Abstract
Background
The impact of antiretroviral therapy (ART) initiation on the vaginal microbiome is unknown. This is of particular importance among women living in sub-Saharan Africa. Understanding this relationship could help elucidate if and how the host immune system interacts with the vaginal microbiome.
Methods
The vaginal microbiome of HIV-1/HSV-2-coinfected women (n = 92) in Uganda was evaluated from self-collected vaginal swabs 1 month pre-ART and at 4 and 6 months post–ART initiation. The vaginal microbiome was characterized by 16S rRNA gene-based sequencing and quantitative polymerase chain reaction. Vaginal community state types (CSTs) were identified using proportional abundance data. Changes in microbiome composition were assessed with permutational analyses of variance (PerMANOVA).
Results
Five vaginal CSTs were identified, which varied significantly by bacterial load (P < .01): CST-1 was characterized by Lactobacillus iners, CST-2 by Gardnerella, CST-3 by Gardnerella and Prevotella, CST-4 by Lactobacillus crispatus, and CST-5 was highly diverse. Vaginal microbiome composition also did not change significantly after ART initiation (P = .985). Immune reconstitution after ART initiation did not affect vaginal microbiome CST assignment (P = .722) or individual-level changes in bacterial load (log response ratio [interquartile range], –0.50 [–2.75 to 0.38] vs –0.29 [–2.03 to 1.42]; P = .40).
Conclusions
The vaginal microbiome of HIV-infected women was not affected by the initiation of ART or immune reconstitution in this observational study. Further research is needed to explore the long-term effects of ART treatment on the vaginal microbiome.
Keywords: antiretroviral therapy, anaerobes, HIV, microbiome, vaginal dysbiosis
The vaginal microbiome is an important factor in women’s health. It is also dynamic, with a bacterial composition that is a function of various demographic and host factors, including race, menarche, pregnancy, and menopause [1–4]. Typically, Lactobacillus-dominated vaginal microbiomes have been associated with better gynecological and reproductive health outcomes [5–7]. In the absence of Lactobacillus dominance, the vaginal microbiome is most often dominated by anaerobes such as Gardnerella and Prevotella [8, 9], which are often associated with bacterial vaginosis (BV) [10]. These BV-associated anaerobes are frequently prevalent in the vaginal microbiome of African and African American women as compared with women from other racial and ethnic backgrounds [11]. Overall, vaginal microbiomes characterized by a low abundance of Lactobacillus spp. and high abundance of anaerobes are associated with negative gynecologic and obstetric outcomes. These outcomes include pre-term birth and sexually transmitted infections, including HIV [12–15]. Although oral pre-exposure prophylaxis (PrEP) for HIV prevention does not appear to be affected by the vaginal microbiome [16], Lactobacillus-deficient vaginal microbiomes may reduce the efficacy of topical HIV PrEP [17]. Among women living with HIV, vaginal microbiomes characterized by a high abundance of anaerobes are associated with vaginal inflammation [18], higher genital HIV viral load [19], and greater HIV shedding [20]. Among women who have not achieved HIV virologic suppression, a high abundance of vaginal anaerobes is associated with an increased risk of sexual transmission of HIV to male partners [21]. Additionally, among women who are virologically suppressed on antiretroviral therapy (ART), the vaginal microbiome is associated with vaginal ART concentration. Both low- and high-diversity microbiomes are associated with reduced vaginal plasma ART concentrations, as compared with intermediate-diversity microbiomes [22].
Sub-Saharan Africa represents an area of notably high HIV prevalence and transmission, where it is particularly important to understand the relationship between the vaginal microbiome and HIV disease [23]. Although there are few studies that have characterized the vaginal microbiome in sub-Saharan African women, existing evidence suggests that anaerobic taxa are commonly present [24, 25]—particularly anaerobic taxa associated with BV [26]. Data on the vaginal microbiome of HIV-positive women of African descent are especially limited. The vaginal microbiome of Tanzanian women living with HIV shows a high prevalence of Gardnerella vaginalis and Lactobacillus iners, and a high proportional abundance of diverse BV-associated anaerobes [27]. A study of predominantly African American and African women living with HIV in the United States and Kenya found a high prevalence of Lactobacillus crispatus and BV-associated anaerobes including Atopobium and Sneathia in both populations [20].
HIV treatment is critical to improving clinical outcomes and reducing HIV transmission [28]. However, the impact of ART initiation on the vaginal microbiome of women living with HIV is unknown. If the prevalence and abundance of vaginal anaerobes are reduced by restoration of CD4 T-cell counts with ART, this may suggest that the host immune system interacts with the microbiome, possibly indicating strategies for HIV transmission prevention. The aim of this study was to characterize how ART initiation and CD4 T-cell reconstitution affect the vaginal microbiome among women living with HIV in Rakai, Uganda.
METHODS
Study Population and Sample Collection
Women aged 18 years and older coinfected with HIV and herpes simplex virus type 2 (HSV-2) with CD4 T-cell counts of 300–400 cells/μL (n = 440) were included in a double-blind randomized placebo-controlled trial examining the effect of acyclovir on HIV disease progression in Rakai, Uganda [29, 30]. ART was initiated in participants with a CD4 T-cell count <250 cells/μL or who developed World Health Organization stage 4 clinical disease, in accordance with the national clinical guidelines at the time of the trial. Women who initiated ART during the trial and who provided at least 1 monthly self-collected vaginal swab before and after initiating ART, from which DNA was successfully extracted, were included in the current study (n = 92). The method for self-collection of vaginal swabs has been described elsewhere [31] and shown to be equivalent to clinically collected swabs for the detection of sexually transmitted infections (STIs) [32].
All participants provided written informed consent. The trial was approved by the Uganda National Council for Science and Technology (Kampala, Uganda), the Uganda Virus Research Institute Research and Ethics Committee, and the National Institute of Allergy and Infectious Diseases Intramural Institutional Review Board. The trial was registered at ClinicalTrials.gov (NCT00405821).
Vaginal Microbiome Characterization
The self-collected vaginal swabs were stored in 1000 µL of Specimen Transport Medium (STM; Roche, Indianapolis IN). Total DNA was extracted from vaginal swabs as previously described [21]. Using the extracted DNA, the vaginal bacterial load was quantified with broad-coverage quantitative PCR (qPCR) targeting the V3-V4 region, measured as 16S rRNA gene copies per swab. The vaginal microbiome composition was characterized by sequencing the V3-V4 region of the bacterial 16S rRNA gene as previously described [33], yielding a total of 519 347 sequences. Taxonomic classification of the resultant sequences was performed using the Ribosomal Database Project Naïve Bayesian Classifier (RDP release 10, update 20) [34]. Lactobacillus species were additionally classified to the species level at 97% confidence using an in-house trained classifier. After filtering for taxa present at >0.025% of the total reads, the vaginal microbiome data set contained 83 genera and 7 Lactobacillus species. The absolute abundance of each vaginal bacterial taxon was calculated as: absolute abundance = vaginal bacterial load × proportional abundance of the given taxon (measured as the number of 16S rRNA gene sequences assigned to a taxon in a given sample, divided by the total number of 16S rRNA sequences obtained for that sample) [33, 35].
Covariate Data
Baseline HSV-2 serostatus was determined using Focus HerpeSelect-2 enzyme immunoassay (EIA; Cypress, CA); HIV-1 serostatus was determined by the Vironostika HIV-1 (Charlotte, NC) and Organon Teknika (Cambridge Biotech, Worcester, MA) EIAs at study enrollment. Discordant EIAs were confirmed with HIV-1 Western blot using Bio-Merieux-Vitek (St. Louis, MO). Plasma HIV viral load copies/mL was determined with the Roche Monitor v1/5 assay (Indianapolis, IN). FACSCalibur (Becton Dickenson, Franklin Lakes, NJ) was used to determine CD4 T cells/μL at the study visit immediately before ART initiation and the study visit closest to 6 months post–ART initiation. Immune reconstitution was defined as a CD4 T-cell count increase of at least 50 cells/μL from pre– to post–ART initiation [36].
Statistical Analysis
Vaginal community state types (CSTs) were identified using proportional abundance data with Bray-Curtis distance and hierarchal clustering by Ward’s method [37]. The CST number was set to 5, consistent with prior work [2]. Data from all time points were used in CST assignment (273 samples), which assumes that the same CST classifications were present in the community at both pre- and post-ART time points. CST assignments for each individual were allowed to vary across time. Alpha diversity was calculated using the Shannon diversity index. Nonmetric multidimensional scaling plots (nMDS) were used to visualize the overall microbiome composition across CSTs. The dominant bacterial taxa for each CST were characterized by prevalence (defined as the percentage of vaginal microbiome samples with a given taxon present within each CST), proportional abundance, and absolute abundance. Overall vaginal bacterial loads were compared across CSTs using Wilcoxon rank-sum tests.
Graphical analyses and Wilcoxon rank-sum tests were used to assess changes in vaginal bacterial load across time for each CST. Differences in vaginal bacterial load over time were tested using the Wilcoxon rank-sum test; resulting P values were adjusted for the false discovery rate of 0.05.
Changes in vaginal bacterial load after ART initiation were evaluated using log response ratios (LRRs), defined as: ln([vaginal bacterial load 6 months post-ART]/[vaginal bacterial load 1 month pre-ART]) [38]. LRRs were compared based on level of CD4 change and by immune reconstitution status using 2-tailed t tests with unequal variance at an α value of 0.05. A sensitivity analysis was conducted using a linear mixed model to examine bacterial load over time since ART initiation. The Shannon diversity index was also examined over time using a linear mixed model. Overall vaginal microbiome compositions were visualized by nMDS and compared using permutational multivariate analysis of variance (PerMANOVA) [39] at each time point, as well as for the post-ART vaginal microbiome by immune reconstitution status.
Data analyses were conducted in R (version 3.5).
RESULTS
Participant Characteristics
The median age at enrollment for the 92 female participants was 34.5 years (Table 1); 45.7% of participants were assigned to the acyclovir trial arm and initiated HSV-2 treatment at study enrollment. Before initiating ART, the median viral load (interquartile range [IQR]) was 96 843 (20 559–251 379) copies per mL, and 42.3% of the participants had a CD4 T-cell count <200 cells/μL. All participants received ART, with the majority of participants (83.7%) receiving zidovudine/lamivudine/nevirapine (AZT/3TC/NVP). After ART initiation, the median CD4 T-cell increase (IQR) was 105 (43–159) cells/μL, with 73.9% of participants having achieved immune reconstitution, defined as an increase of at least 50 cells/μL.
Table 1.
Baseline Characteristics of Female Study Population (n = 92)
| Baseline Characteristics | No. (%) |
|---|---|
| Age at enrollment, y | |
| 20–29 | 23 (25.0) |
| 30–39 | 39 (42.4) |
| 40–49 | 22 (23.9) |
| ≥50 | 8 (8.7) |
| Acyclovir trial arm | |
| Placebo | 50 (54.3) |
| Acyclovir | 42 (45.7) |
| ART regimen | |
| AZT/3TC/NVP | 77 (83.7) |
| CBV/EFV | 5 (5.4) |
| d4T/3TC/NVP | 6 (6.5) |
| TDF/3TC/NVP | 3 (3.3) |
| TVD/EFV | 1 (1.1) |
| Baseline CD4 count, cells/μL | |
| <200 | 38 (42.3) |
| 200–450 | 54 (58.7) |
| Baseline HIV viral load, copies/mL | |
| <10 000 | 14 (15.2) |
| 10 000–99 999 | 46 (50.0) |
| ≥100 000 | 32 (34.8) |
| BMI, kg/m2 | |
| <18.5 | 8 (8.7) |
| 18.5–24.9 | 66 (71.7) |
| ≥25 | 18 (19.6) |
Abbreviations: 3TC, lamivudine; ART, antiretroviral therapy; AZT, zidovudine; BMI, body mass index; CBV, zidovudine-lamivudine; d4T, stavudine; EFV, efavirenz; NVP, nevirapine; TDF, tenofovir disoproxil fumarate; TVD, tenofovir disoproxil furminate-emtricitabine.
The 5 Vaginal Microbiome Community State Types
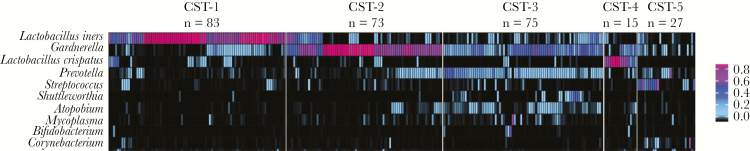
The vaginal microbiome profiles (n = 273 samples) of the 92 participants were clustered into 5 CSTs (Figure 1). Each CST had a distinct microbiome composition (PerMANOVA P = .01) (Supplementary Figure 1) characterized by a unique set of dominant taxa. The most prevalent CST, CST-1 (30.4% of samples), was dominated by L. iners based on both proportional abundance (median [IQR], 78% [64%–92%]) and absolute abundance (median [IQR], 7.30 [5.59–7.08] log10 16S rRNA gene copies per swab) (Table 2). CST-2 and CST-3 had similar prevalence rates (26.7% and 27.5%, respectively), with CST-2 being dominated primarily by Gardnerella and CST-3 being dominated by both Gardnerella and Prevotella. CST-4 and CST-5 were the least prevalent (5.5% and 9.9%, respectively), with CST-4 being uniquely dominated by L. crispatus and CST-5 being highly diverse and not dominated by any specific taxon (Figure 1).
Figure 1.
Pre- and post-ART vaginal microbiome samples (n = 273) from 92 women with HIV cluster into 5 distinct CSTs using proportional abundance data. The full analytic data set (83 genera and 7 Lactobacillus species) was used to cluster samples into CSTs; the 10 most prevalent taxa are shown. Abbreviations: ART, antiretroviral therapy; CST, community state type.
Table 2.
Prevalence and Abundance Characteristics of Key Taxa for Vaginal Microbiome Community State Types 1–5
| CST-1 | CST-2 | CST-3 | CST-4 | CST-5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prev. | PA, % | AA | Prev. | PA, % | AA | Prev. | PA, % | AA | Prev. | PA, % | AA | Prev. | PA, % | AA | |
| Lactobacillus iners | 100 | 78 | 7.30 | 67 | 2 | 5.61 | 88 | 2 | 6.86 | 67 | 0 | 4.51 | 59 | 0 | 2.94 |
| Lactobacillus crispatus | 59 | 0 | 4.38 | 41 | 0 | 0.00 | 20 | 0 | 0.00 | 100 | 66 | 7.23 | 15 | 0 | 0.00 |
| Gardnerella | 76 | 2 | 5.70 | 100 | 71 | 7.61 | 99 | 32 | 8.03 | 100 | 8 | 6.18 | 70 | 3 | 4.44 |
| Prevotella | 78 | 0 | 4.75 | 85 | 0 | 5.76 | 100 | 16 | 7.93 | 73 | 0 | 4.14 | 74 | 2 | 4.36 |
| Sample assignment, No. (%) | 83 (30.4) | 73 (26.7) | 75 (27.5) | 15 (5.5) | 27 (9.9) | ||||||||||
| Shannon diversity | 5.30 | 5.23 | 6.38 | 3.77 | 5.07 | ||||||||||
| Total bacterial loada | 7.38 (6.94–7.93) | 7.90 (7.19–8.22) | 8.65 (8.06–8.99) | 7.45 (6.82–7.90) | 6.49 (5.59–7.08) | ||||||||||
Abbreviations: AA, median log-transformed absolute abundance of taxa; CST, community state type; PA, median proportional abundance of taxa as percentage; Prev., percentage of samples with taxa present.
aTotal bacterial load was measured in log 16S rRNA gene copies. Data are median (interquartile range). Total bacterial load varies significantly by CST assignment (Kruskal-Wallis chi-square, 93.05; P < .0001).
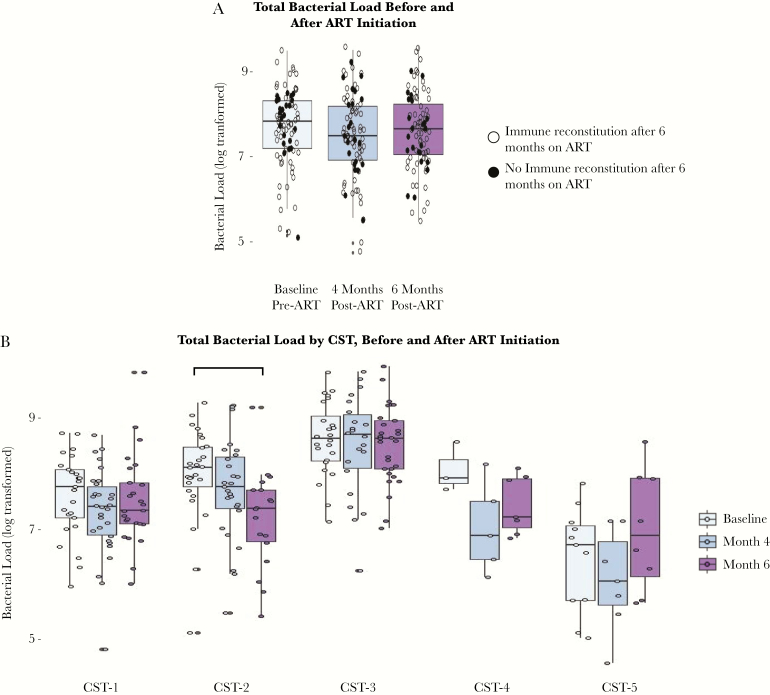
The 5 CSTs also varied significantly in total bacterial load at both baseline and follow-up (P < .01). Specifically, CST-3 had the highest total bacterial load (median [IQR], 8.65 [8.06–8.99] log10 16S rRNA gene copies per swab), and CST-5 had the lowest total bacterial load (median [IQR], 6.49 [5.59–7.08]) (Figure 2).
Figure 2.
Vaginal bacterial load overall and by CST before and after ART initiation. Total bacterial load is log-transformed. Horizontal bars represent significance of P < .05 from pairwise FDR-adjusted Wilcoxon rank-sum tests. Abbreviations: ART, antiretroviral therapy; CST, community state type.
Before ART initiation, vaginal microbiome CST assignment was not significantly associated with the participants’ age, body mass index, CD4 count, or HIV viral load at study baseline (Table 3). It is notable that before ART, all participants with CST-4 had CD4 counts >200 cells/μL, and by contrast, 72.7% of women with CST-5 had CD4 counts <200 cells/μL; however, this difference was not statistically significant, and power was limited by the small sample sizes.
Table 3.
Participant Characteristics by Pre-antiretroviral Therapy Initiation Community State Type Assignment
| CST-1 | CST-2 | CST-3 | CST-4 | CST-5 | ||
|---|---|---|---|---|---|---|
| 26 (28.3) | 28 (30.4) | 24 (26.1) | 3 (3.3) | 11 (12.0) | P | |
| Baseline age, y | .275 | |||||
| 20–29 | 5 (19.2) | 7 (25.0) | 8 (33.3) | 1 (33.3) | 2 (18.2) | |
| 30–39 | 14 (53.8) | 11 (39.3) | 9 (37.5) | 2 (66.7) | 3 (27.3) | |
| 40–49 | 6 (23.1) | 8 (28.6) | 7 (29.2) | 0 (0.0) | 1 (9.1) | |
| 50+ | 1 (3.8) | 2 (7.1) | 0 (0.0) | 0 (0.0) | 5 (45.5) | |
| Baseline BMI, kg/m2 | .645 | |||||
| <18.5 | 3 (11.5) | 3 (10.7) | 0 (0.0) | 0 (0.0) | 2 (18.2) | |
| 18.5–24.9 | 18 (69.2) | 19 (67.9) | 20 (83.3) | 2 (66.7) | 7 (63.6) | |
| 25+ | 5 (19.2) | 6 (21.4) | 4 (16.7) | 1 (33.3) | 2 (18.2) | |
| Baseline CD4 count, cells/μL | .098 | |||||
| <200 | 8 (30.8) | 13 (46.4) | 10 (41.7) | 0 (0.0) | 8 (72.7) | |
| 200–450 | 18 (69.2) | 15 (53.6) | 14 (58.3) | 3 (100.0) | 3 (27.3) | |
| Baseline HIV viral load, copies/mL | .486 | |||||
| <10 000 | 2 (7.7) | 6 (21.4) | 3 (12.5) | 1 (33.3) | 1 (9.1) | |
| 10 000–99 999 | 12 (46.2) | 9 (32.1) | 10 (41.7) | 0 (0.0) | 2 (18.2) | |
| 100 000+ | 12 (46.2) | 13 (46.4) | 11 (45.8) | 2 (66.7) | 8 (72.7) |
Data are presented as No. (%). P values from the Fisher exact test.
Abbreviations: BMI, body mass index; CST, community state type.
Changes in Vaginal Bacteria After ART Initiation and Association With Immune Reconstitution
The majority of women (66.3%) switched from their baseline CST over the 3 time points: 31.5% of women shifted once, and 34.8% shifted twice. Baseline CST assignment was not associated with CST assignment stability. Additionally, acyclovir trial arm status was not associated with overall microbiome composition (PerMANOVA P = .139) (Supplementary Figure 2).
Overall microbiome composition did not vary significantly with ART initiation (PerMANOVA P = .985) (Supplementary Figure 3). ART initiation was not associated with changes in overall bacterial load, regardless of immune reconstitution status at month 6 (Figure 2A). No differences were found in Shannon diversity (beta [95% confidence interval {CI}], 0.02 [–0.08 to 0.09]) over time from pre–ART initiation through to 6 months post–ART initiation. After ART initiation, the variation in total bacterial load across CSTs was attenuated compared with baseline (from 6 significant pairwise differences at baseline to 3 significant pairwise differences at month 6) (Supplementary Figure 4). Although there were no differences in CST prevalence before vs after ART initiation, there were changes in the characteristics of women in specific CSTs post-ART. In particular, the vaginal microbiomes of women assigned to CST-2 at 6 months after ART initiation showed a significantly lower total bacterial load than the vaginal microbiomes of women assigned to CST-2 at baseline (P < .01) (Figure 2B).
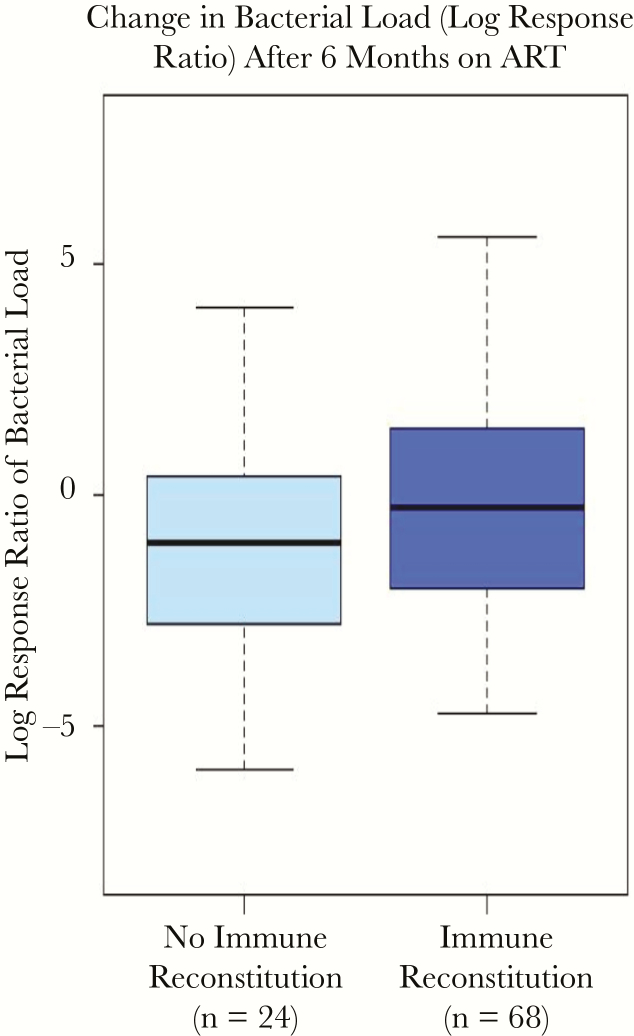
Further, the participants’ immune reconstitution status post-ART was not associated with any changes in vaginal microbiome composition or total bacterial load (P = .40) (Figure 3; Supplementary Figure 5). The participants’ immune reconstitution status post-ART was also not associated with significant changes in the absolute abundance of the dominant vaginal bacterial taxa, including L. iners, L. crispatus, Gardnerella, and Prevotella. When following individuals over time, no statistically significant changes were observed in total vaginal bacterial load (beta [95% CI], –0.15 [–0.35 to 0.05]).
Figure 3.
Associations of CD4 T-cell recovery and immune reconstitution with within-person changes in bacterial load from baseline pre-ART to 6 months post–ART initiation. Log response ratio defined as: ln([bacterial load at month 6]/[bacterial load at baseline]). No significant differences were found between immune reconstitution groups by t test (P = .40). Abbreviations: ART, antiretroviral therapy; CST, community state type.
DISCUSSION
To our knowledge, this is the first study that has examined the vaginal microbiome during ART initiation. In this study, ART initiation and subsequent immune reconstitution did not appear to modify the vaginal microbiome. There was no significant change in the overall vaginal microbiome composition after ART initiation, regardless of CD4 T-cell reconstitution. However, the bacterial load in Gardnerella-dominated CST-2 decreased significantly after ART initiation. Vaginal microbiome characteristics before ART, such as vaginal bacterial load and absolute abundance of specific taxa, were not associated with subsequent immune reconstitution after ART initiation.
Microbiomes at certain body sites, including the gut microbiome, are known to be affected by HIV infection [40–43]. By contrast, the vaginal microbiomes observed in women living with HIV in the present study did not appear to differ vastly from the vaginal microbiomes that have previously been reported in HIV-negative women of African descent [24, 25, 44]. We identified 5 vaginal microbiome community state types among the women living with HIV, each with a distinct dominant taxon/taxa, overall composition, and total bacterial load. The composition of the vaginal microbiomes observed in this present study were similar to those previously reported in women of African descent. A study that examined 1268 vaginal microbiome samples from HIV-negative African American women similarly found 4 vaginal CSTs that are dominated by L. iners, Gardnerella, L. crispatus, and multiple anaerobic taxa [45]. A study of Rwandan female sex workers found high prevalence rates of vaginal microbiomes dominated by L. iners and by multiple anaerobes (eg, Prevotella, Gardnerella, etc.), irrespective of HIV status [19]. However, the difference we observed in vaginal bacterial load by CST within this HIV-positive population has never before been reported.
This study has several limitations. The generalizability of these findings is limited due to all participants being co-infected with HIV and HSV-2. Additionally, the majority of women (83.7%) were administered AZT/3TC/NVP per national clinical guidelines at the time of the trial, which have subsequently changed to include TDF- and DTG-containing regimens [46]. The blood and vaginal samples were collected at separate time points, with CD4 being measured every 6 months, whereas vaginal swabs were collected monthly. However, the median time between pre-ART and final post-ART assessment of CD4 counts and vaginal microbiomes was 6 months. As most women in this study were started on the same ART regimen, microbiome changes associated with different ART regimens could not be assessed. Given the limited follow-up, only the short-term impact of ART could be evaluated. We did not collect data on contraceptive use, vaginal hygiene, sexual behaviors, or menstruation. However, the effect of menstruation in this study is likely to be accounted for as random noise, as there is no reason to believe it would vary in proportion between the visits before and after ART initiation. Lastly, the vaginal microbiome outcome data could not be linked with bacterial vaginosis diagnoses, as the relevant clinical and microscopic data were not collected.
In summary, these observational findings suggest that the vaginal microbiome of women living with HIV were not impacted by the initiation of ART nor by subsequent immune reconstitution. However, further research is needed to explore the long-term impacts of ART and immune reconstitution on the vaginal microbiome.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Financial support. This work was supported in part by extramural funding from the National Institutes of Health (1R01AI120938 and 1R01AI128779 [to A.A.R.T.] and R01A123002-01A1 [to L.B.P.]) and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Gupta S, Kumar N, Singhal N, et al. . Vaginal microflora in postmenopausal women on hormone replacement therapy. Indian J Pathol Microbiol 2006; 49:457–61. [PubMed] [Google Scholar]
- 2. Romero R, Hassan SS, Gajer P, et al. . The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2014; 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borgdorff H, van der Veer C, van Houdt R, et al. . The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS One 2017; 12:e0181135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamamoto T, Zhou X, Williams CJ, et al. . Bacterial populations in the vaginas of healthy adolescent women. J Pediatr Adolesc Gynecol 2009; 22:11–8. [DOI] [PubMed] [Google Scholar]
- 5. Hickey RJ, Zhou X, Pierson JD, et al. . Understanding vaginal microbiome complexity from an ecological perspective. Transl Res 2012; 160:267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown RG, Al-Memar M, Marchesi JR, et al. . Establishment of vaginal microbiota composition in early pregnancy and its association with subsequent preterm prelabor rupture of the fetal membranes. Transl Res 2019; 207:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Champer M, Wong AM, Champer J, et al. . The role of the vaginal microbiome in gynaecological cancer. BJOG 2018; 125:309–15. [DOI] [PubMed] [Google Scholar]
- 8. Shipitsyna E, Roos A, Datcu R, et al. . Composition of the vaginal microbiota in women of reproductive age–sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS One 2013; 8:e60670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdool Karim SS, Passmore JS, Baxter C. The microbiome and HIV prevention strategies in women. Curr Opin HIV AIDS 2018; 13:81–7. [DOI] [PubMed] [Google Scholar]
- 10. Onderdonk AB, Delaney ML, Fichorova RN. The human microbiome during bacterial vaginosis. Clin Microbiol Rev 2016; 29:223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chehoud C, Stieh DJ, Bailey AG, et al. . Associations of the vaginal microbiota with HIV infection, bacterial vaginosis, and demographic factors. AIDS 2017; 31:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J, McCormick J, Bocking A, Reid G. Importance of vaginal microbes in reproductive health. Reprod Sci 2012; 19:235–42. [DOI] [PubMed] [Google Scholar]
- 13. Brotman RM, Bradford LL, Conrad M, et al. . Association between Trichomonas vaginalis and vaginal bacterial community composition among reproductive-age women. Sex Transm Dis 2012; 39:807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McClelland RS, Lingappa JR, Srinivasan S, et al. . Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect Dis 2018; 18:554–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sewankambo N, Gray RH, Wawer MJ, et al. . HIV-1 infection associated with abnormal vaginal flora morphology and bacterial vaginosis. Lancet 1997; 350:546–50. [DOI] [PubMed] [Google Scholar]
- 16. Heffron R, McClelland RS, Balkus JE, et al. ; Partners PrEP Study Team Efficacy of oral pre-exposure prophylaxis (PrEP) for HIV among women with abnormal vaginal microbiota: a post-hoc analysis of the randomised, placebo-controlled partners PrEP study. Lancet HIV 2017; 4:e449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klatt NR, Cheu R, Birse K, et al. . Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 2017; 356:938–45. [DOI] [PubMed] [Google Scholar]
- 18. Gautam R, Borgdorff H, Jespers V, et al. ; Vaginal Biomarkers Study Group Correlates of the molecular vaginal microbiota composition of African women. BMC Infect Dis 2015; 15:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borgdorff H, Tsivtsivadze E, Verhelst R, et al. . Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J 2014; 8:1781–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitchell C, Balkus JE, Fredricks D, et al. . Interaction between lactobacilli, bacterial vaginosis-associated bacteria, and HIV type 1 RNA and DNA genital shedding in U.S. and Kenyan women. AIDS Res Hum Retroviruses 2013; 29:13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu CM, Hungate BA, Tobian AA, et al. . Penile microbiota and female partner bacterial vaginosis in Rakai, Uganda. MBio 2015; 6:e00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donahue Carlson R, Sheth AN, Read TD, et al. . The female genital tract microbiome is associated with vaginal antiretroviral drug concentrations in human immunodeficiency virus-infected women on antiretroviral therapy. J Infect Dis 2017; 216:990–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKinnon LR, Karim QA. Factors driving the HIV epidemic in Southern Africa. Curr HIV/AIDS Rep 2016; 13:158–69. [DOI] [PubMed] [Google Scholar]
- 24. Jespers V, Kyongo J, Joseph S, et al. . A longitudinal analysis of the vaginal microbiota and vaginal immune mediators in women from sub-Saharan Africa. Sci Rep 2017; 7:11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gosmann C, Anahtar MN, Handley SA, et al. . Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity 2017; 46:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bayigga L, Kateete DP, Anderson DJ, et al. . Diversity of vaginal microbiota in sub-Saharan Africa and its effects on HIV transmission and prevention. Am J Obstet Gynecol 2019; 220:155–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hummelen R, Changalucha J, Butamanya NL, et al. . Lactobacillus rhamnosus GR-1 and L. reuteri RC-14 to prevent or cure bacterial vaginosis among women with HIV. Int J Gynaecol Obstet 2010; 111:245–8. [DOI] [PubMed] [Google Scholar]
- 28. Granich R, Gupta S, Hersh B, et al. . Trends in AIDS deaths, new infections and ART coverage in the top 30 countries with the highest AIDS mortality Burden; 1990–2013. PLoS One 2015; 10:e0131353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reynolds SJ, Makumbi F, Newell K, et al. . Effect of daily aciclovir on HIV disease progression in individuals in Rakai, Uganda, co-infected with HIV-1 and herpes simplex virus type 2: a randomised, double-blind placebo-controlled trial. Lancet Infect Dis 2012; 12:441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gianella S, Redd AD, Grabowski MK, et al. . Vaginal cytomegalovirus shedding before and after initiation of antiretroviral therapy in Rakai, Uganda. J Infect Dis 2015; 212:899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tobian AA, Grabowski MK, Serwadda D, et al. ; Rakai Health Sciences Program Reactivation of herpes simplex virus type 2 after initiation of antiretroviral therapy. J Infect Dis 2013; 208:839–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nason MC, Patel EU, Kirkpatrick AR, et al. . Immunological signaling during herpes simplex virus-2 and cytomegalovirus vaginal shedding after initiation of antiretroviral treatment. Open Forum Infect Dis 2016; 3:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu CM, Hungate BA, Tobian AA, et al. . Male circumcision significantly reduces prevalence and load of genital anaerobic bacteria. MBio 2013; 4:e00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cole JR, Wang Q, Cardenas E, et al. . The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009; 37:D141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu CM, Price LB, Hungate BA, et al. . Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv 2015; 1:e1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rositch AF, Gravitt PE, Tobian AA, et al. . Frequent detection of HPV before and after initiation of antiretroviral therapy among HIV/HSV-2 co-infected women in Uganda. PLoS One 2013; 8:e55383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oksanen J, Blanchet FG, Kindt R, et al. . Vegan: community ecology package. R package version 1.15-4. 2011; 117–8. Available at: http://CRAN.R-project.org/package=vegan. Accessed 19 June 2019.
- 38. Lajeunessei MJ. On the meta-analysis of response ratios for studies with correlated and multi-group designs. Ecology 2011; 92:2049–55. [DOI] [PubMed] [Google Scholar]
- 39. Mandal S, Van Treuren W, White RA, et al. . Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 2015; 26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Serrano-Villar S, Rojo D, Martínez-Martínez M, et al. . Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine 2016; 8:203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Noguera-Julian M, Rocafort M, Guillén Y, et al. . Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016; 5:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lozupone CA, Li M, Campbell TB, et al. . Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, et al. . Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol 2015; 8:760–72. [DOI] [PubMed] [Google Scholar]
- 44. Ravel J, Gajer P, Abdo Z, et al. . Vaginal microbiome of reproductive-age women. Proc Nat Acad Sci U S A 2011; 108(Suppl 1):4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fettweis JM, Brooks JP, Serrano MG, et al. . Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 2014; 160:2272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. World Health Organization. Consolidated ARV guidelines, June 2013 Available at: https://www.who.int/hiv/pub/guidelines/arv2013/intro/rag/en/index4.html. Accessed 19 June 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.