Abstract
Objective:
The follicular fluid (FF) of women with polycystic ovary syndrome (PCOS) seems to exhibit a profile different from that of fertile women, which may be related to folliculogenesis disruption in PCOS patients. The aim of this study was to evaluate the differentially expressed proteins in the FF of women with PCOS compared to oocyte donors (ODs).
Methods:
This screening study included thirteen (13) women who underwent in vitro fertilization (IVF) cycles: seven (7) ODs and six (6) PCOS patients. The patients underwent standard ovarian stimulation, and the FF was analysed using ion trap and time-of-flight liquid chromatography-mass spectrometry (LCMS-IT-TOF).
Results:
The FF of the patients was matched to 229 proteins, with 61 proteins exclusive to the PCOS group, 123 proteins exclusive to the ODs, and 45 proteins found in both groups. We highlight fetuin-A and vitamin D ligand protein, which were exclusively expressed in the PCOS group; Complement C3 overexpressed in the PCOS group; and 26S protease only expressed in the OD group. The canonical pathways LXR/RXR activation, FXR/RXR activation, prothrombin activation are directly related to the disrupted metabolism and increased inflammatory status found in PCOS patients.
Conclusions:
The findings of the differentially expressed proteins and matched pathways are associated with folliculogenesis, indicating it relevance to oocyte quality.
Keywords: polycystic ovary syndrome, proteomic, follicular fluid, in vitro fertilization
INTRODUCTION
Polycystic ovary syndrome (PCOS) is characterized by hyperandrogenism, ovulation disorder and polycystic ovaries (PCO) and the exclusion of other endocrinopaties (Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group, 2004). PCOS affects 6-8% of women of reproductive age. Although PCOS was first described eighty years ago (Stein & Leventhal, 1935), its aetiology is not yet fully elucidated, as it is a heterogeneous and complex disorder with metabolic and reproductive implications. PCOS represents the major ovulatory cause of infertility, which leads some PCOS patients to pursue in vitro fertilization (IVF) treatments (Dumesic et al., 2015).
The follicular fluid (FF) that surrounds the cumulus-oocyte complex contains several factors that originate from the blood transudate and are secreted by cumulus cells, such as proteins, steroids, polysaccharides and other metabolites; thus, FF provides a unique microenvironment in which to study oocyte development and maturation (Schweigert et al., 2006; Appasamy et al., 2008)
It is recognized that the FF from women with PCOS is characterized by deregulated expression of several compounds, including anti-Müllerian hormone (AMH), inhibin-B, activin-A, amphiregulin, heparan sulfate proteoglycan 2; tumour necrosis factor (TNF), α-induced protein 6 and plasminogen (Ambekar et al., 2015). Although previous studies have identified molecules in the FF of PCOS patients that are associated with the deregulation of follicle maturation, this process is not completely understood. We aimed to identify putative differences in the FF profiles of PCOS patients and fertile women, represented by egg donors, using mass spectrometric analysis to better understand the mechanisms that lead to deregulated oocyte development.
MATERIAL AND METHODS
Study design
This prospective study evaluated the protein components of FF from oocyte donors (ODs) in comparison to those of FF from infertile women with PCOS who underwent IVF at Huntington Reproductive Medicine Centre and the Reproductive Unit of the Federal University of São Paulo (UNIFESP) from 2012 to 2015. This study protocol was approved by the ethics committee of Federal University of São Paulo (No. 1620/2011), and informed written consent was obtained from each patient.
Casuistic
Thirteen (13) patients were enrolled and divided into two groups: ODs (n=7) and infertile PCOS patients (PCOS; n=6). The ODs were healthy female volunteers under the age of 32 years with body mass indices between 18 and 30 kg/m2, antral follicle counts ≥10, normal karyotypes, and the absence of endometriosis who had been screened and tested for infectious diseases. The PCOS patients were diagnosed with infertility according the Rotterdam criteria (Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group, 2004). All the PCOS patients presented body mass indices (BMI) below 25 kg/m2, basal follicular stimulating hormone (FSH) levels below 15 IU/L, basal oestradiol levels below 50 pg/mL, the presence of both ovaries, and no ongoing infectious diseases or uterine abnormalities, and they had undergone intracytoplasmic sperm injection (ICSI) cycles with ejaculated sperm. For both groups, patients who presented gynaecological bleeding, hydrosalpinx, allergy to gonadotropins or other medications used in the treatment, severe oligo- or azoospermia, abusive use of any medications or ovarian hyperstimulation syndrome (OHSS) during the treatment were excluded.
Ovarian stimulation protocol and sample collection
FF was obtained from women who underwent the standard short protocol of IVF (using a GnRH antagonist - Cetrotide®, Merck, Germany). Controlled ovarian stimulation was performed using recombinant FSH (rFSH - Gonal-F®, Merck, Germany) and was monitored with ultrasound. Ovulation was triggered with a GnRH agonist (aGnRH - Gonapeptyl, Ferring, Germany) when at least two follicles reached 20 mm. The FF was collected from the dominant follicles through aspiration between 34 and 36 h after aGnRH administration, using transvaginal ultrasound guidance. Only clear FF samples, without blood or flushing medium contamination, were processed. The selected FF samples were centrifuged at 1200 rpm for 10 to 15 min to remove cellular debris. The supernatants were stored at −80ºC until purification.
Protein extraction
Before analysis, albumin and immunoglobulins were removed from the FF samples (25 µL) using the Albumin & IgG Depletion SpinTrap (GE Healthcare Life Sciences™) according to the manufacturer’s protocol. The protein concentration in each FF sample was measured in triplicate using a bicinchoninic acid assay (BCA assay) (Smith et al., 1985). Twenty-five to thirty micrograms of albumin/IgG-depleted FF protein was subjected to electrophoresis via 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions (Laemmli, 1970) and stained with Coomassie brilliant blue R-250.
All gels were analysed, and ten bands were cut equally for each sample and processed separately for in-gel digestion according to the protocol described by Westermeier et al. (2002), with slight modifications.
Mass spectrometry analysis
Liquid chromatography-mass spectrometry (LC-MS) analyses were performed using an Electrospray-Ion Trap-Time of Flight system (ESI-IT-TOF) (Shimadzu Co., Japan) equipped with a binary Ultra-Fast Liquid Chromatography system (UFLC) (20A Prominence, Shimadzu) at the Laboratory of Biochemistry and Biophysics of the Butantan Institute (São Paulo, Brazil). First, each band sample was lyophilized, resuspended in 50 µL of 0.1% acetic acid and loaded on a C18 column (Discovery C18, 5 µm; 50 × 2.1 mm) in a binary solvent system: (A2) water/acetic acid (999/1, v/v) and (B2) ACN/water/acetic acid (900/99/1, v/v/v). The column was eluted at a constant flow rate of 0.2 mL.min−1 with a 5 to 70% gradient of solvent B2 over 35 min. The eluates were monitored by a Shimadzu SPD-M20A PDA detector before introduction into the mass spectrometer. The interface voltage was adjusted to 4.5 kV, and the capillary voltage was 1.76 kV at 200ºC. MS spectra were acquired in positive mode and collected in the 80-2000 mass charge (m/z) range. MS/MS spectra were collected in the 50-1950 m/z range. Instrument control, data acquisition, and data processing were performed with LabSolutions (LCMSsolution 3.60.361 version, Shimadzu).
Bioinformatics analysis
Proteomic analysis was performed using the Mascot Server (ion search) in house version (2.4) and Peaks Studio V7 (Bioinformatics Solutions, Inc., Waterloo, Canada). The following parameters were adjusted for the search: parent mass and fragment mass error tolerance: 0.1 Da; enzyme: trypsin; fixed modification: carbamidomethylation; variable modification: methionine oxidation; precursor mass search type: monoisotopic; max missed cleavages: 3; non-specific cleavages: one; database: SwissProt, taxon: Homo sapiens; peptide - 10 lgP: ≥15; and protein - 10 lgP: ≥20. The false discovery rate (FDR) for peptide-spectrum matches was ≤1%.
Although each band was analysed separately in LC-MS, we performed a protein search combining all ten bands obtained from each patient. A protein was considered exclusive when it was detected in the FF of patients in either the OD or PCOS group and was totally absent in all of the samples from the other group; a protein was considered overexpressed when it was detected in both groups, but one group had a mean detected peptide level greater than that of the other group by 50% (greater than 1.5-fold in one group and less than 0.5-fold in the other group).
Protein identification and classification
The identified proteins were classified according to their classes, locations, biological functions and processes using the PANTHER Classification System (Gene Ontology Phylogenetic Annotation Project, Los Angeles, USA) (Mi et al., 2016). System biology analysis was carried out using Ingenuity™ Pathway Analysis software (IPA™, QIAGEN, Redwood, USA). The overexpressed proteins were selected for the analysis of canonical pathways and biological interaction networks. The biological processes were staggered according to the IPA™ Knowledge Base. The associations between the identified proteins and canonical pathways in the database were assessed with Ingenuity™ software using Fisher's exact test (significance of p<0.01).
Statistical analysis
Clinical proteomic studies is a multistage biomarker pipeline that begin with the identification of a large number of proteins in a small set of sample. This screening step, as this is our study, the number of samples included was based on the principle that a minimum number of samples considering biological and technical variation inherent in the experiment. Thus, we included a small number of samples and non-parametric statistic was applied.
The patients’ demographic data were evaluated using descriptive statistics. Normality was evaluated with the Kolmogorov-Smirnov test. Non-paired continuous data were compared using the Mann-Whitney test for means comparisons and paired data were compared using Wilcoxon’s signed-rank test. Data analyses were performed using SPSS 22 (IBM SPSS Software, USA), and significance was accepted for p-values ≤0.05.
RESULTS
The patients’ demographics and clinical outcomes are described in Table 1. The ovarian reserves of the patients in both groups had similar profiles in terms of basal FSH dosages and antral follicle counts. As expected, the OD patients were younger, and the PCOS patients had longer menstrual cycle intervals. The parameters related to ovarian induction (length, serum hormone levels, and mature (metaphase II-MII) oocytes collected) were similar between the groups, except for the amount of gonadotropin administered, which was higher in the OD group. The OD group had a higher number of top-quality embryos (3rd day) than the PCOS group.
Table 1.
Demographic and clinical data for the patients in the PCOS and OD groups
| OD group | PCOS group | p-value | |
|---|---|---|---|
| Number of samples | 7 | 6 | ---- |
| Age (years) | 24.71±3.45 | 33.50±1.38 | 0.0050 |
| BMI (kg/m2) | 24.03±1.15 | 21.37±2.02 | 0.0264 |
| Menstrual cycle interval (days) | 29.29±1.60 | 50.00±21.68 | 0.0033 |
| Antral follicle count | 14.0±4.6 | 14.8±4.8 | 0.7723 |
| Basal FSH (UI/mL) | 5.56±1.41 | 5.97±1.44 | 0.5180 |
| Gonadotropin dose (UI) | 2829.0± 494.9 | 1758.0±1097.0 | 0.0513 |
| Oocyte induction length (days) | 12.0±0.0 | 11.7±1.9 | 0.4282 |
| Oestradiol (E2)* (nmol/L) | 3503±1777 | 2892±2474 | 0.7308 |
| Progesterone (P4)* (nmol/L) | 2.74±1.40 | 2.24±1.278 | 0.4295 |
| MII oocytes collected | 10.86±2.27 | 13.33±3.39 | 0.1277 |
| Top-quality embryos (D3) | 3.4±14.8 | 2.2±2.4 | 0.0343 |
Measured prior to oocyte collection.
The proteomic analysis of the proteins from the FF samples matched 229 proteins in the SwissProt database. Forty-five (45) proteins were detected in both groups. Three of these shared proteins were excluded from analyses, as they were contaminants (trypsins and keratins), resulting in 42 proteins shared between the two groups. There were 61 proteins that were exclusive to the PCOS group, and 123 proteins that were exclusive to the OD group (Supplemental Tables I and II). To refine the SwissProt results, only proteins that were expressed in at least two patients from each group were considered. Five proteins were selected from those exclusively expressed in the PCOS group, and three proteins were selected from those exclusively expressed in the OD group (Table 2).
Table 2.
Proteins that were exclusively detected in at least two FF samples from either the PCOS or OD group
| Group | SwissProt ID | Description | Peptide mean | Average mass (Da) | Patients |
|---|---|---|---|---|---|
| PCOS | Q17R60 | Interphotoreceptor matrix proteoglycan 1 | 1.5 | 89387 | 2 |
| Q8 WXI7 | Mucin-16 | 1.3 | 2.00E+06 | 3 | |
| P02765 | Alpha-2-HS glycoprotein | 1.5 | 39325 | 2 | |
| P02774 | Vitamin D-binding protein | 2.0 | 52964 | 2 | |
| Q15020 | S Squamous cell carcinoma antigen recognized by T-cells 3 | 1.0 | 109935 | 2 | |
| OD | P43686 | 26S protease regulatory subunit 6B | 1.5 | 47366 | 2 |
| Q8IWI9 | MAX gene-associated protein | 1.0 | 331836 | 2 | |
| Q8IWI9 | Transformation/transcription domain - associated protein | 1.0 | 437603 | 2 |
The differentially expressed proteins were rated and selected. Six proteins were the highest occurring peptides in the PCOS group, and ten proteins were the highest occurring peptides in the OD group (Table 3). The most significant proteins, which were expressed in the FF of at least two patients, were the complement C3 protein, which was overexpressed in the PCOS group, and titin, serum albumin, complement C4-A, complement C4-B, alpha-1-acid glycoprotein 1 and alpha-2-macroglobulin, which were overexpressed in the OD group.
Table 3.
Proteins that were differentially expressed in the FF of PCOS and OD patients and for which one group had at least 50% more peptides than the other
| SwissProt ID | Description | Peptide mean | Fold change | |
|---|---|---|---|---|
| OD | PCOS | |||
| P04004 | Vitronectin | 1 | 4 | 4.0 |
| P25311 | Zinc-alpha-2 glycoprotein | 1 | 2 | 2.0 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | 1 | 2 | 2.0 |
| Q6 V0I7 | Protocadherin Fat 4 | 1 | 2 | 2.0 |
| P01834 | Complement factor B | 4 | 7 | 1.8 |
| P01024 | Complement C3 | 5.3 | 8 | 1.5 |
| Q8 WZ42 | Titin | 3 | 2 | 0.6 |
| Q9C0G6 | Dynein heavy chain 6, axonemal | 3 | 2 | 0.6 |
| P02768 | Serum albumin | 16.75 | 10 | 0.6 |
| P0C0 L4 | Complement C4-A | 4 | 2 | 0.5 |
| P0C0 L5 | Complement C4-B | 4 | 2 | 0.5 |
| P02763 | Alpha-1-acid glycoprotein 1 | 2.5 | 1 | 0.4 |
| P01023 | Alpha-2-macroglobulin | 5.75 | 1 | 0.2 |
The proteins that were identified as exclusive or overexpressed were classified according to the Gene Ontology database and analysed with respect to biological pathways with the Ingenuity™ software. Six molecular functions were identified, and four of them were very similar between the two groups (GO:0005488, GO:0004872, GO:0005198 and GO:0003824). The PCOS patients had fewer proteins related to transporter activity (GO:0005215) (7.10% OD vs. 2.30% PCOS). Additionally, translation regulation activity (GO:0045182) was detected only in the OD patients (1.80%) but was represented by only one protein.
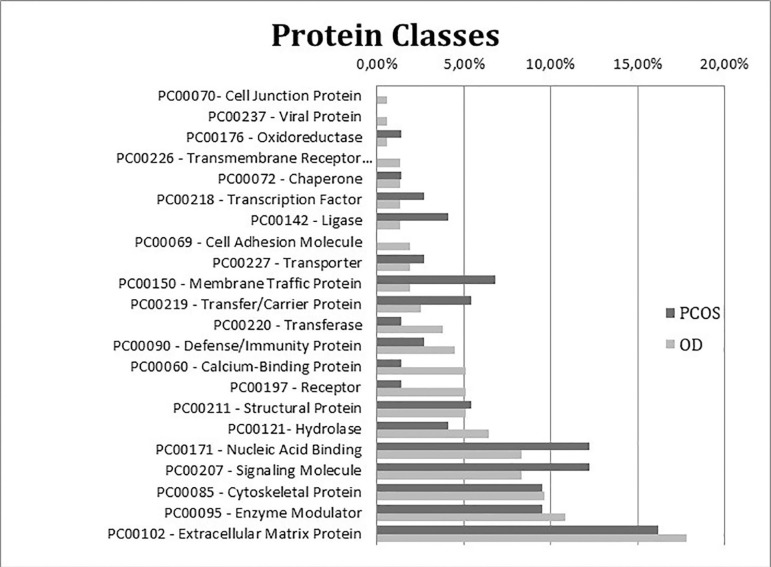
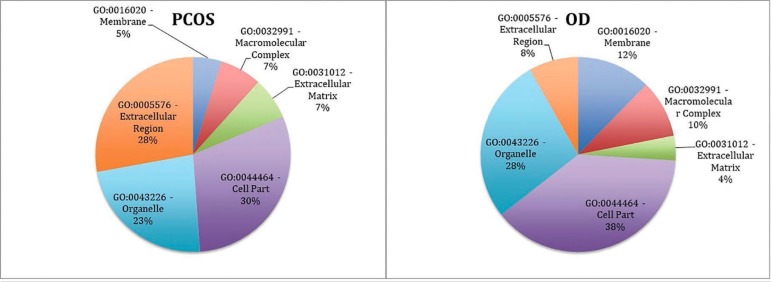
The evaluation of protein classes resulted in nineteen different classes (Figure 1). The most representative classes for the OD group were cell junction, cell adhesion and transmembrane receptor regulatory/adaptor, which were exclusive to this group. The PCOS group presented more proteins related to the oxireductase, membrane traffic protein and ligase classes. The distribution of the protein classes in terms of cellular components differed between the groups: the PCOS group had more extracellular proteins, and the OD group had more membrane and membrane-related proteins (Figure 2).
Figure 1.
Chart indicating the percentages of exclusive and upregulated FF proteins from the PCOS (dark grey) and OD (light grey) groups classified according to protein classes based on the Gene Ontology database
Figure 2.
Chart indicating the percentages of exclusive and upregulated FF proteins from the PCOS and OD groups, classified according to cellular components based on the Gene Ontology database
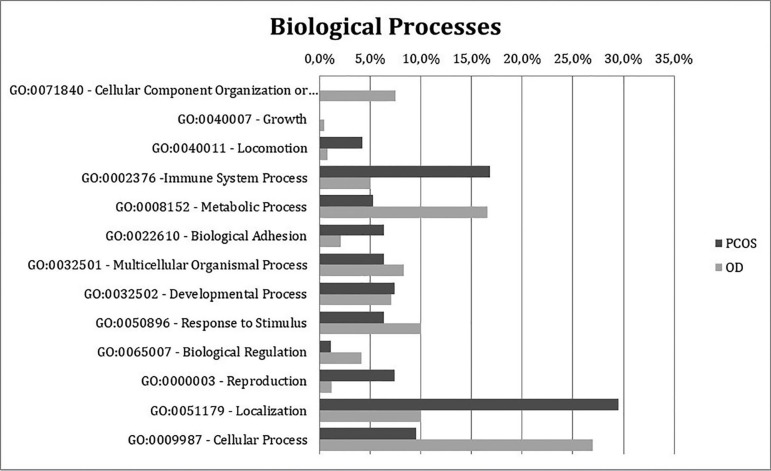
The biological processes associated with the detected proteins differed remarkably between the groups (Figure 3). The PCOS group had more proteins associated with immune process, cell localization and biological adhesion molecules. The OD group had more proteins associated with metabolic processes and cell component organization, suggesting that the OD group was more metabolically active.
Figure 3.
Chart indicating the percentages of exclusive and upregulated FF proteins from the PCOS (dark grey) and OD (light grey) groups classified according to biological processes based on the Gene Ontology database
These results were corroborated by the biological pathway analysis (Table 4), as the proteins identified in the FF of the OD patients were related to cellular assembly and organization and cellular function and maintenance. The PCOS group had fewer proteins matched to cellular assembly and organization. As expected, the proteins of the OD group matched biological functions related to embryo and general organism development; only two of these proteins were detected in the FF of the patients in the PCOS group. The main canonical pathways (Supplemental Table III) found only for the proteins in the FF from the PCOS patients were LXR/RXR activation (p=9.04 E-11, overlap 7%) and FXR/RXR activation (p=1.67 E-10, overlap 6.6%), which are key for the metabolism of lipids, lipoproteins and glucose, reflecting the disrupted metabolism exhibited by PCOS patients. In addition, proteins associated with the intrinsic (p=1.70E-07, overlap 22.2%) and extrinsic (p=1.48E-06, overlap 13.3%) prothrombin activation pathways were identified in the FF from the PCOS group.
Table 4.
Molecular and cellular function and physiological system development and function matched to the proteins detected in the FF from PCOS and OD patients
| PCOS | OD | |||
|---|---|---|---|---|
| p-value | Molecules | p-value | Molecules | |
| Molecular and Cellular Function | ||||
| Carbohydrate Metabolism | 8.43E-03 – 1.94E-06 | 10 | ||
| Lipid Metabolism | 8.43E-03 – 1.94E-06 | 11 | ||
| Small Molecule Biochemistry | 8.43E-03 – 1.94E-06 | 11 | ||
| Cell-To-Cell Signalling and Interaction | 8.43E-03 – 3.52E-06 | 15 | ||
| Cellular Assembly and Organization | 8.43E-03 – 8.38E-06 | 24 | 1.19E-02 – 3.89E-05 | 45 |
| Cellular Function and Maintenance | 1.14E-02 – 3.89E-05 | 38 | ||
| Post-Translational Modification | 8.95E-03 – 9.71E-05 | 3 | ||
| Protein Degradation | 9.71E-05 – 9.71E-05 | 2 | ||
| Protein Synthesis | 8.95E-03 – 9.71E-05 | 2 | ||
| Physiological System Development and Function | ||||
| Embryonic Development | 8.43E-03 – 3.52E-06 | 14 | 1.14E-02 – 1.94E-04 | 29 |
| Haematological System Development and Function | 8.43E-03 – 6.08E-06 | 18 | ||
| Hair and Skin Development and Function | 5.63E-03 – 7.13E-06 | 9 | ||
| Tissue Development | 8.43E-03 - 8.38E-06 | 21 | 1.24E-02 – 1.94E-04 | 33 |
| Renal and Urological System Development and Function | 5.63E-03 – 1.19E-05 | 10 | ||
| Nervous System Development and Function | 1.14E-02 – 1.94E-04 | 22 | ||
| Organ Development | 1.14E-02 – 1.94E-04 | 20 | ||
| Organismal Development | 11.14E-02 – 1.94E-04 | 32 | ||
DISCUSSION
Our findings showed significantly diminished expression of proteins involved in key processes associated with oocyte competence and embryo development in PCOS patients. In addition, overexpression of proteins related to oxidative stress, the immune response and lipid, lipoprotein and carbohydrate metabolism was observed in these patients. Although many proteomics analyses of FF have been published recently, the functional correlations among these proteins are still poorly recognized. We attempted to correlate the differentially expressed proteins in the FF from PCOS patients with physiological pathways. We believe that the observed differences may reflect the PCOS patients’ diminished embryo quality, as this factor is directly reliant on oocyte characteristics.
In our study, the inflammatory pathway represented by complement C3 protein and vitronectin was overexpressed in the FF from the PCOS group. The augmented levels of these proteins in the FF seems to be related to poor oocyte quality, potentially explaining IVF failure (Estes et al., 2009). Additionally, excess complement cascade activation leads to deficiencies in vascular endothelial growth factor (VEGF) activity, which is essential for proper oocyte maturation (Jarkovska et al., 2010). Another marker for the disruption of the inflammatory pathway in PCOS patients is the overexpression of alpha-2-HS-glycoprotein (fetuin-A). This protein is an acute-phase inflammatory regulator that is usually upregulated in OHSS (Jarkovska et al., 2011). As we excluded OHSS patients and applied a GnRH agonist analogue to trigger ovulation, the presence of fetuin-A was not expected and may contribute to the decreased oocyte quality in those patients.
Moreover, the poor oocyte quality and deregulated inflammatory status of PCOS patients may be related to the overexpression of vitamin D-binding protein (VDBP) in their FF. VDBP was another protein found exclusively in the FF of the PCOS group, and according to the literature, this protein may be related to decreased implantation, pregnancy (Estes et al., 2009), and live birth rates (Benkhalifa et al., 2015); VDBP is even more strongly associated with a higher risk of miscarriages (Kushnir et al., 2012) and foetal growth restriction (Wookey et al., 2017).
The overexpressed coagulation pathway found in PCOS-FF, characterized by intrinsic and extrinsic prothrombin activation, is also linked to an inflammatory response; this pathway has important roles in follicle physiology (de Agostini, 2006) and may be associated with poor IVF outcomes (Bianchi et al., 2016).
The exclusive and overexpressed proteins in the OD group, such as 26S protease, alpha-1-acid glycoprotein 1 and alpha-2-macroglobulin, are correlated with a better ovarian stimulation response. The 26S protease is a highly specialized, conserved ribonucleoprotein that facilitates assembly of proteasome complexes; this protein is directly and indirectly involved in the regulation of gene expression (Mittenberg, 2014). Alpha-2-macroglobulin is linked to intrinsic and extrinsic coagulation cascades and is correlated with the complement pathway (Hanrieder et al., 2009). The adequate regulation of coagulation and immune response pathways is essential for the extracellular matrix (ECM) modelling that facilitates follicular growth, ovulation and corpus luteum formation (Kamat et al., 1995; Curry & Smith, 2006), which may be more effective in fertile women.
Furthermore, PCOS is frequently associated with disrupted lipid and carbohydrate metabolism (Dumesic et al., 2015). We found some proteins in the PCOS-FF that were absent in the OD group; these proteins represented metabolic pathways, and their presence corroborated previous findings (Dai & Lu, 2012; Ambekar et al., 2015). Our findings at the FF level suggest that the impairment of lipid and lipoprotein metabolism also occurs within a specific microenvironment, such as that of infertile women with PCOS and a normal BMI. The increased inflammatory status and metabolic disruption observed through the protein composition of the FF from our PCOS patients seem to lead to a worse prognosis for oocyte viability and may affect IVF outcomes. Previous studies of PCOS patients undergoing IVF treatment obtained a great number of oocytes but reported poor fertilization and embryo development rates, an outcome that may be linked to deregulated oocyte activation through a damaged microenvironment (Jungheim et al., 2009).
To find potential markers of oocyte quality, our inclusion and exclusion criteria were very strict to allow us to identify markers that are exclusive to PCOS-FF without overlapping with other pathologies and conditions, such as obesity or OHSS. We hypothesize that the evaluation of fetuin-A, VDBP, complement C3 and 26S protease expression in the FF of PCOS patients undergoing IVF could be associated with oocyte quality. The limitations of these findings include the absence of experimental validation of the candidate markers through other techniques, such as Western blotting; additionally, the differentially expressed proteins must be correlated with the final IVF outcomes to endorse their use in clinical practice.
ACKNOWLEDGEMENTS
The authors are grateful for the contributions of the team from Huntington Reproductive Medicine and the Reproductive Unit of the Federal University of São Paulo (UNIFESP) for their excellent support with patients and procedures.
Supplementary Table I.
List of proteins identified exclusively in the FF from PCOS group in comparison to OD group.
| SwissProt ID | Name | Class | Molecular Function | Cellular compartment | Number of Patients |
|---|---|---|---|---|---|
| Q17R60 | Interphotoreceptor matrix proteoglycan 1 | Glicoproteína de matriz extracelular (PC00102); receptor (PC00100) | receptor activity(GO:0004872) | extracellular region(GO:0005576) | 3 |
| Q8WXI7 | Mucin-16 | NA | NA | NA | 3 |
| PNA1NA42 | Complement C3 | NA | cysteine-type endopeptidase inhibitor activity(GO:0003824) | #N/D | 2 |
| P02765 | Alpha-2-HS-glycoprotein | Inibidor de cisteína protease (PC00095); Glicoproteína de matriz extracelular (PC00191) | peptidase inhibitor activity(GO:0003824) | extracellular space(GO:0005576) | 2 |
| P02774 | Vitamin D-binding protein | NA | NA | NA | 2 |
| Q15020 | Squamous cell carcinoma antigen recognized by T-cells 3 | NA | NA | NA | 2 |
| A6NDX4 | Putative transmembrane protein | NA | NA | #N/D | 1 |
| O14526 | FCH domain only protein 1 | Proteína de citoesqueleto da família das actinas (PC00085); proteína reguladora de trafficking de membrana (PC00041) | NA | NA | 1 |
| O14578 | Citron Rho-interacting kinase | non-receptor serine/threonine protein kinase(PC00220) | protein kinase activity(GO:0003824) | NA | 1 |
| O15020 | Spectrin beta chain, non-erythrocytic 2 | non-motor actin binding protein(PC00085) | actin binding(GO:0005488);structural constituent of cytoskeleton(GO:0005515) | intracellular(GO:0044464) | 1 |
| O15357 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | phosphatase(PC00121) | NA | NA | 1 |
| O75445 | Usherin | extracellular matrix linker protein(PC00102);receptor(PC00101) | receptor activity(GO:0004872) | extracellular matrix(GO:0031012);extracellular region(GO:0005576) | 1 |
| O75691 | Small subunit processome component 20 homolog | NA | NA | intracellular(GO:0044464);nucleolus(GO:0005622);ribonucleoprotein complex(GO:0043226) | 1 |
| P00739 | Haptoglobin-related protein | annexin(PC00060);calmodulin(PC00050);peptide hormone(PC00131);protease inhibitor(PC00061);receptor(PC00207);serine protease(PC00179) | NA | NA | 1 |
| P01008 | Antithrombin-III | serine protease inhibitor(PC00095) | serine-type endopeptidase inhibitor activity(GO:0003824);serine-type peptidase activity(GO:0016787) | extracellular space(GO:0005576) | 1 |
| P01019 | Angiotensinogen | serine protease inhibitor(PC00095) | serine-type endopeptidase inhibitor activity(GO:0003824);serine-type peptidase activity(GO:0016787) | extracellular space(GO:0005576) | 1 |
| P02538 | Keratin, type II cytoskeletal 6A | intermediate filament(PC00085);structural protein(PC00129) | structural constituent of cytoskeleton(GO:0005198) | intermediate filament cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| P02671 | Fibrinogen alpha chain | signaling molecule(PC00207) | receptor binding(GO:0005488) | extracellular region(GO:0005576) | 1 |
| P02675 | Fibrinogen beta chain | signaling molecule(PC00207) | receptor binding(GO:0005488) | extracellular region(GO:0005576) | 1 |
| P02679 | Fibrinogen gamma chain | NA | NA | NA | 1 |
| P02790 | Hemopexin | transfer/carrier protein(PC00219) | NA | extracellular matrix(GO:0031012) | 1 |
| P04259 | Keratin, type II cytoskeletal 6B | intermediate filament(PC00085);structural protein(PC00129) | structural constituent of cytoskeleton(GO:0005198) | intermediate filament cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| P08574 | Cytochrome c1, heme protein, mitochondrial | NA | NA | cytoplasm(GO:0044464);mitochondrion(GO:0005622) | |
| P08603 | Complement factor H | NA | NA | NA | 1 |
| P09529 | Inhibin beta B chain | growth factor(PC00207) | transforming growth factor beta receptor binding(GO:0005488) | extracellular space(GO:0005576) | 1 |
| P13645 | Keratin, type I cytoskeletal 10 | intermediate filament(PC00085);structural protein(PC00129) | structural constituent of cytoskeleton(GO:0005198) | intermediate filament cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| P19827 | Inter-alpha-trypsin inhibitor heavy chain H1 | serine protease inhibitor(PC00095) | protein binding(GO:0005488);serine-type endopeptidase inhibitor activity(GO:0005515) | NA | 1 |
| P20742 | Pregnancy zone protein | complement component(PC00090);cytokine(PC00078);serine protease inhibitor(PC00207) | cytokine activity(GO:0005488);serine-type endopeptidase inhibitor activity(GO:0005515) | NA | 1 |
| P27169 | Serum paraoxonase/arylesterase 1 | NA | hydrolase activity, acting on ester bonds(GO:0003824) | NA | 1 |
| P33991 | DNA replication licensing factor MCM4 | DNA helicase(PC00171);hydrolase(PC00009) | DNA helicase activity(GO:0003824);hydrolase activity(GO:0004386);nucleic acid binding(GO:0003678) | NA | 1 |
| P35908 | Keratin, type II cytoskeletal 2 epidermal | intermediate filament(PC00085);structural protein(PC00129) | structural constituent of cytoskeleton(GO:0005198) | intermediate filament cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| P48668 | Keratin, type II cytoskeletal 6C | intermediate filament(PC00085);structural protein(PC00129) | structural constituent of cytoskeleton(GO:0005198) | intermediate filament cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| P49454 | Centromere protein F | NA | NA | NA | 1 |
| P49589 | Cysteine--tRNA ligase, cytoplasmic | RNA binding protein(PC00171);aminoacyl-tRNA synthetase(PC00031) | aminoacyl-tRNA ligase activity(GO:0003824) | cytosol(GO:0044464) | 1 |
| P49842 | Serine/threonine-protein kinase 19 | NA | NA | NA | 1 |
| P78312 | Protein FAM193A | NA | NA | NA | 1 |
| P98170 | E3 ubiquitin-protein ligase XIAP | protease inhibitor(PC00095) | cysteine-type endopeptidase inhibitor activity(GO:0003824);ubiquitin-protein ligase activity(GO:0016787) | cytoplasm(GO:0044464);microtubule(GO:0005622);nucleus(GO:0005737) | 1 |
| Q12873 | Chromodomain-helicase-DNA-binding protein 3 | DNA helicase(PC00171) | NA | NA | 1 |
| Q13129 | Zinc finger protein Rlf | nuclease(PC00171);transcription factor(PC00170) | nuclease activity(GO:0003824);sequence-specific DNA binding transcription factor activity(GO:0016787) | NA | 1 |
| Q15813 | Tubulin-specific chaperone E | chaperone(PC00072) | NA | NA | 1 |
| Q3KP22 | Uncharacterized protein C11orf85 | NA | NA | NA | 1 |
| Q53GS7 | Nucleoporin GLE1 | nucleic acid binding(PC00171);transfer/carrier protein(PC00219) | nucleic acid binding(GO:0005488) | NA | 1 |
| Q63HN8 | E3 ubiquitin-protein ligase RNF213 | NA | NA | NA | 1 |
| Q6YP21 | Kynurenine--oxoglutarate transaminase 3 | transaminase(PC00220) | transaminase activity(GO:0003824) | NA | 1 |
| Q70CQ2 | Ubiquitin carboxyl-terminal hydrolase 34 | NA | NA | NA | 1 |
| Q7Z6M1 | Rab9 effector protein with kelch motifs | NA | NA | NA | 1 |
| Q86YQ8 | Copine-8 | membrane traffic protein(PC00150) | NA | NA | 1 |
| Q8IWT3 | Cullin-9 | NA | NA | NA | 1 |
| Q8IY50 | Putative thiamine transporter SLC35F3 | NA | NA | NA | 1 |
| Q8N9W7 | Putative transmembrane protein FLJ36131 | NA | #N/D | #N/D | 1 |
| Q8NHM4 | Putative trypsin-6 | serine protease(PC00121) | serine-type peptidase activity(GO:0003824) | extracellular space(GO:0005576) | 1 |
| Q8TDI0 | Chromodomain-helicase-DNA-binding protein 5 | DNA helicase(PC00171) | NA | NA | 1 |
| Q8TF01 | Arginine/serine-rich protein PNISR | NA | NA | NA | 1 |
| Q92665 | 28S ribosomal protein S31, mitochondrial | NA | NA | NA | 1 |
| Q9H1H9 | Kinesin-like protein KIF13A | microtubule binding motor protein(PC00085) | microtubule motor activity(GO:0003824) | cytoskeleton(GO:0043226);intracellular(GO:0005856);protein complex(GO:0044464) | 1 |
| Q9NQ66 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | calcium-binding protein(PC00060);guanyl-nucleotide exchange factor(PC00095);phospholipase(PC00022);signaling molecule(PC00113) | calcium ion binding(GO:0005488);guanyl-nucleotide exchange factor activity(GO:0005509);phospholipase activity(GO:0003824);receptor binding(GO:0030234);small GTPase regulator activity(GO:0005085) | NA | 1 |
| Q9NTG1 | Polycystic kidney disease and receptor for egg jelly-related protein | G-protein modulator(PC00095);ion channel(PC00022);membrane-bound signaling molecule(PC00227) | Cation channel activity(GO:0005215) | NA | 1 |
| Q9P212 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | calcium-binding protein(PC00060);guanyl-nucleotide exchange factor(PC00095);phospholipase(PC00022);signaling molecule(PC00113) | calcium ion binding(GO:0005488);guanyl-nucleotide exchange factor activity(GO:0005509);phospholipase activity(GO:0003824);receptor binding(GO:0030234);small GTPase regulator activity(GO:0005085) | NA | 1 |
| Q9P225 | Dynein heavy chain 2, axonemal | hydrolase(PC00121);microtubule binding motor protein(PC00085) | microtubule motor activity(GO:0003824);structural constituent of cytoskeleton(GO:0016787) | intracellular(GO:0044464);microtubule(GO:0005622) | 1 |
| Q9P2N5 | RNA-binding protein 27 | RNA binding protein(PC00171) | RNA binding(GO:0005488) | NA | 1 |
| Q9UKA4 | RNA-binding protein 27 | kinase modulator(PC00095) | protein binding(GO:0005488) | cytoplasm(GO:0044464) | 1 |
Supplementary Table II.
List of proteins identified exclusively in the FF from OD group in comparison to PCOS group.
| SwissProt ID | Name | Class | Molecular Function | Cellular compartment | Number of Patients |
|---|---|---|---|---|---|
| P43686 | 26S protease regulatory subunit 6B | hydrolase(PC00121) | protein binding(GO:0005488) | cytosol(GO:0044464);nucleus(GO:0005622);protein complex(GO:0005737) | 2 |
| Q8IWI9 | MAX gene-associated protein | nucleic acid binding(PC00171);transcription factor(PC00218) | sequence-specific DNA binding transcription factor activity(GO:0005488) | NA | 2 |
| Q9Y4A5 | Transformation/transcription domain-associated protein | non-receptor serine/threonine protein kinase(PC00220);nucleotide kinase(PC00137) | kinase activity(GO:0003824) | NA | 2 |
| A0AUZ9 | KAT8 regulatory NSL complex subunit 1-like protein | NA | NA | NA | 1 |
| A5PLK6 | Regulator of G-protein signaling protein-like | G-protein modulator(PC00095) | GTPase activity(GO:0003824);enzyme activator activity(GO:0016787);pyrophosphatase activity(GO:0003924) | cytoplasm(GO:0044464);plasma membrane(GO:0005622) | 1 |
| A6NJZ7 | RIMS-binding protein 3C | NA | NA | NA | 1 |
| A6NNM3 | RIMS-binding protein 3B | NA | NA | NA | 1 |
| O00268 | Transcription initiation factor TFIID subunit 4 | NA | NA | NA | 1 |
| O14641 | Segment polarity protein dishevelled homolog DVL-2 | enzyme modulator(PC00095);signaling molecule(PC00207) | receptor binding(GO:0005488) | cytosol(GO:0044464) | 1 |
| O14948 | Transcription factor EC | basic helix-loop-helix transcription factor(PC00218) | NA | NA | 1 |
| O15294 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | glycosyltransferase(PC00220) | transferase activity, transferring glycosyl groups(GO:0003824) | NA | 1 |
| O43187 | Interleukin-1 receptor-associated kinase-like 2 | NA | protein kinase activity(GO:0003824) | cytoplasm(GO:0044464);nucleus(GO:0005622) | 1 |
| O43451 | Maltase-glucoamylase, intestinal | glucosidase(PC00121) | glucosidase activity(GO:0003824) | NA | 1 |
| O60229 | Kalirin | guanyl-nucleotide exchange factor(PC00095);signaling molecule(PC00022) | guanyl-nucleotide exchange factor activity(GO:0003824);receptor binding(GO:0030234);small GTPase regulator activity(GO:0005085) | NA | 1 |
| O60318 | Germinal-center associated nuclear protein | ATP synthase(PC00227);hydrolase(PC00068) | cation transmembrane transporter activity(GO:0005215);hydrogen ion transmembrane transporter activity(GO:0022857);hydrolase activity(GO:0008324) | NA | 1 |
| O60494 | Cubilin | apolipoprotein(PC00219);cell adhesion molecule(PC00052);enzyme modulator(PC00069);extracellular matrix protein(PC00095);membrane-bound signaling molecule(PC00102);metalloprotease(PC00207);oxidase(PC00152);serine protease(PC00121);transporter(PC00190) | enzyme regulator activity(GO:0003824);lipid transporter activity(GO:0030234);metallopeptidase activity(GO:0005215);oxidoreductase activity(GO:0005319);receptor activity(GO:0016787);serine-type peptidase activity(GO:0008233);transmembrane transporter activity(GO:0008237) | extracellular matrix(GO:0031012);extracellular region(GO:0005576) | 1 |
| O75094 | Slit homolog 3 protein | NA | NA | NA | 1 |
| O75901 | Ras association domain-containing protein 9 | membrane traffic protein(PC00150) | NA | NA | 1 |
| O94813 | Slit homolog 2 protein | NA | NA | NA | 1 |
| O95206 | Protocadherin-8 | cadherin(PC00069) | calcium ion binding(GO:0005488) | NA | 1 |
| O95490 | Latrophilin-2 | G-protein coupled receptor(PC00197);antibacterial response protein(PC00021) | receptor activity(GO:0004872) | NA | 1 |
| O95613 | Pericentrin | chromatin/chromatin-binding protein(PC00171);hydrolase(PC00009);kinase modulator(PC00077) | chromatin binding(GO:0005488);hydrolase activity(GO:0003682);kinase regulator activity(GO:0003824);nucleic acid binding(GO:0016787);protein binding(GO:0016740) | NA | 1 |
| O95661 | GTP-binding protein Di-Ras3 | small GTPase(PC00095) | GTPase activity(GO:0003824);protein binding(GO:0016787) | NA | 1 |
| O95835 | Serine/threonine-protein kinase LATS1 | annexin(PC00060);calmodulin(PC00050);non-receptor serine/threonine protein kinase(PC00131);transfer/carrier protein(PC00061) | protein kinase activity(GO:0003824) | intracellular(GO:0044464) | 1 |
| O95947 | T-box transcription factor TBX6 | nucleic acid binding(PC00171);transcription factor(PC00218) | sequence-specific DNA binding transcription factor activity(GO:0005488) | NA | 1 |
| P01857 | Ig gamma-1 chain C region | NA | antigen binding(GO:0005488);receptor binding(GO:0003823) | extracellular space(GO:0005576);immunoglobulin complex(GO:0005615);plasma membrane(GO:0032991) | 1 |
| P06396 | Gelsolin | non-motor actin binding protein(PC00085) | actin binding(GO:0005488);structural constituent of cytoskeleton(GO:0005515) | actin cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| P07333 | Macrophage colony-stimulating factor 1 receptor | NA | NA | NA | 1 |
| P0C7U3 | Probable palmitoyltransferase ZDHHC11B | NA | NA | NA | 1 |
| P13761 | HLA class II histocompatibility antigen, DRB1-7 beta chain | #N/D | #N/D | #N/D | 1 |
| P35527 | Keratin, type I cytoskeletal 9 | intermediate filament(PC00085);structural protein(PC00129) | structural constituent of cytoskeleton(GO:0005198) | intermediate filament cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| P42345 | Serine/threonine-protein kinase mT | non-receptor serine/threonine protein kinase(PC00220);nucleotide kinase(PC00137) | kinase activity(GO:0003824) | NA | 1 |
| P46013 | Antigen KI-67 | NA | NA | NA | 1 |
| P49750 | YLP motif-containing protein 1 | nucleic acid binding(PC00171) | nucleic acid binding(GO:0005488) | NA | 1 |
| P49792 | E3 SUM | G-protein modulator(PC00095) | protein binding(GO:0005488);small GTPase regulator activity(GO:0005515) | NA | 1 |
| P51805 | Plexin-A3 | signaling molecule(PC00207);tyrosine protein kinase receptor(PC00197) | NA | NA | 1 |
| P98161 | Polycystin-1 | G-protein modulator(PC00095);ion channel(PC00022);membrane-bound signaling molecule(PC00227) | cation channel activity(GO:0005215) | NA | 1 |
| Q04637 | Eukaryotic translation initiation factor 4 gamma 1 | translation initiation factor(PC00171) | translation initiation factor activity(GO:0045182) | NA | 1 |
| Q08170 | Serine/arginine-rich splicing factor 4 | NA | NA | NA | 1 |
| Q08AE8 | Protein spire homolog 1 | actin family cytoskeletal protein(PC00085) | structural constituent of cytoskeleton(GO:0005198) | intracellular(GO:0044464) | 1 |
| Q13402 | Unconventional myosin-VIIa | G-protein modulator(PC00095);actin binding motor protein(PC00022);cell junction protein(PC00085) | enzyme regulator activity(GO:0003824);motor activity(GO:0030234);protein binding(GO:0016787);structural constituent of cytoskeleton(GO:0016462) | actin cytoskeleton(GO:0043226);intracellular(GO:0005856);plasma membrane(GO:0015629) | 1 |
| Q13563 | Polycystin-2 | G-protein modulator(PC00095);ion channel(PC00022);membrane-bound signaling molecule(PC00227) | cation channel activity(GO:0005215) | NA | 1 |
| Q14643 | Inositol 1,4,5-trisphosphate receptor type 1 | ligand-gated ion channel(PC00227) | ligand-gated ion channel activity(GO:0005215);receptor activity(GO:0022857) | NA | 1 |
| Q14980 | Nuclear mitotic apparatus protein 1 | NA | NA | NA | 1 |
| Q15149 | Plectin | non-motor actin binding protein(PC00085) | actin binding(GO:0005488);structural constituent of cytoskeleton(GO:0005515) | actin cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| Q15413 | Ryanodine receptor 3 | ligand-gated ion channel(PC00227) | ligand-gated ion channel activity(GO:0005215);receptor activity(GO:0022857) | NA | 1 |
| Q15772 | Striated muscle preferentially expressed protein kinase | G-protein coupled receptor(PC00197);immunoglobulin receptor superfamily(PC00021);immunoglobulin superfamily cell adhesion molecule(PC00090);protein phosphatase(PC00124) | NA | NA | 1 |
| Q16531 | DNA damage-binding protein 1 | damaged DNA-binding protein(PC00171);mRNA polyadenylation factor(PC00009) | damaged DNA binding(GO:0005488) | intracellular(GO:0044464);nucleus(GO:0005622);protein complex(GO:0043226) | 1 |
| Q2LD37 | Uncharacterized protein KIAA1109 | NA | NA | NA | 1 |
| Q3B7T1 | Erythroid differentiation-related factor 1 | NA | NA | NA | 1 |
| Q460N5 | Poly [ADP-ribose] polymerase 14 | nucleic acid binding(PC00171) | nucleic acid binding(GO:0005488) | NA | 1 |
| Q562E7 | WD repeat-containing protein 81 | esterase(PC00121);kinase inhibitor(PC00097);mRNA splicing factor(PC00095) | NA | NA | 1 |
| Q5JSL3 | Dedicator of cytokinesis protein 11 | guanyl-nucleotide exchange factor(PC00095) | guanyl-nucleotide exchange factor activity(GO:0003824);small GTPase regulator activity(GO:0030234) | NA | 1 |
| Q5T5C0 | Syntaxin-binding protein 5 | membrane trafficking regulatory protein(PC00150) | GTPase activity(GO:0003824);enzyme activator activity(GO:0016787);pyrophosphatase activity(GO:0003924) | cytoplasm(GO:0044464);plasma membrane(GO:0005622) | 1 |
| Q5TC82 | Roquin-1 | NA | NA | NA | 1 |
| Q5TZA2 | Rootletin | kinase modulator(PC00095);viral protein(PC00140) | kinase regulator activity(GO:0003824);protein binding(GO:0016740) | NA | 1 |
| Q5UIP0 | Telomere-associated protein RIF1 | NA | NA | NA | 1 |
| Q5VST9 | Obscurin | immunoglobulin receptor superfamily(PC00090);immunoglobulin superfamily cell adhesion molecule(PC00124);protein phosphatase(PC00069) | NA | NA | 1 |
| Q5VT52 | Regulation of nuclear pre-mRNA domain-containing protein 2 | kinase inhibitor(PC00095) | kinase inhibitor activity(GO:0003824);protein binding(GO:0030234) | NA | 1 |
| Q5VUA4 | Zinc finger protein 318 | NA | NA | NA | 1 |
| Q5VYK3 | Proteasome-associated protein ECM29 homolog | kinase modulator(PC00095) | kinase regulator activity(GO:0003824);protein binding(GO:0016740) | NA | 1 |
| Q5VZM2 | Ras-related GTP-binding protein B | small GTPase(PC00095) | GTPase activity(GO:0003824);pyrophosphatase activity(GO:0016787) | cytoplasm(GO:0044464);endosome(GO:0005622);lysosome(GO:0005737);membrane(GO:0043226);protein complex(GO:0005768);vacuole(GO:0005764) | 1 |
| Q66K64 | DDB1- and CUL4-associated factor 15 | NA | NA | NA | 1 |
| Q6P9F0 | Coiled-coil domain-containing protein 62 | #N/D | #N/D | #N/D | 1 |
| Q6ZN55 | Zinc finger protein 574 | KRAB box transcription factor(PC00218) | NA | NA | 1 |
| Q6ZT12 | E3 ubiquitin-protein ligase UBR3 | NA | NA | cytoplasm(GO:0044464);protein complex(GO:0005622) | 1 |
| Q6ZV29 | Patatin-like phospholipase domain-containing protein 7 | esterase(PC00121) | phospholipase activity(GO:0003824) | cytoplasm(GO:0044464);endoplasmic reticulum(GO:0005622) | 1 |
| Q70JA7 | Chondroitin sulfate synthase 3 | glycosyltransferase(PC00220) | transferase activity, transferring glycosyl groups(GO:0003824) | NA | 1 |
| Q7L523 | Ras-related GTP-binding protein A | small GTPase(PC00095) | GTPase activity(GO:0003824);pyrophosphatase activity(GO:0016787) | cytoplasm(GO:0044464);endosome(GO:0005622);lysosome(GO:0005737);membrane(GO:0043226);protein complex(GO:0005768);vacuole(GO:0005764) | 1 |
| Q7Z494 | Nephrocystin-3 | NA | NA | microtubule(GO:0043226) | 1 |
| Q7Z794 | Keratin, type II cytoskeletal 1b | intermediate filament(PC00085);structural protein(PC00129) | structural constituent of cytoskeleton(GO:0005198) | intermediate filament cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
| Q7Z7G8 | Vacuolar protein sorting-associated protein 13B | NA | NA | NA | 1 |
| Q7Z7M0 | Multiple epidermal growth factor-like domains protein 8 | extracellular matrix linker protein(PC00102);receptor(PC00101);small GTPase(PC00197) | GTPase activity(GO:0003824);receptor activity(GO:0016787) | extracellular matrix(GO:0031012);extracellular region(GO:0005576) | 1 |
| Q86TS7 | Putative UPF0730 protein encoded by LINC00643 | NA | NA | NA | 1 |
| Q8IVE3 | Pleckstrin homology domain-containing family H member 2 | NA | NA | NA | 1 |
| Q8IYF3 | Testis-expressed sequence 11 protein | chaperone(PC00072) | NA | NA | 1 |
| Q8IZD9 | Dedicator of cytokinesis protein 3 | guanyl-nucleotide exchange factor(PC00095) | guanyl-nucleotide exchange factor activity(GO:0003824);small GTPase regulator activity(GO:0030234) | NA | 1 |
| Q8N3S3 | Putative homeodomain transcription factor 2 | homeobox transcription factor(PC00218);nucleic acid binding(PC00116) | sequence-specific DNA binding transcription factor activity(GO:0005488) | NA | 1 |
| Q8N4F7 | RING finger protein 175 | ubiquitin-protein ligase(PC00142) | NA | Golgi apparatus(GO:0043226);cytoplasm(GO:0005794);endoplasmic reticulum(GO:0044464);nuclear outer membrane-endoplasmic reticulum membrane network(GO:0005622) | 1 |
| Q8N4S9 | MARVEL domain-containing protein 2 | tight junction(PC00070);transcription cofactor(PC00214) | sequence-specific DNA binding transcription factor activity(GO:0005488);transcription cofactor activity(GO:0003676) | plasma membrane(GO:0016020) | 1 |
| Q8N554 | Zinc finger protein 276 | NA | NA | NA | 1 |
| Q8N8Z8 | Zinc finger protein 441 | NA | NA | NA | 1 |
| Q8NA56 | Tetratricopeptide repeat protein 29 | guanyl-nucleotide exchange factor(PC00095);transmembrane receptor regulatory/adaptor protein(PC00022) | guanyl-nucleotide exchange factor activity(GO:0003824);small GTPase regulator activity(GO:0030234) | NA | 1 |
| Q8NE71 | ATP-binding cassette sub-family F member 1 | ATP-binding cassette (ABC) transporter(PC00227);hydrolase(PC00003);translation elongation factor(PC00121) | ATPase activity, coupled to transmembrane movement of substances(GO:0003824);translation elongation factor activity(GO:0016787);transmembrane transporter activity(GO:0042626) | NA | 1 |
| Q8NEZ4 | Histone-lysine N-methyltransferase 2C | DNA binding protein(PC00171);methyltransferase(PC00009) | NA | NA | 1 |
| Q8NF91 | Nesprin-1 | non-motor actin binding protein(PC00085) | actin binding(GO:0005488);structural constituent of cytoskeleton(GO:0005515) | intracellular(GO:0044464) | 1 |
| Q8NFC6 | Biorientation of chromosomes in cell division protein 1-like 1 | NA | NA | NA | 1 |
| Q8NG31 | Protein CASC5 | NA | NA | NA | 1 |
| Q8NI35 | InaD-like protein | NA | NA | NA | 1 |
| Q8TER5 | Rho guanine nucleotide exchange factor 40 | guanyl-nucleotide exchange factor(PC00095);signaling molecule(PC00022) | guanyl-nucleotide exchange factor activity(GO:0003824);receptor binding(GO:0030234);small GTPase regulator activity(GO:0005085) | NA | 1 |
| Q8WVC0 | RNA polymerase-associated protein LE | DNA-directed RNA polymerase(PC00171) | NA | intracellular(GO:0044464);nucleus(GO:0005622) | 1 |
| Q8WWG9 | Potassium voltage-gated channel subfamily E member 4 | NA | NA | NA | 1 |
| Q92823 | Neuronal cell adhesion molecule | G-protein coupled receptor(PC00197);immunoglobulin receptor superfamily(PC00021);immunoglobulin superfamily cell adhesion molecule(PC00090);protein phosphatase(PC00124) | NA | NA | 1 |
| Q969F9 | Hermansky-Pudlak syndrome 3 protein | NA | NA | NA | 1 |
| Q969V4 | Tektin-1 | NA | NA | NA | 1 |
| Q96B21 | Transmembrane protein 45B | NA | NA | NA | 1 |
| Q96ID5 | Immunoglobulin superfamily member 21 | immunoglobulin receptor superfamily(PC00090);immunoglobulin superfamily cell adhesion molecule(PC00124);protein phosphatase(PC00069) | NA | NA | 1 |
| Q96MB7 | Putative nuclease HARBI1 | NA | NA | NA | 1 |
| Q96NH3 | Protein broad-minded | NA | NA | NA | 1 |
| Q96PX9 | Pleckstrin homology domain-containing family G member 4B | guanyl-nucleotide exchange factor(PC00095);signaling molecule(PC00022) | guanyl-nucleotide exchange factor activity(GO:0003824);receptor binding(GO:0030234);small GTPase regulator activity(GO:0005085) | NA | 1 |
| Q9BQ52 | Zinc phosphodiesterase ELAC protein 2 | NA | endoribonuclease activity(GO:0003824) | NA | 1 |
| Q9BSC4 | Nucleolar protein 10 | NA | NA | NA | 1 |
| Q9GIY3 | HLA class II histocompatibility antigen, DRB1-14 beta chain | #N/D | #N/D | #N/D | 1 |
| Q9GZQ4 | Neuromedin-U receptor 2 | G-protein coupled receptor(PC00197) | receptor activity(GO:0004872) | NA | 1 |
| Q9H0X9 | Oxysterol-binding protein-related protein 5 | NA | NA | NA | 1 |
| Q9H1A4 | Anaphase-promoting complex subunit 1 | ligase(PC00142) | ligase activity(GO:0003824) | NA | 1 |
| Q9H254 | Spectrin beta chain, non-erythrocytic 4 | non-motor actin binding protein(PC00085) | actin binding(GO:0005488);structural constituent of cytoskeleton(GO:0005515) | intracellular(GO:0044464) | 1 |
| Q9H7M6 | Zinc finger SWIM domain-containing protein 4 | NA | NA | NA | 1 |
| Q9NRL2 | Bromodomain adjacent to zinc finger domain protein 1A | acetyltransferase(PC00220);chromatin/chromatin-binding protein(PC00038) | acetyltransferase activity(GO:0003824);chromatin binding(GO:0016740);nucleic acid binding(GO:0016746) | NA | 1 |
| Q9NZC2 | Bromodomain adjacent to zinc finger domain protein 1A | immunoglobulin receptor superfamily(PC00090) | receptor activity(GO:0004872) | NA | 1 |
| Q9P107 | GEM-interacting protein | NA | NA | NA | 1 |
| Q9P227 | Rho GTPase-activating protein 23 | NA | GTPase activity(GO:0003824);enzyme activator activity(GO:0016787);pyrophosphatase activity(GO:0003924) | NA | 1 |
| Q9UFD9 | RIMS-binding protein 3A | NA | NA | NA | 1 |
| Q9UFH2 | Dynein heavy chain 17, axonemal | hydrolase(PC00121);microtubule binding motor protein(PC00085) | microtubule motor activity(GO:0003824);structural constituent of cytoskeleton(GO:0016787) | intracellular(GO:0044464);microtubule(GO:0005622) | 1 |
| Q9UGM6 | Tryptophan--tRNA ligase, mitochondrial | NA | NA | cytoplasm(GO:0044464) | 1 |
| Q9UHT4 | Putative uncharacterized protein PR | #N/D | #N/D | #N/D | 1 |
| Q9UKN7 | Unconventional myosin-XV | G-protein modulator(PC00095);actin binding motor protein(PC00022);cell junction protein(PC00085) | enzyme regulator activity(GO:0003824);motor activity(GO:0030234);protein binding(GO:0016787);structural constituent of cytoskeleton(GO:0016462) | actin cytoskeleton(GO:0043226);intracellular(GO:0005856);plasma membrane(GO:0015629) | 1 |
| Q9ULL8 | Protein Shroom4 | ligand-gated ion channel(PC00227) | ligand-gated ion channel activity(GO:0005215);receptor activity(GO:0022857) | NA | 1 |
| Q9Y2H0 | Disks large-associated protein 4 | transmembrane receptor regulatory/adaptor protein(PC00226) | NA | NA | 1 |
| Q9Y493 | Zonadhesin | cell adhesion molecule(PC00069);extracellular matrix glycoprotein(PC00102) | NA | extracellular matrix(GO:0031012);extracellular region(GO:0005576) | 1 |
| Q9Y5T5 | Ubiquitin carboxyl-terminal hydrolase 16 | #N/D | #N/D | #N/D | 1 |
| Q9Y5Y6 | Suppressor of tumorigenicity 14 protein | annexin(PC00060);calmodulin(PC00050);peptide hormone(PC00131);protease inhibitor(PC00061);receptor(PC00207);serine protease(PC00179) | NA | NA | 1 |
| Q9Y6U3 | Adseverin | non-motor actin binding protein(PC00085) | actin binding(GO:0005488);structural constituent of cytoskeleton(GO:0005515) | actin cytoskeleton(GO:0043226);intracellular(GO:0005856) | 1 |
Supplementary Table III.
Canonical Pathways matched for the unique and increased proteins of the FF from PCOS and OD groups.
| PCOS Group | OD Group | |||||
|---|---|---|---|---|---|---|
| Canonical Pathways | p-value | Overlap | p-value | Overlap | ||
| LXR/RXR Activation | 9,04E-11 | 7.0% | 9/128 | |||
| FXR/RXR Activation | 1,67E-10 | 6.6% | 9/137 | |||
| Extrinsic Prothrombin Activation Pathway | 1,70E-07 | 22.2% | 4/18 | |||
| Acute Phase Response Signaling | 5,21E-07 | 4.1% | 7/171 | 3,06E-03 | 2.9% | 5/171 |
| Intrinsic Prothrombin Activation Pathway | 1,48E-06 | 13.3% | 4/30 | |||
| Allograft Rejection Signaling | 1,29E-02 | 3.5% | 3/85 | |||
| Communication between Innate and Adaptive Immune Cells | 1,50E-02 | 3.3% | 3/90 | |||
| Crosstalk between Dendritic Cells and Natural Killer Cells | 1,50E-02 | 3.3% | 3/90 | |||
| Germ Cell-Sertoli Cell Junction Signaling | 1,86E-02 | 2.3% | 4/176 | |||
Funding Statement
This study was supported by Coordination for the Improvement of Higher Education Personnel (CAPES), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP Proc. No. 2010-51873-6) and Conselho Nacional De Desenvolvimento Científico E Tecnológico (CNPQ. Proc. No. 472930/2010-9).
Congresses where this study was presented: ASRM Meeting 2016 Salt Lake City - USA.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Support
This study was supported by Coordination for the Improvement of Higher Education Personnel (CAPES), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP Proc. No. 2010-51873-6) and Conselho Nacional De Desenvolvimento Científico E Tecnológico (CNPQ. Proc. No. 472930/2010-9).
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
REFERENCES
- Ambekar AS, Kelkar DS, Pinto SM, Sharma R, Hinduja I, Zaveri K, Pandey A, Prasad TS, Gowda H, Mukherjee S. Proteomics of follicular fluid from women with polycystic ovary syndrome suggests molecular defects in follicular development. J Clin Endocrinol Metab. 2015;100:744–753. doi: 10.1210/jc.2014-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appasamy M, Jauniaux E, Serhal P, Al-Qahtani A, Groome NP, Muttukrishna S. Evaluation of the relationship between follicular fluid oxidative stress, ovarian hormones, and response to gonadotropin stimulation. Fertil Steril. 2008;89:912–921. doi: 10.1016/j.fertnstert.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Benkhalifa M, Madkour A, Louanjli N, Bouamoud N, Saadani B, Kaarouch I, Chahine H, Sefrioui O, Merviel P, Copin H. From global proteome profiling to single targeted molecules of follicular fluid and oocyte: contribution to embryo development and IVF outcome. Expert Rev Proteomics. 2015;12:407–423. doi: 10.1586/14789450.2015.1056782. [DOI] [PubMed] [Google Scholar]
- Bianchi L, Gagliardi A, Landi C, Focarelli R, De Leo V, Luddi A, Bini L, Piomboni P. Protein pathways working in human follicular fluid: the future for tailored IVF? Expert Rev Mol Med. 2016;18:e9. doi: 10.1017/erm.2016.4. [DOI] [PubMed] [Google Scholar]
- Curry Jr TE, Smith MF. Impact of extracellular matrix remodeling on ovulation and the folliculo-luteal transition. Semin Reprod Med. 2006;24:228–241. doi: 10.1055/s-2006-948552. [DOI] [PubMed] [Google Scholar]
- Dai G, Lu G. Different protein expression patterns associated with polycystic ovary syndrome in human follicular fluid during controlled ovarian hyperstimulation. Reprod Fertil Dev. 2012;24:893–904. doi: 10.1071/RD11201. [DOI] [PubMed] [Google Scholar]
- de Agostini A. An unexpected role for anticoagulant heparan sulfate proteoglycans in reproduction. Swiss Med Wkly. 2006;136:583–590. doi: 10.4414/smw.2006.11368. [DOI] [PubMed] [Google Scholar]
- Dumesic DA, Oberfield SE, Stener-Victorin E, Marshall JC, Laven JS, Legro RS. Scientific Statement on the Diagnostic Criteria, Epidemiology, Pathophysiology, and Molecular Genetics of Polycystic Ovary Syndrome. Endocr Rev. 2015;36:487–525. doi: 10.1210/er.2015-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes SJ, Ye B, Qiu W, Cramer D, Hornstein MD, Missmer SA. A proteomic analysis of IVF follicular fluid in women <or=32 years old. Fertil Steril. 2009;92:1569–1578. doi: 10.1016/j.fertnstert.2008.08.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrieder J, Zuberovic A, Bergquist J. Surface modified capillary electrophoresis combined with in solution isoelectric focusing and MALDI-TOF/TOF MS: a gel-free multidimensional electrophoresis approach for proteomic profiling--exemplified on human follicular fluid. J Chromatogr A. 2009;1216:3621–3628. doi: 10.1016/j.chroma.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Jarkovska K, Martinkova J, Liskova L, Halada P, Moos J, Rezabek K, Gadher SJ, Kovarova H. Proteome mining of human follicular fluid reveals a crucial role of complement cascade and key biological pathways in women undergoing in vitro fertilization. J Proteome Res. 2010;9:1289–1301. doi: 10.1021/pr900802u. [DOI] [PubMed] [Google Scholar]
- Jarkovska K, Kupcova Skalnikova H, Halada P, Hrabakova R, Moos J, Rezabek K, Gadher SJ, Kovarova H. Development of ovarian hyperstimulation syndrome: interrogation of key proteins and biological processes in human follicular fluid of women undergoing in vitro fertilization. Mol Hum Reprod. 2011;17:679–692. doi: 10.1093/molehr/gar047. [DOI] [PubMed] [Google Scholar]
- Jungheim ES, Lanzendorf SE, Odem RR, Moley KH, Chang AS, Ratts VS. Morbid obesity is associated with lower clinical pregnancy rates after in vitro fertilization in women with polycystic ovary syndrome. Fertil Steril. 2009;92:256–261. doi: 10.1016/j.fertnstert.2008.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat BR, Brown LF, Manseau EJ, Senger DR, Dvorak HF. Expression of vascular permeability factor/vascular endothelial growth factor by human granulosa and theca lutein cells. Role in corpus luteum development. Am J Pathol. 1995;146:157–165. [PMC free article] [PubMed] [Google Scholar]
- Kushnir MM, Naessén T, Wanggren K, Rockwood AL, Crockett DK, Bergquist J. Protein and steroid profiles in follicular fluid after ovarian hyperstimulation as potential biomarkers of IVF outcome. J Proteome Res. 2012;11:5090–5100. doi: 10.1021/pr300535g. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenberg AG. Non-canonical activities of the proteasomes. Tsitologiia. 2014;56:331–339. [PubMed] [Google Scholar]
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Schweigert FJ, Gericke B, Wolfram W, Kaisers U, Dudenhausen JW. Peptide and protein profiles in serum and follicular fluid of women undergoing IVF. Hum Reprod. 2006;21:2960–2968. doi: 10.1093/humrep/del257. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Stein IF, Leventhal ML. Amenorrhea associated with bilateral polycystic ovaries. Am J Obstet Gynecol. 1935;29:181–191. [Google Scholar]
- Westermeier R, Naven T, Höpker HR, editors. Proteomics in Practice: A Guide to Successful Experimental Design. Weinheim: Wiley-VCH Verlag GmbH; 2002. [Google Scholar]
- Wookey AF, Chollangi T, Yong HE, Kalionis B, Brennecke SP, Murthi P, Georgiou HM. Placental Vitamin D-Binding Protein Expression in Human Idiopathic Fetal Growth Restriction. J Pregnancy. 2017;2017:5120267. doi: 10.1155/2017/5120267. [DOI] [PMC free article] [PubMed] [Google Scholar]