HIV, the virus that causes AIDS, is responsible for millions of infections and deaths annually. Despite intense research for the past 25 years, there remains no safe and effective vaccine available. The significance of this work is in identifying the pros and cons of adding a protein boost to an already well-established DNA/MVA HIV vaccine that is currently being tested in the clinic. Characterizing the effects of the protein boost can allow researchers going forward to design vaccines that generate responses that will be more effective against HIV. Our results in rhesus macaques show that boosting with a specific HIV envelope protein does not significantly boost antibody responses that were identified as immune correlates of protection in a moderately successful RV144 HIV vaccine trial in humans and highlight the need for the development of improved HIV envelope immunogens.
KEYWORDS: DNA/MVA, SHIV challenge, V1V2, V2 HS, gp140 protein, human immunodeficiency virus
ABSTRACT
The RV144 human immunodeficiency virus type 1 (HIV-1) vaccine trial showed a strong association between anti-gp70 V1V2 scaffold (V1V2) and anti-V2 hot spot peptide (V2 HS) antibody responses and reduced risk of HIV infection. Accordingly, a primary goal for HIV vaccines is to enhance the magnitude and breadth of V1V2 and V2 HS antibody responses in addition to neutralizing antibodies. Here, we tested the immunogenicity and efficacy of HIV-1 C.1086 gp140 boosts administered sequentially after priming with CD40L-adjuvanted DNA/simian-human immunodeficiency virus (SHIV) and boosting with modified vaccinia virus Ankara (MVA)-SHIV vaccines in rhesus macaques. The DNA/MVA vaccination induced robust vaccine-specific CD4 and CD8 T cell responses with a polyfunctional profile. Two gp140 booster immunizations induced very high levels (∼2 mg/ml) of gp140 binding antibodies in serum, with strong reactivity directed against the homologous (C.1086) V1V2, V2 HS, V3, and gp41 immunodominant (ID) proteins. However, the vaccine-induced antibody showed 10-fold (peak) and 32-fold (prechallenge) weaker binding to the challenge virus (SHIV1157ipd3N4) V1V2 and failed to bind to the challenge virus V2 HS due to a single amino acid change. Point mutations in the immunogen V2 HS to match the V2 HS in the challenge virus significantly diminished the binding of vaccine-elicited antibodies to membrane-anchored gp160. Both vaccines failed to protect from infection following repeated SHIV1157ipd3N4 intrarectal challenges. However, only the protein-boosted animals showed enhanced viral control. These results demonstrate that C.1086 gp140 protein immunizations administered following DNA/MVA vaccination do not significantly boost heterologous V1V2 and V2 HS responses and fail to enhance protection against heterologous SHIV challenge.
IMPORTANCE HIV, the virus that causes AIDS, is responsible for millions of infections and deaths annually. Despite intense research for the past 25 years, there remains no safe and effective vaccine available. The significance of this work is in identifying the pros and cons of adding a protein boost to an already well-established DNA/MVA HIV vaccine that is currently being tested in the clinic. Characterizing the effects of the protein boost can allow researchers going forward to design vaccines that generate responses that will be more effective against HIV. Our results in rhesus macaques show that boosting with a specific HIV envelope protein does not significantly boost antibody responses that were identified as immune correlates of protection in a moderately successful RV144 HIV vaccine trial in humans and highlight the need for the development of improved HIV envelope immunogens.
INTRODUCTION
Human immunodeficiency virus (HIV) infects nearly 2 million individuals annually worldwide. Despite vigorous efforts, there remains no effective vaccine or functional cure (1). Developing a safe and effective vaccine that can prevent HIV transmission remains a high priority to curtail the AIDS epidemic. Of the limited vaccine trials that have been tested for efficacy in humans, the RV144 trial is the only trial that showed a low but significant impact on reducing HIV transmission. At the conclusion of the 3-year trial, the vaccine efficacy was 31.2% (2). Importantly, the RV144 trial identified multiple correlates for reduced risk of infection (3–6). These include higher binding titers to an HIV envelope (Env) V1V2 scaffolded protein and antibody (Ab)-dependent cellular cytotoxicity (ADCC) activity (3, 4). Further analyses of responses from vaccine participants revealed a correlation between V2 loop responses and reduced risk of infection (5). Within the V2 loop, there lies a relatively conserved linear region directly preceding the α4β7 binding site, known as the V2 hot spot (V2 HS). Binding titers against this relatively conserved region within the V2 loop also correlated with reduced risk of infection in vaccine participants (7). Consistent with RV144 results, multiple studies in nonhuman primates (NHPs) showed a strong association between anti-V2 antibody responses and protection (8–11). Accordingly, a major focus of HIV vaccine development is to generate strong and broad anti-V1V2 responses, including those against the V2 HS, and to understand the mechanisms of protection mediated by these antibodies.
The DNA prime and modified vaccinia virus Ankara (MVA) boost approach has been shown to induce a strong T cell and antibody response against simian immunodeficiency virus (SIV) and HIV in NHPs (12–21), and DNA/MVA HIV-1 vaccines have been shown to be safe and highly immunogenic in people (22–33). Our previous studies using DNA/MVA SIVmac239 immunogens showed a significant delay in the acquisition of intrarectal (i.r.) SIVsmE660 infection in rhesus macaques (RMs) (15, 16, 34). We also showed that adjuvanting the DNA/MVA SIVmac239 vaccine with membrane-anchored CD40L further enhanced protection against both SIVsmE660 and SIVmac251 challenges (16, 17). In all of these studies, we observed multiple correlates for a delay in the acquisition of SIV infection, which included mucosal IgG in rectal secretions prechallenge, the avidity of gp160 binding antibody in serum, and antibody effector functions such as antibody-dependent cellular viral inhibition (ADCVI). These DNA/MVA SIV vaccines also induced a strong anti-V2 response (8, 17). Although our SIVmac239-based vaccines induced neutralizing antibodies against SIVsmE660 challenge infection, these responses did not correlate with protection (34–36), suggesting that antibody effector functions significantly contributed to vaccine protection.
Given these highly encouraging results, it is important to further enhance the protection afforded by the DNA/MVA vaccines. In this regard, the addition of Env protein boosts is an excellent choice, as proteins are ideal for inducing strong antibody responses. Thus, it is important to test if the addition of protein boosts to the DNA/MVA vaccine can further boost the protective antibody responses and enhance protection. However, the choice of the protein (gp120 versus gp140), the nature of the adjuvant, and the timing of protein administration with respect to MVA boosts (simultaneous versus sequential) can significantly influence the magnitude, specificity, and quality of the antibody response and, thereby, protection. A recent DNA/MVA HIV-1 clade B vaccine study in macaques tested the influence of gp120 protein in alum delivered simultaneously with MVA boosts (37). The gp120 protein enhanced binding antibody titers by 10-fold, but it failed to boost the anti-V1V2 response. However, the immunogenicity and efficacy of sequential HIV Env protein boosts following DNA/MVA vaccination have not yet been tested in macaques.
In this study, we developed and tested clade C C.1086 Env-based DNA and MVA immunogens that express simian-human immunodeficiency virus (SHIV) virus-like particles (VLPs) (SIVmac239 Gag core with HIV Env VLPs) displaying trimeric C.1086 Env on the membrane. We focused on clade C because HIV infections by these strains are most prevalent worldwide. We chose the C.1086 strain because it was down-selected from a series of clade C Envs by Duke CHAVI based on antigenicity and stability, and C.1086 gp120 protein is being used in the HVTN702 large-scale efficacy trial in South Africa. We adjuvanted the DNA vaccine with membrane-anchored macaque CD40L due to our previous success with this adjuvant (16, 17). Using these immunogens, we compared the immunogenicities and efficacies of sequential C.1086 gp140 boosts following DNA/MVA vaccination. Our results demonstrate that the addition of sequential gp140 protein boosts in Adjuplex adjuvant to the DNA/MVA SHIV vaccine induces a robust binding response to gp140 protein but fails to protect against pathogenic intrarectal SHIV challenge. The majority of the antibodies generated by the Env protein boosts were directed against the V1V2, V2 HS, V3, and gp41 immunodominant (ID) regions. However, these antibodies were poorly cross-reactive with the challenge virus V1V2 and V2 HS. Point mutations in the V2 HS of the immunogen to match the V2 HS in the challenge virus abrogated the binding of vaccine-elicited sera to membrane-anchored HIV Env.
RESULTS
DNA and MVA SHIV VLP immunogens.
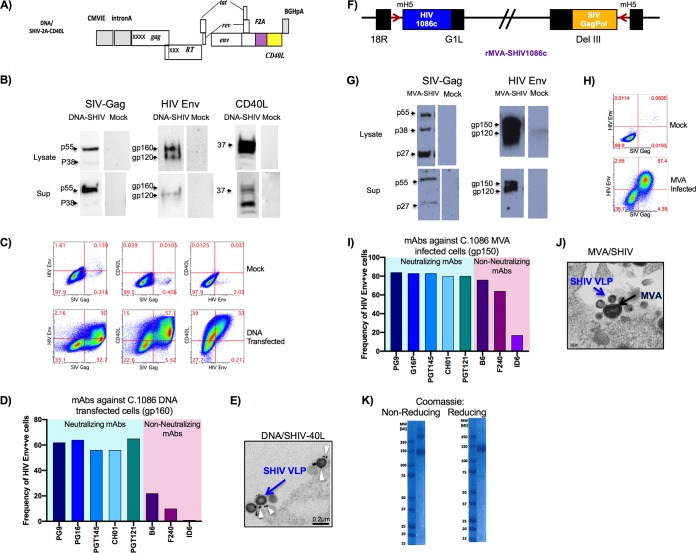
The DNA/SHIV-C.1086-2A-CD40L immunogen expressed SIVmac239 Gag, protease (Prt), and reverse transcriptase (RT); HIV C.1086 Env, Tat, and Rev; and rhesus macaque CD40L linked via an F2A peptide from foot-and-mouth disease virus (38). The F2A peptide was chosen over the traditional internal ribosomal entry site (IRES) method due to the enhanced coexpression of CD40L and HIV Env proteins in preliminary studies (Fig. 1A and data not shown). By Western blot analysis, Gag pr55, Env gp120 and gp160, and CD40L could be detected at the correct sizes in both the cell lysate and supernatant at 48 h posttransfection in 293T cells (Fig. 1B). Coexpression of Env gp160 and CD40L was detectable on the surface of transfected 293T cells, and SIV Gag was detectable by intracellular staining at 48 h posttransfection (Fig. 1C). The Env expressed on DNA transfected bound strongly to multiple broadly neutralizing antibodies (bnAbs), including antibodies that primarily recognize the trimeric form of Env, such as PG16 and PGT145, and bound weakly to nonneutralizing antibodies (nnAbs), suggesting that a significant fraction of Env on the surface exists as a trimer (Fig. 1D). Electron microcopy revealed the formation of SHIV VLPs budding off the surface of transfected 293T cells, and gold labeling using anti-CD40L antibody detected CD40L protein expression on the surface of budding VLPs from DNA-transfected cells (Fig. 1E). Similarly, MVA/SHIVC.1086 also expressed SIVmac239 Gag, Prt, and RT and HIV C.1086 Env gp150 and produced VLPs (Fig. 1F to J). The MVA-infected cells showed strong binding to both bnAbs and nnAbs, suggesting that the gp150 form of C.1086 Env may expose some of the nonneutralizing epitopes (Fig. 1I). The gp140 protein predominantly existed in monomeric and dimeric forms, with a small portion forming trimers (Fig. 1K).
FIG 1.
Characterization of DNA and MVA immunogens. (A) Schematic of C.1086 SHIV DNA. (B) Western blot analysis of the SHIV DNA-CD40L construct expressing SIV Gag, SIV CD40L, and HIV C.1086 Env proteins. Sup, supernatant. (C) Flow cytometry displaying surface coexpression of HIV Env and SIV CD40L on SIV Gag-positive transfected 293T cells. (D) Binding of mAb to C.1086-transfected cells. +ve, positive. (E) Electron microcopy images of SHIV DNA-CD40L VLPs budding from the surface of transfected 293T cells. (F) Schematic of C.1086 SHIV MVA constructs. (G) Western blot analysis of the SHIV MVA construct expressing SIV Gag and HIV C.1086 Env proteins. (H) Flow cytometry of Gag-positive cells expressing HIV Env on infected DF1 cells. (I) Binding of mAbs to SHIV MVA-infected cells. (J) Electron microcopy images of MVA-infected DF1 cells expressing SHIV MVA VLPs and MVA particles. (K) Coomassie gels of C.1086 gp140 protein postpurification. CMVIE, cytomegalovirus immediate early promoter.
DNA immunization induces strong activation of dendritic cells and monocytes in blood.
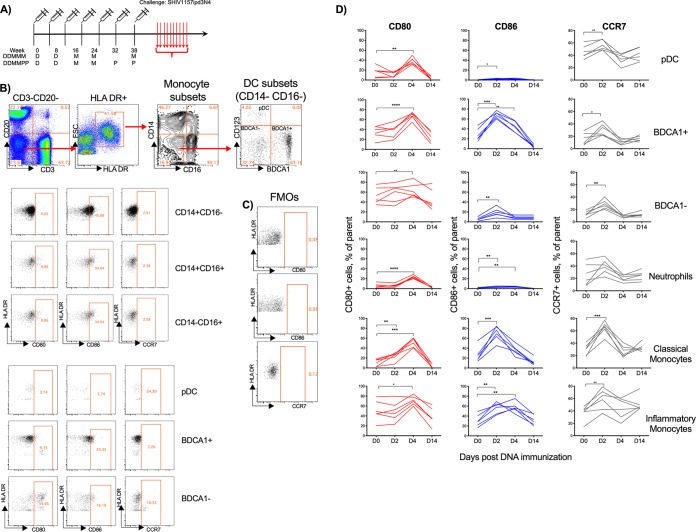
Young adult male (n = 20) and female (n = 10) RMs were vaccinated intramuscularly (i.m.) in the upper thigh at weeks 0 and 8 with 3 mg of DNA/SHIV-C.1086-CD40L (here referred to as SHIV-rhCD40L DNA) and at weeks 16 and 24 with 108 PFU of MVA/SHIV-C.1086 (referred to as SHIV-MVA). At this point, the RMs were divided into two groups. One group (DDMMM; 10 males and 5 females) received a third i.m. MVA immunization at week 38, and the second group (DDMMPP; 10 males and 5 females) received two subcutaneous protein immunizations adjuvanted with Adjuplex at weeks 32 and 38 (Fig. 2A). As mentioned above, rhesus macaque CD40L was included in the DNA construct to serve as a built-in vaccine adjuvant. In assessing the adjuvant properties of CD40L, the expression of CD80, CD86, and CCR7 was monitored on plasmacytoid dendritic cells (pDCs) (lineage CD14− CD16− HLR-DR+ CD123+), BDCA-1+ DCs (lineage CD14− CD16− HLR-DR+ BDCA-1+), monocytes, and neutrophils (more granular cells and CD66+ cells) in blood on days 0, 2, 4, and 14 after the 2nd DNA vaccination. Monocytes were categorized as classical monocytes (CD14+ CD16−), inflammatory monocytes (CD14+ CD16+), and patrolling monocytes (CD14− CD16+) (Fig. 2B). The fluorescence-minus-one (FMO) controls were used for all activation markers at all time points to accurately assess expression levels of all cellular subsets (Fig. 2C). DNA immunization induced strong activation of all innate subsets analyzed based on the expression of either CD80, CD86, or CCR7 (Fig. 2D). CD80 expression peaked on day 4 for all monocytes, DCs, and neutrophils sampled except for BDCA-1− DCs and returned to levels near or below baseline levels by day 14. Expression was mostly observed for BDCA-1+ and BDCA-1− DCs and classical monocytes on day 2, while expression peaked at day 2 and remained elevated at day 4 for inflammatory monocytes (Fig. 2D). CD86 expression was also marginally increased on neutrophils on day 2 and day 4 (Fig. 2D). Similarly, CCR7 expression also peaked on day 2 for most of the cellular subsets studied and dropped below baseline levels by day 4. However, by day 14, the expression of CCR7 returned to baseline levels in all subsets monitored. On day 2 postimmunization, the CCR7 expression level was significantly higher on BDCA-1− and classical monocytes (Fig. 2D). Taken together, these data show that DNA immunization induced high levels of activated DCs, monocyte subsets, and neutrophils with the potential to migrate to lymphoid tissue. However, it is important to note that the contribution of the CD40L adjuvant to this innate activation could not be determined due to the lack of data from an unadjuvanted DNA vaccine group.
FIG 2.
DNA/SHIV-CD40L immunizations activate DC and monocyte subsets. (A) Experimental design of the monkey study. (B) Gating strategy for monocyte subsets, DC subsets, and neutrophils. FSC, forward scatter. (C) Fluorescence-minus-one (FMO) gating strategy for CD80, CD86, and CCR7. (D) Expression of CD80, CD86, and CCR7 by DCs, neutrophils, and monocyte subsets at days 0, 2, 4, and 14 postimmunization in six immunized RMs (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
gp140 protein booster immunizations induce robust homologous Env-specific IgG and IgA binding antibodies and V1V2 antibodies in serum and rectal secretions.
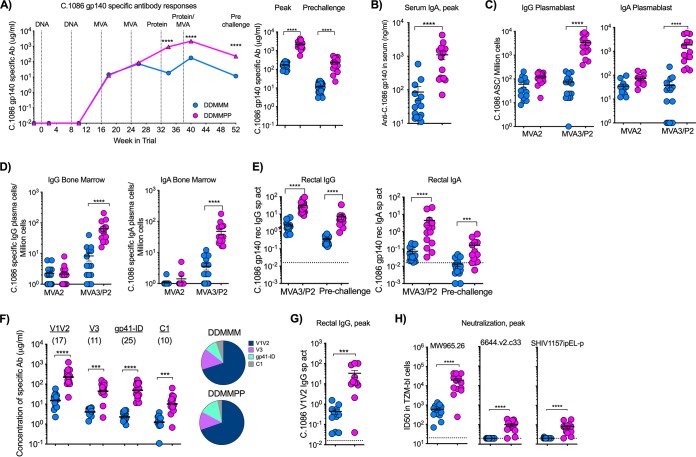
Consistent with our previous studies (16, 17, 39), DNA immunizations did not elicit detectable levels of gp140-specific antibodies in serum. However, the subsequent two MVA immunizations boosted the HIV Env C.1086 gp140-specific IgG response to about 85 μg/ml in both vaccine groups (Fig. 3A). Impressively, in the DDMMPP group, the 1st gp140 protein immunization on week 32 boosted the response nearly 10-fold, reaching a geometric mean titer of 761 μg/ml at week 34, and the 2nd protein immunization on week 38 further boosted these titers by about 2.5-fold to a geometric mean titer of 1,908 μg/ml on week 40. In the DDMMM group, the 3rd MVA vaccination on week 38 only marginally boosted the peak antibody response to a titer of 169 μg/ml on week 40. Compared to the DDMMM group, the addition of gp140 protein immunization was observed in the DDMMPP group to increase the gp140-specific antibody titers by 11-fold (P < 0.0001) at the peak and 17-fold (P < 0.0001) at the prechallenge time point (Fig. 3A). Similarly, DDMMPP-immunized RMs had 15-fold-higher gp140-specific IgA responses in serum than those of DDMMM-immunized RMs (P < 0.0001) (Fig. 3B). The higher-magnitude serum IgG and IgA responses in the DDMMPP group were associated with 10- to 50-fold-higher IgG and IgA plasmablast responses in blood 5 days after the final boost (Fig. 3C) and plasma cells in bone marrow 6 weeks after the final boost (Fig. 3D).
FIG 3.
Humoral immunity following immunization. (A) Kinetics of serum HIV Env gp140 binding Ab titers in DDMMM- and DDMMPP-vaccinated RMs. Data shown are geometric means for the group. Data for individual animals for the peak (week 40) and prechallenge (week 54) time points are shown on the right. (B to E) C.1086 gp140-specific IgA in serum, IgG and IgA plasmablast responses in blood, IgG and IgA plasma cells in bone marrow, and IgG and IgA in rectal secretions at the indicated time points. ASC, antigen-secreting cells. (F) Specificity of serum binding antibody to 1086c immunogen-derived gp70-V1V2 scaffold, V3, gp41 ID, and C1 peptides. Numbers in parentheses indicate fold differences between DDMMM and DDMMPP vaccine groups. (G) C.1086 V1V2-specific response in rectal secretions at the peak. (H) Neutralizing antibody titers against tier 1A and 1B clade C viruses. ID50, 50% infective dose; ***, P < 0.001; ****, P < 0.0001.
gp140 protein booster immunizations also boosted HIV gp140-specific antibody responses in the rectum, the route of infection for this vaccine study (Fig. 3E). In the DDMMPP group, titers of gp140-specific IgG antibodies in rectal secretions were 13-fold higher at the peak (week 40) and 19-fold higher at the prechallenge time point (Fig. 3E). A similar increase was also observed for the rectal IgA level, which was 60-fold higher at the peak and 12-fold higher at the prechallenge time point (Fig. 3E). These increases mirrored those in serum, suggesting that the majority of the IgA, as well as the IgG, in rectal secretions may have been derived from serum. Taken together, these data demonstrate that successive gp140 protein immunizations following DNA/MVA vaccination resulted in a strong boosting of gp140-specific IgG and IgA responses in serum, rectal secretions, and plasma cells in the bone marrow.
We next determined the specificity of these antibodies by measuring reactivity to specific regions of C.1086 Env. DDMMM vaccination induced antibody response against homologous gp70-V1V2, V3, gp41 ID, and constant 1 domain (C1) proteins, with a dominant response to V1V2 (Fig. 3F). DDMMPP animals showed 10- to 25-fold-higher levels of anti-gp70 V1V2-, anti-V3-, anti-gp41 ID-, and anti-C1-specific IgG responses and maintained the same V1V2 dominance (Fig. 3F). Similarly, protein boosting resulted in significantly (P < 0.0003) higher titers of C.1086 V1V2-specific antibodies in rectal secretions (Fig. 3G), which correlated with V1V2 responses in serum (data not shown). With respect to neutralization potential, protein boosts induced higher titers of neutralizing antibodies against MW965.26 (tier 1A; 32-fold), 6644.v2.c33 (tier 1B; 4-fold), and SHIV1157ipEL-p (the tier 1 counterpart to the challenge virus SHIV1157ipd3N4; 4-fold) (Fig. 3H). However, both vaccine regimens failed to generate any neutralizing activity against tier 2 viruses, including the autologous virus C.1086 and challenge virus 1157ipd3N4 (data not shown).
gp140 protein boosts induce strong Env-specific CD4 but not CD8 T cells and skew the T cell response toward a higher vaccine-specific CD4-to-CD8 T cell ratio.
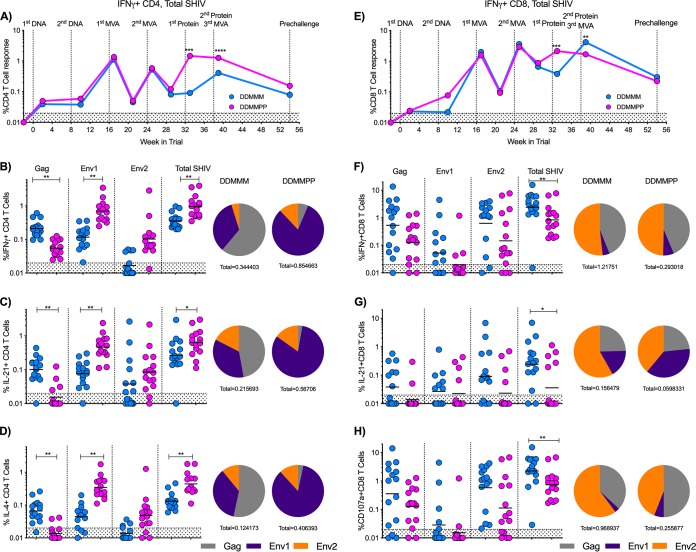
We next assessed the effect of gp140 protein immunization on the CD4 and CD8 T cell responses. Following the first protein boost, the total SHIV (Gag plus Env)-specific CD4 T cell response was recalled at levels similar to those observed at week 1 after the first MVA immunization. The second protein immunization did not further boost the total SHIV-specific interferon gamma-positive (IFN-γ+) CD4 T cell response. However, compared to the third MVA immunization in the DDMMM group, the protein-boosted animals showed significantly higher (P < 0.001) SHIV-specific IFN-γ+ CD4 T cell responses by more than 3-fold at the peak (week 1) and 2-fold at the prechallenge time point (Fig. 4A and B). This was also true for SHIV-specific interleukin 21-positive (IL-21+) and IL-4+ CD4 T cell responses at the peak (P = 0.02 and 0.001, respectively) (Fig. 4C and D). In addition, there was a difference in the relative proportions of Gag- and Env-specific CD4 T cells between groups. The IFN-γ, IL-21, and IL-4 CD4 T cell responses in the DDMMM group were more evenly distributed between the SIV Gag and HIV Env proteins, while the DDMMPP response was predominantly directed against the Env1 peptide pool (first 435 amino acids in gp120) of the HIV Env protein (Fig. 4B to D).
FIG 4.
DDMMPP immunization elicits significantly higher CD4 T cell responses, while DDMMM immunization elicits higher CD8 T cell responses. (A) Longitudinal total SHIV-specific IFN-γ+ CD4 T cell response. Geometric means for the group are shown. (B to D) SHIV-specific IFN-γ+, IL-21+, and IL-4+ CD4 T cell responses 1 week after the final immunization. Pie charts depict the relative distributions of the CD4 T cell responses to Gag, Env1, and Env2 peptide pools. (E) Longitudinal total SHIV-specific IFN-γ+ CD8 T cell response. Geometric means for the group are shown. (F to H) SHIV-specific IFN-γ+, IL-21+, and CD107+ CD8 T cell responses 1 week after the final immunization. Pie charts depict the relative distributions of the CD8 T cell responses to Gag, Env1, and Env2 peptide pools. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
While protein immunizations induced higher CD4 T cell responses at the peak, consecutive MVA immunizations led to 2-fold-higher peak SHIV-specific CD8 T cell responses, although these differences did not persist to the prechallenge time point (Fig. 4E and F). DDMMM immunization also induced higher total SHIV-specific IL-21+ and CD107a+ CD8 T cell responses at the peak (P = 0.02 and 0.007) than in DDMMPP-immunized RMs (Fig. 4G and H). As observed for CD4 T cell responses, DDMMM immunization elicited an evenly distributed SIV Gag and HIV Env (predominantly to the Env2 peptide pool) IFN-γ CD8 T cell response, but the distribution shifted toward Env for IL-21+ and CD107a+ responses (Fig. 4G and H). In contrast to the CD4 T cell responses in DDMMPP animals, CD8 T cells exhibited a more balanced SHIV-specific IFN-γ+ and CD107a+ response (Fig. 4F and H), but IL-21+ CD8 T cells were skewed toward HIV Env (Fig. 4G). Taken together, these data illustrate that incorporation of gp140 into the vaccine regimen shifted the CD4 and CD8 T cell responses toward the HIV Env protein and diminished the SIV Gag response.
gp140 protein boost elicits a polyfunctional Env-specific CD4 T cell response in blood.
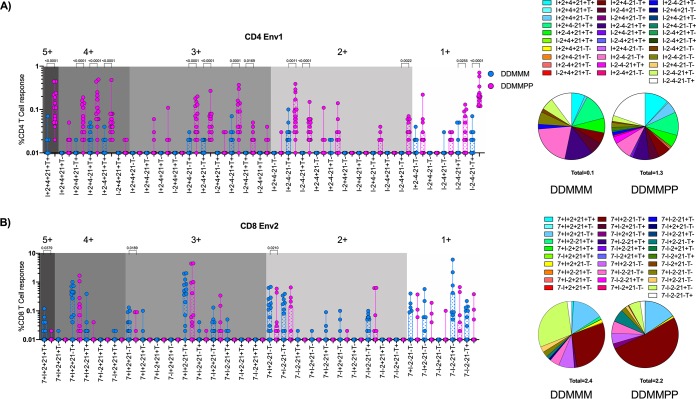
To get a better understanding of the cytokine profile of the CD4 and CD8 T cell responses post-DDMMM and -DDMMPP immunization following the final immunization, we determined cytokine coexpression profiles using Boolean gating (Fig. 5). We focused our analyses on Env-specific CD4 and CD8 T cells since this was the common immunogen in both vaccine groups at this time point. In addition, we used Env1-specific CD4 T cells and Env2-specific CD8 T cells since they showed the strongest immune responses. In the DDMMPP group, the Env1-specific CD4 T cells were distributed across 5+, 4+, 3+, 2+, and 1+ cytokines/functions relatively equally. These responses were significantly greater in the DDMMPP group than in the DDMMM group for all major subsets (Fig. 5A). The Env2-specific CD8 T cells mostly consisted of 4+, 3+, and 2+ cytokines/functions, and some of the responses were higher in the DDMMM group than in the DDMMPP group. The 5+ cells were a minority and were mostly present in the DDMMM group (Fig. 5B). Taken together, these data demonstrate that a gp140 protein boost generates polyfunctional SHIV-specific CD4 T cells capable of providing CD4 help to antigen-specific CD8 T cells and B cells.
FIG 5.
Boolean analysis of vaccine-specific CD4 and CD8 T cell responses. (A) Peak CD4 T cells response to the Env1 peptide pool. (B) Peak CD8 T cell response to the Env2 peptide pool. 7, CD107a; I, IFN-γ; T, TNF-α; 2, IL-2; 21, IL-21; 4, IL-4.
Both the DDMMM and DDMMPP vaccines fail to prevent intrarectal heterologous SHIV1157ipd3N4 infection; however, protein-boosted RMs show enhanced viral control.
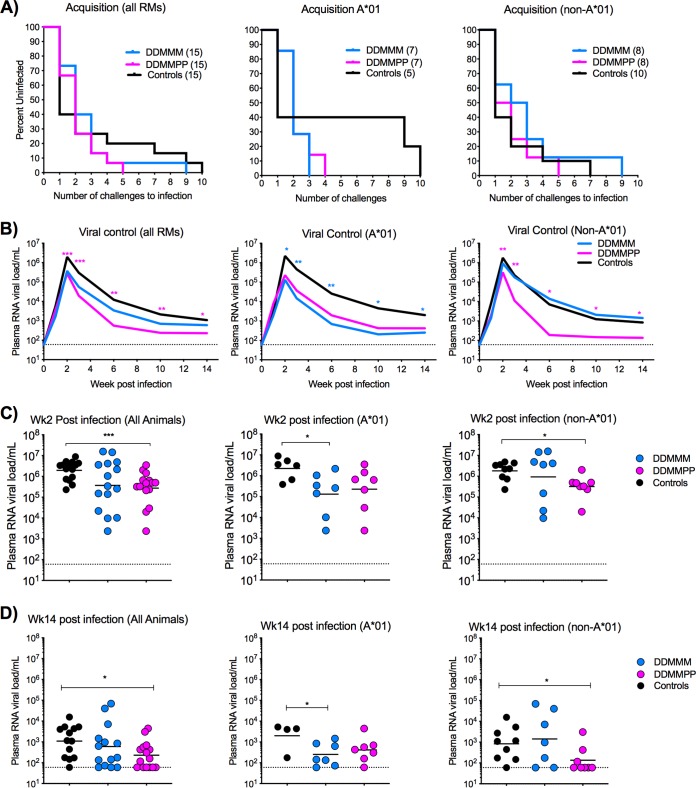
In order to determine the efficacy of the DDMMM and DDMMPP vaccine regimens, RMs were challenged intrarectally (i.r.) weekly with SHIV1157ipd3N4 at a 1:400 dilution. SHIV1157ipd3N4 is a tier 2 clade C SHIV derived from a clinical isolate serially passaged in infant RMs (40). Fifteen age- and sex-matched RMs were added to the study to serve as unvaccinated controls. After the first challenge, 53% of the unvaccinated controls were infected, compared to 27% and 33% of RMs in the DDMMM and DDMMPP groups, respectively. However, by the second challenge, there was no difference in the rate of acquisition between any of the groups, as 60 to 70% of the animals in each group were infected. The remaining animals were infected after 5 to 10 challenges (Fig. 6A). Acquisition of infection was comparable between Mamu-A*01+ and Mamu-A*01-negative (Mamu-A*01−) animals except that there was a trend toward slower acquisition in Mamu-A*01+ controls than in Mamu-A*01− controls (Fig. 6A).
FIG 6.
Acquisition of infection and viral control after SHIV1157ipd3N4 infection. (A) Kaplan-Meier plots depicting the number of challenges required for productive infection. (B) Geometric mean values for plasma RNA viral titers for all, Mamu-A*01+, and Mamu-A*01− animals. Asterisks represent statistical significance between immunized and unvaccinated controls. A magenta asterisk indicates significance between the DDMMPP group and unvaccinated controls. A blue asterisk indicates significance between the DDMMM group and unvaccinated controls. (C) Plasma viral loads for individual animals at week 2 postinfection. (D) Plasma viral loads for individual animals at week 14 postinfection. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To study the influence of vaccination on viral control, we compared plasma viral loads in the vaccinated animals and unvaccinated controls up to 14 weeks. The protein-boosted animals showed significantly lower viral RNA loads at multiple time points than those in unvaccinated controls, including the Mamu-A*01-negative animals (Fig. 6B to D). In contrast, DDMMM-immunized RMs showed lower viral RNA loads only in Mamu-A*01+ animals (Fig. 6B to D). These data demonstrate that the addition of C.1086 gp140 boosts to the DNA/MVA C.1086 vaccine regimen did not improve protection against SHIV1157ipd3N4 rectal challenges, but they enhanced viral control after infection.
Vaccine-elicited antibody and CD8 T cell responses correlate with decreased viral burden postinfection.
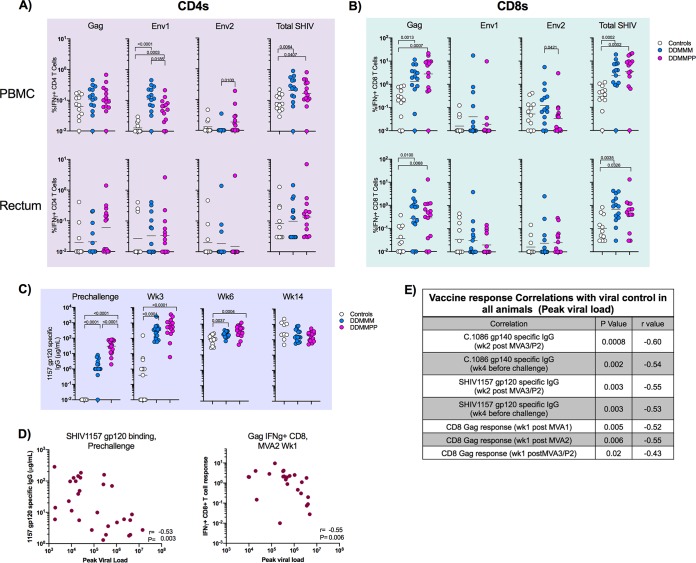
To get a better understanding of what factors were contributing to viral control postinfection, we first examined vaccine-specific CD4 and CD8 T cells at 3 weeks postinfection. We observed a strong anamnestic expansion of SHIV-specific IFN-γ+ CD4 and CD8 T cells in vaccinated animals compared to unvaccinated animals in both blood and rectum (Fig. 7A and B). Specifically, the Env1-specific IFN-γ+ CD4 T cell and Gag-specific IFN-γ+ CD8 T cell responses were significantly higher in the vaccinated animals than in the unvaccinated controls (Fig. 7A and B). Higher SHIV-specific CD8 T cell responses were also observed in the rectum of DDMMM- and DDMMPP-immunized RMs than in unvaccinated controls (Fig. 7B). SHIV1157 gp120-specific antibody responses were measured in the serum both before infection and at weeks 3, 6, and 14 postinfection. Vaccination resulted in significantly higher SHIV1157-specific gp120 antibody titers at weeks 3 and 6 postinfection; however, by week 14 postinfection, there was no difference between vaccinated and unvaccinated RMs (Fig. 7C). The enhanced viral control in the protein-boosted animals also suggested that vaccine-induced antibody would have contributed to viral control. Accordingly, we found a significant inverse association between C.1086 or SHIV1157 gp120 Env-specific binding antibody and the peak viral load (Fig. 7D and E). We also found a significant inverse association between Gag-specific CD8 T cells and the week 3 viral load (Fig. 7D and E). However, none of the postinfection immune responses showed an inverse association with viral load. These results strongly suggested that both vaccine-induced antibody and CD8 T cells contribute to enhanced viral control.
FIG 7.
Postinfection T and B cell responses. (A) C.1086-specific CD4 T cell response in the blood and rectum at 3 weeks postinfection. (B) C.1086-specific CD8 T cell response in the blood and rectum at 3 weeks postinfection. (C) SHIV1157ipd3N4 gp120-specific antibody responses prechallenge and at weeks 3, 6, and 14 postinfection. (D) Correlations between SHIV1157ipd3N4-specific serum antibody responses in the blood at the prechallenge time point and CD8 Gag responses in the blood at week 1 after MVA2 with week 3 postinfection plasma viral loads. (E) Table showing correlations between the vaccine-specific response and week 3 postinfection plasma viral load.
Vaccine-induced V1V2 and V2 HS antibodies poorly cross-react with the challenge virus V1V2 and V2 HS and show diminished binding to membrane-anchored V2 HS-modified Env.
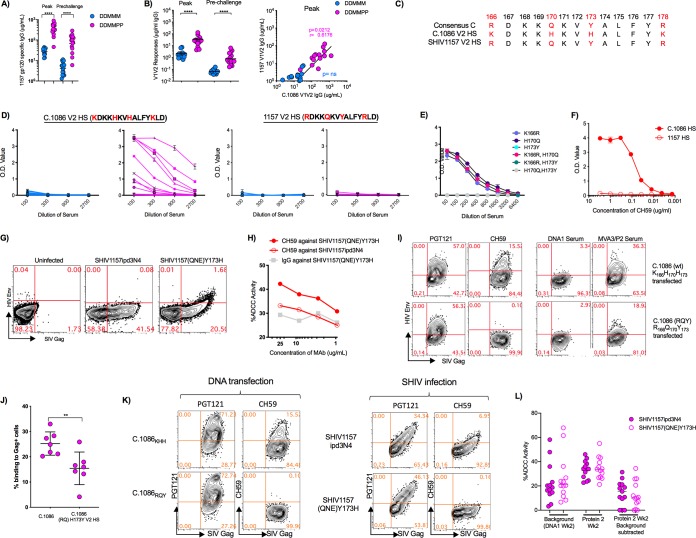
The vaccine-induced antibody showed strong binding to challenge virus gp120, and sera from DDMMPP-vaccinated animals showed 14- and 19-fold-higher titers at the peak and prechallenge time points than those in DDMMM animals (Fig. 8A). However, the vaccine-induced antibody bound very poorly to gp70-V1V2 of the 1157ipd3N4 challenge virus at both the peak and prechallenge time points (Fig. 8B). This was true for both vaccine groups, although the gp140-boosted animals had about 15-fold-higher levels of binding antibodies at the peak and prechallenge time points (Fig. 8B). At the peak, the levels of 1157 V1V2-specific IgG were roughly 10-fold lower than those of C.1086 V1V2-specific IgG, demonstrating that the C.1086 Env immunogen induced antibodies that poorly cross-reacted with the challenge virus V1V2. Peak 1157 gp70-V1V2 responses in serum directly correlated with the C.1086 gp70-V1V2 responses in the DDMMPP group (P = 0.02) but not in the DDMMM immunization group (Fig. 8B).
FIG 8.
Poor cross-reactivity of vaccine-induced antibody to the challenge virus V1V2 and V2 HS. (A) 1157 gp120 IgG responses in the blood at the peak and prechallenge time points in DDMMM- and DDMMPP-immunized RMs. (B) 1157 gp70 V1V2 IgG responses in the blood at the peak and prechallenge time points in DDMMM- and DDMMPP-immunized RMs. Correlations between responses against 1157 V1V2 and C.1086 V1V2 are shown on the right. (C) V2 HS sequences for the clade C consensus sequence, 1086c, and 1157ipd3N4. (D) ELISA data showing binding of individual RM sera to the 1086c peptide (TELKDKKHKVHALFY) and 1157 V2 HS peptide (TELRDKKQKVYALFY) for each immunization group. O.D., optical density. (E) Pooled peak serum from DDMMPP-immunized RMs against C.1086 V2 HS singly or doubly mutated peptides. (F) ELISA data showing binding of mAb CH59 to the 1086c and 1157 V2 HS sequences. (G) Flow cytometry data demonstrating binding of CH59 to CEM NKr cells infected with SHIV1157ipd3N4 and SHIV1157(QNE)Y173H. (H) ADCC activity of CH59 against SHIV1157ipd3N4- and SHIV1157(QNE)Y173H-infected CEM NKr cells. (I) Representative flow data demonstrating binding of PGT121 and CH59 mAbs and serum from DDMMPP animals to DNA-transfected cells. C.1086 is the vaccine DNA without CD40L. C.1086 (RQ)H173Y DNA is identical to C.1086 except for three mutations in the HS, K166R, H170Q, and H173Y. wt, wild type. (J) Binding of DDMMPP sera to DNA-transfected cells. (K) Binding to DNA/SHIV-C.1086-CD40L (KHH and RQY)-transfected (left) and SHIV1157ipd3N4/SHIV1157(QNE)Y173H-infected (right) cells using mAbs PGT121 and CH59. (L) ADCC activity against SHIV1157ipd3N4 and SHIV1157(QNE)Y173H. DNA1, time point used as the background for the assay. **, P < 0.01; ****, P < 0.0001.
We next analyzed binding antibodies to the V2 HS. For this, we synthesized peptides encompassing the V2 HS of C.1086 and 1157 Envs. The V2 HS sequences of C.1086 and 1157 differ at 4 amino acid locations: K166R, H170Q, H173Y, and K178R (Fig. 8C). Serum from DDMMPP but not DDMMM animals showed strong binding to the 1086 V2 HS. However, the same sera did not show any binding to the 1157 V2 HS (Fig. 8D). Single-amino-acid substitutions in the C.1086 V2 HS sequence showed that H173Y completely abolishes binding (Fig. 8E). Collectively, these data demonstrate that neither DDMMM nor DDMMPP immunizations generated antibodies that cross-reacted with the challenge virus V2 HS due to the H173Y amino acid change in the 1157 V2 HS.
To determine if V2 HS-specific antibodies contribute to ADCC activity against virus-infected cells, we used the CH59 monoclonal antibody (mAb) that was isolated from an RV144 trial participant and shown to bind to an epitope in the V2 HS region. The CH59 mAb bound strongly to the C.1086 V2 HS but showed no binding to the 1157 V2 HS (Fig. 8F). Next, we looked at binding to SHIV1157ipd3N4- and SHIV1157(QNE)Y173H-infected cells. SHIV1157(QNE)Y173H is similar to SHIV1157ipd3N4 but has a Y173H mutation in the V2 HS as well as two additional mutations, Q170K and I192R (41). CH59 bound to targets infected with SHIV1157(QNE)Y173H but not SHIV1157ipd3N4, confirming that V2 HS-directed antibodies can recognize HIV Env on infected cells and that this recognition is sensitive to the histidine at position 173 (Fig. 8G). Consistent with the cell binding results, CH59 showed ADCC activity against SHIV1157(QNE)Y173H-infected cells but not against SHIV1157ipd3N4-infected cells (Fig. 8H). These results demonstrate that antibodies directed against the V2 HS recognize membrane-anchored HIV Env and mediate ADCC activity against SHIV-infected cells.
To determine if C.1086 vaccine-induced antibodies recognize Env expressed on the cell membrane, and whether this recognition may be V2 HS sequence dependent, we transfected 293T cells with two DNA constructs expressing SIV Gag and C.1086 HIV Env proteins. The first DNA construct was the exact same DNA used to immunize the macaques in the trial, while the second was mutated in the V2 HS at 3 locations (K166R, H173Q, and H173Y) to match the HIV-1 clade C consensus sequence. PGT121, the positive-control mAb, bound equally well to cells transfected with either of the DNA constructs, while immune serum bound significantly better (P < 0.007) to cells transfected with the homologous DNA construct (Fig. 8I and J). These results suggest that V2 HS-specific antibodies generated by the DNA/MVA C.1086 vaccine contribute to the recognition of membrane-anchored Env on the cell surface and that the heterogeneity between the vaccine and challenge virus V2 HS sequences may render these antibodies less functional against challenge virus-infected cells. Our attempts to measure binding of vaccine sera to SHIV-infected cells with V2 HS mutations did not yield good binding with either (H or Y at position 173) of the V2 HS viruses (data not shown). The mAbs PGT121 and CH59 themselves showed significantly lower binding to SHIV-infected cells than to DNA-transfected cells, indicating lower levels of Env on the cell membrane of the former than of the latter (Fig. 8K). Finally, we measured the ADCC activity against the challenge SHIV and Y173H mutant SHIV using sera from DDMMPP-vaccinated animals. We observed a low level of ADCC activity against both viruses, and the activity was comparable against both viruses (Fig. 8L). These results showed that vaccine-induced antibodies show poor ADCC activity against infected cells, possibly due to the heterologous nature of the Env and poor expression on the cell surface, and the H residue at position 173 does not improve this activity.
DISCUSSION
Here, we tested the ability of a CD40L-adjuvanted clade C DNA/MVA SHIV vaccine with and without gp140 protein boosts to protect against heterologous tier 2 clade C intrarectal SHIV challenges. Our results showed that the CD40L-adjuvanted DNA/MVA vaccine induces robust SHIV-specific CD4 and CD8 T cell responses with a polyfunctional profile, and gp140 protein boosts in Adjuplex adjuvant markedly enhance the IgG and IgA binding antibodies in blood as well as in mucosal secretions. The protein boosts also induced high levels of vaccine-specific gp70-V1V2 and V2 HS antibody responses but failed to induce a neutralizing antibody response against tier 2 HIV Envs, including against the challenge virus. This strong cellular and humoral binding antibody response in the DDMMPP group did not translate to better protection. Importantly, our results showed that the C.1086 gp140-induced anti-V1V2 and -V2 HS antibodies poorly cross-react with the challenge virus V1V2 and V2 HS, and three amino acid changes in the V2 HS of the immunogen to match the challenge virus V2 HS greatly diminish antibody recognition of membrane-anchored HIV Env. This is quite concerning for HIV vaccine approaches that aim to induce a strong and broad anti-V1V2 response. Thus, it is important to develop HIV Env immunogens that can strongly increase both the magnitude and breadth of the anti-V1V2 response. To overcome this limitation, a recent study used pentavalent gp120 protein boosts to enhance the breadth of the anti-V2 antibody response and showed that this improved protection against clade C SHIV (41). Toward this goal, we recently developed a trimeric, cyclically permuted gp120 protein that induces a strong and broad anti-V1V2 response in rabbits and macaques (42). In conclusion, these results demonstrate that sequential immunizations with C.1086-based DNA, MVA, and gp140 protein induce a robust cellular and humoral immune response to HIV, but they do not protect against a heterologous intrarectal clade C SHIV challenge.
Encouragingly, the gp140 protein boosts enhanced the control of SHIV replication at the peak and set point stages of infection. Impressively, this enhanced control was observed in both Mamu-A*01+ and Mamu-A*01− animals. Roughly 30% of gp140 protein-boosted RMs had undetectable viral levels in the blood at 6 weeks postinfection. By 10 weeks postinfection, 40% of gp140-boosted RMs had undetectable virus in the blood relative to DDMMM (30%) and control (13%) RMs. Based on the immune correlate analysis, the higher levels of vaccine-induced antibody responses and SIV Gag-specific CD8 T cells at the prechallenge time point contributed to enhanced viral control in Mamu-A*01− animals. These data indicate that the vaccine-induced antibody response to C.1086 and SHIV1157 contributed significantly to viral control against heterologous infection. The contribution of anti-Env antibody responses to enhanced viral control is consistent with findings from a recent study showing that passive transfer of anti-V1V2 monoclonal antibody reduces cell-associated viral DNA after SHIV infection in macaques (43).
An important finding of our study is that anti-V2 HS antibodies are important for the recognition of membrane-anchored trimeric gp160, and point mutations in the V2 HS significantly diminish the binding of vaccine sera to membrane-anchored trimeric gp160. Although the RV144 antibody analyses revealed that anti-V2 HS binding antibody is a correlate for reduced risk of HIV acquisition (5), the precise mechanisms by which these antibodies may provide protection are still being worked out. There is very little information about the anti-V2 HS antibodies induced by vaccination. Our study shows that gp140 protein immunizations can significantly boost autologous anti-V2 HS antibodies, but the binding of these antibodies to the challenge virus V2 HS can be completely abrogated by a single amino acid change at position 173 from histidine to tyrosine. Serum from unvaccinated controls was tested against the challenge virus V2 HS and found not to bind to the homologous V2 HS, indicating that infection does not induce antibodies to this region. These results strongly suggest that it is important for HIV vaccines to generate high-magnitude anti-V2 HS antibodies with broad reactivity to diverse V2 HS sequences. It is also important to note that the V2 HS of the C.1086 protein is distant from the V2 HS of the clade C consensus sequence, suggesting that C.1086 Env-induced anti-V2 HS antibodies may not cross-react with the V2 HS of most of the primary clade C isolates. Our recent study with cyclically permuted gp120 protein showed that it is possible to generate broad anti-V2 HS antibodies by vaccination in rabbits and macaques (42).
In this study, the C.1086 gp140 protein boosts did not generate either autologous or heterologous tier 2-neutralizing antibody responses. This is not surprising for multiple reasons. First, the gp140 protein that we used is not stabilized to make native-like trimers. The majority of this protein exists as a monomer or dimer. Despite this, we used this protein because the stabilized trimeric version of C.1086 Env protein was not available. Second, not all native-like trimers will induce a strong autologous neutralizing antibody response. It is quite possible that C.1086 trimers may not be good at inducing autologous neutralizing antibody responses. Third, we may need a longer gap between the two protein immunizations. We used 6 weeks; however, a recent study and our ongoing studies with BG505 SOSIP immunizations suggest that an interval of 16 weeks or longer may be better for inducing autologous neutralizing antibody responses. Our hope was to at least induce a strong nonneutralizing antibody with effector function.
In this study, we included a third MVA immunization in the DDMMM group to boost the CD8 T cell response. Given that CD8 T cells can kill virus-infected cells, we hoped that any CD4 T cells infected in the rectum would be eliminated before they could spread the infection to secondary lymphoid tissues. Indeed, the third MVA immunization significantly boosted the CD8 T cell response at the peak. However, by the prechallenge time point, there was no significant difference between the two groups. Despite having more virus-specific CD8 T cells and fewer CD4 T cells at the time of challenge, DDMMM immunization did not delay acquisition compared to unvaccinated controls. However, the greater SHIV-specific CD8 T cell response resulted in lower viremia and better viral control in the Mamu-A01+ RMs than in their DDMMPP counterparts or unvaccinated controls.
In conclusion, our results show that the addition of C.1086 gp140 protein boosts to the DNA/MVA vaccine modality markedly enhanced levels of binding antibodies to gp140, but they failed to induce a strong antibody response against the heterologous anti-V1V2 and clade C consensus V2 HS. They also show that the unstabilized gp140 protein boosts do not induce autologous tier 2-neutralizing antibody responses. These results highlight the need for the development of clade C trimeric Env immunogens that induce broadly cross-reactive anti-V1V2 and anti-V2 HS responses with effector function and neutralization potential.
MATERIALS AND METHODS
Animals and immunizations.
Young Indian male and female rhesus macaques (RMs) from the Yerkes primate breeding colony were selected based on their Mamu-A*01, Mamu-B*08, and Mamu-B*17 alleles and cared for according to the Animal Welfare Act and the National Institutes of Health (NIH) (Bethesda, MD) Guide for the Care and Use of Laboratory Animals (44), using protocols approved by the Emory University Institutional Animal Care and Use Committee (45, 46). Thirty RMs were divided into 2 groups based on weight, sex, and Mamu-A*01 status. Each group contained 5 females and 10 males, of which 7 were Mamu-A*01 positive and 8 were Mamu-A*01 negative. Fifteen age-, sex-, weight-, and Mamu-A*01 status-matched controls were added after vaccinations were finished. All vaccinated animals were primed i.m. with 3 mg of CD40L-adjuvanted SHIV DNA on weeks 0 and 8 and then boosted i.m. with 1 × 108 PFU of SHIV MVA on weeks 16 and 24. On weeks 32 and 38, one group (DDMMPP) was immunized subcutaneously at the base of the neck with 100 μg of C.1086 gp140 HIV Env protein supplemented with Adjuplex adjuvant (Sigma) according to the manufacturer’s protocol. The other group (DDMMM) was given an additional i.m. vaccination with 108 PFU of SHIV MVA on week 38.
Immunogens.
The clade C HIV C.1086 gp140 K160N Env protein (47, 48) was obtained from Barton Haynes (Duke Human Vaccine Institute). The DNA vaccine PGA1/SHIV1086C_2ACD40L was constructed as previously described (16). The PGA1/SHIV1086C_2ACD40L construct was engineered from the pGA1/SHIV89.6 DNA plasmid (15). A single DNA fragment containing HIV Env with a K160N mutation, Tat, and Rev derived from C.1086 sequences was synthesized (GeneArt, Germany) and cloned using the EcoRI and NheI restriction sites by replacing the EcoRI-NheI fragment of the HIV89.6 Env and Tat/Rev genes. A DNA fragment containing an F2A peptide derived from foot-and-mouth disease virus along with monkey CD40L was inserted downstream of the HIV Env gene using a gene infusion kit (Clontech Laboratories, Mountain View, CA). Expression of the proteins by DNA was tested in transiently transfected 293T cells using flow cytometry and Western blotting. After 48 h, the supernatant and cells were collected. The cells were tested by flow cytometry for the expression of SIV Gag using 2F12 antibody (catalog no. 2343; NIH AIDS Reagent Program), HIV Env using ID6 antibody (catalog no. 1610; AIDS Reagent Program), and CD40L using anti-CD40L antibody (catalog no. AF617; R&D Systems). Western blotting with the above-described antibodies was used to detect the expressed proteins in cell supernatants and lysates.
For MVA/SHIV-C.1086, C.1086 gp140 K160N Env (GenBank accession no. FJ444399.1) (49) was codon optimized for vaccinia virus expression, synthesized from GeneArt, and cloned into pLW-73 using the XmaI site under the control of an independent mH5 promoter. This plasmid DNA was subsequently recombined, as described previously (20), into the essential region (DelII) of MVA expressing SIV Gag and Pol genes (kindly provided by B. Moss) between genes I8R and G1L. Recombinant MVA (rMVA) was isolated using standard methods, except that sorting was used during the first round of selection using green fluorescent protein (GFP) and PGT121 antibody to detect cell surface Env expression. Plaques were picked after 7 rounds to obtain GFP-negative rMVA/SHIV, and DNA sequences were confirmed. The recombinants were characterized for Gag and Env expression by flow cytometry and Western blotting. VLP formation was verified by electron microscopy. Viral stocks were purified from lysates of infected DF1 cells using a 36% sucrose cushion.
SHIV challenges.
Vaccinated and control RMs were challenged weekly by the intrarectal route with the tier 2 clade C SHIV1157ipd3N4 virus (40). This virus was titrated and used at a 1:400 dilution of the stock (containing 9.8 × 106 50% tissue culture infective doses [TCID50]/ml and 257 ng/ml of p27) for a maximum of 10 challenges. Animals were considered infected after two consecutive positive plasma viral load readouts.
Quantitation of SHIV RNA plasma load.
The SHIV copy number was determined using quantitative real-time PCR as previously described (12). Briefly, total RNA was extracted from plasma of infected RMs, amplified, and used for quantitative real-time PCR analyses.
Collection and processing of blood and rectal secretions.
Peripheral blood mononuclear cells (PBMCs) were collected and isolated as previously described (12). Rectal secretions were collected using Weck-Cel sponges (Beaver Visitec, Waltham, MA) and eluted as previously described (50).
T cell responses.
Intracellular cytokine staining (ICS) was performed as previously described (16), with a few modifications. Briefly, PBMCs were stimulated with 1 μg/ml of SIVmac239 Gag or HIV C.1086 Env overlapping peptide pools in the presence of 1 μg/ml anti-CD28 and anti-CD49d (BD Pharmingen, San Diego, CA) in complete RPMI 1640 medium (containing 10% fetal bovine serum [FBS], HEPES, gentamycin, and penicillin-streptomycin). The HIV Env overlapping peptides (15-mers overlapping by 11 residues) was divided into two pools, Env1 and Env2. Env1 consists of the first 106 peptides (amino acids 1 to 435 of gp120), and Env2 consists of the last 106 peptides (remaining gp120 and gp41) of the HIV Env protein. After 1.5 h of stimulation at 37°C in 5% CO2, GolgiStop (0.5 μg/ml; BD Pharmingen) and brefeldin A (0.5 μg/ml; BD Pharmingen) were added. After an additional 4.5-h incubation, the cells were placed at 4°C overnight. The following morning, cells were washed in fluorescence-activated cell sorter (FACS) wash buffer (phosphate-buffered saline [PBS] with 2% FBS and 0.05% sodium azide) and then surface stained with CD4-BV650 (clone L200; BD Pharmingen), anti-human CD8-AmCyan (clone SK1; BD Biosciences), and Live/Dead fixable near-infrared (IR) allophycocyanin (APC)-Cy7 stain (Invitrogen, CA) for 20 min at 4°C. Cells were then washed, permeabilized with Cytofix/Cytoperm (BD Biosciences) for 25 min at 4°C, and washed twice with Perm wash buffer (BD Biosciences). Intracellular staining was then done using a mixture of anti-human CD3-BV421 (clone SP34-2; BD Biosciences), anti-human interferon gamma (IFN-γ)-Alexa 700 (clone B27; BD Biosciences), anti-human interleukin 2 (IL-2)-BV605 (clone MQ1-17H12; BD Biosciences), anti-human interleukin 21 (IL-21)-APC (clone 3A3-N2.1; BD Biosciences), anti-human tumor necrosis factor alpha (TNF-α)-phycoerythrin (PE)-CF594 (clone Mab11; BD Biosciences), anti-human IL-4–PE (clone 7A3-3; Miltenyi Biotech, Auburn, CA), and anti-human CD107A-BV786 (clone H4A3; BD Biosciences) for 25 min at 4°C. Cells were washed with Perm wash buffer followed by FACS wash buffer and then resuspended in FACS wash buffer. Cells were acquired on an LSRII instrument (BD Immunocytometry Systems, San Jose, CA) and analyzed using FlowJo software (TreeStar, Ashland, OR).
Measurement of binding antibodies.
Anti-Env antibody responses were measured by an enzyme-linked immunosorbent assay (ELISA) using either the gp140 protein immunogen, murine leukemia virus gp70-scaffolded V1V2 proteins (from Abraham Pinter, Rutgers Medical School), or linear V2 HS peptides captured on Costar high-binding microtiter plates (Corning Life Sciences, Lowell, MA) as previously described (51). Briefly, plates were coated with the antigen at 1 μg/ml in PBS and incubated overnight at 4°C. The following day, the plates were washed, blocked, and incubated for 1 h with 3-fold dilutions of serum. For the standard, known concentrations of purified rhesus IgG (serially diluted) (NHP reagent resource) were captured using anti-rhesus IgG (Rockland). Bound IgG was detected using peroxidase-conjugated anti-monkey IgG (Accurate Chemical and Scientific, Westbury, NY) and the tetramethylbenzidine substrate (KPL, Gaithersburg, MD). The reaction was stopped by adding 100 μl of 2 N H2SO4. Each plate included a standard curve generated using goat anti-monkey IgG and rhesus macaque IgG (both from Accurate).
A customized Luminex binding antibody multiplex assay (BAMA) was used as previously described (8, 52–54) to measure rectal antibodies to the C.1086 gp140 immunogen or gp70-V1V21157ipd3N4 protein and serum antibodies to consensus C peptides representing the gp41 immunodominant (ID) domain (DQQLLGMWGCSGKLIC; GenScript), the gp120 V3 loop (cysteine-bonded CTRPNNNTRKSIRIGPGQTFYATGDIIGDIRQAHC; Neolabs), and the gp120 constant 1 domain (C1) (MHEDIISLWDESLKPCVKLTPLCV; Neolabs). Briefly, protein- or peptide-conjugated beads were mixed overnight at 4°C with 5-fold dilutions of the standard and serum or secretions. The standards were calibrated by an ELISA (54) and consisted of pooled purified IgG or IgG-depleted serum (for IgA assays) from SHIV-vaccinated and -infected macaques (55). Beads were alternatively washed and mixed for 30 min with 100 μl of 20 μg/ml biotinylated anti-monkey IgG or IgA (Rockland Immunochemicals, Pottstown, PA) followed by a 1:400 dilution of neutravidin-phycoerythrin (SouthernBiotech, Birmingham, AL). After measurement of fluorescence in a BioPlex 200 instrument (Bio-Rad, Hercules, CA), the concentrations of antibody were interpolated from standard curves. Antibody concentrations in secretions were divided by the total IgG or IgA concentrations to obtain the specific activity. Total IgG and IgA were measured by an ELISA using goat anti-monkey IgG or IgA antibodies (Rockland) as described previously (50).
ADCC assay.
The ADCC assay was performed as previously described (56). Briefly, CCR5+ CEM NKr cells with a Tat-inducible luciferase promoter (kindly provided by David Evans, University of Wisconsin at Madison) were infected through spinoculation with 250 μl of the SHIV1157ipd3N4 challenge stock virus for 2 h at 1,200 × g at 25°C. Infection was allowed to proceed for 4 days. On day 3, infected cells were checked for the level of infection by flow cytometry after intracellular staining for SIV Gag. On day 4, infected targets were incubated with serum (1:50 dilution) or monoclonal antibodies and rhesus CD16-expressing KHYG1 NK effector cells (from David Evans) at a 10:1 effector-to-target cell ratio for 8 h. A 150-μl volume of the cell mixture was then added to 50 μl of BriteLite Plus luciferase substrate reagent in a black 96-well plate (both from Perkin Elmer, Duluth, GA). Luciferase activity was measured 2 min later. ADCC activity was calculated as the percent reduction in luciferase compared to effector and target cells alone.
Plasmablasts in blood and plasma cells in bone marrow.
IgG and IgA plasmablast responses were measured 5 days after the 2nd and 3rd MVA immunizations (MVA2 and MVA3) and after the 1st and 2nd Env protein immunizations (P1 and P2) as previously described (57). Briefly, 96-well multiscreen HTS filter plates (Millipore, Burlington, MA) were coated overnight at 4°C with 8 μg/ml of goat anti-monkey IgG or IgA antibody (Rockland) or with 10 μg/ml of C.1086 gp140. The plates were washed and blocked with complete RPMI 1640 medium for 2 h at 37°C in 5% CO2. Threefold serial dilutions of PBMCs from each RM were added and incubated overnight at 37°C. The next day, the PBMCs were discarded, and the plates were washed. They were then developed using biotinylated anti-monkey IgG or IgA (Rockland), streptavidin-conjugated horseradish peroxidase (Vector Laboratories), and nitroblue tetrazolium (NBT)–5-bromo-4-chloro-3-indolylphosphate (BCIP) (Thermo Fisher Scientific). Water was used to stop the reaction after spots became visible. Plates were allowed to dry, and spots were counted the following day. For measuring plasma cells in bone marrow, we followed the same procedure except that cells from bone marrow were used in place of PBMCs.
Measurement of neutralizing antibody.
SHIV-specific neutralization was measured as a reduction in luciferase expression after a single round of infection in TZM-bl cells as described previously (58).
Statistical analysis.
GraphPad Prism v7.0 was used to perform all nonparametric two-tailed statistical analyses. For comparisons between DDMMM and DDMMPP vaccination groups, a Mann-Whitney rank sum test was used. For comparisons between time points within the same group, the Wilcoxon matched-pairs test was used. The Spearman rank test was used for correlations. Statistical significance was defined as a P value of <0.05.
ACKNOWLEDGMENTS
We thank the veterinary and animal care staff of Yerkes National Primate Research Center for their assistance in the care of animals and for sample collection; the Emory CFAR virology core, Benton Lawson, and Melon Tekola Nega for viral load assays; the NIH AIDS Research and Reference Reagent Program for the provision of peptides and cell lines; and Ruth Ruprecht and the Dana-Farber Cancer Institute for the generous donation of the SHIV1157ipd3N4 virus to the repository.
This work was supported in part by National Institutes of Health grants PO1 AI088575 (to R.R.A.), U19AI096187 (to R.R.A.), and U19AI109633 (to R.R.A.), Emory University CFAR grant P30 AI050409, NCRR/NIH base grant P30 RR00165, and the Emory Vaccinology Training Program (T32AI074492). R.R.A. is a coinventor of DNA/MVA vaccine technology that has been licensed to Geovax Inc. by Emory University.
REFERENCES
- 1.Fauci AS, Folkers GK, Marston HD. 2014. Ending the global HIV/AIDS pandemic: the critical role of an HIV vaccine. Clin Infect Dis 59(Suppl 2):S80–S84. doi: 10.1093/cid/ciu420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH-TAVEG Investigators. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 3.Kim JH, Excler JL, Michael NL. 2015. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu Rev Med 66:423–437. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- 4.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zolla-Pazner S, deCamp AC, Cardozo T, Karasavvas N, Gottardo R, Williams C, Morris DE, Tomaras G, Rao M, Billings E, Berman P, Shen X, Andrews C, O’Connell RJ, Ngauy V, Nitayaphan S, de Souza M, Korber B, Koup R, Bailer RT, Mascola JR, Pinter A, Montefiori D, Haynes BF, Robb ML, Rerks-Ngarm S, Michael NL, Gilbert PB, Kim JH. 2013. Analysis of V2 antibody responses induced in vaccinees in the ALVAC/AIDSVAX HIV-1 vaccine efficacy trial. PLoS One 8:e53629. doi: 10.1371/journal.pone.0053629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Zhang L, Janes H, Frahm N, Isaacs A, Kim JH, Montefiori D, McElrath MJ, Tomaras GD, Gilbert PB. 2017. Predictors of durable immune responses six months after the last vaccination in preventive HIV vaccine trials. Vaccine 35:1184–1193. doi: 10.1016/j.vaccine.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tassaneetrithep B, Tivon D, Swetnam J, Karasavvas N, Michael NL, Kim JH, Marovich M, Cardozo T. 2014. Cryptic determinant of alpha4beta7 binding in the V2 loop of HIV-1 gp120. PLoS One 9:e108446. doi: 10.1371/journal.pone.0108446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer SS, Gangadhara S, Victor B, Shen X, Chen X, Nabi R, Kasturi SP, Sabula MJ, Labranche CC, Reddy PB, Tomaras GD, Montefiori DC, Moss B, Spearman P, Pulendran B, Kozlowski PA, Amara RR. 2016. Virus-like particles displaying trimeric simian immunodeficiency virus (SIV) envelope gp160 enhance the breadth of DNA/modified vaccinia virus Ankara SIV vaccine-induced antibody responses in rhesus macaques. J Virol 90:8842–8854. doi: 10.1128/JVI.01163-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NPM, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, Rao M, Chung AW, Dowell KG, Bailey-Kellogg C, Brown EP, Ackerman ME, Vargas-Inchaustegui DA, Whitney S, Doster MN, Binello N, Pegu P, Montefiori DC, Foulds K, Quinn DS, Donaldson M, Liang F, Loré K, Roederer M, Koup RA, McDermott A, Ma Z-M, Miller CJ, Phan TB, Forthal DN, Blackburn M, Caccuri F, Bissa M, Ferrari G, Kalyanaraman V, Ferrari MG, Thompson D, Robert-Guroff M, Ratto-Kim S, Kim JH, Michael NL, Phogat S, Barnett SW, Tartaglia J, Venzon D, Stablein DM, et al. . 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22:762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaccari M, Fourati S, Gordon SN, Brown DR, Bissa M, Schifanella L, Silva de Castro I, Doster MN, Galli V, Omsland M, Fujikawa D, Gorini G, Liyanage NPM, Trinh HV, McKinnon KM, Foulds KE, Keele BF, Roederer M, Koup RA, Shen X, Tomaras GD, Wong MP, Munoz KJ, Gach JS, Forthal DN, Montefiori DC, Venzon DJ, Felber BK, Rosati M, Pavlakis GN, Rao M, Sekaly RP, Franchini G. 2018. HIV vaccine candidate activation of hypoxia and the inflammasome in CD14(+) monocytes is associated with a decreased risk of SIVmac251 acquisition. Nat Med 24:847–856. doi: 10.1038/s41591-018-0025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, Forthal DN, Ourmanov I, Hirsch VM, Carville A, Mansfield KG, Stablein D, Pau MG, Schuitemaker H, Sadoff JC, Billings EA, Rao M, Robb ML, Kim JH, Marovich MA, Goudsmit J, Michael NL. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, Candido MA, Kozyr NL, Earl PL, Smith JM, Ma HL, Grimm BD, Hulsey ML, Miller J, McClure HM, McNicholl JM, Moss B, Robinson HL. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- 13.Smith JM, Amara RR, McClure HM, Patel M, Sharma S, Yi H, Chennareddi L, Herndon JG, Butera ST, Heneine W, Ellenberger DL, Parekh B, Earl PL, Wyatt LS, Moss B, Robinson HL. 2004. Multiprotein HIV type 1 clade B DNA/MVA vaccine: construction, safety, and immunogenicity in macaques. AIDS Res Hum Retroviruses 20:654–665. doi: 10.1089/0889222041217419. [DOI] [PubMed] [Google Scholar]
- 14.Lai L, Vodros D, Kozlowski PA, Montefiori DC, Wilson RL, Akerstrom VL, Chennareddi L, Yu T, Kannanganat S, Ofielu L, Villinger F, Wyatt LS, Moss B, Amara RR, Robinson HL. 2007. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology 369:153–167. doi: 10.1016/j.virol.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai L, Kwa S, Kozlowski PA, Montefiori DC, Ferrari G, Johnson WE, Hirsch V, Villinger F, Chennareddi L, Earl PL, Moss B, Amara RR, Robinson HL. 2011. Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. J Infect Dis 204:164–173. doi: 10.1093/infdis/jir199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwa S, Lai L, Gangadhara S, Siddiqui M, Pillai VB, Labranche C, Yu T, Moss B, Montefiori DC, Robinson HL, Kozlowski PA, Amara RR. 2014. CD40L-adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus SIV239 vaccine enhances SIV-specific humoral and cellular immunity and improves protection against a heterologous SIVE660 mucosal challenge. J Virol 88:9579–9589. doi: 10.1128/JVI.00975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwa S, Sadagopal S, Shen X, Hong JJ, Gangadhara S, Basu R, Victor B, Iyer SS, LaBranche CC, Montefiori DC, Tomaras GD, Villinger F, Moss B, Kozlowski PA, Amara RR. 2015. CD40L-adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus (SIV) vaccine enhances protection against neutralization-resistant mucosal SIV infection. J Virol 89:4690–4695. doi: 10.1128/JVI.03527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chea LS, Amara RR. 2017. Immunogenicity and efficacy of DNA/MVA HIV vaccines in rhesus macaque models. Expert Rev Vaccines 16:973–985. doi: 10.1080/14760584.2017.1371594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JM, Amara RR, Campbell D, Xu Y, Patel M, Sharma S, Butera ST, Ellenberger DL, Yi H, Chennareddi L, Herndon JG, Wyatt LS, Montefiori D, Moss B, McClure HM, Robinson HL. 2004. DNA/MVA vaccine for HIV type 1: effects of codon-optimization and the expression of aggregates or virus-like particles on the immunogenicity of the DNA prime. AIDS Res Hum Retroviruses 20:1335–1347. doi: 10.1089/aid.2004.20.1335. [DOI] [PubMed] [Google Scholar]
- 20.Wyatt LS, Earl PL, Liu JY, Smith JM, Montefiori DC, Robinson HL, Moss B. 2004. Multiprotein HIV type 1 clade B DNA and MVA vaccines: construction, expression, and immunogenicity in rodents of the MVA component. AIDS Res Hum Retroviruses 20:645–653. doi: 10.1089/0889222041217428. [DOI] [PubMed] [Google Scholar]
- 21.Manrique M, Kozlowski PA, Cobo-Molinos A, Wang S-W, Wilson RL, Martinez-Viedma MDP, Montefiori DC, Carville A, Aldovini A. 2014. Resistance to infection, early and persistent suppression of simian immunodeficiency virus SIVmac251 viremia, and significant reduction of tissue viral burden after mucosal vaccination in female rhesus macaques. J Virol 88:212–224. doi: 10.1128/JVI.02523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Shinawi M, Mohamed HT, El-Ghonaimy EA, Tantawy M, Younis A, Schneider RJ, Mohamed MM. 2013. Human cytomegalovirus infection enhances NF-kappaB/p65 signaling in inflammatory breast cancer patients. PLoS One 8:e55755. doi: 10.1371/journal.pone.0055755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goonetilleke N, Moore S, Dally L, Winstone N, Cebere I, Mahmoud A, Pinheiro S, Gillespie G, Brown D, Loach V, Roberts J, Guimaraes-Walker A, Hayes P, Loughran K, Smith C, De Bont J, Verlinde C, Vooijs D, Schmidt C, Boaz M, Gilmour J, Fast P, Dorrell L, Hanke T, McMichael AJ. 2006. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J Virol 80:4717–4728. doi: 10.1128/JVI.80.10.4717-4728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes P, Gilmour J, von Lieven A, Gill D, Clark L, Kopycinski J, Cheeseman H, Chung A, Alter G, Dally L, Zachariah D, Lombardo A, Ackland J, Sayeed E, Jackson A, Boffito M, Gazzard B, Fast PE, Cox JH, Laufer D. 2013. Safety and immunogenicity of DNA prime and modified vaccinia Ankara virus-HIV subtype C vaccine boost in healthy adults. Clin Vaccine Immunol 20:397–408. doi: 10.1128/CVI.00637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer SS, Amara RR. 2014. DNA/MVA vaccines for HIV/AIDS. Vaccines (Basel) 2:160–178. doi: 10.3390/vaccines2010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramanathan VD, Kumar M, Mahalingam J, Sathyamoorthy P, Narayanan PR, Solomon S, Panicali D, Chakrabarty S, Cox J, Sayeed E, Ackland J, Verlinde C, Vooijs D, Loughran K, Barin B, Lombardo A, Gilmour J, Stevens G, Smith MS, Tarragona-Fiol T, Hayes P, Kochhar S, Excler JL, Fast P. 2009. A phase 1 study to evaluate the safety and immunogenicity of a recombinant HIV type 1 subtype C-modified vaccinia Ankara virus vaccine candidate in Indian volunteers. AIDS Res Hum Retroviruses 25:1107–1116. doi: 10.1089/aid.2009.0096. [DOI] [PubMed] [Google Scholar]
- 27.Sandstrom E, Nilsson C, Hejdeman B, Brave A, Bratt G, Robb M, Cox J, Vancott T, Marovich M, Stout R, Aboud S, Bakari M, Pallangyo K, Ljungberg K, Moss B, Earl P, Michael N, Birx D, Mhalu F, Wahren B, Biberfeld G, HIV Immunogenicity Study 01/02 Team. 2008. Broad immunogenicity of a multigene, multiclade HIV-1 DNA vaccine boosted with heterologous HIV-1 recombinant modified vaccinia virus Ankara. J Infect Dis 198:1482–1490. doi: 10.1086/592507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cebere I, Dorrell L, McShane H, Simmons A, McCormack S, Schmidt C, Smith C, Brooks M, Roberts JE, Darwin SC, Fast PE, Conlon C, Rowland-Jones S, McMichael AJ, Hanke T. 2006. Phase I clinical trial safety of DNA- and modified virus Ankara-vectored human immunodeficiency virus type 1 (HIV-1) vaccines administered alone and in a prime-boost regime to healthy HIV-1-uninfected volunteers. Vaccine 24:417–425. doi: 10.1016/j.vaccine.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 29.Hanke T, Goonetilleke N, McMichael AJ, Dorrell L. 2007. Clinical experience with plasmid DNA- and modified vaccinia virus Ankara-vectored human immunodeficiency virus type 1 clade A vaccine focusing on T-cell induction. J Gen Virol 88:1–12. doi: 10.1099/vir.0.82493-0. [DOI] [PubMed] [Google Scholar]
- 30.Guimaraes-Walker A, Mackie N, McCormack S, Hanke T, Schmidt C, Gilmour J, Barin B, McMichael A, Weber J, Legg K, Babiker A, Hayes P, Gotch F, Smith C, Dally L, Dorrell L, Cebere I, Kay R, Winstone N, Moore S, Goonetilleke N, Fast P, IAVI-006 Study Group. 2008. Lessons from IAVI-006, a phase I clinical trial to evaluate the safety and immunogenicity of the pTHr.HIVA DNA and MVA.HIVA vaccines in a prime-boost strategy to induce HIV-1 specific T-cell responses in healthy volunteers. Vaccine 26:6671–6677. doi: 10.1016/j.vaccine.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Aboud S, Nilsson C, Karlen K, Marovich M, Wahren B, Sandstrom E, Gaines H, Biberfeld G, Godoy-Ramirez K. 2010. Strong HIV-specific CD4+ and CD8+ T-lymphocyte proliferative responses in healthy individuals immunized with an HIV-1 DNA vaccine and boosted with recombinant modified vaccinia virus Ankara expressing HIV-1 genes. Clin Vaccine Immunol 17:1124–1131. doi: 10.1128/CVI.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goepfert PA, Elizaga ML, Sato A, Qin L, Cardinali M, Hay CM, Hural J, DeRosa SC, DeFawe OD, Tomaras GD, Montefiori DC, Xu Y, Lai L, Kalams SA, Baden LR, Frey SE, Blattner WA, Wyatt LS, Moss B, Robinson HL, National Institute of Allergy and Infectious Diseases HIV Vaccine Trials Network. 2011. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis 203:610–619. doi: 10.1093/infdis/jiq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsson C, Godoy-Ramirez K, Hejdeman B, Brave A, Gudmundsdotter L, Hallengard D, Currier JR, Wieczorek L, Hasselrot K, Earl PL, Polonis VR, Marovich MA, Robb ML, Sandstrom E, Wahren B, Biberfeld G. 2014. Broad and potent cellular and humoral immune responses after a second late HIV-modified vaccinia virus Ankara vaccination in HIV-DNA-primed and HIV-modified vaccinia virus Ankara-boosted Swedish vaccinees. AIDS Res Hum Retroviruses 30:299–311. doi: 10.1089/AID.2013.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chamcha V, Kannanganat S, Gangadhara S, Nabi R, Kozlowski PA, Montefiori DC, LaBranche CC, Wrammert J, Keele BF, Balachandran H, Sahu S, Lifton M, Santra S, Basu R, Moss B, Robinson HL, Amara RR. 2016. Strong, but age-dependent, protection elicited by a DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. Open Forum Infect Dis 3:ofw034. doi: 10.1093/ofid/ofw034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burton SL, Kilgore KM, Smith SA, Reddy S, Hunter E, Robinson HL, Silvestri G, Amara RR, Derdeyn CA. 2015. Breakthrough of SIV strain smE660 challenge in SIV strain mac239-vaccinated rhesus macaques despite potent autologous neutralizing antibody responses. Proc Natl Acad Sci U S A 112:10780–10785. doi: 10.1073/pnas.1509731112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilgore KM, Murphy MK, Burton SL, Wetzel KS, Smith SA, Xiao P, Reddy S, Francella N, Sodora DL, Silvestri G, Cole KS, Villinger F, Robinson JE, Pulendran B, Hunter E, Collman RG, Amara RR, Derdeyn CA. 2015. Characterization and implementation of a diverse simian immunodeficiency virus SIVsm envelope panel in the assessment of neutralizing antibody breadth elicited in rhesus macaques by multimodal vaccines expressing the SIVmac239 envelope. J Virol 89:8130–8151. doi: 10.1128/JVI.01221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen X, Basu R, Sawant S, Beaumont D, Kwa SF, LaBranche C, Seaton KE, Yates NL, Montefiori DC, Ferrari G, Wyatt LS, Moss B, Alam SM, Haynes BF, Tomaras GD, Robinson HL. 2017. HIV-1 gp120 and modified vaccinia virus Ankara (MVA) gp140 boost immunogens increase immunogenicity of a DNA/MVA HIV-1 vaccine. J Virol 91:e01077-17. doi: 10.1128/JVI.01077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, Rapolu M, Kumar S, Gupta M, Liang Z, Han Z, Williams P, Su WW. 2017. Coordinated protein co-expression in plants by harnessing the synergy between an intein and a viral 2A peptide. Plant Biotechnol J 15:718–728. doi: 10.1111/pbi.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer SS, Gangadhara S, Victor B, Gomez R, Basu R, Hong JJ, Labranche C, Montefiori DC, Villinger F, Moss B, Amara RR. 2015. Codelivery of envelope protein in alum with MVA vaccine induces CXCR3-biased CXCR5+ and CXCR5− CD4 T cell responses in rhesus macaques. J Immunol 195:994–1005. doi: 10.4049/jimmunol.1500083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol 80:8729–8738. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradley T, Pollara J, Santra S, Vandergrift N, Pittala S, Bailey-Kellogg C, Shen X, Parks R, Goodman D, Eaton A, Balachandran H, Mach LV, Saunders KO, Weiner JA, Scearce R, Sutherland LL, Phogat S, Tartaglia J, Reed SG, Hu SL, Theis JF, Pinter A, Montefiori DC, Kepler TB, Peachman KK, Rao M, Michael NL, Suscovich TJ, Alter G, Ackerman ME, Moody MA, Liao HX, Tomaras G, Ferrari G, Korber BT, Haynes BF. 2017. Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge. Nat Commun 8:15711. doi: 10.1038/ncomms15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones AT, Chamcha V, Kesavardhana S, Shen X, Beaumont D, Das R, Wyatt LS, LaBranche CC, Stanfield-Oakley S, Ferrari G, Montefiori DC, Moss B, Tomaras GD, Varadarajan R, Amara RR. 2017. A trimeric HIV-1 envelope gp120 immunogen induces potent and broad anti-V1V2 loop antibodies against HIV-1 in rabbits and rhesus macaques. J Virol 92:e01796-17. doi: 10.1128/JVI.01796-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hessell AJ, Shapiro MB, Powell R, Malherbe DC, McBurney SP, Pandey S, Cheever T, Sutton WF, Kahl C, Park B, Zolla-Pazner S, Haigwood NL. 2018. Reduced cell-associated DNA and improved viral control in macaques following passive transfer of a single anti-V2 monoclonal antibody and repeated simian/human immunodeficiency virus challenges. J Virol 92:e02198-17. doi: 10.1128/JVI.02198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 45.Lai L, Kwa SF, Kozlowski PA, Montefiori DC, Nolen TL, Hudgens MG, Johnson WE, Ferrari G, Hirsch VM, Felber BK, Pavlakis GN, Earl PL, Moss B, Amara RR, Robinson HL. 2012. SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine 30:1737–1745. doi: 10.1016/j.vaccine.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kannanganant S, Gangadhara S, Lai L, Lawson B, Kozlowski PA, Robinson HL, Amara RR. 2014. Local control of repeated-dose rectal challenges in DNA/MVA-vaccinated macaques protected against a first series of simian immunodeficiency virus challenges. J Virol 88:5864–5869. doi: 10.1128/JVI.00145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. 2013. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burton S, Spicer LM, Charles TP, Gangadhara S, Reddy PB, Styles TM, Velu V, Kasturi SP, Legere T, Hunter E, Pulendran B, Amara R, Hraber P, Derdeyn CA. 2019. Clade C HIV-1 envelope vaccination regimens differ in their ability to elicit antibodies with moderate neutralization breadth against genetically diverse tier 2 HIV-1 envelope variants. J Virol 93:e01846-18. doi: 10.1128/JVI.01846-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O’Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozlowski PA, Lynch RM, Patterson RR, Cu-Uvin S, Flanigan TP, Neutra MR. 2000. Modified wick method using Weck-Cel sponges for collection of human rectal secretions and analysis of mucosal HIV antibody. J Acquir Immune Defic Syndr 24:297–309. doi: 10.1097/00042560-200008010-00001. [DOI] [PubMed] [Google Scholar]
- 51.Pegu P, Vaccari M, Gordon S, Keele BF, Doster M, Guan Y, Ferrari G, Pal R, Ferrari MG, Whitney S, Hudacik L, Billings E, Rao M, Montefiori D, Tomaras G, Alam SM, Fenizia C, Lifson JD, Stablein D, Tartaglia J, Michael N, Kim J, Venzon D, Franchini G. 2013. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J Virol 87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol 82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trama AM, Moody MA, Alam SM, Jaeger FH, Lockwood B, Parks R, Lloyd KE, Stolarchuk C, Scearce R, Foulger A, Marshall DJ, Whitesides JF, Jeffries TL Jr, Wiehe K, Morris L, Lambson B, Soderberg K, Hwang KK, Tomaras GD, Vandergrift N, Jackson KJL, Roskin KM, Boyd SD, Kepler TB, Liao HX, Haynes BF. 2014. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe 16:215–226. doi: 10.1016/j.chom.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nabi R, Moldoveanu Z, Wei Q, Golub ET, Durkin HG, Greenblatt RM, Herold BC, Nowicki MJ, Kassaye S, Cho MW, Pinter A, Landay AL, Mestecky J, Kozlowski PA. 2017. Differences in serum IgA responses to HIV-1 gp41 in elite controllers compared to viral suppressors on highly active antiretroviral therapy. PLoS One 12:e0180245. doi: 10.1371/journal.pone.0180245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao J, Lai L, Amara RR, Montefiori DC, Villinger F, Chennareddi L, Wyatt LS, Moss B, Robinson HL. 2009. Preclinical studies of human immunodeficiency virus/AIDS vaccines: inverse correlation between avidity of anti-Env antibodies and peak postchallenge viremia. J Virol 83:4102–4111. doi: 10.1128/JVI.02173-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alpert MD, Heyer LN, Williams DE, Harvey JD, Greenough T, Allhorn M, Evans DT. 2012. A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J Virol 86:12039–12052. doi: 10.1128/JVI.01650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasturi SP, Kozlowski PA, Nakaya HI, Burger MC, Russo P, Pham M, Kovalenkov Y, Silveira EL, Havenar-Daughton C, Burton SL, Kilgore KM, Johnson MJ, Nabi R, Legere T, Sher ZJ, Chen X, Amara RR, Hunter E, Bosinger SE, Spearman P, Crotty S, Villinger F, Derdeyn CA, Wrammert J, Pulendran B. 2017. Adjuvanting a simian immunodeficiency virus vaccine with Toll-like receptor ligands encapsulated in nanoparticles induces persistent antibody responses and enhanced protection in TRIM5alpha restrictive macaques. J Virol 91:e01844-16. doi: 10.1128/JVI.01844-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter12:Unit 12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]