Abstract
Background
Studies have shown that HSP20 (heat-shock protein 20) genes play important roles in regulating plant growth, development, and stress response. However, the grape HSP20 gene family has not been well studied.
Results
A total of 48 VvHSP20 genes were identified from the grape genome, which were divided into 11 subfamilies (CI, CII, CIII, CV, CVI, CVII, MI, MII, ER, CP and PX/Po) based on a phylogenetic analysis and subcellular localization. Further structural analysis showed that most of the VvHSP20 genes (93.8%) had no intron or only one intron, while genes that clustered together based on a phylogenetic tree had similar motifs and evolutionarily conserved structures. The HSP20s share a conservedα-crystalline domain (ACD) and the different components of the ACD domain suggest the functional diversity of VvHSP20s. In addition, the 48 VvHSP20 genes were distributed on 12 grape chromosomes and the majority of VvHSP20 genes were located at the proximal or distal ends of chromosomes. Chromosome mapping indicated that four groups of VvHSP20 genes were identified as tandem duplication genes. Phytohormone responsive, abiotic and biotic stress-responsive, and plant development-related cis-elements were identified from the cis-regulatory elements analysis of VvHSP20s. The expression profiles of VvHSP20s genes (VvHSP20–1, 11, 14, 17, 18, 19, 20, 24, 25, 28, 31, 39, 42, and 43) were largely similar between RNA-Seq and qRT-PCR analysis after hydrogen peroxide (H2O2) treatment. The results showed that most VvHSP20s were down-regulated by H2O2 treatment during fruit development. VvHSP20s genes were indeed found to be involved in the grape berry development and differences in their transcriptional levels may be the result of functional differentiation during evolution.
Conclusions
Our results provide valuable information on the evolutionary relationship of genes in the VvHSP20 family, which is useful for future studies on the functional characteristics of VvHSP20 genes in grape.
Keywords: Grape, HSP20, Gene family, Genome-wide analysis, H2O2
Background
As one of the most important cultivated fruit crops in the world, grape has high economic value. ‘Kyoho’ is a tetraploid interspecific hybrid and mid-late ripening grape cultivar derived from a cross between Vitis vinifera x Vitis labrusca, which is widely cultivated in China. Our previous studies on ‘Kyoho’ have shown that hydrogen peroxide (H2O2) treatment could promote the early ripening of ‘Kyoho’ grape, causing it to ripen 20 days earlier than the control [1, 2]. Other studies in tomato [3] and pear [4] have also demonstrated that H2O2 is associated with fruit development. H2O2 is an early component of the thermal signal pathway, which is a necessary condition for the activation of heat-shock protein 20 (HSP20) synthesis [5]. In addition, the response of HSP20s to H2O2 has also been revealed in tomato and rice, where H2O2 was shown to induce the expression of mitochondrial HSP22 and chloroplast HSP26, respectively [6, 7]. It has been reported that HSP21 could protect photosystem II (PSII) from oxidative stress, promote color change during fruit ripening, and play a key role in the transformation of chloroplasts to pigment mother cells during fruit ripening [8].
The expression of HSPs is activated or increased under high temperature stress. According to molecular weight and sequence homology, HSPs can be divided into five families, which include HSP100, HSP90, HSP70, HSP60, and HSP20 [9, 10]. Among them, the molecular weights of HSP20 proteins are between 15 and 42 kDa, and are thus considered small HSPs. In some plant tissues, HSP20s comprise the largest proportion of HSPs [9]. HSP20s possess a typical conserved domain, known as the α-crystalline domain (ACD), which contains a conserved 80–100 amino acid sequence, a compact β-strand structure, and two conserved regions (CRs): CR I with β2, β3, β4, and β5; and CR II with β7, β8, β9, and a β6 loop [11]. HSP20s can prevent the damage of proteins caused by environmental stress and help them to fold or degrade [12, 13]. Thus, HSP20s are the important parts of cellular molecular chaperones.
In plants, HSP20 genes are involved in many developmental processes and responses to abiotic stresses [14, 15]. Under heat stress, HSP20s can prevent the aggregation and irreversible denaturation of heat-denatured proteins, which ensures that other proteins can perform normal functions at high temperature, providing a strong basis for improving the heat resistance of plant organs. HSP20s have been shown to be located in mitochondria, cytoplasm, and endoplasmic reticulum [16].
The number of HSP20 genes in plants is about four times greater than that in animals [17]. For example, 19, 35, 39, 42, 44, 51 members of the HSP20 gene family were respectively investigated in Arabidopsis (Arabidopsis thaliana) [11], pepper (Capsicum annuum L.) [18], rice (Oryza sativa) [19], tomato (Solanum lycopersicum) [20], watermelon (Citrullus lanatus L.) [21], and soybean (Glycine max) [22]. To date, HSP20 gene family members in grape have not been identified. Therefore, this study aims to elucidate the composition, gene structure, evolution, and expression of the grape HSP20 gene family, in an attempt to characterize structural and functional features, and to establish a foundation for further utilization of plant HSPs.
Results
Genome-wide identification of VvHSP20 gene family in grape
A total of 61 VvHSP20 genes were obtained by Hidden Markov Model (HMM) analysis. The presence of an ACD domain was confirmed by submitting the protein sequences to CDD, Pfam, and SMART database. The sequences without the typical ACD domain were discarded. A total of 48 sequences were retained and confirmed as grape HSP20 after removing the sequences with a molecular weight beyond the 15–42 kDa. Detailed information on physicochemical properties of these HSP20s are listed in Table 1. The length of the VvHSP20 proteins varied from 136 (VvHSP20–47 and VvHSP20–48) to 365 amino acids (VvHSP20–41); the molecular weights of VvHSP20s were from 15.27 kDa (VvHSP20–30) to 40.59 kDa (VvHSP20–41). The predicted pI values of VvHSP20s ranged from 4.68 (VvHSP20–41) to 9.48 (VvHSP20–20).
Table 1.
Features of VvHSP20 genes identified in grape
| Gene name | Sequence ID | ORF Length (bp) | Chr | Chromosome Position | Length (aa) | MW (KDA) | pI | ProtComp |
|---|---|---|---|---|---|---|---|---|
| HSP20–1 | VIT_01s0010g02290.t01 | 549 | 1 | 19,257,410–19,258,339 | 182 | 20.74 | 5.42 | Chloroplast |
| HSP20–2 | VIT_02s0154g00480.t01 | 606 | 2 | 5,248,542–5,250,041 | 201 | 22.45 | 9.24 | Mitochondrial |
| HSP20–3 | VIT_02s0154g00490.t01 | 606 | 2 | 5,255,141–5,256,154 | 201 | 22.55 | 9.11 | Mitochondrial |
| HSP20–4 | VIT_04s0008g01490.t01 | 471 | 4 | 1,219,942–1,220,465 | 156 | 17.34 | 5.94 | Cytoplasmic |
| HSP20–5 | VIT_04s0008g01500.t01 | 459 | 4 | 1,221,915–1,222,579 | 1521 | 16.69 | 6.84 | Cytoplasmic |
| HSP20–6 | VIT_04s0008g01510.t01 | 471 | 4 | 1,223,278–1,224,822 | 156 | 17.40 | 5.77 | Cytoplasmic |
| HSP20–7 | VIT_04s0008g01520.t01 | 471 | 4 | 1,224,823–1,225,354 | 156 | 17.58 | 5.58 | Cytoplasmic |
| HSP20–8 | VIT_04s0008g01550.t01 | 471 | 4 | 1,237,576–1,240,002 | 156 | 17.41 | 5.94 | Cytoplasmic |
| HSP20–9 | VIT_04s0008g01570.t01 | 501 | 4 | 1,244,737–1,246,530 | 166 | 18.60 | 5.95 | Cytoplasmic |
| HSP20–10 | VIT_04s0008g01580.t01 | 471 | 4 | 1,248,569–1,249,249 | 156 | 17.42 | 6.62 | Cytoplasmic |
| HSP20–11 | VIT_04s0008g01590.t01 | 468 | 4 | 1,251,984–1,252,699 | 155 | 17.29 | 5.94 | Cytoplasmic |
| HSP20–12 | VIT_04s0008g01610.t01 | 477 | 4 | 1,255,490–1,256,222 | 158 | 18.14 | 6.33 | Cytoplasmic |
| HSP20–13 | VIT_04s0008g01620.t01 | 480 | 4 | 1,257,262–1,257,741 | 159 | 18.42 | 8.46 | Cytoplasmic |
| HSP20–14 | VIT_06s0004g05770.t01 | 435 | 6 | 6,524,201–6,524,870 | 144 | 16.31 | 6.93 | Cytoplasmic |
| HSP20–15 | VIT_06s0009g01090.t01 | 948 | 6 | 12,540,124–12,542,550 | 315 | 34.98 | 8.58 | – |
| HSP20–16 | VIT_08s0032g00100.t01 | 579 | 8 | 3,034,519–3,035,221 | 192 | 21.52 | 8.45 | – |
| HSP20–17 | VIT_08s0058g00210.t01 | 447 | 8 | 8,901,763–8,902,216 | 148 | 16.88 | 5.81 | Nuclear |
| HSP20–18 | VIT_09s0002g00640.t01 | 483 | 9 | 440,684–441,359 | 160 | 17.89 | 6.3 | Cytoplasmic |
| HSP20–19 | VIT_09s0002g06790.t01 | 702 | 9 | 6,710,140–6,711,133 | 233 | 26.31 | 7.78 | Mitochondrial |
| HSP20–20 | VIT_12s0028g01390.t01 | 780 | 12 | 2,044,541–2,046,498 | 259 | 28.71 | 9.48 | – |
| HSP20–21 | VIT_12s0035g01910.t01 | 753 | 12 | 22,228,715–22,229,897 | 250 | 28.39 | 7.94 | Endoplasmic reticulum |
| HSP20–22 | VIT_13s0019g00860.t01 | 429 | 13 | 2,689,279–2,690,178 | 142 | 15.81 | 6.75 | Peroxisomal |
| HSP20–23 | VIT_13s0019g02740.t01 | 456 | 13 | 3,999,388–4,000,084 | 151 | 17.17 | 5.81 | Nuclear |
| HSP20–24 | VIT_13s0019g02760.t01 | 423 | 13 | 4,003,325–4,003,954 | 140 | 15.8 | 6.77 | Cytoplasmic |
| HSP20–25 | VIT_13s0019g02770.t01 | 456 | 13 | 4,006,363–4,007,091 | 151 | 17.1 | 5.81 | Nuclear |
| HSP20–26 | VIT_13s0019g02780.t01 | 456 | 13 | 4,015,394–4,016,080 | 151 | 17.02 | 5.8 | Nuclear |
| HSP20–27 | VIT_13s0019g02820.t01 | 456 | 13 | 4,036,190–4,036,907 | 151 | 17.12 | 5.81 | Nuclear |
| HSP20–28 | VIT_13s0019g02840.t01 | 456 | 13 | 4,043,383–4,044,010 | 151 | 17.09 | 5.54 | Nuclear |
| HSP20–29 | VIT_13s0019g02850.t01 | 456 | 13 | 4,048,636–4,049,360 | 151 | 17.05 | 5.8 | Nuclear |
| HSP20–30 | VIT_13s0019g02920.t01 | 411 | 13 | 4,108,657–4,109,160 | 136 | 15.27 | 5.7 | Cytoplasmic |
| HSP20–31 | VIT_13s0019g02930.t01 | 483 | 13 | 4,112,675–4,113,430 | 160 | 18.17 | 6.78 | Cytoplasmic |
| HSP20–32 | VIT_13s0019g03000.t01 | 483 | 13 | 4,149,244–4,149,995 | 160 | 18.15 | 7.93 | Cytoplasmic |
| HSP20–33 | VIT_13s0019g03010.t01 | 435 | 13 | 4,151,427–4,155,706 | 144 | 16.37 | 9.21 | Cytoplasmic |
| HSP20–34 | VIT_13s0019g03050.t01 | 498 | 13 | 4,180,057–4,183,444 | 165 | 19.23 | 6.46 | Cytoplasmic |
| HSP20–35 | VIT_13s0019g03090.t01 | 483 | 13 | 4,195,524–4,196,187 | 160 | 18.17 | 5.43 | Cytoplasmic |
| HSP20–36 | VIT_13s0019g03160.t01 | 483 | 13 | 4,227,250–4,227,937 | 160 | 18.02 | 7.94 | Cytoplasmic |
| HSP20–37 | VIT_13s0019g03170.t01 | 480 | 13 | 4,234,111–4,234,852 | 159 | 18.19 | 6.17 | Nuclear |
| HSP20–38 | VIT_14s0128g00280.t01 | 750 | 14 | 2,945,472–2,949,352 | 249 | 26.84 | 5.82 | – |
| HSP20–39 | VIT_16s0022g00510.t01 | 627 | 16 | 11,604,847–11,606,213 | 208 | 23.74 | 5.61 | Mitochondrial |
| HSP20–40 | VIT_16s0098g01060.t01 | 684 | 16 | 21,339,160–21,340,093 | 227 | 25.03 | 6.35 | Chloroplast |
| HSP20–41 | VIT_18s0072g00490.t01 | 1098 | 18 | 19,691,814–19,692,987 | 365 | 40.59 | 4.68 | – |
| HSP20–42 | VIT_18s0089g01270.t01 | 561 | 18 | 29,188,982–29,189,738 | 186 | 21.13 | 5.89 | Cytoplasmic |
| HSP20–43 | VIT_19s0014g05050.t01 | 579 | 19 | 5,376,784–5,377,821 | 192 | 22.39 | 5.35 | Cytoplasmic |
| HSP20–44 | VIT_19s0085g01050.t01 | 441 | 19 | 23,631,036–23,631,743 | 146 | 16.44 | 5.9 | Cytoplasmic |
| HSP20–45 | VIT_18s0001g01570.t01 | 492 | 18_random | 2,135,300–2,136,089 | 163 | 18.28 | 6.33 | Nuclear |
| HSP20–46 | VIT_18s0001g01610.t01 | 480 | 18_random | 2,183,110–2,184,273 | 159 | 18 | 5.74 | Nuclear |
| HSP20–47 | VIT_00s0707g00010.t01 | 411 | unknow | 34,510,489–34,511,388 | 136 | 15.69 | 4.89 | Nuclear |
| HSP20–48 | VIT_00s0992g00020.t01 | 411 | unknow | 37,397,842–37,398,790 | 136 | 15.7 | 5.01 | Nuclear |
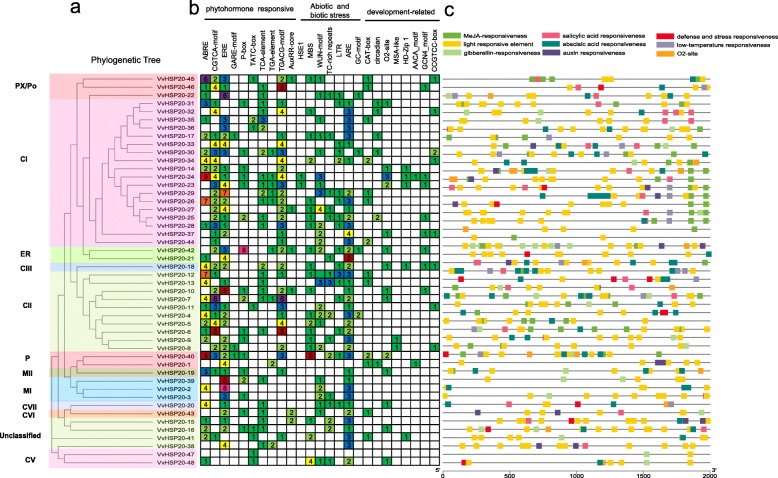
Phylogenetic analysis of VvHSP20 genes
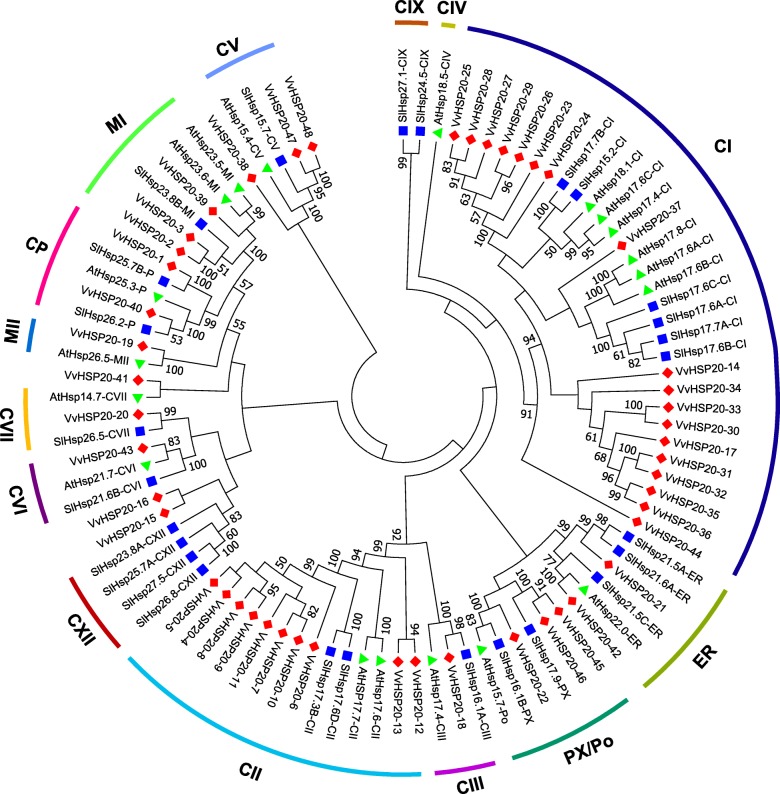
An unrooted Neighbor-Joining (NJ) phylogenetic tree was constructed based on the alignment of amino acid sequences of HSP20 from grape, Arabidopsis, tomato (Fig. 1). In total, 19 sequences from Arabidopsis, 26 sequences from tomato, and 48 sequences from grape were assessed in the phylogenetic tree. According to the phylogenetic and the subcellular localization analysis, the grape HSP20 protein are divided into 11 subfamilies (CI, CII, CIII, CV, CVI, CVII, MI, MII, ER, CP, and PX/Po) (Fig. 1, Table 1). Clustering of the subfamilies in grape is largely consistent with the subcellular localization, i.e., the proteins in the same cluster were located in the same subcellular sites. Specifically, six HSP20 subfamilies (CI-CVI), MTI and MTII subfamilies, CP, ER and PX /Po localize to the cytoplasm/nucleus, mitochondria, chloroplast, endoplasmic reticulum and peroxisome, respectively. The 93 HSP20s were classified into 14 distinct subfamilies, except for the unclassified VvHSP20s (VvHSP20–15, VvHSP20–16, VvHSP20–38, and VvHSP20–41), the subcellular localization of which could not be predicted using the online tool Protcomp. Most of the VvHSP20s, including 33 out of 44, were classified into CI–CVII, which suggested that the cytosol may be the primary functional site of plant HSP20s.
Fig. 1.
Phylogenetic tree of HSP20 proteins from grape and other plants. Phylogenetic tree of HSP20 proteins from grape and other plants including Arabidopsis thaliana and Solanum lycopersicum was constructed using MEGA7.0 based on the NJ method; bootstrap was 10,000 replicates. Percentage bootstrap scores of > 50% were displayed
Characterization of the amino acid sequences and gene structure of VvHSP20s
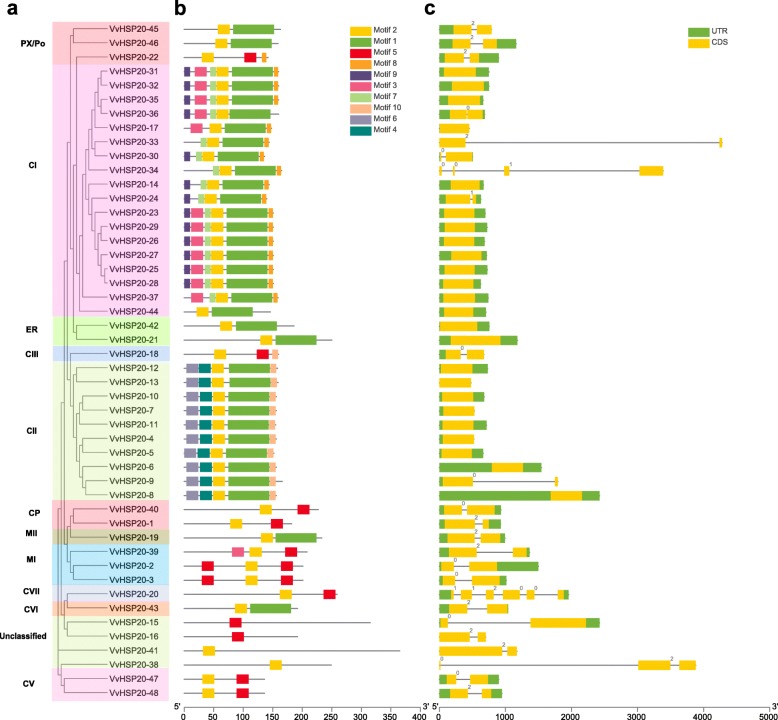
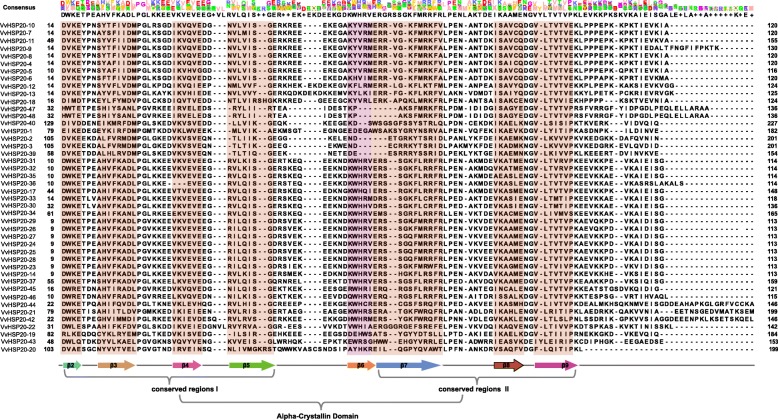
As shown in Fig. 2a, 48 VvHSP20s were divided into 11 subgroups, except for the unclassified HSP20 (VvHSP20–15, VvHSP20–16, VvHSP20–38 and VvHSP20–41). Ten conserved motifs of VvHSP20 proteins were identified by the MEME website and listed in Table 2. The lengths of these conserved motifs ranged from 6 to 60 amino acids (Fig. 2b, Table 2). ACD consists of two conserved regions, CRI of β2, β3 and β4, and CRI of β7, β8 and β9, separated by a variable length hydrophilic region β6 loop (Fig. 3). VvHSP20–2, 3, 39, 40, 47 and 48 lacked the β6-loop. VvHSP20–36 lacked the β-strands 4. The different components of the ACD domain suggest functional diversity among VvHSP20s. The same group of VvHSP20 proteins in the phylogenetic tree had the same motif, which indicated that they were highly conserved.
Fig. 2.
Phylogenetic tree, gene structure and domain analyses of VvHSP20s. a Phylogenetic tree of VvHSP20s was constructed with clustalx software. b Domain analyses of VvHSP20 proteins. Different color boxes represented the different types of motifs. c Gene structure of VvHSP20s. CDS sequences are represented by yellow round-corner rectangles and introns by grey lines, UTRs are shown with green boxes
Table 2.
Motif sequences identified by MEME tools
| Motif | Length (aa) | Sequence |
|---|---|---|
| 1 | 70 | VEEGRILQISGDRSVEKEEKNDKWHRVERSSGKFMRRFRLPENVKVDEVKAAMENGVLTVTVPKAEVQKP |
| 2 | 21 | DWKETPEAHVFKADLPGLKKE |
| 3 | 21 | NNMFDLWDPFQDFPFTGGALS |
| 4 | 21 | KSVSAPTRTYVRDAKAMAATP |
| 5 | 21 | MIDIDGISAGYEDGVLTVTVP |
| 6 | 21 | MMGFDSPLFSALQHMLDATDD |
| 7 | 11 | GETSAFANTRI |
| 8 | 8 | VKAIDISG |
| 9 | 11 | MSLIPSFFGGR |
| 10 | 11 | KKPKTIEVKIA |
Fig. 3.
The alignment of ACDs of HSP20s in grape. Names of all members are shown on the left side of the figure. Each predicted β-plated sheet is shown for shadow. The primary structure of the ACD, including the conserved regions I (CRI), II (CRII), and β 6-loop, is shown at the bottom of the figure
Next, we analyzed gene structure in order to better understand HSP20s. Among the VvHSP20s genes, 24 (50.0%) were intronless, and 21 genes (43.8%) possessed one intron. VvHSP20–38 (2 introns), VvHSP20–34 (3 introns), and VvHSP20–20 (5 introns) had two or more introns (Fig. 2c). Genes of the same subgroup had the same intron phase, which indicated that the structure was quite conserved over evolution.
Chromosomal location and gene duplication of VvHSP20
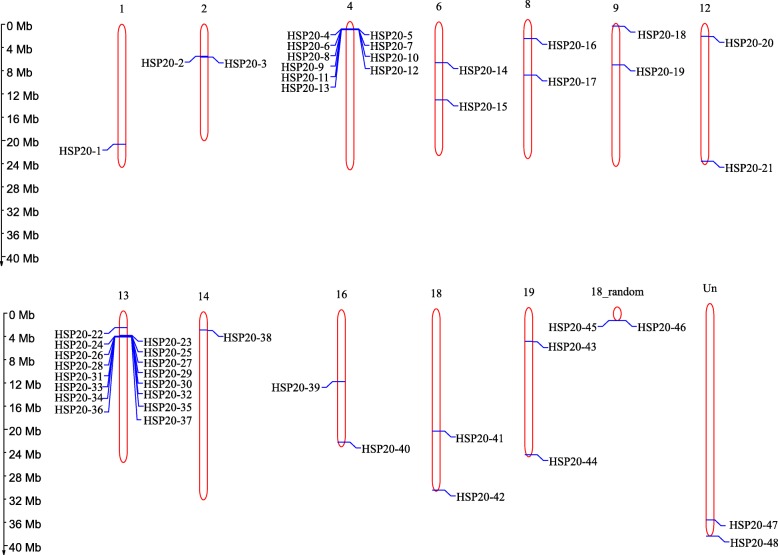
The 48 VvHSP20 genes were distributed on 12 grape chromosomes (Fig. 4). Most of the VvHSP20 genes were present on chromosome 4 (10 genes) and chromosome 13 (16 genes), while each of the remaining 10 chromosomes had one or two genes. Both tandem and segmental duplication contribute to the production of gene families during the process of evolution. Thus, potential duplication events of VvHSP20 genes were analyzed. In total, four groups of VvHSP20 genes (VvHSP20–2, 3; VvHSP20–4, 5, 6, 7, 8, 9, 10, 11; VvHSP20–23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 35, 36 and VvHSP20–47, 48) were identified as tandem duplication genes (Additional file 1: Figure S1). Furthermore, none of the genes were suggested to be products of segmental duplication. Based on the above results, we inferred that tandem duplication played an important role in the expansion of the VvHSP20 family in grape.
Fig. 4.
Chromosomal locations of VvHSP20 genes on grape chromosomes. Blue lines indicated gene position
Analysis of cis-element in VvHSP20 gene promoters
To understand the possible role of cis-regulatory elements of VvHSP20, the promoter sequences (comprising − 2000 bp upstream of the translation start site) of 48 VvHSP20 genes were submitted into PlantCARE to detect the cis-elements. Three categories of cis-elements, including phytohormone responsive, abiotic and biotic stress-responsive, and plant development-related cis-elements were identified and are shown in Fig. 5. Among the three categories of cis-elements, the phytohormone responsive category accounts for the highest proportion. In this category, cis-acting elements were widely present in the promoter region, including auxin responsive (TGA-element and AuxRR-core), gibberellin-responsive elements (GARE-motif, P-box, and TATC-box), ethylene-responsive (ERE), MeJA-responsive (TGACG-motif and CGTCA-motif), abscisic acid-responsive (ABRE), and salicylic acid-responsive (TCA-element). Among these elements, ABRE and ERE accounted for the largest part of the phytohormone responsive category. In the abiotic and biotic stress-responsive category, stress response-related cis-elements, such as HSE1 (heat stress), WUN motif (wound-responsive element), TC-rich repeats (stress response), LTR (low temperature-responsive), ARE (anaerobic induction), and GC-motif (anoxia) were detected. In the last category, plant development-related elements, including meristem expression (CCGTCC-box and CAT-box), circadian, zein metabolism regulation (O2-site), cell cycle regulation (MSA-like), differentiation of the palisade mesophyll cells (HD-Zip 1), and endosperm expression (AACA_motif and GCN4_motif) were identified. In addition, most of the VvHSP20 genes possessed W boxes and MYB binding sites, including CCAAT-boxes.
Fig. 5.
Investigation of cis-acting element numbers in VvHSP20 genes. a The phylogenetic tree of VvHSP20s was constructed using Muscle module and neighbor-joining (NJ) method with 1000 bootstrap replicates implemented in MEGA7. Displaying of different background colors to distinguish the clades and subclades. b The different colors and numbers of the grid indicated the numbers of different promoter elements in these VvHSP20 genes. c The different colored block represented the different types of cis-acting elements and their locations in each VvHSP20 gene
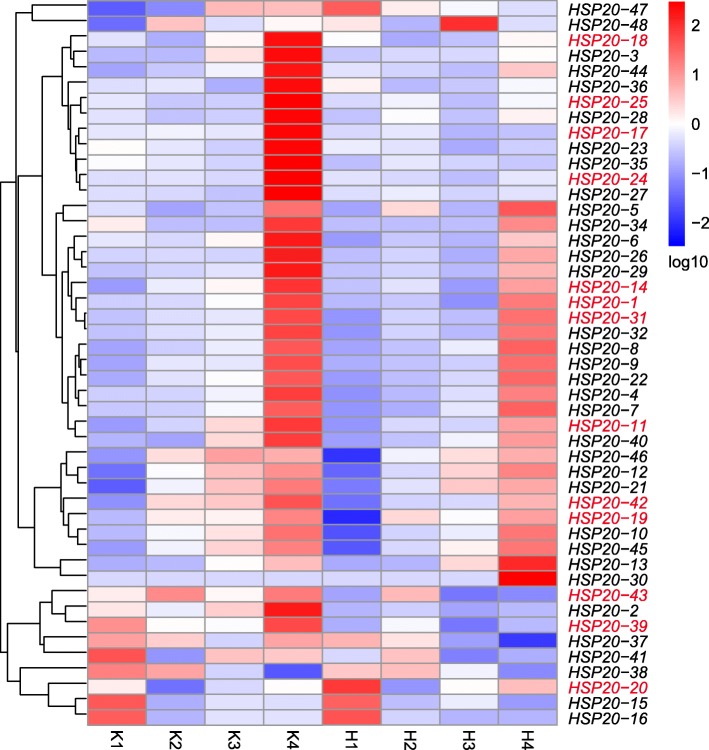
Expression patterns of VvHSP20s in response to H2O2 treatment
There is a close relationship between gene expression and function. To determine the functions of VvHSP20s in grape, a heatmap of 47 VvHSP20 genes was constructed using FPKM values from RNA-Seq data in control and H2O2-treated berries of ‘Kyoho’ (Fig. 6, sampling period is described in Materials and Methods and Table 3). The expression level of HSP20–33 was extremely low and not detected by RNA-Seq analysis during fruit development. Most VvHSP20s were down-regulated after treatment, especially at the fourth period. However, the opposite trend was also observed for a few genes, including HSP20–13, HSP20–20, and HSP20–30. These results indicated that most of the VvHSP20 genes responded to H2O2 treatment, and the response mechanisms of different VvHSP20 genes to H2O2 were different.
Fig. 6.
Relative transcriptional expression levels of VvHSP20 from RNA-seq data in the H2O2 treatment and the control. Data were plotted after the Z-score normalization across the row based on absolute FPKM values of each gene at different development stages. The colors vary from blue to red representing the scale of the relative expression levels. ID in Red indicate genes selected for qRT-PCR analysis
Table 3.
The sampling date of grape berries in this study
| Development stage (dpa) | Sampling time | Code | |
|---|---|---|---|
| control | Treated | ||
| 35 | 6.12 | K1 | H1 |
| 45 | 6.21 | K2 | H2 |
| 55 | 7.1 | K3 | H3 |
| 65 | 7.12 | K4 | H4 |
dpa: Days post anthesis
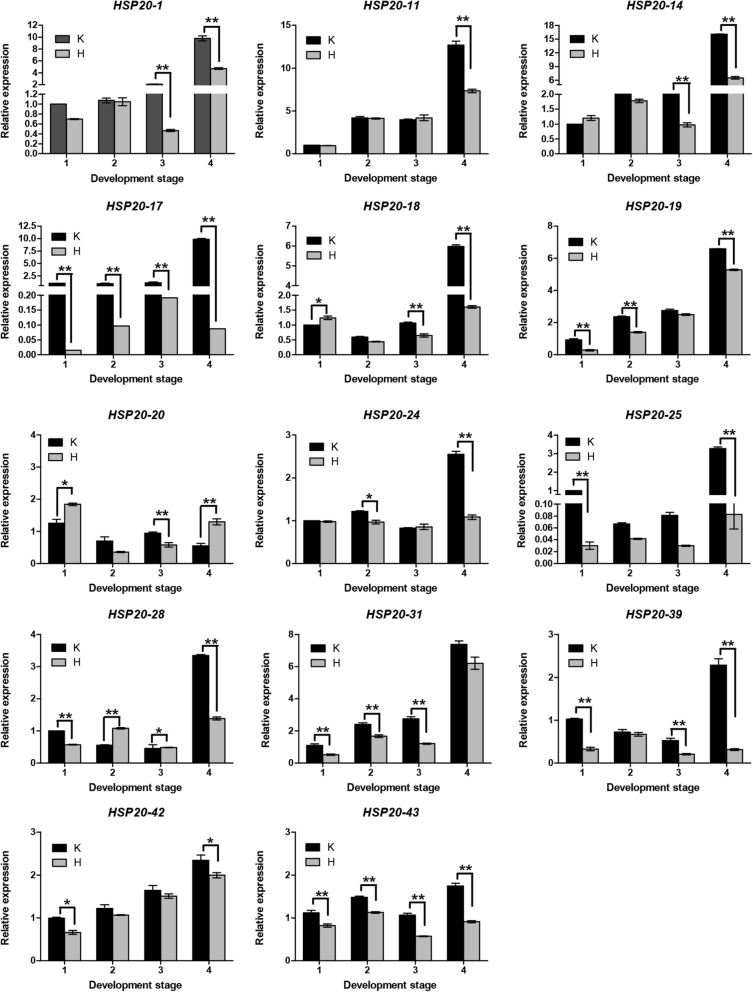
Based on the statistical significance of the gene expression levels from the RNA-Seq analysis and the partitioning of the clusters of genes from the phylogenetic analysis, 14 differentially expressed VvHSP20 genes were selected to be further validated by qRT-PCR in response to control and H2O2 treatment (Fig. 7). Consistent with the RNA-Seq data, the expression level of most genes decreased after the treatment. Besides HSP20–31, the relative expression levels of the remaining 13 genes were extremely down-regulated at the fourth period. It is worth noting that VvHSP20–17 and VvHSP20–25 were hardly expressed after treatment. Similar expression patterns were revealed within the tandem duplicated gene groups (VvHSP20–25 and VvHSP20–28). The similar expression patterns indicated that the tandem duplicated VvHSP20 genes had similar functions and structures. Members of the CI subgroup (VvHSP20–24, VvHSP20–25, VvHSP20–28, and VvHSP20–31) had similar expression patterns after the treatment, which suggested that they had similar functions in response to H2O2 treatment.
Fig. 7.
Expression profiles of VvHSP20s from qRT-PCR in the H2O2 treatment and the control. The x-axis represented different sampling date, while relative expression levels for the y-axis. Data represented the mean of three biological replicates. Error bars represented standard deviations from three independent technical replicates. And the expression level of K1 was used as the calibrator. The asterisks indicate the significant level (*P < 0.05, **P < 0.01) based on a Duncan’s multiple range test
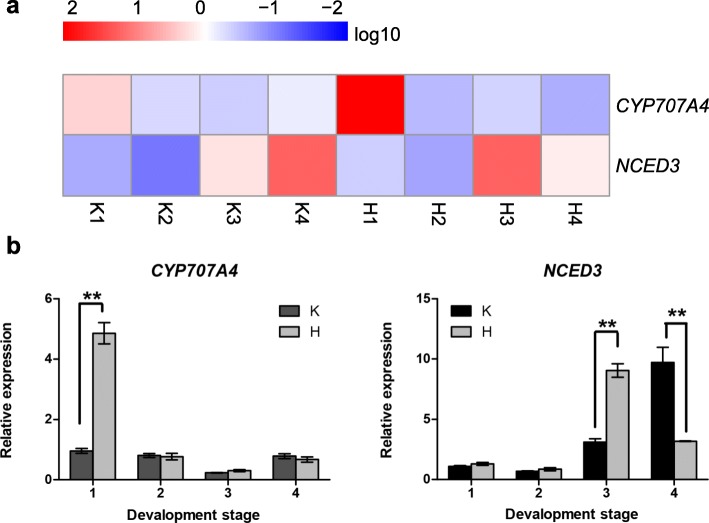
Expression patterns of ABA-related genes in response to H2O2 treatment
It is well known that ABA plays an important role in grape [23, 24]. In the previous study [1], H2O2 treatment was shown to promote the early fruit ripening of ‘Kyoho’. To further explore the role of ABA in this process, RNA-Seq and qRT-PCR were performed to examine the expression analysis of ABA-related genes. As shown in Fig. 8, the expression patterns of the ABA synthesis-related gene (NCED3) and degradation-related gene (CYP707A4) were different following H2O2 treatment. Compared with the control, the expression level of NCED3 reached the highest level at veraison (H3 stage), then decreased at the H4 stage. On the contrary, the expression level of the CYP707A4 gene increased rapidly after treatment and reached its lowest level at veraison. The changes in the expression levels of ABA-related genes indicated that H2O2 may regulate fruit development possibly through control of ABA catabolism and biosynthesis.
Fig. 8.
Expression profiles of ABA-related genes in the H2O2 treatment and the control. a The expression pattern of ABA-related genes from RNA-seq data. b Expression patterns of ABA-related genes from qRT-PCR. The x axis represented different sampling dates, while the y axis indicated relative expression levels. The data represent the average of three biological replicates. The error bar represents the standard deviation of three independent techniques. The expression quantity of K1 was used as calibrator. The asterisk indicates the significance level based on the Duncan multiple range test (*P < 0.05, **P < 0.01)
Discussion
Fruit ripening is known to be regulated by a balance between reactive oxygen species (ROS) formation and detoxification by antioxidant enzymes [25, 26]. ROS causes senescence by accumulation of superoxide anion (O2.-) and hydrogen peroxide (H2O2) during fruit ripening [3]. H2O2 not only acts as a stress inducing factor, but also as a signaling molecule. Imbalance between ROS generation and removal can lead to oxidative stress in aerobic organisms [27, 28]. Previous studies on H2O2 signaling have identified a number of genes that are regulated by H2O2 levels [29, 30]. Among H2O2-inducible genes, HSPs are related to defense or stress responses [5]. However, the relationship between H2O2 and HSP20 in grape berry development is not clear. Therefore, a preliminary study on this issue was conducted.
HSP20 proteins as molecular chaperones play an important role in plant growth and development, and deter or reduce the irreversible aggregation of denatured proteins under stress [14, 15]. Although HSP20s block the aggregation and stabilization of non-natural proteins in an ATP-independent manner [17], HSP20s themselves could not refold non-native proteins. Pea Hsp18.1 had to work with the hsp70 system to refold thermally-modified proteins [31]. In recent years, due to the availability of whole genome sequences, HSP20 families have been identified from plants, such as Arabidopsis [11], tomato [32], rice [19], and soybean [22]. However, there are few studies on the HSP20 family in grapes.
Following an integrated approach to detect HSP20s in grape, 48 putative VvHSP20 genes were identified. These genes were divided into 11 subgroups (CI, CII, CIII, CV, CVI, CVII, MI, MII, ER, CP, and PX/Po). Previous research showed that 12 HSP20 gene subgroups were identified from Arabidopsis (CI–CVII, MI, MII, ER, CP, and PX/Po) [11, 33]. Likewise, four new nuclear subgroups from rice (CVIII, CIX, CX, and CXI) were reported [9]. However, several subgroups including CIV, CVIII, CIX, CX, and CXI of rice were not identified from the VvHSP20 genes of grape. One study demonstrated that the CIV subgroup may be involved in coping with diverse stress conditions and may be developmentally regulated [33]. Under normal growth conditions, members of the CVIII subgroup may be heat-induced, while the CX subgroup of genes may be related to specific housekeeping functions [9]. Interestingly, in pepper plants, the HSP20 CIV, CV, CVIII, CIX, CX, and CXI subgroups were found to be absent [18]. In addition, the HSP20 family of rice lacked CIV and CVII subgroups [9]. Therefore, it was easy to see that gene acquisition and loss events are widespread in plant species. The absence of subgroups may be due to the loss of genes during the evolution of HSP20 genes.
Gene structure plays a crucial role in the evolution of multiple gene families. Our results showed that most of the VvHSP20 genes (93.8%) had no intron or only one intron of short length. Plants tend to retain genes without introns or with shorter introns [34]. This is consistent with previous reports from pepper [18] and tomato [32], where 97.14 and 83.33% of HSP20 genes, respectively, have no or one short intron. Most VvHSP20s in the CII and ER subgroups had no intron, which is consistent with orthologs in pepper, rice, and soybean [18, 19, 22], but the gene structure (exon-intron) of the CI group in grape was different from those in these species, indicating that the intron pattern might not be well preserved among different species. In addition, the stability index of most VvHSP20 proteins was greater than or equal to 40, indicating that most of them were unstable proteins. Instability is believed to be a common feature of stress-responsive proteins, and may also reflect the rapid induction of VvHSP20 genes [35].
The expression of heat-shock proteins (HSPs) is activated or increased under hight termperature stress, a condition in which HSP20s play important roles in protecting against protein aggregation [14]. HSP20s could be induced not only by environmental stresses, including heat, cold, drought, and salinity, but also by various developmental processes, such as embryogenesis, seed germination, and fruit ripening [22, 36–38]. In this study, the expression of VvHSP20s was down-regulated by H2O2 treatment during fruit development (Fig. 7), in line with our previous research showing hydrogen peroxide can promote the early ripening of ‘Kyoho’ grape [1]. Similarly, FaHSP17.4 was highly expressed in leaves and flower organs of ‘Fengxiang’ strawberry, but the expression decreased gradually during fruit development [36]. In addition, HSP expression is induced at specific developmental stages in plants. HSP20s were highly expressed in the development stages of zygotic embryonic tissues, and during pollen maturation in rice and tomato [9, 39]. The NJJS4 gene is a type of HSP20-coding gene, which accumulates in strawberry fruit (Fragaria x ananassa cv, receptacle) during ripening [40]. Class II sHSP17.4 is expressed at almost all stages of fruit development, and maintained at a high level at the later stage of fruit ripening, while Class II sHSP17.6 reached a peak at the turning stage, and Class I HSP17.7 reached a high level at the pink stage [41]. Four differentially expressed HSP20 genes were revealed from the RNA-Seq results of tomato fruit (Heize 1706), which were considered to play an important role in fruit development [42]. These observations indicate that HSP20s are associated with fruit development.
ABA plays an important role in promoting fruit ripening. In non-climacteric grape berries, ABA is considered to be the main signal that triggers the onset of maturation-related processes as it peaks at version, accompanied by the beginning of berry softening and skin coloration [43]. ABA content is determined by the dynamic balance of endogenous ABA biosynthesis and catabolism [44]. A previous study showed that 9-cis-epoxycarotenoid dioxygenase (NCED) is a key enzyme involved in ABA biosynthesis [45] and CYP707A (an key ABA degradation enzyme) plays a predominant role in ABA catabolism in vivo in strawberry [46, 47]. NCED plays an important role in the ABA-mediated signaling pathway [45, 48]. In order to further understand the relationship between hydrogen peroxide and ABA during grape development, we analyzed the expression of ABA synthesis and degradation-related genes after H2O2 treatment (Fig. 8). In this study, NCED3 was found to have low expression at the early stages of fruit development, but to rapidly increase at the K4 stage in the control. However, it reached peak levels at veraison then rapidly decreased at H4 stage. This is consistent with changes in ABA during fruit development, whereby ABA reaches peak levels at the veraison stage and decreases after that [49, 50]. ABA catabolism and biosynthesis are closely linked through feedback and feedforward loops to limit the amount of ABA needed for fruit growth and to rapidly increase the amount of ABA before fruit ripening [47]. The CYP707A4 gene is highly induced at the H1 stage, then gradully decreases, and finally reaches the lowest values at veraison after H2O2 treatment. It was previously shown that the expression level of FveCYP707A4a was higher in the early stages of fruit development in woodland strawberry [47]. This may be due to a high level of ABA inhibiting early fruit growth [47] and accelerated ABA degradation following hydrogen peroxide treatment.
We propose a model for HSP20s and ABA, H2O2, fruit development, and high temperature (Fig. 9). Under high temperature, HSP20s are activated or increased [51]. In our study, the expression levels of most HSP20s were down-regulated during fruit development after H2O2 treatment (Fig. 7) and H2O2 treatment promoted early ripening of ‘Kyoho’ grape [1]. In addition, ABA play significant roles in promoting fruit ripening and it is considered that ABA is the main signal triggering the beginning of maturation-related processes. ABA synthesis and metabolism were also affected by H2O2 (Fig. 8). Interestingly, other studies have shown that ABA induces H2O2 formation [52]. However, the role of HSP20s in this process needs to be further explored.
Fig. 9.
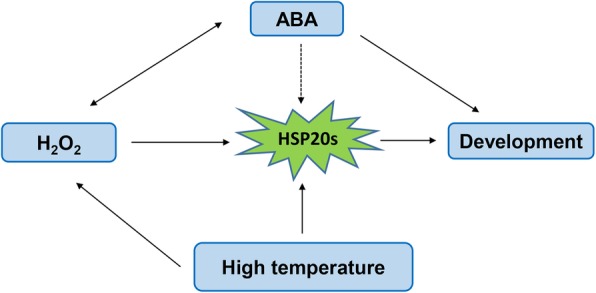
Proposed Model for HSP20 and ABA, H2O2, high temperature, fruit development
Conclusion
In this study, the HSP20 gene family of grape was comprehensively identified. The phylogenetic relationships, gene structures, conserved motifs, and cis-acting elements of 48 VvHSP20 genes were analyzed, while the expression levels were explored by RNA-Seq and qRT-PCR analysis. A total of 48 HSP20 were divided into 11 subfamilies according to the phylogenetic tree and subcellular localization. The expression levels of HSP20 genes in grape under H2O2 treatment were verified by qRT-PCR analysis, providing a basis for further study on the functional analysis of HSP20 genes during fruit development. Finally, the expression levels of ABA-related genes were verified. We confirmed that H2O2 indeed affected ABA metabolism and the expression of HSP20 genes to promote fruit development and ripening.
Methods
Identification of HSP20 genes in grape genome
We downloaded the grapevine reference genome assembly and protein sequences from Ensembl Plants Database (http://plants.ensembl.org/index.html). The grape HSP20 candidates were identified based on the HMM profile of HSP20 (PF00011). The CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), Pfam and SMART (http://smart.embl-heidelberg.de/) were used to further confirm the conserved HSP20 domain. Finally, 48 HSP20s were identified after removing the redundant sequence without the conserved ACD domain of HSP20 and with the molecular weight outside the range of 15–42 kDa. The Protparam online tools (https://web.expasy.org/protparam/) were used to predict physicochemical properties of HSP20 proteins. The online tool Protcomp (http://linux1.softberry.com/) was used to perform the subcellular localization prediction. The identified VvHSP20 genes were named according to their positions on pseudomolecules [19].
Phylogenetic analysis of HSP20 genes in plants
The amino acid sequences of HSP20s derived from Arabidopsis and tomato and newly identified VvHSP20s were used for phylogenetic analysis. The neighbor joining phylogenetic tree was constructed with the default parameters based on the multiple sequence alignments of the HSP20s amino acid sequences by MEGA 7.0 software.
Characterization of the amino acid sequences and gene structure of VvHSP20s
The conserved motifs of VvHSP20s were identified using MEME program (version 4.11.2, http://alternate.meme-suite.org/tools/meme), and the parameters were as follows: optimum motif width ranges from 6 to 200 amino acid residues and maximum of 10 misfits. The structures of VvHSP20 genes in grape was identified using TBtools software [53].
Chromosomal location and gene duplication of HSP20 genes
Chromosomal localization information of VvHSP20 genes was obtained from Ensembl Plants Database (http://plants.ensembl.org/index.html) and the chromosome location images were generated using the MapDraw V2.1 tool (http://mg2c.iask.in/mg2c_v2.0/). The definition of CaHSP20 gene replication is based on the previous research [54]. The duplication events and syntenic analysis of VvHSP20 genes were determined using MCScanX (Multiple Collinearity Scan) [55] and Circos software, respectively.
Analysis of cis-elements in VvHSP20 gene promoters
The cis-elements were identified from the upstream 2 kb promoter sequences of the VvHSP20 genes which were submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [56].
Plant material
Plant samples were collected from the farm of Henan University of Science & Technology, Luoyang, China in 2017. ‘Kyoho’ grape treated with distilled water (containing 0.03% silicon wet-77 surfactant) was naturally grown for 6 years as a control and treated twice with 300 mmol/L H2O2. The first spraying was conducted at 25 days post anthesis (dpa) in 2017 and the second was 35 dpa. Samples were taken 35 days after flowering and every 10 days until the treated fruits were ripe (Table 2). In addition, the treated berries reached the veraison at 55 dpa. Representative pest-free samples were collected from 5 individual vines of ‘Kyoho’. Thirty samples were randomly selected from each tree to record the phenological data of fruit development.
RNA extraction and quantitative real-time PCR (qRT-PCR) data analysis
The RNAprep Pure Plant Kit (TIANGEN, Beijing China) was used to isolate total RNA. cDNAs were obtained by total RNA reverse transcription using HiScript® II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing China). Primers for the VvHSP20 genes were designed by Primer Premier 5.0 software and listed in Additional file 2: Table S1. The grape ubiquitin1 gene was used as the reference gene [57, 58] and the expression level of K1 was used as the calibrator. Quantitative real-time PCR was conducted with a total volume of 10 μL of TransStart Top Green qPCR SuperMix kit (TRANSGEN, Beijing China) in CFX96 Real-Time PCR Detection System (Bio-Rad). The relative expression changes of VvHSP20s genes were calculated using the 2-ΔΔCt method from three independent replicates [59]. SPSS version 21.0 was employed to analyze the statistical significant differences of the gene expression levels by ANOVA with Duncan’s multiple range test.
The FPKM values of VvHSP20 genes were from the RNA-Seq data (Accession codes, SRA: PRJNA541089). The average FPKM value of each repetition was converted to log10. Pheatmap (R package) was used to generate the heatmap.
Supplementary information
Additional file 1: Figure S1. Syntenic relationships among VvHSP20s genes. Different colors represent different chromosomes. Lines of different colors represent different tandem duplication genes.
Additional file 2: Table S1. Primers used for the qRT-PCR reactions.
Acknowledgements
We would like to thank all the colleagues in our laboratory for providing useful discussions and technical assistance. We are very grateful to the editors and reviewers for their critical evaluation of the manuscript and for constructive comments on the improvement of the manuscript.
Abbreviations
- ABA
Amino acid
- ACD
Alpha-crystallin domain
- CRI
Conserved region I
- CRII
Conserved region II
- HSP20s
Small heat shock proteins
- PSII
Photosystem II
Authors’ contributions
D-LG and X-RJ conceived and designed the experiments; X-RJ conduct the bioinformatics analysis and wrote the manuscript draft. X-RJ and P-YN examined the expression of genes by qRT-PCR. D-LG, G-HZ and Y-HY reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by Natural Science Foundation of China (NSFC: 31672106) and Zhongyuan Science and Technology Innovation Leaders (194200510007), China. The Funding bodies were not involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
In this study, the grape genome sequence used to identify HSP20 genes were downloaded from Ensembl Plants Database (http://plants.ensembl.org/index.html). Expression data of VvHSP20 genes in grape used in this study can be accessed via the NCBI SRA database with accession numbers of PRJNA541089 from 5th May 2020 onwards, as until then there is an embargo due to a complementary manuscript. Until then, these sequences are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiao-Ru Ji, Email: wushangdaya962464@163.com.
Yi-He Yu, Email: yuyihe@haust.edu.cn.
Pei-Yi Ni, Email: npy15637921287@163.com.
Guo-Hai Zhang, Email: hnzhanggh@163.com.
Da-Long Guo, Email: guodalong@haust.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-019-2031-4.
References
- 1.Guo D-L, Wang Z-G, Li Q, Gu S-C, Zhang G-H, Yu Y-H. Hydrogen peroxide treatment promotes early ripening of Kyoho grape. Aust J Grape Wine Res. 2019;25:357–362. doi: 10.1111/ajgw.12399. [DOI] [Google Scholar]
- 2.Xi F-F, Guo L-L, Yu Y-H, Wang Y, Li Q, Zhao H-L, et al. Comparison of reactive oxygen species metabolism during grape berry development between 'Kyoho' and its early ripening bud mutant 'Fengzao'. Plant Physiol Biochem. 2017;118:634–642. doi: 10.1016/j.plaphy.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Jimenez A, Creissen G, Kular B, Firmin J, Robinson S, Verhoeyen M, et al. Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta. 2002;214:751–758. doi: 10.1007/s004250100667. [DOI] [PubMed] [Google Scholar]
- 4.Chiriboga M, Giné Bordonaba J, Schotsmans W-C, Larrigaudière C, Recasens I. Antioxidant potential of ‘conference’ pears during cold storage and shelf life in response to 1-methylcyclopropene. Lwt-Food Sci Technol. 2013;51:170–176. doi: 10.1016/j.lwt.2012.10.023. [DOI] [Google Scholar]
- 5.Königshofer H, Tromballa H-W, Löppert H-G. Early events in signalling high-temperature stress in tobacco BY2 cells involve alterations in membrane fluidity and enhanced hydrogen peroxide production. Plant Cell Environ. 2008;31:1771–1780. doi: 10.1111/j.1365-3040.2008.01880.x. [DOI] [PubMed] [Google Scholar]
- 6.Lee B-H, Lee H-S, Won S-H, Miyao M, Chung W-I, Kim I-J, et al. Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene. 2000;245:283–290. doi: 10.1016/S0378-1119(00)00043-3. [DOI] [PubMed] [Google Scholar]
- 7.Banzet N, Richaud C, Deveaux Y, Kazmaier M, Gagnon J, Triantaphylidès C. Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J. 1998;13:519–527. doi: 10.1046/j.1365-313X.1998.00056.x. [DOI] [PubMed] [Google Scholar]
- 8.Neta-Sharir I, Isaacson T, Lurie S, Weiss D. Dual role for tomato heat shock protein 21: protecting photosystem II from pxidative ptress and promoting color changes during fruit maturation. Plant Cell. 2005;17:1829–1838. doi: 10.1105/tpc.105.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar N-K, Kim Y, Grover A. Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics. 2009;10:393–411. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao P, Wang D, Wang R, Kong N, Zhang C, Yang C, et al. Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics. 2018;19:1–13. doi: 10.1186/s12864-017-4368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scharf K, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:TEFOAT>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feder M-E, Hofmann G-E. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 13.Wallace E-W-J, Kear-Scott J-L, Pilipenko E-V, Schwartz M-H, Laskowski P-R, Rojek A-E, et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 2015;162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters E-R. The evolution, function, structure, and expression of the plant sHSPs. J Exp Bot. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- 15.Mogk A, Bukau B. Role of sHsps in organizing cytosolic protein aggregation and disaggregation. Cell Stress Chaperones. 2017;22:493–502. doi: 10.1007/s12192-017-0762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mani N, Ramakrishna K, Suguna K. Characterization of rice small heat shock proteins targeted to different cellular organelles. Cell Stress Chaperones. 2015;20:451–460. doi: 10.1007/s12192-015-0570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters E-R, Lee G-J, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–338. doi: 10.1093/jxb/47.3.325. [DOI] [Google Scholar]
- 18.Guo M, Liu J, Lu J, Zhai Y, Wang H, Gong Z, et al. Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front Plant Sci. 2015;6:806. doi: 10.3389/fpls.2015.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouyang Y, Chen J, Xie W, Wang L, Zhang Q. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol Biol. 2009;70:341–357. doi: 10.1007/s11103-009-9477-y. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, Cheng Y, Feng K, Ruan M, Ye Q, Wang R, et al. Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Front Plant Sci. 2016;7:1-14. [DOI] [PMC free article] [PubMed]
- 21.He Y, Fan M, Sun Y, Li L. Genome-wide analysis of watermelon HSP20s and their expression profiles and subcellular locations under stresses. Int J Mol Sci. 2019;20:12. doi: 10.3390/ijms20010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes-Caitar V-S, de Carvalho M-C, Darben L-M, Kuwahara M-K, Nepomuceno A-L, Dias W-P, et al. Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics. 2013;14:577–594. doi: 10.1186/1471-2164-14-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Liu Q, Xi B, Dai H. Study on the regulation of anthocyanin biosynthesis by exogenous abscisic acid in grapevine. Sci Hortic-Amsterdam. 2019;250:294–301. doi: 10.1016/j.scienta.2019.02.054. [DOI] [Google Scholar]
- 24.Koyama K, Sadamatsu K, Goto-Yamamoto N. Abscisic acid stimulated ripening and gene expression in berry skins of the cabernet sauvignon grape. Funct Integr Genomics. 2010;10:367–381. doi: 10.1007/s10142-009-0145-8. [DOI] [PubMed] [Google Scholar]
- 25.Kumar V, Irfan M, Ghosh S, Chakraborty N, Chakraborty S, Datta A. Fruit ripening mutants reveal cell metabolism and redox state during ripening. Protoplasma. 2016;253:581–594. doi: 10.1007/s00709-015-0836-z. [DOI] [PubMed] [Google Scholar]
- 26.Pilati S, Brazzale D, Guella G, Milli A, Ruberti C, Biasioli F, et al. The onset of grapevine berry ripening is characterized by ROS accumulation and lipoxygenase-mediated membrane peroxidation in the skin. BMC Plant Biol. 2014;14:87. doi: 10.1186/1471-2229-14-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandenabeele S, Katrien V-D-K, Dat J, Gadjev I, Boonefaes T, Morsa S, et al. A comprehensive analysis of hydrogen peroxide-induced gene eExpression in tobacco. Proc Natl Acad Sci U S A. 2003;100:16113–16118. doi: 10.1073/pnas.2136610100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desikan R, Mackerness AH. S, Hancock J-T, Neill S-J. regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Guo Y, Jia L, Chu H, Zhou S, Chen K, et al. Hydrogen peroxide acts upstream of nitric oxide in the heat shock pathway in Arabidopsis seedlings. Plant Physiol. 2014;164:2184–2196. doi: 10.1104/pp.113.229369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desikan R, Clarke A, Hancock J-T, Neill S-J. H2O2 activates a MAP kinase-like enzyme in Arabidopsis thaliana suspension cultures. J Exp Bot. 1999;50:1863–1866. [Google Scholar]
- 31.Lee G-J, Vierling E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 2000;122:189–198. doi: 10.1104/pp.122.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Cheng Y, Feng K, Ruan M, Ye Q, Wang R, et al. Genome-wide identification and expression profiling of tomato Hsp20 gene family in response to biotic and abiotic stresses. Front Plant Sci. 2016;7:1–14. doi: 10.3389/fpls.2016.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddique M, Gernhard S, von Koskull-Döring P, Vierling E, Scharf K. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones. 2008;13:183–197. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattick J-S, Gagen M-J. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- 35.Rao P-K, Roxas B-A-P, Li Q. Determination of global protein turnover in stressed mycobacterium cells using hybrid-linear ion trap-fourier transform mass spectrometry. Anal Chem. 2008;80:396–406. doi: 10.1021/ac701690d. [DOI] [PubMed] [Google Scholar]
- 36.Gang L, Xia Z, Lu Z, Houcheng Z. Cloning and expression analysis of small heat shock protein (FaHSP17.4) gene from strawberry fruit (Fragaria ananassa) Mol Plant Breed. 2016;14:328–336. [Google Scholar]
- 37.Reddy P-S, Kishor P-B-K, Seiler C, Kuhlmann M, Eschen-Lippold L, Lee J, et al. Unraveling regulation of the small heat shock proteins by the heat shock factor HvHsfB2c in barley: its implications in drought stress response and seed development. PLoS One. 2014;9:e89125. doi: 10.1371/journal.pone.0089125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srivastava A, Gupta A-K, Datsenka T, Mattoo A-K, Avtar HK. Maturity and ripening-stage specific modulation of tomato (Solanum lycopersicum) fruit transcriptome. GM Crops. 2010;1:237–249. doi: 10.4161/gmcr.1.4.13737. [DOI] [PubMed] [Google Scholar]
- 39.Wehmeyer N, Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 2000;122:1099–1108. doi: 10.1104/pp.122.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medina-Escobar N, Cárdenas J, Muñoz-Blanco J, Caballero J-L. Cloning and molecular characterization of a strawberry fruit ripening-related cDNA corresponding a mRNA for a low-molecular-weight heat-shock protein. Plant Mol Biol. 1998;36:33–42. doi: 10.1023/A:1005994800671. [DOI] [PubMed] [Google Scholar]
- 41.Ding C, Wang C-Y, Gross K-C, Smith D-L. Reduction of chilling injury and transcript accumulation of heat shock proteins in tomato fruit by methyl jasmonate and methyl salicylate. Plant Sci. 2001;161:1153–1159. doi: 10.1016/S0168-9452(01)00521-0. [DOI] [Google Scholar]
- 42.Arce D-P, Krsticevic F-J, Bertolaccini M-R, Ezpeleta J, Ponce S-D, Tapia E. Analysis of small heat shock protein gene family expression (RNA-Seq) during the tomato fruit maturation. IFMBE Proc. 2015;49:679–682. doi: 10.1007/978-3-319-13117-7_173. [DOI] [Google Scholar]
- 43.Kuhn N, Guan L, Dai Z-W, Wu B-H, Lauvergeat V, Gomès E, et al. Berry ripening: recently heard through the grapevine. J Exp Bot. 2014;65:4543–4559. doi: 10.1093/jxb/ert395. [DOI] [PubMed] [Google Scholar]
- 44.Leng P, Yuan B, Guo Y-D. The role of abscisic acid in fruit ripening and responses to abiotic stress. J Exp Bot. 2014;65:4577–4588. doi: 10.1093/jxb/eru204. [DOI] [PubMed] [Google Scholar]
- 45.Qin X., Zeevaart J. A. D. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proceedings of the National Academy of Sciences. 1999;96(26):15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, et al. The Arabidopsis cytochrome P450 CYP707A encodes ABA 80-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao X, Li M, Liu B, Yan M, Yu X, Zi H, et al. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc Natl Acad Sci U S A. 2018;115:E11542–E11550. doi: 10.1073/pnas.1812575115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujita Y, Yoshida T, Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013;147:15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 49.Davies C, BOSS P-K, Robinson S-P. Treatment of crape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol. 1997;115:1155–1161. doi: 10.1104/pp.115.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wheeler S, Loveys B, Ford C, Davies C. The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust J Grape Wine Res. 2009;15:195–204. doi: 10.1111/j.1755-0238.2008.00045.x. [DOI] [Google Scholar]
- 51.Zhao P, Wang D-D, Wang R-Q, Kong N-N, Zhang C, Yang C-H, et al. Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics. 2018;19:61. doi: 10.1186/s12864-018-4443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ni L, Fu X-P, Zhang H, Li X, Cai X, Zhang P-P, et al. Abscisic acid inhibits rice protein phosphatase PP45 via H2O2 and relieves epression of the Ca2+/CaM-dependent protein kinase DMI3. Plant Cell. 2019;31:128–152. doi: 10.1105/tpc.18.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, Chen H, He Y, Xia R. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv [Preprint]. 2018;289660. 10.1101/289660.
- 54.Gu Z, Cavalcanti A, Chen F, Bouman P, Li W. Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol Biol Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Tang H, DeBarry J-D, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Downey M-O, Harvey J-S, Robinson S-P. Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of shiraz and chardonnay (Vitis vinifera L.) Aust J Grape Wine R. 2003;9:110–121. doi: 10.1111/j.1755-0238.2003.tb00261.x. [DOI] [Google Scholar]
- 58.Guo D-L, Xi F-F, Yu Y-H, Zhang X-Y, Zhang G-H, Zhong G-Y. Comparative RNA-Seq profiling of berry development between table grape 'Kyoho' and its early-ripening mutant 'Fengzao. BMC Genomics. 2016;17:795–812. doi: 10.1186/s12864-016-3051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmittgen T-D, Livak K-J. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Syntenic relationships among VvHSP20s genes. Different colors represent different chromosomes. Lines of different colors represent different tandem duplication genes.
Additional file 2: Table S1. Primers used for the qRT-PCR reactions.
Data Availability Statement
In this study, the grape genome sequence used to identify HSP20 genes were downloaded from Ensembl Plants Database (http://plants.ensembl.org/index.html). Expression data of VvHSP20 genes in grape used in this study can be accessed via the NCBI SRA database with accession numbers of PRJNA541089 from 5th May 2020 onwards, as until then there is an embargo due to a complementary manuscript. Until then, these sequences are available from the corresponding author upon reasonable request.