Abstract
Objective
Neural network–based representations (“embeddings”) have dramatically advanced natural language processing (NLP) tasks, including clinical NLP tasks such as concept extraction. Recently, however, more advanced embedding methods and representations (eg, ELMo, BERT) have further pushed the state of the art in NLP, yet there are no common best practices for how to integrate these representations into clinical tasks. The purpose of this study, then, is to explore the space of possible options in utilizing these new models for clinical concept extraction, including comparing these to traditional word embedding methods (word2vec, GloVe, fastText).
Materials and Methods
Both off-the-shelf, open-domain embeddings and pretrained clinical embeddings from MIMIC-III (Medical Information Mart for Intensive Care III) are evaluated. We explore a battery of embedding methods consisting of traditional word embeddings and contextual embeddings and compare these on 4 concept extraction corpora: i2b2 2010, i2b2 2012, SemEval 2014, and SemEval 2015. We also analyze the impact of the pretraining time of a large language model like ELMo or BERT on the extraction performance. Last, we present an intuitive way to understand the semantic information encoded by contextual embeddings.
Results
Contextual embeddings pretrained on a large clinical corpus achieves new state-of-the-art performances across all concept extraction tasks. The best-performing model outperforms all state-of-the-art methods with respective F1-measures of 90.25, 93.18 (partial), 80.74, and 81.65.
Conclusions
We demonstrate the potential of contextual embeddings through the state-of-the-art performance these methods achieve on clinical concept extraction. Additionally, we demonstrate that contextual embeddings encode valuable semantic information not accounted for in traditional word representations.
Keywords: clinical concept extraction, contextual embeddings, language model
INTRODUCTION
Concept extraction is the most common clinical natural language processing (NLP) task1–4 and a precursor to downstream tasks such as relations,5 frame parsing,6 co-reference,7 and phenotyping.8,9 Corpora such as those from Informatics for Integrating Biology and the Bedside (i2b2),10–12 ShARe/CLEF,13,14 and SemEval15–17 act as evaluation benchmarks and datasets for training machine learning (ML) models. Meanwhile, neural network–based representations continue to advance nearly all areas of NLP, from question answering18 to named entity recognition (a close analogue of concept extraction).3,19–22 Recent advances in contextual representations, including ELMo23 and BERT,24 have pushed performance even further. These have demonstrated that relatively simple downstream models using contextual embeddings can outperform complex models25 using embeddings such as word2vec26 and GloVe.27
In this article, we aim to explore the potential impact these representations have on clinical concept extraction. Our contributions include the following:
An evaluation exploring numerous embedding methods: word2vec,26 GloVe,27 fastText,28 ELMo,23 and BERT.24
An analysis covering 4 clinical concept corpora, demonstrating the generalizability of these methods.
A performance increase for clinical concept extraction that achieves state-of-the-art results on all 4 corpora.
A demonstration of the effect of pretraining on clinical corpora vs larger open-domain corpora, an important trade-off in clinical NLP.29
A detailed analysis of the effect of pretraining time when starting from prebuilt open-domain models, which is important due to the long pretraining time of methods such as ELMo and BERT.
In the following sections, we introduce the theoretical knowledge that supports the shift from word-level embeddings to contextual embeddings.
Word embedding models
Word-level vector representation methods learn a real-valued vector to represent a single word. One of the most prominent methods for word-level representation is word2vec.26 So far, word2vec has widely established its effectiveness for achieving state-of-the-art performances in a variety of clinical NLP tasks.30 GloVe27 is another unsupervised learning approach to obtain a vector representation for a single word. Unlike word2vec, GloVe is a statistical model that aggregates both a global matrix factorization and a local context window. The learning relies on dimensionality reduction on the co-occurrence count matrix based on how frequently a word appears in a context. fastText28 is also an established library for word representations. Unlike word2vec and GloVe, fastText considers individual words as character n-grams. For instance, cold is made of the n-grams c, co, col, cold, o, ol, old, l, ld, and d. This approach enables handling of infrequent words that are not present in the training vocabulary, alleviating some out-of-vocabulary issues.
However, the effectiveness of word-level representations is hindered by the limitation that they conflate all possible meanings of a word into a single representation and so the embedding is not adjusted to the surrounding context. In order to tackle these deficiencies, advanced approaches have attempted to directly model the word's context into the vector representation. Figure 1 illustrates this with the word cold, in which a traditional word embedding assigns all senses of the word cold with a single vector, whereas a contextual representation varies the vector based on its meaning in context (eg, cold temperature, medical symptom or condition, an unfriendly disposition). Although a fictional figure is shown here, we later demonstrate this on real data.
Figure 1.
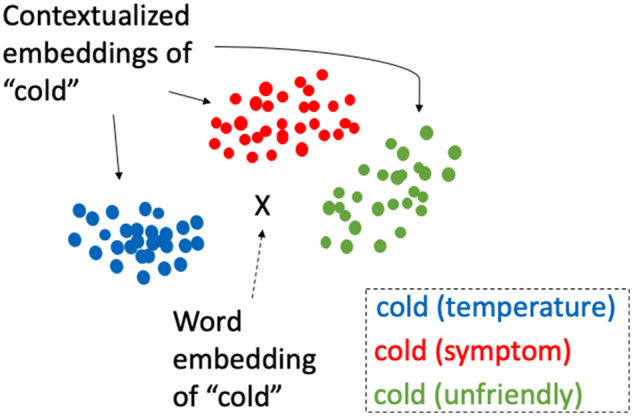
Fictional embedding vector points and clusters of cold.
The first contextual word representation that we consider to overcome this issue is ELMo.23 Unlike the previously mentioned traditional word embeddings that constitute a single vector for each word and the vector remains stable in downstream tasks, this contextual word representation can capture the context information and dynamically alter a multilayer representation. At training time, a language model objective is used to learn the context-sensitive embeddings from a large text corpus. The training step of learning these context-sensitive embeddings is known as pretraining. After pretraining, the context-sensitive embedding of each word will be fed into the sentences for downstream tasks. The downstream task learns the shared weights of the inner state of pretrained language model by optimizing the loss on the downstream task.
BERT24 is also a contextual word representation model, and, similar to ELMo, pretraining on an unlabeled corpus with a language model objective. Compared to ELMo, BERT is deeper in how it handles contextual information due to a deep bidirectional transformer for encoding sentences. It is based on a transformer architecture employing self-attention.31 The deep bidirectional transformer is equipped with multiheaded self-attention to prevent locality bias and to achieve long-distance context comprehension. Additionally, in terms of the strategy for how to incorporate these models into the downstream task, ELMo is a feature-based language representation while BERT is a fine-tuning approach. The feature-based strategy is similar to traditional word embedding methods that considers the embedding as input features for the downstream task. The fine-tuning approach, on the other hand, adjusts the entire language model on the downstream task to achieve a task-specific architecture. So while the ELMo embeddings may be used as the input of a downstream model, with the BERT fine-tuning method, the entire BERT model is integrated into the downstream task. This fine-tuning strategy is more likely to make use of the encoded information in the pretrained language models.
Clinical concept extraction
Clinical concept extraction is the task of identifying medical concepts (eg, problem, test, treatment) from clinical notes. This is typically considered as a sequence tagging problem to be solved with machine learning–based models (eg, conditional random field [CRF]) using hand-engineered clinical domain knowledge as features.4,32 Recent advances have demonstrated the effectiveness of deep learning–based models with word embeddings as input. Up to now, the most prominent model for clinical concept extraction is a bidirectional long short-term memory (Bi-LSTM) with CRF architecture.19,22,33 The bidirectional LSTM-based recurrent neural network captures both forward and backward information in the sentence and the CRF layer considers sequential output correlations in the decoding layer using the Viterbi algorithm.
Most similar to this article, several recent works have applied contextual embedding methods to concept extraction, both for clinical text and biomedical literature. For instance, ELMo has shown excellent performance on clinical concept extraction.34 BioBERT35 applied BERT primarily to literature concept extraction, pretraining on MEDLINE abstracts and PubMed Central articles, but also applied this model to the i2b2 2010 corpus10 without clinical pretraining (we include BioBERT in our experiments). A recent preprint by Alsentzer et al36 pretrains on MIMIC-III (Medical Information Mart for Intensive Care III), similar to our work, but achieves lower performance on the 2 tasks in common, i2b2 2010 and 2012.11 Their work does suggest potential value in only pretraining on MIMIC-III discharge summaries, as opposed to all notes, as well as combining clinical pretraining with literature pretraining. Finally, another recent preprint proposes the use of BERT not for concept extraction, but rather for clinical prediction tasks such as 30-day readmission prediction.37
MATERIALS AND METHODS
In this article, we consider both off-the-shelf embeddings from the open domain as well as pretraining clinical domain embeddings on clinical notes from MIMIC-III,38 which is a public database of intensive care unit patients.
For the traditional word-embedding experiments, the static embeddings are fed into a Bi-LSTM CRF architecture. All words that occur at least 5 times in the corpus are included and infrequent words are denoted as UNK. To compensate for the loss due of those unknown words, character embeddings for each word are included.
For ELMo, the context-independent embeddings with trainable weights are used to form context-dependent embeddings, which are then fed into the downstream task. Specifically, the context-dependent embedding is obtained through a low-dimensional projection and a highway connection after a stacked layer of a character-based convolutional neural network (char-CNN) and a 2-layer Bi-LSTM language model (bi-LM). Thus, the contextual word embedding is formed with a trainable aggregation of highly-connected bi-LM. Because the context-independent embeddings already consider representation of characters, it is not necessary to learn a character embedding input for the Bi-LSTM in concept extraction. Finally, the contextual word embedding for each word is fed into the prior state-of-the-art sequence labeling architecture, Bi-LSTM CRF, to predict the label for each token.
For BERT, both the BERTBASE and BERTLARGE off-the-shelf models are used with additional Bi-LSTM layers at the top of the BERT architecture, which we refer to as BERTBASE(General) and BERTLARGE(General), respectively. For background, the BERT authors released 2 off-the-shelf cased models: BERTBASE and BERTLARGE, with 110 million and 340 million total parameters, respectively. BERTBASE has 12 layers of transformer blocks, 768 hidden units, and 12 self-attention heads, while BERTLARGE has 24 layers of transformer blocks, 1024 hidden units, and 16 self-attention heads. So BERTLARGE is both “wider” and “deeper” in model structure, but is otherwise essentially the same architecture. The models initiated from BERTBASE(General) and BERTLARGE(General) are fine-tuned on the downstream task (ie, clinical concept recognition in our case). Because BERT integrates sufficient label-correlation information, the CRF layer is abandoned and only a Bi-LSTM architecture is used for sequence labeling. Additionally, 2 clinical domain embedding models are pretrained on MIMIC-III, initiated from the BERTBASE and BERTLARGE checkpoints, which we refer to as BERTBASE(MIMIC) and BERTLARGE(MIMIC), respectively.
Datasets
Our experiments are performed on 4 widely-studied shared tasks, the 2010 i2b2/VA challenge,10 the 2012 i2b2 challenge,11 the SemEval 2014 Task 7,15 and the SemEval 2015 Task 14.16 The descriptive statistics for the datasets are shown in Table 1. The 2010 i2b2/VA challenge data contain a total of 349 training and 477 testing reports with clinical concept types: PROBLEM, TEST, and TREATMENT. The 2012 i2b2 challenge data contain 190 training and 120 testing discharge summaries, with 6 clinical concept types: PROBLEM, TEST, TREATMENT, CLINICAL DEPARTMENT, EVIDENTIAL and OCCURRENCE. The SemEval 2014 Task 7 data contain 199 training and 99 testing reports with the concept type: DISEASE DISORDER. The SemEval 2015 Task 14 data consists of 298 training and 133 testing reports with the concept type: DISEASE DISORDER. For the 2 SemEval tasks, the disjoint concepts are handled with “BIOHD” tagging schema.39
Table 1.
Descriptive statistics for concept extraction datasets
| Dataset | Subset | Notes | Entities |
|---|---|---|---|
| i2b2 2010 | Train | 349 | 27 837 |
| Test | 477 | 45 009 | |
| i2b2 2012 | Train | 190 | 16 468 |
| Test | 120 | 13 594 | |
| SemEval 2014 Task 7 | Train | 199 | 5816 |
| Test | 99 | 5351 | |
| SemEval 2015 Task 14 | Train | 298 | 11 167 |
| Test | 133 | 7998 |
i2b2: Informatics for Integrating Biology and the Bedside.
The clinical embeddings are trained on MIMIC-III,38 which consists of almost 2 million clinical notes. Notes that have an ERROR tag are first removed, resulting in 1 908 359 notes with 786 414 528 tokens and a vocabulary of size 712 286. For pretraining traditional word embeddings, words are lowercased, as is standard practice. For pretraining ELMo and BERT, casing is preserved.
Experimental settings
Concept extraction
Concept extraction is based on the model proposed in Lample et al,40 a Bi-LSTM CRF architecture. For traditional embedding methods and ELMo embeddings, we use the same hyperparameters setting: hidden unit dimension at 512, dropout probability at 0.5, learning rate at 0.001, learning rate decay at 0.9, and Adam as the optimization algorithm. Early stopping of training is set to 5 epochs without improvement to prevent overfitting.
Pretraining of clinical embeddings
Across embedding methods, 2 different scenarios of pretraining are investigated and compared:
Off-the-shelf embeddings from the official release, referred to as the General model.
Pretrained embeddings on MIMIC-III, referred to as the MIMIC model.
In the first scenario, more details related to the embedding models are shown in in Table 2. We also apply BioBERT,35 which is the most recent pretrained model on biomedical literature initiated from BERTBASE.
Table 2.
Resources of off-the-shelf embeddings from the open domain
| Method | Resource (tokens / vocab) | Size | Language model |
|---|---|---|---|
| word2vec | Google News (100B / 3M) | 300 | NA |
| Glove | Gigaword5 + Wikipedia2014 (6B / 0.4M) | 300 | NA |
| fastText | Wikipedia 2017 + UMBC corpus + statmt.org news (16B / 1M) | 300 | NA |
| ELMo | WMT 2008-2012 + Wikipedia (5.5B / 0.7M) | 512 | 2-layer, 4096-hidden,93.6M parameters |
| BERTBASE | BooksCorpus + English Wikipedia (3.3B / 0.03Ma) | 768 | 12-layer, 768-hidden, 12 heads, 110M parameters |
| BERTLARGE | BooksCorpus + English Wikipedia (3.3B / 0.03Ma) | 1024 | 24-layer, 1024-hidden, 16-heads, 340M parameters |
B: billion; M: million; NA: Not Applicable.
aVocabulary size calculated after wordpiece tokenization.
In the second scenario, for all the traditional embedding methods, we pretrain 300 dimension embeddings from MIMIC-III clinical notes. We apply the following hyperparameter settings for all 3 traditional embedding methods including word2vec, GloVe, and fastText: window size of 15, minimum word count of 5, 15 iterations, and embedding size of 300 to match the off-the-shelf embeddings.
For ELMo, the hyperparameter setting for pretraining follows the default in Peters et al.23 Specifically, a char-CNN embedding layer is applied with 16-dimension character embeddings, filter widths of [1, 2, 3, 4, 5, 6, 7] with respective [32, 32, 64, 128, 256, 512, 1024] number of filters. After that, a 2-layer Bi-LSTM with 4096 hidden units in each layer is added. The output of the final bi-LM language model is projected to 512 dimensions with a highway connection. The total number of tokens in pretraining on MIMIC-III was 786 414 528. MIMIC-III was split into a training corpus (80%) for pretraining and a held-out testing corpus (20%) for evaluating perplexity. The pretraining step is performed on the training corpus for 15 epochs. The average perplexity on the testing corpus is 9.929.
For BERT, 2 clinical-domain models initialized from BERTBASE and BERTLARGE are pretrained. Unless specified, we follow the authors’ detailed instructions to set up the pretraining parameters, as other options were tested and it has been concluded that this is a useful recipe when pretraining from their released model (eg, poor model convergence). The vocabulary list consisting of 28 996 word-pieced tokens applied in BERTBASE and BERTLARGE is adopted. According to their article, the performance on the downstream tasks decrease as the training steps increase, thus we decide to save the intermediate checkpoint (every 20 000 steps) and report the performance of intermediate models on the downstream task.
Fine-tuning BERT
Fine-tuning the BERT clinical models on the downstream task requires some adjustments. First, instead of randomly initializing the Bi-LSTM output weights, Xavier initialization is utilized, without which the BERT fine-tuning failed to converge (this was not necessary for ELMo). Second, early stopping of fine-tuning is set to 800 steps without improvement to prevent overfitting. Finally, postprocessing steps are conducted to align the BERT output with the concept gold standard, including handling truncated sentences and word-pieced tokenization.
Evaluation and computational costs
A total of 10% of the official training set is used as a development set and the official test set is used to report performance. The specific performance metrics are precision, recall, and F1 measure for exact matching. The pretraining BERT experiments are implemented in TensorFlow41 on a NVidia Tesla V100 GPU (32G) (NVIDIA, Santa Clara, CA), other experiments are performed in TensorFlow on a NVIDIA Quadro M5000 (8G). The time for pretraining ELMo, BERTBASE, and BERTLARGE for every 20 000 checkpoint is 4.83 hours, 3.25 hours, and 5.16 hours, respectively. These three models were run until manually set to stop at 320 000 iterations (82.66 hours [roughly 3.4 days]), 700 000 iterations (113.75 hours [roughly 4.7 days]), and 700,000 iterations (180.83 hours [roughly 7.5 days]), respectively.
RESULTS
Performance comparison
The performance on the respective test sets for the embedding methods on the 4 clinical concept extraction tasks are reported in Table 3. The performance is evaluated in exact matching F1. In general, embeddings pretrained on the clinical corpus performed better than the same method pretrained on an open-domain corpus.
Table 3.
Test set comparison in exact F1 of embedding methods across tasks
| Method | i2b2 2010 |
i2b2 2012 |
SemEval 2014 Task 7 |
SemEval 2015 Task 14 |
||||
|---|---|---|---|---|---|---|---|---|
| General | MIMIC | General | MIMIC | General | MIMIC | General | MIMIC | |
| word2vec | 80.38 | 84.32 | 71.07 | 75.09 | 72.2 | 77.48 | 73.09 | 76.42 |
| GloVe | 84.08 | 85.07 | 74.95 | 75.27 | 70.22 | 77.73 | 72.13 | 76.68 |
| fastText | 83.46 | 84.19 | 73.24 | 74.83 | 69.87 | 76.47 | 72.67 | 77.85 |
| ELMo | 83.83 | 87.8 | 76.61 | 80.5 | 72.27 | 78.58 | 75.15 | 80.46 |
| BERTBASE | 84.33 | 89.55 | 76.62 | 80.34 | 76.76 | 80.07 | 77.57 | 80.67 |
| BERTLARGE | 85.48 | 90.25 b | 78.14 | 80.91 b | 78.75 | 80.74 b | 77.97 | 81.65 b |
| BioBERT | 84.76 | – | 77.77 | – | 77.91 | – | 79.97 | – |
| Prior SOTA | 88.6034 | a42 | 80.339 | 81.343 | ||||
i2b2: Informatics for Integrating Biology and the Bedside; MIMIC: Medical Information Mart for Intensive Care; SOTA: state-of-the-art.
aThe SOTA on the i2b2 2012 task is only reported in partial-matching F1. That result, 92.29,42 is below the equivalent we achieve on partial-matching F1 with BERTLARGE(MIMIC), 93.18.
bThe best performing result in the respective task.
For i2b2 2010, the best performance is achieved by BERTLARGE(MIMIC), with an F1 of 90.25. It improves the performance by 5.18 over the best performance of the traditional embeddings achieved by GloVe(MIMIC), with an F1 of 85.07. As expected, both ELMo and BERT clinical embeddings outperform the off-the-shelf embeddings with relative increase up to 10%.
The best performance on the i2b2 2012 task is achieved by BERTLARGE(MIMIC), with an F1 of 80.91 across all the alternative methods. It increases F1 by 5.64 over GloVe(MIMIC), which obtains the best score (75.27) among the traditional embedding methods. As expected, ELMo and BERT with pretrained clinical embedddings exceed the off-the-shelf open-domain models.
The most effective model for SemEval 2014 task achieved an exact matching F1 of 80.74 by BERTLARGE(MIMIC). Notably, traditional embedding models pretrained on the clinical corpus such as GloVe(MIMIC) obtained a higher performance than contextual embedding model trained on open domain, namely ELMo(General).
For the SemEval 2015 task, as the experiments are performed only in concept extraction, the models are evaluated using the official evaluation script from the SemEval 2014 task. Note that the training set (298 notes) for the SemEval 2015 task is the training (199 notes) and test set (99 notes) combined for the SemEval 2014 task. The best performance on the 2015 task is achieved by BERTLARGE(MIMIC) with an F1 of 81.65.
The detailed performance for each entity category including PROBLEM, TEST, and TREATMENT on the 2010 task is shown in Table 4. Both ELMo and BERT show improvements to all 3 categories, with ELMo outperforming the traditional embeddings on all 3, and BERT outperforming ELMo on all 3. One notable aspect with BERT is that Treatments see a larger jump: Treatment is the lowest-performing category for ELMo and the traditional embeddings despite there being slightly more Treatments than Tests in the data, but for BERT the Treatments category outperforms Tests.
Table 4.
Performance of each label category with pretrained MIMIC models on i2b2 2010 task.
| word2vec | GloVe | fastText | ELMo | BERTBASE | BERTLARGE | |
|---|---|---|---|---|---|---|
| PROBLEM | 84.16 | 85.08 | 84.32 | 88.76 | 89.61 a | 89.26 |
| TEST | 85.93 | 84.96 | 84.01 | 87.39 | 88.09 | 88.8 a |
| TREATMENT | 83.14 | 84.73 | 83.89 | 86.98 | 88.3 | 89.14 a |
i2b2: Informatics for Integrating Biology and the Bedside; MIMIC: Medical Information Mart for Intensive Care;
aThe best performing result in the respective task.
Table 5 shows the results for each event type on the 2012 task with embeddings pretrained from MIMIC-III. Generally, the biggest improvement by the contextual embeddings over the traditional embeddings is achieved on the PROBLEM type (BERTLARGE: 86.1, GloVe: 77.83). This is reasonable because in clinical notes, diseases and conditions normally appear in certain types of surrounding context with similar grammar structures. Thus, it is necessarily important to take advantage of contextual representations to capture the surrounding context for that particular concept. Interestingly, ELMo outperforms both BERT models for CLINICAL DEPT and OCCURRENCE.
Table 5.
Performance of each label category with pretrained MIMIC models on i2b2 2012 task
| word2vec | GloVe | fastText | ELMo | BERTBASE | BERTLARGE | |
|---|---|---|---|---|---|---|
| PROBLEM | 76.49 | 77.83 | 75.35 | 84.1 | 85.91 | 86.1 a |
| TEST | 78.12 | 81.26 | 76.94 | 84.76 | 86.88 a | 86.56 |
| TREATMENT | 76.22 | 78.52 | 76.88 | 83.9 | 84.27 | 85.09 a |
| CLINICAL DEPT | 78.18 | 77.92 | 77.27 | 83.71 a | 77.92 | 78.23 |
| EVIDENTIAL | 73.14 | 74.26 | 72.94 | 72.95 | 74.21 | 74.96 a |
| OCCURRENCE | 64.77 | 64.19 | 61.02 | 66.27 a | 62.36 | 65.65 |
MIMIC: Medical Information Mart for Intensive Care;
aThe best performing result in the respective task.
Pretraining evaluation
The efficiency of pretrained ELMo and BERT models are investigated by reporting the loss during pretraining steps and by evaluating the intermediate checkpoints on downstream tasks. It is observed for both ELMo and BERT at their pretraining stages, the train perplexity or loss decreases as the steps increase, indicating that the language model is actually adapting to the clinical corpus. If there is no intervention to stop the pretraining process, it will lead to a very small loss value (see Supplementary Figure 1). However, this will ultimately cause overfitting on the pretraining corpus.
Using i2b2 2010 as the downstream task, the final performance at each intermediate checkpoint of the pretrained model is shown in Figure 2. For ELMo, as the pretraining proceeds, the performance of the downstream task remains stable after a certain number of iterations (the maximum F1 reaches 87.80 at step 280 000). For BERTBASE, the performance on the downstream task is less steady and tends to decrease after achieving its optimal model, with the maximum F1 89.55 at step 340 000. We theorize that this is due to initializing the MIMIC model with the open-domain BERT model: over many iterations on the MIMIC data, the information learned from the large open corpus (3.3 billion words) is lost and would eventually converge on a model similar to one initialized from scratch. Thus, limiting pretraining on a clinical corpus to a certain number of iterations provides a useful trade-off, balancing the benefits of a large open-domain corpus while still learning much from a clinical corpus. We hope that this is a practical piece of guidance for the clinical NLP community when they intend to generate their own pretrained model from a clinical corpus.
Figure 2.
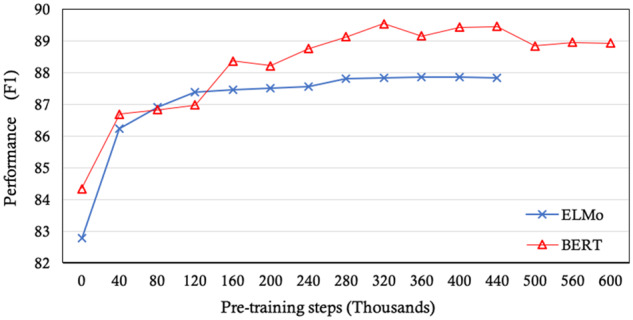
Performances on the Informatics for Integrating Biology and the Bedside (i2b2) 2010 task governed by the steps of pretraining epochs on ELMo(MIMIC) and BERT-BASE(MIMIC). MIMIC: Medical Information Mart for Intensive Care.
DISCUSSION
This study explores the effects of numerous embedding methods on 4 clinical concept extraction tasks. Unsurprisingly, domain-specific embedding models outperform open-domain embedding models. All types of embeddings enable consistent gains in concept extraction tasks when pretrained on a clinical domain corpus. Further, the contextual embeddings outperform traditional embeddings in performance. Specifically, large improvements can be achieved by pretraining a deep language model from a large corpus, followed by a task-specific fine tuning.
State-of-the-art comparison
Among the 4 clinical concept extraction corpora, the i2b2 2012 task reports the partial matching F1 as the organizers reported in Sun et al.11 and the other 3 tasks report the exact matching F1. Currently, the state-of-the-art models for i2b2 2010, i2b2 2012, SemEval 2014 task 7, and SemEval 2015 Task 14 are reported with F1 of 88.60,34 92.29,42 80.3,39 and 81.3,43 respectively. With the most advanced language model representation method pretrained on a large clinical corpus, namely BERTLARGE(MIMIC), we achieved new state-of-the-art performances across all tasks. BERTLARGE(MIMIC) outperform the state-of-the-art models on all 4 tasks with respective F measures of 90.25, 93.18 (partial F1), 80.74, and 81.65.
Semantic information from contextual embeddings
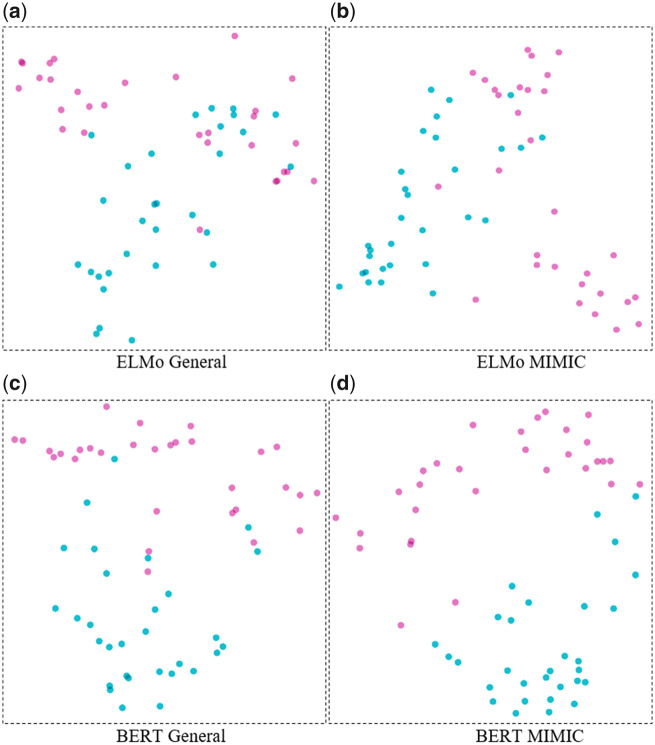
Here, we explore the semantic information captured by the contextual representation and infer that the contextual embedding can encode information that a single word vector fails to. We select 30 sentences from both web texts and clinical notes in which the word cold appears (The actual sentences can be found in Supplement Table 1). The embedding vectors of cold in 30 sentences from 4 embedding models, ELMo(General), ELMo(MIMIC), BERTLARGE(General), and BERTLARGE (MIMIC), were derived. This results in 120 vectors for the same word across 4 embeddings. For each embedding method, Principal Component Analysis (PCA) is performed to reduce the dimensionality to 2.
The PCA visualizations are shown in Figure 3. As expected, the vectors of cold generated by ELMo(General) are mixed within 2 different meaning labels. The vectors generated by BERTLARGE(General) and BERTLARGE(MIMIC) are more clearly clustered into 2 groups. ELMo(General) is unable to discriminate the different meanings of the word cold, specifically between temperature and symptom. The visualization result is also consistent with the performance on the concept extract tasks where ELMo(General) tends to get a poorer performance compared with the other 3 models.
Figure 3.
Principal Component Analysis (PCA) visualizations using embedding vectors of cold from embedding models (purple: cold as temperature meaning; red: cold as symptom). (a) ELMo General (b) ELMo MIMIC (c) BERT General (d) BERT MIMIC
Traditional word embeddings are commonly evaluated using lexical similarity tasks, such as those by Pakhomov et al,44,45 which compare 2 words outside any sentence-level context. While not entirely appropriate for comparing contextual embeddings such as ELMo and BERT because the centroid of the embedding clusters are not necessarily meaningful, such lexical similarity tasks do provide motivation for investigating the clustering effects of lexically similar (and dissimilar) words. In Supplementary Figure 2, we compare 4 words from Pakhomov et al45: tylenol, motrin, pain, and herpes based on 50-sentence samples from MIMIC-III and the same 2-dimension PCA visualization technique. One would expect tylenol (acetaminophen) and motrin (ibuprofen) to be similar, and in fact the clusters overlap almost completely. Meanwhile, pain is a nearby cluster, while herpes is quite distant. So while contextual embeddings are not well-suited to context-free lexical similarity tasks, the aggregate effects (clusters) still demonstrate similar spatial relationships as traditional word embeddings.
Lexical segmentation in BERT
One important notable difference between BERT and both ELMo and the traditional word embeddings is that BERT breaks words down into subword tokens, referred to as wordpieces.46 This is accomplished via statistical analysis on a large corpus, as opposed to using a morphological lexicon. The concern for clinical NLP, then, is if a different word piece tokenization method is appropriate for clinical text as opposed to general text (ie, books and Wikipedia for the pretrained BERT models). Supplementary Table 2 shows the word piece tokenization for the medical words from the lexical similarity corpus developed by Pakhomov et al.45 The results do not exactly conform to traditional medical term morphology (eg, appendicitis is broken into app, -end, -icit, -is, as opposed to having the suffix -itis). Note that this isn't necessary a bad segmentation: it is possible this would outperform a word piece tokenization based on the SPECIALIST lexicon.47 What is not in dispute, however, is that further experimentation is required, such as determining word pieces from MIMIC-III. Note this is not as simple as it at first seems. The primary issue is that the BERT models we use in this article were first pretrained on a 3.3 billion word open-domain corpus, then pretrained further on MIMIC-III. Performing word piece tokenization on MIMIC-III would at a minimum require repeating the pretraining process on the open-domain corpus (with the clinical word pieces) to get comparable embedding models. Given the range of experimentation necessary to determine the best word piece strategy, we leave this experimentation to future work.
CONCLUSION
In this article, we present an analysis of different word embedding methods and investigate their effectiveness on 4 clinical concept extraction tasks. We compare between traditional word representation methods as well as the advanced contextual representation methods. We also compare pretrained contextual embeddings using a large clinical corpus against the performance of off-the-shelf pretrained models on open-domain data. Primarily, the efficacy of contextual embeddings over traditional word vector representations are highlighted by comparing the performances on clinical concept extraction. Contextual embeddings also provide interesting semantic information that is not accounted for in traditional word representations. Further, our results highlight the benefits of embeddings through unsupervised pretraining on clinical text corpora, which achieve higher performance than off-the-shelf embedding models and result in new state-of-the-art performance across all tasks.
FUNDING
This work was supported by the U.S. National Institutes of Health (NIH) and the Cancer Prevention and Research Institute of Texas (CPRIT). Specifically, NIH support comes from the National Library of Medicine (NLM) under award R00LM012104 (to KR) and R01LM010681 (to HX), as well as the National Cancer Institute under award U24CA194215 (to HX). CPRIT support for computational resources was provided under awards RP170668 (W. Jim Zheng) and RR180012 (Xiaoqian Jiang).
AUTHOR CONTRIBUTIONS
YS performed the experiments and drafted the initial manuscript. KR conceived of the study and oversaw the design. JW and HX contributed to the experiment design. All authors reviewed and approved the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement
Dr. Xu, Mr. Wang, and The University of Texas Health Science Center at Houston have research-related financial interests in Melax Technologies, Inc.
Supplementary Material
REFERENCES
- 1. Tang B, Cao H, Wu Y, et al. Recognizing clinical entities in hospital discharge summaries using Structural Support Vector Machines with word representation features. BMC Med Inform Decis Mak 2013; 13 (suppl 1): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kundeti SR, Vijayananda J, Mujjiga S, et al. Clinical named entity recognition: challenges and opportunities. Proceedings of the IEEE International Conference on Big Data (Big Data); 2016: 1937–45.
- 3. Unanue IJ, Borzeshi EZ, Piccardi M.. Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition. J Biomed Inform 2017; 76: 102–9. [DOI] [PubMed] [Google Scholar]
- 4. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rink B, Harabagiu S, Roberts K.. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc 2011; 185: 594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Si Y, Roberts K.. A Frame-Based NLP System for Cancer-Related Information Extraction. AMIA Annu Symp Proc 2018; 2018: 1524–33. [PMC free article] [PubMed] [Google Scholar]
- 7. Lee H, Peirsman Y, Chang A, et al. Stanford’s multi-pass sieve coreference resolution system at the CoNLL-2011 shared task. Proceedings of the Fifteenth Conference on Computational Natural Language Learning: Shared Task; 2011: 28–34.
- 8. Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. In: AMIA Annu Symp Proc. 2011; 2011: 1564–72.. [PMC free article] [PubMed] [Google Scholar]
- 9. Velupillai S, Suominen H, Liakata M, et al. Using clinical Natural Language Processing for health outcomes research: overview and actionable suggestions for future advances. J Biomed Inform 2018; 88: 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 185: 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun W, Rumshisky A, Uzuner O.. Evaluating temporal relations in clinical text: 2012 i2b2 challenge. J Am Med Inform Assoc 2013; 205: 806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stubbs A, Kotfila C, Uzuner Ö.. Automated systems for the de-identification of longitudinal clinical narratives: overview of 2014 i2b2/UTHealth shared task Track 1. J Biomed Inform 2015; 58: S11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suominen H, Salanterä S, Velupillai S, et al. Overview of the ShARe/CLEF eHealth evaluation lab 2013. Proceedings of the International Conference of the Cross-Language Evaluation Forum for European Languages; 2013: 212–231.
- 14. Kelly L, Goeuriot L, Suominen H, et al. Overview of the ShARe/CLEF eHealth Evaluation Lab 2014. Proceedings of the International Conference of the Cross-Language Evaluation Forum for European Languages; 2014: 172–91.
- 15. Pradhan S, Elhadad N, Chapman W, et al. Semeval-2014 Task 7: Analysis of Clinical Text. Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014); 2014: 54–62.
- 16. Elhadad N, Pradhan S, Gorman S, et al. SemEval-2015 Task 14: Analysis of Clinical Text. Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015); 2015: 303–10.
- 17. Bethard S, Savova G, Chen W-T, et al. Semeval-2016 task 12: clinical tempeval. Proceedings of the 10th International Workshop on Semantic Evaluation (SemEval-2016); 2016: 1052–62.
- 18. Shen Y, Rong W, Jiang N, et al. Word embedding based correlation model for question/answer matching. Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence; 2017: 3511–7.
- 19. Habibi M, Weber L, Neves M, Wiegandt DL, Leser U.. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 2017; 3314: i37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang F, Guo J, Xu W, et al. Application of word embeddings in biomedical named entity recognition tasks. J Digit Inf Manag 2015; 135: 321–7. [Google Scholar]
- 21. Wu Y, Xu J, Jiang M, Zhang Y, Xu H.. A study of neural word embeddings for named entity recognition in clinical text. AMIA Annu Symp Proc; 2015: 1326. [PMC free article] [PubMed] [Google Scholar]
- 22. Florez E, Precioso F, Riveill M, et al. Named entity recognition using neural networks for clinical notes. Proceedings of the International Workshop on Medication and Adverse Drug Event Detection; 2018: 7–15.
- 23. Peters ME, Neumann M, Iyyer M., et al. Deep contextualized word representations. Proceedings of NAACL-HLT; 2018: 2227–2237.
- 24. Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of NAACL-HLT; 2019: 4171–4186.
- 25. Seo M, Kembhavi A, Farhadi A, Hajishirzi H. Bidirectional Attention Flow for Machine Comprehension. Proceedings of the International Conference on Learning Representations; 2017.
- 26. Mikolov T, Sutskever I, Chen K, et al. Distributed representations of words and phrases and their compositionality. Advances in Neural Information Processing Systems; 2013: 3111–9.
- 27. Pennington J, Socher R, Manning C. Glove: global vectors for word representation. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014: 1532–43.
- 28. Bojanowski P, Grave E, Joulin A, Mikolov T. Enriching Word Vectors with Subword Information. Transactions of the Association for Computational Linguistics 2017; 5:135–146.
- 29. Roberts K. Assessing the corpus size vs. similarity trade-off for word embeddings in clinical NLP. Proceedings of the Clinical Natural Language Processing Workshop (ClinicalNLP); 2016: 54–63.
- 30. Wang Y, Liu S, Afzal N, et al. A comparison of word embeddings for the biomedical natural language processing. J Biomed Inform 2018; 87: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaswani A, Shazeer N, Parmar N, et al. Attention is All You Need. Advances in Neural Information Processing Systems; 2017: 5998–6008. [Google Scholar]
- 32. de Bruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc 2011; 185: 557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chalapathy R, Borzeshi EZ, Piccardi M. Bidirectional LSTM-CRF for clinical concept extraction. Proceedings of the Clinical Natural Language Processing Workshop; 2016: 7–12.
- 34. Zhu H, Paschalidis IC, Tahmasebi A. Clinical concept extraction with contextual word embedding. Proceedings of the Machine Learning for Health (ML4H) Workshop; 2018.
- 35. Lee J, Yoon W, Kim S, et al. BioBERT: pre-trained biomedical language representation model for biomedical text mining. arXiv 2019. Feb 3 [E-pub ahead of print]. [DOI] [PMC free article] [PubMed]
- 36. Alsentzer E, Murphy JR, Boag W, et al. Publicly Available Clinical BERT Embeddings. Proceedings of the Clinical Natural Language Processing workshop; 2019: 72–78.
- 37. Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. arXiv 2019. Apr 16 [E-pub ahead of print].
- 38. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang B, Chen Q, Wang X, et al. Recognizing disjoint clinical concepts in clinical text using machine learning-based methods. AMIA Annu Symp Proc.; 2015: 1184–93. [PMC free article] [PubMed] [Google Scholar]
- 40. Lample G, Ballesteros M, Subramanian S, et al. Neural Architectures for Named Entity Recognition. Proceedings of NAACL-HLT; 2016: 260–270.
- 41. Abadi M, Barham P, Chen J, et al. Tensorflow: a system for large-scale machine learning. Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation; 2016: 265–83. [Google Scholar]
- 42. Liu Z, Yang M, Wang X, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak 2017; 17: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y, Wang J, Tang B, et al. UTH_CCB: a report for semeval 2014–task 7 analysis of clinical text. In: Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014); 2014: 802–6.
- 44. Pakhomov S, McInnes B, Adam T, et al. Semantic similarity and relatedness between clinical terms: an experimental study. AMIA Annu Symp Proc; 2010: 572–6. [PMC free article] [PubMed] [Google Scholar]
- 45. Pakhomov SVS, Finley G, McEwan R, et al. Corpus domain effects on distributional semantic modeling of medical terms. Bioinformatics 2016; 32: 3635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuster M, Nakajima K. Japanese and Korean voice search. Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) Kyoto, Japan: IEEE; 2012: 5149–52.
- 47. Browne AC, McCray AT, Srinivasan S.. The Specialist Lexicon. Washington, DC: National Library of Medicine Technical Reports; 2000: 18–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.