Although APOBEC3 proteins are known to be important antiviral restriction factors in both mice and humans, their roles in the restriction of endogenous retroviruses (ERV) have been limited to in vitro studies. Here, we report that human APOBEC3G expressed as a transgene in mice prevents the emergence of infectious ERV from endogenous loci. This study reveals that APOBEC3G can powerfully restrict active retrotransposons in vivo and demonstrates how transgenic mice can be used to investigate host mechanisms that inhibit retrotransposons and reinforce genomic integrity.
KEYWORDS: APOBEC, innate immunity, retroviruses, Toll-like receptors
ABSTRACT
Endogenous retroviruses (ERV) are found throughout vertebrate genomes, and failure to silence their activation can have deleterious consequences on the host. Mutation and subsequent disruption of ERV loci is therefore an indispensable component of the cell-intrinsic defenses that maintain the integrity of the host genome. Abundant in vitro and in silico evidence have revealed that APOBEC3 cytidine-deaminases, including human APOBEC3G (hA3G), can potently restrict retrotransposition; yet, in vivo data demonstrating such activity is lacking, since no replication-competent human ERV have been identified. In mice deficient for Toll-like receptor 7 (TLR7), transcribed ERV loci can recombine and generate infectious ERV. In this study, we show that ectopic expression of hA3G can prevent the emergence of replication-competent, infectious ERV in Tlr7−/− mice. Mice encode one copy of Apobec3 in their genome. ERV reactivation in Tlr7−/− mice was comparable in the presence or absence of Apobec3. In contrast, expression of a human APOBEC3G transgene abrogated emergence of infectious ERV in the Tlr7−/− background. No ERV RNA was detected in the plasma of hA3G+ Apobec3−/− Tlr7−/− mice, and infectious ERV virions could not be amplified through coculture with permissive cells. These data reveal that hA3G can potently restrict active ERV in vivo and suggest that expansion of the APOBEC3 locus in primates may have helped to provide for the continued restraint of ERV in the human genome.
IMPORTANCE Although APOBEC3 proteins are known to be important antiviral restriction factors in both mice and humans, their roles in the restriction of endogenous retroviruses (ERV) have been limited to in vitro studies. Here, we report that human APOBEC3G expressed as a transgene in mice prevents the emergence of infectious ERV from endogenous loci. This study reveals that APOBEC3G can powerfully restrict active retrotransposons in vivo and demonstrates how transgenic mice can be used to investigate host mechanisms that inhibit retrotransposons and reinforce genomic integrity.
INTRODUCTION
Roughly 8 to 10% of both human and murine genomes is composed of endogenous retroviruses (ERV), the endogenized counterparts of ancient retroviruses that invaded the germ line and became fixed within these genomes (1, 2). The provirus-like ERV present in the genomes of common laboratory mouse strains (3, 4) formed following infection by exogenous murine leukemia virus (MLV) (5), and these ERV loci are actively transcribed and translated. Although wild-type C57BL/6 mice do not contain a proviral ERV locus capable of independently generating replication-competent ERV (6–9), infectious ERV virions readily emerge when B-cell-dependent humoral control is compromised or when Toll-like receptor 7 (TLR7) signaling is deficient (10, 11). In addition to antibody- and TLR7-mediated control, transcriptional silencing, and stochastic recombination events that remove ERV from the genome (12, 13), mutagenesis of retroelement sequences by apolipoprotein B editing complex 3 (APOBEC3) proteins is an important component of this innate defense against ERV.
Present throughout vertebrate genomes, APOBEC3 proteins are zinc-dependent cytosine deaminases that act on single-stranded DNA (ssDNA) to cause cytosine-to-uracil mutations in targeted sequences (14, 15). Although mouse genomes encode a single Apobec3 gene, mA3, expansion of this locus in primates has given rise to seven APOBEC3 genes, APOBEC3A, -3B, -3D/E, -3F, -3G, and -3H (16, 17). APOBEC3 proteins, and particularly the human APOBEC3G (hA3G), have long been appreciated for their potent restriction of exogenous retroviruses (18–20). Originally characterized for its activity against human immunodeficiency virus 1 (HIV-1) (21), hA3G is a restriction factor that is packaged into retroviral virions, which upon entry into a target cell hypermutates reverse transcribed viral ssDNA through its deaminase domain (22–27). hA3G also inhibits reverse transcriptase in a deaminase-independent manner (28–30). Just as HIV-1 is inhibited by hA3G, MLV is potently restricted by both mA3 and hA3G (22, 25, 31, 32), with hA3G capable of blocking primary infection with exogenous MLV when expressed as a transgene in mice (33).
In addition, multiple studies have demonstrated that human APOBEC3 proteins can act to inhibit the in vitro retrotransposition of retroelements, including the yeast retrotransposon Ty1, murine intracisternal A particles (IAP) and MusD ERV elements, and human long interspersed element 1 (LINE1) and Alu elements (34–40). Although hA3G was first shown to possess activity against Ty1 (38, 41), it has since been demonstrated that hA3G restricts MusD and IAP elements when overexpressed in in vitro reporter assays (39, 40) and can hypermutate human ERV (HERV) sequences (42, 43). This in vitro evidence is also supported by in silico data showing that mA3 and hA3 family members have targeted ERV and HERV genomic loci (44), respectively, including those encoding the proviral ERV capable of emergence (45). However, the extent to which hA3G restricts ERV and other retroelements in vivo remains unclear, particularly since replication-competent HERV have not been identified in the human genome (25) and identification of A3-restricted retrotransposons is complicated by the high copy number and repetitive nature of the retroelements themselves.
In C57BL/6 mice, the backbone of the recombinant infectious ERV that emerge is formed by a single proviral ERV, Emv2 (10). Because this locus is unique, its increased expression serves as an indicator of ERV emergence. We therefore took advantage of this phenomenon and available hA3G transgenic mice on the mA3 knockout (mA3−/−) background (33) to investigate whether ectopic expression of hA3G protein is able to prevent or impede the emergence of replication-competent ERV from Tlr7−/− mice in vivo.
(This article was submitted to an online preprint archive [46]. )
RESULTS AND DISCUSSION
To investigate the role of hA3G in the restriction of ERV, we first investigated whether mice deficient in mA3 demonstrate spontaneous ERV emergence. As with emerged ERV from recombination-activating gene 1-deficient (Rag1−/−) mice (10), we observed that infectious ERV in Tlr7−/− mice result from recombination between several endogenous retroviral loci (Fig. 1A). Thus, as with exogenous retroviruses (47), TLR7-dependent immunity is essential to limit the transcription of ERV and their potential for recombination and emergence. In contrast to Tlr7−/− (48) and Myd88−/− (49) mice (Fig. 1B), which lack TLR7 and its signaling adapter MyD88, respectively, and are unable to prevent emergence of infectious ERV, mA3−/− mice retain control of ERV (Fig. 1B). This reveals that endogenous mA3 is not required to prevent ERV emergence.
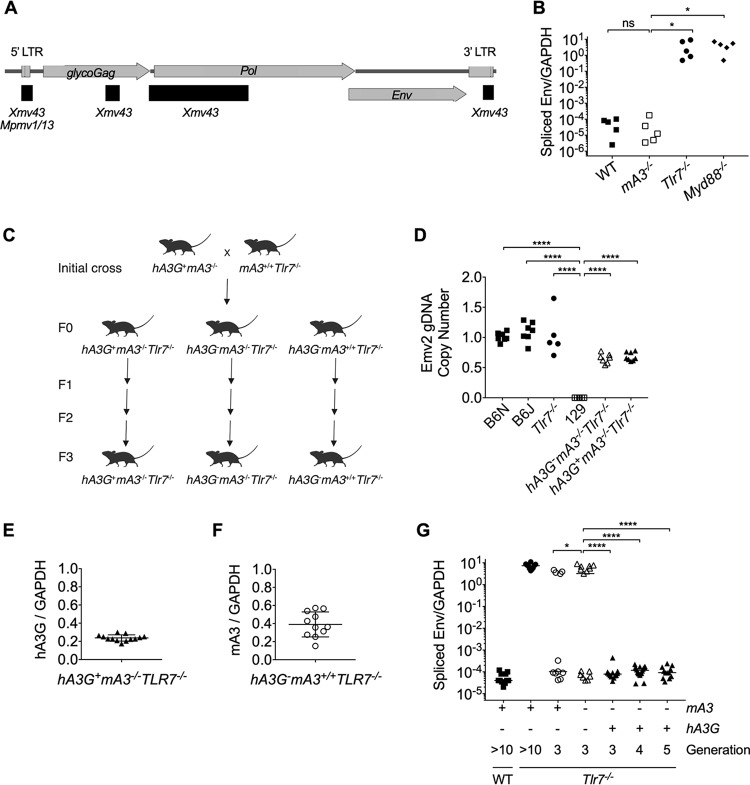
FIG 1.
Human APOBEC3G, but not murine APOBEC3, expression prevents the emergence of infectious ERV in Tlr7−/− mice. (A) Schematic of the structure and open reading frames of the Emv2-based ERV genome isolated from virions amplified through coculture with Tlr7−/− splenocytes. Recombined regions are denoted by black horizontal bars, and the ERV locus that contributed sequence is listed. (B) RT-qPCR of spliced Emv2 envelope expression from peripheral blood of C57BL/6N (n = 5), mA3−/− (n = 5), Tlr7−/− (n = 5), and Myd88−/− (n = 5) mice. (C) Breeding scheme used in this study. The initial cross used to generate the experimental lines is shown. Once the desired homozygous genotypes were obtained (F0), the separate lines were individually bred to the third generation (F3). (D) Quantification of Emv2 copy number by qPCR of gDNA isolated from ear punches of wild-type (WT) C57BL6/N (n = 7), WT C57BL/6J (n = 7), Tlr7−/− (n = 5), 129S1/SvImJ (n = 5), hA3G– mA3−/− Tlr7−/− (n = 7), and hA3G+ mA3−/− Tlr7−/− (n = 8). A primer set amplifying the envelope region of Emv2 was used. The fold copy number over the mean WT C57BL/6N value, normalized to the telomerase reverse transcriptase (Tert) copy number, is plotted. (E) RT-qPCR of hA3G expression from peripheral blood of hA3G+ mA3−/− Tlr7−/− (n = 13) mice. (F) RT-qPCR of mA3 expression from peripheral blood of hA3G– mA3+/+ Tlr7−/− (n = 11) mice. (G) RT-qPCR of spliced Emv2 envelope expression from peripheral blood of C57BL/6N (n = 13), Tlr7−/− (n = 11), F3 hA3G– mA3+/+ Tlr7−/− (n = 11), F3 hA3G-mA3−/− Tlr7–/– (n = 15), F3 hA3G+ mA3−/− Tlr7−/− (n = 13), F4 hA3G+ mA3−/− Tlr7−/− (n = 18), and F5 hA3G+ mA3−/− Tlr7−/− (n = 12) mice. Adjusted P values in Fig. 1B were calculated for one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test comparing normalized spliced Emv2 expression values to those of the mA3−/− mice. Adjusted P values in Fig. 1D were calculated for one-way ANOVA with Dunnett’s multiple-comparison test comparing normalized Emv2 copy number values to those of the 129 controls, which lack the Emv2 locus. Adjusted P values in Fig. 1G were calculated for one-way ANOVA with Dunnett’s multiple-comparison test comparing spliced Emv2 expression values to those of the F3 hA3G– mA3−/− Tlr7−/− controls. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
We next crossed hA3G+ mice lacking mA3 (hA3G+ mA3−/−) to Tlr7−/− mice to generate a first generation (F0) of transgene-positive and -negative mice with homozygous loss of TLR7 (hA3G+ mA3−/− Tlr7−/− and hA3G− mA3−/− Tlr7−/−) (Fig. 1C). In the transgene-positive mice, hA3G expression is driven by the chicken beta-actin promoter, resulting in high expression levels that are comparable across tissues, and comparable to the level found in resting human peripheral blood mononuclear cells (33). We also bred mice that maintained mA3 expression in the absence of TLR7 (hA3G− mA3+/+ Tlr7−/−). These three strains were then bred out for several generations and their gDNA was screened by quantitative PCR (qPCR) to assess the Emv2 copy number in the genome, since the mA3−/− mice, although backcrossed to C57BL/6N for multiple generations, were originally generated using 129 embryonic stem cells, whose genome does not contain the Emv2 locus (50). In this way, we ensured that all mice with a contribution from the mA3−/− genome nevertheless possessed copies of the Emv2 locus (Fig. 1D). These strains were then screened for hA3G transgene expression (Fig. 1E) or mA3 expression (Fig. 1F) and ERV emergence (Fig. 1G) by reverse transcription qPCR (RT-qPCR) from peripheral blood samples.
We first observed ERV emergence in hA3G− mA3+/+ Tlr7−/− and hA3G− mA3−/− Tlr7−/− mice by the third generation (F3) of breeding between homozygous knockouts (Fig. 1G). More than half (8/15) of the hA3G− mA3−/− Tlr7−/− mice were ERV positive, as were 4 of the 11 hA3G− mA3+/+ Tlr7−/− controls. Our finding that the emergence of recombined infectious ERV occurs in TLR7−/− mice is consistent with previously published data (10, 11) that implicate B cells and TLR3/7/9 signaling in the control of ERV. It has also been shown that infectious ERV are generated through recombination between ERV loci (5). Our data (Fig. 1A) are consistent with previously published findings that the generation of replication-competent ERV requires multiple recombination events to restore polymerase function and endow the Emv2-based virus with a nonrestricted capsid (7, 51). We and others (10) observe that several generations of breeding are required for this infectious ERV emergence to occur. We hypothesize that more than one recombination event, and potentially multiple successful reintegration events by the same Emv2-derived sequence, is required to initially generate the infectious ERV.
The previous studies (10, 11) that investigated control of ERV were all performed in mice with intact mA3. Further, ERV emergence in the absence of cytosolic retroviral sensors has not been reported. Our data indicated that mA3 is neither required to prevent ERV emergence nor sufficient to prevent the emergence of ERV in Tlr7−/− mice, although its presence appears to delay this emergence (Fig. 1G). Like most MLV, ERV express glycosylated Gag (Fig. 1A), a longer, glycosylated variant of Gag protein that counters restriction by mA3 (18). The dominant ERV that emerges in our TLR7−/− mice encodes Pr80 and has 84% amino acid identity and 89% amino acid similarity to Moloney MLV Pr80 with no gaps in alignment. Given this high degree of homology, we believe it likely that ERV Pr80 also retains the anti-mA3 activity of Moloney MLV Pr80. If so, ERV glycosylated Gag antagonism of mA3 may underlie the failure of mA3 to prevent ERV emergence.
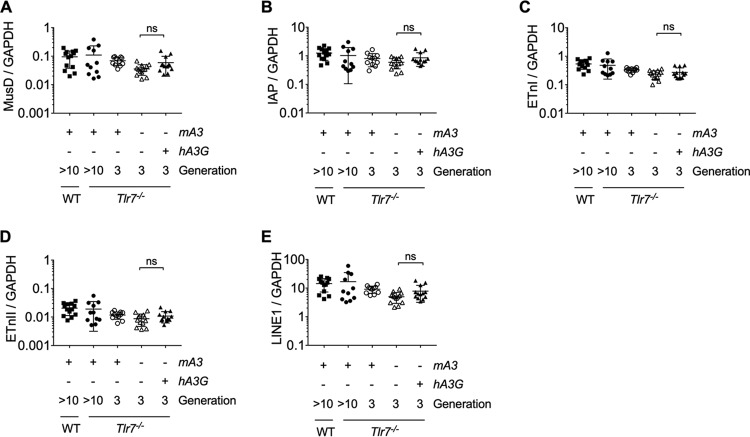
In stark comparison to the effect of mA3, hA3G expression in the Tlr7−/− background entirely abrogated ERV emergence (Fig. 1G). In the F3 offspring, all 13 hA3G+ mA3−/− Tlr7−/− mice retained control of ERV, and this impressive capacity to prevent ERV emergence extended to the fourth (F4) and to the fifth (F5) generations, where all hA3G+ mA3−/− Tlr7−/− mice remained infectious ERV negative. Previous work (39, 40) implicates hA3G in the repression of ERV elements, since ectopic expression of hA3G in cell culture leads to both a decrease in the number of transposed MusD and IAP elements and the deamination of newly transposed cDNA sequences, with no difference in the quantity of intermediary MusD or IAP RNA produced. However, these in vitro data were generated using transposable element reporter constructs in cell culture, and it is not known whether in vitro ectopic hA3G expression recapitulates its in vivo effects upon transposable element expression. We therefore compared global transcription levels of IAP, MusD, early transposons I and II (ETnI and ETnII), or LINE1 elements in hA3G+ mA3−/− Tlr7−/− and hA3G− mA3−/− Tlr7−/− mice. Consistent with in vitro data (39, 40), we did not observe differences in global expression levels of these transposable element families between any of the genotypes by RT-qPCR from peripheral blood (Fig. 2A to E). Thus, our data suggest that neither transgene expression itself nor the mechanism of hA3G restriction involves widespread suppression of long terminal repeat (LTR) retroelement or LINE1 transcription.
FIG 2.
LTR and LINE1 retroelements are not suppressed by the transgenic expression of human APOBEC3G in Tlr7−/− mice. (A to E) Expression of select LTR retrotransposon families and LINE1 families via RT-qPCR using RNA isolated from peripheral blood of C57BL/6N (n = 13), Tlr7−/− (n = 11), F3 hA3G− mA3+/+ Tlr7−/− (n = 11), F3 hA3G− mA3−/− Tlr7–/– (n = 13), and F3 hA3G+ mA3−/− Tlr7−/− (n = 13) mice. Primers amplify the gag or polymerase regions of IAP, MusD, and ETn elements (31) or LINE1 ORFp1. All values are normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression. Means and standard deviations are plotted. The adjusted P values in Fig. 2 were calculated for multiple t tests (two-tailed) comparing transposable element expression values of hA3G− mA3−/− Tlr7−/− and hA3G+ mA3−/− Tlr7−/− mice. All adjusted P values were corrected for the five independent hypotheses tested in Fig. 2 using the Holm-Šidák method with an alpha value of 0.05 for the entire family of comparisons. ns, not significant.
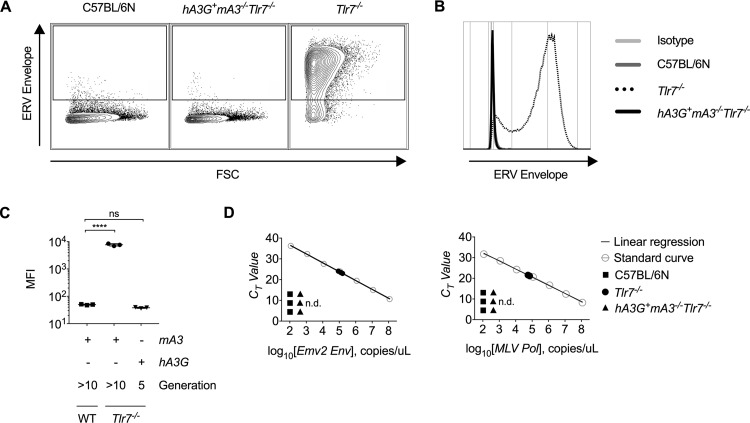
The in vivo restriction of MLV by hA3G occurs by hA3G-mediated deamination of the viral RNA and by inhibition of viral reverse transcriptase (33). We hypothesize that restriction of ERV by hA3G includes hypermutation of partially or fully recombinant ERV transcripts that inhibits their subsequent infectivity. Infectious ERV from hA3G− mA3+/+ Tlr7−/− splenocytes was amplified in coculture with permissive cell lines (Fig. 3A to C), and circulating ERV RNA was detected in plasma from these mice (Fig. 3D). However, we were unable to detect or amplify infectious ERV from splenocytes from hA3G+ mA3−/− Tlr7−/− mice (Fig. 3A to C). Similarly, we did not detect ERV RNA in the plasma of hA3G+ mA3−/− Tlr7−/− mice (Fig. 3D). Because we could not isolate and sequence emerged ERV from these mice, we are unable to ascertain the extent to which deamination contributes to restriction in transgene-positive mice. It is also possible that, as with MLV and HIV-1 infection, hA3G inhibits the reverse transcriptase of polymerase-restored ERV or otherwise impairs subsequent integration (52, 53) during the initial stages of emergence, before infectious virions are reconstituted. To study how hA3G specifically restricts ERV emergence would require the development of a new in vitro system that accurately recapitulates the events that give rise to emergence.
FIG 3.
Infectious ERV cannot be detected in the plasma or isolated through splenocyte coculture from hA3G+ mA3−/− Tlr7−/− mice. (A to C) Representative flow cytometry plots (A), histograms (B), and calculated mean fluorescence intensities (C) of ERV envelope expression on live, CD45.2-negative DFJ8 cells after 7 days of coculture with C57BL/6N, Tlr7−/−, or F5 hA3G+ mA3−/− Tlr7−/− splenocytes (n = 3 mice per group). (D) Absolute quantification of the number of polymerase or unspliced ERV envelope RNA copies per microliter of cDNA generated from plasma of 16-week-old C57BL/6N, Tlr7−/−, and hA3G+ mA3−/− Tlr7−/− mice. Plots are representative of three independent experiments. P values in Fig. 1C were calculated using one-way ANOVA with Dunnett’s multiple-comparison test comparing values to those of the WT control. ****, P < 0.0001; ns, not significant.
In this in vivo system, the ectopic expression of hA3G in murine cells was driven by a chicken beta-actin promoter, yielding constitutive expression across tissue subtypes. This basal and global expression enabled us to capture the effect of hA3G expression upon ERV transcripts prior to their recombination and emergence, independent of the breeding generation or tissue subtype within which the recombination events occurred. However, while hA3G is endogenously expressed in unstimulated human hematopoietic cells and testis (54), its expression is rapidly induced across cell types in response to interferon stimulation (18, 19). Because of this difference in expression pattern, endogenous hA3G may not act upon transcribed HERV in an entirely analogous manner. Similarly, although hA3G restricts both murine and human retroviruses through conserved mechanisms, it is possible that its anti-ERV and anti-HERV activities differ.
In this study, we have demonstrated that hA3G, which has previously been shown to restrict exogenous retrovirus infection, powerfully restricts emergence of endogenous retroviruses in TLR7-deficient mice in vivo. These data extend our understanding of the function of this protein and reveal an important layer of host defense that reinforces genomic integrity. Indeed, the expansion of the APOBEC3 locus and the presence of hA3G may contribute to the mechanisms that prevent reconstitution of replication-competent HERV in humans. While the sequence of events that are required for restriction of ERV emergence by hA3G have yet to be characterized, this study demonstrates that transgenic mice can serve as a powerful tool to investigate how proteins such as hA3G impact retroelements and restrict their movement within the genome.
MATERIALS AND METHODS
Mice.
C57BL/6N mice (strain code 027) were obtained from Charles River Laboratories and bred in-house. C57BL/6J mice (stock number 000664) and 129S1/SvImJ mice (stock number 002448) were obtained from The Jackson Laboratory. Tlr7−/− (C57BL/6N) mice (48) were bred in-house. High-expression-level transgenic hA3G+ mA3−/− mice and hA3G− mA3−/− mice (33) were maintained by breeding between transgene-positive and/or -negative mA3 knockout mice. Myd88−/− mice (49) were kindly provided by Doug Golenbock. All non-wild-type mouse strains were backcrossed at least six times to the C57BL/6N genome and were positive for at least two genomic copies of the Emv2 locus by qPCR (Fig. 1D). All mice were housed under specific-pathogen-free conditions, and care was provided in accordance with Yale University IACUC guidelines (protocol 10365). The hA3G− mA3−/− Tlr7−/− mice were maintained as a separate line from the transgene-negative littermates of hA3G+ mA3−/− Tlr7−/− crosses to ensure that the ERV transcripts and genomic loci in the hA3G− mA3−/− Tlr7–/– genome were not subject to effects of hA3G expression.
Genotyping.
Genomic DNA was obtained from ear punches by boiling the tissues in lysis buffer (25 mM NaOH, 2 mM EDTA [pH 8.0]) for 30 min and neutralizing in equal volume of neutralizing buffer (40 mM Tris-HCl [pH 8.0]). PCR was performed as 20-μl reactions using TopTaq Master Mix (Qiagen) and cycling conditions as follows: 94°C for 3 min, 30 × 94°C for 30 s and 53°C for 30 s and 72°C for 30 s, followed by a final extension of 72°C for 5 min. The primer sets used for genotyping were as follows: murine Apobec3, forward (5′-CCCAGGACAACATCCACGC-3′) and reverse (5′-GCTCTGCACATTCGAAACAGGG-3′); human APOBEC3G (33), forward (5′-GGGACCCAGATTACCAGGAG-3′) and reverse (5′-GCAGATTATTCCAAGGCTCAA-3′); and murine Tlr7, KO forward (5′-TCATTCTCAGTATTGTTTTGCC-3′), WT forward (5′-AGGGTATGCCGCCAAATCTAAAG-3′), and reverse (5′-ACCTTTGTGTGCTCCTGGAC-3′).
gDNA Emv2 copy number analysis.
To quantitate genomic Emv2 copy number, primers amplifying the envelope region of Emv2 were used. Genomic DNA (gDNA) lysates were obtained as described above. Real-time quantitative PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) in 10-μl reactions in triplicate using 5 to 30 ng of cDNA per reaction. Primers were used at a final concentration of 0.225 μM. Fold copy number over the WT C57BL/6N mean value, normalized to Tert copy number, was calculated. The primer sets used for copy number analysis were as follows: murine Emv2 Env (10, 55), forward (5′-AGGCTGTTCCAGAGATTGTG-3′) and reverse (5′-TTCTGGACCACCACACGAC-3′); and murine Tert, forward (5′-GCCACTTAGGTGGGCATGCTA-3′) and reverse (5′-CTGTCCCTGGATCGTGAGGT-3′).
Peripheral blood isolation.
Mice were anesthetized, and blood was obtained via a retro-orbital bleed. Blood was collected with heparinized Natelson tubes (Fisher Scientific) into 8 mM EDTA in phosphate-buffered saline (PBS). For cellular RNA isolation, red blood cells were lysed with ACK lysis buffer (150 mM NH4Cl, 1 M KHCO3, 0.1 mM EDTA [pH 7.4]), and cells were washed twice with PBS before the addition of RLT buffer (Qiagen). Samples were stored at –80°C prior to RNA isolation.
Reverse transcription-quantitative PCR.
RNA was isolated from peripheral blood using the RNeasy kit (Qiagen) and cDNA was synthesized using an iScript cDNA Synthesis kit (Bio-Rad). Quantitative PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) in 10-μl reactions in triplicate using 5 to 30 ng of cDNA per reaction. Primers were used at a final concentration of 0.225 μM. The primer sequences were as follows: murine Apobec3, forward (5′-CTGCCTGCTAAGCGAGAAAGGC-3′) and reverse (5′-CTTTTGAATGCCGCCAGTTGCC-3′); human APOBEC3G, forward (5′-CCGAGGACCCGAAGGTTAC-3′) and reverse (5′-TCCAACAGTGCTGAAATTCG-3′); spliced Emv2 Env (56), forward (5′-CCAGGGACCACCGACCCACCGT-3′) and reverse (5′-TAGTCGGTCCCGGTAGGCCTCG-3′); MLV Pol (57), forward (5′-CACTTTGAGGGATCAGGAGCC-3′) and reverse (5′-CTTCTAGGTTTAGGGTCAACACCTGT-3′); unspliced Emv2 Env (55), forward (5′-AGGCTGTTCCAGAGATTGTG-3′) and reverse (5′-TTCTGGACCACCACATGAC-3′); murine Gapdh, forward (5′-GAAGGTCGGTGTGAACGGA-3′) and reverse (5′-GTTAGTGGGGTCTCGCTCCT-3′); IAP (58), forward (5′-AAGCAGCAATCACCCACTTTGG-3′) and reverse (5′-CAATCATTAGATGTGGCTGCCAAG-3′); MusD (58), forward (5′-GTGGTATCTCAGGAGGAGTGCC-3′) and reverse (5′-GGGCAGCTCCTCTATCTGAGTG-3′); ETnI (59), forward (5′-TGAGAAACGGCAAAGGATTTTTGGA-3′) and reverse (5′-ATTACCCAGCTCCTCACTGCTGA-3′); ETnII (59), forward (5′-GTGCTAACCCAACGCTGGTTC-3′) and reverse (5′-ACTGGGGCAATCCGCCTATTC-3′); and LINE1 ORFp, forward (5′-AAGCCTACAGAACTCCAAATAG-3′) and reverse (5′-AGGCTTGCCTTTATATGTTACT-3′).
Plasma RNA isolation and cDNA synthesis.
Peripheral blood was isolated from 16-week-old mice and centrifuged at 14,000 rpm for 15 min at 4°C and 200 μl of plasma was removed to a new Eppendorf tube. Plasma was homogenized with 1 ml of TRIzol and 200 μl of chloroform, and the aqueous layer was isolated by centrifugation for 15 min at 12,000 × g at 4°C. The aqueous layer was combined with 500 μl of isopropanol and 90 μg/ml glycerol and frozen for 1 h at –80°C. The RNA was then pelleted by centrifugation for 10 min at 12,000 × g at 4°C and washed twice with cold 75% ethanol before resuspending in 10 μl of RNase-free water. cDNA was synthesized using the Superscript III Cells Direct cDNA synthesis kit (Invitrogen), and qPCR was performed as described above, using sequenced ERV plasmid (described below) to generate a standard curve for absolute quantification using unspliced Emv2 Env and MLV Pol primers.
Splenocyte isolation and coculture.
The day before coculture, 100,000 DFJ8 avian fibroblasts were plated in 1 ml of Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Gibco) in a 12-well tissue culture-treated dish. On the day of coculture, spleens were isolated and dissociated through a 40-μm-pore size filter in RPMI medium (Gibco), and red blood cells were lysed with ACK lysis buffer. The splenocytes were then washed and passed through a 70-μm-pore size filter prior to counting, and 5 million splenocytes from each mouse were added to a corresponding well of DFJ8 cells, supplemented with an additional 1 ml of medium. Four days later, each coculture (supernatant plus cells) was moved to a 60-mm dish using 2 mM EDTA in PBS to dissociate the adherent cells, with a final culture volume of 4 ml. On day 7 of coculture, the adherent cells were stained for ERV envelope expression by flow cytometry.
Flow cytometry.
Hybridoma supernatant containing monoclonal antibody 573 was kindly provided by Leonard Evans (60). This antibody recognizes the envelope of all MLV, including ecotropic classes. Cells were washed twice with PBS and stained with mAb573 diluted 1:1 with PBS and then washed twice again and stained with anti-mouse CD45.2-FITC (BioLegend, catalog no. 109805), anti-mouse IgM-APC (Jackson, catalog no. 115-136-075), and 7-AAD viability staining solution (eBioscience, catalog no. 00-6993-50). Prior to analysis, cells were fixed in 1% paraformaldehyde in PBS. All incubations were performed at a final volume of 30 μl for 15 to 20 min at 4°C. Flow cytometry was performed on a BD LSRII Green cytometer, and the data were analyzed using FlowJo.
ERV isolation and sequencing.
Individual ERV-infected DFJ8 cells from coculture with TLR7−/− splenocytes were seeded in a 96-well plate and expanded until confluent in 12-well dishes. These monoclonal cultures were analyzed for ERV envelope expression by flow cytometry (as described above), and a single infected clone (D61) was selected. Two million D61 cells were plated in T-175 flasks in 30 ml of medium and grown for 1 week, after which the supernatant was harvested. Cell debris was removed by centrifuging the sample at 1500 rpm for 5 min, and the resulting supernatant was clarified through a 0.45-μm-pore size filter. The clarified supernatant was underlaid with a 1.12 g/μl sucrose cushion and ultracentrifuged at 23,000 rpm for 2 h at 4°C. The resulting viral pellet was resuspended in Opti-MEM (Gibco) and stored at –20°C. ERV RNA was isolated from these viral stocks using TRIzol/chloroform extraction and random hexamer cDNA synthesis (described above). Using the Emv2 mm10 genomic sequence, primers to highly conserved regions of the Emv2 backbone were used to amplify overlapping segments of the viral genome, which was assembled using Gibson Assembly (NEB) and cloned into the pUC19 vector for sequencing.
ERV recombination analysis.
A moving, overlapping window of size 50 bp was used to extract fragment sequences from the Emv2 ERV sequence. The step of the moving window is 1 bp. The fragment sequences were used as queries to search the GRCm38 reference genome by BLAT with the “-fastMap” option. An overused tile file with tile size 11 was used in the search. For each query sequence, all hits were sorted based on score = (% identity × alignment length), chromosomes and positions; only hits that had a maximum score (including ties) were kept. Each hit on this hit list was searched in both directions to expand the hit to maximum length, and the chromosome position with the maximum length was used as the mapped position of the Emv2 ERV sequence in the GRCm38 genome. Regions of the ERV sequence that mapped to non-Emv2 hits and were >10 bp were considered to have recombined with Emv2. Any recombined positions corresponding to unique ERV xenotropic (Xmv), polytropic (Pmv), or modified polytropic (Mpmv) loci (45) were identified and are shown in Fig. 1A.
ACKNOWLEDGMENTS
We thank Huiping Dong for maintaining the mouse strains used in this study. We also thank Leonard Evans for sharing the 573 hybridoma supernatant.
This study was supported in part by the Howard Hughes Medical Institute (to A.I.) and by NIH awards R01 AI054359 and R01 AI127429 (to A.I.). R.S.T. was supported by NIH training grant 5-T32-GM00720540 and by grant F30 (5-F30-AI129265-02).
R.S.T., M.T., and A.I. designed the experiments. R.S.T., M.T., H.D., and K.S.-B. performed the experiments. Y.K. analyzed ERV sequence data. R.S.T., S.R.R., and A.I. analyzed data. R.S.T. and A.I. prepared the manuscript.
We declare no competing interests.
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, et al. . 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, et al. . 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins NA, Copeland NG, Taylor BA, Lee BK. 1982. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol 43:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoye JP, Coffin JM. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol 61:2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak CA. 2014. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. doi: 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young GR, Kassiotis G, Stoye JP. 2012. Emv2, the only endogenous ecotropic murine leukemia virus of C57BL/6J mice. Retrovirology 9:23. doi: 10.1186/1742-4690-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozak CA. 2012. Viewpoint on Emv2, the only endogenous ecotropic murine leukemia virus of C57BL/6 mice. Retrovirology 9:25–25. doi: 10.1186/1742-4690-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak CA, Rowe WP. 1982. Genetic mapping of ecotropic murine leukemia virus-inducing loci in six inbred strains. J Exp Med 155:524–534. doi: 10.1084/jem.155.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King SR, Berson BJ, Risser R. 1988. Mechanism of interaction between endogenous ecotropic murine leukemia viruses in (BALB/c × C57BL/6) hybrid cells. Virology 162:1–11. doi: 10.1016/0042-6822(88)90388-1. [DOI] [PubMed] [Google Scholar]
- 10.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. 2012. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu P, Lubben W, Slomka H, Gebler J, Konert M, Cai C, Neubrandt L, Prazeres da Costa O, Paul S, Dehnert S, Dohne K, Thanisch M, Storsberg S, Wiegand L, Kaufmann A, Nain M, Quintanilla-Martinez L, Bettio S, Schnierle B, Kolesnikova L, Becker S, Schnare M, Bauer S. 2012. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity 37:867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Goodier JL. 2016. Restricting retrotransposons: a review. Mob DNA 7:16. doi: 10.1186/s13100-016-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mager DL, Stoye JP. 2015. Mammalian endogenous retroviruses. Microbiol Spectr 3:Mdna3-0009-2014. [DOI] [PubMed] [Google Scholar]
- 14.Conticello SG. 2008. The AID/APOBEC family of nucleic acid mutators. Genome Biol 9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. 2012. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol 23:258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. 2002. An anthropoid-specific locus of orphan C-to-U RNA-editing enzymes on chromosome 22. Genomics 79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 17.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 18.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stavrou S, Ross SR. 2015. APOBEC3 proteins in viral immunity. J Immunol 195:4565–4570. doi: 10.4049/jimmunol.1501504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koito A, Ikeda T. 2012. Apolipoprotein B mRNA-editing, catalytic polypeptide cytidine deaminases and retroviral restriction. Wiley Interdiscip Rev RNA 3:529–541. doi: 10.1002/wrna.1117. [DOI] [PubMed] [Google Scholar]
- 21.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 22.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 23.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 26.Zennou V, Perez-Caballero D, Göttlinger H, Bieniasz PD. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol 78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafer A, Bogerd HP, Cullen BR. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Guo F, Cen S, Niu M, Saadatmand J, Kleiman L. 2006. Inhibition of tRNA(3)(Lys)-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J Virol 80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo F, Cen S, Niu M, Yang Y, Gorelick RJ, Kleiman L. 2007. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J Virol 81:11322–11331. doi: 10.1128/JVI.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollpeter D, Parsons M, Sobala AE, Coxhead S, Lang RD, Bruns AM, Papaioannou S, McDonnell JM, Apolonia L, Chowdhury JA, Horvath CM, Malim MH. 2018. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat Microbiol 3:220–233. doi: 10.1038/s41564-017-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Low A, Okeoma CM, Lovsin N, de las Heras M, Taylor TH, Peterlin BM, Ross SR, Fan H. 2009. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology 385:455–463. doi: 10.1016/j.virol.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stavrou S, Blouch K, Kotla S, Bass A, Ross SR. 2015. Nucleic acid recognition orchestrates the anti-viral response to retroviruses. Cell Host Microbe 17:478–488. doi: 10.1016/j.chom.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stavrou S, Crawford D, Blouch K, Browne EP, Kohli RM, Ross SR. 2014. Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo. PLoS Pathog 10:e1004145. doi: 10.1371/journal.ppat.1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol 16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 36.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res 34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O’Shea KS, Moran JV, Cullen BR. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A 103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dutko JA, Schafer A, Kenny AE, Cullen BR, Curcio MJ. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr Biol 15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esnault C, Millet J, Schwartz O, Heidmann T. 2006. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res 34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 41.Schumacher AJ, Nissley DV, Harris RS. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc Natl Acad Sci U S A 102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YN, Malim MH, Bieniasz PD. 2008. Hypermutation of an ancient human retrovirus by APOBEC3G. J Virol 82:8762–8770. doi: 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esnault C, Priet S, Ribet D, Heidmann O, Heidmann T. 2008. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology 5:75. doi: 10.1186/1742-4690-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anwar F, Davenport MP, Ebrahimi D. 2013. Footprint of APOBEC3 on the genome of human retroelements. J Virol 87:8195–8204. doi: 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jern P, Stoye JP, Coffin JM. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet 3:e183. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treger RS, Tokuyama M, Dong H, Ross SR, Kong Y, Iwasaki A. 2019. Human APOBEC3G prevents emergence of infectious endogenous retrovirus in mice. bioRxiv 10.1101/457937. [DOI] [PMC free article] [PubMed]
- 47.Browne EP. 2013. Toll-like receptor 7 inhibits early acute retroviral infection through rapid lymphocyte responses. J Virol 87:7357–7366. doi: 10.1128/JVI.00788-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. 2004. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143–150. doi: 10.1016/S1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 50.Chattopadhyay SK, Lander MR, Rands E, Lowy DR. 1980. Structure of endogenous murine leukemia virus DNA in mouse genomes. Proc Natl Acad Sci U S A 77:5774–5778. doi: 10.1073/pnas.77.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bamunusinghe D, Liu Q, Plishka R, Dolan MA, Skorski M, Oler AJ, Yedavalli VRK, Buckler-White A, Hartley JW, Kozak CA. 2017. Recombinant origins of pathogenic and nonpathogenic mouse gammaretroviruses with polytropic host range. J Virol 91:e00855-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu X-F. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J Virol 81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol 81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papatheodorou I, Fonseca NA, Keays M, Tang YA, Barrera E, Bazant W, Burke M, Füllgrabe A, Fuentes A-P, George N, Huerta L, Koskinen S, Mohammed S, Geniza M, Preece J, Jaiswal P, Jarnuczak AF, Huber W, Stegle O, Vizcaino JA, Brazma A, Petryszak R. 2018. Expression atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res 46:D246–D251. doi: 10.1093/nar/gkx1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshinobu K, Baudino L, Santiago-Raber ML, Morito N, Dunand-Sauthier I, Morley BJ, Evans LH, Izui S. 2009. Selective up-regulation of intact, but not defective env RNAs of endogenous modified polytropic retrovirus by the Sgp3 locus of lupus-prone mice. J Immunol 182:8094–8103. doi: 10.4049/jimmunol.0900263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Young GR, Ploquin MJ, Eksmond U, Wadwa M, Stoye JP, Kassiotis G. 2012. Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog 8:e1002709. doi: 10.1371/journal.ppat.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lötscher M, Recher M, Lang KS, Navarini A, Hunziker L, Santimaria R, Glatzel M, Schwarz P, Böni J, Zinkernagel RM. 2007. Induced prion protein controls immune-activated retroviruses in the mouse spleen. PLoS One 2:e1158. doi: 10.1371/journal.pone.0001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 59.Maksakova IA, Mager DL, Reiss D. 2008. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci 65:3329–3347. doi: 10.1007/s00018-008-8494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans LH, Morrison RP, Malik FG, Portis J, Britt WJ. 1990. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol 64:6176–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]