Clinically, Zika virus (ZIKV) infection can lead to developmental defects in the cortex of the fetal brain. How ZIKV triggers this event in developing neural cells is not well understood at a molecular level and likely requires many contributing factors. ZIKV efficiently infects human neural progenitor cells (hNPCs) and leads to growth arrest of these cells, which are critical for brain development. Here, we demonstrate that infection with ZIKV, but not dengue virus, disrupts the cell cycle of hNPCs by halting DNA replication during S phase and inducing DNA damage. We further show that ZIKV infection activates the ATM/Chk2 checkpoint but prevents the activation of another checkpoint, the ATR/Chk1 pathway. These results unravel an intriguing mechanism by which an RNA virus interrupts host DNA replication. Finally, by mimicking virus-induced S-phase arrest, we show that ZIKV manipulates the cell cycle to benefit viral replication.
KEYWORDS: cell cycle, DNA damage, DNA damage checkpoints, DNA damage response, DNA replication, S phase, Zika virus, neural progenitors
ABSTRACT
Zika virus (ZIKV) infection attenuates the growth of human neural progenitor cells (hNPCs). As these hNPCs generate the cortical neurons during early brain development, the ZIKV-mediated growth retardation potentially contributes to the neurodevelopmental defects of the congenital Zika syndrome. Here, we investigate the mechanism by which ZIKV manipulates the cell cycle in hNPCs and the functional consequence of cell cycle perturbation on the replication of ZIKV and related flaviviruses. We demonstrate that ZIKV, but not dengue virus (DENV), induces DNA double-strand breaks (DSBs), triggering the DNA damage response through the ATM/Chk2 signaling pathway while suppressing the ATR/Chk1 signaling pathway. Furthermore, ZIKV infection impedes the progression of cells through S phase, thereby preventing the completion of host DNA replication. Recapitulation of the S-phase arrest state with inhibitors led to an increase in ZIKV replication, but not of West Nile virus or DENV. Our data identify ZIKV’s ability to induce DSBs and suppress host DNA replication, which results in a cellular environment favorable for its replication.
IMPORTANCE Clinically, Zika virus (ZIKV) infection can lead to developmental defects in the cortex of the fetal brain. How ZIKV triggers this event in developing neural cells is not well understood at a molecular level and likely requires many contributing factors. ZIKV efficiently infects human neural progenitor cells (hNPCs) and leads to growth arrest of these cells, which are critical for brain development. Here, we demonstrate that infection with ZIKV, but not dengue virus, disrupts the cell cycle of hNPCs by halting DNA replication during S phase and inducing DNA damage. We further show that ZIKV infection activates the ATM/Chk2 checkpoint but prevents the activation of another checkpoint, the ATR/Chk1 pathway. These results unravel an intriguing mechanism by which an RNA virus interrupts host DNA replication. Finally, by mimicking virus-induced S-phase arrest, we show that ZIKV manipulates the cell cycle to benefit viral replication.
INTRODUCTION
Zika virus (ZIKV) is a mosquito-borne flavivirus of significant public health concern, closely related to dengue virus (DENV), West Nile virus (WNV), and yellow fever virus (YFV). These single-stranded, positive-sense RNA viruses share similar genome characteristics and replication strategies, yet infection with each of these viruses can lead to very different clinical outcomes, suggesting that distinct cellular and molecular mechanisms underlie their pathogenesis. ZIKV’s preferential targeting of neural progenitor cells (1), which give rise to the fetal cortex during brain development, appears to be unique among these flaviviruses and likely contributes to the severe neurological complications, such as microcephaly, observed in congenitally infected fetuses and newborns (2, 3).
In response to the devastating effects of ZIKV on fetal neural development brought to light by the 2015 outbreak in Brazil, significant research efforts have been undertaken to understand the mechanisms of Zika pathogenesis. We previously demonstrated that ZIKV causes cell cycle perturbation and apoptosis in human cortical neural progenitor cells (hNPCs) (1). These findings were recapitulated in three-dimensional (3D) cerebral organoids (4–6) and the developing fetal brain in pregnant mice (7, 8). Additionally, global transcriptome analyses of ZIKV-infected hNPCs and murine fetal brain tissues revealed a dysregulation of cell-cycle-related genes (1, 7, 8), further highlighting the involvement of cell cycle pathways in ZIKV biology.
Progression through the cell cycle is tightly controlled by an array of cellular regulators and effectors. This control can be exerted through the activation of signaling pathways that halt the cell cycle and facilitate the repair of damaged DNA, collectively referred to as the DNA damage response (DDR) (9–11). The DDR is mediated by members of the phosphoinositide 3 (PI-3) kinase family, primarily ataxia telangiectasia mutated (ATM) and ATM and Rad3 related (ATR), whose checkpoint signals are relayed to downstream effector proteins by the Chk2 and Chk1 transducer kinases, respectively (12, 13). Typically, the ATM/Chk2 signaling pathway is activated upon the detection of double-strand breaks (DSBs) in DNA, while the ATR/Chk1 signaling pathway is initiated by the presence of accumulated single-strand DNA (ssDNA) at stalled replication forks (14). Activation of either of these protein kinase cascades can ultimately result in widespread effects on cell cycle progression, DNA repair, and the induction of apoptosis (15).
Following the detection of DNA damage, a central function of the DDR is to arrest cell cycle progression. The ATM/ATR kinases mediate activation of three major cell cycle checkpoints: the G1/S checkpoint ensures that DNA replication or S phase is not initiated in the presence of DNA damage, the intra-S-phase checkpoint activates in response to replication stress, and the G2/M checkpoint prevents the onset of cell division in the presence of DNA damage or unreplicated DNA, thereby inhibiting the segregation of damaged chromosomes (9, 16–18). DDR-mediated arrest of the cell cycle occurs to allow time for the damaged DNA to be repaired prior to reentry into the cell cycle. An early event that occurs in response to DNA damage is the phosphorylation of S139 on histone variant H2A.X (γH2A.X) by the ATM/ATR kinases at sites of damaged DNA, which serves as a sensitive marker for DNA DSBs (19, 20). This event leads to the recruitment of numerous DNA repair proteins, such as the p53-binding protein 1 (53BP1) (21, 22). Recently, it was demonstrated that ZIKV infection leads to increased γH2A.X signals (7, 23, 24); however, neither direct evidence of DNA breaks during ZIKV infection, nor the consequences of this damage on cell cycle progression or viral replication have been shown.
Here, we report that ZIKV infection activates the ATM/Chk2 signaling pathway, while suppressing the ATR/Chk1 signaling pathway, and causes a dramatic inhibition of DNA replication and progression through S phase. Additionally, we observe increased levels of phosphorylated H2A.X and 53BP1 protein and detect physical breaks in DNA. Together, these data suggest that ZIKV infection triggers an ATM-dependent DDR in hNPCs by inducing DNA DSBs. We also demonstrate that aphidicolin/thymidine-mediated S-phase arrest leads to an increase of ZIKV replication. Lastly, we show that while DENV and WNV both efficiently infect hNPCs, these viruses do not block host DNA replication, and their infections are not enhanced by S-phase arrest.
RESULTS
ZIKV infection causes DNA damage and activation of the DDR.
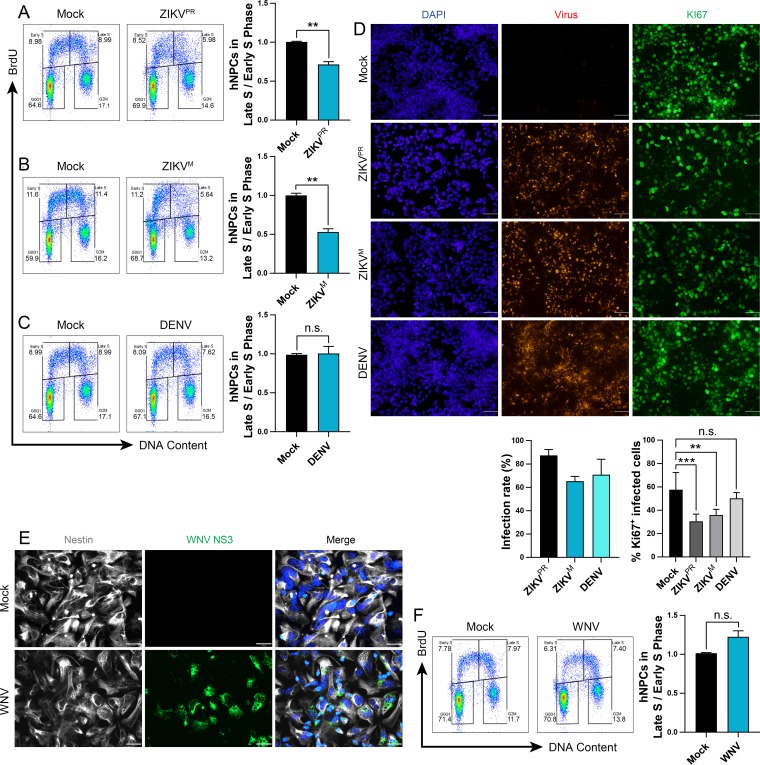
We have previously shown that infection of hNPCs with the ZIKV MR766 (ZIKVM) Uganda strain led to a perturbation of the host cell cycle as measured in DNA content using propidium iodide (PI) staining (1). To determine if infection by an epidemic strain of ZIKV interferes with active DNA replication and proliferation, we infected hNPCs for 48 h with ZIKV PRVABC59 (ZIKVPR) or ZIKVM (multiplicity of infection [MOI] of 0.4) and then labeled these cells with a nucleoside analogue, bromodeoxyuridine (BrdU), to identify cells undergoing DNA replication. Infection of hNPCs with either strain of ZIKV significantly decreased the number of cells positive for BrdU and with a near-4C DNA content (i.e., late-S-phase cells). This resulted in a reduced ratio of cells in late S versus early S phase to 50 to 60% of that in mock-infected hNPCs (Fig. 1A and B). In contrast, DENV infection (MOI of 0.4) did not significantly alter the ratio of cells in late S versus early S phase compared to mock-infected hNPCs (Fig. 1C). Immunofluorescence analysis showed comparable infection efficiencies for ZIKVPR, ZIKVM, and DENV, with over 60% of hNPCs infected in each population at 48 h (Fig. 1D). We also observed a significant attenuation of the cell proliferation marker KI67 in hNPCs infected with ZIKV, but not for DENV (Fig. 1D), consistent with previous studies in ZIKV-infected mice and brain organoids (6, 8).
FIG 1.
ZIKV infection disrupts DNA replication of hNPCs. (A to F) hNPCs were infected with (A to D) ZIKVM (MOI of 0.4), ZIKVPR (MOI of 0.4), or DENV (MOI of 0.4) for 48 h or (E and F) WNV (MOI of 0.5) for 24 h. (A to C and F) Representative cell cycle profiles and quantifications of BrdU-pulse-labeled hNPCs. The same mock-infected cells were used for the ZIKVPR and DENV representative images. Error bars are mean ± SD, representing the average from two biological replicates. (D) Representative immunofluorescence images and quantifications of viral infection of hNPCs using anti-flavivirus envelope (4G2) and anti-KI67 staining 48 h postinfection. KI67+ cell quantification is the percentage of infected cells. Scale bars are 50 μm. Error bars are mean ± SD, representing the average from two biological replicates (n > 500 cells per treatment). (E) Representative immunofluorescence images of hNPCs infected with WNV (MOI of 0.5) for 24 h and stained with anti-WNV NS3 or antinestin. Scale bars are 26 μm. In panels A to D and F, ** indicates P ≤ 0.01 and *** indicates P ≤ 0.001. (A to C and F) Unpaired t test. (D) One-way ANOVA.
We next determined whether the ability of ZIKV to perturb the cell cycle in hNPCs was shared by another neurotropic flavivirus, WNV. We first tested the permissiveness of hNPCs to the WNV NY99 strain (MOI of 0.5) and observed efficient infection, with approximately 60% of hNPCs infected at 24 h (Fig. 1E). In contrast to ZIKV, WNV-infected hNPCs displayed no significant difference in either the number of late-S-phase cells or the ratio of cells in late S versus early S phase compared to mock-infected hNPCs at 24 h postinfection (Fig. 1F).
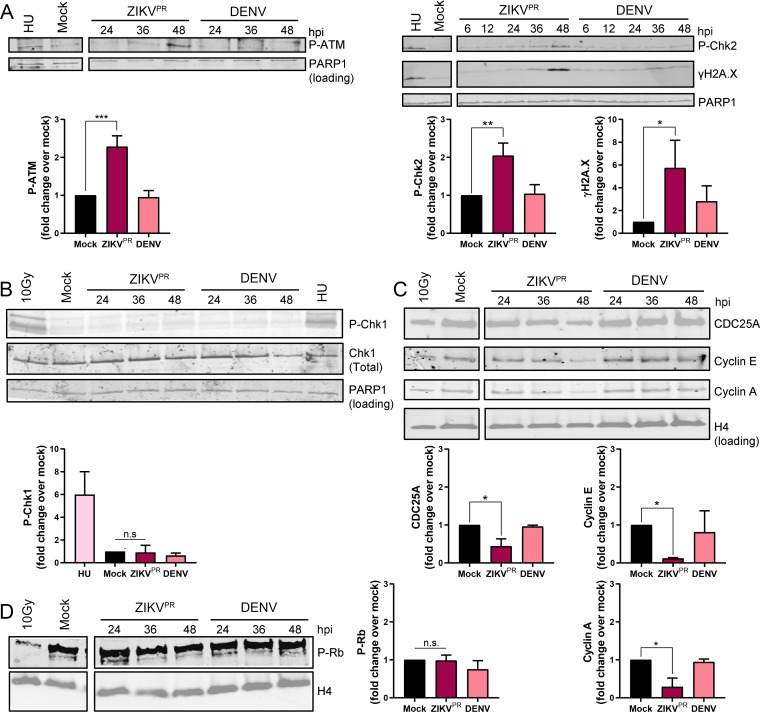
To delineate the molecular mechanism of S-phase disruption by ZIKV, we analyzed the phosphorylation state of key cell cycle checkpoint molecules in mock-, ZIKV-, or DENV-infected hNPCs. We detected increased phosphorylated ATM and its substrate Chk2 in ZIKV-infected hNPCs by 48 h postinfection (Fig. 2A), suggesting activation of the ATM/Chk2 checkpoint, but did not observe Chk1 phosphorylation in ZIKV-infected hNPCs (Fig. 2B). We next detected reduced protein levels of several downstream targets of the ATM/Chk2 signaling pathway, including the cell cycle regulators CDC25A, cyclin E, and cyclin A, in ZIKV-infected hNPCs (Fig. 2C). In contrast to ZIKV, we did not detect checkpoint activation in DENV-infected hNPCs (Fig. 2A and B). Of note, similar levels of hyperphosphorylated retinoblastoma (Rb) protein were observed in virus-infected and mock-infected hNPCs (Fig. 2D), suggesting that these cells are not restricted at the G1/S transition.
FIG 2.
ZIKV infection activates the ATM/Chk2 checkpoint in hNPCs. (A to C) Representative Western blot images and quantifications of DNA damage response signaling pathway protein expression in hNPCs infected with ZIKVPR (MOI of 0.4) or DENV (MOI of 0.4) analyzed over the time course shown (hours postinfection [hpi]). Positive controls include cells treated with 1 mM hydroxyurea (HU) for 22 h and 10-Gy-irradiated cells. Quantifications are of 48-h time points. Error bars are mean ± SD, representing the average from (A and B) three or (C) two biological replicates. (D) Representative Western blot image and quantification of phosphorylated Rb in hNPCs infected with ZIKVPR (MOI of 0.4) or DENV (MOI of 0.4) analyzed over the time course shown (hpi). The positive control is 10-Gy-irradiated cells. Quantifications are of 48-h time points. Error bars are mean ± SD, representing the average from three biological replicates. In panels A to D, * indicates P ≤ 0.05, ** indicates P ≤ 0.01, and *** indicates P ≤ 0.001 (one-way ANOVA).
We next investigated what triggers ATM/Chk2 checkpoint activation upon ZIKV infection. As part of the DDR, checkpoint proteins such as ATM and Chk2 are activated in response to DNA DSBs, which are marked by the presence of γH2A.X. To determine if γH2A.X signals are increased during ZIKV infection, we infected hNPCs with ZIKV or DENV and collected cell lysates over a time course for Western blot analysis. We observed a higher γH2A.X signal from hNPCs infected with ZIKV than for those from mock- or DENV-infected hNPCs (Fig. 2A). We confirmed these results with an immunofluorescence assay that allowed for the quantification of the number of the nuclear γH2A.X foci at 48 h after infection and observed an approximately 4-fold increase in number of γH2A.X foci in ZIKV-infected compared to mock-infected hNPCs (Fig. 3A). As DNA damage can also occur during virus-induced apoptosis, we excluded cells expressing cleaved caspase-3 and detected an approximately 3-fold increase in γH2A.X foci in ZIKV-infected compared to mock-infected hNPCs (Fig. 3B). Downstream of γH2A.X signaling, the 53BP1 protein is phosphorylated at damaged DNA foci as part of the DSB repair process. We observed an approximately 5-fold increase in phosphorylated 53BP1 foci in ZIKV-infected compared to mock-infected hNPCs 48 h postinfection (Fig. 3C). Finally, to directly visualize DSBs in DNA, we utilized the neutral comet assay, which is a sensitive method that combines gel electrophoresis with fluorescence microscopy to directly visualize and measure DNA DSBs in individual eukaryotic cells (25). We detected a significant increase in mean tail moment in ZIKV-infected compared to mock- and DENV-infected hNPCs 48 h postinfection (Fig. 3D and E).
FIG 3.
ZIKV infection induces DNA damage in hNPCs, while simultaneously restricting activation of the ATR/Chk1 checkpoint. (A) Representative immunofluorescence images and quantification of γH2A.X foci in hNPCs infected for 48 h with ZIKVM (MOI of 0.4) or DENV (MOI of 0.4). Cells were stained with anti-γH2A.X, and nuclear foci were counted. DAPI is represented in gray; γH2A.X is represented in green. The scale bar is 20 μm. Error bars are mean ± SD (n > 500 cells per treatment). (B) Quantification of γH2A.X foci in cleaved caspase-3-negative (c-Cas3−) hNPCs infected for 48 h with ZIKVPR (MOI of 0.4) or DENV (MOI of 0.4). Cells were stained with anti-γH2A.X, and anti-cleaved caspase-3, and nuclear foci were counted in cells negative for caspase-3 staining. Error bars are mean ± SD (n > 500 cells per treatment,). (C and D) Representative immunofluorescence images and quantification of (C) phosphorylated 53BP1 foci or (D and E) tail moment in hNPCs infected for 48 h with ZIKVPR (MOI of 0.4) or DENV (MOI of 0.4). (C) Cells were stained with anti-phospho-53BP1, and nuclear foci were counted. DAPI is represented in gray; phospho-53BP1 is represented in green. Scale bars are 20 μm. Error bars are mean ± SD for images representative of two biological replicates (n > 500 cells per treatment). (D) Immunofluorescence images and (E) cluster plot analysis of the neutral comet assay. Following neutral comet assay lysis and single-cell electrophoresis, infected hNPCs were stained with SYBR gold, and tail moment was measured. Scale bars are 50 μm. Error bars are mean ± SD, representing the average from three biological replicates (immunofluorescence, n > 50 comets per replicate; cluster plot, n = 160 comets per treatment). (F) Representative Western blot image and quantification of phosphorylated Chk1 in hNPCs infected with ZIKVPR (MOI of 0.4) or DENV (MOI of 0.4) for 44 or 26 h and then treated with 1 mM HU for 4 or 22 h prior to collection. Error bars are mean ± SD, representing the average from two biological replicates. In panels A to E, * indicates P ≤ 0.05, ** indicates P ≤ 0.01, *** indicates P ≤ 0.001, and **** indicates P ≤ 0.0001 (one-way ANOVA).
Our BrdU analysis results (Fig. 1A and B) strongly indicate a disruption in S phase in hNPCs during ZIKV infection. Yet, while the ATR/Chk1 signaling pathway is typically activated at stalled replication forks, Chk1 phosphorylation was not observed in ZIKV-infected hNPCs (Fig. 2B), suggesting that the ATR/Chk1 signaling pathway is not initiated upon ZIKV infection. Thus, we investigated the intriguing possibility that ZIKV infection suppresses ATR activation. We infected hNPCs with ZIKV or DENV for 44 or 26 h and then treated the cells with 1 mM hydroxyurea (HU) for an additional 4 or 22 h to activate the ATR/Chk1 signaling pathway. As we previously noted, Chk1 phosphorylation was not observed in the mock- or virus-infected untreated hNPCs; however, there was a significant increase in Chk1 phosphorylation in the mock- and DENV-infected hNPCs following treatment with HU (Fig. 3F). In contrast, ZIKV-infected hNPCs showed only minimal phosphorylation of Chk1 following HU treatment (Fig. 3F), suggesting that ZIKV does indeed suppress activation of the ATR/Chk1 signaling pathway. Collectively, these findings demonstrate that ZIKV, but not DENV, infection of hNPCs leads to DSBs in DNA and activates the DDR through the ATM/Chk2 signaling pathway, while suppressing the ATR/Chk1 signaling pathway.
ZIKV infection arrests the progression of DNA replication.
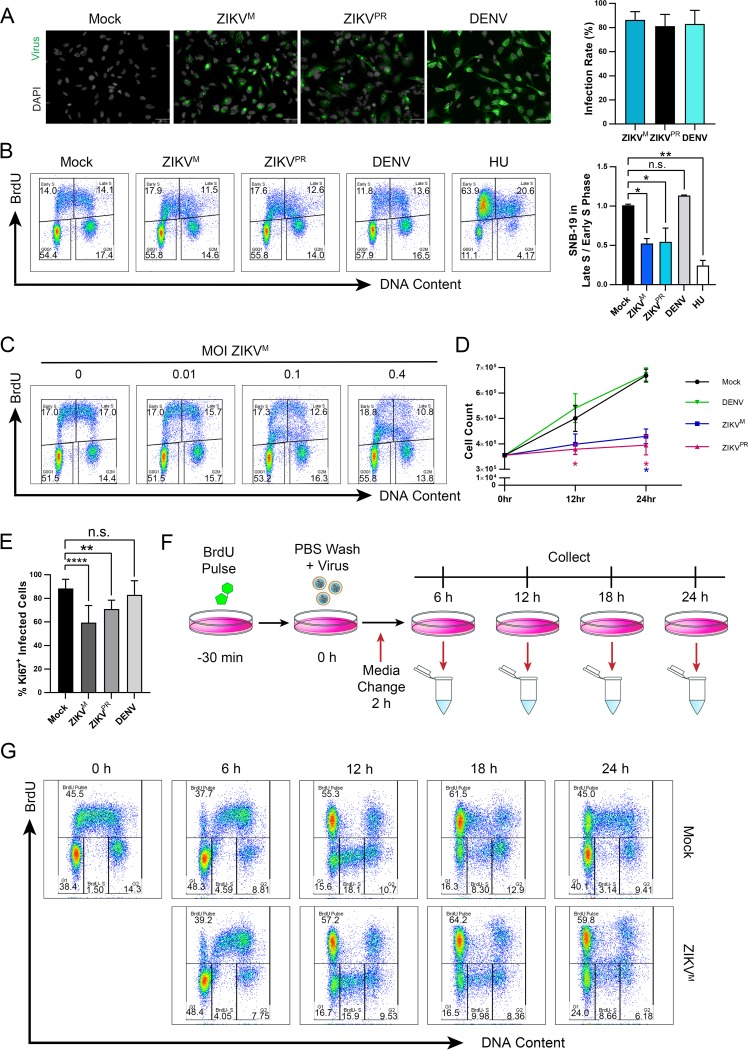
Next, we analyzed the kinetics of ZIKV-mediated perturbation of DNA replication in a highly permissive human glioblastoma cell line, SNB-19, that is more amenable to synchronization in vitro than hNPCs. Infection of SNB-19 cells by ZIKV (MOI of 1.0) or DENV (MOI of 1.0) resulted in greater than 80% of cells infected at 24 h (Fig. 4A). ZIKV, but not DENV, infection significantly decreased the ratio of cells in late S versus early S phase (Fig. 4B), similar to that observed in hNPCs, and this decrease was inversely correlated with the ZIKV MOI (Fig. 4C). Additionally, these cells exhibited significantly reduced cell proliferation (Fig. 4D) and attenuation of KI67 staining (Fig. 4E) in populations infected with ZIKV, but not DENV, comparable to infected hNPCs. To determine the timing of this ZIKV-induced cell cycle disruption, we labeled cells with BrdU prior to virus inoculation to follow the progress of the S-phase pulse-labeled cells during infection (Fig. 4F). A difference in cell cycle profiles between mock- and ZIKV-infected cells became detectable at around 18 h postinfection (Fig. 4G); thus, this time was used as a reference point to track the progression of these perturbed cells in subsequent experiments.
FIG 4.
ZIKV infection alters the proliferation and cell cycle progression of SNB-19 cells. (A, B, D, and E) SNB-19 cells were infected with ZIKVM (MOI of 1.0), ZIKVPR (MOI of 1.0), or DENV (MOI of 1.0) for (A and B) 24 h or (D) 12 to 24 h prior to analysis. (A) Representative immunofluorescence images and quantification of infected SNB-19 cells stained with anti-flavivirus envelope (4G2). Scale bars are 50 μm. Error bars are mean ± SD (n > 500 cells per treatment). (B) Representative cell cycle profiles and quantifications of BrdU-pulse-labeled SNB-19 cells following infection or treatment with 1 mM HU for 22 h. Error bars are mean ± SD, representing the average from two biological replicates. (C) Cell cycle profile of SNB-19 cells infected with increasing MOI of ZIKVM at 24 h prior to BrdU pulse-labeling. (D) Time course of SNB-19 cell proliferation for infected or mock-infected cells. Error bars are mean ± SD, representing the average from three biological replicates. (E) Quantification of KI67-positive SNB-19 cells in the infected population. Cells were stained with anti-KI67 and anti-flavivirus envelope (4G2) 24 h postinfection and counted for double-positive cells. Error bars are mean ± SD (n > 500 per treatment). (F) Schematic of SNB-19 BrdU pulse-labeling for cell cycle profile analysis of DNA replication progression. SNB-19 cells were initially pulsed with BrdU for 30 min and then infected for 2 h with ZIKVM (MOI of 1.0). Samples were collected at 6, 12, 18, and 24 h postinfection for analysis by flow cytometry. (G) Cell cycle progression of BrdU-pulse-labeled SNB-19 cells throughout ZIKV infection as indicated in panel F. Analyzed gates include the BrdU-labeled cells (BrdU Pulse), and all BrdU-negative cells divided into the G1, BrdU− S, and G2 DNA content groups. The cell cycle profiles shown are representative of three biological replicates. In panels B, D, and E, * indicates P ≤ 0.05, ** indicates P ≤ 0.01, *** indicates P ≤ 0.001, and **** indicates P ≤ 0.0001 (one-way ANOVA).
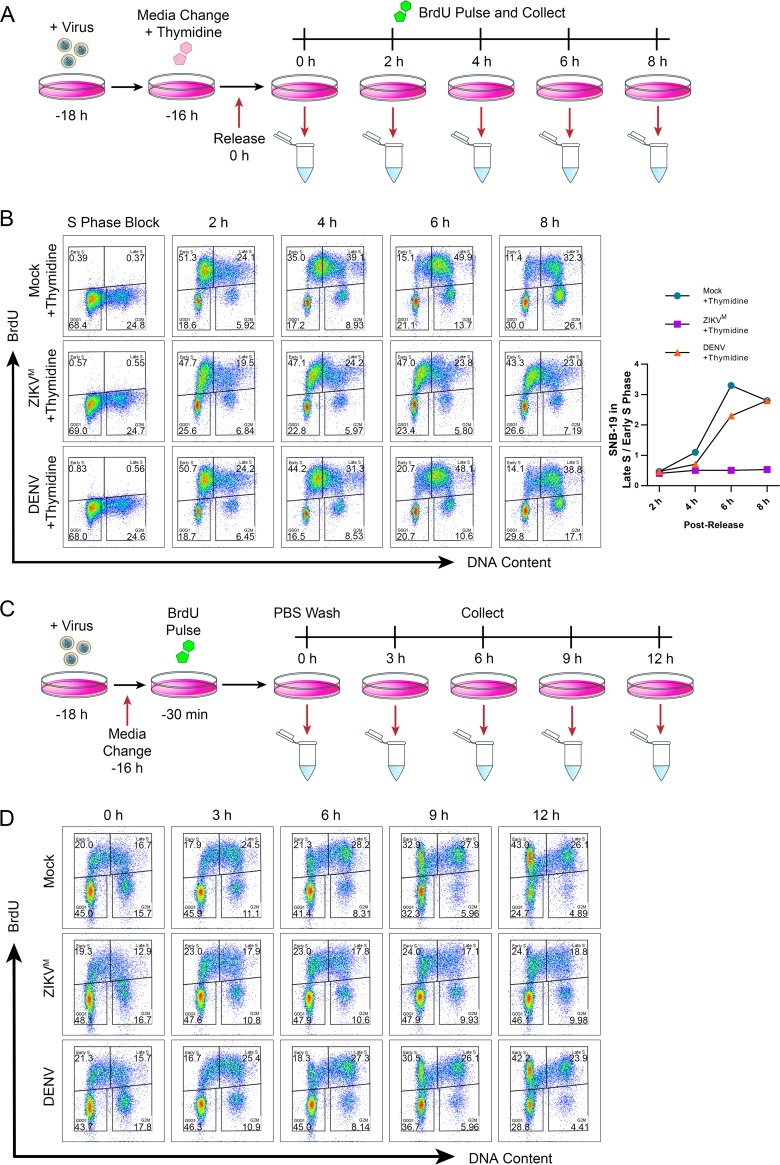
A specific reduction of late-S-phase cells by ZIKV infection suggests an impediment of S-phase progression, which can be visualized more clearly in synchronized cells. We treated infected cells for 16 h with thymidine, which blocks DNA replication in a reversible fashion, to obtain a synchronized cell population and then detected cell progression through S phase following thymidine removal (Fig. 5A). After thymidine treatment, but before release, the BrdU profiles were similar for both infected and uninfected cells (Fig. 5B). After release from the block, the uninfected and DENV-infected cells moved as a synchronized population through S phase, reaching late S phase around 8 h postrelease. In contrast, the ZIKV-infected cells entered early S phase but were unable to progress through to late S phase, with progression effectively stopped by 4 h postrelease (Fig. 5B).
FIG 5.
ZIKV infection disrupts progression of S phase and causes an S-phase arrest in SNB-19 cells. (A) Schematic of SNB-19 S-phase synchronization and BrdU pulse-labeling for cell cycle profile analysis of S-phase progression. SNB-19 cells were initially infected for 2 h with ZIKVM (MOI of 1.0) or DENV (MOI of 1.0) followed by treatment with 2 mM thymidine for 16 h. At 18 h postinfection, 0-h samples were BrdU pulse-labeled for 30 min and collected, while all other cells were released from thymidine block. At 2, 4, 6, and 8 h postrelease, cells were BrdU pulse-labeled for 30 min and collected for analysis by flow cytometry. (B) Cell cycle synchronization and progression of ZIKVM- or DENV-infected SNB-19 cells through S phase treated as indicated in panel A. Shown is quantification of representative profile. The cell cycle profiles shown are representative of three biological replicates. (C) Schematic of SNB-19 BrdU pulse-labeling for cell cycle profile analysis of S-phase progression in infected cells. SNB-19 cells were initially infected for 2 h with ZIKVM (MOI of 1.0) or DENV (MOI of 1.0). At 18 h postinfection, samples were BrdU pulse-labeled for 30 min and collected at 0, 3, 6, 9, and 12 h post-BrdU labeling for analysis by flow cytometry. (D) Cell cycle progression of an asynchronous population of BrdU-pulse-labeled SNB-19 cells during infection as indicated in panel C. The cell cycle profiles shown are representative of three biological replicates.
To follow the natural progression of S-phase cells without synchronization, we pulse-labeled mock- and virus-infected SNB-19 cells with BrdU and then chased them for up to 12 h (Fig. 5C). The early-S-phase BrdU-labeled cells in the uninfected and DENV-infected populations progressed from a 2C to a 4C DNA content and then accumulated with a 2C DNA content (following a division) by 12 h (Fig. 5D), suggesting that these cells had cycled through back to the G1 phase. In contrast, the early-S-phase BrdU-labeled cells in the ZIKV-infected population were less efficient at increasing their DNA content, while a small subset of the late-S-phase BrdU-labeled cells appeared to be able to progress to a 2C DNA content normally (Fig. 5D). Together, our data demonstrate that ZIKV, but not DENV, infection induces an S-phase arrest, thereby disrupting the progression of DNA replication and preventing the completion of S phase.
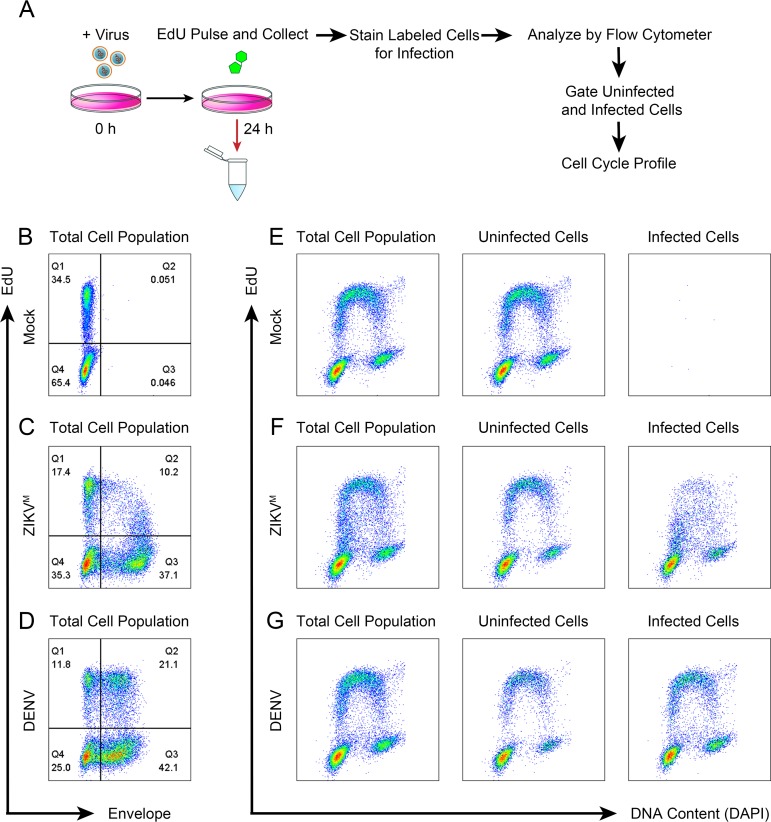
We then confirmed that the S-phase arrest was induced only in ZIKV-infected cells but not in uninfected cells by simultaneously analyzing infection and the cell cycle profile. For this, we utilized 5-ethynyl-2′-deoxyuridine (EdU) to measure DNA replication alongside detection of the viral envelope protein (Fig. 6A). Infection of SNB-19 cells with ZIKV (MOI of 0.5) or DENV (MOI of 0.5) resulted in approximately 50 to 60% of the cells positive for viral envelope (Fig. 6B to D), permitting the comparison of the cell cycle profiles of the individually gated infected and uninfected subpopulations in the same sample (Fig. 6E to G). For both ZIKV and DENV, the cell cycle profiles of the uninfected subpopulations closely resembled that of mock-infected cells (Fig. 6E to G, left and middle panels). However, the ZIKV-positive subpopulation exhibited a clear reduction in EdU labeling compared to the uninfected subpopulation, suggesting inhibition of DNA replication (Fig. 6F, right panel). In contrast, the DENV-positive subpopulation did not show this reduction, and there was no difference in DNA replication between the infected and uninfected subpopulations (Fig. 6G, middle and right panel). These results indicate that the S-phase arrest induced by ZIKV infection is limited to infected cells.
FIG 6.
ZIKV-induced S-phase arrest is specific to infected cells. (A) Schematic of EdU pulse-labeling for cell cycle profile analysis of infected versus uninfected cells. SNB-19 cells were infected with ZIKVM (MOI of 0.5) or DENV (MOI of 0.5) for 24 h. Samples were pulse-labeled with 100 μM EdU for 30 min, collected, and then stained with anti-flavivirus envelope (4G2) prior to Click-iT chemistry and staining with DAPI. (B to D) Representative 2D analysis of EdU-labeled cells and envelope protein staining of SNB-19 cells treated as indicated in panel A. (E to G) Corresponding cell cycle profiles of panels B to D. The “Uninfected Cells” plot displays cells gated from Q1 and Q4 of panels B to D. The “Infected Cells” plot displays cells gated from Q2 and Q3 of panels B to D. (B to G) The cell cycle profiles shown are representative of three biological replicates.
Induced S-phase arrest enhances ZIKV replication.
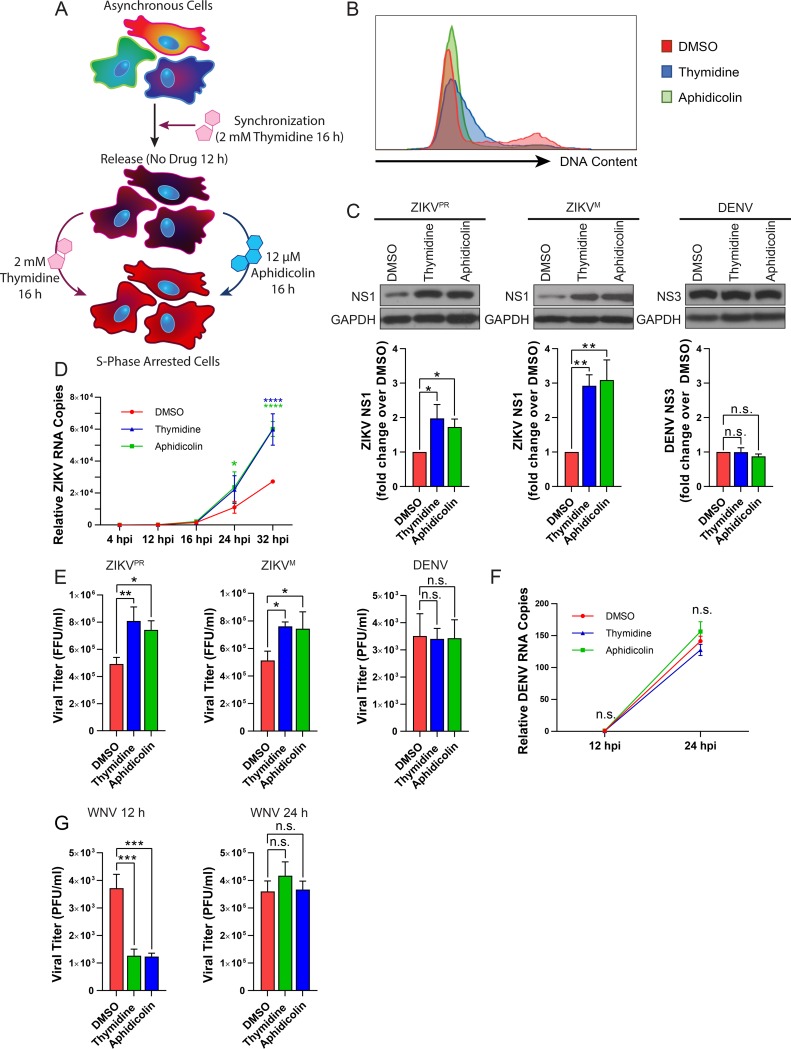
We next determined if ZIKV-induced S-phase arrest was beneficial for viral replication. To mimic the S-phase arrest observed during ZIKV infection, we used two treatment strategies in SNB-19 cells: a double thymidine block or a thymidine block followed by treatment with a DNA polymerase inhibitor, aphidicolin (APH) (Fig. 7A). PI analysis revealed that both treatments arrested cells in early S phase (Fig. 7B).
FIG 7.
Induced S-phase arrest of SNB-19 cells enhances ZIKV, but not DENV or WNV replication. (A) Schematic of two treatment conditions used to synchronize SNB-19 cells in S phase. Cells were initially treated with 2 mM thymidine for 16 h and then were released back into the cell cycle for 12 h. Following release, the cells were treated with either 2 mM thymidine or 12 μM aphidicolin for an additional 16 h to induce S-phase arrest prior to infection. (B) Representative flow cytometry analysis of SNB-19 cells treated as indicated in panel A. Samples were stained with propidium iodide (PI) and analyzed by flow cytometry. (C, D, and F) SNB-19 cells were infected for (C) 24 h, (D) 4 to 32 h, or (F) 12 to 24 h with ZIKVPR (MOI of 0.5), ZIKVM (MOI of 0.5), or DENV (MOI of 0.5). (C) Western blot images and quantification of intracellular viral protein expression. Viral protein was detected by anti-ZIKV NS1 or anti-DENV NS3 for each respective sample. Viral protein band intensities were normalized to the DMSO control, representing the mean from three biological replicates ± SD. (D and F) Time course of relative intracellular viral RNA copies in (D) ZIKVM- or (F) DENV-infected SNB-19 cells as measured by qPCR. Error bars are the mean ± SD from three biological replicates. (E and G) Infectivity titers of culture supernatants at (E) 48 h postinfection as measured by focus-forming units (ZIKVPR and ZIKVM [MOI of 0.5]) and at 36 h postinfection as measured by PFU (DENV [MOI of 0.5]) or (G) at 12 and 24 h postinfection as measured by PFU (WNV [MOI of 0.5]) on (E) Vero cells or (F) BHK cells. Error bars are the mean ± SD from three biological replicates. In panels C to G, * indicates P ≤ 0.05, ** indicates P ≤ 0.01, *** indicates P ≤ 0.001, and **** indicates P ≤ 0.0001. (C, E, and G) One-way ANOVA. (D and F) Two-way ANOVA.
With either S-phase inhibitor treatment strategy, intracellular ZIKV protein levels were significantly enhanced by 24 h after infection compared to the levels in dimethyl sulfoxide (DMSO)-treated cells (Fig. 7C). An infection time course analysis revealed that ZIKV RNA remained at similar levels between 4 and 16 h and then increased by 24 h for the S-phase inhibitor-treated samples only, indicating that detectable enhancement occurred later in the virus life cycle (Fig. 7D), consistent with our previous time course (Fig. 4G). Although there was no significant increase in ZIKV infectious titers at 24 h after infection, by 48 h, there was a significant increase in ZIKV yields from S-phase inhibitor-treated cells compared to DMSO-treated cells (Fig. 7E). In contrast, these drug treatments did not significantly alter DENV protein (Fig. 7C) or RNA (Fig. 7F) levels or infectious titers (Fig. 7E). We also tested the potential effect of S-phase arrest on WNV infectious particle production. Interestingly, both S-phase inhibitor treatment strategies resulted in a significant decrease of WNV infectious titers at 12 h—a difference that was no longer apparent by 24 h—compared to DMSO-treated cells (Fig. 7G). These results suggest that an S-phase arrest environment does not benefit WNV, but instead delays its replication kinetics.
To confirm that the enhancement of ZIKV replication was specific to S-phase arrest, we cultured SNB-19 cells in reduced serum for 24 h to accumulate cells in G0/G1 phase prior to infection. Propidium iodide staining confirmed that the reduced serum medium led to an increase in G0/G1-phase cells (Fig. 8A). In contrast to the results in S-phase-arrested cells, the reduced serum conditions markedly decreased ZIKV, but not DENV, protein expression (Fig. 8B) and infectious titers (Fig. 8C). Because reduced serum in culture medium may deplete serum proteins required for efficient viral entry (26, 27), we infected cells with ZIKV in standard serum medium to allow entry before adding the reduced serum medium and observed a similar, albeit less dramatic, decrease in ZIKV intracellular protein levels (Fig. 8D). Overall, these results indicate that an S-phase arrest environment, during which cellular DNA replication is inhibited, fosters ZIKV infection, while a G0/G1 arrest environment is detrimental.
FIG 8.
G0/G1-phase arrest of SNB-19 cells does not promote ZIKV replication. (A) Flow cytometry analysis of SNB-19 cells cultured for 24 h in 10, 1, and 0.1% serum media, respectively. Samples were stained with propidium iodide (PI) and analyzed by flow cytometry. (B and C) SNB-19 cells were infected for (B) 24 h or (C) 36 h with ZIKVPR (MOI of 0.5) or ZIKVM (MOI of 0.5) or 48 h with DENV (MOI of 0.5). (B) Western blot images of intracellular viral protein expression in serum-starved SNB-19 cells 24 h postinfection. Following culture for 24 h in standard (10% FBS) or reduced (1% or 0.1% FBS) serum medium, cells were infected by direct addition of virus culture to the existing culture medium. Viral protein was detected by anti-ZIKV NS1 or anti-DENV NS3 for each respective sample. Viral protein band intensities were normalized to the 10% FBS control, representing the mean ± SD from three biological replicates. (C) Infectivity titers of culture supernatants as measured by FFU (ZIKVM and ZIKVPR) or PFU (DENV) on Vero cells. Error bars are the mean ± SD from three biological replicates. (D) Western blot images of intracellular viral protein expression in SNB-19 cells 24 h postinfection. Following a 2-h infection in standard serum medium (10% FBS) with ZIKVPR (MOI of 2.0), ZIKVM (MOI of 2.0), or DENV (MOI of 2.0), the medium was changed to standard serum medium (10% FBS) or reduced serum medium (1% or 0.1% FBS) for 24 h prior to collection. Viral protein was detected by anti-ZIKV NS1 or anti-DENV NS3 for each respective sample. Viral protein band intensities were normalized to the 10% FBS control, representing the mean ± SD from three biological replicates. In panels B to D, * indicates P ≤ 0.05, ** indicates P ≤ 0.01, *** indicates P ≤ 0.001, and **** indicates P ≤ 0.0001 (one-way ANOVA).
DISCUSSION
Here, we report that ZIKV activates an ATM/Chk2-dependent DDR pathway in response to ZIKV-induced DNA breaks in hNPCs while simultaneously suppressing the ATR/Chk1 signaling pathway, ultimately culminating in an S-phase arrest and the halting of host DNA replication (Fig. 9). Furthermore, our data suggest that this S-phase arrest state provides a beneficial environment for ZIKV replication in proliferating neural cells (Fig. 9).
FIG 9.
Model depicting the effects of ZIKV infection on cell cycle progression in hNPCs. In this study, we demonstrate that ZIKV infection of neural progenitors causes DNA double-strand breaks (DSBs) and suppresses activation of the ATR/Chk1 signaling pathway, resulting in induction of the DNA damage response (DDR) via the ATM/Chk2 signaling pathway. Activation of ATM and the subsequent Chk2-mediated signaling cascade leads to the phosphorylation of the DNA damage repair proteins H2A.X and 53BP1 and degradation of the S-phase regulatory and effector proteins CDC25A, cyclin A, and cyclin E, culminating in an S-phase arrest environment that fosters productive ZIKV replication.
Many viruses capable of modulating the host cell cycle employ the strategy of DDR induction; however, the mechanisms of DDR activation differ in their requirements for DNA damage. Some viral protein kinases can directly bind to components of the DDR pathway to activate downstream signaling cascades (28, 29), while other viruses trigger the DDR by inducing physical breaks in host DNA (i.e., nicks and DSBs) or replication stress (30–32). Our work presented here suggests that ZIKV activates the DDR through the ATM/Chk2 checkpoint pathway by causing DSBs in DNA, revealing the mechanism by which ZIKV induces cell cycle perturbation and restricts the growth of hNPCs. It is also possible that other as-yet-undiscovered events exist that provide additional mechanisms for DDR induction during ZIKV infection. Recently, it was shown that DENV infection causes leakage of mitochondrial DNA (mtDNA) into the cytoplasm (33). Although ZIKV infection may similarly cause mtDNA leakage, ATM activation by cytoplasmic DNA has not been reported (34, 35).
Physical breaks in DNA can occur by indirect or direct mechanisms. Viruses can trigger the production of reactive oxygen species (ROS), which are a known cause of nicks or DSBs in host DNA (36, 37). While ZIKV can transcriptionally upregulate ROS-related genes, such as NOX1 (38, 39), and DENV and WNV have been shown to induce ROS, these viruses simultaneously upregulate antioxidative responses, negating DNA-damaging redox imbalances in the cell (40–42).
As a direct mechanism of inducing DNA breaks, a virus can impede the progression of DNA replication forks by disrupting a critical interaction or by depleting a DNA replication or repair factor (43, 44). Failure to remove the stress can result in collapsed replication forks and subsequent DSBs (45), a process that is accelerated in the absence of ATR signaling (46) and leads to ATM activation (9). Our BrdU labeling results suggest that ZIKV-infected cells can enter early S phase but then are unable to further replicate their DNA, while a small subset of cells in late S phase can complete DNA replication. These data, complemented by our previous findings that ZIKV infection of hNPCs led to a general downregulation of numerous cell-cycle-related genes (1, 39), are consistent with a mechanism of virus-mediated depletion of a crucial DNA replication factor(s). In this scenario, we would typically expect activation of the ATR checkpoint; however, our results suggest that ZIKV suppresses activation of the ATR/Chk1 signaling pathway. Collectively, ZIKV-mediated depletion of a vital DNA replication factor(s) in conjunction with suppression of the ATR/Chk1 signaling pathway would likely accelerate fork collapse and subsequent DSB formation. It is also possible that ZIKV suppression of ATR activation alone, in the absence of a depleted DNA replication factor(s), would increase DSBs in DNA as hNPCs have been shown to be particularly susceptible to endogenous stresses such as replication stress (47). Either possibility would almost certainly result in activation of the ATM/Chk2 signaling pathway and an arrest of cell cycle progression in S phase, consistent with our observations in hNPCs.
During DNA virus infection, DDR factors are recruited to nuclear virus replication centers (48), thereby shifting host cellular resources to facilitate efficient viral lytic replication (49). For RNA viruses that replicate exclusively in the cytoplasm, how DDR activation or the resulting cell cycle arrest benefits viral replication is less clear. We show that the enhancement of ZIKV infection is specific to the S-phase arrest state, as serum starvation to induce G0/G1 arrest negatively impacted ZIKV replication, and that this enhancement is unique to ZIKV among the flaviviruses tested. Thus, it is possible that ZIKV requires DDR factors or some other aspects of the host DNA replication machinery for efficient replication.
The enhancement of viral replication in an arrest state has also been reported for other RNA viruses (32, 50); however, the specific mechanisms by which this enhancement occurs remain unclear. While we have demonstrated that induction of an S-phase arrest state leads to increased ZIKV protein expression, RNA levels, and titers, we have not yet explicitly determined at which stage of the viral life cycle this enhancement occurs. It is of note that the induced S-phase arrest state led to increased viral protein production at 24 h while the viral RNA levels and titers were not increased until later time points. This temporal discrepancy suggests that the S-phase arrest state either increases viral protein stability or production prior to any detectable benefit to the viral RNA level or that the arrest state is not immediately beneficial to RNA replication.
We observed that an accumulation of ZIKV proteins at viral replication factories coincided with the appearance of the cell cycle arrest, suggesting that this phenotype may be dependent upon a certain threshold of a triggering ZIKV protein or RNA, which remains unidentified. In future studies, it would be interesting to map the viral sequence variations responsible for the observed differences between ZIKV and the other two flaviviruses we tested, DENV and WNV, in their effect on the cell cycle.
While the effect of ZIKV-induced S-phase arrest benefits viral replication in neural cells, viral particles released from the fetal brain likely play a limited role in viral transmission: thus, any evolutionary advantage of a fetal brain-specific effect mediated by ZIKV remains unclear. Furthermore, future research is needed to determine if the enhancement of ZIKV replication by cell cycle perturbation is specific to cells of neural origin.
It is likely that the impact of ZIKV infection on multiple cell types in the central nervous system contributes to the collective mechanism by which ZIKV disrupts fetal neural development and causes the large assortment of phenotypes characteristic of congenital Zika syndrome. Our findings provide a specific mechanistic link between ZIKV infection of hNPCs and growth retardation of these cells, which likely plays an important role in ZIKV-induced microcephaly. Furthermore, we identify an aspect of ZIKV’s unique biology among other flaviviruses, such as DENV and WNV, that provides a handle for dissecting virus-specific disease mechanisms.
MATERIALS AND METHODS
Cell lines.
The human glioblastoma SNB-19 cell line (from the NCI-60 collection) was a kind gift from David Meckes (Florida State University, Tallahassee, FL). SNB-19 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS [Invitrogen]) and incubated at 37°C in 5% CO2. Aedes albopictus C6/36 cells (ATCC) were cultured in Eagle’s minimum essential medium (EMEM) supplemented with 10% FBS and incubated at 28°C in 5% CO2. Vero cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS and incubated at 37°C in 5% CO2. BHK-21 strain WI2 cells were obtained from Tadeusz Wiktor (Wistar Institute, Philadelphia, PA), maintained in minimum essential medium (MEM) supplemented with 2.5% fetal calf serum (FCS) and 10 μg/ml gentamicin, and incubated at 37°C in 5% CO2.
Culture of hiPSCs and differentiation into cortical neural progenitor cells.
Human induced pluripotent stem cell (hiPSC) lines (C1-2 and C3-1) were generated in Guo-Li Ming’s lab (1, 51) and were differentiated into forebrain-specific hNPCs following previous protocols (51). Briefly, hiPSC colonies were detached from the feeder layer with 1-mg/ml collagenase treatment for 1 h and suspended in embryonic body (EB) medium, consisting of FGF-2-free iPSC medium supplemented with 2 μM dorsomorphin and 2 μM A-83, in nontreated polystyrene plates for 4 days with a daily medium change. After 4 days, EB medium was replaced by neural induction medium (NPC medium) consisting of DMEM–Ham’s F-12, N2 supplement, NEAA, 2 μg/ml heparin, and 2 μM cyclopamine. The floating EBs were then transferred to Matrigel-coated 6-well plates at day 7 to form neural tube-like rosettes. The attached rosettes were kept for 15 days with NPC medium change every other day. On day 22, the rosettes were picked mechanically and transferred to low-attachment plates (Corning) to form neurospheres in NPC medium containing B27. The neurospheres were then dissociated with Accutase at 37°C for 10 min and placed onto Matrigel-coated 6-well plates at day 24 to form monolayer NPCs in NPC medium containing B27. These hNPCs expressed markers for forebrain-specific progenitor markers, including nestin, PAX6, EMX-1, FOXG1, and OTX2 (51).
Virus production and infection.
ZIKV MR766 was obtained in the form of culture fluid from ZeptoMetrix (Buffalo, NY). ZIKV PRVABC59 was obtained from ATCC (Manassas, VA). Dengue virus type 2—16681 was a kind gift from Qianjun Li (University of Alabama, Birmingham). WNV NY99 was obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA [Galveston, TX]). ZIKV and DENV stocks were amplified in C6/36 cells (ATCC) at 28°C. Briefly, C6/36 cells cultured in T-75 flasks were incubated with ZIKV and DENV viral inoculum for 1 h at room temperature, then fresh medium was added, and cells were cultured for 7 days, filtered with a 0.45-μm-pore filter, aliquoted, and stored at –80°C. For mock infections, an equal amount of spent uninfected C6/36 supernatant was used. A stock of WNV strain NY99 was prepared by infecting BHK cell monolayers at an MOI of 0.1 and harvesting culture fluid at 32 hpi. Clarified culture fluid was aliquoted and stored at –80°C. The titers of amplified viruses were determined by focus-forming unit (FFU) or PFU assays as described in the following section.
Virus infectivity titration assays.
ZIKV titers were determined by FFU assay, and DENV titers were determined by PFU assay on Vero cells. For the PFU assay, culture supernatants were collected, serially diluted, and incubated with a confluent monolayer of Vero cells for 2 h at 37°C prior to addition of a 2% methylcellulose overlay mixed 1:1 with 2× DMEM. After 48 to 72 h of incubation, cells were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde, stained with a solution of 0.5% crystal violet for 20 min, and rinsed with double-distilled water (ddH2O). For the FFU assay, cells were blocked in PBTG, which is PBT (PBS plus 0.5% Tween 20 and 1% bovine serum albumin [BSA]) plus 0.2% BSA and 5% normal goat serum. The cells were then incubated overnight at 4°C with anti-flavivirus envelope (clone D1-4G2-4-15, produced from hybridoma ATCC HB-112). The following day, the cells were washed three times with PBS prior to incubation with goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) (Santa Cruz Biotechnology; catalog no. sc-2005) for 1 h at room temperature and then washed three times with PBS prior to incubation with 3,3′-diaminobenzidine (DAB) peroxidase substrate (Vector Labs; catalog no. SK-4100) for 10 min. WNV titers were determined using a PFU assay on BHK cells. Culture supernatants were collected, serially diluted, and incubated with a confluent monolayer of cells for 1 h at 37°C, prior to addition of a 1% molten Sea ME agarose (Lonza) overlay mixed 1:1 with 2× MEM. After 72 h, the agarose plug was removed, and the cells were stained with 0.05% crystal violet in 10% ethanol.
BrdU/EdU pulse-labeling and analysis by flow cytometry.
Cells were pulse-labeled with 100 μM BrdU (Sigma-Aldrich; catalog no. B5002) for 30 min at 37°C and protected from light for the remainder of the procedure. Cells were then rinsed two times in ice-cold PBS, and the cell pellet was collected. After suspension in ice-cold PBS–1% FBS, ice-cold 100% ethanol was added dropwise while mixing to fix the cells at a final concentration of 75% ethanol at −20°C overnight. For analysis by 2D flow cytometry, the samples were denatured by treatment with 2N HCl–0.5% Triton X-100 and incubated in the dark for 30 min prior to pH neutralization with 0.1 M sodium tetraborate. Cells were then incubated with 0.15 μg anti-BrdU (BD Pharmingen; catalog no. 555627) in a PBS solution containing 0.5% Tween 20 and 1% BSA (PBT) for 1 h. Samples were washed with PBT solution and incubated with 1 μg goat anti-mouse IgG conjugated with fluorescein isothiocyanate (FITC [Sigma-Aldrich; catalog no. F0257]) in PBT for 30 min. After washing with PBT, samples were resuspended in PI staining solution (Abcam; catalog no. ab139418) and incubated for 30 min at 37°C.
Cells were pulse-labeled with 100 μM EdU (Invitrogen; catalog no. C10632) for 30 min at 37°C and protected from light for the remainder of the procedure. Cells were then rinsed two times in ice-cold PBS, and the cell pellet was collected. Pellets were washed an additional time with PBS and resuspended in 4% paraformaldehyde at room temperature for 10 min. Samples were washed with PBS containing 1% BSA and incubated with 2.5 μg anti-flavivirus envelope (clone D1-4G2-4-15, produced from hybridoma ATCC HB-112) in Click-iT saponin-based permeabilization and wash reagent (SPW) (Invitrogen; catalog no. C10632) for 1 h. Samples were washed with PBS–1% BSA and incubated with 1 μg goat anti-mouse IgG–Cy3 (Invitrogen; catalog no. A10521) in SPW for 30 min. After washing with PBS–1% BSA, Click-iT chemistry was performed according to the manufacturer’s protocol (Invitrogen; catalog no. C10632) with Alexa Fluor 488 picolyl azide for 30 min. After washing with PBS–1% BSA, samples were resuspended in FoxP3 permeabilization buffer (Biolegend; catalog no. 421402) with a final concentration of 6 μM DAPI (4′,6-diamidino-2-phenylindole [Biolegend; catalog no. 422801]) for 15 min at room temperature prior to analysis. All samples were analyzed using a BD FACSCanto machine (BD Biosciences). Cell cycle plots were generated using FlowJo software (FlowJo, LLC).
Western blotting for hNPCs.
Samples were washed with ice-cold PBS and then directly lysed in Laemmli sample buffer containing 3× Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific; catalog no. 78440) and 10 μM MG132 (Calbiochem; catalog no. 474790). The lysates were gently collected, immediately flash-frozen in liquid nitrogen, and stored at –80°C until analysis. Proteins in the lysates were resolved either on 4 to 12% precast gradient gels (Bio-Rad) using MES (morpholineethanesulfonic acid) running buffer or on SDS–18% polyacrylamide gels optimized for the resolution of histone proteins. Proteins were transferred to 0.2-μm-pore nitrocellulose membranes prior to blocking with 5% milk in Tris-buffered saline containing 0.1% Tween 20 (TBST). Blots were incubated overnight in TBST plus 0.5% milk with the appropriate antibodies before washing them four times for 4 min each with TBST. The blots were incubated for 45 min with the appropriate fluorescently labeled secondary antibodies (LiCor) at a 1:15,000 dilution in TBST. Then the blots were washed four times with TBST followed by two washes in TBS prior to detection of the fluorescent signals using a LiCor Odyssey infrared imager. The protein bands were quantified using ImageJ (https://imagej.nih.gov/ij/). Quantifications were determined relative to the indicated loading controls prior to normalization to the uninfected sample (value set to 1). The following primary antibodies were obtained commercially and used at a 1:1,000 dilution: cyclin A (Santa Cruz Biotechnology; catalog no. sc-596), cyclin E (Santa Cruz Biotechnology; catalog no. sc-481), CDC25A (Santa Cruz Biotechnology; catalog no. sc-097), Chk1–phospho-S345 (Cell Signaling; catalog no. 13303), Chk2–phospho-T68 (Cell Signaling; catalog no. C13C1), Rb–phospho-T356 (Genetex; catalog no. GTX10922), ATM–phospho-S1981 (Abcam; catalog no. ab81292), γH2A.X–phospho-S139 (Millipore; catalog no. 05-636), PARP1 (Bethyl Laboratories; catalog no. A301-376A), Chk1 (Bethyl Laboratories; catalog no. A300-298A), and H2A.X (Abcam; catalog no. ab20669). Rabbit polyclonal antibodies against histone H4 were developed in the laboratory and have been described previously (52).
Immunofluorescence staining.
Cells were seeded on coverslips and fixed with 4% paraformaldehyde, permeabilized with PBT (0.2% Triton X-100 in PBS), and then blocked in either PBTG (PBT with 0.2% BSA and 5% normal goat serum) (ZIKV and DENV) or PBS with 5% normal horse serum (WNV). After blocking, cells were stained solely or in combination with the following antibodies: anti-phospho-53BP1 (Ser25) (Thermo Fisher Scientific; catalog no. PA5-38465), anti-phospho-H2A.X (Ser140) (Thermo Fisher Scientific; catalog no. MA1-2022), anti-KI67 (Abcam; catalog no. ab15580), anti-flavivirus envelope (clone D1-4G2-4-15, produced from hybridoma ATCC HB-112), anti-WNV NS3 (R&D Systems; catalog no. AF2907), antinestin (Millipore; catalog no. MAB5326), or anti-cleaved caspase-3 (Cell Signaling; catalog no. 9661). Slides were washed three times in PBS and then stained with one or a combination of the following secondary antibodies: goat anti-mouse IgG–FITC (Sigma-Aldrich; catalog no. F0257), goat anti-rabbit IgG–FITC (Sigma-Aldrich; catalog no. F0382), goat anti-mouse IgG–Cy3 (Invitrogen; catalog no. A10521), goat anti-rabbit IgG–Cy3 (Invitrogen; catalog no. A10520), mouse anti-goat–Alexa Fluor 488, or goat anti-mouse–Alexa Fluor 647. Slides were washed three times in PBS and mounted using Vectashield mounting medium with DAPI (Vector Laboratories; H-1200) (ZIKV, DENV). For WNV staining, slides were simultaneously stained with secondary antibody and Hoechst 33342 to stain the nucleus and mounted using Prolong Gold antifade mountant (Thermo Fisher Scientific; catalog no. p36930). ZIKV- or DENV-infected cells and KI67-stained cells were imaged using an Olympus BX61 fluorescence microscope (20× objective). Phospho-H2A.X and phospho-53BP1-stained slides were imaged using a Zeiss LSM 880 confocal microscope (63× objective). Collected images were analyzed and quantified using ImageJ (https://imagej.nih.gov/ij/). WNV-infected cells and nestin-stained cells were imaged using a Zeiss Axio Observer Z1 fluorescence microscope and processed using Volocity 64 software (Perkin Elmer).
Comet assay.
The neutral lysis comet assay was performed as previously described (25). Briefly, 8 × 103 hNPCs resuspended in 400 μl Dulbecco’s PBS (DPBS [VWR; catalog no. 02-0119-1000]) were mixed with 1.2 ml molten 1% low-gelling-temperature agarose (Sigma-Aldrich; catalog no. A4018), and 1.2 ml of the mixture was overlaid onto slides scored and precoated with 1% low-gelling-temperature agarose. Slides were immersed in 4°C neutral lysis buffer (2% Sarkosyl, 0.5 M Na2EDTA, 0.5 mg/ml proteinase K, pH 8.0) and incubated at 37°C for 18 to 20 h. After lysis, slides were submerged for 30 min three times in neutral electrophoresis buffer (90 mM Tris, 90 mM boric acid, 2 mM Na2EDTA, pH 8.5) prior to electrophoresis for 25 min at 0.6 V/cm. Slides were briefly rinsed in ddH2O and stained with 1× SYBR gold (Thermo Fisher Scientific; catalog no. S11494) for 30 min. Excess stain was rinsed away with ddH2O, and slides were imaged using an Olympus BX61 microscope (10× objective). Comet tail moment was quantified using CASPlab software (http://casplab.com).
S-phase cell synchronization.
A total of 1.0 × 105 SNB-19 cells were seeded into 12-well plates. The next day, 2 mM thymidine (Sigma-Aldrich; catalog no. T1895) was added in fresh medium for 16 h to synchronize cells in S phase. Cells were washed with PBS two times, and fresh medium was added for 12 h to release cells back into the cell cycle. Following release, either 2 mM thymidine or 12 μM aphidicolin (Sigma-Aldrich; catalog no. A4487) was added in fresh medium for 16 h to block cells in early S phase. Nuclear DNA was stained with propidium iodine (PI) solution (Abcam; catalog no. ab139418) to confirm synchronization of cells in S phase.
Western blotting for SNB-19 cells.
Mock-, ZIKV-, or DENV- infected SNB-19 cells were trypsinized and collected. Pellets were lysed in Laemmli sample buffer and immediately boiled for 10 min. Proteins in the samples were separated by 12% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The membranes were blocked in 10% milk in PBST and then incubated either at room temperature for 1 h or overnight at 4°C with the following primary antibodies: anti-ZIKV NS1 (BioFront Technologies; catalog no. BF-1225-36), anti-DENV NS3 (Genetex; catalog no. GTX124252), or anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase [Santa Cruz Biotechnology; catalog no. sc-25778]). Membranes were washed three times in PBST and then probed with HRP-conjugated secondary antibodies for 1 h at room temperature. After three PBST washes, a chemiluminescent HRP substrate (Millipore) was applied to detect protein. The protein bands were quantified using ImageJ (https://imagej.nih.gov/ij/). Quantifications were determined relative to loading controls (GAPDH) prior to normalization to the experimental control (value set to 1).
qRT-PCR.
Total cellular RNA was purified from mock-, ZIKVM-, or DENV-infected SNB-19 cells using an RNeasy Plus kit (Qiagen) per the manufacturer’s instructions. cDNA was synthesized from purified RNA using random hexamers and a SuperScript III First-Strand kit (Invitrogen). For viral RNA quantification, quantitative real-time PCR (qRT-PCR) was performed using SYBR green qPCR master mix (Thermo Fisher Scientific) per the manufacturer’s protocol and genome-specific primers for ZIKV MR766 (forward, CAAGGAGTGGGAAGCGGAG; reverse, CCATGTGATGTCACCTGCTCT) or DENV-2 16881 (forward, TAGCCAACCAAGCCACAGTGT; reverse, TTGACTTGTGAGTAGCATCCAATG). All qRT-PCR results were normalized to GAPDH mRNA in the same sample, and relative viral RNA copies were calculated using the threshold cycle (ΔΔCT) method.
Serum starvation.
A total of 1 × 105 SNB-19 cells were seeded in standard culture medium (10% FBS, RPMI), and the next day, the medium was replaced with fresh standard (10% FBS) or reduced (1% or 0.1% FBS) serum medium. After 24 h, cultures were infected with each respective virus by direct inoculation into the existing culture medium. At 24 h postinfection, culture supernatants were collected for viral titer analysis, and samples were trypsinized and collected for Western blot analysis. In a second experiment, 1 × 105 SNB-19 cells were seeded in standard culture medium (10% FBS, RPMI) and the next day infected with each respective virus. After 2 h, the cells were washed with PBS, and the medium was replaced with standard full (10% FBS) or reduced (1% or 0.1% FBS) serum medium. At 24 h postinfection, cells were trypsinized and collected for Western blot analysis.
Statistical analysis.
Quantified data are reported as means ± standard deviations (SD). Experiments were completed in either biological duplicate or triplicate, as indicated in the corresponding figure legend. All statistical analyses were carried out in GraphPad Prism 7. Unless otherwise specified, data were statistically analyzed for the P value using one-way analysis of variance (ANOVA) with Bonferroni’s correction. P value significance is indicated as follows: n.s., not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; and ****, P ≤ 0.0001.
ACKNOWLEDGMENTS
We thank Alyssa Poe, Farrah Karimipour, Brian Washburn, Kristina Poduch, and Ashley Tang for technical assistance.
This study was supported by Public Health Service grants U19 AI131130 and R21 AI119530 to H.T., R01 AI45135 and U19AI131130 to M.A.B., U19 AI131130 to Z.W., and R01 GM083337 to D.M.G. Partial support for J.C.M. was provided by a Georgia State University Molecular Basis of Disease fellowship.
C.H., S.C.O., and H.T. conceived of the research, designed the study, interpreted data, and wrote the manuscript. C.H. and S.C.O. performed experiments and contributed equally to this study. J.C.M. conducted the WNV experiments. A.M. conducted the serum starvation experiments. E.P., Y.S., A.C., and S.G. performed and interpreted the cell cycle protein experiments. R.A.D. contributed to the acquisition and analysis of the cell cycle profiles. C.X. and Z.W. performed hNPC differentiation and designed the initial hNPC analysis. D.M.G. and A.G. contributed to conception of the cell cycle experiments, and A.G. analyzed data. M.A.B. conceived of the West Nile virus experiments. H.S. and G.M. interpreted the hNPC data. H.T. directed the research.
REFERENCES
- 1.Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, Didier RA, Jin P, Song H, Ming GL. 2016. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, Pižem J, Petrovec M, Avšič Županc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 3.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016. Zika virus and birth defects—reviewing the evidence for causality. N Engl J Med 374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 4.Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. 2016. Zika virus impairs growth in human neurospheres and brain organoids. Science 352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 5.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL. 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandão WN, Rossato C, Andrade DG, Faria DP, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrão-Braga PC. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, Xu Z. 2016. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19:672–677. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 8.Wu K-Y, Zuo G-L, Li X-F, Ye Q, Deng Y-Q, Huang X-Y, Cao W-C, Qin C-F, Luo Z-G. 2016. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res 26:645–654. doi: 10.1038/cr.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastan MB, Bartek J. 2004. Cell-cycle checkpoints and cancer. Nature 432:316. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 11.Zhou B-B, Elledge SJ. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 12.Abraham RT. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 13.Bartek J, Lukas J. 2003. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3:421–429. doi: 10.1016/S1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 14.Marechal A, Zou L. 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber AM, Ryan AJ. 2015. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther 149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev 12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C. 2001. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol 154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 19.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. 2008. γH2AX and cancer. Nat Rev Cancer 8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 21.Huyen Y, Zgheib O, DiTullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 22.Panier S, Boulton SJ. 2014. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 23.El Ghouzzi V, Bianchi FT, Molineris I, Mounce BC, Berto GE, Rak M, Lebon S, Aubry L, Tocco C, Gai M, Chiotto AM, Sgrò F, Pallavicini G, Simon-Loriere E, Passemard S, Vignuzzi M, Gressens P, Di Cunto F. 2016. ZIKA virus elicits P53 activation and genotoxic stress in human neural progenitors similar to mutations involved in severe forms of genetic microcephaly and p53. Cell Death Dis 7:e2440. doi: 10.1038/cddis.2016.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devhare P, Meyer K, Steele R, Ray RB, Ray R. 2017. Zika virus infection dysregulates human neural stem cell growth and inhibits differentiation into neuroprogenitor cells. Cell Death Dis 8:e3106. doi: 10.1038/cddis.2017.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olive PL, Banáth JP. 2006. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 26.Meertens L, Labeau A, Dejarnac O, Cipriani S, Sinigaglia L, Bonnet-Madin L, Le Charpentier T, Hafirassou ML, Zamborlini A, Cao-Lormeau VM, Coulpier M, Missé D, Jouvenet N, Tabibiazar R, Gressens P, Schwartz O, Amara A. 2017. Axl mediates Zika virus entry in human glial cells and modulates innate immune responses. Cell Rep 18:324–333. doi: 10.1016/j.celrep.2016.12.045. [DOI] [PubMed] [Google Scholar]
- 27.Richard AS, Shim B-S, Kwon Y-C, Zhang R, Otsuka Y, Schmitt K, Berri F, Diamond MS, Choe H. 2017. AXL-dependent infection of human fetal endothelial cells distinguishes Zika virus from other pathogenic flaviviruses. Proc Natl Acad Sci U S A 114:2024–2029. doi: 10.1073/pnas.1620558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, Zhu J, Xie Z, Liao G, Liu J, Chen M, Hu S, Woodard C, Lin J, Taverna SD, Desai P, Ambinder RF, Hayward GS, Qian J, Zhu H, Hayward SD. 2011. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 10:390–400. doi: 10.1016/j.chom.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarakanova VL, Leung-Pineda V, Hwang S, Yang C-W, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HW IV, 2007. Gammaherpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1:275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijaya Lakshmi A, Ramana M, Vijayashree B, Ahuja Y, Sharma G. 1999. Detection of influenza virus induced DNA damage by comet assay. Mutat Res Toxicol Environ Mutagen 442:53–58. doi: 10.1016/S1383-5718(99)00058-3. [DOI] [PubMed] [Google Scholar]
- 31.Li N, Parrish M, Chan TK, Yin L, Rai P, Yoshiyuki Y, Abolhassani N, Tan KB, Kiraly O, Chow VTK, Engelward BP. 2015. Influenza infection induces host DNA damage and dynamic DNA damage responses during tissue regeneration. Cell Mol Life Sci 72:2973–2988. doi: 10.1007/s00018-015-1879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu LH, Huang M, Fang SG, Liu DX. 2011. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase delta. J Biol Chem 286:39546–39559. doi: 10.1074/jbc.M111.242206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, Webb LG, Bernal-Rubio D, Solovyov A, Greenbaum B, Simon V, Basler CF, Mulder LCF, García-Sastre A, Fernandez-Sesma A. 2017. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2:1–11. doi: 10.1038/nmicrobiol.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakad R, Schumacher B. 2016. DNA damage response and immune defense: links and mechanisms. Front Genet 7:147. doi: 10.3389/fgene.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paludan SR, Bowie AG. 2013. Immune sensing of DNA. Immunity 38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. 2011. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res 711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 37.Reshi ML, Su Y-C, Hong J-R. 2014. RNA viruses: ROS-mediated cell death. Int J Cell Biol 2014:467452. doi: 10.1155/2014/467452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aid M, Abbink P, Larocca RA, Boyd M, Nityanandam R, Nanayakkara O, Martinot AJ, Moseley ET, Blass E, Borducchi EN, Chandrashekar A, Brinkman AL, Molloy K, Jetton D, Tartaglia LJ, Liu J, Best K, Perelson AS, La Barrera RAD, Lewis MG, Barouch DH. 2017. Zika virus persistence in the central nervous system and lymph nodes of rhesus monkeys. Cell 169:610–620. doi: 10.1016/j.cell.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F, Hammack C, Ogden SC, Cheng Y, Lee EM, Wen Z, Qian X, Nguyen HN, Li Y, Yao B, Xu M, Xu T, Chen L, Wang Z, Feng H, Huang W, Yoon K, Shan C, Huang L, Qin Z, Christian KM, Shi P, Xu M, Xia M, Zheng W, Wu H, Song H, Tang H, Ming G-L, Jin P. 2016. Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res 44:8610–8620. doi: 10.1093/nar/gkw765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basu M, Courtney SC, Brinton MA. 2017. Arsenite-induced stress granule formation is inhibited by elevated levels of reduced glutathione in West Nile virus-infected cells. PLoS Pathog 13:e1006240. doi: 10.1371/journal.ppat.1006240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olagnier D, Peri S, Steel C, Van Montfoort N, Chiang C, Beljanski V, Slifker M, He Z, Nichols CN, Lin R, Balachandran S, Hiscott J. 2014. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 10:e1004566. doi: 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Chen Y, Gao N, Wang Y, Tian Y, Wu J, Zhang J, Zhu J, Fan D, An J. 2013. Inhibitory effect of glutathione on oxidative liver injury induced by dengue virus serotype 2 infections in mice. PLoS One 8:e55407. doi: 10.1371/journal.pone.0055407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berti M, Vindigni A. 2016. Replication stress: getting back on track. Nat Struct Mol Biol 23:103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. 2009. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem 284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B. 2016. Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell 164:644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmid M, Speiseder T, Dobner T, Gonzalez RA. 2014. DNA virus replication compartments. J Virol 88:1404–1420. doi: 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luftig MA. 2014. Viruses and the DNA damage response: activation and antagonism. Annu Rev Virol 1:605–625. doi: 10.1146/annurev-virology-031413-085548. [DOI] [PubMed] [Google Scholar]
- 50.Yu J, Zhang L, Ren P, Zhong T, Li Z, Wang Z, Li J, Liu X, Zhao K, Zhang W, Yu XF. 2015. Enterovirus 71 mediates cell cycle arrest in S phase through non-structural protein 3D. Cell Cycle 14:425. doi: 10.4161/15384101.2014.980631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim N-S, Yoon K-J, Shin J, Zhang C, Makri G, Nauen D, Yu H, Guzman E, Chiang C-H, Yoritomo N, Kaibuchi K, Zou J, Christian KM, Cheng L, Ross CA, Margolis RL, Chen G, Kosik KS, Song H, Ming G. 2014. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh RK, Kabbaj MHM, Paik J, Gunjan A. 2009. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol 11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]