Abstract
Objective
Our objective is to develop algorithms for encoding clinical text into representations that can be used for a variety of phenotyping tasks.
Materials and Methods
Obtaining large datasets to take advantage of highly expressive deep learning methods is difficult in clinical natural language processing (NLP). We address this difficulty by pretraining a clinical text encoder on billing code data, which is typically available in abundance. We explore several neural encoder architectures and deploy the text representations obtained from these encoders in the context of clinical text classification tasks. While our ultimate goal is learning a universal clinical text encoder, we also experiment with training a phenotype-specific encoder. A universal encoder would be more practical, but a phenotype-specific encoder could perform better for a specific task.
Results
We successfully train several clinical text encoders, establish a new state-of-the-art on comorbidity data, and observe good performance gains on substance misuse data.
Discussion
We find that pretraining using billing codes is a promising research direction. The representations generated by this type of pretraining have universal properties, as they are highly beneficial for many phenotyping tasks. Phenotype-specific pretraining is a viable route for trading the generality of the pretrained encoder for better performance on a specific phenotyping task.
Conclusions
We successfully applied our approach to many phenotyping tasks. We conclude by discussing potential limitations of our approach.
Keywords: natural language processing, biomedical informatics, phenotyping
INTRODUCTION
Recent neural network models have shown state-of-the-art results on a number of natural language processing (NLP) benchmarks and even human-level performance on several narrowly defined tasks such as question answering1 and machine translation.2 Yet, this success required tens or hundreds of thousands of labeled examples. Procuring annotated datasets of this size is not feasible for most tasks in clinical NLP due to the high cost of manual labeling. The well-known clinical NLP benchmarks such as Integrating Biology and the Bedside (i2b2) obesity comorbidity recognition, i2b2 smoking status detection, and the recent National NLP Clinical Challenges (https://n2c2.dbmi.hms.harvard.edu) have only hundreds of examples per phenotype, making it difficult to take advantage of highly expressive deep learning methods.
This problem of insufficient training data is addressed in computer vision by means of pretraining classifiers on massive datasets such as ImageNet3,4 and subsequently refining them on a more specialized classification task. The idea is that the large datasets train early layers in a deep network to recognize universal vision primitives that should apply across tasks (lines of various orientations, edges, basic shapes, etc.) In NLP, researchers have exploited large unlabeled corpora to pretrain shallow representations of such units of meaning as words and short phrases using techniques such as word2vec5 and GloVe.6 However, these methods focus only on the bottom layer in the hierarchy of language, with new ways to encode higher level language representations needed for most NLP tasks.
More recently, word-level approaches have been extended to sentences.7 Last year saw a number of successful attempts to continue to move up this hierarchy of language complexity by pretraining text encoders using language modeling objectives8–11; text encoding methods such as BERT10 and ULMfit9 can represent sentences or even paragraph-size units. As a result of these works, language modeling is now viewed as NLP's equivalent of ImageNet pretraining. We observe that the analogy with ImageNet is far from perfect because ImageNet has a true source of supervision and language modeling is self-supervised (ie, trained without manual labels; a computer vision analogy would have to involve predicting artificially removed pixels of an image). In this work, we find evidence that clinical NLP is uniquely positioned to investigate a much closer analogy to ImageNet pretraining, which is manifested in billing code prediction. Because billing codes are available in abundance in healthcare institutions and are linked to document-level entities, such as clinical encounters, we are able to move beyond sentence-level encoding to document level by means of supervised pretraining. Prior to this work, representation learning research focused on sentence or paragraph-length sized units, with little research on encoding larger units such as documents and encounters.
In our previous work,12 we introduced a simple text encoder that takes Unified Medical Language System (UMLS) concepts as input and is trained using a billing code prediction objective. The encoder is subsequently used to generate patient representations that succinctly captured patient information. In this follow-up work, we refine this idea, extend it to raw text, and introduce a novel phenotype-specific encoder that makes it possible to trade the generality of the resulting text representations for better performance on a specific phenotyping task.
There are currently 2 strategies for deploying pretrained models: feature extraction and fine-tuning. In feature extraction (eg, ELMo,8 Flair11) one divides learning into 2 independent processes: learning (1) a general model of how language works to encode sentences or documents (pretraining) and (2) how to classify documents encoded with such a model. In concrete terms, the weights of the pretraining network are frozen before learning how to do the downstream task. In fine-tuning (eg, BERT,10 GPT,13 ULMFit9), pretraining works the same way, but training for the downstream task can update the way that the network thinks language works. In other words, the pretraining network weights are allowed to update during the second phase. This work focuses on feature extraction, as our preliminary work finds fine-tuning to be less replicable (for more details, see Discussion).
Our work can also be viewed as learning patient representations, which are the output of our text encoder. Most of the recent work in clinical informatics focused on using structured EHR data, such as International Classification of Diseases (ICD) codes, procedure codes, and medication orders for learning patient representations.14–19 One of the few patient representation learning systems to focus on EHR text is DeepPatient,17 which not only operates on a variety of features including structured EHR information, but also uses topic modeling as a way to represent text. To learn patient representations, they use a model consisting of stacked denoising autoencoders. The learned representations are used to predict ICD codes occurring in the next 30, 60, 90, and 180 days. In contrast to the previous works, Sushil et al20 focuses exclusively on EHR text to learn patient representations using unsupervised methods, such as stacked denoising autoencoders and doc2vec.7 They find that the learned representations outperform traditional bag-of-words representations when few training examples are available and that the target task does not rely on strong lexical features. Like Sushil et al,20 our work uses text variables only.
Existing work on encoding patient representations has focused on predictions of convenience (tasks for which coded data is available), such as mortality prediction or future billing codes. We evaluate our encoder on several phenotyping tasks using labeled datasets. First, in the interests of reproducibility, we evaluate on the publicly available i2b2 comorbidity challenge data, establishing a new state of the art. We also apply our encoder to 2 novel and high-impact substance misuse tasks, predicting opioid and alcohol misuse in trauma patients.
MATERIALS AND METHODS
Document-level clinical text encoder
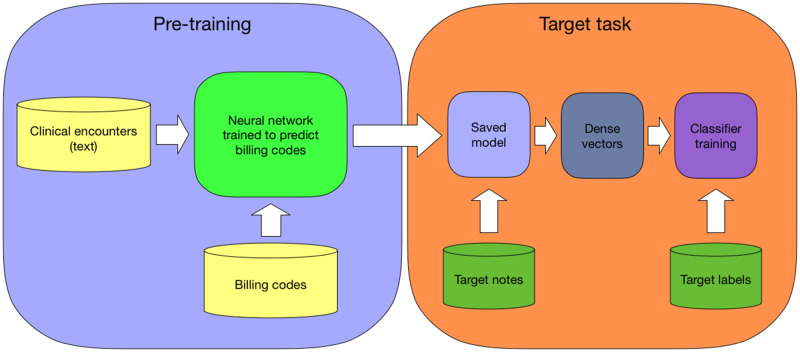
With the advent of electronic health records (EHR), massive amounts of patient data have become available at healthcare institutions. EHR consists of 2 distinct types of data: (1) structured data such as lab results, billing codes, and medication orders and (2) unstructured data such as clinical notes. Our goal is training a clinical text encoder and we observe that structured data potentially presents a good source of supervision. An encoder that learns how to map notes text to structured data, when trained on massive amounts of data, could capture key elements of the information present in the notes text. Text representations derived from this encoder, when used for downstream machine learning tasks such as automatic phenotyping (Figure 1), will likely benefit classifier performance because they have representational power of a large dataset. The methods we discuss can be viewed as representation learning. In this work, we focus on using the billing codes as a source of supervision, which are typically available in abundance in a healthcare institution.
Figure 1.
We train a neural network to predict billing codes given the text of clinical encounters. After the training is finished, we save the model. We use the saved model as a text encoder to create dense representations for the notes in a target task. These representations can be used to train a classifier.
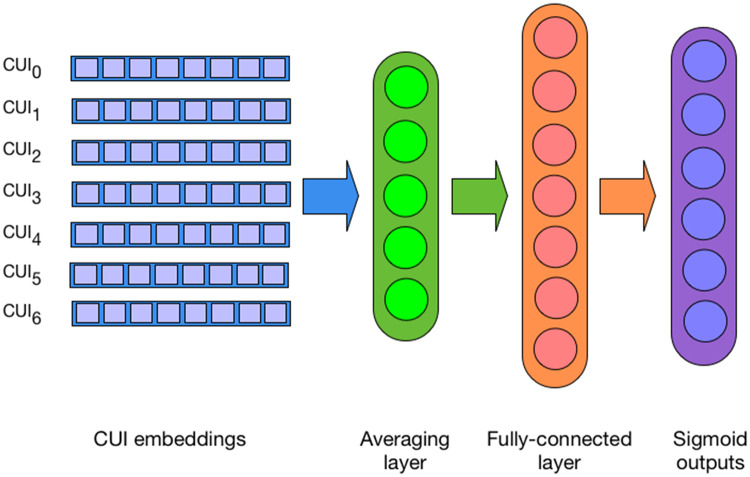
We explore several neural architectures that work directly with text and simple named entity features automatically extracted from text. The first encoder is similar to the one used in our previous work, which is a deep averaging network (DAN) that takes a set of UMLS concept unique identifiers (CUIs), maps them to their 300-dimensional embeddings, averages them, and projects them to the penultimate fully connected hidden layer, essentially encoding the input as a fixed-sized dense vector. During pretraining, the final (output) network layer consists of n sigmoid units, each representing a unique billing code (Figure 2). The architecture presented in Dligach and Miller12 is trained at the patient level, which is suboptimal because billing codes are assigned at the encounter level; the unit of classification in this work is a single encounter. CUIs are extracted from notes by mapping spans of clinically relevant text (eg, shortness of breath, appendectomy, MRI [magnetic resonance imaging]) to entries in the UMLS Metathesaurus. CUIs can be easily extracted by existing tools such as Apache clinical Text Analysis Knowledge Extraction System (cTAKES) (http://ctakes.apache.org). The advantage of this architecture is extremely fast training, which facilitates efficient exploration of the hyperparameter space.
Figure 2.
Deep averaging network that takes as input concept unique identifier (CUI) embeddings and is trained to predict billing codes.
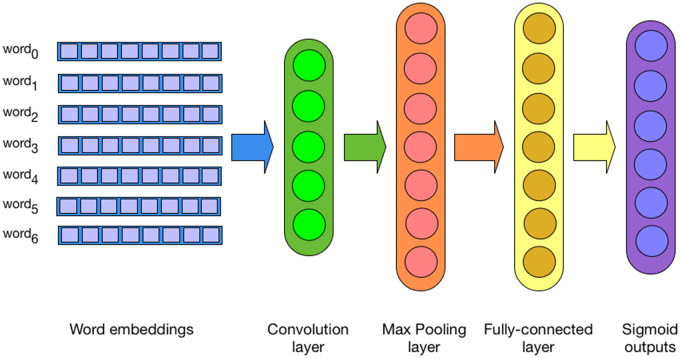
The second encoder is a convolutional neural network (CNN) that operates directly on the text of the notes. The embedding layer is followed by a convolutional layer, a max pooling layer, and a fully connected layer. The output layer is identical to the DAN architecture mentioned previously (Figure 3). In preliminary work, we also experimented with Recurrent Neural Network (RNN)-based architectures, but their performance was subpar both in terms of accuracy and speed, likely due to the difficulty capturing long-distance dependencies.
Figure 3.
Convolutional neural network that takes as input word embeddings and is trained to predict billing codes.
Both encoders are trained using binary cross-entropy loss function and RMSProp optimizer to jointly predict billing codes. To use the patient encoder as a feature extractor, we freeze the network weights, push the text of the notes through the network, and collect the computed values of the hidden layer nodes, thus obtaining a dense vector representing the input text, that can be used as input for any machine learning task (eg, to train a supervised classifier).
As noted, our text encoders are trained to jointly predict all billing codes associated with a clinical encounter. An encoder trained this way, given sufficient amount of data, should capture a broad spectrum of clinical information that exists in the input text, making the representations that the encoder generates appropriate for predicting a wide range of outcomes. While our ultimate goal is learning a universal clinical text encoder, we also observe that it is possible to train a phenotype-specific encoder by restricting the billing code prediction targets to a set that is relevant to a specific phenotype. While a universal encoder could be more practical, as it needs to be pretrained only once, a phenotype-specific encoder could potentially perform better for a specific medical condition. We train 2 phenotype-specific text encoders for detecting substance misuse status, by modifying the encoder's training objective to predict only the codes associated with 2 substance misuse scenarios: alcohol and opioid misuse. This approach can be viewed as a kind of transfer learning21 because the model learns to encode the knowledge obtained from large amounts of data of a source task (billing code prediction) to subsequently apply it to a target task (substance misuse).
Data
We pretrain all text encoders using the Medical Information Mart for Intensive Care III (MIMIC III) corpus.22 MIMIC III contains notes and structured data for over 40 000 Beth Israel Deaconess Medical Center critical care patients. Because billing codes are assigned at the encounter level, we use a patient encounter as a unit of classification when training an encoder; this is different from our previous approach that worked at the patient level. Our unit of classification is thus all notes in an encounter concatenated into a single document that the encoder learns to map to ICD-9 and Current Procedural Terminology code targets. We process these documents with cTAKES to extract UMLS CUIs. cTAKES is an open-source library for processing clinical texts with an efficient dictionary lookup component for identifying mentions of clinically relevant spans of text.
To speed up the training of the encoders, we limit the maximum length of input, set a threshold on the minimum number of examples required for a billing code to be used as a prediction target, and collapse billing codes to their general categories. This last step is currently necessary to make the training viable because there are thousands of unique billing codes. Specifically, for the DAN and CNN encoders, we (1) collapse all ICD-9 and Current Procedural Terminology codes to their more general category (eg, first 3 digits for ICD-9 diagnostic codes), (2) discard all tokens that appear fewer than 100 times, (3) discard encounters that have more than 25 000 tokens, and (4) discard all collapsed billing codes that have fewer than 500 examples. This preprocessing results in a dataset of 58 011 encounters mapped to 276 categories total. For the phenotype-specific encoders, we obtain ICD-9 codes for alcohol misuse (28 codes) and ICD-9 codes for opioid misuse (21 codes) and use them as prediction targets. The ICD code groups for both alcohol and opioid misuse were based on the Agency for Healthcare Research and Quality disease category classifications.23,24 We randomly split this dataset into a training set (80%) and a validation set (20%) for tuning model hyperparameters.
For evaluation, we use a publicly available dataset from the i2b2 obesity challenge,25 which consists of 1237 discharge summaries from the Partners HealthCare annotated with respect to obesity and its 15 most common comorbidities. Each patient was thus labeled for 16 different categories. We focus on the more challenging intuitive task, containing 3 label types (present, absent, and questionable). The diagnosis was annotated as present if it could be inferred even in cases when it was not explicitly mentioned in the text, requiring complex decision making and inferencing, and making this task particularly difficult. In this evaluation, our encoders are evaluated in 16 different classification tasks.
In addition, we use 2 in-house substance misuse datasets developed at the Loyola University Medical Center. The opioid misuse dataset is a part of a larger effort to create manually annotated substance misuse data. The dataset was annotated by trained substance use reviewers and in accordance with the National Survey on Drug Use and Health criteria for nonmedical opioid use (patients taking an opioid for reasons other than prescribed).26 At the time of the experiments described here, 413 patients (208 positive cases and 205 negative cases) annotated with respect to opioid misuse were available. The alcohol misuse dataset comprised 1423 patients (329 positive and 1094 negative cases). All patients completed an Alcohol Use Disorders Identification Test, a validated screening tool for misuse, and labeled cases met criteria if scores of ≥ 5 for women and ≥ 8 for men were met.27 Both datasets were split into training (80%) and test (20%) sets. Note that the prediction targets for both datasets are not ICD codes: for the opioid data, the labels were assigned manually by trained reviewers, while the alcohol data used a patient survey to derive the labels.28
We emphasize that the patient data we use in our evaluation originates from healthcare institutions (Partners HealthCare and Loyola) that are different from the one on which the encoders were trained (Beth Israel). This evaluation is challenging, yet it presents a true test of robustness of the proposed methods.
Experiments
Pre training: We begin by pretraining 2 document-level clinical text encoders: a DAN, which takes CUIs as input, and a CNN with max pooling, which takes words as input. The hyperparameters of both encoders are tuned on the validation set by optimizing the macro F1 score using random search.29 Importantly, the encoders are all tuned independently from the datasets on which we evaluate them; ie, we tune the encoders using MIMIC data but evaluate them on i2b2 comorbidity and Loyola substance misuse datasets. Note that our goal is neither to achieve the best possible billing code prediction performance on MIMIC nor to formally evaluate the performance on the billing code prediction task. Thus we are not allocating separate validation and test sets. Once the encoders achieve an acceptable level of performance, we combine the training and the validation sets and retrain them.
We train the DAN encoder with 5000 hidden units for 16 epochs with a learning rate of 0.001 and a batch size of 16 as determined by random search. We train the CNN encoder with 500 hidden units and 1024 filters of size 5 for 8 epochs with a learning rate of 0.001 and a batch size of 8 using AdaDelta optimizer, also as determined by random search.
Encoder evaluation: To evaluate the quality of an encoder, we deploy it as a feature extractor to generate text representations we can use as input to a linear support vector machine (SVM) classifier. To obtain a vector representing a set of notes, we freeze the network weights and push the notes text through the encoder, harvesting the computed values of the units of one of the intermediate network layers. For the DAN encoder, this is the hidden layer containing 5000 units. For the CNN encoder, we experiment with using either the max pooling layer (1024 units) or the hidden fully connected layer (500 units) to encode the text of the notes. In addition, we evaluate the representations obtained from the encoders in combination with traditional sparse bag-of-words and bag-of-CUIs representations, hypothesizing that the dense representations contain the information about the patient as a whole, while the sparse features may contain the explicit signal. To this end, we concatenate the dense encoder-derived vectors with sparse bag-of-words or bag-of-CUIs vectors. We then train an SVM classifier using these vectors as input.
For example, to run the evaluation on the i2b2 comorbidity data, we obtain patient text representations from the CNN encoder by feeding the text of the notes of a patient into the encoder. Rather than reading the classifier's code predictions, we collect the hidden layer node values, forming a 500-dimensional dense vector. We then train a multiclass SVM classifier for each disease in the comorbidity data, building 16 classifiers. Following the i2b2 obesity challenge, the models are evaluated using macro precision, recall, and F1 scores.25 We report the average macro precision, recall, and F1 across all 16 diseases for each system.
We compare all models to a baseline SVM classifier that we train for each phenotype with bag-of-cui features. We use 10-fold cross-validation on the training set to tune classifier parameters before we evaluate on the test set.
Phenotype-specific pre training: In addition to training an encoder to predict all billing codes associated with an encounter in the MIMIC corpus, we also evaluate the effectiveness of phenotype-specific pretraining by restricting the set of target ICD-9 codes only to the ones associated with the target phenotype. We identify ICD-9 codes for alcohol and opioid misuse and train a DAN encoder for each of these conditions. These encoders are then treated as feature extractors and evaluated as described before using the opioid and alcohol misuse datasets in terms of area under the receiver-operating characteristic curve.
RESULTS
Linear classifier performance for our first evaluation task, i2b2 comorbidity challenge, is in Table 1. Line 1 shows the performance of a traditional baseline – a linear SVM classifier trained with bag-of-CUIs features. Line 2 (DANprev) is our previous system12 that uses a DAN-based patient-level encoder (included for comparison). Lines 3-8 all use various encounter-level neural network encoders.
Table 1.
Average SVM classifier performance on 16 Integrating Biology and the Bedside comorbidity challenge phenotyping tasks
| Encoder | Encoder input | SVM input | Macro P | Macro R | Macro F1 |
|---|---|---|---|---|---|
| none | none | bag-of-CUIs | 0.733 | 0.65 | 0.675 |
| DANprev | CUIs | DAN hidden layer | 0.709 | 0.725 | 0.715 |
| CNN | words | CNN hidden layer | 0.719 | 0.723 | 0.718 |
| CNN | words | CNN hidden layer + bag-of-CUIs | 0.719 | 0.723 | 0.718 |
| CNN | words | CNN max pooling layer | 0.737 | 0.726 | 0.729 |
| CNN | words | CNN max pooling layer + bag-of-words | 0.737 | 0.726 | 0.729 |
| DAN | CUIs | DAN hidden layer | 0.752 | 0.751 | 0.746 |
| DAN | CUIs | DAN hidden layer + bag-of-CUIs | 0.784 | 0.744 | 0.755 |
Performance is compared with SVM trained on a bag-of-CUIs representation of input notes (baseline) vs the representations derived from encoders pretrained on billing code prediction tasks.
CUIs: concept unique identifiers; DAN: deep averaging network; SVM: support vector machine.
In Tables 2 and 3, we show the performance of an SVM classifier on the alcohol and opioid misuse datasets. The first line in these tables show the performance of an SVM classifier trained using bag-of-CUIs representation of the input notes (baseline). The subsequent lines show the performance of an SVM classifier that uses input note representations obtained from a DAN-based encoder pretrained on different billing code prediction tasks. CUIs are used as input to the encoder. Lines 2 and 3 show the performance when the encoder was pretrained on all billing codes. The last line shows the performance of a phenotype-specific encoder, ie, when the encoder was pretrained on opioid misuse billing codes only.
Table 2.
Comparison of different input representations on the performance of an SVM classifier on the opioid misuse data
| Encoder | Pretraining targets | SVM input | ROC AUC |
|---|---|---|---|
| none | none | bag-of-CUIs | 0.838 |
| DAN | all billing codes | DAN hidden layer | 0.889 |
| DAN | all billing codes | DAN hidden layer + bag-of-CUIs | 0.916 |
| DAN | opioid-specific billing codes | DAN hidden layer | 0.951 |
Bag-of-CUIs input (baseline) is compared with the performance of the input obtained from a deep averaging network encoder pretrained on different billing code prediction tasks.
AUC: area under the curve; CUIs: concept unique identifiers; DAN: deep averaging network; ROC: receiver-operating characteristic curve; SVM: support vector machine.
Table 3.
Comparison of different input representations on the performance of an SVM classifier on alcohol misuse data.
| Encoder | Pretraining targets | SVM input | ROC AUC |
|---|---|---|---|
| none | none | bag-of-CUIs | 0.714 |
| DAN | all billing codes | encoder hidden layer | 0.725 |
| DAN | all billing codes | encoder hidden layer + bag-of- CUIs | 0.723 |
| DAN | alcohol-specific billing codes | encoder hidden layer | 0.730 |
Bag-of-CUIs input (baseline) is compared with the performance of the input obtained from a deep averaging network encoder pretrained on different billing code prediction tasks.
AUC: area under the curve; CUIs: concept unique identifiers; ROC: receiver-operating characteristic curve.
DISCUSSION AND CONCLUSION
Our CNN-based clinical text encoder outperformed the bag-of-CUIs baseline by a wide margin and showed approximately the same performance as our previous DAN-based encoder. Concatenating the encoder-generated representations with sparse bag-of-CUIs vectors did not lead to improvements over the dense representations only scenario, likely because the CNN encoder already captures explicit strong features. Using CNN max pooling layer as a text representation improved the performance further, indicating that the CNN-generated feature map already contained the necessary signal and no benefits could be obtained by capturing feature interactions in an additional fully connected layer.
Our DAN-based encoder outperformed the bag-of-CUIs baseline and our previous encoder by a wide margin. Adding additional bag-of-CUIs features to the DAN-generated representations helped to improve the performance further, establishing the new state-of-the art on i2b2 comorbidity data. Prior to this work, to the best of our knowledge, the state-of-the-art on the i2b2 obesity challenge data is presented in Yao et al,30 who report the macro F1 score of 0.677 (precision and recall not reported). In all, utilizing our text encoder improved over the performance of the bag-of-CUIs features baseline by 8 points and by more than 7 points over the previous state-of-the-art.
Our DAN-based encoder showed the best performance on the comorbidity data and we proceeded by evaluating it on the opioid misuse data, where it helped to improve the classifier performance by more than 5 points. Combining sparse bag-of-CUIs with the encoder generated representations improved the performance further. Finally, when the encoder was pretrained on the opioid-specific billing codes only, we obtained further improvements, outperforming the bag-of-CUIs baseline by over eleven points.
Similarly to the opioid misuse task, we find that the use of our text encoder helps to improve the classifier performance on the alcohol misuse detection task, although the size of the improvements is more modest. This is likely due to the fact that alcohol misuse prediction relies on the detection of only a handful of strong lexical features which are captured well by a bag-of-CUIs baseline.28
In general, we find that pretraining using billing codes is a viable route for pretraining. The representations generated by jointly predicting the billing codes associated with a patient encounter have properties of universal patient representations as they were beneficial for all the phenotyping tasks reported here. While phenotype-specific pretraining is beneficial, it is less practical because it requires additional effort tuning the encoder to a specific set of billing codes. Nevertheless, we find that this is a viable route for trading the generality of the pretrained encoder for better performance on a specific phenotyping task.
As mentioned in the Introduction, an alternative to using pretraining for feature extraction (as in our methods described above) is an approach known as fine-tuning. In a fine-tuning approach, a new task is added as an additional output layer to a pretrained network. The task labels are then given to the network, whose entire set of weights can be updated while learning to predict the labels for the new task. While fine-tuning sounds better in theory, we find that it is difficult in practice. Fine-tuning requires optimizing an order of magnitude more hyperparameters, including the learning rate, the dropout rate, and number of training epochs, batch size, the optimizer parameters, not to mention the choices related to the training schedule to deal with issues like catastrophic forgetting.31 Howard and Ruder9 discuss a number of fine-tuning methods such as discriminative fine-tuning, which tunes the learning rate for each layer, and gradual unfreezing, which “thaws” one layer at a time for training. These methods amount to useful heuristics but the accumulated scientific knowledge about how fine-tuning works seems to be insufficiently precise to allow for reliable use. We made some preliminary attempts to fine-tune our pretrained encoder using heuristic approaches, but reverted to using our model in feature extraction as a more practical alternative. Future work will continue investigating the fine-tuning approach.
Prior to our work, methods for text encoding, such as BERT,10 BioBERT,32 and ELMo,8 focused on encoding sentence or paragraph-sized units of text. In this work, we target larger units of text such as individual clinical notes or collections of notes for a patient. While it may be possible to combine sentence-level representations derived from models like BERT into a document-level representation, we leave this investigation for future work. While our ultimate goal is developing a universal patient encoder, which captures most essential information represented in the text of the notes, we acknowledge that using only ICD codes as pretraining targets has limitations. It is likely that extending our methods to include other structured variables, such as medication orders, primary diagnosis, demographic information, and readmission status, could be the next step toward building a universal encoder, leading to even more robust document representations.
The approach we describe here has now been successfully applied to many separate phenotyping tasks, but it is worth thinking about the limitations of using billing codes for pretraining. Because billing codes often describe existing diagnoses, it is possible that the tasks we describe here are successful because they have strong relations to several billing codes. In that case, one might expect that classifiers for nondisease target variables, such as smoking status or specific symptoms, may not benefit from the pretraining regimen described here. In such cases it may be necessary to augment the source of supervision with other types of labels. Future work will explore potential limitations of billing codes as a source of supervision and additional feasible sources of supervision; for example, combining billing code and language modeling objectives could lead to a truly universal clinical text encoder.
FUNDING
This work was supported by National Institutes of Health/National Library of Medicine grant number R01LM012973 (Timothy Miller (PI), Dmitriy Dligach (Co-I)) and National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism grant number K23AA024503 (Majid Afshar (PI)).
AUTHOR CONTRIBUTIONS
DD and TM contributed to the design, experiments, analysis, and writing the manuscript. MA provided the substance misuse data and contributed to the analysis and writing the manuscript.
Conflict of interest statement
None declared.
References
- 1. Rajpurkar P, Jia R, Liang P. Know what you don’t know: unanswerable questions for SQuAD. Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics. 2018: 784–789.
- 2. Hassan H, Aue A, Chen C, et al. Achieving human parity on automatic Chinese to English news translation [published online ahead of print Mar 15]. arXiv2018.
- 3. Long M, Cao Y, Wang J, et al. Learning transferable features with deep adaptation networks. Proceedings of the 32nd International Conference on International Conference on Machine Learning. 2015;37:97–105. JMLR. org. [Google Scholar]
- 4. Razavian AS, Azizpour H, Sullivan J, et al. CNN features off-the-shelf: an astounding baseline for recognition. In: IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops. 2014. doi: 10.1109/CVPRW.2014.131.
- 5. Mikolov T, Corrado G, Chen K, et al. Efficient estimation of word representations in vector space. In: Proceedings of the International Conference on Learning Representations (ICLR 2013). 2013.
- 6. Pennington J, Socher R, Manning CD. GloVe: global vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing. 2014: 1532–43. doi: 10.3115/v1/D14-1162.
- 7. Le Q, Mikolov T. Distributed representations of sentences and documents. In: International Conference on Machine Learning—ICML 2014. 2014: 1188–96.
- 8. Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations. Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers). 2018: 2227–2237.
- 9. Howard J, Ruder S. Universal language model fine-tuning for text classification. Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). 2018:328–339.
- 10. Devlin J, Chang M-W, Lee K, et al. Bert: pre-training of deep bidirectional transformers for language understanding [published online ahead of print Oct 11]. arXiv2018.
- 11. Akbik A, Blythe D, Vollgraf R. Contextual string embeddings for sequence labeling. In: Proceedings of COLING 2018. 2018. doi: 10.1007/s11517-008-0365-4.
- 12. Dligach D, Miller T. Learning patient representations from text. In: Proceedings of the Seventh Joint Conference on Lexical and Computational Semantics. 2018: 119–23.
- 13. Radford A, Narasimhan K, Salimans T, et al. Improving language understanding with unsupervised learning. 2018. Technical report, OpenAI.
- 14. Choi E, Bahadori MT, Song L, et al. GRAM: graph-based attention model for healthcare representation learning. In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2017: 787–95. [DOI] [PMC free article] [PubMed]
- 15. Choi E, Bahadori MT, Schuetz A, et al. Doctor ai: predicting clinical events via recurrent neural networks. In: Machine Learning for Healthcare Conference. 2016: 301–18. [PMC free article] [PubMed]
- 16. Lipton ZC, Kale D, Elkan C, et al. Learning to diagnose with LSTM recurrent neural networks [published online ahead of print Mar 21]. arXiv2017.
- 17. Miotto R, Li L, Kidd BA, et al. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016; 6: 26094.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen P, Tran T, Wickramasinghe N, et al. Deepr: a convolutional net for medical records. IEEE J Biomed Health Inform2017; 21 (1): 22–30. doi: 10.1109/JBHI.2016.2633963. [DOI] [PubMed]
- 19. Pham T, Tran T, Phung D, et al. Deepcare: a deep dynamic memory model for predictive medicine. In: Pacific-Asia Conference on Knowledge Discovery and Data Mining. 2016: 30–41.
- 20. Sushil M, Šuster S, Luyckx K, et al. Patient representation learning and interpretable evaluation using clinical notes. J Biomed Inform2018; 84: 103–13. doi: 10.1016/j.jbi.2018.06.016. [DOI] [PubMed]
- 21. Pan SJ, Yang Q.. A survey on transfer learning. IEEE Trans Knowl Data Eng 2010; 2210: 1345–59. [Google Scholar]
- 22. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharabiani MTA, Aylin P, Bottle A.. Systematic review of comorbidity indices for administrative data. Med Care 2012; 5012: 1109–18. doi: 10.1097/MLR.0b013e31825f64d0. [DOI] [PubMed] [Google Scholar]
- 24. Weiss AJ, Bailey MK, O’Malley L, et al. Patient characteristics of opioid-related inpatient stays and emergency department visits nationally and by state, 2014. HCUP Statistical Brief, 2017. [PubMed]
- 25. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 185: 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Substance Use and Mental Health Services Administration. 2015 National Survey on Drug Use and Health Rockville, MD: Center for Behavioral Health Statistics and Quality; 2016. doi: 10.2186/jjps.49.498.
- 27. Saunders JB, Aasland OG, Babor TF, et al. Development of the alcohol use disorders identification test (AUDIT): WHO collaborative project on early detection of persons with harmful alcohol consumption‐II. Addiction1993; 88 (6): 791–804. doi: 10.1111/j.1360-0443.1993.tb02093.x. [DOI] [PubMed]
- 28. Afshar M, Phillips A, Karnik N, et al. Natural language processing and machine learning to identify alcohol misuse from the electronic health record in trauma patients: development and internal validation. J Am Med Inform Assoc 2019; 263: 254–61. doi: 10.1093/jamia/ocy166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergstra J, Bengio Y.. Random search for hyper-parameter optimization. J Mach Learn Res 2012; 13: 281–305. [Google Scholar]
- 30. Yao L, Mao C, Luo Y. Clinical text classification with rule-based features and knowledge-guided convolutional neural networks. In: 2018 IEEE International Conference on Healthcare Informatics Workshop (ICHI-W). 2018: 70–1. [DOI] [PMC free article] [PubMed]
- 31. French RM. Catastrophic forgetting in connectionist networks. Trends Cogn Sci 1999; 34: 128–35. doi: 10.1016/S1364-6613(99)01294-2. [DOI] [PubMed] [Google Scholar]
- 32. Lee J, Yoon W, Kim S, et al. BioBERT: pre-trained biomedical language representation model for biomedical text mining [published online ahead of print Feb 3]. arXiv2019. [DOI] [PMC free article] [PubMed]