Abstract
Objective
This article presents a novel method of semisupervised learning using convolutional autoencoders for optical endomicroscopic images. Optical endomicroscopy (OE) is a newly emerged biomedical imaging modality that can support real-time clinical decisions for the grade of dysplasia. To enable real-time decision making, computer-aided diagnosis (CAD) is essential for its high speed and objectivity. However, traditional supervised CAD requires a large amount of training data. Compared with the limited number of labeled images, we can collect a larger number of unlabeled images. To utilize these unlabeled images, we have developed a Convolutional AutoEncoder based Semi-supervised Network (CAESNet) for improving the classification performance.
Materials and Methods
We applied our method to an OE dataset collected from patients undergoing endoscope-based confocal laser endomicroscopy procedures for Barrett’s esophagus at Emory Hospital, which consists of 429 labeled images and 2826 unlabeled images. Our CAESNet consists of an encoder with 5 convolutional layers, a decoder with 5 transposed convolutional layers, and a classification network with 2 fully connected layers and a softmax layer. In the unsupervised stage, we first update the encoder and decoder with both labeled and unlabeled images to learn an efficient feature representation. In the supervised stage, we further update the encoder and the classification network with only labeled images for multiclass classification of the OE images.
Results
Our proposed semisupervised method CAESNet achieves the best average performance for multiclass classification of OE images, which surpasses the performance of supervised methods including standard convolutional networks and convolutional autoencoder network.
Conclusions
Our semisupervised CAESNet can efficiently utilize the unlabeled OE images, which improves the diagnosis and decision making for patients with Barrett’s esophagus.
Keywords: endomicroscopy, Barrett’s esophagus, semisupervised learning, convolutional autoencoders
INTRODUCTION
The computer-aided diagnosis (CAD) system utilizes digital imaging processing and machine learning for medical imaging analytics. CAD systems aim at assisting doctors in the interpretation of medical images to speed up the diagnosis and reduce the human biases. A typical CAD system consists of image quality control, feature extraction, predictive modeling, and model visualization, which enables automatic decision making with reliable and reproducible performance.1 With the ability to quantitatively represent medical images, the CAD system is a helpful tool to detect rare events and subtle changes that may be extremely challenging for human observers. Thus, researchers have developed CAD systems for multiple imaging modalities including computed tomography,2 magnetic resonance imaging,3 and whole-slide images,4 which have significantly improved the diagnosis of various diseases.
Optical endomicroscopy (OE) is a newly emerged endoscopic imaging based on confocal microscopy, spectroscopy-based imaging, or optical coherence tomography.5 By combining endoscopy and microscopy, OE can function as “optical biopsies,” which enables real-time in situ biopsy instead of the conventional biopsy and histopathology. Using an OE technique, the physician can make real-time clinical decisions about the grade of dysplasia, if present, and potentially treat the patient during the same endoscopic session. Thus, this novel technique can significantly decrease the waiting time from the time of diagnosis to time for endoscopic treatment.
The newly emerged OE technique provides gastrointestinal endoscopists the opportunity to evaluate the esophageal lining and mucosa in real time through optical biopsy. Compared with the conventional biopsy through endoscope plus microscopic examination afterward, OE can potentially improve the clinical care for patients with gastrointestinal conditions like Barrett’s esophagus (BE). BE is a disorder defined as abnormal changes from normal squamous epithelium to the columnar epithelium. The abnormal changes usually happen in the lower portion of the esophagus.6 BE is a well-known risk factor for esophageal adenocarcinoma (EAC). Once BE is diagnosed, doctors will perform a biopsy and use the histologic severity of the targeted BE tissue to determine the cancer prevention surveillance intervals and treatment recommendations. With the limited amount of tissue collected during a biopsy and the time delay between biopsy and actual diagnosis, OE is a promising novel technique for real-time diagnosis using optical biopsy. Clinical trials have demonstrated that OE could achieve improved clinical care quality for patients with BE.7–9 However, a large number of microscopic images are generated during an OE session. Human examination of all microscopic images in real time through an OE session can be demanding and prone to errors. Thus, a fast and reliable CAD system to automatically process these microscopic images is essential to enable real-time diagnosis for BE patients using OE techniques.
To build a reliable CAD system, we typically need substantial labeled training data to select the optimal feature subsets and train robust classification models through supervised learning. However, accumulating a large labeled dataset for OE images can be expensive and time consuming, and there are no public OE datasets available yet. On the other hand, it is relatively easy to collect a significant number of unlabeled images through each OE session. Thus, considering the lack of labeled OE images and the easy access to unlabeled OE images, we propose to improve the classification of OE images by utilizing the unlabeled images through semisupervised learning. In the previous study, we have applied handcrafted feature extraction and label propagation methods for semisupervised learning.10 With the rapid development of deep learning and their massive success in natural image processing, we propose to improve our previous method by exploiting the convolutional autoencoders (CAEs) and developing a semisupervised deep neural network Convolutional AutoEncoder based Semi-supervised Network (CAESNet) for improving the multiclass classification of BE. With extensive experiments on the OE dataset collected at Emory University, we have demonstrated the superior performance of our semisupervised CAESNet compared with all baselines.
Optical endomicroscopy
OE is a novel optical technology integrating endoscopy with microscopy for in situ diagnosis. It enables real-time diagnosis and treatment, in contrast to the delay in treatment due to the inherent time needed to obtain a final diagnosis by histopathology.
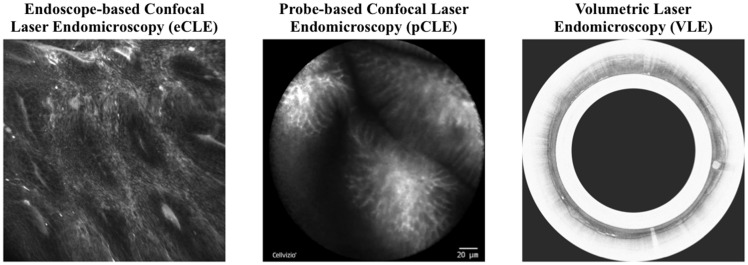
Currently, 3 types of commercial OE systems are available for clinical use: endoscope-based confocal laser endomicroscopy (eCLE), probe-based CLE, and volumetric laser endomicroscopy (Figure 1),5 which have been developed and approved in 2004, 2006, and 2013, respectively. Table 1 compares specifications of the 3 technologies. They are significantly different in resolution, acquisition position, acquisition speed, and microscopic imaging presentation, and we refer the readers to these articles for detailed comparison.5,11–13 As clinical trials suggest that eCLE has better performance than probe-based CLE in the diagnosis of esophageal diseases,14 we focus on developing a semisupervised classification method for eCLE images, and we believe that this method can be readily generalized to other OE modalities.
Figure 1.
Example images of three types of commercial optical endomicroscopy systems. Left: endoscope-based confocal laser endomicroscopy (eCLE); middle: probe-based confocal laser endomicroscopy (pCLE); right: volumetric laser endomicroscopy (VLE).
Table 1.
Comparison of specifications for 3 optical endomicroscopy technologies
| eCLE | pCLE | VLE | |
|---|---|---|---|
| Company | Pentax, Tokyo, Japan; and Optiscan, Victoria, Australia | Mauna Kea Technologies, Paris, France | NinePoint Medical, Cambridge, MA |
| Model | Pentax ISC 1000 | Cellvizio 100 Series | Nvision VLE |
| Data Format | Surface images | Surface image video | Helical scan video |
| Axial Information | Multiple z-planes | Single z-plane | Multiple z-planes |
| Axial resolution, µm | 7 | NA | 7 |
| Lateral resolution, µm | 0.7 | 3.5 | NA |
| Z-depth, µm | 0–250 | 60 | 0–3000 |
| Frame size | 600-µm diameter | 6-cm scan acquisition length | |
| Image size | NA | ||
| Speed | 0.8-1.2 frames/s | 12 frames/s | 1200 slices/90 s |
| Intravenous fluorescein required? | Yes | Yes | No |
eCLE: endoscope-based confocal laser endomicroscopy; NA: not applicable; pCLE: probe-based confocal laser endomicroscopy; VLE: volumetric laser endomicroscopy.
Barrett’s esophagus
BE, a well-known risk factor for EAC,15 is characterized by the abnormal changes from normal squamous epithelium to the columnar epithelium at the lower portion of the esophagus. The population incidence of EAC in the United States has an estimated rise of 300%-350% since the 1970s,16,17 making up 60% of the new esophageal cancer diagnosis in the United States in 2009,18 with only a 5-year survival rate of 15%-20%.19 The progression from BE to adenocarcinoma involves multiple stages, namely nondysplastic BE, low-grade dysplasia, high-grade dysplasia, and finally adenocarcinoma.20
The impact of BE on the mortality from EAC is still unclear, which makes directly screening for BE controversial.21 Thus, physicians identify patients with BE through either subjective selectively screening22 or an upper endoscopy performed for an unrelated reason. The rapid development of OE may provide another mechanism to help detect the neoplastic changes earlier than conventional endoscopy and improve the efficiency of cancer surveillance.14
CAD for BE classification
The CAD-based BE classification using OE has become an emerging field of research due to the rapidly increasing population incidence of EAC, as it can realize real-time diagnosis, improve the patients’ prognosis by early detection, and reduce the physicians’ workload.
Conventional CAD pipeline includes image quality control, feature extraction, predictive modeling, and model visualization. For example, Grisan et al23 applied support vector machine to identify gastric metaplasia vs intestinal metaplasia using rotation invariant local binary pattern features. Veronese et al24 improved the approach via a 2-stage classification pipeline, and Ghatwary et al25 applied image enhancement before feature extraction for increased overall accuracy.
More recently, deep learning has also been applied to the classification of BE using OE images. For example, Mendel et al26 have implemented deep convolutional neural networks for binary classification of patients into EAC and non-EAC, realizing a sensitivity of 0.94 and a specificity of 0.88 with leave-1-out cross-validation. Hong et al27 have also applied convolutional neural networks for 3-class classification for BE and neoplasia using endomicroscopic images with an accuracy of 80.77%.
However, the performance and generalization capability of CAD systems is still largely constrained by the scarcity of annotated images. Besides collecting more annotated data, researchers incorporate unsupervised, semisupervised, and weakly supervised learning to make the most of the data available.
One major application of semisupervised learning is image segmentation. Papandreou et al28 are among the first in studying weakly and semisupervised learning for semantic image segmentation. The authors designed an expectation maximization algorithm for iteratively updating the prediction for pixel-level annotations and the parameters of segmentation neural networks. Jia et al29 approached the weakly supervised segmentation with multiple instance learning by aggregating the pixel-level annotations and designed a constrained optimization process when additional supervision information is available. More recently, Li et al30 studied weakly supervised segmentation for the prostate cancer pathological images by utilizing the prior knowledge about the epithelium-stroma distribution.
Besides image segmentation, semisupervised learning has also been applied for image classification. For example, our prior work applies semisupervised learning to eCLE images by propagating labels to unlabeled images.10 However, handcrafted feature extraction is a major bottleneck for further performance improvement. The unsupervised nature of CAE makes it a popular choice for automated image feature extraction,31 which can naturally utilize the unlabeled data for improved feature representation. Thus, in this article, we adopt the CAE to build a semisupervised deep neural network called CAESNet for improving the classification of BE status using OE images.
Materials and METHODS
Data
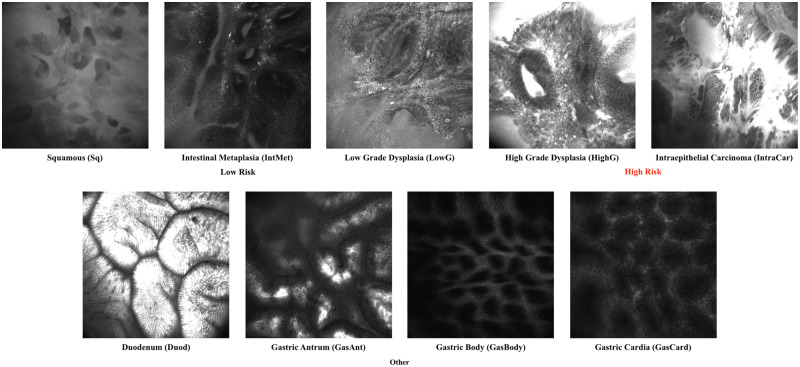
The data we used were images collected from patients undergoing eCLE procedures for BE at Emory Hospital. The dataset consists of 429 images labeled as one of the nine classes by an expert gastrointestinal endomicroscopist and 2826 unlabeled images for semisupervised learning. The statistics of these images are summarized in Table 2. Example images for each class are shown in Figure 2.
Table 2.
Statistics of the Barrett’s esophagus dataset
| Categories | Subclass | Images |
|---|---|---|
| Low risk | Squamous (Sq) | 41 |
| Intestinal metaplasia (IntMet) | 153 | |
| Low-grade dysplasia (LowG) | 23 | |
| High risk | High-grade dysplasia (HighG) | 4 |
| Intraepithelial carcinoma (IntraCar) | 43 | |
| Other | Duodenum (Duod) | 48 |
| Gastric antrum (GasAnt) | 60 | |
| Gastric body (GasBody) | 28 | |
| Gastric cardia (GasCard) | 29 | |
| Total | 429 | |
| Unlabeled | 2826 | |
Figure 2.
Example images of the endoscope-based confocal laser endomicroscopy (eCLE) dataset we used in this article. The images can be classified into nine categories including squamous (Sq), intestinal metaplasia (IneMet), low-grade dysplasia (LowG), high-grade dysplasia (HighG), intraepithelial carcinoma (IntraCar), duodenum (Duod), gastric antrum (GasAnt), gastric body (GasBody), and gastric cardia (GasCard).
Image preprocessing and data augmentation
The original size of our eCLE images is Pixels. Because of the limited number of training data, we apply image augmentation to increase the number of instances for training, which is expected to improve the performance of image reconstruction and classification. For each batch of training images, we apply a data augmentation consisting of random rotation, zooming in, and flipping (Supplementary Note 1). The augmented images are then scaled into a smaller size of to reduce the number of parameters. Examples of the augmented images are shown in Supplementary Figures S1 and S2.
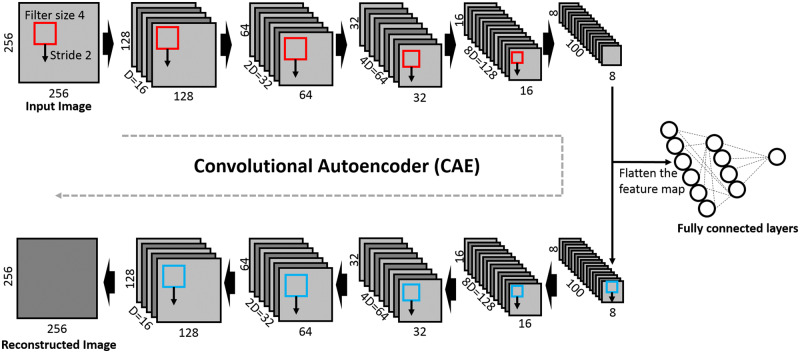
CAESNet: stacked CAEs for semisupervised learning
The structure of CAESNet is shown in Figure 3, which consists of a stacked convolutional autoencoder for unsupervised feature representation and fully connected layers for image classification. The encoder consists of 5 convolution layers, each with a filter of size 4 and stride 2, resulting in encoded hidden codes of size . Similarly, the decoder consists of 5 deconvolution (transposed convolution) layers with the same filter size and stride as that of encoders. The depth of each layer of the encoder is 16, 32, 64, 128, and 100, respectively, with the last layer as the bottleneck layer. The bottleneck layer with dimensions of is first flattened into a vector of length 6400 and then connected to the second fully connected layers through a rectified linear unit activation function, a dropout layer, and batch normalization layer. Finally, the label is predicted from the second fully connected layer with a softmax function. The dropout layer is applied during training stage and turned off during test stage.
Figure 3.
Visualization of the proposed convolutional autoencoders based semisupervised learning model Convolutional AutoEncoder based Semi-supervised Network. The original images are first encoded into hidden codes through 5 layers of convolutional layers with a filter size of 4 × 4 and a stride of 2. The hidden codes can be either fed into fully connected layers for classification or into the 5 layers of deconvolutional layers for decoding into original images.
There are two loss functions in this network, namely reconstruction loss and classification loss. In the unsupervised stage, we use the reconstruction loss to train the encoder-decoder with both labeled and unlabeled images. In the supervised stage, we use the classification loss to train the encoder-classifier with only labeled images. The reconstruction loss is a measure of the differences between the input images and the reconstructed images, measured by the mean squared errors:
where is the number of images in the batch, is the number of pixels of each input image, is the original value of the th pixel of the th image, and is the value of the th pixel of the th reconstructed image.
We use the cross-entropy function as classification loss to measure of the differences between the real labels and the predicted labels of the images:
where is the number of images in the batch, is the real label of the th image in the batch, and is the predicted probability of the th image. The is a vector of one-hot coded labels, which equals 1 for the real label and 0 for other labels.
We construct four models utilizing all or parts of this network structure with or without the unlabeled images. The major components of these four models are summarized in Table 3.
Table 3.
The major components of 4 models
| Components | Loss function | Train set | Endpoint | |
|---|---|---|---|---|
| Model 1 | Encoder, decoder | Reconstruction loss | Labeled / unlabeled | Reconstruction |
| Model 2 | Encoder, fully connected | Classification loss | Labeled | Classification |
| Model 3 | Encoder, decoder, fully connected | Reconstruction loss | Labeled | Classification |
| Classification loss | ||||
| Model 4 (CAESNet) | Encoder, decoder, fully connected | Reconstruction loss | Labeled and unlabeled | Classification |
| Classification loss |
Algorithm 2: Training schemes for four models.
Def Model_1(UnlabeledImages):
while modelConverged() do:
unsupervisedTrain(UnlabeledImages)
end while
Def Model_2({LabeledImages, Labels}):
while modelConverged() do:
supervisedTrain({LabeledImages, Labels})
end while
Def Model_3({LabeledImages, Labels}):
while modelConverged() do:
unsupervisedTrain(LabeledImages)
supervisedTrain({LabeledImages, Labels})
end while
Def Model_4(UnlabeledImages, {LabeledImages, Labels}):
while modelConverged() do:
unsupervisedTrain(UnlabeledImages+LabeledImages)
if LabeledImages:
supervisedTrain({LabeledImages, Labels})
end if
end while
In model 1, we implemented a stacked CAE, which is an unsupervised feature representation network. Each input image is encoded by an encoder with 5 convolution layers into hidden codes. Then the input images are reconstructed by a decoder with 5 deconvolution layers (transposed convolution layers) from the hidden codes. The network is trained by minimizing the reconstruction loss (Algorithm 1 unsupervisedTrain).
Algorithm 1: Unsupervised and supervised learning.
Def unsupervisedTrain(Image):
while unsupervisedTraining() do
end while
Def supervisedTrain(Image, Label):
while supervisedTraining() do
end while
In model 2, we use only the stacked convolutional encoders and the fully connected layers for image classification, which is a standard implementation for image classification. Each input image is first encoded and then classified into multiple categories. The network is trained by minimizing the classification loss using only the labeled images (Algorithm 1 supervisedTrain).
In model 3, we use both the stacked CAE for image reconstruction and the fully connected layers for image classification, but only using the labeled images. In each training step, we first update the encoder and decoder by reconstruction loss, then we update the encoder and fully connected layers by classification loss. All images are firstly encoded into hidden codes. After encoding, we reconstruct the input images and update the encoder and decoder by taking the gradient of reconstruction loss. Then, we use the updated encoder to regenerate the hidden codes and pass them through fully connected layers for image classification. We update the encoder and fully connected layers by taking the gradient of the classification loss.
In model 4 (CAESNet), we use the same structure as that in model 3, but utilize both labeled and unlabeled images for training. For each image, if it is labeled, we first update the encoder and decoder, then the encoder and fully connected layers; on the other hand, if it is unlabeled, we only update the encoder and decoder by minimizing the reconstruction loss. The implementation details of model 4 are presented in Supplementary Note 2.
Model evaluation and classification metrics
The three classification pipelines are evaluated using stratified 4-fold cross-validation (Supplementary Note 3.1). In each fold of the cross-validation, we split the labeled data into training, validation, and test datasets. We initiate the weights of our networks with multiple random seeds and select the best model based on the classification loss on the validation set. Data augmentation has been applied only on the training datasets. To enable statistical test, we repeat the 4-fold cross-validation for three times.
We evaluate the multiclass classification results using four metrics, namely accuracy, precision, F1 score, and Cohen’s kappa score (Supplementary Note 3.2). We do not include recall because it is the same as accuracy for multiclass classification if we weighted the recall by the number of samples in each class.
Experiment configuration
We run all data processing pipelines and models on a single server with multiple CPU cores and two NVIDIA Tesla K80 GPUs (NVIDIA, Santa Clara, CA). The image augmentation is implemented with Python Library Augmentor32 with random rotation, scaling, and flipping (Supplementary Note 1.3). The CAESNet and baseline deep learning models are implemented with PyTorch Version 0.4.0. The classification results are evaluated with scikit-learn.33 For the training of semisupervised networks, we use a batch size of 20, epochs of 200, and a learning rate of 0.0002. We apply different depths (ie, 16, 32, and 64) to each convolutional layer to see the influence on the classification and reconstruction performance. We also compare the results with our previous semisupervised method,10 which is also briefly described in Supplementary Note 4.
RESULTS
Improved image reconstruction performance by extra training with unlabeled images
The image reconstruction quality increases as the number of training images increases. Figure 4 shows the original images and the reconstructed images from the test set for models 1, 3 and 4. The reconstructed image quality of model 3 is the worst compared with models 1 and 4. However, model 4 can achieve similar reconstruction performance as model 1 even with the extra task of classification. The poor reconstruction performance of model 3 may result from the trade-off between image reconstruction and classification, and the limited number of labeled images.
Figure 4.
The original images (blue rectangle) and the corresponding reconstructed images (red rectangle) by autoencoders in 3 different models. Model 1: an autoencoder using labeled images. Model 3: an autoencoder + a classifier (Clf) using only labeled images. Model 4 (Convolutional AutoEncoder based Semi-supervised Network): an autoencoder + a classifier using both labeled and unlabeled images. Models 1 and 4 (Convolutional AutoEncoder based Semi-supervised Network) achieve better reconstruction results compared with those of model 3.
Improved classification performance by semisupervised learning with unlabeled images
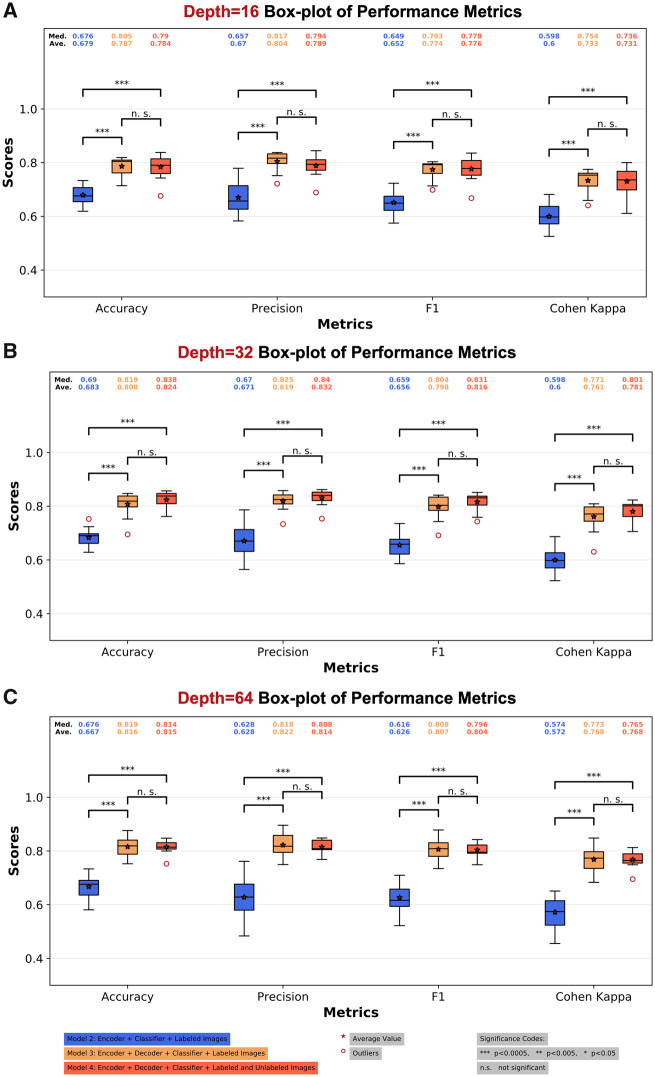
The classification performance of 3 models is shown in Figure 5 and Table 4. Based on Table 4, the performance of model 2 is inferior compared with the best baseline model (label spreading). However, models 3 and 4 consistently achieve better performance compared with the baseline models at all network depths, likely resulting from utilizing the reconstruction loss for regularizing the network. Thus, when trained with the same amount of labeled data, only model 2 suffers from underfitting. Models 3 and 4 achieve similar prediction performance, where model 4 at depth 32 achieves the best average performance (0.824 ± 0.0329). We have also performed the pairwise 2-sample t test for the prediction performance of all models in Table 4 (Supplementary Note 5). Model 3 and model 4 significantly outperforms model 2. However, no significant difference has been identified between models 3 and 4. In Figure 5, we visualized the significance levels among models 2, 3, and 4 at the same network depth. We have also visualized the performance of all models with boxplots and their pairwise 2-sample t-test results in Supplementary Note 5. The training losses of models 2, 3, and 4 are visualized in Supplementary Figure S10.
Figure 5.
Boxplot of the classification performance of various models and depths. (A) The classification performance of three models using depth 16; (B) the performance of three models using depth 32; (C) the performance of three models using depth of 64. Models 3 and 4 (Convolutional AutoEncoder based Semi-supervised Network) achieve similar prediction performance and consistently outperforms model 2. Model 4 (Convolutional AutoEncoder based Semi-supervised Network) at depth 32 achieves the best average performance (0.824 ± 0.0329). The asterisk indicates an average value and a red circle indicates outliers. ***P < .005. Ave.: average; Med.: median; n.s.: not significant.
Table 4.
Classification performance of models with various implementations
| Category | Implementation | Accuracy | Precision | F1 | Cohen’s kappa |
|---|---|---|---|---|---|
| Graph based (baseline) | LP | 0.734 ± 0.0175 | 0.763 ± 0.0479 | 0.674 ± 0.0269 | 0.642 ± 0.0241 |
| Randomized LP (20) | 0.652 ± 0.0465 | 0.709 ± 0.0277 | 0.584 ± 0.0453 | 0.528 ± 0.0741 | |
| Randomized LP (90) | 0.686 ± 0.0318 | 0.702 ± 0.0519 | 0.624 ± 0.0394 | 0.577 ± 0.0477 | |
| Model 2 (supervised) | Network depth 16 | 0.679 ± 0.0375 | 0.67 ± 0.0623 | 0.652 ± 0.046 | 0.6 ± 0.0491 |
| Network depth 32 | 0.683 ± 0.0368 | 0.671 ± 0.0661 | 0.656 ± 0.0458 | 0.6 ± 0.0486 | |
| Network depth 64 | 0.667 ± 0.0431 | 0.628 ± 0.0784 | 0.626 ± 0.0554 | 0.572 ± 0.0601 | |
| Model 3 (supervised) | Network depth 16 | 0.787 ± 0.0369 | 0.804 ± 0.0376 | 0.774 ± 0.0354 | 0.733 ± 0.0439 |
| Network depth 32 | 0.808 ± 0.045 | 0.819 ± 0.034 | 0.798 ± 0.0448 | 0.761 ± 0.052 | |
| Network depth 64 | 0.816 ± 0.0384 | 0.822 ± 0.0446 | 0.807 ± 0.0425 | 0.768 ± 0.0498 | |
| Model 4 (semisupervised) | Network depth 16 | 0.784 ± 0.046 | 0.789 ± 0.0419 | 0.776 ± 0.0458 | 0.731 ± 0.0542 |
| Network depth 32a | 0.824 ± 0.0329 | 0.832 ± 0.0302 | 0.816 ± 0.0342 | 0.781 ± 0.04 | |
| Network depth 64 | 0.815 ± 0.0252 | 0.814 ± 0.0265 | 0.804 ± 0.0262 | 0.768 ± 0.0306 |
LP = label spreading.
Best performing model.
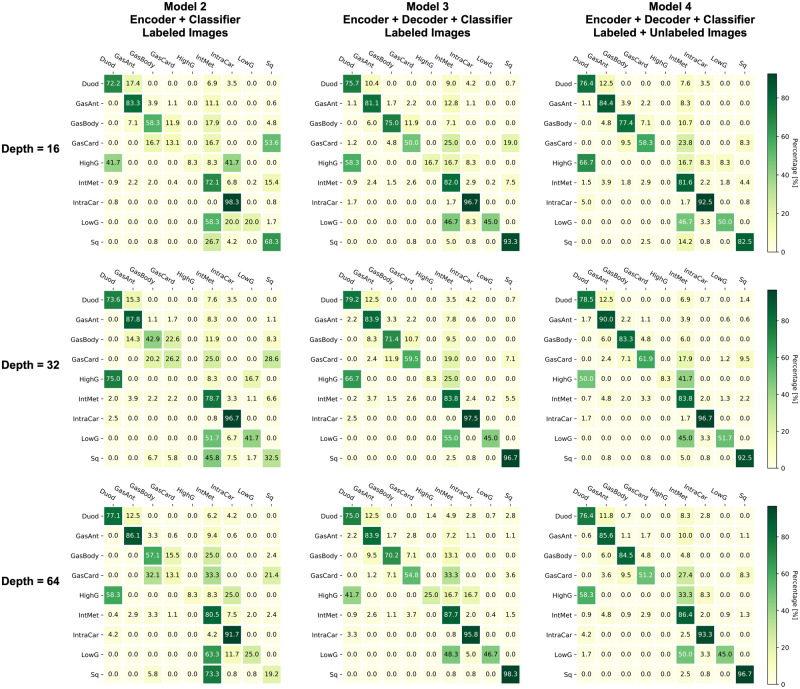
The confusion matrices of three models with various depths are shown in Figure 6. An ideal classification should achieve the diagonal pattern in the confusion matrix. Model 3 and model 4 concentrate more on the diagonal cells compared with model 2, which is consistent with their overall performance. However, both models 3 and 4 makes a relatively poor classification for gastric cardia (GasCard), low grade dysplasia (LowG), and high grade dysplasia (HighG). These three classes are tended to be misclassified as the intestinal metaplasia (IntMet). We suspect that this misclassification could be caused by the limited numbers of samples for GasCard, LowG, and HighG, where we only have 29, 23, and 4 samples, respectively, of the 429 labeled images. On the contrary, we have 153 IntMet samples, which might dominate the classification decisions.
Figure 6.
Confusion matrices of various models and depths. Each confusion matrix is color-coded as a heatmap for visualization purpose. Models 3 and 4 (Convolutional AutoEncoder based Semi-supervised Network) consistently achieves better performance compared with model 2 at all network depths. Duod: duodenum; GasAnt: gastric antrum; GasBody: gastric body; GasCard: gastric cardia; HighG: high-grade dysplasia; IneMet: intestinal metaplasia; IntraCar: intraepithelial carcinoma; LowG: low-grade dysplasia; Sq: squamous.
Fluctuated performance with the number of unlabeled images
After confirming that utilizing unlabeled images to help the regularization of the encoder can improve the prediction performance, we want to investigate the influence of the number of unlabeled images utilized for the model training. With the 2826 unlabeled images, we feed the model in an accumulative fashion. For ratio increasing from 0 to 1.0 with a step size of 0.1, we always use the first proportions of the unlabeled images. For example, when ratio equals to 0.1, we use the first 10% of unlabeled images; when ratio equals to 0.2, we use the same first 10% of unlabeled images plus the subsequent 10% of unlabeled images. There are fluctuations in the prediction performance along the number of unlabeled images applied (Supplementary Figure S11 and Supplementary Table S5). We have also investigated the influence of data augmentation on classification performance using model 2 (Supplementary Note 8), where similar performance fluctuations have been observed.
CONCLUSION
In this article, we developed CAESNet, a CAE-based semisupervised learning framework for multiclass classification of endomicroscopic images. Based on the extensive experiments, we conclude that the stacked CAE is an effective deep learning method to extract informative features from the eCLE images. The CAE network allows us to not only add a regularization for the classification loss but also allows us to utilize the unlabeled images to optimize the encoder in a semisupervised learning fashion.
DISCUSSION
When utilizing a different number of unlabeled images, the performance does not follow a monotonic increasing pattern but fluctuates as the number of unlabeled images increased. There are multiple potential explanations. First, the results could be caused by the training configurations in which we apply the same hyperparameters for experiments with different numbers of unlabeled images. To solve this issue, optimized configurations may need to be searched for each model and dataset. Second, the performance may be related to the quality and underlined labels of the unlabeled images. If the unlabeled images are less relevant with the labeled images, we may experience an adverse effect when utilizing these unrelated unlabeled images. We also suspect that if most unlabeled images are from a specific class, they may preoccupy the autoencoder and make it overfit images from that class. One potential way to solve this problem is to introduce another coefficient to balance the reconstruction loss and classification loss. When we have a larger number of unlabeled images, the reconstruction loss typically drops much faster compared with classification loss. Thus, we can assign larger weights to classification loss so that we can balance the training of image classification as well as the image reconstruction.
There are multiple future directions for our semisupervised model. One future direction is to improve the unsupervised feature representation by applying adversarial autoencoders (AAEs). AAE is a probabilistic autoencoder by matching the aggregated posterior of the hidden code vector of the autoencoder with an arbitrary prior distribution.34 As a result, the encoder tends to generate more meaningful hidden codes. Currently, we only use the unlabeled data to improve the unsupervised feature representation with simple handcrafted data augmentation. With the AAE framework, we can additionally autoaugment the labeled images from a few labeled images and the unlabeled images.35 In this method, we learn a generator for sequences of incremental, black-box transformation functions from the unlabeled images and then apply the learned transformation function generator to the labeled images for augmentation of realistic labeled images. This advanced autoaugmentation for labeled images should be able to improve the classification performance.
Introducing interpretation for the deep model we build is another direction to enable the clinical translation of our method. By introducing techniques like the attention to visualize the model, we want to identify the features of the original images that contribute most to the right classification. This process can, in turn, serve as a parameter tuning or diagnosis step for the model. By examining whether the neural network is truly picking up the disease-relevant structures for the prediction, we can differentiate models fitting the data noise from the truly effective models.
FUNDING
This work was supported by the grants from the National Institutes of Health (National Cancer Institute Transformative R01 CA163256), the National Center for Advancing Translational Sciences (UL1TR000454), Microsoft Research, and Hewlett Packard for LT, HW, and MDW. This work was also supported in part by a scholarship from China Scholarship Council (CSC 201406010343) for LT. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or China Scholarship Council.
AUTHOR CONTRIBUTIONS
All authors listed are justifiably credited with authorship. In detail: LT was involved in conception, design, analysis, and interpretation of data, and drafting of the manuscript. HW was involved in conception, design, analysis, and interpretation of data, and drafting of the manuscript. MDW was involved in conception, design, and interpretation of data, and drafting of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Kevin E. Woods for providing the dataset analyzed in this article and the help throughout the project.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kothari S, Phan JH, Stokes TH, Wang MD.. Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc 2013; 206: 1099–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheng JZ, Ni D, Chou YH, et al. Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci Rep 2016; 6: 24454.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bron EE, Smits M, van der Flier WM, et al. Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: the CADDementia challenge. Neuroimage 2015; 111: 562–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cooper LA, Kong J, Gutman DA, et al. Integrated morphologic analysis for the identification and characterization of disease subtypes. J Am Med Inform Assoc 2012; 192: 317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carignan CS, Yagi Y.. Optical endomicroscopy and the road to real-time, in vivo pathology: present and future. Diagn Pathol 2012; 7: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spechler SJ, Souza RF.. Barrett's esophagus. N Engl J Med 2014; 3719: 836–45. [DOI] [PubMed] [Google Scholar]
- 7. Dunbar KB, Okolo P 3rd, Montgomery E, Canto MI.. Confocal laser endomicroscopy in Barrett's esophagus and endoscopically inapparent Barrett's neoplasia: a prospective, randomized, double-blind, controlled, crossover trial. Gastrointest Endosc 2009; 704: 645–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canto MI, Anandasabapathy S, Brugge W, et al. In vivo endomicroscopy improves detection of Barrett's esophagus-related neoplasia: a multicenter international randomized controlled trial (with video). Gastrointest Endosc 2014; 792: 211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sharma P, Meining AR, Coron E, et al. Real-time increased detection of neoplastic tissue in Barrett's esophagus with probe-based confocal laser endomicroscopy: final results of an international multicenter, prospective, randomized, controlled trial. Gastrointest Endosc 2011; 743: 465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu H, Tong L, Wang MD. Improving multi-class classification for endomicroscopic images by semi-supervised learning. In: 2017 IEEE EMBS International Conference on Biomedical & Health Informatics; 2017: 5–8 [DOI] [PMC free article] [PubMed]
- 11. Sturm MB, Wang TD.. Emerging optical methods for surveillance of Barrett's oesophagus. Gut 2015; 6411: 1816–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leggett CL, Gorospe EC, Chan DK, et al. Comparative diagnostic performance of volumetric laser endomicroscopy and confocal laser endomicroscopy in the detection of dysplasia associated with Barrett's esophagus. Gastrointest Endosc 2016; 835: 880–8 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kang D, Schlachter SC, Carruth RW, et al. Comprehensive confocal endomicroscopy of the esophagus in vivo. EIO 2014; 2 (3): E135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li CQ, Guo J, Zhang JY, Liu JW, Li YQ. Sa1492 A Paralleled Comparison Between Two Sets of Confocal LASER Endomicroscopy in Gastrointestinal Tract. Gastrointestinal Endoscopy. 2014; 79(5):AB233. [Google Scholar]
- 15. Sharma P. Barrett's esophagus. N Engl J Med 2009; 36126: 2548–56. [DOI] [PubMed] [Google Scholar]
- 16. Devesa SS, Blot WJ, Fraumeni JF Jr.. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer 1998; 8310: 2049–53. [PubMed] [Google Scholar]
- 17. Chang JT, Katzka DA.. Gastroesophageal reflux disease, Barrett esophagus, and esophageal adenocarcinoma. Arch Intern Med 2004; 16414: 1482–8. [DOI] [PubMed] [Google Scholar]
- 18. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ.. Cancer statistics, 2009. CA Cancer J Clin 2009; 594: 225–49. [DOI] [PubMed] [Google Scholar]
- 19. Sharma P, McQuaid K, Dent J, et al. A critical review of the diagnosis and management of Barrett's esophagus: the AGA Chicago Workshop. Gastroenterology 2004; 1271: 310–30. [DOI] [PubMed] [Google Scholar]
- 20. Anaparthy R, Sharma P.. Progression of Barrett oesophagus: role of endoscopic and histological predictors. Nat Rev Gastroenterol Hepatol 2014; 119: 525–34. [DOI] [PubMed] [Google Scholar]
- 21. Wang KK, Sampliner RE; Practice Parameters Committee of the American College of Gastroenterology. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett's esophagus. Am J Gastroenterol 2008; 1033: 788–97. [DOI] [PubMed] [Google Scholar]
- 22. Gill RS, Singh R.. Endoscopic imaging in Barrett's esophagus: current practice and future applications. Ann Gastroenterol 2012; 252: 89–95. [PMC free article] [PubMed] [Google Scholar]
- 23. Grisan E, Veronese E, Diamantis G, Trovato C, Crosta C, Battaglia G.. Computer aided diagnosis of barrett's esophagus using confocal laser endomicroscopy: preliminary data. Gastrointest Endosc 2012; 754: 126–26. [Google Scholar]
- 24. Veronese E, Grisan E, Diamantis G, Battaglia G, Crosta C, Trovato C.. Hybrid patch-based and image-wide classification of confocal laser endomicroscopy images in Barrett's esophagus surveillance. I S Biomed Imaging 2013: 362–65. [Google Scholar]
- 25. Ghatwary N, Ahmed A, Ye XJ, Jalab H.. Automatic grade classification of Barretts esophagus through feature enhancement. Proc SPIE 2017: 1013433. [Google Scholar]
- 26. Mendel R, Ebigbo A, Probst A, Messmann H, Palm C.. Barrett’s esophagus analysis using convolutional neural networks In: Maier-Hein KH, Deserno TM, Handels H, Tolxdorff T, eds. Bildverarbeitung für die Medizin 2017. Berlin, Germany: Springer; 2017: 80–5. [Google Scholar]
- 27. Hong J, Park B-Y, Park H.. Convolutional neural network classifier for distinguishing Barrett's esophagus and neoplasia endomicroscopy images. In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE; 2017. [DOI] [PubMed]
- 28. Papandreou G, Chen LC, Murphy KP, Yuille AL.. Weakly- and semi-supervised learning of a deep convolutional network for semantic image segmentation. IEEE I Conf Comp Vis 2015: 1742–50. [Google Scholar]
- 29. Jia ZP, Huang XY, Chang EIC, Xu Y.. Constrained deep weak supervision for histopathology image segmentation. IEEE Trans Med Imaging 2017; 3611: 2376–88. [DOI] [PubMed] [Google Scholar]
- 30. Li JY, Speier W, Ho KC, et al. An EM-based semi-supervised deep learning approach for semantic segmentation of histopathological images from radical prostatectomies. Comput Med Imag Grap 2018; 69: 125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Masci J, Meier U, Ciresan D, Schmidhuber J.. Stacked convolutional auto-encoders for hierarchical feature extraction. Lect Notes Comput Sc 2011; 6791: 52–59. [Google Scholar]
- 32. Bloice MD, Stocker C, Holzinger A. Augmentor: An Image Augmentation Library for Machine Learning. arXiv e-prints 2017. https://ui.adsabs.harvard.edu/\#abs/2017arXiv170804680B. Accessed August 1, 2017.
- 33. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011; 12: 2825–30. [Google Scholar]
- 34. Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B. Adversarial autoencoders. arXiv2015. May 25 [E-pub ahead of print].
- 35. Ratner AJ, Ehrenberg HR, Hussain Z, Dunnmon J, Re C.. Learning to compose domain-specific transformations for data augmentation. Adv Neural Inf Process Syst 2017; 30: 3239–49. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.