil-borne wheat mosaic virus and the possible role of the bottlenecks in speeding up selection of variations in trans-acting genes or elements. J Virol 84:1828–1837. doi: 10.1128/JVI.01890-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatineni S, French R. 2016. The coat protein and NIa protease of two Potyviridae family members independently confer superinfection exclusion. J Virol 90:10886–10905. doi: 10.1128/JVI.01697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang XF, Sun R, Guo Q, Zhang S, Meulia T, Halfmann R, Li D, Qu F. 2017. A self-perpetuating repressive state of a viral replication protein blocks superinfection by the same virus. PLoS Pathog 13:e1006253. doi: 10.1371/journal.ppat.1006253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall JS, French R, Hein GL, Morris TJ, Stenger DC. 2001. Three distinct mechanisms facilitate genetic isolation of sympatric wheat streak mosaic virus lineages. Virology 282:230–236. doi: 10.1006/viro.2001.0841. [DOI] [PubMed] [Google Scholar]
- 35.Ziebell H, Carr JP. 2009. Effects of dicer-like endoribonucleases 2 and 4 on infection of Arabidopsis thaliana by cucumber mosaic virus and a mutant virus lacking the 2b counter-defence protein gene. J Gen Virol 90:2288–2292. doi: 10.1099/vir.0.012070-0. [DOI] [PubMed] [Google Scholar]
- 36.Ziebell H, Payne T, Berry JO, Walsh JA, Carr JP. 2007. A cucumber mosaic virus mutant lacking the 2b counter-defence protein gene provides protection against wild-type strains. J Virol 88:2862–2871. doi: 10.1099/vir.0.83138-0. [DOI] [PubMed] [Google Scholar]
- 37.Folimonova SY, Robertson CJ, Shilts T, Folimonov AS, Hilf ME, Garnsey SM, Dawson WO. 2010. Infection with strains of citrus tristeza virus does not exclude superinfection by other strains of the virus. J Virol 84:1314–1325. doi: 10.1128/JVI.02075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folimonova SY. 2012. Superinfection exclusion is an active virus-controlled function that requires a specific viral protein. J Virol 86:5554–5561. doi: 10.1128/JVI.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergua M, Zwart MP, El-Mohtar C, Shilts T, Elena SF, Folimonova SY. 2014. A viral protein mediates superinfection exclusion at the whole-organism level but is not required for exclusion at the cellular level. J Virol 88:11327–11338. doi: 10.1128/JVI.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergua M, Kang SH, Folimonova SY. 2016. Understanding superinfection exclusion by complex populations of citrus tristeza virus. Virology 499:331–339. doi: 10.1016/j.virol.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Zhang XF, Guo J, Zhang X, Meulia T, Paul P, Madden LV, Li D, Qu F. 2015. Random plant viral variants attain temporal advantages during systemic infections and in turn resist other variants of the same virus. Sci Rep 5:15346. doi: 10.1038/srep15346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beachy RN. 1999. Coat-protein-mediated resistance to tobacco mosaic virus: discovery mechanisms and exploitation. Philos Trans R Soc Lond B Biol Sci 354:659–664. doi: 10.1098/rstb.1999.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valkonen JP, Rajamaki ML, Kekarainen T. 2002. Mapping of viral genomic regions important in cross-protection between strains of a potyvirus. Mol Plant Microbe Interact 15:683–692. doi: 10.1094/MPMI.2002.15.7.683. [DOI] [PubMed] [Google Scholar]
- 44.Atallah OO, Kang SH, El-Mohtar CA, Shilts T, Bergua M, Folimonova SY. 2016. A 5′-proximal region of the citrus tristeza virus genome encoding two leader proteases is involved in virus superinfection exclusion. Virology 489:108–115. doi: 10.1016/j.virol.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 45.Lindbo JA, Falk BW. 2017. The impact of “coat protein-mediated virus resistance” in applied plant pathology and basic research. Phytopathology 107:624–634. doi: 10.1094/PHYTO-12-16-0442-RVW. [DOI] [PubMed] [Google Scholar]
- 46.Klas FE, Fuchs M, Gonsalves D. 2006. Comparative spatial spread overtime of zucchini yellow mosaic virus (ZYMV) and watermelon mosaic virus (WMV) in fields of transgenic squash expressing the coat protein genes of ZYMV and WMV, and in fields of nontransgenic squash. Transgenic Res 15:527–541. doi: 10.1007/s11248-006-9001-y. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S, Sun R, Guo Q, Zhang XF, Qu F. 2019. Repression of turnip crinkle virus replication by its replication protein p88. Virology 526:165–172. doi: 10.1016/j.virol.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 48.Heaton LA, Zuidema D, Jackson AO. 1987. Structure of the M2 protein gene of sonchus yellow net virus. Virology 161:234–241. doi: 10.1016/0042-6822(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 49.Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. 2005. Biology of plant rhabdoviruses. Annu Rev Phytopathol 43:623–660. doi: 10.1146/annurev.phyto.43.011205.141136. [DOI] [PubMed] [Google Scholar]
- 50.Scholthof KB, Hillman BI, Modrell B, Heaton LA, Jackson AO. 1994. Characterization and detection of sc4: a sixth gene encoded by sonchus yellow net virus. Virology 204:279–288. doi: 10.1006/viro.1994.1532. [DOI] [PubMed] [Google Scholar]
- 51.Wang Q, Ma X, Qian S, Zhou X, Sun K, Chen X, Zhou X, Jackson AO, Li Z. 2015. Rescue of a plant negative-strand RNA virus from cloned cDNA: Insights into enveloped plant virus movement and morphogenesis. PLoS Pathog 11:e1005223. doi: 10.1371/journal.ppat.1005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson AO, Li Z. 2016. Developments in plant negative-strand RNA virus reverse genetics. Annu Rev Phytopathol 54:469–498. doi: 10.1146/annurev-phyto-080615-095909. [DOI] [PubMed] [Google Scholar]
- 53.Goodin MM, Chakrabarty R, Yelton S, Martin K, Clark A, Brooks R. 2007. Membrane and protein dynamics in live plant nuclei infected with sonchus yellow net virus, a plant-adapted rhabdovirus. J Gen Virol 88:1810–1820. doi: 10.1099/vir.0.82698-0. [DOI] [PubMed] [Google Scholar]
- 54.Sun K, Zhou X, Lin W, Zhou X, Jackson AO, Li Z. 2018. Matrix-glycoprotein interactions required for budding of a plant nucleorhabdovirus and induction of inner nuclear membrane invagination. Mol Plant Pathol 19:2288–2301. doi: 10.1111/mpp.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clinton GM, Little SP, Hagen FS, Huang AS. 1978. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell 15:1455–1462. doi: 10.1016/0092-8674(78)90069-7. [DOI] [PubMed] [Google Scholar]
- 56.Carroll AR, Wagner RR. 1979. Role of the membrane (M) protein in endogenous inhibition of in vitro transcription by vesicular stomatitis virus. J Virol 29:134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De BP, Thornton GB, Luk D, Banerjee AK. 1982. Purified matrix protein of vesicular stomatitis virus blocks viral transcription in vitro. Proc Natl Acad Sci U S A 79:7137–7141. doi: 10.1073/pnas.79.23.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finke S, Conzelmann KK. 2003. Dissociation of rabies virus matrix protein functions in regulation of viral RNA synthesis and virus assembly. J Virol 77:12074–12082. doi: 10.1128/jvi.77.22.12074-12082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganesan U, Bragg JN, Deng M, Marr S, Lee MY, Qian S, Shi M, Kappel J, Peters C, Lee Y, Goodin MM, Dietzgen RG, Li Z, Jackson AO. 2013. Construction of a sonchus yellow net virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses. J Virol 87:10598–10611. doi: 10.1128/JVI.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodin MM, Austin J, Tobias R, Fujita M, Morales C, Jackson AO. 2001. Interactions and nuclear import of the N and P proteins of sonchus yellow net virus, a plant nucleorhabdovirus. J Virol 75:9393–9406. doi: 10.1128/JVI.75.19.9393-9406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang XF, Zhang S, Guo Q, Sun R, Wei T, Qu F. 2018. A new mechanistic model for viral cross protection and superinfection exclusion. Front Plant Sci 9:40. doi: 10.3389/fpls.2018.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziebell H, Macdiarmid R. 2017. Prospects for engineering and improvement of cross-protective virus strains. Curr Opin Virol 26:8–14. doi: 10.1016/j.coviro.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Whitaker-Dowling P, Youngner JS, Widnell CC, Wilcox DK. 1983. Superinfection exclusion by vesicular stomatitis virus. Virology 131:137–143. doi: 10.1016/0042-6822(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 64.Simon KO, Cardamone JJ Jr, Whitaker-Dowling PA, Youngner JS, Widnell CC. 1990. Cellular mechanisms in the superinfection exclusion of vesicular stomatitis virus. Virology 177:375–379. doi: 10.1016/0042-6822(90)90494-C. [DOI] [PubMed] [Google Scholar]
- 65.Tatineni S, Robertson CJ, Garnsey SM, Bar-Joseph M, Gowda S, Dawson WO. 2008. Three genes of citrus tristeza virus are dispensable for infection and movement throughout some varieties of citrus trees. Virology 376:297–307. doi: 10.1016/j.virol.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 66.Zakowski JJ, Wagner RR. 1980. Localization of membrane-associated proteins in vesicular stomatitis virus by use of hydrophobic membrane probes and cross-linking reagents. J Virol 36:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lenard J, Vanderoef R. 1990. Localization of the membrane-associated region of vesicular stomatitis virus M protein at the N terminus, using the hydrophobic, photoreactive probe 125I-TID. J Virol 64:3486–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mebatsion T, Weiland F, Conzelmann KK. 1999. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol 73:242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jackson AO. 1978. Partial characterization of the structural proteins of sonchus yellow net virus. Virology 87:172–181. doi: 10.1016/0042-6822(78)90169-1. [DOI] [PubMed] [Google Scholar]
- 70.Newcomb WW, Brown JC. 1981. Role of the vesicular stomatitis virus matrix protein in maintaining the viral nucleocapsid in the condensed form found in native virions. J Virol 39:295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newcomb WW, Tobin GJ, McGowan JJ, Brown JC. 1982. In vitro reassembly of vesicular stomatitis virus skeletons. J Virol 41:1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lyles DS, McKenzie MO. 1998. Reversible and irreversible steps in assembly and disassembly of vesicular stomatitis virus: equilibria and kinetics of dissociation of nucleocapsid-M protein complexes assembled in vivo. Biochemistry 37:439–450. doi: 10.1021/bi971812j. [DOI] [PubMed] [Google Scholar]
- 73.Lyles DS, McKenzie MO, Ahmed M, Woolwine SC. 1996. Potency of wild-type and temperature-sensitive vesicular stomatitis virus matrix protein in the inhibition of host-directed gene expression. Virology 225:172–180. doi: 10.1006/viro.1996.0585. [DOI] [PubMed] [Google Scholar]
- 74.McCreedy BJ Jr, McKinnon KP, Lyles DS. 1990. Solubility of vesicular stomatitis virus M protein in the cytosol of infected cells or isolated from virions. J Virol 64:902–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gaudin Y, Barge A, Ebel C, Ruigrok R. 1995. Aggregation of VSV M protein is reversible and mediated by nucleation sites: implications for viral assembly. Virology 206:28–37. doi: 10.1016/S0042-6822(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 76.Min BE, Martin K, Wang R, Tafelmeyer P, Bridges M, Goodin M. 2010. A host-factor interaction and localization map for a plant-adapted rhabdovirus implicates cytoplasm-tethered transcription activators in cell-to-cell movement. Mol Plant Microbe Interact 23:1420–1432. doi: 10.1094/MPMI-04-10-0097. [DOI] [PubMed] [Google Scholar]
- 77.Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol 283:61–119. [DOI] [PubMed] [Google Scholar]
- 78.Sherwood JL, Fulton RW. 1982. The specific involvement of coat protein in tobacco mosaic virus cross protection. Virology 119:150–158. doi: 10.1016/0042-6822(82)90072-1. [DOI] [PubMed] [Google Scholar]
- 79.Bendahmane M, Chen I, Asurmendi S, Bazzini AA, Szecsi J, Beachy RN. 2007. Coat protein-mediated resistance to TMV infection of Nicotiana tabacum involves multiple modes of interference by coat protein. Virology 366:107–116. doi: 10.1016/j.virol.2007.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Asurmendi S, Berg RH, Koo JC, Beachy RN. 2004. Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc Natl Acad Sci U S A 101:1415–1420. doi: 10.1073/pnas.0307778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bendahmane M, Fitchen JH, Zhang G, Beachy RN. 1997. Studies of coat protein-mediated resistance to tobacco mosaic tobamovirus: correlation between assembly of mutant coat proteins and resistance. J Virol 71:7942–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu B, Stubbs G, Culver JN. 1998. Coat protein interactions involved in tobacco mosaic tobamovirus cross-protection. Virology 248:188–198. doi: 10.1006/viro.1998.9280. [DOI] [PubMed] [Google Scholar]
- 83.Asurmendi S, Berg RH, Smith TJ, Bendahmane M, Beachy RN. 2007. Aggregation of TMV CP plays a role in CP functions and in coat-protein-mediated resistance. Virology 366:98–106. doi: 10.1016/j.virol.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Makinen K, Hafren A. 2014. Intracellular coordination of potyviral RNA functions in infection. Front Plant Sci 5:110. doi: 10.3389/fpls.2014.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Wezel R, Liu H, Tien P, Stanley J, Hong Y. 2001. Gene C2 of the monopartite geminivirus tomato yellow leaf curl virus-China encodes a pathogenicity determinant that is localized in the nucleus. Mol Plant Microbe Interact 14:1125–1128. doi: 10.1094/MPMI.2001.14.9.1125. [DOI] [PubMed] [Google Scholar]
- 86.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. 2002. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31:375–383. doi: 10.1046/j.1365-313X.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- 87.Yang X, Baliji S, Buchmann RC, Wang H, Lindbo JA, Sunter G, Bisaro DM. 2007. Functional modulation of the geminivirus AL2 transcription factor and silencing suppressor by self-interaction. J Virol 81:11972–11981. doi: 10.1128/JVI.00617-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qian S, Chen X, Sun K, Zhang Y, Li Z. 2017. Capped antigenomic RNA transcript facilitates rescue of a plant rhabdovirus. Virol J 14:113. doi: 10.1186/s12985-017-0776-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jackson AO, Wagner JD. 1998. Procedures for plant rhabdovirus purification, polyribosome isolation, and replicase extraction. Methods Mol Biol 81:77–97. doi: 10.1385/0-89603-385-6:77. [DOI] [PubMed] [Google Scholar]
- 90.Jackson AO, Christie SR. 1977. Purification and some physicochemical properties of sonchus yellow net virus. Virology 77:344–355. doi: 10.1016/0042-6822(77)90431-7. [DOI] [PubMed] [Google Scholar]
- 91.Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]