Abstract
Existing US epidemiological data demonstrate that consumption of smokeless tobacco, particularly moist snuff, is less harmful than cigarette smoking. However, the molecular and biochemical changes due to moist snuff consumption relative to smoking remain incompletely understood. We previously reported that smokers (SMK) exhibit elevated oxidative stress and inflammation relative to moist snuff consumers (MSC) and non-tobacco consumers (NTC), based on metabolomic profiling data of saliva, plasma, and urine from MSC, SMK, and NTC. In this study, we investigated the effects of tobacco consumption on additional metabolic pathways using pathway-based analysis tools. To this end, metabolic pathway enrichment analysis and topology analysis were performed through pair-wise comparisons of global metabolomic profiles of SMK, MSC, and NTC. The analyses identified >8 significantly perturbed metabolic pathways in SMK compared with NTC and MSC in all 3 matrices. Among these differentially enriched pathways, perturbations of caffeine metabolism, energy metabolism, and arginine metabolism were mostly observed. In comparison, fewer enriched metabolic pathways were identified in MSC compared with NTC (5 in plasma, none in urine and saliva). This is consistent with our transcriptomics profiling results that show no significant differences in peripheral blood mononuclear cell gene expression between MSC and NTC. These findings, taken together with our previous biochemical, metabolomic, and transcriptomic analysis results, provide a better understanding of the relative changes in healthy tobacco consumers, and demonstrate that chronic cigarette smoking, relative to the use of smokeless tobacco, results in more pronounced biological changes, which could culminate in smoking-related diseases.
Keywords: Biomarker, metabolomics, moist snuff, smoking, pathway analysis
Introduction
Cigarette smoking is an important risk factor for many diseases including lung cancer, chronic obstructive pulmonary disease (COPD), cardiovascular disease (CVD), and oral cancer.1 Cigarette smoke is a dynamic and complex aerosol containing non-volatile compounds in liquid droplet form (termed the “particulate phase”) and volatile constituents in the gas phase. More than 8000 chemical compounds have been identified in the particulate and gas phase of cigarette smoke,2 including many well-known toxicants. For example, 93 cigarette smoke constituents have been classified by the Food and Drug Administration (FDA) as harmful and potentially harmful constituents (HPHCs), which are further classified as carcinogens, respiratory toxicants, reproductive or developmental toxicants, cardiovascular toxicants, and/or addictive constituents.3
While cigarette smoking is the predominant form of tobacco consumption in the United States and other countries, consumption of smokeless tobacco products (STPs) is also common.4 Existing US and Swedish epidemiological data indicate that consumption of STPs is less harmful compared with cigarette smoking and the health risks associated with STPs use are lower than those with smoking.5-8 Although the health effects of cigarette smoking and knowledge of smoking-related biomarkers have been extensively documented,9 relatively less information exists on the effects of STP consumption. Consequently, very few biomarkers inform the biological effects (BioEff) of STP use.
To gain a better understanding of the physiological effects of smoking and STP use, R. J. Reynolds Tobacco Company (RJRT) has conducted several clinical studies to evaluate biomarkers of tobacco exposure (BioExp) and BioEff.10-14 Among these are 2 cross-sectional studies that include a CVD biomarker study13,14 and a biomarker discovery study11 consisting of 3 cohorts (non-tobacco consumers [NTC], smokers [SMK], and moist snuff consumers [MSC]). Both studies consistently showed that biomarkers of combustible toxicants were substantially higher in SMK compared with MSC. As expected, NTC exhibited the lowest levels of BioExp among these cohorts. Moreover, SMK showed higher levels of BioEff associated with oxidative stress (urinary isoprostanes and leukotriene E4), inflammation (white blood cell count), platelet activation (thromboxane metabolites), and lipid metabolism (apolipoprotein B100 and oxidized low-density lipoprotein), relative to NTC and MSC.11 Principal components analysis of serum CVD BioEff suggests that interleukin (IL)-12, soluble intracellular adhesion molecule (sICAM)-1, and IL-8 are potential BioEff that differentiate SMK, MSC, and NTC.14
Untargeted metabolomics profiling technologies such as high-resolution nuclear magnetic resonance spectroscopy and mass spectrometry (MS) have been used for discovery of metabolic biomarkers to evaluate health effects of short-term and long-term tobacco usage.15-22 Untargeted metabolomics enables high-throughput measurements of hundreds or even thousands of small molecules without prior knowledge and thus leads to identification of novel potential biomarkers. In the biomarker discovery study discussed above, Prasad et al12 applied gas chromatography (GC)–MS and liquid chromatography (LC)–MS/MS-based metabolomics to identify metabolomic biomarkers from plasma, urine, and saliva collected from clinical trial participants. Statistical analysis of the global metabolomics profiling suggests that the overall biochemical profile of SMK is distinct from that of MSC and NTC. Fewer metabolic differences in both number and magnitude of biochemical compounds were observed between MSC and NTC, compared with differences between SMK and NTC. Several metabolites associated with oxidative stress and inflammatory pathways were identified as potential metabolic BioEff.
Metabolic pathway analysis identifies clusters of metabolites related to key cellular signaling and metabolic networks, which provides mechanistic insight into the underlying biology of differentially expressed metabolites.23 In this study, we used metabolomic profiles from these 3 cohorts (NTC, SMK, and MSC) and an integrated metabolic pathway analysis approach that included pathway enrichment analysis and pathway topology analysis to assess the impact of smoking and consumption of moist snuff on human physiology. Perturbations in caffeine metabolism, energy metabolism, and arginine metabolism distinguished the 3 different cohorts. These findings, taken together with our previous investigations,11-13,24 provide a more comprehensive understanding of biochemical and physiological changes induced by consumption of various tobacco products.
Materials and Methods
Clinical study conduct
A single-blinded, cross-sectional clinical study was conducted at the High Point Clinical Trails Center, High Point, North Carolina. The clinical conduct and sample collection have been described elsewhere.11 Briefly, 40 healthy, male participants (age 35-60 years) were enrolled into 1 of 3 consumer group cohorts (SMK, MSC, or NTC) after they provided informed consent. The participants fasted overnight and refrained from tobacco use prior to sample collection. Plasma was collected into tubes containing ethylenediaminetetraacetic acid (EDTA). Unstimulated saliva was collected into tubes containing protease and phosphatase inhibitors. The 24 hour urine samples were collected under ambulatory conditions and stored at −80°C. Aliquots of the plasma, urine, and saliva samples were analyzed using global metabolomic profiling at Metabolon Inc. (Durham, North Carolina). The clinical conduct of the study was approved by the Independent Investigational Review Board, Inc (Plantation, Florida) and registered at ClinicialTrials.gov (ClinicalTrials.gov Identifier: NCT01923402).
Metabolomic profiling
Metabolomic profiling was performed using saliva, plasma, and urine samples collected from the participants.25-27 Briefly, all samples were extracted with a methanol solution and split into equal parts for analysis by GC-MS and LC-MS/MS.28 Two separate ultrahigh performance LC-MS/MS injections were optimized for basic and acidic species. After chromatographic separation using separate acid/base 2.1 × 100 mm Waters BEH C18 1.7 µm particle LC columns (or 20 m × 0.18 mm GC column with 0.18 µm film phase), full-scan MS was conducted to record and quantify all detectable ions formed after molecule fragmentation. Metabolites were identified by matching the ion’s chromatographic retention index, nominal mass, and spectral fragmentation signatures to Metabolon’s in-house reference library, which was generated from standard metabolites under similar analytical procedures as the experimental samples. Once identified, metabolite ions were quantified by integration of their corresponding peak area.29
Statistical analysis of individual metabolites
Raw data were imputed with minimum values, mean-normalized, and then log-transformed. A 2-sample unequal variances t test was used to compare individual plasma, urine, and saliva metabolomic profiles obtained from the 3 cohorts. Statistical significance was defined as P ⩽ .05 and q ⩽ .1. False discovery rates (FDRs), estimated by q values, were used to account for multiple comparisons.
Metabolic pathway analysis
Metabolic pathway enrichment analysis and pathway topology analysis were conducted using MetaboAnalyst 3.0 computational platform to better understand the functional impact of tobacco use on human metabolism.23 Pathway enrichment analysis computes a single P value for each metabolic pathway (a group of functional-associated metabolites), as opposed to the t test, which calculates statistical significance of the difference between individual metabolites. Pathway topology analysis applies graph theory to measure a given experimentally identified metabolite’s importance in a pre-defined metabolic pathway. Measurements were computed using centrality, a common metric used in graph theory to estimate the relative importance of individual nodes to the overall network. Similarly, the importance measures for other unidentified metabolites in the pathway were computed. A “pathway impact score” was then computed as the sum of the importance measures of identified metabolites divided by the total sum of the importance measures of all the identified and unidentified metabolites in the pathway. The pathway impact score represents an objective estimate of the importance of a given pathway relative to a global metabolic network.
In step 1 of the analysis, we created a 2-column data input file comprising an individual metabolite’s relative abundance and Human Metabolome Database entry number. Data on nicotine and its metabolites were excluded as these analyses were focused on identifying distinguishing BioEff as opposed to BioExp. In addition, unnamed structures from global metabolomic profiles were excluded from the data input file. The data input files were uploaded to the MetaboAnalyst 3.0 web server. Data pre-processing, such as normalization and scaling, and metabolic pathway analysis were performed using the following parameters: (1) enrichment analysis was performed using the global test method,30 (2) centrality was measured using Relative Betweenness, and (3) 80 human metabolic pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used as reference metabolic pathways. Details of the algorithms for enrichment pathway analysis and topology analysis were previously described.31
Classical FDR approach was used to control for false positives for metabolite enrichment analysis. A large FDR threshold, similar to gene set enrichment analysis (GSEA),32 was applied to identify significant “enriched” pathways (in our case, 0.22-0.52 and in GSEA, 0.25). The optimal FDR threshold identified is 0.32, as described in the “Results” section. The cut-off value of 0.1 for pathway impact score was used consistently across multiple comparisons to filter less important pathways, similar to previous work.33
Results
Pathway enrichment analysis and topology analysis were performed to analyze metabolomic profiles measured from plasma, urine, and saliva samples. Pair-wise comparisons were conducted among 3 cohorts (SMK, NTC, and MSC), namely, between SMK and NTC (SMK vs NTC), between MSC and NTC (MSC vs NTC), and between SMK and MSC (SMK vs MSC). Enriched functional pathways were identified and their impact scores were measured.
Optimal FDR threshold
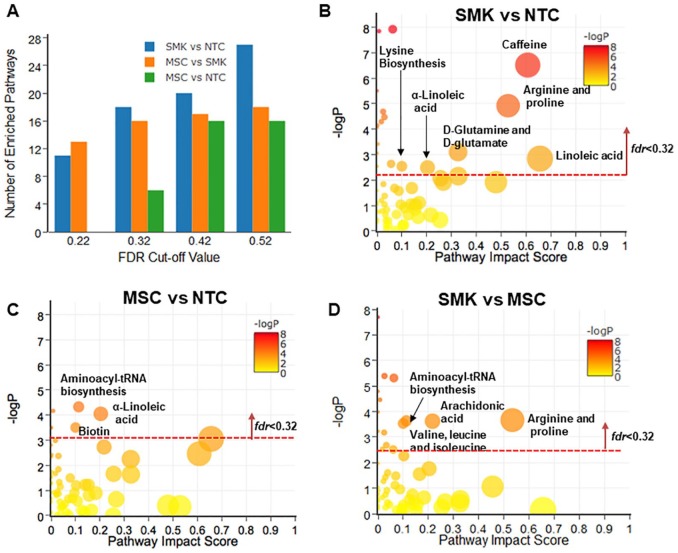
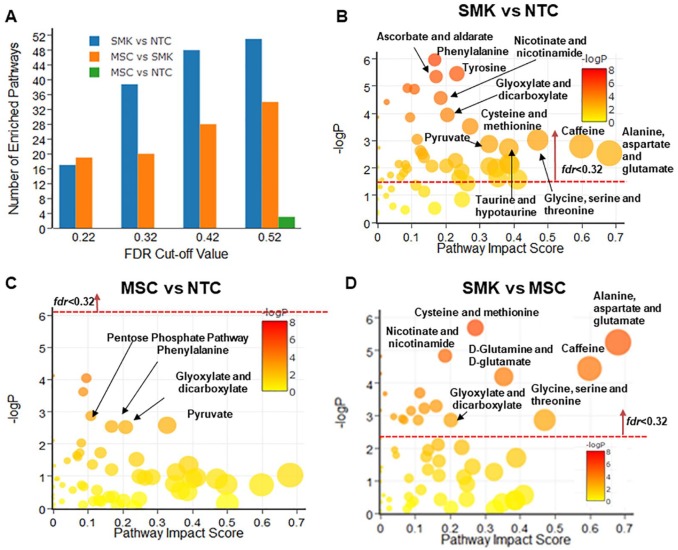
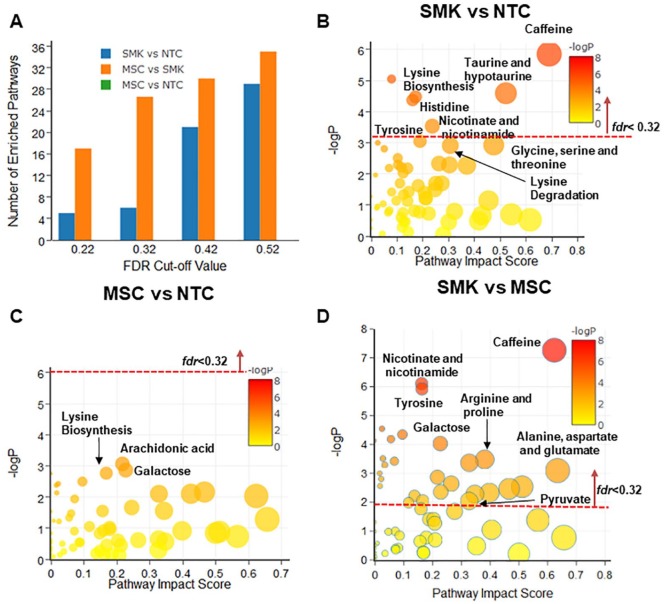
Different FDR threshold values were explored for pathway analysis of plasma metabolomics data (Figure 1A). When the FDR cut-off value (fdrcut-off) was 0.22, 11 and 13 significantly enriched metabolic pathways were observed between SMK and NTC, and between MSC and SMK, respectively. However, no significantly enriched pathways were found when MSC were compared with NTC. When the fdrcut-off was 0.32, 6 metabolic pathways were identified as significantly different between MSC and NTC, 18 pathways between SMK and NTC, and 16 between SMK and NTC (Figure 1A). However, there was no appreciable increase in the number of significantly enriched pathways between MSC and SMK when the fdrcut-off was increased to either 0.42 or 0.52. Thus, 0.32 was used as an optimal fdrcut-off for analyzing plasma metabolomics data. For urine, the number of significantly enriched pathways between MSC and NTC remained zero until fdrcut-off reached 0.52 (Figure 2A). In the case of saliva, none of the significantly enriched pathways between MSC and NTC were found when fdrcut-off increased from 0.22 to 0.52 (Figure 3A). For consistency with plasma analysis, fdrcut-off 0.32 was used as the threshold for analyzing urine and saliva metabolomics data.
Figure 1.
Pathway enrichment and topology analysis of plasma metabolomics data from 3 cohorts including SMK, MSC, and NTC. The most enriched pathways were identified when (A) SMK were compared with NTC (SMK vs NTC), (B) MSC versus NTC, and (C) SMK versus MSC. “Pathway Impact Score” in x-axis represents the impact of these enriched pathways computed from topology analysis. “–log P” in y-axis refers to negative natural logarithmic value of the original P value from statistical analysis of pathway difference between 2 cohorts. (D) The number of enriched pathways was computed when different fdrcut-off values were used. When fdrcut-off = 0.32, 18, 5, and 16 pathways were considered for the case of SMK versus NTC, MSC versus NTC, and SMK versus MSC, respectively. MSC indicates moist snuff consumers; NTC, non-tobacco consumers; SMK, cigarette smokers.
Figure 2.
Pathway enrichment and topology analysis of urinary metabolomics data from 3 cohorts including SMK, MSC, and NTC. The most enriched pathways were identified when (A) SMK were compared with NTC (SMK vs NTC); (B) MSC versus NTC; and (C) SMK versus MSC. “Pathway Impact Score” in x-axis represents the impact of these enriched pathways computed from topology analysis. “–log P” in y-axis refers to negative natural logarithmic value of the original P value from statistical analysis of pathway difference between 2 cohorts. (D) The number of enriched pathways was computed when different fdrcut-off values were used. None of the enriched pathways in the case of MSC versus NTC were identified as significantly different, except fdrcut-off = 0.52, respectively. MSC indicates moist snuff consumers; NTC, non-tobacco consumers; SMK, cigarette smokers.
Figure 3.
Pathway enrichment and topology analysis of saliva metabolomics data from 3 cohorts including SMK, MSC, and NTC. The most enriched pathways were identified when (A) SMK were compared with NTC (SMK vs NTC), (B) MSC versus NTC, and (C) SMK versus MSC. “Pathway Impact Score” in x-axis represents the impact of these enriched pathways computed from topology analysis. “–log P” in y-axis refers to negative natural logarithmic value of the original P value from statistical analysis of pathway difference between 2 cohorts. (D) The number of enriched pathways was computed when a number of fdrcut-off values were used. None of enriched pathways in the case of MSC versus NTC were identified as significantly different when fdrcut-off varies from 0.22 to 0.52. MSC indicates moist snuff consumers; NTC, non-tobacco consumers; SMK, cigarette smokers.
Plasma metabolomic pathway analysis
The enriched pathways and their impact scores identified from plasma metabolomics data are visualized in Figure 1. When SMK metabolomics data were compared with NTC (SMK vs NTC), 18 pathways were identified as significantly enriched, among which 6 pathways had relatively large impact scores (>0.1) (Figure 1B). These pathways included linoleic acid (LA) metabolism, alpha linolenic (α-LA) metabolism, caffeine metabolism, arginine and proline metabolism, lysine biosynthesis, and D-glutamine and D-glutamate metabolism. In the case of MSC versus NTC, only 6 enriched pathways were identified (Figure 1C); 3 of them had higher-impact scores (>0.1) including α-LA metabolism, aminoacyl-tRNA biosynthesis, and biotin metabolism, suggesting different biological pathways were affected by moist snuff consumption from cigarette smoking. In the comparison between SMK and MSC, aminoacyl-tRNA biosynthesis, arachidonic acid metabolism, valine/leucine/isoleucine metabolism, and arginine and proline metabolism were identified as enriched pathways with an impact score of >0.1 (Figure 1D).
Urine metabolomic pathway analysis
Comparative analysis of urine metabolomics data (Figure 2) revealed a greater number of differentially regulated pathways in SMK than those found in plasma (Figure 1). Relative to NTC, 38 differentially regulated pathways were detected in SMK (Figure 2B), whereas 20 pathways were enriched compared with MSC (Figure 2D). No enriched pathways were found between MSC and NTC (Figure 2C).
In SMK versus NTC, 28 enriched metabolic pathways with impact score >0.1 were found. They belong to a wide range of metabolic pathways, including carbohydrate metabolism (pyruvate metabolism, ascorbate and aldarate metabolism), metabolism of cofactors and vitamins (nicotinate and nicotinamide metabolism), amino acid (glycine, serine, and threonine; alanine, aspartate, and glutamate) metabolism, biosynthesis of other secondary metabolites (caffeine metabolism), and lipid (glycerophospholipid) metabolism (Figure 2B). Thus, urine appears to provide richer metabolic information compared with plasma. In SMK versus MSC, 11 out of 20 enriched pathways were identified with impact scores >0.1. They include caffeine metabolism, amino acid–related metabolisms (such as alanine, aspartate, and glutamate metabolism, and cysteine and methionine metabolism), nicotinate and nicotinamide metabolism, and others (Figure 2D).
Saliva metabolomic pathway analysis
Similar to urine, but not plasma, pathway analysis of the saliva metabolomic data revealed no significant difference in enriched pathways between MSC and NTC (Figure 3A and C). Six enriched pathways were identified between SMK and NTC in saliva (Figure 3B) as compared with plasma (18 pathways) and urine (38 pathways). Among these enriched pathways, 5 have >0.1 impact scores including caffeine metabolism, taurine and hypotaurine metabolism, histidine metabolism, and nicotinate and nicotinamide metabolism.
In contrast, 26 enriched pathways were observed between SMK and MSC in saliva (Figure 3D). Among them, 18 pathways were scored with an impact score higher than 0.10. Interestingly, they included not only the metabolic pathways previously identified (e.g., caffeine metabolism, arginine and proline metabolism, nicotinate and nicotinamide metabolism), but also the pathways of tricarboxylic acid (TCA) cycle (impact score = 0.25) and sphingolipid metabolism (impact score = 0.39). Thus, analyses of saliva data revealed several enriched pathways between SMK and MSC, which were not identified in plasma or urine.
Enriched metabolic pathways
Among the enriched pathways identified, several were consistently enriched in SMK when compared with both NTC and MSC in urine, plasma, or saliva and therefore were selected for further evaluation, namely, caffeine metabolism, amino acid metabolism (e.g., arginine and proline metabolism), and energy metabolism (e.g., pyruvate metabolism).
Caffeine metabolism
Caffeine metabolism, which involves the Phase I liver detoxification enzyme cytochrome P-450 1A2 (CYP1A2), was identified as a significantly enriched pathway when comparing SMK versus NTC and SMK versus MSC in plasma, urine, and saliva (Figures 1-3). A number of the 14 possible caffeine metabolites were detected in urine (13), plasma (8), and saliva (6) (Table 1).
Table 1.
Fold changes for the metabolites involved in caffeine metabolism among SMK, MSC, and NTC.
| Biochemical name | Fold changes |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Plasma |
Urine |
Saliva |
|||||||
| MSC/NTC | SMK/NTC | SMK/MSC | MSC/NTC | SMK/NTC | SMK/MSC | MSC/NTC | SMK/NTC | SMK/MSC | |
| Caffeine | 1.79 | 0.52 | 0.29 | 1.47 | 1.35 | 0.92 | 1.55 | 0.42 | 0.27 |
| Paraxanthine | 1.19 | 0.82 | 0.69 | 1.09 | 1.48 | 1.37 | 1.15 | 0.86 | 0.74 |
| Theobromine | 1.06 | 0.43 | 0.41 | 1.05 | 0.74 | 0.70 | 0.79 | 0.44 | 0.56 |
| 1-methylurate | ND | ND | ND | 1.02 | 1.34 | 1.32 | ND | ND | ND |
| 1,3-dimethylurate | ND | ND | ND | 0.97 | 1.71 | 1.75 | ND | ND | ND |
| 1,7-dimethylurate | 1.24 | 0.89 | 0.70 | 1.19 | 1.26 | 1.06 | 1.1 | 1.05 | 0.95 |
| 3,7-dimethylurate | ND | ND | ND | 0.88 | 0.62 | 0.71 | ND | ND | ND |
| 1,3,7-trimethylurate | ND | ND | ND | 1.13 | 1.11 | 0.98 | ND | ND | ND |
| 1-methylxanthine | 1.03 | 1.23 | 1.20 | 1.03 | 1.64 | 1.59 | ND | ND | ND |
| 3-methylxanthine | 0.90 | 0.81 | 0.90 | 0.98 | 0.88 | 0.89 | ND | ND | ND |
| 7-methylxanthine | 0.91 | 0.81 | 0.88 | 0.90 | 0.94 | 1.05 | 0.69 | 0.91 | 1.32 |
| 5-acetylamino-6-amino-3-methyluracil | ND | ND | ND | 0.97 | 1.93 | 2.00 | ND | ND | ND |
| 5-acetylamino-6-formylamino-3-methyluracil | ND | ND | ND | 0.94 | 2.07 | 2.20 | ND | ND | ND |
| Theophylline | 1.24 | 0.91 | 0.74 | ND | ND | ND | 1.23 | 0.89 | 0.72 |
The statistical significance P < .05 is highlighted by shaded and bolded “fold change” values; bolded fold of change values indicate .05 < P < .1. Abbreviations: MSC, moist snuff consumers; ND, not detected; NTC, non-tobacco consumers; SMK, cigarette smokers.
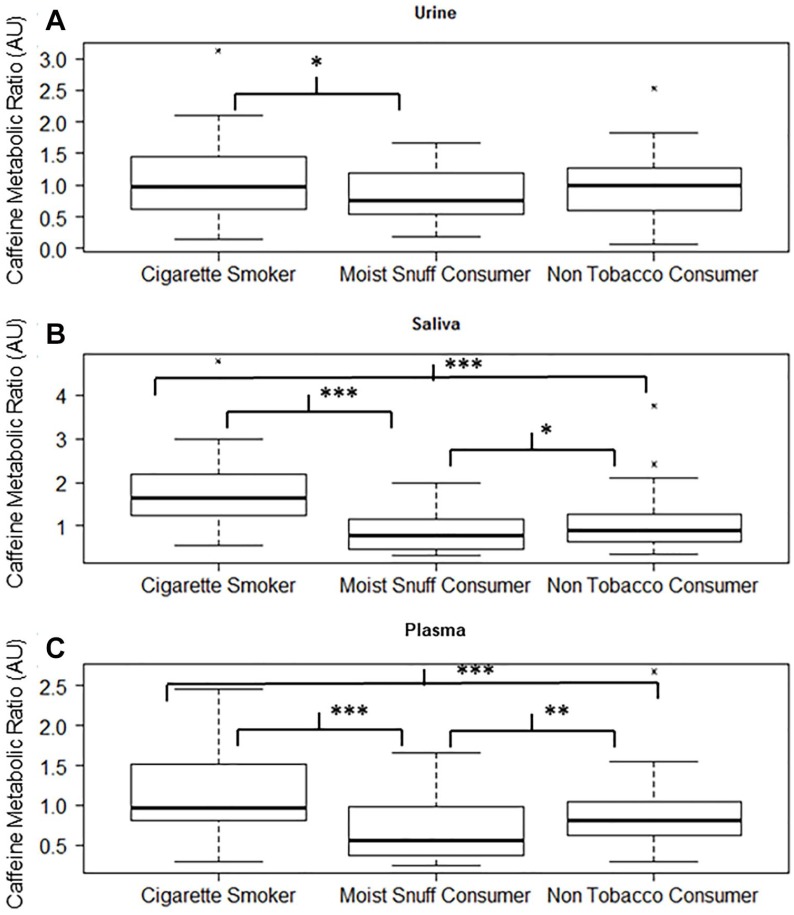
First, the level of caffeine in urine of SMK was higher relative to NTC (fold change [FC]: 1.35) although it did not achieve statistical significance. In contrast, the level of caffeine was significantly lower in both plasma and saliva of SMK relative to NTC. Relative to NTC, MSC exhibited a high level of caffeine in plasma, urine, and saliva (FC: 1.79, 1.47, and 1.55), respectively. Second, 1-methylurate and 1,3-dimethylurate were significantly higher in the urine of SMK relative to NTC and MSC. However, they were not detected in plasma or saliva of SMK, NTC, and MSC. Third, caffeine metabolic ratios (CMR = Cparaxanthine / Ccaffeine) were computed for each participant and compared with different cohorts in each matrix (Figure 4). In urine, no significant differences were observed between MSC and NTC, and between SMK and NTC, although there was a marginal significant difference between SMK and MSC (P = .093). In both saliva and plasma, significant differences in CMR were observed between SMK and NTC (P < .001), and between SMK and MSC (P < .001). The P value for the comparison between MSC and NTC in saliva was between .1 and .05, but in plasma was less than .05.
Figure 4.
Caffeine metabolic ratio of SMK, MSC, and NTC in urine, saliva, and plasma. Box plots of caffeine metabolic ratios (computed using scaled intensity of paraxanthine divided by caffeine) in (A) urine, (B) saliva, and (C) plasma show altered caffeine metabolic ratio in saliva and plasma. MSC indicates moist snuff consumers; NTC, non-tobacco consumers; SMK, smokers. *indicates .05 < P ⩽ 0.1, **indicates .001 < P ⩽ .05, and ***indicates P ⩽ .001.
Energy metabolism
Among many metabolic pathways that contribute to providing energy for cellular and physiological functions, nicotinate and nicotinamide metabolism, pyruvate metabolism, and the TCA cycle were identified to be significantly enriched in urine or saliva of SMK compared with NTC and MSC. Among the metabolites involved in these metabolic pathways, levels of 1,5-anhydroglucitol (1,5-AG), a naturally occurring monosaccharide, were different in the study cohorts (Table 2). The levels of 1,5-AG were significantly lower in the plasma, but higher in urine of SMK compared with NTC (Table 2).
Table 2.
SMK had indications of disrupted energy metabolism.
| Biochemical name | Fold of change
(plasma) |
Fold of change
(urine) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMK |
P value | SMK |
P value | MSC |
P value | SMK |
P value | SMK |
P value | MSC |
P value | |
| NTC | MSC | NTC | NTC | MSC | NTC | |||||||
| Glycolysis | ||||||||||||
| 1,5-anhydroglucitol (1,5-AG) | 0.9 | .034 | 0.93 | .184 | 0.97 | .312 | 1.32 | .026 | 1.18 | .268 | 1.12 | .199 |
| Pyruvate | 1.04 | .856 | 1.03 | .850 | 1.01 | .991 | 0.77 | .021 | 0.72 | .003 | 1.08 | .501 |
| Lactate | 1.09 | .148 | 1.04 | .409 | 1.04 | .625 | 1.62 | .046 | 1.1 | .750 | 1.47 | .006 |
| Glucose | 1.07 | .503 | 0.97 | .459 | 1.1 | .067 | 1.76 | .169 | 0.96 | .720 | 1.84 | .061 |
| TCA cycle | ||||||||||||
| Citrate | 0.9 | .018 | 0.97 | .691 | 0.92 | .077 | 0.9 | .255 | 0.96 | .858 | 0.94 | .379 |
| Succinylcarnitine | 0.94 | .390 | 1.01 | .943 | 0.93 | .329 | 0.82 | .004 | 0.83 | .028 | 0.99 | .688 |
| 2-methylcitrate | 0.78 | .006 | 0.84 | .078 | 0.93 | .365 | ||||||
| Itaconate | 0.84 | .044 | 0.82 | .064 | 1.03 | .995 | ||||||
| Succinate | 0.92 | .292 | 0.87 | .112 | 1.06 | .570 | 0.71 | .099 | 0.7 | .028 | 1.01 | .698 |
| Malate | 0.83 | .094 | 0.94 | .917 | 0.88 | .151 | 1.21 | .560 | 0.94 | .257 | 1.29 | .057 |
| NADH/NADPH metabolism | ||||||||||||
| Nicotinate | 2.04 | .002 | 2.04 | .001 | 1 | .724 | ||||||
| Quinolinate | 0.77 | .012 | 0.77 | .125 | 1 | .503 | ||||||
| Trigonelline (N’-methylnicotinate) | 1.25 | .293 | 1.4 | .049 | 0.89 | .319 | 1.74 | .019 | 1.7 | .006 | 1.02 | .617 |
| Nicotinamide | 0.93 | .690 | 0.92 | .417 | 1.01 | .721 | 0.86 | .099 | 0.82 | .014 | 1.05 | .508 |
The statistical significance P < .05 is highlighted by shaded fold of change values; bolded fold of change values indicates .05 < P < .1. Abbreviations: MSC, moist snuff consumers; NADH, nicotinamide adenine dinucleotide; NADPH, nicotinamide adenine dinucleotide phosphate; NTC, non-tobacco consumers; SMK, cigarette smokers; TCA cycle, tricarboxylic acid cycle.
In addition, urinary pyruvate levels were significantly lower in SMK, compared with NTC and MSC. In contrast, compared with NTC, only lactate was higher in the urine of MSC, while malate trended higher but was not statistically significant.
Among the TCA cycle metabolites, circulating plasma citrate levels were lower in SMK compared with NTC. Other indications of altered energy metabolism that were also significantly lower in SMK relative to NTC, include urinary succinylcarnitine, 2-methylcitrate, and itaconate. While a similar trend was evident in SMK compared with MSC, only the urinary succinylcarnitine was significantly lower.
The urine of SMK, compared with NTC, also had significantly elevated nicotinate and trigonelline levels along with decreased levels of quinolinate, a biosynthetic intermediate in NAD+ biosynthesis (Table 2). Similarly, SMK exhibited elevated levels of nicotinate and trigonelline compared with MSC. Smokers also exhibited decreased urinary nicotinamide relative to NTC and MSC.
Arginine and proline metabolism
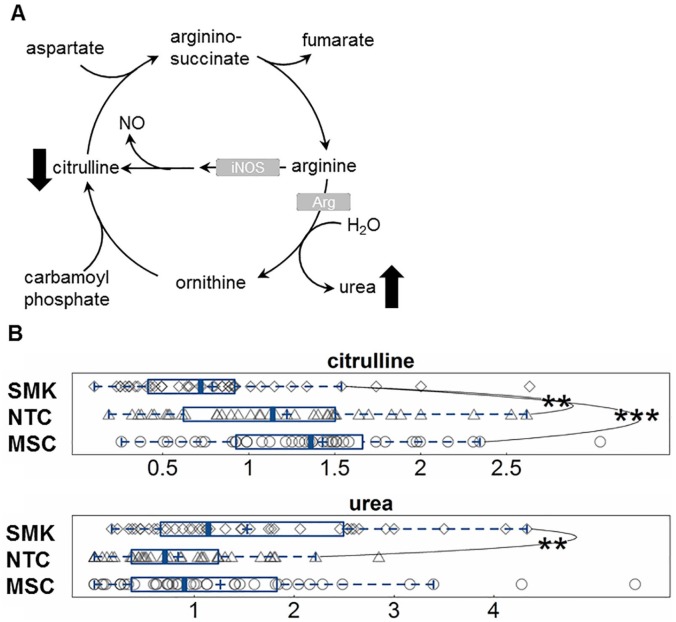
Arginine and proline metabolism was identified as a commonly enriched pathway affected by tobacco consumption (Figures 1 and 3). Arginine serves as a building block for the synthesis of proteins and as a precursor for many other small molecules including nitric oxide, urea, and polyamines (Figure 5A). The levels of citrulline, which is formed by the deamination of arginine, were significantly lower in the saliva of SMK relative to NTC and MSC (Figure 5B). The mean value of salivary citrulline levels was higher in MSC compared with NTC but the difference is not statistically significant. The levels of salivary urea were elevated, but not statistically higher (.05 < P < .1), in MSC and SMK, relative to NTC.
Figure 5.
SMK have altered arginine metabolism in saliva. The increased urea and decreased citrulline in saliva are consistent with up-regulation of arginase and decreased nitric oxide synthase. (A) Pathway diagram of urea cycle and (B) boxplots showing altered urea cycle metabolites in SMK. MSC indicates moist snuff consumers; NTC, non-tobacco consumers; SMK, smokers. Vertical lines in the boxplots represent the median; boxed areas represent 50% of the distribution; whiskers represent the maximum and minimum values excluding outliers, and the plus sign represents the mean. **indicates .001 < P ⩽ .05 and ***indicates P ⩽ .001.
Discussion
To better understand metabolism in tobacco product consumers, pathway enrichment and topology analyses methods were applied to evaluate the metabolomic profiles of 3 biological matrices obtained from SMK, MSC, and NTC. The metabolomic profiles were generated using an untargeted metabolomics platform in our previous effort to identify metabolic biomarkers for MSC and SMK in urine, plasma, and saliva.12 We reported previously that many differentially expressed metabolites in SMK, relative to NTC and MSC, were indicative of higher levels of oxidative stress and inflammation.12 In the current work, as opposed to focusing on individual metabolites, we analyzed the metabolomic data using knowledge-based pathway analysis approaches and identified significantly affected metabolic pathways in SMK and MSC. Key findings of this work are (1) SMK exhibit more pronounced and extensive metabolic pathway changes relative to MSC and NTC and (2) energy metabolism, caffeine metabolism, and arginine and proline metabolism are prominently enriched in SMK, but not in MSC, when compared with NTC.
In this study, a relatively large fdrcut-off (0.32) was used to identify enriched metabolic networks. Previously, a conventional fdrcut-off of 0.05 was used to identify differentiating metabolites in the tobacco consumers.12 In the global network analyses, application of a conventional 0.05 cut-off did not yield any differentially enriched pathways among cohorts (data not shown). This could be due to the fact that these data were generated from generally healthy tobacco consumers (SMK and MSC). Thus, we explored multiple FDR cut-off values in 0.02 increments. Using this approach, a 0.32 fdrcut-off was found to provide a separation for plasma metabolomic profiles between MSC and NTC (Figure 1A); however, when a more stringent cut-off of 0.22 was applied, no differences were detected between MSC and NTC in plasma (Figure 1A). Hence, for consistency across comparisons and other matrices, a 0.32 FDR cut-off was used in this study (Figures 2A and 3A).
Using the 0.32 fdrcut-off, our pathway analysis of urinary, plasma, and saliva metabolomic profiles suggested that a diverse range of metabolic pathways, including carbohydrate, amino acid, lipid, vitamin, and nucleotide metabolism, were perturbed in SMK relative to NTC (Table S1 and Figure S1 in the Supplementary Material). First, two enriched pathways including caffeine metabolism and ascorbate and aldarate metabolism were consistently seen in urine, plasma, and saliva of SMK. Second, three carbohydrate metabolic pathways (amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, galactose metabolism), three amino acid metabolic pathways (arginine and proline metabolism, glutathione metabolism, and D-glutamine and D-glutamate metabolism), and one nucleotide metabolic pathway were enriched in plasma and urine of SMK. Third, lysine biosynthesis was enriched in plasma and saliva of SMK. Fourth, one metabolic pathway concerning cofactors and vitamins (nicotinate and nicotinamide metabolism) and two amino acid metabolic pathways (histidine metabolism, taurine and hypotaurine metabolism) were enriched in urine and saliva of SMK. Finally, eight metabolic pathways were uniquely enriched in plasma of SMK including steroid hormone biosynthesis; 30 uniquely enriched metabolic pathways were identified in urine and zero in saliva (Table S1 in the Supplementary Material). Multiple enriched lipid metabolic pathways including glycerolipid metabolism, sphingolipid metabolism, and glycerophospholipid metabolism were identified in urine only.
We further compared the enriched pathways in SMK and MSC with NTC, and found only two pathways (lysine biosynthesis and α-LA metabolism) were enriched in plasma of both SMK and MSC. Given the similarity between urinary and saliva metabolomes of MSC and NTC, no enriched pathways were detected in MSC.
The enrichment of pyruvate metabolism, the TCA cycle, glycolysis, and gluconeogenesis pathways in urine of SMK, but not in urine of MSC, suggests a perturbed energy metabolism in SMK, but not in MSC. Such finding is also indirectly supported by enriched nicotinate and nicotinamide metabolism in urine and saliva of SMK, but not MSC, as well as many enriched carbohydrate and amino acid metabolic pathways shown above. This finding is not surprising as these pathways are closely linked to energy metabolism. In addition, the increased urinary lactate in SMK suggests that the rate of pyruvate production through glycolysis exceeded the rate of pyruvate consumption through mitochondrial oxidative phosphorylation. We also observed that circulating citrate was decreased in plasma, and multiple metabolites related to TCA cycle intermediates were decreased in SMK urine. Although lactate was also elevated in the urine of MSC, we did not observe other signs of energy metabolism impairment. Our findings are consistent with the well-known smoking-related energy metabolism perturbations. For example, cigarette smoke exposure to cultured cells causes structural and functional abnormalities in the mitochondria34,35; the levels of several TCA cycle intermediates, malate, fumarate and succinate, were decreased in the urine of SMK.36
Caffeine metabolism was prominently altered in tobacco consumers. Our analysis showed that caffeine metabolism was profoundly altered in SMK, compared with NTC, and was less pronounced in MSC. Caffeine is a naturally occurring stimulant found in coffee, tea, chocolate, and many other beverages. CYP1A2 is the major enzyme responsible for the metabolism of caffeine37; 95% of ingested caffeine is metabolized by CYP1A2 to paraxanthine as the primary intermediate product. Our metabolomics analysis identified that SMK exhibit marked changes in caffeine metabolism. This observation is consistent with the CMR measured in saliva and plasma of SMK relative to NTC (Figure 4), suggesting CYP1A2 enzyme activity is up-regulated by cigarette smoking, as reported previously.38 In our study, several caffeine metabolites were significantly elevated in urine of SMK, but not in MSC, compared with NTC (Table 1). However, this could be attributed to higher coffee consumption in SMK compared with NTC and MSC cohorts (unpublished data), which is consistent with a published report.39
Our analysis also suggested enrichment of arginine and proline metabolism in plasma and urine of SMK, but not MSC. The finding that urea production in the saliva of SMK was elevated indicates the up-regulation of arginine metabolism in SMK. Interestingly, this BioEff was not apparent in MSC’s saliva, SMK’s urine, and plasma compared with NTC (Figure 5). Our results are consistent with a report of increased salivary urea, which may account for dental implant failure in SMK.40 In asthmatic patients, arginase activity in airway endothelial cells is up-regulated with smoking41 and may impair nitrous oxide production.42
In addition to the aforementioned metabolic pathway perturbations, we also observed that glutathione metabolism was significantly enriched in plasma and urine of SMK. Glutathione metabolism plays an important role in alleviating oxidative stress,43 and this finding is consistent with our previous observation from single metabolite analyses12 that SMK exhibited higher levels of oxidative stress. In addition, SMK exhibited elevated levels of 1,5-AG in urine, relative to MSC and NTC. The 1,5-anhydroglucitol, a clinically established marker for hyperglycemia, is a non-metabolized food component and its concentration remains relatively stable in the blood. It is filtered and reabsorbed by the kidney and a small amount is excreted in the urine. During hyperglycemia, glucose competes with 1,5-AG for re-absorption. Thus, hyperglycemia leads to elevated 1,5-AG urinary levels and a decrease in plasma levels. Elevated levels of 1,5-AG indicate a hyperglycemic state, which is a risk factor for diabetes.44 Thus, our finding suggests that SMK are relatively hyperglycemic, which is consistent with the reported association between smoking and impaired glucose control.45
Our results demonstrate that pathway-based analysis approach is a useful means of overcoming the limitations imposed by univariate analysis of metabolomics data, and it offers a methodology for uncovering the biologically plausible pathways affected by tobacco product consumption. Many of these identified enriched biochemical networks such as oxidative stress response, and arginine and proline metabolism, have also been reported to be involved with smoking-associated diseases like COPD and lung cancer. For example, oxidative stress response, and glycolysis and gluconeogenesis pathways were identified as two common underlying pathogenic pathways of lung cancer and COPD diseases from proteomics analysis of the bronchoalveolar lavage fluid in lung cancer and COPD patients.46 Serum metabolomics analysis of lung cancer patients identified sphingolipid metabolism, glycine, serine, and threonine metabolism, arginine and proline metabolism, and LA metabolism as commonly altered pathways.47 Taken together, the smoking-related metabolic perturbations in biological pathways may drive the progression to disease phenotypes such as COPD and lung cancer.
Although the current study has provided important insights into biochemical perturbations in tobacco consumers, particularly SMK, several limitations exist. First, although KEGG pathway databases have been widely used in enrichment analysis as reference databases, high-quality and high-resolution annotation of condition- and cell-specific metabolites remains challenging.48 Second, the enrichment analysis method used in the current study assumes that each pathway in the database is independent of other pathways; it does not account for the interactions between different pathways.49 Third, the current analyses do not consider all identified metabolites due to a lack of accurate KEGG annotation of metabolites identified from Metabolon’s MS platform. Nevertheless, our analyses provide an unbiased qualitative and semi-quantitative approach to compare the significance and the importance of a given biological pathway perturbed across different conditions.
Conclusions
In summary, combined with our previous findings of BioExp and BioEff,11-13,24 pathway analysis of the plasma, urinary, and saliva metabolomic profiles from SMK, MSC, and NTC provides additional insights into the biochemical changes of tobacco consumers. We show that SMK, but not MSC, exhibit prominent changes in caffeine, energy, and arginine metabolism relative to NTC. Collectively, our findings suggest cigarette smoking, but not moist snuff consumption, is a prominent modifier of human physiology.
Supplemental Material
Supplemental material, MPA_suppl_resub_clean_xyz259589b262172 for Pathway Analysis of Global Metabolomic Profiles Identified Enrichment of Caffeine, Energy, and Arginine Metabolism in Smokers but Not Moist Snuff Consumers by Gang Liu, Douglas P Lee, Eckhardt Schmidt and GL Prasad in Bioinformatics and Biology Insights
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by RAI Services Company.
Declaration of Conflicting Interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: G.L., E.S., and G.L.P. are full-time employees of RAI Services Company. RAI Services Company is a wholly owned subsidiary of Reynolds American Inc., which is a wholly owned subsidiary of British American Tobacco plc. D.P.L. is a full-time employee of Omic Insight Inc. None of the authors have any other conflict of interest to declare.
Author Contributions: GL and GLP conceived and designed the study; GL performed the analysis; GL, DPL, ES and GLP interpreted the results; GL, DPL and GLP wrote the manuscript. All authors edited and approved the final manuscript.
Data Availability: The metabolomics data used to support the findings of this study were previously published as supplemental files S4-S6 in Prasad et al.12
ORCID iD: Gang Liu  https://orcid.org/0000-0002-8758-1797
https://orcid.org/0000-0002-8758-1797
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Jamal A, Agaku IT, O’Connor E, King BA, Kenemer JB, Neff L. Current cigarette smoking among adults—United States, 2005-2013. MMWR Morb Mortal Wkly Rep. 2014;63:1108-1112. [PMC free article] [PubMed] [Google Scholar]
- 2. Perfetti TA, Rodgman A. The complexity of tobacco and tobacco smoke. Beitr Tabakforsch. 2011;25:215-232. [Google Scholar]
- 3. U.S. Food and Drug Administration. Harmful and Potentially Harmful Constituents in Tobacco Products and Tobacco Smoke: Established List. Silver Spring, MD: Center for Tobacco Products; 2012. [Google Scholar]
- 4. Levy DT, Mays D, Boyle RG, Tam J, Chaloupka FJ. The effect of tobacco control policies on US smokeless tobacco use: a structured review. Nicotine Tob Res. 2017;20:3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang F, Pedersen NL, Ye W, et al. Moist smokeless tobacco (Snus) use and risk of Parkinson’s disease. Int J Epidemiol. 2017;46:872-880. [DOI] [PubMed] [Google Scholar]
- 6. Asthana S, Labani S, Kailash U, Sinha DN, Mehrotra R. Association of smokeless tobacco use and oral cancer: a systematic global review and meta-analysis. Nicotine Tob Res. 2019;21:1162-1171. [DOI] [PubMed] [Google Scholar]
- 7. Lee PN, Hamling J. Systematic review of the relation between smokeless tobacco and cancer in Europe and North America. BMC Med. 2009;7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sponsiello-Wang Z, Weitkunat R, Lee PN. Systematic review of the relation between smokeless tobacco and cancer of the pancreas in Europe and North America. BMC Cancer. 2008;8:356-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yanbaeva DG, Dentener MA, Creutzberg EC, Wesseling G, Wouters EF. Systemic effects of smoking. Chest. 2007;131:1557-1566. [DOI] [PubMed] [Google Scholar]
- 10. Ogden MW, Marano KM, Jones BA, Morgan WT, Stiles MF. Switching from usual brand cigarettes to a tobacco-heating cigarette or snus: part 2. Biomarkers of exposure. Biomarkers. 2015;20:391-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prasad GL, Jones BA, Chen P, Gregg EO. A cross-sectional study of biomarkers of exposure and effect in smokers and moist snuff consumers. Clin Chem Lab Med. 2016;54:633-642. [DOI] [PubMed] [Google Scholar]
- 12. Prasad GL, Jones BA, Schmidt E, Chen P, Kennedy AD. Global metabolomic profiles reveal differences in oxidative stress and inflammation pathways in smokers and moist snuff consumers. J Metabol. 2015;1:2. [Google Scholar]
- 13. Campbell LR, Brown BG, Jones BA, Marano KM, Borgerding MF. Study of cardiovascular disease biomarkers among tobacco consumers, part 1: biomarkers of exposure. Inhal Toxicol. 2015;27:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nordskog BK, Brown BG, Marano KM, Campell LR, Jones BA, Borgerding MF. Study of cardiovascular disease biomarkers among tobacco consumers, part 2: biomarkers of biological effect. Inhal Toxicol. 2015;27:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsu PC, Zhou B, Zhao Y, et al. Feasibility of identifying the tobacco-related global metabolome in blood by UPLC-QTOF-MS. J Proteome Res. 2013;12:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hsu PC, Lan RS, Brasky TM, et al. Menthol smokers: metabolomic profiling and smoking behavior. Cancer Epidemiol Biomarkers Prev. 2017;26:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gu F, Derkach A, Freedman ND, et al. Cigarette smoking behaviour and blood metabolomics. Int J Epidemiol. 2016;45:1421-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spitale RC, Cheng MY, Chun KA, et al. Differential effects of dietary supplements on metabolomic profile of smokers versus non-smokers. Genome Med. 2012;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muller DC, Degen C, Scherer G, Jahreis G, Niessner R, Scherer M. Metabolomics using GC-TOF-MS followed by subsequent GC-FID and HILIC-MS/MS analysis revealed significantly altered fatty acid and phospholipid species profiles in plasma of smokers. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;966:117-126. [DOI] [PubMed] [Google Scholar]
- 20. Mueller DC, Piller M, Niessner R, Scherer M, Scherer G. Untargeted metabolomic profiling in saliva of smokers and nonsmokers by a validated GC-TOF-MS method. J Proteome Res. 2013;13:1602-1613. [DOI] [PubMed] [Google Scholar]
- 21. Kaluarachchi MR, Boulange CL, Garcia-Perez I, Lindon JC, Minet EF. Multiplatform serum metabolic phenotyping combined with pathway mapping to identify biochemical differences in smokers. Bioanalysis. 2016;8: 2023-2043. [DOI] [PubMed] [Google Scholar]
- 22. Garcia-Perez I, Lindon JC, Minet E. Application of CE-MS to a metabonomics study of human urine from cigarette smokers and non-smokers. Bioanalysis. 2014;6:2733-2749. [DOI] [PubMed] [Google Scholar]
- 23. Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251-W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arimilli S, Madahian B, Chen P, Marano K, Prasad GL. Gene expression profiles associated with cigarette smoking and moist snuff consumption. BMC Genomics. 2017;18:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barnes VM, Kennedy AD, Panagakos F, et al. Global metabolomic analysis of human saliva and plasma from healthy and diabetic subjects, with and without periodontal disease. PLoS ONE. 2014;9:e105181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gall WE, Beebe K, Lawton KA, et al. Alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose tolerance in a nondiabetic population. PLoS ONE. 2010;5:e10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ganti S, Taylor SL, Kim K, et al. Urinary acylcarnitines are altered in human kidney cancer. Int J Cancer. 2012;130:2791-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evans A, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81: 6656-6667. [DOI] [PubMed] [Google Scholar]
- 29. Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2: 9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93-99. [DOI] [PubMed] [Google Scholar]
- 31. Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342-2344. [DOI] [PubMed] [Google Scholar]
- 32. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X, Gao J, Chen J, et al. Identification of metabolic biomarkers in patients with type 2 diabetic coronary heart diseases based on metabolomic approach. Sci Rep. 2016;6:30785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffmann RF, Zarrintan S, Brandenburg SM, et al. Prolonged cigarette smoke exposure alters mitochondrial structure and function in airway epithelial cells. Respir Res. 2013;14:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aravamudan B, Kiel A, Freeman M, et al. Cigarette smoke-induced mitochondrial fragmentation and dysfunction in human airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2014;306:L840-L854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salek RM, Maguire ML, Bentley E, et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics. 2007;29: 99-108. [DOI] [PubMed] [Google Scholar]
- 37. Nehlig A. Interindividual differences in caffeine metabolism and factors driving caffeine consumption. Pharmacol Rev. 2018;70:384-411. [DOI] [PubMed] [Google Scholar]
- 38. Kalow W, Tang BK. Caffeine as a metabolic probe: exploration of the enzyme-inducing effect of cigarette smoking. Clin Pharmacol Ther. 1991;49:44-48. [DOI] [PubMed] [Google Scholar]
- 39. Bjorngaard JH, Nordestgaard AT, Taylor AE, et al. Heavier smoking increases coffee consumption: findings from a Mendelian randomization analysis. Int J Epidemiol. 2017;46:1958-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Queiroz DA, Cortelli JR, Holzhausen M, Rodrigues E, Aquino DR, Saad WA. Smoking increases salivary arginase activity in patients with dental implants. Clin Oral Investig. 2009;13:263-267. [DOI] [PubMed] [Google Scholar]
- 41. Bergeron C, Boulet LP, Page N, et al. Influence of cigarette smoke on the arginine pathway in asthmatic airways: increased expression of arginase I. J Allergy Clin Immunol. 2007;119:391-397. [DOI] [PubMed] [Google Scholar]
- 42. Sikka G, Pandey D, Bhuniya AK, et al. Contribution of arginase activation to vascular dysfunction in cigarette smoking. Atherosclerosis. 2013;231:91-94. [DOI] [PubMed] [Google Scholar]
- 43. Wu G, Fang Y-Z, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489-492. [DOI] [PubMed] [Google Scholar]
- 44. McGill JB, Cole TG, Nowatzke W, et al. Circulating 1,5-anhydroglucitol levels in adult patients with diabetes reflect longitudinal changes of glycemia: a U.S. trial of the GlycoMark assay. Diabetes Care. 2004;27:1859-1865. [DOI] [PubMed] [Google Scholar]
- 45. Gerber PA, Locher R, Schmid B, Spinas GA, Lehmann R. Smoking is associated with impaired long-term glucose metabolism in patients with type 1 diabetes mellitus. Nutr Metab Cardiovasc Dis. 2013;23:102-108. [DOI] [PubMed] [Google Scholar]
- 46. Pastor MD, Nogal A, Molina-Pinelo S, et al. Identification of proteomic signatures associated with lung cancer and COPD. J Proteomics. 2013;89:227-237. [DOI] [PubMed] [Google Scholar]
- 47. Chen Y, Ma Z, Min L, et al. Biomarker identification and pathway analysis by serum metabolomics of lung cancer. Biomed Res Int. 2015;2015:183624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garcia-Campos MA, Espinal-Enriquez J, Hernandez-Lemus E. Pathway analysis: state of the art. Front Physiol. 2015;6:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, MPA_suppl_resub_clean_xyz259589b262172 for Pathway Analysis of Global Metabolomic Profiles Identified Enrichment of Caffeine, Energy, and Arginine Metabolism in Smokers but Not Moist Snuff Consumers by Gang Liu, Douglas P Lee, Eckhardt Schmidt and GL Prasad in Bioinformatics and Biology Insights