While species A rotavirus is commonly associated with diarrheal disease in young children, species B rotavirus has caused sporadic outbreaks of adult diarrheal disease. A major genetic difference between species A and B rotaviruses is the NSP1 gene, which encodes two proteins for species B rotavirus. We demonstrate that the smaller of these proteins, NSP1-1, can mediate fusion of cultured human cells. Comparison with viral proteins of similar function provides insight into NSP1-1 domain organization and fusion mechanism. These comparisons suggest that there is a fatty acid modification at the amino terminus of the protein, and our results show that an intact amino terminus is required for NSP1-1-mediated fusion. NSP1-1 from a human virus mediates fusion of human cells, but not hamster cells, and enhances species A rotavirus replication in culture. These findings suggest potential, but currently untested, roles for NSP1-1 in RVB host species tropism, immune evasion, and pathogenesis.
KEYWORDS: FAST, NSP1-1, RVB, Reoviridae, fusion, rotavirus, syncytia
ABSTRACT
Rotavirus is an important cause of diarrheal disease in young mammals. Rotavirus species A (RVA) causes most human rotavirus diarrheal disease and primarily affects infants and young children. Rotavirus species B (RVB) has been associated with sporadic outbreaks of human adult diarrheal disease. RVA and RVB are predicted to encode mostly homologous proteins but differ significantly in the proteins encoded by the NSP1 gene. In the case of RVB, the NSP1 gene encodes two putative protein products of unknown function, NSP1-1 and NSP1-2. We demonstrate that human RVB NSP1-1 mediates syncytium formation in cultured human cells. Based on sequence alignment, NSP1-1 proteins from species B, G, and I contain features consistent with fusion-associated small transmembrane (FAST) proteins, which have previously been identified in other genera of the Reoviridae family. Like some other FAST proteins, RVB NSP1-1 is predicted to have an N-terminal myristoyl modification. Addition of an N-terminal FLAG peptide disrupts NSP1-1-mediated fusion. NSP1-1 from a human RVB mediates fusion of human cells but not hamster cells and, thus, may serve as a species tropism determinant. NSP1-1 also can enhance RVA replication in human cells, both in single-cycle infection studies and during a multicycle time course in the presence of fetal bovine serum, which inhibits rotavirus spread. These findings suggest potential yet untested roles for NSP1-1 in RVB species tropism, immune evasion, and pathogenesis.
IMPORTANCE While species A rotavirus is commonly associated with diarrheal disease in young children, species B rotavirus has caused sporadic outbreaks of adult diarrheal disease. A major genetic difference between species A and B rotaviruses is the NSP1 gene, which encodes two proteins for species B rotavirus. We demonstrate that the smaller of these proteins, NSP1-1, can mediate fusion of cultured human cells. Comparison with viral proteins of similar function provides insight into NSP1-1 domain organization and fusion mechanism. These comparisons suggest that there is a fatty acid modification at the amino terminus of the protein, and our results show that an intact amino terminus is required for NSP1-1-mediated fusion. NSP1-1 from a human virus mediates fusion of human cells, but not hamster cells, and enhances species A rotavirus replication in culture. These findings suggest potential, but currently untested, roles for NSP1-1 in RVB host species tropism, immune evasion, and pathogenesis.
INTRODUCTION
Rotaviruses are members of the Reoviridae family of nonenveloped viruses with segmented double-stranded RNA (dsRNA) genomes and causative agents of diarrheal disease in many young mammals, including humans. Adults are often resistant to rotavirus diarrheal disease. Acquired immunity, particularly local and systemic antibodies, plays an important role in protection from rotavirus disease, and immunity appears to increase with repeated infection or immunization (1). However, innate immunity also may contribute to rotavirus disease, and rotaviruses have been shown to antagonize innate signaling pathways using multiple distinct mechanisms (1–3).
Rotaviruses are currently classified into nine species, A to I, which further resolve into two major clades (https://talk.ictvonline.org/) (4–6). One clade contains species A, C, D, and F, and the other contains species B, G, H, and I. A tentatively proposed 10th species, rotavirus species J (RVJ), clusters phylogenetically with RVH (7). The majority of human rotavirus diarrheal disease occurs in infants and young children and is associated with rotavirus species A (RVA) infection (1). RVA also causes diarrheal disease in birds and numerous mammals though subsets of RVA genotypes are associated with specific hosts (8). RVB, RVC, RVH, and RVI have been detected mostly in domesticated mammals, while RVD, RVF, and RVG have been detected in birds (9, 10). Tentative species RVJ was detected in Schreiber’s bats (7). However, instances of zoonotic transmission of rotaviruses and their gene segments, particularly between humans and domesticated animals, have been documented (11, 12). Although some factors, such as lack of appropriate attachment and entry machinery or adaptive immune cross-protection, are known to impose barriers, factors permitting or limiting rotavirus zoonotic transmission remain incompletely understood.
Evidence of RVB infection has been commonly detected in diarrheic pigs (13–15), and RVB has been associated with sporadic outbreaks of diarrheal disease in humans (16–19). The first reported human RVB outbreak occurred in China from 1982 to 1983, ultimately affecting more than a million people with cholera-like diarrhea (20–24). While RVB disease symptoms resemble those of RVA gastroenteritis, RVB causes disease primarily in adults rather than pediatric populations (25). Studies suggest there is low-level RVB seroprevalence in humans (26–28). RVB outbreaks in humans are not thought to be caused by viruses directly transmitted from animals; rather, phylogenetic analysis of RVB sequences suggests that viruses affecting humans and other animals are distinct (29). Factors contributing to the capacity of these viruses to cause disease in adults remain unknown.
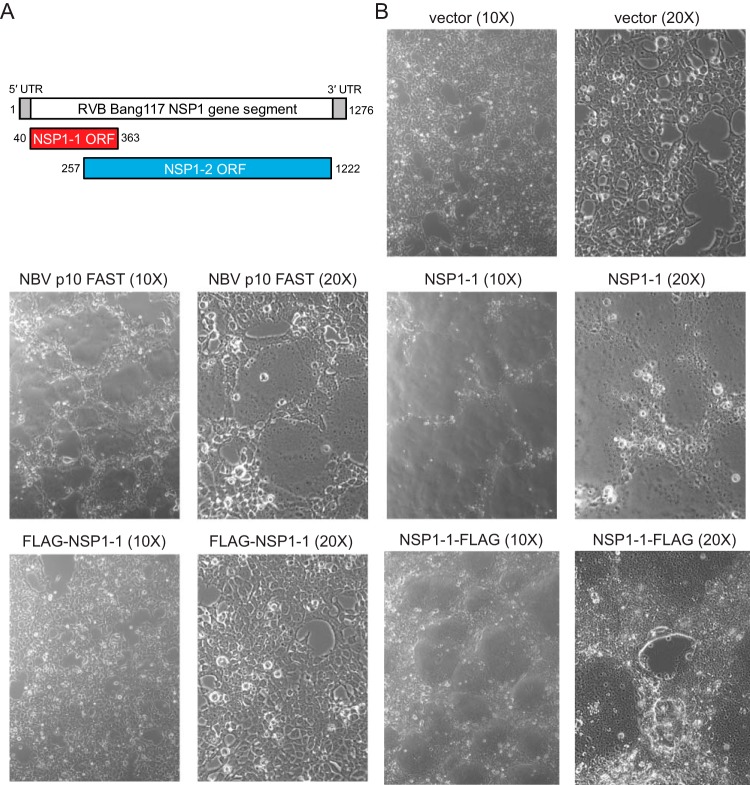
Like RVA, RVB has a genome containing 11 segments of dsRNA. Based on sequence alignment and structure prediction, 10 of the 11 RVB segments encode proteins with RVA homologs (30, 31). However, the segment encoding the RVA innate immune antagonist NSP1 differs significantly. For most rotaviruses in the clade containing RVB, including RVG and RVI, the NSP1 gene segment contains two overlapping open reading frames (ORFs) whose encoded protein products have little predicted sequence or structural homology with known proteins (32). The first RVB ORF, NSP1-1, is predicted to encode a product approximately 100 amino acids in length (Fig. 1A) though the protein product has not been shown to be expressed (32, 33). The length and predicted structural features of NSP1-1 are reminiscent of fusion-associated small transmembrane (FAST) proteins.
FIG 1.
RVB Bang117 NSP1-1 mediates syncytium formation in 293T cells. (A) Schematic showing the organization of RVB Bang117 NSP1 gene segment, including untranslated regions (UTRs) and two putative ORFs (32, 33). (B) Differential interference contrast images of 293T cells transfected with vector alone or plasmids encoding NBV p10 FAST or RVB Bang117 NSP1-1 in its untagged form or with an N- or C-terminal FLAG tag. Representative images are shown. Plasmids used for transfection and lens objective magnification are indicated.
FAST proteins are a family of small, bitopic plasma membrane proteins that mediate cell-cell fusion and syncytium formation (reviewed in references 34 to 36). These nonstructural viral proteins are encoded by fusogenic members of the Aquareovirus and Orthoreovirus genera of the nonenveloped Reoviridae family. There are multiple types of FAST proteins, and they range in size from approximately 90 to 200 amino acids. Each contains three modular domains: a small, N-terminal extracellular domain that is often acylated, a transmembrane domain, and a C-terminal cytoplasmic tail containing a polybasic motif. Additional putative functional motifs have been identified and vary among FAST protein family members. FAST proteins are nonstructural proteins that are expressed following virus entry, viral mRNA transcription, and translation. Membrane fusion occurs in closely apposed cells. FAST protein clustering and interactions with the opposing lipid bilayer, including insertion of fatty acid moieties or hydrophobic residues, favors lipid mixing and membrane curvature, leading to pore formation. Following pore formation, cellular proteins, including annexin 1 and actin, promote pore expansion and, thereby, syncytium formation.
In the current publication, we provide evidence that RVB NSP1-1 is a FAST protein that is capable of mediating syncytium formation in some, but not all, mammalian cell types. Based on sequence alignment, we suggest that other rotavirus species, RVG and RVI, may also encode functional FAST proteins. We demonstrate that the N terminus is required for NSP1-1-mediated fusion and provide experimental support for a role of NSP1-1 in viral replication and possibly in spread. These findings have potential implications for the role of NSP1-1 in host immune evasion and human RVB disease.
(This article was submitted to an online preprint archive [37]).
RESULTS
The N terminus is required for RVB NSP1-1-mediated fusion in 293T cells.
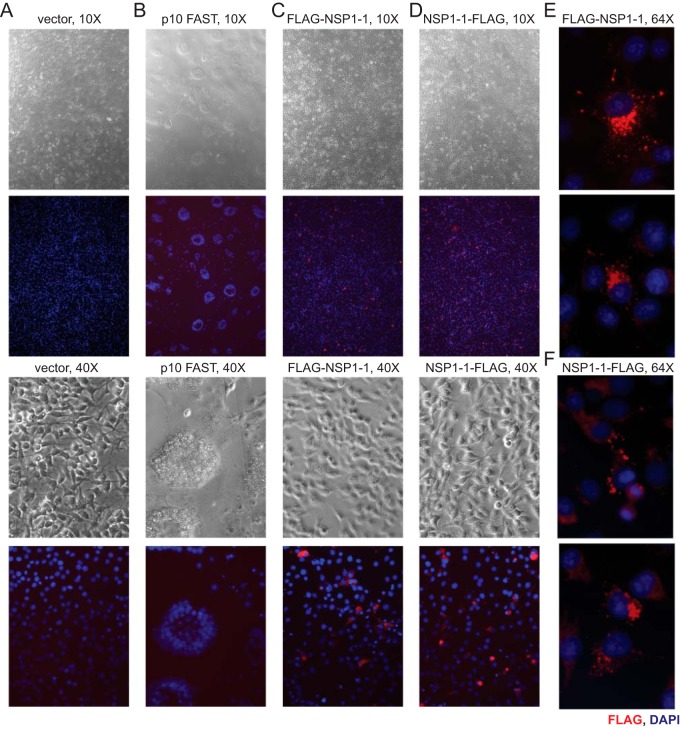
To test the hypothesis that NSP1-1 is a FAST protein, we first transfected human embryonic kidney 293T cells with a pCAGGS vector, pCAGGS encoding the fusogenic Nelson Bay orthoreovirus (NBV) p10 FAST protein (38), or pCAGGS encoding human RVB Bang117 NSP1-1 (33) and then permitted expression for 24 h and examined cell morphology using differential interference contrast microscopy. Vector-transfected cells were indistinguishable from mock-transfected or untransfected cells. Transfection with NBV p10, a known FAST protein (38–42), or with RVB NSP1-1 changed the cell morphology from individually distinct cells to a monolayer pockmarked by smooth oval-shaped regions lacking defined cell edges, which likely represent syncytia (Fig. 1B). These observations suggest that, like NBV p10, RVB NSP1-1 can mediate cell-cell fusion.
To enable detection of RVB NSP1-1, we engineered a FLAG peptide at the N or C terminus. Following transfection of 293T cells with pCAGGS encoding tagged forms of RVB NSP1-1, we found that C-terminally tagged NSP1-1 (NSP1-1–FLAG) mediated morphological changes resembling syncytia in the cell monolayer (Fig. 1B). Cells transfected with plasmids encoding N-terminally tagged NSP1-1 (FLAG–NSP1-1), however, were morphologically indistinguishable from vector-transfected cells. This finding suggests that the N terminus plays an important role in RVB NSP1-1-mediated cell morphological changes.
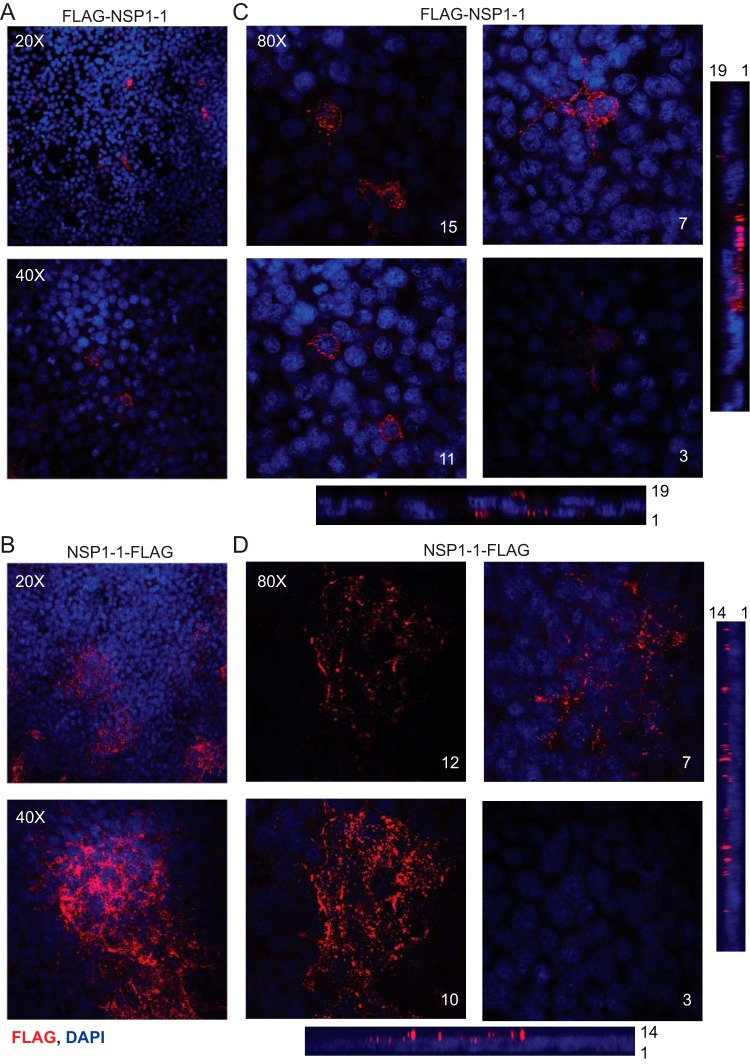
To gain insight into RVB NSP1-1 localization and cell morphological changes, we transfected 293T cells with pCAGGS encoding FLAG-tagged NSP1-1, waited 24 h to permit protein expression, fixed and stained the cells to detect FLAG and nuclei, and imaged them using confocal microscopy. FLAG–NSP1-1 was typically expressed in the cytoplasm of individual, or sometimes adjacent, cells (Fig. 2A). In z-stacks, FLAG–NSP1-1 was detected in the cytoplasm at the level of the nucleus, and individual stained cells were distinct (Fig. 2C). In striking contrast to FLAG–NSP1-1, NSP1-1–FLAG was detected in clusters containing many nuclei (Fig. 2B). Observed through a z-stack, NSP1-1–FLAG was detected at and above the level of the nucleus, consistent with cellular and plasma membrane localization, and the edges of individual stained cells were indistinguishable (Fig. 2D).
FIG 2.
RVB Bang117 NSP1-1 localization in 293T cells. 293T cells were transfected with plasmids encoding RVB Bang117 NSP1-1 with an N- or C-terminal FLAG tag. Cells were fixed and stained with antibodies to detect FLAG (red) or nuclei (blue) and imaged using confocal microscopy. (A and B) Images from a single confocal plane of 293T cells transfected with plasmids encoding FLAG–NSP1-1 or NSP1-1–FLAG, as indicated, taken using a 20× or 40× objective, as indicated. (C and D) Confocal images from comparable focal planes in z-stacks taken using the 80× objective for 293T cells transfected with plasmids encoding FLAG–NSP1-1 or NSP1-1–FLAG, as indicated. The z-section number is indicated; numbers increase coincident with distance from the adherent surface of the monolayer. Bars at the bottom and right side represent expanded orthogonal views through the z-stack, approximately at the center of the images, with the order of z-sections indicated.
Shared features of RVB NSP1-1 and Reoviridae FAST proteins.
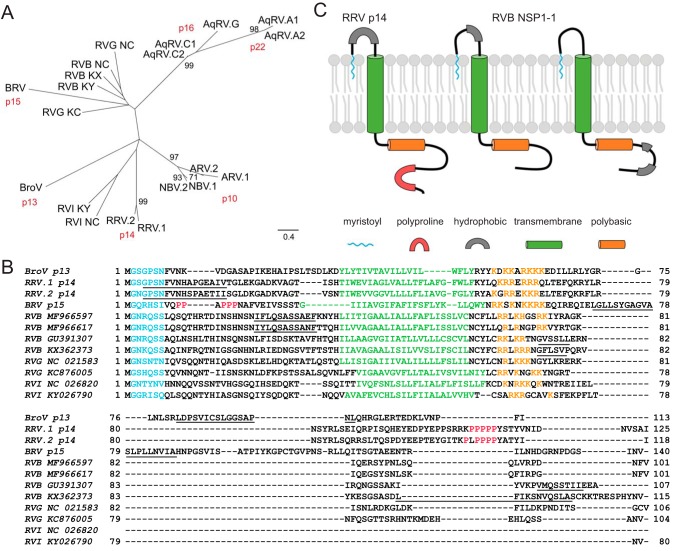
The similarity in protein size and cell morphological changes induced upon RVB NSP1-1 expression suggested that it may be a FAST protein. To gain insight into the relationships between rotavirus NSP1-1 proteins and Reoviridae FAST proteins, we constructed a maximum likelihood (ML) tree using the sequences of representative RVB, RVG, and RVI NSP1-1 proteins and orthoreovirus and aquareovirus FAST proteins (Fig. 3A). We found that aquareovirus and avian reovirus/NBV FAST proteins each formed a clade supported by strong bootstrap values that clustered distinctly from the rotavirus NSP1-1 proteins and baboon orthoreovirus (BRV) p15, Broome orthoreovirus (BroV) p13, and reptilian reovirus (RRV) p14 FAST proteins. While they did not cluster together strongly, RVB and RVG NSP1-1 proteins clustered most closely with BRV p15, whereas RVI NSP1-1 proteins clustered more closely with RRV p14.
FIG 3.
Conserved features of Reoviridae FAST proteins. (A) Maximum likelihood tree showing relationships among Reoviridae FAST and rotavirus NSP1-1 proteins. Abbreviations are as follows: AqRV.A1, Atlantic salmon aquareovirus; AqRV.A2, turbot aquareovirus; AqRV.G, American grass carp aquareovirus; AqRV.C1, golden shiner aquareovirus; AqRV.C2, grass carp aquareovirus; ARV.1, avian orthoreovirus 176; ARV.2, psittacine orthoreovirus; NBV.1, Nelson Bay orthoreovirus; NBV.2, Melaka orthoreovirus; BroV, Broome orthoreovirus; RRV.1, python orthoreovirus; RRV.2, green bush viper orthoreovirus; BRV, baboon orthoreovirus; RVB KY, rotavirus B strain RVB/Goat-wt/USA/Minnesota-1/2016; RVB KX, rotavirus B strain RVB/Pig-wt/VNM/12089_7; RVB NC, human rotavirus B strain Bang373; RVG KC, rotavirus G pigeon/HK18; RVG NC, rotavirus G chicken/03V0567/DEU/2003; RVI NC, rotavirus I strain KE135/2012; RVI KY, rotavirus I cat. Scale, in amino acid substitutions per site, is indicated. Each bootstrap value greater than 70% is shown adjacent to the supported node. (B) Alignment of selected Reoviridae FAST and rotavirus NSP1-1 proteins. Abbreviations for FAST proteins are as given in panel A. Rotavirus species (RVB, RVG, or RVI) and accession number are indicated. Predicted N-myristoylation motifs are shown in cyan, transmembrane helices are in green, polybasic regions are in orange, polyproline regions are in red, and hydrophobic regions are underlined. (C) Cartoon models highlighting the predicted features and membrane topology for the RRV p14 FAST protein and for RVB NSP1-1. The leftmost RVB model exemplifies RVB MF966597 and MF966617 listed in panel B, which contain a hydrophobic region in the predicted extracellular N-terminal domain, whereas the rightmost RVB model exemplifies RVB GU391307 and KX362373 from panel B, which contain hydrophobic regions in the predicted C-terminal cytoplasmic tail. Features and models of BroV p13, RRV p14, and BRV p15 shown in panels B and C are based on previously published work (35).
To gain insight into sequence and structural features of rotavirus NSP1-1 proteins, we used software to scan for sequence motifs and constructed amino acid alignments with the most closely clustering FAST proteins from the ML tree (Fig. 3A). Based on the PROSITE definition (entry PDOC00008), an N-myristoylation site was predicted at amino acids 2 to 7 in RVB NSP1-1 (Fig. 3B). Although there is a high false-positive prediction rate for N-myristoylation motifs, prediction at this precise location for every complete RVB, RVG, and RVI NSP1-1 sequence deposited in GenBank (as of 12/4/2018) (Fig. 4) provides confidence in its legitimacy. BroV, RRV, and BRV FAST proteins are also known or predicted to be N-myristoylated (43–46). Using the TMHMM server, we identified predicted transmembrane (TM) helices in RVB, RVG, and RVI sequences (Fig. 3B). In each case, the N terminus was predicted to be extracellular, while the C terminus was predicted to be cytoplasmic. For RVB Bang117 NSP1-1, the TM region was predicted to span amino acids 39 to 61. Like the BroV, RRV, and BRV FAST proteins, each of the NSP1-1 proteins contained multiple basic residues shortly after the TM domain (Fig. 3B). Some RVB sequences contained short stretches of hydrophobic residues in the N-terminal domain, while others contained two short hydrophobic regions in the C-terminal domain (Fig. 3B). Analyzed RVG and RVI NSP1-1 proteins lacked strong hydrophobic signatures outside the predicted transmembrane domain. NSP1-1 proteins were typically shorter than FAST proteins, by up to 36 amino acids, with most of the difference in length residing C terminal to the polybasic region (Fig. 3B). The motifs identified by sequence alignment and analysis suggest models of RVB NSP1-1 in which the extracellular, myristoylated N-terminal domain precedes a single transmembrane domain and a short cytoplasmic tail containing a polybasic region, with some RVB NSP1 proteins containing a single hydrophobic region N terminal to the transmembrane domain and others containing two hydrophobic regions C terminal to the polybasic region (Fig. 3C).
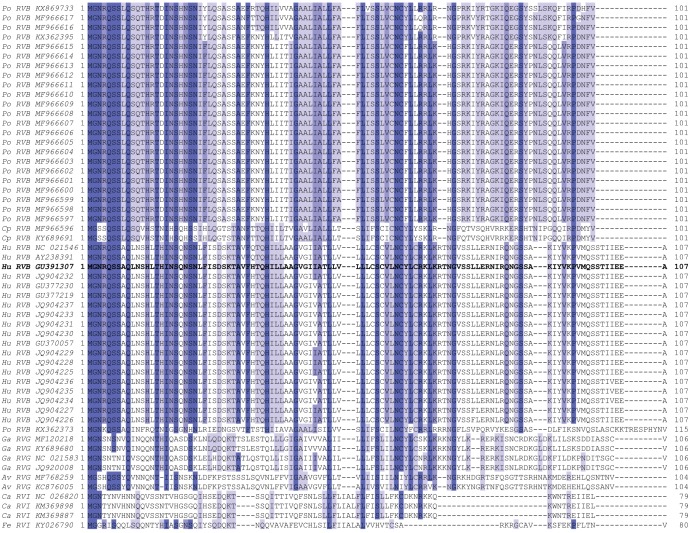
FIG 4.
Alignment of complete RVB, RVG, and RVI NSP1-1 sequences, colored based on amino acid identity, with darker purple indicating higher identity at a given position. RVB Bang117 NSP1-1 is shown in bold text. For each sequence, host origin, rotavirus species, and GenBank accession number are indicated. Av, avian; Ca, canine; Cp, caprine; Fe, feline; Ga, gallinaceous; Hu, human; Po, porcine.
RVB NSP1-1 mediates syncytium formation in Caco-2 cells.
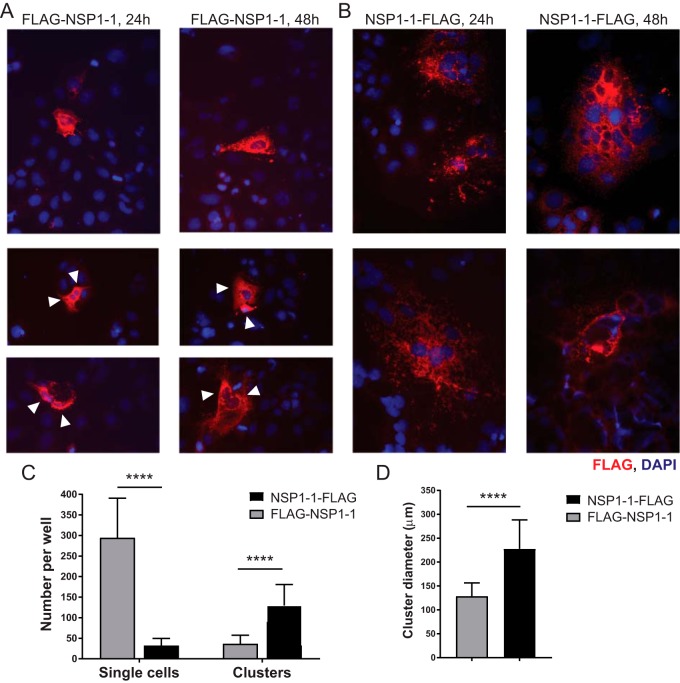
RVA infects enterocytes in the human intestine (1). Similarly, in animal models, RVB infects enterocytes at the tips of small intestinal villi (47–50). To determine whether RVB Bang117 NSP1-1 could mediate syncytium formation in a more biologically relevant cell type, we transfected human epithelial colorectal adenocarcinoma Caco-2 cells with pCAGGS encoding FLAG-tagged RVB NSP1-1. These cells can form polarized monolayers and exhibit many properties typical of absorptive enterocytes in the small intestine. To achieve reasonable transfection efficiency, we transfected subconfluent Caco-2 monolayers, waited 24 or 48 h to permit protein expression, fixed and stained the cells to detect FLAG and nuclei, and then imaged them using indirect immunofluorescence microscopy. Similar to observations made in 293T cells (Fig. 2A), we found that FLAG–NSP1-1 was primarily expressed in the cytoplasm of individual Caco-2 cells or occasionally in groups of 2 to 4 adjacent cells with distinct cell edges, whereas NSP1-1–FLAG was expressed mostly in the cytoplasm of clusters of cells containing many nuclei and lacking distinct cell boundaries (Fig. 5A and B). We quantified the numbers of single cells and clusters, which we defined as at least three immediately adjacent FLAG-positive cells, present in wells of transfected Caco-2 cells at 24 h posttransfection. Consistent with a role for the N terminus in cell-cell fusion, we found significantly more FLAG-positive single cells in FLAG–NSP1-1-transfected than NSP1-1–FLAG-transfected wells (∼9-fold) and significantly more clusters in NSP1-1–FLAG-transfected cells than in FLAG–NSP1-1-transfected wells (∼3.5-fold) (Fig. 5C). Often, groups of FLAG–NSP1-1-transfected cells we identified as “clusters” appeared to be groups of three or four adjacent singly transfected cells, whereas FLAG-positive “clusters” in NSP1-1–FLAG-transfected cells appeared to contain more nuclei per cluster. To quantify differences in cluster size between FLAG–NSP1-1-transfected and NSP1-1–FLAG-transfected Caco-2 cells, we measured cluster diameters and found that NSP1-1–FLAG clusters were significantly larger than FLAG–NSP1-1 clusters (Fig. 5D). Based on FLAG staining pattern, number of nuclei per cluster, and cluster diameter (Fig. 5), we anticipate that FLAG–NSP1-1-transfected cell clusters represent adjacent cells in a well that were simultaneously yet independently transfected or transfected cells that underwent division during the assay, whereas NSP1-1–FLAG-transfected cell clusters are syncytia. Together, these findings suggest that C-terminally tagged RVB Bang117 NSP1-1 can mediate fusion of a cell type similar to that targeted during RVB infection in animal models (47–50).
FIG 5.
RVB Bang117 FAST mediates syncytium formation in Caco-2 cells. (A and B) Caco-2 cells in 24-well plates were transfected with plasmids encoding RVB Bang117 NSP1-1 with an N- or C-terminal FLAG tag. Cells were fixed and stained with antibodies to detect FLAG (red) or nuclei (blue) and imaged at 24 and 48 h posttransfection using immunofluorescence microscopy. Representative images are shown for FLAG–NSP1-1-transfected cells and NSP1-1–FLAG-transfected cells, as indicated. White triangles indicate adjacent FLAG-positive FLAG–NSP1-1-transfected cells. (C) The numbers of FLAG-positive clusters (3 or more adjacent FLAG-positive cells) and single FLAG-positive cells per well were quantified in three wells per experiment in four independent experiments (n = 12). (D) The diameters of 20 FLAG-positive clusters per experiment in four independent experiments were quantified (n = 80). ****, P < 0.0001, by unpaired t test.
RVB NSP1-1 fails to mediate fusion in BHK cells.
We next wanted to test RVB NSP1-1 function in the context of viral infection. NBV p10 enhances reovirus and rotavirus replication in baby hamster kidney cells expressing T7 RNA polymerase (BHK-T7 cells) (38). The reverse genetics system for simian rotavirus strain SA11 involves transfection of BHK-T7 cells with plasmids encoding the 11 rotavirus RNAs under the control of the T7 promoter, along with pCAGGS plasmids encoding viral capping enzymes to enhance viral protein translation and NBV p10 to enhance rotavirus replication and spread. We hypothesized that if RVB NSP1-1 could mediate syncytium formation in BHK-T7 cells, it could functionally replace NBV p10 in rotavirus reverse genetics experiments. As a first step toward this goal, we transfected BHK-T7 cells with pCAGGS alone or pCAGGS expressing NBV p10, FLAG–NSP1-1, or NSP1-1–FLAG, waited 24 h to permit protein expression, fixed and stained the cells to detect FLAG and nuclei, and imaged them using differential interference contrast and indirect immunofluorescence microscopy. Transfection with pCAGGS encoding NBV p10 resulted in the appearance of balls or ring-like clusters of nuclei surrounded by regions of smooth cytoplasm nearly devoid of nuclei (Fig. 6B). Although many transfected cells detectably expressed NSP1-1–FLAG or FLAG–NSP1-1, all cells had distinct borders, and no differences in morphology were observed in comparison to vector-transfected cells (Fig. 6A, C, and D). A similar lack of morphological change was observed following transfection of BHK-T7 cells with untagged RVB NSP1-1 (data not shown). Although localization was not examined in depth, NSP1-1–FLAG staining did not often diffusely fill the cytoplasm but appeared most strongly in perinuclear puncta, suggesting potential mislocalization (Fig. 6F). These findings suggest that RVB NSP1-1 is incapable of mediating cell-cell fusion in all mammalian cell lines.
FIG 6.
RVB NSP1-1 fails to mediate fusion in BHK cells. BHK-T7 cells were transfected with vector alone (A) or plasmids encoding NBV p10 FAST (B) or RVB Bang117 NSP1-1 with an N-terminal (C and E) or C-terminal (D and F) FLAG tag. Cells were fixed and stained with antibodies to detect FLAG (red) or nuclei (blue) and imaged using both differential interference contrast and immunofluorescence microscopy. Representative images are shown. Plasmids used for transfection and lens objective magnification are indicated. In panels E and F, the field of view was cropped and enlarged to highlight FLAG localization in individual cells.
RVB NSP1-1 mediates enhanced RVA replication in human cells.
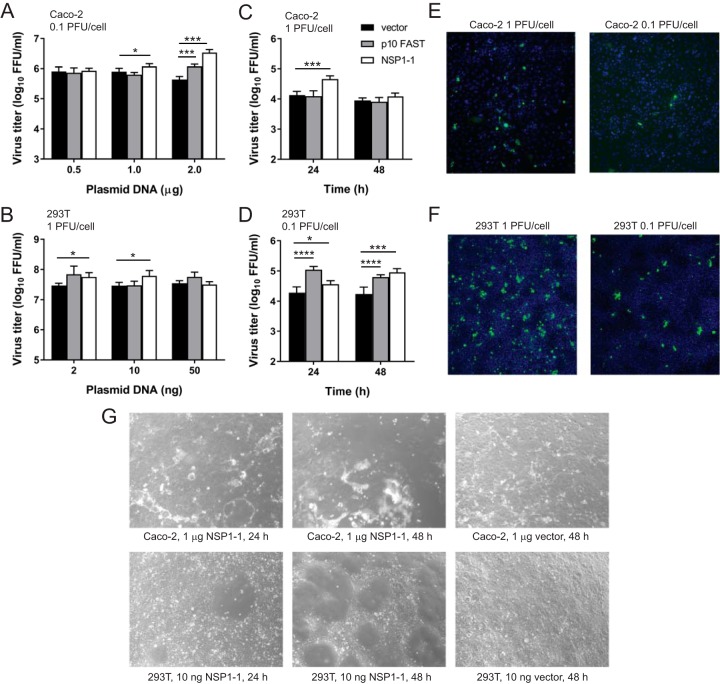
Since NSP1-1 did not mediate cell-cell fusion in BHK-T7 cells, we sought to test RVB NSP1-1 function in the context of viral infection in Caco-2 and 293T cells, which can support RVA replication and RVB NSP1-1-mediated fusion. Although the precise mechanism is unknown, NBV p10 has been shown to enhance replication of simian RVA strain SA11 in cells adsorbed at low multiplicity of infection (MOI) during a 16-h time course, which is less than the time required to complete a round of replication and initiate infection of a new subset of cells (38). Trypsin, which cleaves viral attachment protein VP4, activates RVA for optimal infectivity (1). To determine whether NBV p10 or RBV NSP1-1 affected viral replication in Caco-2 or 293T cells, we transfected these cells with increasing concentrations of pCAGGS alone or pCAGGS encoding NBV p10 or RVB NSP1-1 and then infected them with 0.1 PFU/cell (Caco-2) or 1 PFU/cell (293T) of trypsin-activated RVA strain SA11. The MOI was calculated by titration on MA104 cell monolayers. In the cell lines used for the experiment, these MOIs result in infection of approximately 0.1% (Caco-2) or 10% (293T) of cells (Fig. 7E and F). At 16 h postinfection, we lysed the cells and quantified viral titers. We found that SA11 titer was enhanced in Caco-2 cells transfected with 2 μg of NBV p10 (∼3-fold) or 1 or 2 μg of NSP1-1 (∼2-fold or ∼8-fold, respectively), in comparison to SA11 titers in vector-transfected cells (Fig. 7A). A modest but statistically significant enhancement of SA11 titer (∼2-fold) was detected in 293T cells transfected with 2 or 10 ng of pCAGGS expressing RVB NSP1-1 in comparison to the titer in vector-transfected cells (Fig. 7B) but was not detected at a lower MOI. While the detected differences are smaller than the ∼10-fold or 40-fold increase in RVA titer following transfection of BHK-T7 cells, respectively, with 1 or 2 μg of pCAGGS encoding NBV p10 (38), these findings suggest a potential role for RVB NSP1-1 in enhancing rotavirus replication in human cells during a single infectious cycle.
FIG 7.
RVB Bang117 FAST mediates enhanced rotavirus replication in human cells. (A and B) Short-term infection. Caco-2 cells (A) or 293T cells (B) were transfected with the indicated amount of plasmid DNA. At 4 h posttransfection, cells were adsorbed with trypsin-activated SA11 rotavirus, washed, incubated in serum-free medium containing trypsin at 37°C for 16 h, and lysed. Virus titers in cell lysates were quantified by FFA. (C and D) Rotavirus replication and spread in the presence of FBS. Caco-2 cells were transfected with 1 μg of plasmid DNA, or 293T cells were transfected with 10 ng of plasmid DNA. At 4 h posttransfection, cells were adsorbed with trypsin-activated SA11 rotavirus, washed, incubated in MEM containing 20% (Caco-2) or 10% (293T) FBS for 0, 24, or 48 h, and lysed. Virus titers in cell lysates were quantified by FFA. The MOI, as titrated on MA104 cells, is indicated. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001, in comparison to results with vector alone, by unpaired t test (n = 3). (E and F) Confluent monolayers of Caco-2 or 293T cells were adsorbed with the indicated MOI of SA11 rotavirus, as titrated on MA104 cells, and incubated for 24 h in medium containing FBS. Cells were fixed and stained to detect nuclei (blue) and rotavirus antigen (green) and imaged by indirect immunofluorescence to provide information about SA11 infectivity in experiments shown in panels A to D. (G) Cytopathic effects in transfected cells. Caco-2 or 293T cells were transfected with the indicated concentrations of plasmids and imaged using bright-field microscopy at 24 or 48 h posttransfection to reveal gross morphological changes in the monolayer.
Rotavirus spreads poorly in cultured cells in the presence of fetal bovine serum (FBS), likely due to cleavage inhibition of the viral attachment protein. To determine whether RVB NSP1-1 could facilitate rotavirus spread in the presence of FBS, we transfected Caco-2 or 293T cells with pCAGGS alone or pCAGGS encoding NBV p10 or RVB NSP1-1, infected the cells with RVA strain SA11 at a MOI of 1 PFU/cell (Caco-2) or 0.1 PFU/cell (293T), and then incubated them in standard culture medium containing 20% FBS (Caco-2) or 10% FBS (293T). These MOIs result in the infection of approximately 1% of cells (Fig. 7E and F). At 24 h and 48 h posttransfection, we lysed the cells and quantified the viral titer. While we anticipated an increase in titer between 0 and 24 h, we did not anticipate that the viral titer would increase between 24 and 48 h postadsorption unless viral spread was facilitated by direct cell-cell fusion since FBS should inhibit activation of the SA11 attachment protein and infection of a new round of cells. In Caco-2 cells, we found a modest increase in SA11 titer (<10-fold) at 24 h postinfection in RVB NSP1-1-transfected cells in comparison to the titer in vector-transfected cells (Fig. 7C). No significant difference in titers was detected at 48 h postinfection. However, by this time, RVB NSP1-1-transfected Caco-2 monolayers displayed evidence of significant cytopathic effects, including cell rounding and lifting, which may indicate poor cell viability (Fig. 7G). In 293T cells, we found that SA11 titers were significantly enhanced for both NBV p10- and RVB NSP1-1-transfected cells in comparison to the titer in vector-transfected cells at 24 and 48 h postinfection (Fig. 7D). Notably, the titer increased from 24 to 48 h in NSP1-1-transfected but not vector-transfected 293T cells. Transfection of 293T cells with similar amounts of pCAGGS expressing RVB NSP1-1 resulted in modest (24 h) to significant (48 h) visible syncytium formation within the monolayer, without complete monolayer disruption and cell lifting (Fig. 7G). Together, these findings suggest that RVB NSP1-1 can enhance rotavirus replication during multicycle infection, potentially by enabling direct cell-to-cell spread.
DISCUSSION
Based on our findings, we propose that RVB encodes functional FAST proteins that contain a myristoyl moiety at the N terminus, an extracellular N-terminal loop, a central transmembrane helix, and a relatively short cytoplasmic tail containing a region of approximately four basic residues (Fig. 3C). Short stretches of hydrophobic residues also are predicted in either the N- or C-terminal region of the protein. The morphological changes induced in cultured cells following NSP1-1 expression, which include the appearance of smooth patches lacking distinct cell membranes and resemble those induced by NBV p10, suggest that RVB NSP1-1 can mediate syncytium formation in human 293T cells (Fig. 1B). The detection of FLAG-positive multinucleated clusters in NSP1-1–FLAG-transfected 293T and Caco-2 cells suggests that the cells expressing NSP1-1 are fusing to one another, and addition of a peptide to the C terminus does not disrupt fusion activity (Fig. 2 and 5). The appearance and reduced number of clusters in FLAG–NSP1-1-transfected 293T and Caco-2 cells suggest that the N terminus plays an important role in fusion (Fig. 2, 3, and 5).
While the proposed sequence and structural features of RVB NSP1-1 remain to be biochemically and structurally validated, sequence alignment and analysis support our model of RVB NSP1-1 organization (Fig. 3). For example, the presence of a predicted myristoylation motif at the N terminus of every RVB, RVG, and RVI sequence in GenBank (Fig. 3B and 4) provides support for the presence of this fatty acid modification. Conservation of a myristoylation motif at the N terminus and the polybasic motif in the cytoplasmic tail in all NSP1-1 proteins, as well as clusters of hydrophobic residues in either the N- or C-terminal domain, suggests that these motifs may be critical for fusion (Fig. 3B and C and Fig. 4). The myristoylated N terminus and hydrophobic patch residues of FAST proteins induce lipid mixing between liposomal membranes (44, 51). While insertion of N-terminal fatty acid and hydrophobic moieties may promote membrane merger, cytoplasmic hydrophobic patches may promote pore formation by inducing or stabilizing membrane curvature in the inner leaflet at the rim of a newly formed fusion pore (35). Fewer basic residues are present in NSP1-1 proteins (4 to 5 residues) than in some FAST protein polybasic regions (6 to 7 residues), but the p10 FAST protein has only three to four basic residues in the polybasic motif, and the p14 FAST protein requires only three of its six basic residues to mediate syncytium formation (52, 53). Sequence alignments suggest that RVG and RVI also may encode FAST proteins (Fig. 3B) though their functionality remains to be tested directly. If functional, RVI NSP1-1 will represent the smallest known FAST protein and may help define the minimal requirements for cell-cell fusion.
Many viruses exhibit tropism for certain animal species or cell types. Based on the observation that NSP1-1 can mediate fusion in human cells but not hamster cells, it may serve as a viral tropism determinant though this hypothesis remains to be thoroughly tested. Support for influence on host species preference comes from the observation that NSP1 gene sequences of RVBs from murine, human, ovine, bovine, and porcine hosts cluster in distinct phylogenetic groups, with low identities between them and the greatest diversity detected among porcine RVBs (29). Why NBV p10 FAST is capable of mediating fusion in BHK-T7 cells while RVB NSP1-1 is not currently is unclear. While RVB NSP1-1 is predicted to be myristoylated at the N terminus, p10 FAST proteins form an extracellular cysteine loop and are palmitoylated at a membrane-proximal site in the N terminus (40, 54–56). These differences in acylation may affect functionality. Based on apparent mislocalization of NSP1-1, it is possible that signals that mediate trafficking from the endoplasmic reticulum, where FAST proteins are translated, to the plasma membrane via the secretory pathway (45, 52, 53, 57, 58) fail to function appropriately in some nonhomologous hosts. Chimeric FAST proteins containing individual domain exchanges between RVB Bang117 NSP1-1 and NBV p10, similar to those engineered for other FAST proteins (34, 35, 59), may provide insight into protein domains responsible for the species-specific fusion activity of RVB Bang117 NSP1-1.
Expression of NSP1-1 during infection has not been shown, but indirect evidence suggests that it is expressed. NSP1-1 ORF products failed to be immunoprecipitated using convalescent rat serum following in vitro transcription and translation from full-length IDIR gene 7 (32). NSP1-2 translation is predicted to be more efficient than NSP1-1 translation based on nucleotide sequences flanking the START codons (32). However, villous epithelial syncytial cell formation has been observed in the ileum and jejunum of RVB IDIR-infected neonatal rats, with syncytial cells reported to contain viral particles (47, 48). Additionally, an ovine RVB strain produced RVB-positive syncytia on MA104 monolayers (60). In numerous studies of RVB in pigs and cows, researchers have failed to note detection of syncytia, and in a rodent model of infection with a fusogenic pteropine orthoreovirus (PRV), authors failed to detect syncytia in infected lung tissue when specifically looking for these cells (39). It has been suggested, however, that syncytia are rapidly sloughed from the intestinal epithelium and therefore easily missed (60). While low levels of expression and cytotoxic effects may render this protein, and the syncytia whose formation it mediates, difficult to detect, these observations suggest that NSP1-1 is expressed during RVB infection.
Our results with cells transiently transfected with plasmids expressing RVB NSP1-1 and infected with RVA SA11 suggest potential functions for NSP1-1 in rotavirus replication and spread (Fig. 7). Since only a single successful in vitro culture system has been published for RVB (61), with no follow-up studies, we used RVA to study NSP1-1 function in the context of viral infection. Our experimental results support a role for RVB NSP1-1 in enhancing rotavirus replication in the presence of trypsin at a time point less than the length of a single infectious cycle (Fig. 7A and B) and in the presence of inhibitory FBS at time points that would permit multiple rounds of replication (Fig. 7C and D). The latter result is consistent with enhancement of replication by permitting the virus to spread from cell to cell without having to initiate infection at the plasma membrane, whereas the former result suggests another mechanism of replication enhancement. A major drawback to our experiments using RVA is the inability to ensure that NSP1-1 expression and viral infection occurred in the same cell. With a small percentage of cells infected and only a subset of cells transfected with plasmid DNA, it is likely that only a fraction of infected cells also fused to adjacent cells to mediate virus spread. In our system, we also were unable to modulate NSP1-1 expression levels, and the CAG promoter may have mediated significantly higher levels of protein expression than occur during natural infection, fusing cells too rapidly and resulting in cell death (Fig. 7G). Ultimately, these preliminary observations will need to be validated in a more biologically relevant system. Nonetheless, our results are consistent with those from other published studies showing that FAST proteins can enhance replication of dsRNA viruses on sub-single-cycle and multicycle time scales (38, 39). In one study, the authors detected enhancement of viral RNA synthesis in the presence of FAST proteins as early as 5 h postinfection, and they hypothesized that cell-to-cell fusion provides access to additional substrates for viral transcription, such as nucleotide triphosphates and S-adenosyl methionine (39). Since replication enhancement conferred by FAST proteins was detected even at a high MOI, the authors suggested that enhancement is not mediated by cell-to-cell spread. Mechanisms by which cell fusion could enhance viral replication at a high MOI remain unclear as fusion would not be anticipated to provide access to new sources of material for building progeny virions under these conditions.
What is the biological function of RVB NSP1-1 during infection? Syncytia formed between epithelial cells may increase rates of cell-to-cell spread and enhance viral replication and shedding within the infected host. This hypothesis is supported by the previous detection of syncytial cells, which were reported to contain the majority of virus particles, at the tips of jejunal and ileal villi during RVB infection of neonatal rats (47, 48). Close cellular apposition, mediated by adherens junctions, facilitates FAST protein-mediated fusion (62). Thus, enterocytes, which form close contacts via tight junctions, are ideal candidate cells for FAST protein-mediated syncytium formation. While the hypothesis remains to be tested, our findings (Fig. 7C and D) suggest the possibility that cell-to-cell fusion induced by NSP1-1 aids in the spread of RVB strains following introduction into the gastrointestinal tract. Such a mechanism could potentially enable immune evasion within the host by permitting viral spread in the presence of neutralizing antibodies. A report of dairy cows involved in a 2002 RVB diarrhea outbreak shedding a highly similar strain of RVB during a 2005 outbreak suggests that animals are not completely protected after the initial infection (63) though RVA reinfection with reduced disease severity also occurs (1, 64). Regardless of the mechanism, there is now published evidence supporting a biological role for a FAST protein in vivo (39). In a rodent model, two PRVs that are isogenic except in the capacity to express p10 FAST exhibited significant differences in pathogenesis, with animals infected with PRV containing an intact FAST protein exhibiting reduced body weight and survival and enhanced viral titer and lung pathology compared to levels in animals infected with PRV lacking p10 FAST expression (39). While we currently lack a system in which to directly test its function, these observations suggest a potential role for RVB NSP1-1 and other FAST proteins in viral replication and pathogenesis in vivo.
Although it is reasonable to anticipate that NSP1-1 permits evasion of adaptive immune responses by promoting direct rotavirus cell-to-cell spread within the host, it is unclear how RVB and other viruses in its clade (RVG, RVH, and RVI) evade innate immune signaling in the absence of an RVA NSP1 homolog. RVA NSP1 has been shown to promote degradation of innate signaling molecules, including interferon regulatory factors and β-TrCP (3, 65). In some cases, RVA NSP1 function is host species specific. Perhaps cell-cell fusion permits rotavirus to evade some innate immune mechanisms, or perhaps NSP1-2, whose function remains unknown, obviates the need for an RVA-like NSP1 protein. The evolutionary mechanisms through which FAST proteins became incorporated into rotavirus genomes or into Reoviridae genomes in general and the consequences of the lack of an NSP1-1 ORF for RVH and potentially RVJ viruses also are unclear (66). Future studies using new animal models and technologies, such as reverse genetics and human intestinal organoid culture (38, 67), may permit insights into differences in host interactions among the rotavirus species.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Human embryonic kidney 293T cells were grown in Dulbecco’s modified Eagle’s minimal essential medium (Corning) supplemented to contain 10% fetal bovine serum (FBS) (Gibco) and 2 mM l-glutamine. Human colonic epithelial Caco-2 cells were grown in Eagle’s minimum essential medium (MEM) with Earle’s salts and l-glutamine (Corning) supplemented to contain 20% FBS, 1× MEM nonessential amino acids (Sigma), 10 mM HEPES (Corning), and 1 mM sodium pyruvate (Gibco). Monkey kidney epithelial MA104 cells were grown in MEM with Earle’s salts and l-glutamine (Corning) supplemented to contain 5% FBS. Baby hamster kidney cells expressing T7 RNA polymerase under control of a cytomegalovirus promoter (BHK-T7) (68) were grown in Dulbecco’s modified Eagle’s minimal essential medium (Corning) supplemented to contain 5% fetal bovine serum, 2 mM l-glutamine, and 10% tryptose phosphate broth (Gibco). These cells were propagated in the presence of 1 mg/ml G418 (Invitrogen) during alternate passages.
Simian rotavirus laboratory strain SA11 was propagated in MA104 cells, and viral titers were determined by fluorescent focus assay (FFA) using MA104 cells (69).
Monoclonal mouse anti-FLAG antibody (Sigma), sheep polyclonal rotavirus antiserum (Invitrogen), Alexa Fluor 546-conjugated anti-mouse IgG (Invitrogen), and Alexa Fluor 488-conjugated anti-sheep IgG (Invitrogen) are commercially available.
Plasmids.
NBV p10 in pCAGGS has been described previously (38). pLIC8 was constructed by engineering a ligation-independent cloning site in mammalian expression plasmid pGL4.74. pLIC6 was constructed by engineering a ligation-independent cloning site into mammalian expression plasmid pCAGGS. RVB Bang117 was sequenced from a specimen obtained in 2002 from a 32-year-old male with severe diarrhea in Bangladesh (33). RVB Bang117 NSP1-1 was synthesized (GenScript) and cloned into pLIC8 and pLIC6 using ligation-independent cloning following PCR amplification with appropriate primers and T4 DNA polymerase treatment. Tagged versions of NSP1-1 were engineered using round-the-horn PCR (https://openwetware.org/wiki/%27Round-the-horn_site-directed_mutagenesis). Briefly, a pair of primers, each encoding half of the FLAG peptide (DYKDDDDK), was used to amplify NSP1-1 in pLIC8 (primer sequences available upon request). The resulting PCR fragment was purified and ligated to form a complete tag inserted at the N or C terminus of the ORF. After the sequences of plasmid clones were verified, tagged versions of NSP1-1 were transferred into pLIC6 using ligation-independent cloning, and nucleotide sequences were verified by Sanger sequencing.
Cell transfection and imaging.
For differential interference contrast imaging, 293T cells (∼5 × 105 per well) in 12-well plates were transfected with 0.2 μg of plasmid DNA per well using LyoVec transfection reagent (InvivoGen), according to manufacturer’s instructions, incubated at 37°C for 24 h, and imaged using a Zeiss Axiovert 200 inverted microscope. For confocal imaging, glass coverslips were sterilized, coated with poly-l-lysine (Sigma), rinsed, and dried. 293T cells (1.25 × 105 per well) were seeded onto coverslips in 24-well plates and incubated at 37°C 1 day prior to transfection with 0.1 μg of plasmid DNA per well using LyoVec. At 24 h posttransfection, cells were fixed with cold methanol and blocked with phosphate-buffered saline (PBS) containing 1% FBS. FLAG peptides were detected with mouse anti-FLAG, diluted 1:500, and Alexa Fluor 546-conjugated anti-mouse IgG, diluted 1:1,000, and nuclei were detected using 300 nM 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen), with washes in PBS containing 0.5% Triton X-100 (Fisher Scientific). Stained coverslips were mounted on glass slides using ProLong Gold antifade mountant (Invitrogen) and dried prior to imaging with an Olympus FV-1000 Inverted confocal microscope.
BHK-T7 cells (∼2 × 105 per well) in 24-well plates were transfected with 0.5 μg of plasmid DNA per well using TransIT-LT1 transfection reagent (Mirus Bio) in Opti-MEM (Gibco), according to the manufacturer’s instructions, and incubated at 37°C for 24 h prior to fixing and staining. Cells were fixed with cold methanol and blocked with PBS containing 1% FBS. Staining to detect FLAG and nuclei was performed as described above. Stained cells were imaged using a Zeiss Axiovert 200 inverted microscope equipped with an HBO 100 mercury arc lamp.
Caco-2 cells (∼1 × 105 per well) in 24-well plates were transfected with 1 μg per well of plasmid DNA using TransIT-LT1 transfection reagent in Opti-MEM, according to the manufacturer’s instructions, and incubated at 37°C for 24 h prior to fixing and staining. Cells were fixed with 10% neutral buffered formalin and blocked with PBS containing 0.5% Triton X-100 and 5% FBS. Staining to detect FLAG and nuclei was performed as described above. Stained cells were imaged using a Zeiss Axiovert 200 inverted microscope equipped with an HBO 100 mercury arc lamp.
Quantitation of single cells, clusters, and cluster diameter.
Caco-2 cells in 24-well plates were transfected with 1 μg per well of plasmids encoding RVB Bang117 NSP1-1–FLAG or RVB Bang117 FLAG–NSP1-1 and stained to detect FLAG and nuclei as described for imaging studies. Clusters were defined as groupings of at least three FLAG-positive cells in contact with one another. To quantify single cells and clusters, entire wells of transfected cells were visually analyzed using a Zeiss Axiovert 200 inverted microscope. The person analyzing the wells was not the person who performed the transfections and in most cases was blinded to the identity of the samples. Four independent experiments, each containing three technical replicates, were analyzed. To quantify cluster diameter, entire wells of transfected cells from the four independent experiments just described were imaged using an ImageXpress Micro XL automated microscope imager (Molecular Devices). Diameters of 20 cell clusters per plasmid construct per experiment were quantified using MetaXpress image analysis software (version 6.5; Molecular Devices). To compare numbers of clusters, single cells, and cluster diameters, statistical analyses were performed using unpaired t tests with GraphPad Prism, version 7 (GraphPad).
Transfection-infection experiments in Caco-2 cells.
For short-term rotavirus transfection-infection experiments, Caco-2 cells (∼1 × 105 per well) in 24-well plates were transfected with 0.5, 1, or 2 μg of plasmid DNA per well using TransIT-LT1 transfection reagent in Opti-MEM and incubated at 37°C for 3 h. Medium was removed from the transfected cells and replaced with serum-free MEM for 1 h prior to virus adsorption. SA11 rotavirus was activated by incubation with 1 μg/ml trypsin at 37°C for 1 h. Medium was removed from the cells, and they were adsorbed with activated SA11 rotavirus diluted in 0.1 ml of serum-free MEM per well to an MOI of 0.1 PFU/cell at 37°C for 1 h. After adsorption, cells were washed and then incubated in serum-free MEM containing 0.5 μg/ml of trypsin at 37°C for 16 h. Cell lysates were harvested after three rounds of freezing and thawing, and virus in the resultant lysates was quantified by FFA.
For longer-term rotavirus transfection-infection experiments, Caco-2 cells (∼1 × 105 per well) in 24-well plates were transfected with 1 μg of plasmid DNA per well using TransIT-LT1 transfection reagent in Opti-MEM and incubated at 37°C for 3 h. Medium was removed from the transfected cells and replaced with serum-free MEM for 1 h prior to virus adsorption. SA11 rotavirus was activated by incubation with 1 μg/ml trypsin at 37°C for 1 h. Medium was removed from the cells, and they were adsorbed with activated SA11 rotavirus diluted in 0.1 ml of serum-free MEM per well to an MOI of 1 PFU/cell at 37°C for 1 h. After adsorption, cells were washed and then incubated in MEM containing 20% FBS at 37°C for 24 or 48 h. Cell lysates were harvested after three rounds of freezing and thawing, and virus in the resultant lysates was quantified by FFA.
Transfection-infection experiments in 293T cells.
For short-term rotavirus transfection-infection experiments, 293T cells (∼2.5 × 105 per well) in 24-well plates were transfected with 50, 10, or 2 ng of plasmid DNA per well using LyoVec and incubated at 37°C for 3 h. Medium was removed from the transfected cells and replaced with serum-free Dulbecco’s modified Eagle’s medium (DMEM) for 1 h prior to virus adsorption. SA11 rotavirus was activated by incubation with 1 μg/ml trypsin at 37°C for 1 h. Medium was removed from the cells, and they were adsorbed with activated SA11 rotavirus diluted in 0.1 ml of serum-free DMEM per well to an MOI of 1 PFU/cell at 37°C for 1 h. After adsorption, cells were washed and then incubated in serum-free DMEM containing 0.5 μg/ml trypsin at 37°C for 16 h. Cell lysates were harvested after three rounds of freezing and thawing, and virus in the resultant lysates was quantified by FFA.
For longer-term rotavirus transfection-infection experiments, 293T cells (∼2.5 × 105 per well) in 24-well plates were transfected with 6 ng of plasmid DNA per well using LyoVec and incubated at 37°C for 3 h. Medium was removed from the transfected cells and replaced with serum-free DMEM for 1 h prior to virus adsorption. SA11 rotavirus was activated by incubation with 1 μg/ml trypsin at 37°C for 1 h. Medium was removed from the cells, and they were adsorbed with activated SA11 rotavirus diluted in 0.1 ml of serum-free DMEM per well to an MOI of 0.1 PFU/cell at 37°C for 1 h. After adsorption, cells were washed and then incubated in DMEM containing 10% FBS at 37°C for 24 or 48 h. Cell lysates were harvested after three rounds of freezing and thawing, and virus in the resultant lysates was quantified by FFA.
Fluorescent focus assay.
MA104 cells (4 × 105 per well) were seeded in black-wall 96-well plates and incubated overnight until near confluency. Infected cell lysates were activated with 1 μg/ml trypsin for at 37°C for 1 h and then serially diluted in serum-free MEM. Medium was removed from MA104 cells; they were washed twice in serum-free MEM and adsorbed with serial virus dilutions at 37°C for 1 h. Inocula were removed, and cells were washed with serum-free MEM and then incubated in fresh medium at 37°C for 14 to 18 h. Cells were fixed with cold methanol, and rotavirus proteins were detected by incubation with sheep polyclonal rotavirus antiserum at a 1:500 dilution in PBS containing 0.5% Triton X-100 at 37°C, followed by incubation with Alexa Fluor 488-conjugated anti-sheep IgG diluted 1:1,000 and 300 nM DAPI. Images were captured for four fields of view per well using an ImageXpress Micro XL automated microscope imager (Molecular Devices). Total and percent infected cells were quantified using MetaXpress high-content image acquisition and analysis software (Molecular Devices). Fluorescent foci from four fields of view in duplicate wells for each sample were quantified. Statistical analyses were performed using GraphPad Prism, version 7 (GraphPad). Virus titers in cells transfected with plasmids expressing NBV p10 or RBV NSP1-1 were compared to those in vector-transfected cells using an unpaired t test.
Amino acid alignments and phylogenetic analysis.
Sequences of NSP1-1 were obtained from GenBank. Accession numbers for FAST sequences analyzed for the ML tree shown in Fig. 3A and alignment in Fig. 3B are the following: ACN38055, ADZ31982, ABV01045, AAM92750, AAM92738, AAF45151, ABY78878, AAF45157.1, ABM67655, ACU68609, AAP03134, AHL26969, and AAL01373. Accession numbers for rotavirus segment 5 source sequences for NSP1-1 proteins analyzed for the ML tree shown in Fig. 3A are the following: KY689691, NC_021546, KX362373, KC876005, NC_021583, NC_026820, and KY026790. In each case, the smaller ORF sequence was analyzed. Accession numbers for rotavirus segment 5 source sequences for NSP1-1 proteins are indicated in Fig. 3B and 4.
For phylogenetic analysis, amino acid sequences were aligned using the MUSCLE algorithm in MEGA, version 7.0 (70). The Le Gascuel 2008 model (71) was selected as the best-fit model by Modeltest and used in maximum likelihood (ML) phylogeny construction with 1,000 bootstrap replicates (in MEGA, version 7.0). The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a Jones-Taylor-Thornton (JTT) model and then selecting the topology with superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 8.9228]). The tree with the highest log likelihood is shown in Fig. 3A. The ML tree was visualized using FigTree, version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
For Fig. 3B and 4, amino acid alignments were constructed with MAFFT, version 7.2 (72), using the E-INS-I strategy. N-myristylation motifs were defined using ScanProsite (73). Transmembrane helices were predicted with the TMHMM server, version 2.0 (www.cbs.dtu.dk/services/TMHMM/). Hydrophobic, polybasic, and polyproline regions for FAST proteins were identified previously (35). Hydrophobic patches for NSP1-1 proteins were identified using ProtScale, with a window size of nine amino acids (74). Polybasic regions for NSP1-1 proteins were identified visually.
ACKNOWLEDGEMENTS
We thank Takeshi Kobayashi for NBV p10 in pCAGGS and Marco Morelli for pLIC6 and pLIC8 plasmids.
This research was supported in part by a CTSA award (no. UL1 TR002243) from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
REFERENCES
- 1.Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Rancaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Ding S, Zhu S, Ren L, Feng N, Song Y, Ge X, Li B, Flavell RA, Greenberg HB. 2018. Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells. Elife 7:e39494. doi: 10.7554/eLife.39494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morelli M, Ogden KM, Patton JT. 2015. Silencing the alarms: Innate immune antagonism by rotavirus NSP1 and VP3. Virology 479-480:75–84. doi: 10.1016/j.virol.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, Chappell JD, Ciarlet M, del Vas M, Dermody TS, Dormitzer PR, Duncan R, Fcang Q, Graham R, Guglielmi KM, Harding RM, Hillman B, Makkay A, Marzachì C, Matthijnssens J, Milne RG, Mohd Jaafar F, Mori H, Noordeloos AA, Omura T, Patton JT, Rao S, Maan M, Stoltz D, Suzuki N, Upadhyaya NM, Wei C, Zhou H. 2012. Family Reoviridae, p 541–637. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 5.Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol 157:1177–1182. doi: 10.1007/s00705-012-1273-3. [DOI] [PubMed] [Google Scholar]
- 6.Ogden KM, Johne R, Patton JT. 2012. Rotavirus RNA polymerases resolve into two phylogenetically distinct classes that differ in their mechanism of template recognition. Virology 431:50–57. doi: 10.1016/j.virol.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banyai K, Kemenesi G, Budinski I, Foldes F, Zana B, Marton S, Varga-Kugler R, Oldal M, Kurucz K, Jakab F. 2017. Candidate new rotavirus species in Schreiber’s bats, Serbia. Infect Genet Evol 48:19–26. doi: 10.1016/j.meegid.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhama K, Chauhan RS, Mahendran M, Malik SV. 2009. Rotavirus diarrhea in bovines and other domestic animals. Vet Res Commun 33:1–23. doi: 10.1007/s11259-008-9070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kindler E, Trojnar E, Heckel G, Otto PH, Johne R. 2013. Analysis of rotavirus species diversity and evolution including the newly determined full-length genome sequences of rotavirus F and G. Infect Genet Evol 14:58–67. doi: 10.1016/j.meegid.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Otto PH, Ahmed MU, Hotzel H, Machnowska P, Reetz J, Roth B, Trojnar E, Johne R. 2012. Detection of avian rotaviruses of groups A, D, F and G in diseased chickens and turkeys from Europe and Bangladesh. Vet Microbiol 156:8–15. doi: 10.1016/j.vetmic.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martella V, Banyai K, Matthijnssens J, Buonavoglia C, Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet Microbiol 140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 12.Doro R, Farkas SL, Martella V, Banyai K. 2015. Zoonotic transmission of rotavirus: surveillance and control. Expert Rev Anti Infect Ther 13:1337–1350. doi: 10.1586/14787210.2015.1089171. [DOI] [PubMed] [Google Scholar]
- 13.Brown DW, Beards GM, Chen GM, Flewett TH. 1987. Prevalence of antibody to group B (atypical) rotavirus in humans and animals. J Clin Microbiol 25:316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marthaler D, Rossow K, Culhane M, Goyal S, Collins J, Matthijnssens J, Nelson M, Ciarlet M. 2014. Widespread rotavirus H in commercially raised pigs, United States. Emerg Infect Dis 20:1195–1198. doi: 10.3201/eid2007.140034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marthaler D, Rossow K, Gramer M, Collins J, Goyal S, Tsunemitsu H, Kuga K, Suzuki T, Ciarlet M, Matthijnssens J. 2012. Detection of substantial porcine group B rotavirus genetic diversity in the United States, resulting in a modified classification proposal for G genotypes. Virology 433:85–96. doi: 10.1016/j.virol.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi MS, Ganorkar NN, Ranshing SS, Basu A, Chavan NA, Gopalkrishna V. 2017. Identification of group B rotavirus as an etiological agent in the gastroenteritis outbreak in Maharashtra, India. J Med Virol 89:2244–2248. doi: 10.1002/jmv.24901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan T, Sen A, Choudhury JS, Das S, Naik TN, Bhattacharya SK. 1999. Emergence of adult diarrhoea rotavirus in Calcutta, India. Lancet 353:380–381. doi: 10.1016/s0140-6736(05)74954-0. [DOI] [PubMed] [Google Scholar]
- 18.Lahon A, Maniya NH, Tambe GU, Chinchole PR, Purwar S, Jacob G, Chitambar SD. 2013. Group B rotavirus infection in patients with acute gastroenteritis from India: 1994–1995 and 2004–2010. Epidemiol Infect 141:969–975. doi: 10.1017/S0950268812001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanekata T, Ahmed MU, Kader A, Taniguchi K, Kobayashi N. 2003. Human group B rotavirus infections cause severe diarrhea in children and adults in Bangladesh. J Clin Microbiol 41:2187–2190. doi: 10.1128/jcm.41.5.2187-2190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CM, Hung T, Bridger JC, McCrae MA. 1985. Chinese adult rotavirus is a group B rotavirus. Lancet 2:1123–1124. doi: 10.1016/s0140-6736(85)90710-x. [DOI] [PubMed] [Google Scholar]
- 21.Eiden J, Vonderfecht S, Yolken RH. 1985. Evidence that a novel rotavirus-like agent of rats can cause gastroenteritis in man. Lancet 2:8–11. doi: 10.1016/s0140-6736(85)90057-1. [DOI] [PubMed] [Google Scholar]
- 22.Hung T, Chen GM, Wang CG, Chou ZY, Chao TX, Ye WW, Yao HL, Meng KH. 1983. Rotavirus-like agent in adult non-bacterial diarrhoea in China. Lancet 2:1078–1079. [PubMed] [Google Scholar]
- 23.Hung T, Wang CG, Fang ZY, Chou ZY, Chang XJ, Liong X, Chen G, Yao H, Chao T, Ye W, Den S, Chang W. 1984. Waterborne outbreak of rotavirus diarrhoea in adults in China caused by a novel rotavirus. Lancet i:1139–1142. [PubMed] [Google Scholar]
- 24.Sen A, Kobayashi N, Das S, Krishnan T, Bhattacharya SK, Naik TN. 2001. The evolution of human group B rotaviruses. Lancet 357:198–199. doi: 10.1016/S0140-6736(00)03596-0. [DOI] [PubMed] [Google Scholar]
- 25.Saiada F, Rahman HN, Moni S, Karim MM, Pourkarim MR, Azim T, Rahman M. 2011. Clinical presentation and molecular characterization of group B rotaviruses in diarrhoea patients in Bangladesh. J Med Microbiol 60:529–536. doi: 10.1099/jmm.0.025981-0. [DOI] [PubMed] [Google Scholar]
- 26.Alam MM, Pun SB, Gauchan P, Yokoo M, Doan YH, Tran TN, Nakagomi T, Nakagomi O, Pandey BD. 2013. The first identification of rotavirus B from children and adults with acute diarrhoea in Kathmandu, Nepal. Trop Med Health 41:129–134. doi: 10.2149/tmh.2013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiden JJ, Mouzinho A, Lindsay DA, Glass RI, Fang ZY, Taylor JL. 1994. Serum antibody response to recombinant major inner capsid protein following human infection with group B rotavirus. J Clin Microbiol 32:1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakata S, Estes MK, Graham DY, Wang SS, Gary GW, Melnick JL. 1987. Detection of antibody to group B adult diarrhea rotaviruses in humans. J Clin Microbiol 25:812–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki T, Kuga K, Miyazaki A, Tsunemitsu H. 2011. Genetic divergence and classification of non-structural protein 1 among porcine rotaviruses of species B. J Gen Virol 92:2922–2929. doi: 10.1099/vir.0.036426-0. [DOI] [PubMed] [Google Scholar]
- 30.Fang ZY, Glass RI, Penaranda M, Dong H, Monroe SS, Wen L, Estes MK, Eiden J, Yolken RH, Saif L. 1989. Purification and characterization of adult diarrhea rotavirus: identification of viral structural proteins. J Virol 63:2191–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang ZY, Monroe SS, Dong H, Penaranda M, Wen L, Gouvea V, Allen JR, Hung T, Glass RI. 1992. Coding assignments of the genome of adult diarrhea rotavirus. Arch Virol 125:53–69. doi: 10.1007/BF01309628. [DOI] [PubMed] [Google Scholar]
- 32.Eiden JJ. 1994. Expression and sequence analysis of gene 7 of the IDIR agent (group B rotavirus): similarity with NS53 of group A rotavirus. Virology 199:212–218. doi: 10.1006/viro.1994.1113. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto D, Ghosh S, Ganesh B, Krishnan T, Chawla-Sarkar M, Alam MM, Aung TS, Kobayashi N. 2010. Analysis of genetic diversity and molecular evolution of human group B rotaviruses based on whole genome segments. J Gen Virol 91:1772–1781. doi: 10.1099/vir.0.020412-0. [DOI] [PubMed] [Google Scholar]
- 34.Boutilier J, Duncan R. 2011. The reovirus fusion-associated small transmembrane (FAST) proteins: virus-encoded cellular fusogens. Curr Top Membr 68:107–140. doi: 10.1016/B978-0-12-385891-7.00005-2. [DOI] [PubMed] [Google Scholar]
- 35.Ciechonska M, Duncan R. 2014. Reovirus FAST proteins: virus-encoded cellular fusogens. Trends Microbiol 22:715–724. doi: 10.1016/j.tim.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Duncan R. 5 July 2019. Fusogenic reoviruses and their fusion-associated small transmembrane (FAST) proteins. Annu Rev Virol. doi: 10.1146/annurev-virology-092818-015523. [DOI] [PubMed] [Google Scholar]
- 37.Diller JR, Parrington HM, Patton JT, Ogden KM. 2019. Group B rotavirus encodes a functional fusion-associated small transmembrane (FAST) protein. bioRxiv 10.1101/639070. [DOI] [PMC free article] [PubMed]
- 38.Kanai Y, Komoto S, Kawagishi T, Nouda R, Nagasawa N, Onishi M, Matsuura Y, Taniguchi K, Kobayashi T. 2017. Entirely plasmid-based reverse genetics system for rotaviruses. Proc Natl Acad Sci U S A 114:2349–2354. doi: 10.1073/pnas.1618424114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanai Y, Kawagishi T, Sakai Y, Nouda R, Shimojima M, Saijo M, Matsuura Y, Kobayashi T. 2019. Cell-cell fusion induced by reovirus FAST proteins enhances replication and pathogenicity of non-enveloped dsRNA viruses. PLoS Pathog 15:e1007675. doi: 10.1371/journal.ppat.1007675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Key T, Duncan R. 2014. A compact, multifunctional fusion module directs cholesterol-dependent homomultimerization and syncytiogenic efficiency of reovirus p10 FAST proteins. PLoS Pathog 10:e1004023. doi: 10.1371/journal.ppat.1004023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salsman J, Top D, Boutilier J, Duncan R. 2005. Extensive syncytium formation mediated by the reovirus FAST proteins triggers apoptosis-induced membrane instability. J Virol 79:8090–8100. doi: 10.1128/JVI.79.13.8090-8100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanai Y, Kawagishi T, Nouda R, Onishi M, Pannacha P, Nurdin JA, Nomura K, Matsuura Y, Kobayashi T. 2019. Development of stable rotavirus reporter expression systems. J Virol 93:e01774-18. doi: 10.1128/JVI.01774-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corcoran JA, Duncan R. 2004. Reptilian reovirus utilizes a small type III protein with an external myristylated amino terminus to mediate cell-cell fusion. J Virol 78:4342–4351. doi: 10.1128/jvi.78.8.4342-4351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corcoran JA, Syvitski R, Top D, Epand RM, Epand RF, Jakeman D, Duncan R. 2004. Myristoylation, a protruding loop, and structural plasticity are essential features of a nonenveloped virus fusion peptide motif. J Biol Chem 279:51386–51394. doi: 10.1074/jbc.M406990200. [DOI] [PubMed] [Google Scholar]
- 45.Dawe S, Corcoran JA, Clancy EK, Salsman J, Duncan R. 2005. Unusual topological arrangement of structural motifs in the baboon reovirus fusion-associated small transmembrane protein. J Virol 79:6216–6226. doi: 10.1128/JVI.79.10.6216-6226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dawe S, Duncan R. 2002. The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein. J Virol 76:2131–2140. doi: 10.1128/jvi.76.5.2131-2140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vonderfecht SL, Huber AC, Eiden J, Mader LC, Yolken RH. 1984. Infectious diarrhea of infant rats produced by a rotavirus-like agent. J Virol 52:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salim AF, Phillips AD, Walker-Smith JA, Farthing MJ. 1995. Sequential changes in small intestinal structure and function during rotavirus infection in neonatal rats. Gut 36:231–238. doi: 10.1136/gut.36.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theil KW, Saif LJ, Moorhead PD, Whitmoyer RE. 1985. Porcine rotavirus-like virus (group B rotavirus): characterization and pathogenicity for gnotobiotic pigs. J Clin Microbiol 21:340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huber AC, Yolken RH, Mader LC, Strandberg JD, Vonderfecht SL. 1989. Pathology of infectious diarrhea of infant rats (IDIR) induced by an antigenically distinct rotavirus. Vet Pathol 26:376–385. doi: 10.1177/030098588902600503. [DOI] [PubMed] [Google Scholar]
- 51.Shmulevitz M, Epand RF, Epand RM, Duncan R. 2004. Structural and functional properties of an unusual internal fusion peptide in a nonenveloped virus membrane fusion protein. J Virol 78:2808–2818. doi: 10.1128/JVI.78.6.2808-2818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shmulevitz M, Duncan R. 2000. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the non-enveloped fusogenic reoviruses. EMBO J 19:902–912. doi: 10.1093/emboj/19.5.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parmar HB, Barry C, Kai F, Duncan R. 2014. Golgi complex-plasma membrane trafficking directed by an autonomous, tribasic Golgi export signal. Mol Biol Cell 25:866–878. doi: 10.1091/mbc.E13-07-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barry C, Key T, Haddad R, Duncan R. 2010. Features of a spatially constrained cystine loop in the p10 FAST protein ectodomain define a new class of viral fusion peptides. J Biol Chem 285:16424–16433. doi: 10.1074/jbc.M110.118232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Key T, Sarker M, de Antueno R, Rainey JK, Duncan R. 2015. The p10 FAST protein fusion peptide functions as a cystine noose to induce cholesterol-dependent liposome fusion without liposome tubulation. Biochim Biophys Acta 1848:408–416. doi: 10.1016/j.bbamem.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 56.Shmulevitz M, Salsman J, Duncan R. 2003. Palmitoylation, membrane-proximal basic residues, and transmembrane glycine residues in the reovirus p10 protein are essential for syncytium formation. J Virol 77:9769–9779. doi: 10.1128/jvi.77.18.9769-9779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parmar HB, Barry C, Duncan R. 2014. Polybasic trafficking signal mediates Golgi export, ER retention or ER export and retrieval based on membrane-proximity. PLoS One 9:e94194. doi: 10.1371/journal.pone.0094194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parmar HB, Duncan R. 2016. A novel tribasic Golgi export signal directs cargo protein interaction with activated Rab11 and AP-1-dependent Golgi-plasma membrane trafficking. Mol Biol Cell 27:1320–1331. doi: 10.1091/mbc.E15-12-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clancy EK, Duncan R. 2009. Reovirus FAST protein transmembrane domains function in a modular, primary sequence-independent manner to mediate cell-cell membrane fusion. J Virol 83:2941–2950. doi: 10.1128/JVI.01869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theil KW, Grooms DL, McCloskey CM, Redman DR. 1995. Group B rotavirus associated with an outbreak of neonatal lamb diarrhea. J Vet Diagn Invest 7:148–150. doi: 10.1177/104063879500700124. [DOI] [PubMed] [Google Scholar]
- 61.Sanekata T, Kuwamoto Y, Akamatsu S, Sakon N, Oseto M, Taniguchi K, Nakata S, Estes MK. 1996. Isolation of group B porcine rotavirus in cell culture. J Clin Microbiol 34:759–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salsman J, Top D, Barry C, Duncan R. 2008. A virus-encoded cell-cell fusion machine dependent on surrogate adhesins. PLoS Pathog 4:e1000016. doi: 10.1371/journal.ppat.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayashi M, Murakami T, Kuroda Y, Takai H, Ide H, Awang A, Suzuki T, Miyazaki A, Nagai M, Tsunemitsu H. 2016. Reinfection of adult cattle with rotavirus B during repeated outbreaks of epidemic diarrhea. Can J Vet Res 80:189–196. [PMC free article] [PubMed] [Google Scholar]
- 64.Velazquez FR, Matson DO, Calva JJ, Guerrero L, Morrow AL, Carter-Campbell S, Glass RI, Estes MK, Pickering LK, Ruiz-Palacios GM. 1996. Rotavirus infection in infants as protection against subsequent infections. N Engl J Med 335:1022–1028. doi: 10.1056/NEJM199610033351404. [DOI] [PubMed] [Google Scholar]
- 65.Arnold MM. 2016. The rotavirus interferon antagonist NSP1: many targets, many questions. J Virol 90:5212–5215. doi: 10.1128/JVI.03068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nibert ML, Duncan R. 2013. Bioinformatics of recent aqua- and orthoreovirus isolates from fish: evolutionary gain or loss of FAST and fiber proteins and taxonomic implications. PLoS One 8:e68607. doi: 10.1371/journal.pone.0068607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saxena K, Blutt SE, Ettayebi K, Zeng XL, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, Graham DY, Qureshi W, Sherman V, Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Donowitz M, Estes MK. 2016. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J Virol 90:43–56. doi: 10.1128/JVI.01930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi T, Ooms LS, Ikizler M, Chappell JD, Dermody TS. 2010. An improved reverse genetics system for mammalian orthoreoviruses. Virology 398:194–200. doi: 10.1016/j.virol.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnold M, Patton JT, McDonald SM. 2009. Culturing, storage, and quantification of rotaviruses. Curr Protoc Microbiol Chapter 15:Unit 15C.3. doi: 10.1002/9780471729259.mc15c03s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 72.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]