Abstract
Objective
The goal of the 2018 n2c2 shared task on cohort selection for clinical trials (track 1) is to identify which patients meet the selection criteria for clinical trials. Cohort selection is a particularly demanding task to which natural language processing and deep learning can make a valuable contribution. Our goal is to evaluate several deep learning architectures to deal with this task.
Materials and Methods
Cohort selection can be formulated as a multilabeling problem whose goal is to determine which criteria are met for each patient record. We explore several deep learning architectures such as a simple convolutional neural network (CNN), a deep CNN, a recurrent neural network (RNN), and CNN-RNN hybrid architecture. Although our architectures are similar to those proposed in existing deep learning systems for text classification, our research also studies the impact of using a fully connected feedforward layer on the performance of these architectures.
Results
The RNN and hybrid models provide the best results, though without statistical significance. The use of the fully connected feedforward layer improves the results for all the architectures, except for the hybrid architecture.
Conclusions
Despite the limited size of the dataset, deep learning methods show promising results in learning useful features for the task of cohort selection. Therefore, they can be used as a previous filter for cohort selection for any clinical trial with a minimum of human intervention, thus reducing the cost and time of clinical trials significantly.
Keywords: cohort selection, deep learning, multilabel text classification, convolutional neural network, recurrent neural network
INTRODUCTION
In biomedical research, a cohort is a group of patients who share a set of desired characteristics for a specific study. The fast and accurate selection of cohort patients is critical to the success of research studies, such as clinical trials or epidemiological studies.
The cohort definition is a very time-consuming task because of the large number of patient records that have to be manually reviewed by the researchers. This process is a very challenging problem due to multiple variations of how the information is recorded, medical coding mistakes, sparse data, or missing details, among other issues. Thus, a robust cohort definition requires careful reading of the patient records in order not to miss potential subjects for the study.
Automated natural language processing (NLP) methods for cohort selection can alleviate the manual workload burden for researchers by providing faster access to the relevant information in patient records. Therefore, NLP would facilitate tremendous advances in clinical research. Although there are some rule-based algorithms1,2 capable of identifying patients that satisfy specific criteria relevant to a study, they cannot be reused in other clinical trials. Classical machine learning classifiers can automatically learn rules to identify these patients. However, they still require human expertise to define the most informative feature set for the task. Consequently, there is a need to explore novel methods that can select the most suitable set of patients for any clinical trial, independently of the criteria used and with minimal human intervention. One of the most important advantages of deep learning is that it can automatically learn the most appropriate features from the raw text, without the need for manual work in the feature engineering task.
In this article, we present a comparative study of various popular deep learning architectures applied to the challenging task of cohort selection, posed as a multilabel text classification problem. A significant benefit of the deep learning architectures is that they require little domain knowledge, and thereby, they could be easily applied to identify the patient cohort of any clinical trial or study. Although our deep learning architectures are similar to those proposed in existing deep learning systems for text classification,3–6 we also study if the use of a deep fully connected feedforward (FFF) layer before the prediction layer could improve their results. We also compare random initialization and pretrained word embeddings to initialize our models.
In this section, we discuss the main works based on NLP for cohort selection. Segura-Bedmar et al7 aimed to develop new technologies for supporting epidemiological studies. In particular, they applied different classical machine learning classifiers and a convolutional neural network (CNN) network3 for the automatic detection of anaphylaxis (a severe allergic reaction) cases from a collection of 219 902 clinical records, of which <1% are anaphylaxis cases. Although the CNN model achieves a very high performance (F1 = 95.6%), a linear support vector machine provides the highest performance (F1 = 95.8%), with far less computational complexity.
Glicksberg et al8 used electronic phenotyping algorithms from the PheKB database2 for the automatic cohort selection of 5 diseases: dementia, herpes zoster, sickle cell, type 2 diabetes and attention-deficit/hyperactivity disorder. These algorithms consist of a set of rules based on concepts from terminologies such as the International Statistical Classification of Diseases-Ninth Revision (ICD-9) and can be considered as gold standard selection methods to perform cohort selection for these diseases. The authors also exploited an approach based on word embeddings. Concretely, each patient record was represented as an average of its word embeddings. Then, a similarity measure such as the cosine distance was used to propose the patient records more related to a given disease. The experimental results show that the approach based on word embeddings provides better performance than the phenotyping algorithms.
More recently, Antunes et al9 proposed 3 different approaches for the 2018 n2c2 track 1 on cohort selection: 1) a set of handcrafted rules to identify keywords specific to the criteria; 2) several classical machine learning classifiers (eg, AdaBoostClassifier, BaggingClassifier, DecisionTreeClassifier, GradientBoostingClassifier, XGBClassifier) on the clinical records, which are represented using the ICD-9 codes related to the criteria; and 3) 2 different deep learning methods: a neural network and a CNN, in which the clinical records are represented as matrices of word embeddings. The rule-based approach provides the best results for some criteria, but GradientBoostingClassifier obtains the best overall performance, with a micro-F1 of 0.8356 and a macro-F1 of 0.6517. Deep learning methods provide worse performance than classical classifiers. In particular, the CNN model yields a micro-F1 of 0.7676 and a macro-F1 of 0.4949. Although the best results were obtained by the rule-based method and the classical classifiers trained using ICD-9 codes, the creation of these rules as well as the selection of the most suitable ICD-9 codes for representing each of the selection criteria require a high level of medical expertise to analyze the clinical records. Moreover, these rules and ICD-9 codes can be only applied to the specific criteria defined for this shared task.
In this article, we explore several deep learning architectures, which have proved successful in the task of multilabel text classification, such as a simple CNN,3 a deep CNN,4 a recurrent neural network (RNN),5 and a hybrid architecture combining CNN and RNN.6 To the best of our knowledge, the simple CNN is the only deep learning architecture that has been used in the cohort selection task so far.
MATERIALS AND METHODS
Dataset
We used the dataset of the 2018 n2c2 shared task (track 1), focused on cohort selection for clinical trials. It consists of 311 patient records, which were manually labeled by experts to indicate whether a patient meets a possible criterion from a list of 13 criteria. Some of the 13 selection criteria are the following: Major-diabetes (which indicates whether the patient has a complication related to major-diabetes), Drug-abuse (whether the patient has a drug abuse problem), or Abdominal (whether the patient has a history of intra-abdominal surgery, small or large intestine resection or small bowel obstruction). The organizers split the dataset into 2 parts: training (203 records) and test (108 records) sets. Figure 1 shows the distribution of 13 criteria on the training and test datasets. As can be seen, both datasets are very unbalanced. It also shows that the test dataset has a very similar criteria distribution to the training dataset. Some criteria such as Mi-6Mos (myocardial infarction in the past 6 months), Keto-1yr (diagnosis of ketoacidosis in the past year), Alcohol-abuse (current alcohol use over weekly recommended limit), or Drug-abuse present a very small number of training instances, which makes extremely difficult to learn a model capable of accurately identifying these criteria in the patients records. The most common criteria are Make-decision (patient makes their own medical decisions), English (patient speaks English), and Asp-for-mi (use of aspirin to prevent myocardial infarction), with more than 150 instances in the training dataset.
Figure 1.
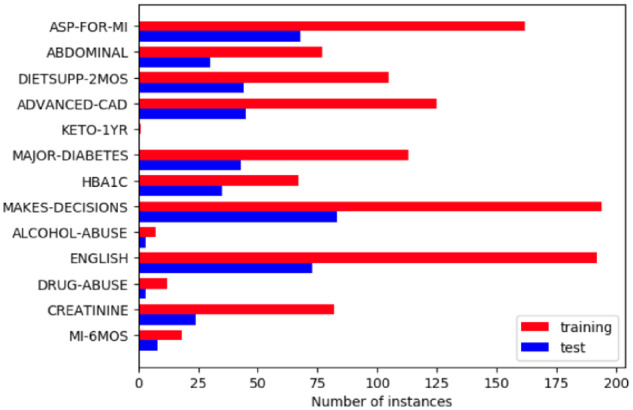
Criteria distribution.
Methods
Cohort selection can be modeled as a multilabel text classification problem, in which the goal is to identify the set of criteria (labels) that are met for each patient record. Compared with multiclass text classification, where each text is classified with a single class label, multilabel text classification is more difficult because there are many possible label combinations.
In this section, we describe several deep learning architectures to classify patient records according to the selection criteria defined in the track 1 of the 2018 n2c2 shared task. The 4 architectures proposed are 1) a simple CNN,3 2) a deep CNN consisting of 3 convolutional blocks,4 3) an RNN with gated recurrent units (GRUs),5 and 4) a hybrid model that comprises a CNN followed by a GRU RNN.6 These architectures have already shown promising results for text classification. To represent the patient records, we explore both random initialization and pretrained word embeddings. In particular, we use a model trained on a large corpus of biomedical and general-domain texts.10
Unlike the original architectures, we also explore the use of a FFF layer before the classification layer. This layer allows us to map the features into a higher-order feature space, which is more easily separable into the different labels. As our goal is to predict multiple labels (selection criteria) for each patient record, the output layer of all our architectures uses the sigmoid activation function, which gives us independent probabilities for each label (criterion). Thus, the output layer has 1 sigmoid output unit per label.
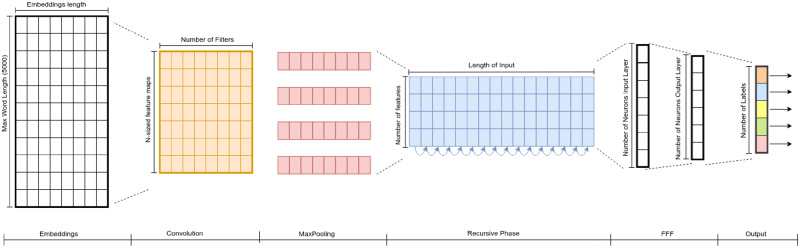
The first of our 4 deep learning approaches is an extension of the CNN architecture3 for sentence classification. This architecture proved to be successful in discovering discriminative and meaningful phrases in a text, which could be useful for the selection criteria task. Each patient record is represented as a matrix of word embeddings. As each record has a different number of words, we studied the distribution of the number of words in the dataset and saw that only 10% of records have more than 5000 words. Thus, we only consider the 5000 first tokens for each cleaned record and pad the shorter records. Then, each record is represented as a matrix of word embeddings with dimension n x k, where n is the number of words (n = 5000) and k is the dimension size of word embeddings. In our case, k is set to 200, which is the dimension of the pretrained word embeddings used in this article. The first layer after the embeddings is a convolutional layer that applies a series of filters of different sizes on the input matrix. The filters slide over the input matrix, producing feature vectors, which are the input of the max-pooling layer. This layer takes the maximum values from the different filters, thus capturing the most relevant features. As said above, our CNN architecture also adds a FFF layer before the last layer, which takes as input the feature vector produced by the max-pooling layer.
As a second approach, we use the deep CNN architecture,4 which applies multiple sequential convolutional layers. Our goal is to study if a deeper CNN model can provide better performance for the cohort selection task than the simple CNN architecture, described previously. Owing to our computational limitations, we could only build a deep CNN model consisting of 3 convolutional blocks with filter sizes of 64, 128, and 256, respectively. In turn, each block contains 2 consecutive convolutional layers and is followed by a max-pooling layer. The deep CNN also integrates an FFF layer before the prediction layer.
Our third approach is based on an RNN, which processes the input text token by token, storing the semantics of the previous tokens in a hidden layer. RNN is capable of capturing contextual information and long-term dependencies,5 which is very important in our case since the clues about the different selection criteria could appear anywhere in a patient record. There are several types of RNN, such as long short-term memory networks (LSTMs)11 or GRUs.12 Both networks use several gates to decide what information should be passed to the output. In this way, the networks can remove irrelevant information, but keep the discriminatory information for the final prediction. While LSTMs have 3 gates (input, output, and forget gates), GRUs only use 2 gates (update and reset gate). In this work, we use GRUs because LSTM units are not appropriate for processing whole long documents,13 as is our case. Moreover, GRUs have shown to achieve better performance on smaller datasets.12 We also add an FFF layer before the last layer.
Our last approach is a hybrid CNN-RNN architecture based on the system proposed by Chen et al.6 CNN is used to learn the features to represent the patient records. Then, these features are the input of an RNN, which is applied to multilabel prediction. However, instead of using LSTM units, we apply an RNN with GRUs. Figure 2 shows this hybrid architecture extended with the FFF layer before the prediction layer.
Figure 2.
Convolutional neural network-recurrent neural network hybrid architecture. FFF: fully connected feedforward.
Our source code is publicly available (https://github.com/PRaezUC3M/cohort_selection/) to enable the reproducibility of our experiments.
RESULTS
Our experiments aim to answer 3 questions: 1) do the pretrained word embeddings overcome the random initialization? 2) does the use of an FFF layer in our deep learning architectures improve the results? and 3) what is the best deep learning architecture for the task of cohort selection?
Precision, recall, and F1 scores are standard metrics to evaluate the performance of a binary classification problem. These metrics can be extended to a multilabeling setting by calculating their macro-averaged and micro-averaged versions. In the macro-average method, labels are equally treated because we compute the metric for each label and then calculate the average. In the micro-average method, we count all true positives, false negatives, and false positives for all labels and then compute the metrics using these values. This method is more suitable for unbalanced datasets, as in our case. Since some criteria (labels) in the dataset are very unbalanced (see Figure 1), the micro-averaged F1 is the most appropriate metric for comparing the different models among them. Moreover, we use the Friedman test to determine if there are significant differences among the models studied in this article.
We used cross-validation for model hyperparameter tuning and performed a grid search study to determine the best parameters of each one of the deep learning architectures studied in this article. Table 1 shows a summary of the parameters used in each architecture. Loss functions are used to measure the error between the output and the predicted output of a model to learn the model parameters in the training phase. To train our models, we apply binary cross-entropy as the loss function because it is more suitable for multilabel problems.13 We also use Adam optimizer for faster convergence.14
Table 1.
Summary of best parameters for our deep learning models
| Method | Hyperparameters |
|---|---|
| CNN |
|
| Deep CNN |
|
| RNN (GRU) |
|
| CNN+RNN |
|
CNN: convolutional neural network; FFF: fully connected feedforward; GRU: gated recurrent unit; RNN: recurrent neural network.
To answer question 1, we compare the effect of random initialization with the pretrained word embeddings. Moreover, to answer question 2, we also study the impact of using an FFF layer in the 4 architectures. Table 2 shows the scores for the simple and deep CNN models. Table 3 shows the results for the RNN and hybrid models.
Table 2.
F1 scores for the simple and deep CNN architectures.
| CNN |
Deep CNN |
|||||||
|---|---|---|---|---|---|---|---|---|
| Without FFF |
With FFF |
Without FFF |
With FFF |
|||||
| Random | Pretrained | Random | Pretrained | Random | Pretrained | Random | Pretrained | |
| Abdominal | 0.5486 | 0.5764a | 0.5764a | 0.4444 | 0.3617 | 0.4886 | 0.3878 | 0.3878 |
| Advanced-cad | 0.3844 | 0.3182 | 0.6411a | 0.3333 | 0.4223 | 0.3182 | 0.3478 | 0.3478 |
| Alcohol-abuse | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 |
| Asp-for-mi | 0.434 | 0.434 | 0.434 | 0.434 | 0.434 | 0.434 | 0.434 | 0.434 |
| Creatinine | 0.5111 | 0.5238a | 0.5111 | 0.4976 | 0.3878 | 0.4118 | 0.4118 | 0.4118 |
| Dietsupp-2mos | 0.4643 | 0.3973 | 0.4258 | 0.4327 | 0.457 | 0.4643 | 0.4712a | 0.4667 |
| Drug-abuse | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 |
| English | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 |
| Hba1c | 0.5581a | 0.4991 | 0.4 | 0.5342 | 0.4 | 0.4 | 0.4 | 0.4 |
| Keto-1yr | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Major-diabetes | 0.4505 | 0.5694a | 0.55 | 0.4444 | 0.4171 | 0.4976 | 0.3478 | 0.3478 |
| Makes-decisions | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 |
| Mi-6mos | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 |
| Overall(micro) | 0.7527 | 0.7515 | 0.7721a | 0.7372 | 0.7347 | 0.7514 | 0.7611 | 0.76 |
| Overall(macro) | 0.4819 | 0.4794 | 0.4963a | 0.4642 | 0.4456 | 0.456 | 0.4395 | 0.4392 |
Random means that random initialization was used. Pretrained means that pretrained word embeddings were used.
CNN: convolutional neural network; FFF: fully connected feedforward.
Best score.
Table 3.
F1 scores for the RNN and hybrid architectures.
| RNN |
CNN+RNN |
|||||||
|---|---|---|---|---|---|---|---|---|
| Without FFF |
With FFF |
Without FFF |
With FFF |
|||||
| Random | Pretrained | Random | Pretrained | Random | Pretrained | Random | Pretrained | |
| Abdominal | 0.3878 | 0.3878 | 0.4792a | 0.3878 | 0.3878 | 0.4792a | 0.3878 | 0.3878 |
| Advanced-cad | 0.3478 | 0.4222 | 0.3478 | 0.3478 | 0.4994a | 0.3478 | 0.4034 | 0.3844 |
| Alcohol-abuse | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 | 0.4915 |
| Asp-for-mi | 0.434 | 0.434 | 0.434 | 0.434 | 0.434 | 0.434 | 0.434 | 0.5238a |
| Creatinine | 0.5249 | 0.4118 | 0.6104 | 0.4118 | 0.6889a | 0.5581 | 0.5833 | 0.5833 |
| Dietsupp-2mos | 0.4886 | 0.4857 | 0.6296 | 0.7333a | 0.5662 | 0.6703 | 0.5982 | 0.4976 |
| Drug-abuse | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 |
| English | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 | 0.4737 |
| Hba1c | 0.4 | 0.4 | 0.4 | 0.4 | 0.4792 | 0.4 | 0.4 | 0.4 |
| Keto-1yr | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Major-diabetes | 0.3478 | 0.5543 | 0.4665 | 0.3478 | 0.6122 | 0.4665 | 0.6296a | 0.4994 |
| Makes-decisions | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 |
| Mi-6mos | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 | 0.4828 |
| Overall(micro) | 0.7623 | 0.7639 | 0.7843 | 0.7811 | 0.7827 | 0.7856a | 0.779 | 0.7233 |
| Overall(macro) | 0.4496 | 0.4622 | 0.4831 | 0.4597 | 0.5062a | 0.4823 | 0.4884 | 0.4761 |
Random means that random initialization was used. Pretrained means that pretrained word embeddings were used.
CNN: convolutional neural network; FFF: fully connected feedforward; RNN: recurrent neural network.
Best score.
DISCUSSION
We first discuss the results for the simple CNN architecture (see Table 2). If the FFF layer is not used, random initialization and the pretrained word embeddings show very similar results without statistically significant differences (see Table 4). However, when the simple CNN architecture includes an FFF layer, the pretrained word embeddings provide worse overall scores than random initialization, with a 3.4% decrease of micro-F1 and 3.2% in macro-F1. Although there are no statistical differences among all the simple CNN models (see Table 4), the results show that the simple CNN architecture extended with the FFF layer and trained with random initialization obtain the best micro-F1 (77.21%) and macro-F1 (49.68%) compared with the other simple CNN models.
Table 4.
P-values for all the deep learning models
| CNN + pretrained | CNN + random + FFF | CNN + pretrained + FFF | DeepCNN + random | DeepCNN + pretrained | DeepCNN + random + FFF | DeepCNN + pretrained + FFF | RNN + random | RNN + pretrained | RNN + random + FFF | RNN + pretrained + FFF | Hybrid + random | Hybrid + pre | Hybrid + random + FFF | Hybrid + pretrained + FFF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CNN + random | .18924 | .07019 | .25179 | .03824a | .02272a | .34299 | .03767a | .7925 | .26414 | .68716 | .83793 | .53839 | .5492 | .2787 | .72477 |
| CNN + pretrained | .09341 | .06587 | .16563 | .09075 | .55969 | .43064 | .33058 | .43149 | .12645 | .08502 | .1452 | .26052 | .15295 | .63016 | |
| CNN + random + FFF | .30045 | .08186 | .17892 | .85835 | .48875 | .34678 | .47327 | .36786 | .19774 | .55937 | .29144 | .44913 | .90165 | ||
| CNN + pretrained + FFF | .45383 | .24682 | .59507 | .25383 | .62239 | .76786 | .57572 | .4946 | .16991 | .45395 | .07955 | .40593 | |||
| DeepCNN + random | .03394a | .74826 | .38392 | .7515 | .68382 | .85001 | .77073 | .67344 | .4473 | .74103 | .68865 | ||||
| DeepCNN + pretrained | .2581 | .02051 | .60709 | .47757 | .23961 | .29475 | .73865 | .85756 | .58568 | .69499 | |||||
| DeepCNN + random + FFF | .02415a | .30862 | .16687 | .23987 | .5289 | .33919 | .27965 | .43141 | .38983 | ||||||
| DeepCNN + pretrained + FFF | .73404 | .22673 | .34101 | .37431 | .41331 | .52991 | .26371 | .48985 | |||||||
| RNN + random | .14826 | .04582a | .22634 | .09698 | .39048 | .22868 | .23598 | ||||||||
| RNN + pretrained | .17518 | .39877 | .22112 | .33279 | .12925 | .21353 | |||||||||
| RNN + random + FFF | .03359a | .46653 | .444 | .22266 | .26722 | ||||||||||
| RNN + pretrained + FFF | .50063 | .27616 | .50124 | .28188 | |||||||||||
| Hybrid + random | .15912 | .03474a | .06604 | ||||||||||||
| Hybrid + pre | .08122 | .1824 | |||||||||||||
| Hybrid + random + FFF | .13658 |
CNN: convolutional neural network; FFF: fully connected feedforward; RNN: recurrent neural network.
Statistically different at a level of significance of 0.05.
If the simple CNN architecture does not uses the FFF layer, the pretrained word embeddings obtain significant improvements in scores for some criteria such as Abdominal, Creatinine, and Major-diabetes, with increases of 2.7%, 1.2%, and 11.8% on F1, respectively. On the other hand, some criteria such as Advanced-cad, Dietsupp-2mos, and Hba1c show better results when the simple CNN model without the FFF layer was trained with random initialization (see Table 2), showing increases of 6.6%, 6.7%, and 5.9% in F1, respectively. If the simple CNN architecture is extended with the FFF layer, random initialization provides statistically significant better scores than the pretrained word embeddings for some criteria such as Abdominal, Advanced-Cad, Creatinine, and Major diabetes, with improvements of 13.2%, 30.7%, 1.3%, and 10.5% in F1, respectively. On the other hand, the pretrained word embeddings only increase the performance for Hba1c (13.4%) and achieve a marginal improvement of 0.69% for Dietsupp-2mos. For the rest of the criteria, the differences are not statistically significant (see Supplementary Appendix Tables 3–8). Therefore, the combination of the pretrained word embeddings and the FFF layer produces a negative impact on the scores for the most criteria. More research needs to be done to understand why some criteria perform better with random initialization. A possible cause could be that these criteria may be described with words that are not presented in the pretrained word embedding model used in this work.
We now analyze the results provided by the deep CNN models. The use of the FFF layer provides better performance than without using it (see Table 2). Using this layer, random initialization and the pretrained word embeddings show very similar results. When the model uses random initialization, this layer obtains an increase of 2.5% on micro-F1, but with a marginal decrease of 0.6% on macro-F1. When the model uses the pretrained word embeddings, the FFF layer provides a slight improvement of only 0.8% on micro-F1, but with a decrease of 1.6% on macro-F1. If the FFF layer is not used, the pretrained word embeddings provide a significant improvement of 1.6% on micro-F1 and 1.4% on macro-F1 compared with random initialization. Comparing the 4 models of the deep CNN architecture, the best micro-F1 (76.11%) is obtained by the model trained with random initialization and using the FFF layer. The deep CNN without the FFF layer provides the best macro-F1 (45.6%) when it is trained using the pretrained word embeddings.
The RNN architecture with the FFF layer shows improvements in the overall scores (see Table 3), with an increase of 2.2% on micro-F1 and 3.3% on macro-F1 for random initialization. The same positive effect is obtained for the pretrained word embeddings, with an improvement of 1.72% on micro-F1, but with a marginal drop of 0.25% on macro-F1. The RNN architecture extended with the FFF layer and trained with random initialization provides the best overall micro-F1 (78.43%) and macro-F1 (48.31%). For the RNN architecture without using the FFF layer, random initialization and the pretrained word embeddings show very similar micro-F1, but with an increase of 1.2% on macro-F1 when the pretrained word embeddings are used. They also obtain significant improvements for the criteria Major-diabetes and Advanced-cad, with increases of 20.6% and 7.4% on F1, respectively. On the other hand, Creatinine shows a significant drop of 11.3% in F1. Moreover, when the architecture uses the FFF layer, random initialization provides better performance for some criteria such as Abdominal, Creatinine, and Major-diabetes, with significant improvements of 9.1%, 19.8%, and 11.8% in F1, respectively.
Finally, we discuss the results provided by the hybrid architecture combining a CNN and an RNN (see Table 3). When the FFF layer is not used, random initialization and the pretrained word embeddings show very similar overall micro-F1 (around 78%), but with an increase of 2.3% on macro-F1 when using random initialization. Some criteria such as Abdominal and Dietsupp-2mos show significant improvements of 9, 1% and 10.4% respectively when using the pretrained word embeddings. On the other hand, random initialization achieves significantly better performance for the Advanced-cad, Creatinine, and Major-diabetes with increases of 15.1%, 13%, and 14.5%, respectively, over the scores given by the pretrained word embeddings. Unlike the previous architectures, the use of the FFF layer has a negative impact on the performance of the hybrid architecture, leading to a decrease of 6.2% on micro-F1 and 1% on macro-F1 when the pretrained word embeddings are used. However, the reduction is smaller for random initialization (0.3% on micro-F1 and 1.7% on macro-F1). The combination of pretrained word embeddings plus the FFF layer shows a substantial decrease in performance compared with the other hybrid models. The overall results indicate that the best hybrid model (micro-F1 = 78.27% and macro-F1 = 50.62%) is trained with random initialization and does not use the FFF layer.
As usual, the macro-F1 scores are lower than their corresponding micro-F1 for all models. Classifiers tend to perform worse in the classes with few instances. Therefore, their F1 scores negatively impact the overall score. We can see that the hybrid approach gives the top macro-F1 (50.62%).
All the models show low performance for the minor criteria such as Alcohol-abuse, Drug-abuse, and Mi-6Mos, which may be due to the high imbalance between their positive and negative instances. Contrary to what was expected, the models do not work well for the criteria that are present in most patient records such as Make-decisions, English, or Asp-for-mi. More research needs to be done to find out its cause. On the other hand, as expected, the balanced criteria such as Advanced-cad (advanced cardiovascular disease), Dietsupp-2mos (taken a dietary supplement in the past 2 months), or Major-diabetes often obtain the best results.
We now answer question 3. The RNN and hybrid architectures provide the best overall results, with micro-F1 around 78% and macro-F1 around 49%. The simple CNN and deep CNN provide slightly lower overall results. Although CNN can identify more discriminative contextual features through its convolutional and max-pooling layers, small filters in the convolutional layer could produce the loss of long-distance patterns, while large filters could cause data scarcity. The use of more than 2 convolutional layers in the deep CNN not only improves the results, but also increases the training time. On the other hand, in terms of time complexity, a CNN with a single convolutional layer is more efficient than the other deep learning models, because it requires a lot less training time.
As was shown previously, Antunes et al9 also exploited a CNN with a word embeddings model trained on the MIMIC-III (Medical Information Mart for Intensive Care III) database, with around 2 billion clinical records. However, this system reported a micro-F1 of 76.76% on the test dataset of the n2c2 cohort selection. Our simple CNN architecture extended with the FFF layer and trained with random initialization obtains a micro-F1 of 77.21%. Likewise, the hybrid and RNN architectures also provide better performance (with micro-F1 around 78%) than the CNN model proposed by Antunes et al.9
We use the Friedman test at 95% to determine if there is statistically difference in results among the models studied in this article. Some observations we can draw from Table 4 include the following:
There are no statistical differences for most of the models.
There are no statistical differences between the 4 models based on the simple CNN architecture. Therefore, neither the FFF layer nor the pretrained word embeddings improve the performance of the simple CNN architecture significantly.
The simple CNN trained with random initialization and without using the FFF layer (FFF) has significant differences with all the deep CNN models, except that trained with random initialization and without the FFF layer. The simple CNN does not present statistical differences with the rest of the models.
If the deep CNN model does not use the FFF layer, the pretrained word embeddings provide significantly better results than random initialization.
In the RNN architecture with random initialization, the use of the FFF layer shows significant better results than those provided by this architecture without using this layer.
If the RNN architecture adds the FFF layer, random initialization provides a significant improvement in macro-F1 compared with that provided using the pretrained word embeddings.
In the hybrid model using random initialization, there are statistical differences when the FFF layer is used.
CONCLUSION
The success of epidemiological studies and clinical trials depends on the selection of the right patients. The medical researchers must perform this selection carefully by the careful analysis of an enormous amount of information from different sources. This process is an expensive and time-consuming task. Rules-based methods or machine learning classifiers exploiting the ICD-9 codes related to the selection criteria can be applied to alleviate the burden of medical researchers in the cohort selection. However, the creation of these rules or the selection of the most representative ICD-9 codes related to the selection criteria still requires extensive additional work involving experts. Thus, in this work, we have explored several deep learning architectures because they can automatically learn the most appropriate features by themselves without any human intervention and prior knowledge.
In this article, we have proposed an extension of 4 deep learning architectures, which have already proved successful for text classification. In particular, we have added a fully connected feed forward layer before their prediction layers. The results show that this layer provides better results for all the architectures, except for the CNN-RNN hybrid architecture. We have also compared random initialization and pretrained word embeddings to initialize our models. Only the deep CNN architecture obtains a significant improvement when pretrained word embeddings are used. Comparing the 4 architectures studied, the hybrid architecture obtains the best results, closely followed by RNN.
To the best of our knowledge, deep learning methods have not been applied to this task, except the CNN model proposed by Antunes et al.9 Our hybrid architecture provides an improvement of almost 2% on micro-F1 compared with this system. However, our results are worse than those provided by the traditional machine learning classifiers proposed in that work. This may be due to the tiny size of the dataset. Indeed, the main limitation of this study is the limited size of the dataset. With a larger dataset, the performance of the deep learning methods would likely be improved significantly.
As future work, we plan to explore semisupervised deep learning techniques to overcome the lack of a sufficient number of training examples. A notable shortcoming of the classical approaches for multilabel text classification is that labels are considered as independent units.3 However, they usually can present strong dependencies among them, especially in the context of clinical trials, in which some patient conditions can be strongly related. For example, criteria such as smoker and lung cancer often occur together in the patient records. Therefore, we also plan to perform a study about if deep learning methods can detect the label dependencies.
FUNDING
This work was supported by the Research Program of the Ministry of Economy and Competitiveness, Government of Spain, grant number TIN2017-87548-C2-1-R (DeepEMR project).
AUTHOR CONTRIBUTIONS
ISB provided the original idea, led the study, conducted the study design, conducted the literature review, helped with some developing tasks, conducted results analysis, and drafted the manuscript. PR developed the models, performed the experiments, helped with the literature review, added critical discussion points, and edited the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement
None declared.
Supplementary Material
REFERENCES
- 1. Gottesman O, Kuivaniemi H, Tromp G, et al. The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet Med 2013; 1510: 761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23: 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim Y. Convolutional neural networks for sentence classification. In: 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014: 1746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conneau A, Schwenk H, Barrault L, Lecun Y.. Very deep convolutional networks for text classification. In: 15th Conference of the European Chapter of the Association for Computational Linguistics; 2017: 1107–16. [Google Scholar]
- 5. Lai S, Xu L, Liu K, Zhao J.. Recurrent convolutional neural networks for text classification. In: Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence; 2015: 2267–73. [Google Scholar]
- 6. Chen G, Ye D, Xing Z, Chen J, Cambria E.. Ensemble application of convolutional and recurrent neural networks for multi-label text categorization. In: 2017 International Joint Conference on Neural Networks (IJCNN); 2017: 2377–83. [Google Scholar]
- 7. Segura-Bedmar I, Colón-Ruíz C, Tejedor-Alonso MA, Moro-Moro M.. Predicting of anaphylaxis in big data EMR by exploring machine learning approaches. J Biomed Inform 2018; 87: 50–9. [DOI] [PubMed] [Google Scholar]
- 8. Glicksberg BS, Miotto R, Johnson KW, Shameer K, Li L, Dudley Chen R. Automated disease cohort selection using word embeddings from electronic health records. Pac Symp Biocomput 2018; 23: 145–56. [PMC free article] [PubMed] [Google Scholar]
- 9. Antunes R, Figueira Silva J, Pereira A, Matos S.. Rule-based and machine learning hybrid system for patient cohort selection. In: 12th International Joint Conference on Biomedical Engineering Systems and Technologies BIOSTEC; 2019. [Google Scholar]
- 10. Pyysalo S, Ginter F, Moen H, Salakoski T, Ananiadou S.. Distributional semantics resources for biomedical text processing. In: Proceedings of LBM; 2013: 39–44. [Google Scholar]
- 11. Hochreiter S, Schmidhuber J.. Long short-term memory. Neural Comput 1997; 98: 1735–80. [DOI] [PubMed] [Google Scholar]
- 12. Cho K, Van Merriënboer B, Gulcehre C, et al. Learning phrase representations using RNN encoder–decoder for statistical machine translation. In: Empirical Methods in Natural Language Processing (EMNLP); 2014: 1724–34. [Google Scholar]
- 13. Dai AM, Qv L.. Semi-supervised sequence learning. In: Advances in Neural Information Processing Systems 28 (NIPS 2015); 2015: 3079–87. [Google Scholar]
- 14.Kingma DP, Ba J. Adam: A method for stochastic optimization. In: International Conference on Learning Representations (ICLR 2015); 2015:1–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.