Abstract
Background
We review genealogical relationships, biogeographic patterns and broad historical drivers of speciation within the Bathyergidae, a group of endemic African rodents, as well as identify key taxa which need further research.
Methods
We sourced comparable cytochrome b sequence data (comparable data available for all members for the Family) and geographic information for all six genera of the African subterranean rodent. This information was combined into the most comprehensive and geographically representative evolutionary study for the Bathyergidae to date.
Results
Species richness within the Bathyergidae appears to be underestimated, with undescribed taxa in five of the six genera. Biogeographic patterns suggest large historical distributions, which were repeatedly fragmented by major landscape changes (especially rifting, uplift and drainage evolution) since the Miocene. Aside from vicariant events, other factors (ecological specialization, population-level responses and climatic change) may have been instrumental in driving divergences in the Bathyergidae. As such, adaptive differences may exist among both populations and species across their discrete ranges, driving independent evolutionary trajectories among taxa. In addition, highly fragmented distributions of divergent (and often relict) lineages indicates the possibility of narrow endemics restricted to diminishing suitable habitats. From this, it is clear that a systematic revision of the Bathyergidae is necessary; such a revision should include comprehensive sampling of all putative taxa, the addition of genomic information to assess adaptive differences, as well as ecological information.
Keywords: African mole-rats, Bathyergidae, Biogeography, Extra-limital, Species richness, Phylogeography
Introduction
Biodiversity is being lost at unprecedented rates, with extinction risk being underestimated for a large number of species in the absence of explicit geospatial and/or population data (Ceballos et al., 2015; Ocampo-Peñuela et al., 2016). It is further clear that species are not ubiquitously distributed across their ranges; habitat specialists are limited to areas of suitable habitat, leading to the possibility of undescribed taxa (Pimm et al., 2014). Accurate and detailed information on species’ distributions and evolutionary histories fill an important void in optimal conservation planning, with DNA evidence aiding in the uncovering of undescribed taxa, and elucidating biogeographic patterns and the drivers thereof.
A case in hand concerns the Bathyergidae, a group of subterranean rodents endemic to sub-Saharan Africa (Faulkes et al., 2010; Ingram, Burda & Honeycutt, 2004; Honeycutt, 2017). The family currently comprises six monophyletic genera (Allard & Honeycutt, 1992; Walton, Nedbal & Honeycutt, 2000; Huchon & Douzery, 2001; Faulkes et al., 1997, 2004; Ingram, Burda & Honeycutt, 2004) with largely disjunct distributions across their ranges. Heterocephalus and Heliophobius are restricted to East Africa, Bathyergus and Georychus to South Africa, with Cryptomys and Fukomys having wide distributions across southern and south-central Africa (see Fig. 1; Honeycutt et al., 1991).
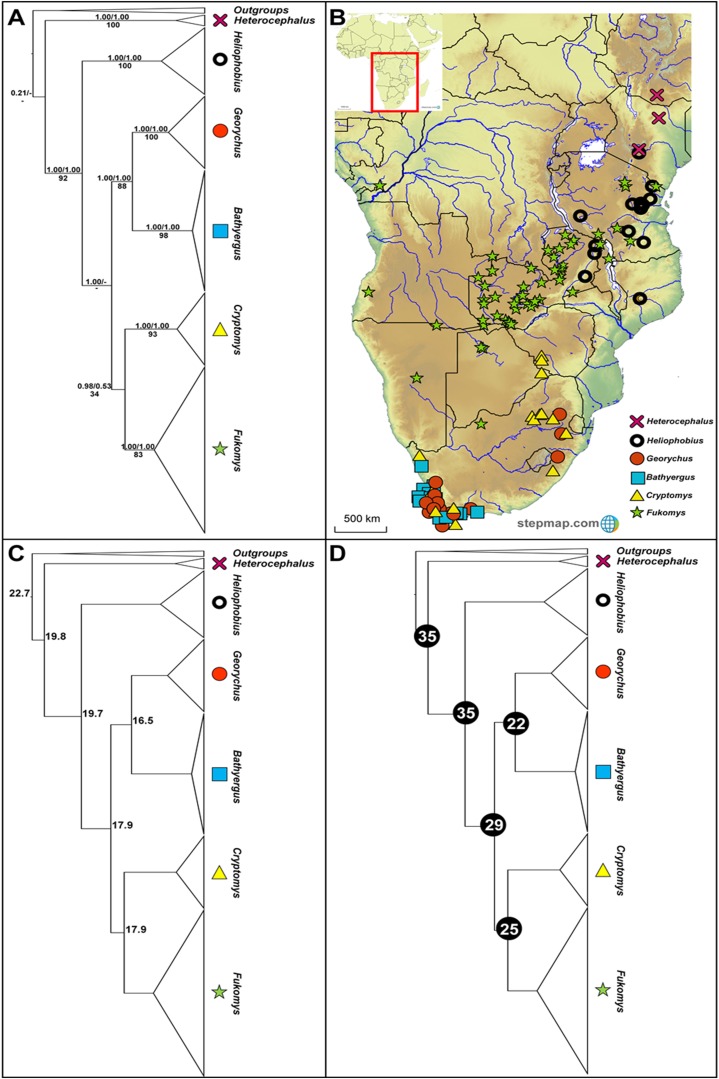
Figure 1. Phylogeny, geographic distributions, sequence divergences and divergence dates of the six bathyergid genera.
(A) Phylogeny of all six bathyergid genera based on the cytochrome b data. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML. A “-” indicates that the node was not retrieved by the particular analysis. The geographic distribution of these genera across sub-Saharan Africa is portrayed in (B) with the uncorrected sequence divergences (%) at the various nodes shown in (C). The divergence date estimates (in Mya) for each node is indicated within the black circles in (D).
The social systems within this family range from solitary (Heliophobius, Georychus and Bathyergus) to eusocial (Heterocephalus and Fukomys; Ingram, Burda & Honeycutt, 2004). Because of their eusocial nature (relatively uncommon amongst mammals), Heterocephalus and Fukomys have received much attention (Bennett & Jarvis, 1988; Nevo & Reig, 1990; Sherman, Jarvis & Alexander, 1991; Jarvis & Bennett, 1993; Nevo, 1999; Bennett & Faulkes, 2000; Lacey, Patton & Cameron, 2000; Faulkes & Bennett, 2007, 2009), leaving the other genera largely under-investigated by comparison (but see Honeycutt et al., 1987; Allard & Honeycutt, 1992; Faulkes et al., 1997; Walton, Nedbal & Honeycutt, 2000; Huchon & Douzery, 2001). Recent systematic/phylogeographic studies have hinted that diversity within the family is notably under-represented (Faulkes et al., 2004; Ingram, Burda & Honeycutt, 2004; Visser, Bennett & Jansen van Vuuren, 2014, 2018, 2019) and as a result, several species have recently been described (Honeycutt et al., 1991; Aguilar, 1993; Macholan et al., 1998; Burda et al., 1999; Chitaukali, Burda & Kock, 2001; Gippoliti & Amori, 2011; Van Daele et al., 2013; Faulkes et al., 2017), with further suggestions of possibly undescribed species in Heliophobius (Faulkes et al., 2011), Georychus, Bathyergus and Cryptomys (Visser, Bennett & Jansen van Vuuren, 2019).
Given the spate of phylogeographic investigations across wide geographic areas (Faulkes et al., 2004, 2011, 2017; Van Daele et al., 2007b; Visser, Bennett & Jansen van Vuuren, 2014, 2018, 2019), sufficient information now exist to synthesize a comprehensive systematic and biogeographic review for the Bathyergidae based on comprehensive geographic coverage and including all the bathyergid genera. In doing so, we construct a comprehensive phylogeny for the group to review genealogical relationships and timing of diversification events, and identify and review biogeographic patterns and broad historical processes driving speciation. This study therefore represents the most comprehensive and geographically representative study for the Bathyergidae to date, encompassing some 740 individuals representing all six genera. We use this information to assess the integrity of current species, as well as highlight possible undescribed cryptic species. To this end, we base inferences on consistent and well-supported monophyly together with sequence divergence estimates (with reference to other intra-generic divergences). Finally, we comment on future directions for research into this group.
Materials and Methods
Specimens
The cytochrome b gene region has been the predominant marker of choice in phylogeographic investigations within the bathyergid genera, and a representative dataset of this marker is available for a large number of species and lineages within the Bathyergidae including a comprehensive coverage of geographic localities. Comparable cytochrome b sequence data were sourced from public databases for all six mole-rat genera (Heterocephalus, Heliophobius, Georychus, Bathyergus, Cryptomys and Fukomys, see Fig.1; Table S1). These data were aligned using ClustalW as implemented in Geneious Pro™ 7.0 software (Biomatters Ltd, Auckland, New Zealand). Sequences shorter than 735 bp were removed to ensure a dataset with minimal missing data (see Joly, Stevens & van Vuuren, 2007 for a discussion of the impact of missing data).
Because of the large sample size (see below) and to reduce computational time, analyses are based on haplotypes only; alignments of the data were deconstructed into haplotypes using TCS 1.21 (Clement, Posada & Crandall, 2000). In total, 740 sequences (see Table S2 for species names and authorities) were included (Heterocephalus: n = 7 from three localities; Heliophobius: n = 60 from 19 localities; Georychus: n = 265 from 15 localities; Bathyergus: n = 190 from 15 localities; Cryptomys: n = 76 from 22 localities; Fukomys: n = 142 from 60 localities) which collapsed into 289 haplotypes (Heterocephalus: n = 7; Heliophobius: n = 38; Georychus: n = 41; Bathyergus: n = 68; Cryptomys: n = 41; Fukomys: n = 94). In addition, outgroup taxa of successive relatedness, and representing the Hystricognathi, were included to root topologies. These included the porcupine (Hystrix africaeaustralis), the dassie rat (Petromus typicus) and the cane rat (Thryonomys swinderianus).
Genealogical and molecular dating analyses
Prior to genealogical analyses, we used the Translate Tool in ExPASy (SIB Bioinformatics Resource Portal) to find the open reading frame in the data, and Split Codons (Sequence Manipulation Suite, www.Bioinformatics.org) to remove each codon position as a separate dataset (1st, 2nd and 3rd). Thereafter, the best-fit substitution model for each codon (1st position: TN93+G; 2nd position: GTR+G; 3rd position: HKY85+G+I) was selected through Modeltest version 2.1 (Guindon & Gascuel, 2003; Darriba et al., 2012) using the Akaike information criterion (Akaike, 1973). Phylogenetic reconstructions followed Bayesian inference methods and maximum likelihood (ML) with nodal support determined through posterior probabilities and bootstrapping respectively. Posterior probabilities ≥ 0.90 and bootstraps ≥ 70% were considered acceptable support.
Bayesian inference trees were constructed in MrBayes version 3.2.7 (Ronquist et al., 2012) by running 5 × 106 generations with sampling every 1,000 generations. To assess adequate convergence of the Markov chain Monte Carlo (MCMC) chain, the effective sample size of parameters was verified in Tracer version 1.5 (Rambaut & Drummond, 2003). After discarding the first 25% of the trees as burnin, a majority rule consensus tree with posterior probabilities was constructed and visualized in Figtree version 1.4.2 (available at: http://tree.bio.ed.ac.uk/software/figtree/).
Maximum likelihood analyses were performed in RAxML version 7.0.4 (Stamakis, 2014), which consisted of a 1,000 bootstrap replicates followed by a ML search. A majority rule consensus was constructed and visualized using Dendroscope version 3.5.8 (Huson & Scornavacca, 2012).
To obtain estimates of divergence times for the various clades, an uncorrelated relaxed molecular clock model with a Yule tree prior (most suitable for inter-specific comparisons) and an UPGMA starting tree was adopted in BEAST version 1.6.1 (Drummond et al., 2012). A fossil calibration point for Heliophobius (19 ± 0.5 Mya; Lavocat, 1973) was included in the analysis. Runs were continued for 30 × 106 generations with sampling every 1,000 generations. Results were visualized in Tracer version 1.5 (Rambaut & Drummond, 2003) to assess sufficient mixing and convergence of the MCMC chains, and samples from the posterior distribution of trees were summarized in TreeAnnotator version 1.6.1 (Drummond et al., 2012) with burnin set to 25%. Finally, a consensus tree was constructed using Figtree version 1.4.4 (available at: http://tree.bio.ed.ac.uk/software/figtree/).
In the final phylogenetic tree, nomenclature follows existing species (as listed on public databases) within the Bathyergidae, and the genetic affinity of sequences from unnamed specimens to the existing species (see Table S1). Possibly undescribed species are identified as lineages which are consistently retrieved as monophyletic and with acceptable nodal support (posterior probabilities ≥ 0.90 and bootstraps ≥ 70%) among the different analyses. Furthermore, these possibly undescribed species were identified on the basis of substantial sequence divergence (divergences that are higher than between described species within that genus, or the intrageneric mean). Calculation of uncorrected sequence divergences were performed in DnaSP version 5.10.01 (Librado & Rozas, 2009).
Results
Genealogical- and biogeographic patterns
Genera
The six genera form well-supported monophyletic groups in all the genealogical analyses (see Fig. 1A). Heterocephalus groups separately in the phylogeny relative to the remaining Bathyergidae; supporting the recognition of two subfamilies Bathyerginae and Heterocephalinae (none of our analyses recovered strong support for the monophyly of the Bathyergidae, although other genes and lines of evidence do support the monophyly of the mole-rats; see Davies et al., 2015). Within the Bathyerginae, Heliophobius occupies a basal position relative to the other four genera. The South African endemic genera Georychus and Bathyergus are well-supported sister genera with the sister relationship between the widespread genera Cryptomys and Fukomys retrieved, but not supported by two of the analyses (MrBayes and RAxML).
Generic ranges appear, to some extent, discrete for three of the genera (Heterocephalus, Heliophobius and Fukomys), but with some degree of overlap between Heliophobius and Fukomys, and extensive overlap between the southern African genera Georychus, Bathyergus and Cryptomys (Fig. 1B). Sequence divergences among the genera are generally in the same order of magnitude (17.7– 25.5%; Fig. 1C; Table S3) with divergences between Georychus and Bathyergus, and Cryptomys and Fukomys, largely contemporaneous during the Oligocene (Fig. 1D). Divergences among these generic complexes, and the remaining members of the Bathyergidae, appear more ancient and in the Eocene (Fig. 1D).
Heterocephalus
Within Heterocephalus no major clades are evident; however, there appears to be some separation between animals from Ethiopia and Kenya (see Fig. S1).
Heliophobius
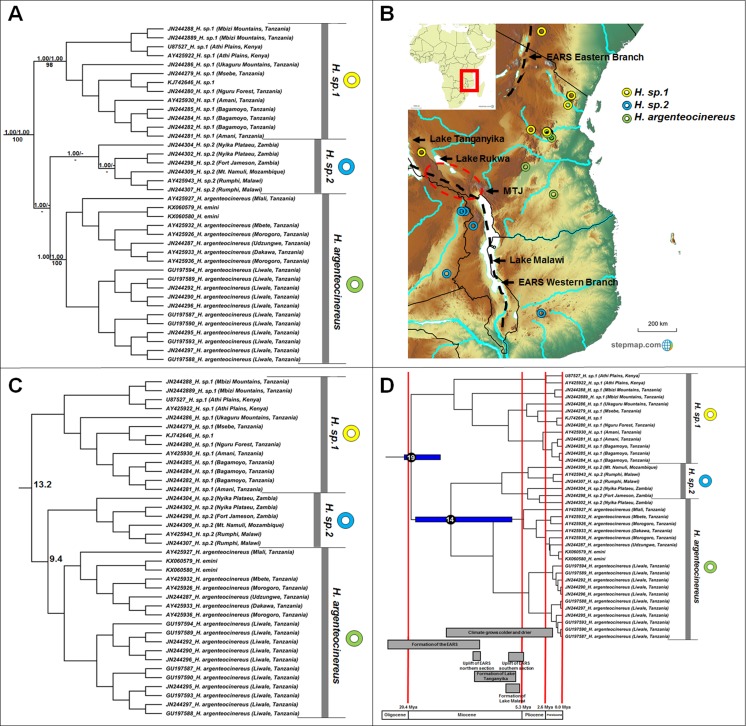
Heliophobius comprises three major clades (Fig. 2A), but with some variations among the sister taxon relationships retrieved by the different analyses. In the BEAST phylogeny (shown), H. sp. 2 is sister to Heliophobius argenteocinereus, while both MrBayes and RAxML groups H. sp. 1 and Heliophobius argenteocinereus as sister taxa with strong support. Heliophobius argenteocinereus and H. sp. 1 both occur to the east of the East African Rift System, whereas H. sp. 2 occupies areas to the west of this region (Fig. 2B). The sister relationships between these three lineages are not clear, with sequence divergence values supporting H. sp. 2 and Heliophobius argenteocinereus as closer related (9.4%; Fig. 2C; Table S4). Divergence times among the Heliophobius species also indicate a more recent middle Miocene divergence between H. sp. 2 and Heliophobius argenteocinereus (Fig. 2D).
Figure 2. Phylogeny, geographic distributions, sequence divergences and divergence dates of species within the genus Heliophobius.
(A) Phylogeny of the genus Heliophobius based on the cytochrome b data and indicating the different species included and identified. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML. A “-” indicates that the node was not retrieved by the particular analysis. The geographic location (point locality for the specimen) of the recovered species is portrayed in (B) with the uncorrected sequence divergences (%) at the various nodes shown in (C). The divergence date estimates (in Mya) for each node is indicated within the black circles in (D), with the blue bars representing the temporal range of the divergence (derived from the dating analysis in BEAST). The red bars represent the temporal boundaries among the epochs, with the timing and temporal extent of major landscape changes which occurred across the distribution of the genus indicated at the bottom.
Georychus
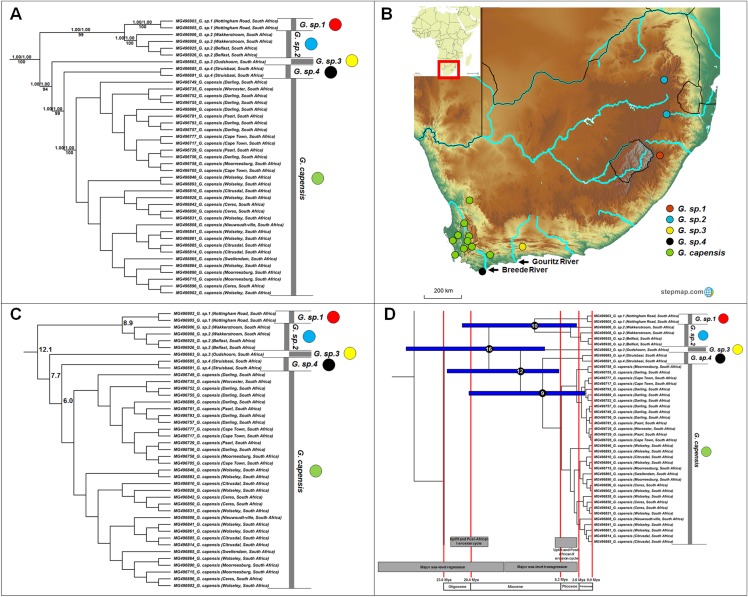
Georychus includes five well-supported, geographically discreet, clades (Fig. 3A) which follow a north-east to south-west clinal pattern across South Africa (Fig. 3B). Georychus sp. 1 and G. sp. 2 appear highly divergent from each other (8.9%) in spite of their geographic proximity (Mpumalanga and KwaZulu-Natal provinces, both in the north eastern part of South Africa), as well as from their congenerics in the Western Cape Province of South Africa (9.9–12.5%; Fig. 3C; Table S5). The times of divergences among the five species span the Miocene (Fig. 3D).
Figure 3. Phylogeny, geographic distributions, sequence divergences and divergence dates of species within the genus Georychus.
(A) Phylogeny of the genus Georychus based on the cytochrome b data and indicating the different species included and identified. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML. The geographic location (point locality for the specimen) of the recovered species is portrayed in (B) with the uncorrected sequence divergences (%) at the various nodes shown in (C). The divergence date estimates (in Mya) for each node is indicated within the black circles in (D), with the blue bars representing the temporal range of the divergence (derived from the dating analysis in BEAST). The red bars represent the temporal boundaries among the epochs, with the timing and temporal extent of major landscape changes which occurred across the distribution of the genus indicated at the bottom.
Bathyergus
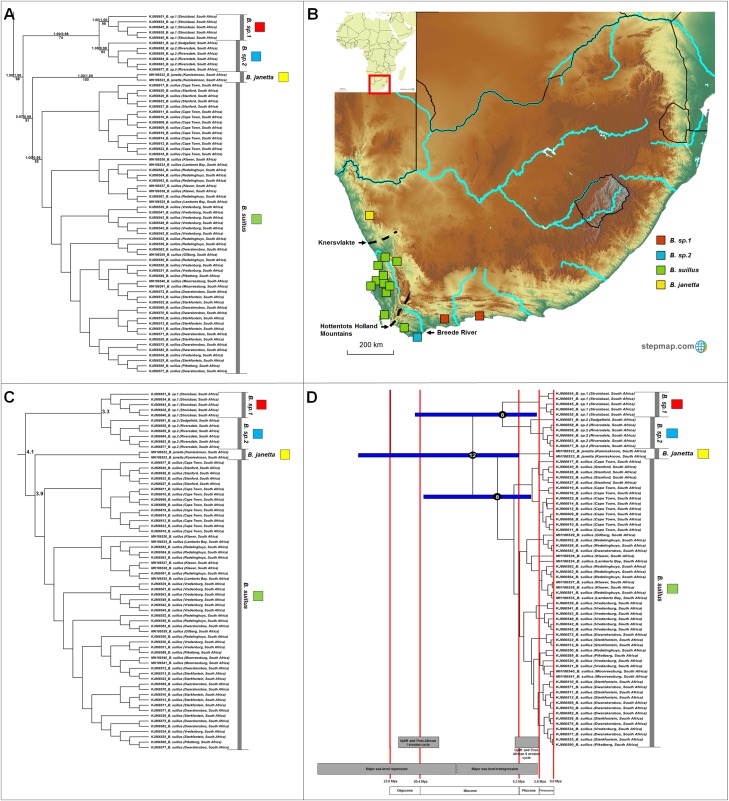
Four well-supported major clades are evident within Bathyergus (Fig. 4A) which occupy non-overlapping ranges across South Africa (Fig. 4B). Sequence divergence values between major clades within this genus is lower compared to those seen in the other genera (3.3–5.3%; Fig. 4C; Table S6), resulting is slightly more recent divergence date estimates (Fig. 4D).
Figure 4. Phylogeny, geographic distributions, sequence divergences and divergence dates of species within the genus Bathyergus.
(A) Phylogeny of the genus Bathyergus based on the cytochrome b data and indicating the different species included and identified. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML. The geographic location (point locality for the specimen) of the recovered species is portrayed in (B) with the uncorrected sequence divergences (%) at the various nodes shown in (C). The divergence date estimates (in Mya) for each node is indicated within the black circles in (D), with the blue bars representing the temporal range of the divergence (derived from the dating analysis in BEAST). The red bars represent the temporal boundaries among the epochs, with the timing and temporal extent of major landscape changes which occurred across the distribution of the genus indicated at the bottom.
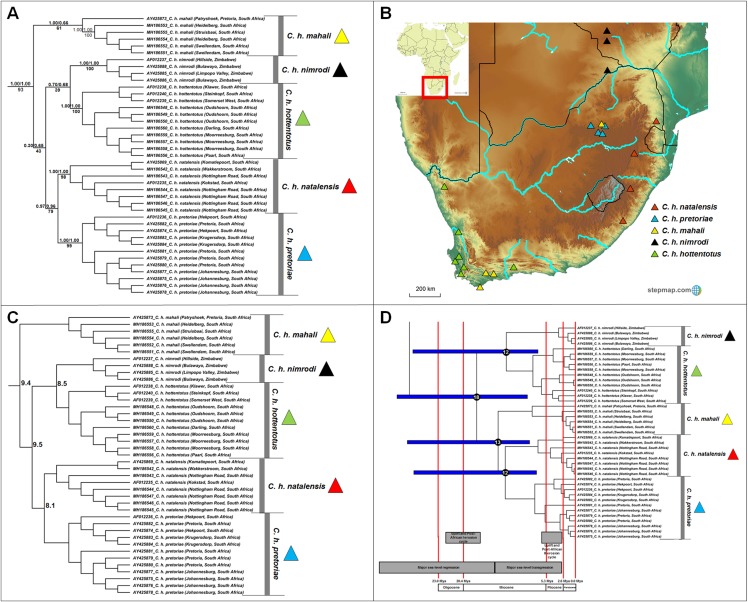
Cryptomys
Five major clades, which corresponds to the described subspecies, were retrieved within Cryptomys (Fig. 5A); four of these clades received strong nodal support from all genealogical analyses (Cryptomys h. nimrodi, C. h. natalensis, C. h. pretoriae and C. h. hottentotus), with the exception of C. h. mahali which was only well-supported in one analysis (BEAST). The putative range covered by this subspecies therefore extends across the interior of South Africa, and includes animals collected in the Western Cape (which group together with strong support; sourced from Visser, Bennett & Jansen van Vuuren (2019)) with an animal from Pretoria grouping basal and with lower support (sourced from Faulkes et al. (2004)). It is therefore possible that distinct genetic clades may exist within this subspecies. In general, intra-generic relationships were not consistently retrieved (with exception of the divergence between C. h. natalensis and C. h. pretoriae) and may indicate rapid and near contemporaneous divergences of groups. A degree of overlap exists in the ranges of C. h. mahali and C. h. hottentotus in the southern coastal parts of South Africa, as well as between C. h. mahali and C. h. pretoriae in the north-eastern interior of the country (Fig. 5B). Sequence divergence among the subspecies appear high and similar in values (8.1–10.3%; Fig. 5C; Table S7), with divergences largely contemporaneous during the Miocene (Fig. 5D).
Figure 5. Phylogeny, geographic distributions, sequence divergences and divergence dates of species within the genus Cryptomys.
(A) Phylogeny of the genus Cryptomys based on the cytochrome b data and indicating the different species included and identified. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML. Values in normal font show support for species which were recovered with the exclusion of a particular specimen. The geographic location (point locality for the specimen) of the recovered species is portrayed in (B) with the uncorrected sequence divergences (%) at the various nodes shown in (C). The divergence date estimates (in Mya) for each node is indicated within the black circles in (D), with the blue bars representing the temporal range of the divergence (derived from the dating analysis in BEAST). The red bars represent the temporal boundaries among the epochs, with the timing and temporal extent of major landscape changes which occurred across the distribution of the genus indicated at the bottom.
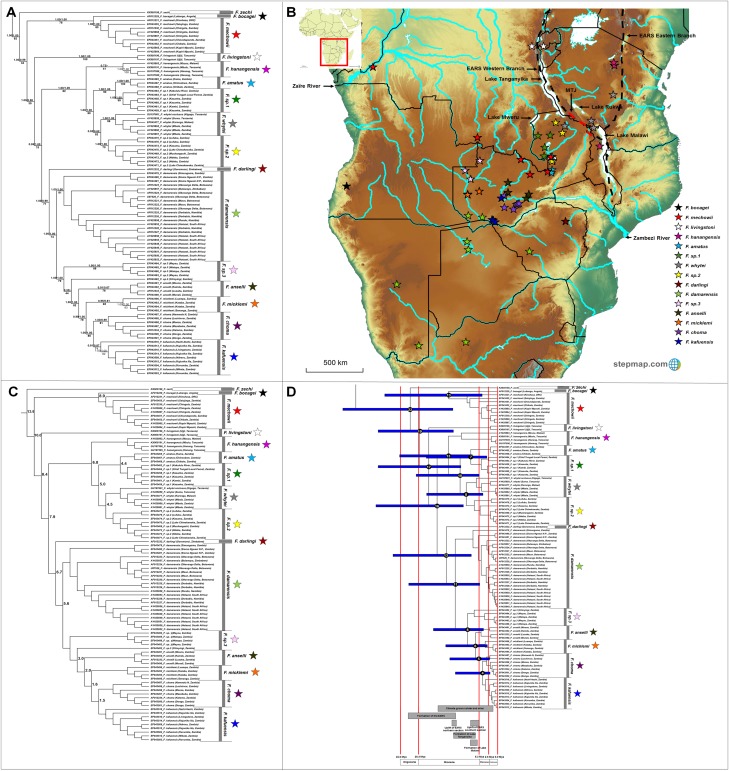
Fukomys
The genus Fukomys occupies a large range, with 16 major clades evident of which 12 are well-supported by all the analyses (Fig. 6A). Among the remaining clades, Fukomys hanagensis, F. micklemi and F. kafuensis also form well-supported monophyletic clades, when excluding sequences AY425863 (F. hanagensis from Mzuzu, Malawi), EF043496 (F. micklemi from Luampa, Zambia) and EF043516 (F. kafuensis from Itezhi-Ithezi, Zambia) from the analyses. The F. anselli clade received poor support irrespective method of analyses.
Figure 6. Phylogeny, geographic distributions, sequence divergences and divergence dates of species within the genus Fukomys.
(A) Phylogeny of the genus Fukomys based on the cytochrome b data and indicating the different species included and identified. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML. A “-” indicates that the node was not retrieved by the particular analysis, while values in normal font show support for species which were recovered with the exclusion of a particular specimen. The geographic location (point locality for the specimen) of the recovered species is portrayed in (B) with the uncorrected sequence divergences (%) at the various nodes shown in (C). The divergence date estimates (in Mya) for each node is indicated within the black circles in (D), with the blue bars representing the temporal range of the divergence (derived from the dating analysis in BEAST). The red bars represent the temporal boundaries among the epochs, with the timing and temporal extent of major landscape changes which occurred across the distribution of the genus indicated at the bottom.
Fukomys zechi (the Ghana mole-rat) is the oldest extant lineage, followed by a number of species complexes and putatively new species distributed across Central, East and Southern Africa (Fig. 6A). Species distributions appear generally discrete, but a degree of overlap exists between the ranges of F. mechowii, F. sp. 1 and F. sp. 2 (Fig. 6B). Sequence divergences within Fukomys vary widely (1.5–16.2%; Fig. 6C; Table S8) resulting in a range of divergence times spanning the Oligocene to the more recent Plio/Pleistocene, but with the majority of events placed in the Miocene (Fig. 6D).
Discussion
Biogeographic patterns and species diversity
The Family Bathyergidae represent an interesting, unique, and highly successful group of sub-Saharan African subterranean mammals, with a range of unique adaptations to their fossorial lifestyle. Although much additional work needs to be done on the group to fully appreciate their evolution, it is clear that the Bathyergidae is characterized by a range of physiological adaptations, karyotype differences and genomic expressions (Deuve et al., 2008; Davies et al., 2015). Here, for the first time, we compile an inclusive phylogeny for the group, and attempt to unravel aspects of the group’s evolutionary and biogeographic histories. For this, we used the mitochondrial cytochrome b gene for which comparable data were available for all taxa. While the cytochrome b data were adequate at resolving intra-generic relationships and highlighting a number of putatively new species, deeper nodes within the phylogenetic tree (inter-generic divergences) remain less well resolved (but still in line with published data; see Allard & Honeycutt, 1992; Faulkes et al., 1997, 2004; Walton, Nedbal & Honeycutt, 2000; Ingram, Burda & Honeycutt, 2004; Deuve et al., 2008).
Inter-generic patterns
The evolutionary placement of Heterocephalus outside the remaining five genera (Heliophobius, Georychus, Bathyergus, Cryptomys and Fukomys) offers strong support for the recognition of two subfamilies namely Heterocephalinae (for Heterocephalus) and Bathyerginae (for the remaining genera) (Ellerman, Morrison-Scott & Hayman, 1953). Within the Bathyerginae, Heliophobius is consistently retrieved as the oldest taxon (also see Allard & Honeycutt, 1992; Janecek et al., 1992; Faulkes et al., 1997, 2011; Walton, Nedbal & Honeycutt, 2000; Ingram, Burda & Honeycutt, 2004; Deuve et al., 2008 for a similar finding). The time of divergence of this genus from the other members of the subfamily is near-contemporaneous with the divergence of the two subfamilies (at approximately 35 Mya). These two oldest genera (Heterocephalus and Heliophobius) predominantly occur to the east of the Eastern Arc Rift System (see Fig. 1B).
Among the remaining genera, the sister relationship between the South African endemics Georychus and Bathyergus is consistently recovered (also see Faulkes et al., 1997, 2004; Walton, Nedbal & Honeycutt, 2000; Ingram, Burda & Honeycutt, 2004; Visser, Bennett & Jansen van Vuuren, 2019) and also the sister relationship between Cryptomys and Fukomys (also see Faulkes et al., 1997, 2004; Walton, Nedbal & Honeycutt, 2000; Ingram, Burda & Honeycutt, 2004). Although Fukomys largely occurs to the west of the Eastern Arc Rift System, some of its member-species straddle this barrier. A notable degree of range overlap exists between Georychus and Bathyergus in the south and south west of South Africa, although two Georychus species occur in the north-eastern parts of the country. Cryptomys is largely co-distributed with these genera across South Africa.
Intra-generic patterns
The intra-generic taxonomy and evolutionary relationships within a number of mole-rat groups have always been contentious; largely as a result of the inconspicuous nature of the animals, and the difficulties of obtaining comprehensive samples across their ranges. For example, Ellerman (1940) proposed nine taxa in Heliophobius, but concluded that these constitute races of Heliophobius argenteocinereus rather than distinct species (with the exception of Heliophobius spalax). However, extensive karyotypic variation (George, 1979; Scharff, Macholan & Burda, 2001) and high levels of mitochondrial sequence divergences (Faulkes et al., 2004, 2011; Ingram, Burda & Honeycutt, 2004), led Faulkes et al. (2011) to propose species statuses for six geographic clades. We recover three distinct and well-supported genetic clades within this genus, although several specimens within each of these clades are highly divergent (e.g., JN244304 from the Nyika Plateau, as well as specimens from the Mbizi Mountains and Athi Plains) and may in fact represent putative species. Although the methods of analyses were not consistent in the relationships between these three clades, it would seem that the oldest species within Heliophobius (H. sp. 2) occurs to the east of the Eastern Arc Rift System (Fig. 2A). Heliophobius sp. 1 is widespread across the Athi Plains and between the Eastern and Western Rifts in the Mbizi Mountains (Tanzania). Indications are that this may be a highland species, restricted to areas of higher elevation (Fig. 2B). In contrast, Heliophobius argenteocinereus occupies a coastal/lowland distribution across Tanzania. Heliophobius sp. 2 occurs to the west of the Eastern Arc Rift System (although a single specimen was found east of the Eastern Arc Rift System in Mozambique), but similarly to Heliophobius argenteocinereus, follows an area of lower elevation along a north-south axis along, and crossing the Eastern Arc Rift System in an east-west axis in the southern part.
A single species is currently recognized for Georychus (Georychus capensis; type locality included in this study). However, our work suggests that there may be as many as four additionally undescribed species within Georychus (Fig. 3A). These unique genetic lineages geographically follow a clinal pattern: G. sp. 1 and G. sp. 2 are restricted to the north-eastern interior of South Africa and is found at higher elevations, G. sp. 3 occurs in a single valley in the southern interior below the Great Escarpment, while G. sp. 4 is located in the south-western coastal region (see Fig. 3B). G. capensis occurs further west, occupying a wide distribution in the interior (below the Great Escarpment) and lowland regions of south-western South Africa.
Bathyergus contains at least four species (Fig. 4A), two of which are the described species (Bathyergus suillus and B. janetta; type localities included in this study). All species are restricted to the southern and western seaboard of South Africa, with the exception of B. janetta which is found further north in the Namaqualand region and southern Namibia (Fig. 4B). Bathyergus sp. 1 and B. sp. 2 are restricted to the southern coastal margins and B. suillus to the western coastal margins.
Based on multi-locus genetic information, Visser, Bennett & Jansen van Vuuren (2019) recently proposed that the genus Cryptomys may be in need of a taxonomic revision, and suggested that C. h. mahali, C. h. hottentotus and C. h. natalensis should be elevated to species level. Considering all the currently described subspecies in this genus, the current study supports this (Fig. 5A, see also Faulkes et al., 2004; Ingram, Burda & Honeycutt, 2004). Although the relationships among the current subspecies within Cryptomys are not consistently retrieved (with the exception of the sister relationship between C. h. natalensis and C. h. pretoriae), the subspecies appear geographically exclusive. The sister clades C. h. natalensis and C. h. pretoriae are restricted to the eastern highland parts of South Africa (also see Ingram, Burda & Honeycutt, 2004), with C. h. natalensis occurring along the margins of the Great Escarpment and C. h. pretoriae further north-west in the Gauteng Province (see Fig. 5B). C. h. hottentotus occupies the coastal margins of south-western and north-western South Africa (also see Aguilar, 1993; Faulkes et al., 1997; Ingram, Burda & Honeycutt, 2004) with C. h. nimrodi found at higher elevations and the furthest north of all the current subspecies in Zimbabwe (also see Faulkes et al., 1997; Van Daele et al., 2004). C. h. mahali was collected in one area of high elevation around Pretoria (Faulkes et al., 1997, 2004), but also occurs in the low-lying coastal range of C. h. hottentotus. Although this would suggest a wide range for this group across South Africa, the genetic divergence between the Pretoria specimen and those from the coast suggest that these may be two highly divergent populations (possible subspecies). Additional sampling across the range is necessary to fully untangle the evolutionary relationships within the mahali group.
Fukomys includes a divergent and widespread collection of distinct genetic lineages (also see Faulkes et al., 1997, 2004, 2017; Ingram, Burda & Honeycutt, 2004; Van Daele et al., 2007b; Deuve et al., 2008) consisting of several species complexes of various evolutionary ages and with various degrees of divergence (Fig. 6; also see Van Daele et al., 2007b; Faulkes et al., 2017). The evolutionary progression within this groups suggests a West African origin, with subsequent migrations to the east and south (Fig. 6B). Specifically, F. zechi occupies the Sudanian savannah (Ghana) at the far north-western edge of the generic distribution, the sister species F. bocagei and F. mechowii are found to the far west (Angola) and along a northern band of the generic range respectively, and F. livingstoni occurs in the far north-east (in Tanzania, and east of the Eastern Arc Rift System). Similarly, F. hanangensis occurs in the far north-east of the distribution (in Tanzania and east of the Eastern Arc Rift System), albeit a single population is found west of the Eastern Arc Rift System (in Malawi). F. darlingi occupies the far south-east of the generic range in Zimbabwe.
Species of intermediate age (e.g., F. amatus, F. sp. 1, F. whytei and F. sp. 2) are located in a single geographic area to the west of the Eastern Arc Rift System (Fig. 6B), enclosed by Lakes Tanganyika, -Mweru Wantipa, -Mweru, -Bangweulu and -Malawi, as well as a single tributary of the Zaïre River system. Among these species, F. whytei occupies a range on both side of the Eastern Arc Rift System, with a distribution centered on and crossing the Mbeya Triple Junction, a volcanic area that formed 2 to 2.5 Mya (Ebinger et al., 1993; Delvaux et al., 1998; Macgregor, 2015) and forms an intersection between the two adjacent basins at the northern end of Lake Malawi (Fig. 6B; Faulkes et al., 2011). F. damarensis has a large distribution in the southern part of the generic distribution and to the south of the Zambezi River, while the most derived chromosomal races/species in Fukomys (F. anselli, F. micklemi, F. choma, F. kafuensis and the undescribed F. sp. 3) occur in south-central Zambia around tributaries of the Zambezi River (Fig. 6B).
Major drivers of inter- and intra-generic divergences
Mole-rats are intimately linked to their subterranean niche, and it is not surprising that biogeographic patterns reviewed here suggest abrupt species turnover across various landscape features. It follows that evolutionary patterns within the Bathyergidae are heavily dependent on landscape structure, and the spatiotemporal variability thereof. Divergences among the genera, as well as among species within these genera, therefore coincide with prominent changes in landscape structure over evolutionary time.
The oldest inter-generic divergences probably coincide with early Miocene landscape changes. Climatically, warm and wet conditions prevailed during the early Miocene, which was interrupted by a progressively colder and drier phase during the middle Miocene (15.5–14 Mya, Siesser, 1980; Van Zinderen Bakker & Mercer, 1986; Zachos et al., 2001; Diekmann, Falker & Kuhn, 2003; Krammer, Baumann & Henrich, 2006). This favored the expansion of arid-adapted lineages (Axelrod & Raven, 1978; Coetzee, 1978, 1983; Bonnefille, 1984; Van Zinderen Bakker & Mercer, 1986), following a corridor of fluctuating aridity and a forest/woodland savannah mosaic across eastern, south-central and southern Africa (Van Zinderen Bakker, 1967; Van Couvering & Van Couvering, 1976; Kortlandt, 1983). Under these conditions ancestral bathyergids likely attained large distributions across sub-Saharan Africa, as supported by fossil bathyergids from the early Miocene deposits from East Africa and Namibia (Lavocat, 1973, 1978; also see Honeycutt et al., 1991; Faulkes et al., 2004). These wide distributions of the early bathyergids likely became fragmented through subsequent upheavals in the climate and geology of sub-Saharan Africa, driving spatial isolation and divergence within the group as a whole.
Major sculpting of the East African landscape began with the formation of the Eastern (Kenya) Rift at 23–11 Mya, with uplift of the Western Branch (Albertine Rift) of the Eastern Arc Rift System occurring at 12–11 Mya in the northern section and 7–5 Mya in the southern section (Van Couvering & Van Couvering, 1976; Ebinger, 1989; Ebinger et al., 1993). Tectonic activity continued into the Pliocene/Pleistocene (Ebinger, 1989; Ebinger et al., 1993; Bauer et al., 2010) and was instrumental in the formation of mountainous areas (e.g., the Eastern Arc Mountains at around 11–5.3 Mya) and volcanoes (see Chorowicz, 2005). This rifting process also drove the formation of the Great Lakes (Lake Tanganyika 12–6 Mya, Lake Malawi 7.2–5.3 Mya, Cohen, Soreghan & Scholz, 1993; Delvaux, 1995; Delvaux et al., 1998; Macgregor, 2015).
The early divergence of the East African Heterocephalus and Heliophobius appears independent of rifting (also see Faulkes et al., 2017), while the divergence between the two generic complexes of Georychus/Bathyergus and Cryptomys/Fukomys may have followed the spatial separation of a common ancestor via different routes of dispersal from East Africa. Within the Georychus/Bathyergus complex, a widespread common ancestor likely diverged parapatrically, with Georychus evolving in the South African interior and Bathyergus diverging within sandy areas of south and south-western South Africa. Divergence between Cryptomys and Fukomys follows the Zambezian and the Kalahari-Highveld phytochoria and was likely driven by the pattern of flow of the paleo-Zambezi River (the upper Zambezi River was linked to the Orange- or Limpopo Rivers; Thomas & Shaw, 1988). This flow pattern isolated ancestors of the two genera, with Cryptomys radiating southward while Fukomys spread north and west to attain a large distribution in south-central and West Africa (also see Ingram, Burda & Honeycutt, 2004; Faulkes et al., 2017).
The major historical landscape changes across East Africa coincide with intra-generic speciation patterns within the genera Heliophobius and Fukomys (Figs. 2D and 6D). The formation of the Eastern Arc Rift System and the Great Lakes likely fragmented ancestral Heliophobius populations, subsequently leading to the divergence of H. sp. 1 and H. sp. 2/Heliophobius argenteocinereus across this newly formed barrier. Elevation differences created through uplift (e.g., of the Eastern Arc Mountains) across this tectonically active region also likely restricted individual Heliophobius species to either highland (H. sp. 1) or lowland (H. sp. 2/Heliophobius argenteocinereus) habitats. Formation of Lakes Rukwa and Malawi also appear instrumental in the divergence between F. whytei and F. sp. 2, from where F. whytei subsequently expanded its range eastward via the Mbeya Triple Junction following this divergence (F. whytei lineages follow a geographically clinal pattern in an eastward direction; Fig. 6A). In contrast, F. hanangensis appears to have originated on the western side of the Eastern Arc Rift System, and has subsequently colonized the eastern side in Tanzania, also likely following the Mbeya Triple Junction (the oldest F. hanangensis lineage is found west of the Eastern Arc Rift System; Fig. 6A).
Aside from impact of rifting in driving divergence among the above Fukomys species, the wider distribution of this genus has been subjected to the consequences of uplift across central and eastern Africa. This undoubtedly influenced drainage evolution in the paleo-Zambezi and paleo-Zaïre River watersheds. Initial drainage evolution of the paleo-Zaïre River watershed (specifically the paleo-Kafue River and Luapula River), along with the formation of lakes (Lakes Tanganyika, -Mweru Wantipa, -Mweru, -Bangweulu and -Malawi), appear to have driven speciation within the F. hanangensis, F. amatus, F. sp. 1, F. sp. 2 species complex. This pattern of isolation is also mirrored in extra-limital species of a similar evolutionary age (F. darlingi and F. damarensis) around the Zambezi River watershed. Continued drainage evolution of the Zambezi River watershed across central Zambia along with geomorphological repatterning of the area, appear to have driven speciation among the younger Fukomys species (e.g., F. sp. 3, F. anselli, F. micklemi, F. choma and F. kafuensis) which are separated by branches of the Zambezi River system. Furthermore, these species exhibit extensive chromosomal rearrangements, possibly following ecological specialization to separate areas in this ecogeographically heterogeneous region (Van Daele et al., 2007b).
Aside from the East African genera, speciation within the three southern African genera Georychus, Bathyergus and Cryptomys exclusively follow geological and climatic changes across this sub-region. In South Africa, early Miocene uplift initiated the Post-African I erosion cycle (Partridge & Maud, 1987) with a second and more pronounced uplift event in the late Miocene/early Pliocene leading to the Post-African II erosion cycle (Partridge & Maud, 1987, 2000). Cold and dry episodes dominated the climate since the middle Miocene (Van Zinderen Bakker & Mercer, 1986) and late Miocene (Shackleton & Kennett, 1975a, 1975b; Van Zinderen Bakker & Mercer, 1986; Tyson & Partridge, 2000), with a cold period at the Miocene/Pliocene boundary (5 Mya) leading to the abolishment of subtropical rainforests in south-western South Africa (Van Zinderen Bakker & Mercer, 1986). Following glacial cycles, oceanic fluctuations exposed or inundated large areas of the coastal shelf (more than 500 km) during oceanic regression and transgression phases respectively (Dingle & Rogers, 1972; Siesser & Dingle, 1981; Maud & Partridge, 1987; Rogers, 1987; Hallam, 1992). A major regression lasted from the early Oligocene up until the end of the early Miocene (low-stand of more than 500 m below present during the Oligocene/Miocene boundary), while a major regression occurred from the middle Miocene lasting until the Pliocene (high-stand of up to 300 m above present during the late Miocene/early Pliocene; Siesser & Dingle, 1981).
Radiations within Georychus, Bathyergus and Cryptomys followed the Post-African I erosion cycle. Prior to this uplift event, the topography across southern Africa would have been more uniform. During this period, a major sea-level regression was also underway (Dingle & Rogers, 1972; Siesser & Dingle, 1981). Under these condition, ancestral lineages in all three genera were probably widespread across the interior as well as along the coastal margins of southern Africa. These wide distributions would have been bisected by the first major uplift event which initiated the Post-African I erosion cycle. This uplift raised the interior plateau of southern Africa (250–300 m in the east and 150–200 m in the west), creating a rugged and sloping topography as well as mountainous areas across the sub-region, consequently leading to the incision of deep river valleys (Partridge & Maud, 1987; Cowling, Procheş & Partridge, 2009).
As a result, divergences in the largely co-distributed genera Georychus and Bathyergus (and possibly Cryptomys) follow elevation differences, mountain barriers and drainage evolution of the major river systems. The influence of elevation differences is evident in the occurrence of several highland species which occupy the southern African interior (e.g., G. sp. 1, G. sp. 2, C. h. pretoriae, C. h. natalensis and possibly C. h. mahali), in contrast to several lowland/coastal species (e.g., G. sp. 3, G. sp. 4, G. capensis and C. h. hottentotus). Mountainous areas such as the Hottentots Holland Mountains also forms a major geographic barrier between B. suillus and B. sp. 1/B. sp. 2. Similar to Fukomys, the influence of drainage evolution as an isolating factor is apparent, with the Breede and Gouritz rivers forming major geographic barriers between B. sp. 1 and B. sp. 2, and G. sp. 3, G. sp. 4 and G. capensis, and drainage evolution of the Limpopo River likely leading to the divergence of C. h. nimrodi. Given that a large part of the distributions of Georychus and Bathyergus are exclusively coastal, the major sea-level transgression which covered the coastal shelf during the late Miocene/early Pliocene also appears instrumental in driving/enforcing isolation of the lowland species such as B. sp. 1 and B. sp. 2, B. suillus and B. janetta, and G. sp. 4.
Possible other factors driving speciation within the Bathyergidae
Although physical landscape changes were undoubtedly instrumental in driving radiations within the Bathyergidae, these events alone cannot explain all the divergences within the family. Historical climatic changes, intrinsic population-level responses or ecological specialization of species undoubtedly contributed (Faulkes et al., 2004, 2010; Van Daele et al., 2007b; Visser, Bennett & Jansen van Vuuren, 2019) although these drivers are frequently overlooked in phylogeoraphic/systematic studies on the Bathyergidae (but see Van Daele et al., 2007b).
Given the ecogeographical turnover across the African landscape, isolation in fringe habitat indicates the possibility that geographically distant species represent narrow endemics which are adapted to localized and diminishing habitat types. In contrast, the majority of younger or less divergent species occupy a central distribution, where suitable habitat (similar ecological circumstances) is likely abundant and divergences largely follow vicariant events. Understanding and incorporating species-specific responses/attributes (e.g., ecological and demographic information) may therefore be critical during phylogeographic/systematic endeavors in the Bathyergidae—a paradigm shift which is becoming increasingly important when interpreting genetic patterns across landscapes (Papadopouloua & Knowles, 2016).
Indeed, the geographic ranges of a large number of mole-rat lineages are spatially limited, and secondary contact is uncommon even in areas without apparent physical barriers. Given the heterogeneous geology, microclimate and phytogeography of Africa across various spatial scales, it is reasonable to assume that ecological and/or adaptive differences exist among species across their respective ranges, and that other intrinsic and extrinsic processes may be of importance in driving or enforcing isolation. As such, the Bathyergidae may include a multitude of habitat specialist taxa. These specialists may be prone to extinction through stochastic population processes, or modern perturbations which diminish preferred habitat (e.g., current levels of climate change).
Highly divergent and geographically restricted species within three of the genera is found at the edges of their respective generic ranges. No obvious link to physical landscape evolution explains the origin or distribution of the oldest species within Fukomys (e.g., F. zechi, F. bocagei, F. livingstoni). These species are separated from other members of the genus by large geographic distances, and likely occupy diminishing fringe habitats in the generic range. Similarly, large distances separate species within the genus Georychus. It is entirely possible that these (or at least G. sp. 1 and 2) species represent relic populations, isolated in pockets of suitable habitat. Similarly, F. zechi was likely isolated form congenerics by the formation of the tropical rainforest in the Congo Basin (Ingram, Burda & Honeycutt, 2004; Van Daele et al., 2007a; Faulkes et al., 2017).
Bathyergid systematics—historical trends and future directions
The data presented here confirms the notion that allopatric speciation is prevalent in taxa which inhabit the fossorial/subterranean niche (Fitzpatrick & Turelli, 2006). Phylogeographic/spatial genetic studies have consistently recovered patterns of intimate co-evolution between fossorial taxa and the landscape, invariably finding genetically discrete lineages or species in separate geographic regions (Albert, Zardoya & García-París, 2007; Opazo et al., 2008; Tang et al., 2010).
While the patterns recovered within fossorial taxa have remained broadly similar, the choice of markers for inferring genetic isolation has grown by leaps and bounds over the past four decades. Initially, inferences of genetically discrete groups were based on karyotypic and allozyme information (Nevo et al., 1974; Patton & Yang, 1977; Hafner et al., 1987). With the advent of more refined genetic sequencing techniques, mitochondrial DNA sequence data dominated fossorial phylogeography and systematics (Albert, Zardoya & García-París, 2007; Opazo et al., 2008; Tang et al., 2010) while more recently, the addition of nuclear markers has aided in the discovery of cryptic species and discrete lineages (Bannikova et al., 2015; Salvi et al., 2018).
Aside from these developments in the types and amount of data generated in systematic studies, there have also been developments in linking genetic patterns (especially adaptive genetic patterns) with the ecogeographic characteristics of the landscape. Adaptive differences (ecological, physiological, behavioral, morphological and genetic) in the fossorial genus Spalax have been intensively studied over the past three decades in four parapatric chromosomal species distributed across an ecogeographical and climatic gradient in Israel (Nevo & Bar-El, 1976; Nevo, 1985; Nevo et al., 1994, 2000). To some extent, similar patterns have been demonstrated in the fossorial genera Nanospalax (Savić, Ćirović & Bugarski-Stanojević, 2017) and Geomys (Heaney & Timm, 1985). Based on advances in genomic sequencing abilities, more recent studies have successfully employed data on adaptive genes and gene expression (whole mitochondrial and nuclear DNA genomes, transcriptomes, DNA methylation, micro RNA and codon usage) to show adaptive ecological speciation between two Spalax species which inhabit adjacent areas contrasting sharply in geology, edaphic attributes, vegetation and ecology (Hadid et al., 2012; Lövy et al., 2015; Li et al., 2015, 2016; Zhao et al., 2016). In these studies, it was shown that natural selection has acted on >300 genes to create adaptive complexes in these sharply contrasting ecogeographic areas.
By comparison, systematic revisions within the Bathyergidae have kept pace with the development of approaches. These revisions were, however, not consistent in the use of specific data types with some studies based on karyotype (Aguilar, 1993; Kawalika, Burda & Brüggert, 2001; Van Daele et al., 2004; Burda et al., 2005; Deuve et al., 2008) or allozyme data (Janecek et al., 1992; Filipucci et al., 1997) or mitochondrial DNA sequence data (Allard & Honeycutt, 1992; Faulkes et al., 1997, 2004, 2010, 2011, 2017; Van Daele et al., 2007b). Only in single instances (and more recently) have information from nuclear markers been included (Walton, Nedbal & Honeycutt, 2000; Huchon & Douzery, 2001; Ingram, Burda & Honeycutt, 2004; Kundu & Faulkes, 2004; Visser, Bennett & Jansen van Vuuren, 2014, 2018, 2019).
A significant advance in bathyergid systematics entailed the investigation of adaptive genes and gene complexes among the six bathyergid genera (Davies et al., 2015). Here, more than 700 genes were identified which convey some form of adaptive function. Based upon this information, Visser, Bennett & Jansen van Vuuren (2019) selected three genetic markers (in addition to mitochondrial DNA sequence data) to investigate genetic exclusiveness across the distributions of the two South African bathyergid genera Bathyergus and Georychus. The authors also demonstrated differences in the ecology (and to some extent the mating system in Georyhcus) among the recovered genetically and geographically discrete groups. More importantly, the discrete groups were recovered using both the adaptive nuclear data, as well as the cytochrome b data, indicating that this marker may be adequate for exploring the existence of highly divergent lineages.
From the cytochrome b and distributional data reviewed here, the possibility is highlighted that several cryptic and yet undescribed taxa exist within the Bathyergidae. In addition, these and many other bathyergid taxa may represent narrow endemics, frequently occupying diminishing habitats. An urgent and thorough systematic revision of the family may therefore be a necessity, especially given the rate of extinction linked to environmental change (Ceballos et al., 2015; Ocampo-Peñuela et al., 2016). The phylogeny presented here may serve as an adequate starting point for future taxonomic endeavors. Readers who have access to the material used in this study are urged to sequence additional and variable or adaptive nuclear markers to enable formal species descriptions. Additional sampling may also be required across wider geographic ranges and including larger sample sizes and more localities, so as to define species distributions- and relationships. A thorough systematic investigation of the Bathyergidae should also aim to include ecological and/or demographic data when defining species integrities. As such, this may aid in the identification of narrow endemics, as well as in conservation initiatives which aim to conserve the suitable habitat which harbors these species.
Conclusions
Here, we presented for the first time a comprehensive phylogeny for the Bathyergidae based on inclusive sampling across the ranges of genera and species, and place our phylogeny within the context of the species’ biologies and Africa’s complex geological and climatic history. We confirm the East African origin for the Bathyergidae (Faulkes et al., 2004, 2011, 2017) but suggest that earlier radiations were independent of rifting. Biogeographic patterns of the genera bear a signature of wide historical distributions (likely following open and savannah type habitats) which were fragmented by Miocene landscape changes. Importantly, the effects of climatic changes, distribution of suitable habitat, habitat specialization and population-level processes remain largely unexplored, but may have acted either unattended or in concert with vicariant events to influence speciation patterns.
Although largely exploratory in nature, it appears that several undescribed species may exist within the Bathyergidae, with five of the six genera harboring multiple species and possibly even undescribed cryptic species—many of which may be narrow endemics. As such, three species are recovered in Heliophobius (Heliophobius argenteocinereus, H. sp. 1 and H. sp. 2), five species in Georychus (G. capensis, G. sp. 1, G. sp. 2, G. sp. 3 and G. sp. 4), four species in Bathyergus (B. suillus, B. janetta, B. sp. 1 and B. sp. 2), five species in Cryptomys (C. h. mahali, C. h. nimrodi, C. h. hottentotus, C. h. natalensis and C. h. preoriae) and 16 species in Fukomys (F. zechi, F. bocagei, F. mechowii, F. livingstoni, F. hanangensis, F. amatus, F. whytei, F. darlingi, F. damarensis, F. anselli, F. micklemi, F. choma, F. kafuensis, and F. sp. 1, F. sp. 2 and F. sp. 3). Given the current rate of loss in biodiversity worldwide, a thorough systematic review of the Bathyergidae is crucial to discover and conserve the patterns and drivers of biodiversity in the family. Such a review would require intensive sampling, large sample sizes, multiple genetic markers and the addition of ecological and/or demographic data.
Supplemental Information
Phylogeny of the Bathyergidae constructed through Bayesian analysis (in BEAST) and based on the cytochrome b sequence data for all the available sample haplotypes listed in Table S1. The various mole-rat species included and identified within each genus are indicated. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML.
Accession numbers of the cytochrome b haplotypes used in this study, also indicating the species (as designated on the public database), reference to the original study in which the sequence was generated, the species designation in the original study, the species designation in the current study and geographic information and coordinates of the sampling locality from where the species originates.
Pairwise estimates of uncorrected sequence divergence among the six bathyergid genera.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Heliophobius.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Georychus.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Bathyergus.
Pairwise estimates of uncorrected sequence divergence among the various species (subspecies) included and identified within the genus Cryptomys.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Fukomys.
Acknowledgments
We would like to thank the University of Johannesburg for supplying the infrastructure to make this project possible. Furthermore, our manuscript benefited greatly from the input of the Editor, Gabriela Parra Olea, as well as from the reviewers Rodney Honeycutt, Mirna Garcia-Castillo and one anonymous reviewer.
Funding Statement
The project was funded through a South African National Research Foundation (NRF) grant to Bettine Jansen van Vuuren. Jacobus H. Visser was financially supported (in the form of a doctoral bursary) by a South African Research Chairs Initiative (SARChI) grant awarded to Nigel C. Bennett. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Jacobus H. Visser conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Nigel C. Bennett conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Bettine Jansen van Vuuren conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The data used in this study are available from public databases (EMBL and Genbank). Their accession numbers are available in the Supplemental File.
References
- Aguilar (1993).Aguilar GH. The karyotype and taxonomic status of Cryptomys hottentotus darlingi (Rodentia: Bathyergidae) South African Journal of Zoology. 1993;28(4):201–204. doi: 10.1080/02541858.1993.11448319. [DOI] [Google Scholar]
- Akaike (1973).Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Second International Symposium on Information theory. Budapest: Akademia Kiado; 1973. pp. 267–281. [Google Scholar]
- Albert, Zardoya & García-París (2007).Albert EM, Zardoya R, García-París M. Phylogeographical and speciation patterns in subterranean worm lizards of the genus Blanus (Amphisbaenia: Blanidae) Molecular Ecology. 2007;16(7):1519–1531. doi: 10.1111/j.1365-294X.2007.03248.x. [DOI] [PubMed] [Google Scholar]
- Allard & Honeycutt (1992).Allard MW, Honeycutt RL. Nucleotide sequence variation in the mitochondrial 12S rRNA gene and the phylogeny of African mole-rats (Rodentia: Bathyergidae) Molecular Biology and Evolution. 1992;9:27–40. doi: 10.1093/oxfordjournals.molbev.a040706. [DOI] [PubMed] [Google Scholar]
- Axelrod & Raven (1978).Axelrod DI, Raven PH. Late Cretaceous and tertiary vegetation history of Africa. In: Werger MJA, editor. Biogeography and Ecology of Southern Africa. Vol. 1. The Hague: Junk; 1978. pp. 77–130. [Google Scholar]
- Bannikova et al. (2015).Bannikova AA, Zemlemerova ED, Colangelo P, Sözen M, Sevindik M, Kidov AA, Dzuev RI, Kryštufek B, Lebedev VS. An underground burst of diversity—a new look at the phylogeny and taxonomy of the genus Talpa Linnaeus, 1758 (Mammalia: Talpidae) as revealed by nuclear and mitochondrial genes. Zoological Journal of the Linnean Society. 2015;175(4):930–948. doi: 10.1111/zoj.12298. [DOI] [Google Scholar]
- Bauer et al. (2010).Bauer FU, Glasmacher UA, Ring U, Schumann A, Nagudi B. Thermal and exhumation history of the central Rwenzori Mountains, Western Rift of the East African Rift System, Uganda. International Journal of Earth Sciences. 2010;99(7):1575–1597. doi: 10.1007/s00531-010-0549-7. [DOI] [Google Scholar]
- Bennett & Faulkes (2000).Bennett NG, Faulkes CG. African mole-rats: ecology and eusociality. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- Bennett & Jarvis (1988).Bennett NC, Jarvis JUM. The social structure and reproductive biology of colonies of the mole-rat Cryptomys damarensis (Rodentia, Bathyergidae) Journal of Mammology. 1988;69(2):293–302. doi: 10.2307/1381379. [DOI] [Google Scholar]
- Bonnefille (1984).Bonnefille R. Cenozoic vegetation and environments of early hominids in East Africa. In: Whyte RO, editor. The Evolution of the East Asian Environment. Vol. 2. Hong Kong: University of Hong Kong; 1984. pp. 579–612. [Google Scholar]
- Burda et al. (2005).Burda H, Šumbera R, Chitaukali WN, Dryden GL. Taxonomic status and remarks on ecology of the Malawian mole-rat Cryptomys whytei (Rodentia, Bathyergidae) Acta Theriologica. 2005;50(4):529–536. doi: 10.1007/BF03192646. [DOI] [Google Scholar]
- Burda et al. (1999).Burda H, Zima J, Scharff A, Macholan M, Kawalika M. The karyotypes of Cryptomys anselli sp nova and Cryptomys kafuensis sp nova: new species of the common mole-rat from Zambia (Rodentia, Bathyergidae) International Journal of Mammalian Biology. 1999;64:36–50. [Google Scholar]
- Ceballos et al. (2015).Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. Accelerated modern human–induced species losses: entering the sixth mass extinction. Science Advances. 2015;1(5):e1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitaukali, Burda & Kock (2001).Chitaukali WN, Burda H, Kock D. On small mammals of the Nyika Plateau, Malawi. Paris: IRD Editions, Collection colloques et seminaires; 2001. [Google Scholar]
- Chorowicz (2005).Chorowicz J. The East African rift system. Journal of African Earth Sciences. 2005;43(1–3):379–410. doi: 10.1016/j.jafrearsci.2005.07.019. [DOI] [Google Scholar]
- Clement, Posada & Crandall (2000).Clement MD, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Coetzee (1978).Coetzee JA. Climatic and biological changes in south-western Africa during the Late Cainozoic. In: Coetzee A, Van Zinderen Bakker EM, editors. Palaeoecology of Africa. Vol. 10. Rotterdam: Balkema; 1978. pp. 13–29. [Google Scholar]
- Coetzee (1983).Coetzee JA. Intimations on the Tertiary vegetation of southern Africa. Bothalia. 1983;14(3/4):345–354. doi: 10.4102/abc.v14i3/4.1179. [DOI] [Google Scholar]
- Cohen, Soreghan & Scholz (1993).Cohen AS, Soreghan MJ, Scholz CA. Estimating the age of formation of lakes: an example from Lake Tanganyika, East-African Rift System. Geology. 1993;21:511–514. [Google Scholar]
- Cowling, Procheş & Partridge (2009).Cowling RM, Procheş S, Partridge TC. Explaining the uniqueness of the Cape flora: Incorporating geomorphic evolution as a factor for explaining its diversification. Molecular Phylogenetics and Evolution. 2009;51(1):64–74. doi: 10.1016/j.ympev.2008.05.034. [DOI] [PubMed] [Google Scholar]
- Darriba et al. (2012).Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies et al. (2015).Davies KTJ, Bennett NC, Tsagkogeorga G, Rossiter SJ, Faulkes CG. Family wide molecular adaptations to underground life in the African mole-rats revealed by phylogenomic analysis. Molecular Biology and Evolution. 2015;32:3089–3107. doi: 10.1093/molbev/msv175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvaux (1995).Delvaux D. Age of Lake Malawi (Nyasa) and water level fluctuations. Musée royal de l’Afrique Centrale (Tervuren) Département de Géologie et Minéralogie Rapport Annuel. 1995;1993:99–108. [Google Scholar]
- Delvaux et al. (1998).Delvaux D, Kervyn F, Vittori E, Kajara RSA, Kilembe E. Late Quaternary tectonic activity and lake level change in the Rukwa Rift Basin. Journal of African Earth Sciences. 1998;26(3):397–421. doi: 10.1016/S0899-5362(98)00023-2. [DOI] [Google Scholar]
- Deuve et al. (2008).Deuve JL, Bennett NC, Britton-Davidian J, Robinson TJ. Chromosomal phylogeny and evolution of the African mole-rats (Bathyergidae) Chromosome Research. 2008;16(1):57–74. doi: 10.1007/s10577-007-1200-8. [DOI] [PubMed] [Google Scholar]
- Diekmann, Falker & Kuhn (2003).Diekmann B, Falker M, Kuhn G. Environmental history of the south-eastern South Atlantic since the Middle Miocene: evidence from the sedimentological records of ODP sites 1088 and 1092. Sedimentology. 2003;50(3):511–529. doi: 10.1046/j.1365-3091.2003.00562.x. [DOI] [Google Scholar]
- Dingle & Rogers (1972).Dingle RV, Rogers J. Effects of sea-level changes on the Pleistocene palaeoecology of the Agulhas Bank. Paleoecology of Africa. 1972;6:55–58. [Google Scholar]
- Drummond et al. (2012).Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinger (1989).Ebinger CJ. Tectonic development of the western branch of the east African rift system. Geological Society of America Bulletin. 1989;1:885–903. doi: 10.1130/0016-7606(1989)101<0885:TDOTWB>2.3.CO;2. [DOI] [Google Scholar]
- Ebinger et al. (1993).Ebinger CJ, Deino AL, Tesha AL, Becker T, Ring U. Tectonic controls on Rift Basin morphology: evolution of the northern Malawi (Nyasa) Rift. Journal of Geophysical Research. 1993;17:821–836. doi: 10.1029/93jb01392. [DOI] [Google Scholar]
- Ellerman (1940).Ellerman JR. The families and genera of living rodents. Vol. 1. London: Trustees of the British Museum (Natural History); 1940. [Google Scholar]
- Ellerman, Morrison-Scott & Hayman (1953).Ellerman JR, Morrison-Scott TCS, Hayman RW. Southern African mammals, 1758-1951: a reclassification. London: Trustees of the British Museum (Natural History); 1953. [Google Scholar]
- Faulkes & Bennett (2007).Faulkes CG, Bennett NC. African mole-rats: behavioral and ecological diversity. In: Wolff J, Sherman PW, editors. Rodent Societies: An Ecological and Evolutionary Perspective. Chicago: The University of Chicago Press; 2007. pp. 427–437. [Google Scholar]
- Faulkes & Bennett (2009).Faulkes CG, Bennett NC. Reproductive skew in African mole-rats: behavioural and physiological mechanisms to maintain high skew. In: Hager R, Jones CB, editors. Reproductive Skew in Vertebrates: Proximate and Ultimate Causes. Cambridge: Cambridge University Press; 2009. pp. 369–396. [Google Scholar]
- Faulkes et al. (1997).Faulkes CG, Bennett NC, Bruford MW, O’Brien HP, Aguilar GH, Jarvis JUM. Ecological constraints drive social evolution in the African mole–rats. Proceedings of the Royal Society of London. Series B: Biological Sciences. 1997;264(1388):1619–1627. doi: 10.1098/rspb.1997.0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkes et al. (2011).Faulkes CG, Bennett NC, Cotterill FPD, Stanley W, Mgode GF, Verheyen E. Phylogeography and cryptic diversity of the solitary-dwelling silvery mole-rat, genus Heliophobius (Family: Bathyergidae) Journal of Zoology. 2011;285(4):324–338. doi: 10.1111/j.1469-7998.2011.00863.x. [DOI] [Google Scholar]
- Faulkes et al. (2017).Faulkes CG, Mgode GF, Archer EK, Bennett NC. Relic populations of Fukomys mole-rats in Tanzania: description of two new species F. livingstoni sp. nov and F. hanangensis sp. nov. PeerJ. 2017;5(1994):e3214. doi: 10.7717/peerj.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkes et al. (2010).Faulkes CG, Mgode GF, Le Comber SC, Bennett NC. Cladogenesis and endemism in Tanzanian mole-rats, genus Fukomys (Family: Bathyergidae): a role for tectonics? Biological Journal of the Linnean Society. 2010;100(2):337–352. doi: 10.1111/j.1095-8312.2010.01418.x. [DOI] [Google Scholar]
- Faulkes et al. (2004).Faulkes CG, Verheyen E, Verheyen W, Jarvis JUM, Bennett NC. Phylogeographical patterns of genetic divergence and speciation in African mole-rats (Family: Bathyergidae) Molecular Ecology. 2004;13(3):613–629. doi: 10.1046/j.1365-294X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- Filipucci et al. (1997).Filipucci MG, Kawalika M, Macholan M, Scharff A, Burda H. Allozyme differentiation and systematic relationship of Zambian Giant mole-rats, Cryptomys mechowi (Bathyergidae, Rodentia) Zeitschrift für Säugetierkunde. 1997;62:172–178. [Google Scholar]
- Fitzpatrick & Turelli (2006).Fitzpatrick BM, Turelli M. The geography of mammalian speciation: mixed signals from phylogenies and range maps. Evolution. 2006;60(3):601–615. doi: 10.1111/j.0014-3820.2006.tb01140.x. [DOI] [PubMed] [Google Scholar]
- George (1979).George W. Conservatism in the karyotypes of two African mole rats (Rodentia, Bathyergidae) Zeitschrift für Säugetierkunde. 1979;44:278–285. [Google Scholar]
- Gippoliti & Amori (2011).Gippoliti S, Amori G. A new species of mole-rat (Rodentia, Bathyergidae) from the Horn of Africa. Zootaxa. 2011;2918(1):39–46. doi: 10.11646/zootaxa.2918.1.4. [DOI] [Google Scholar]
- Guindon & Gascuel (2003).Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hadid et al. (2012).Hadid Y, Tzur S, Pavlíček T, Šumbera R, Šklíba J, Lövy M, Fragman-Sapir O, Beiles A, Arieli R, Raz S, Nevo E. Possible incipient sympatric ecological speciation in blind mole rats (Spalax) Proceedings of the National Academy of Sciences of the United States of America. 2012;110(7):2587–2592. doi: 10.1073/pnas.1222588110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner et al. (1987).Hafner MS, Hafner JC, Patton JL, Smith MF. Macrogeographic patterns of genetic differentiation in the pocket gopher Thomomys umbrinus. Systematic Zoology. 1987;36(1):18–34. doi: 10.2307/2413305. [DOI] [Google Scholar]
- Hallam (1992).Hallam A. Phanerozoic sea-level changes. New York: Columbia University Press; 1992. [Google Scholar]
- Heaney & Timm (1985).Heaney LR, Timm RM. Morphology, genetics, and ecology of pocket gophers (genus Geomys) in a narrow hybrid zone. Biological Journal of the Linnean Society. 1985;25(4):301–317. doi: 10.1111/j.1095-8312.1985.tb00397.x. [DOI] [Google Scholar]
- Honeycutt (2017).Honeycutt RL. Bathyergidae. In: Wilson DE, Lacher TE, Mittermeier RA, editors. Handbook of Mammals of the World, Lagomorphs and Rodents. Vol. 6. Barcelona: Lynx Edicions; 2017. pp. 352–370. [Google Scholar]
- Honeycutt et al. (1991).Honeycutt RL, Allard MW, Edwards SV, Schlitter DA. Systematics and evolution of the family Bathyergidae. In: Sherman PW, Jarvis JUM, Alexander RD, editors. The Biology of the Naked Mole-Rat. Princeton: Princeton University Press; 1991. pp. 45–65. [Google Scholar]
- Honeycutt et al. (1987).Honeycutt RL, Edwards SV, Nelson K, Nevo E. Mitochondrial-DNA variation and the phylogeny of African mole-rats (Rodentia, Bathyergidae) Systematic Zoology. 1987;36(3):280–292. doi: 10.2307/2413067. [DOI] [Google Scholar]
- Huchon & Douzery (2001).Huchon D, Douzery EJP. From the old world to the new world: a molecular chronicle of the phylogeny and biogeography of hystricognath rodents. Molecular Phylogenetics and Evolution. 2001;20(2):238–251. doi: 10.1006/mpev.2001.0961. [DOI] [PubMed] [Google Scholar]
- Huson & Scornavacca (2012).Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Systematic Biology. 2012;61(6):1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Ingram, Burda & Honeycutt (2004).Ingram CM, Burda H, Honeycutt RL. Molecular phylogenetics and taxonomy of the African mole-rats, genus Cryptomys and the new genus Coetomys Gray, 1864. Molecular Phylogenetics and Evolution. 2004;31(3):997–1014. doi: 10.1016/j.ympev.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Janecek et al. (1992).Janecek LL, Honeycutt RL, Rautenbach L, Erasmus BH, Reig S, Schlitter DA. Allozyme variation and systematics of African mole-rats (Rodentia: Bathyergidae) Biochemical Systematics and Ecology. 1992;20(5):401–416. doi: 10.1016/0305-1978(92)90081-N. [DOI] [Google Scholar]
- Jarvis & Bennett (1993).Jarvis JUM, Bennett NC. Eusociality has evolved independently in two genera of bathyergid mole-rats—but occurs in no other subterranean mammal. Behavioral Ecology and Sociobiology. 1993;33(4):253–260. doi: 10.1007/BF02027122. [DOI] [Google Scholar]
- Joly, Stevens & van Vuuren (2007).Joly S, Stevens MI, van Vuuren B. Haplotype networks can be misleading in the presence of missing data. Systematic Biology. 2007;56(5):857–862. doi: 10.1080/10635150701633153. [DOI] [PubMed] [Google Scholar]
- Kawalika, Burda & Brüggert (2001).Kawalika M, Burda H, Brüggert D. Was Zambia a cradle of the genus Cryptomys (Bathyergidae, Rodentia)? A further new ancestral (?) species of Cryptomys from Zambia. In: Denys C, Laurent G, Alain P, editors. African Small Mammals. Vol. 8. Paris: Symposium International sur les Petits Mammifères Africains; 2001. pp. 253–261. [Google Scholar]
- Kortlandt (1983).Kortlandt A. Facts and fallacies concerning Miocene ape habitats. In: Ciochon RL, Corruccini RS, editors. New Interpretations of Ape and Human Ancestry. New York: Plenum; 1983. pp. 465–514. [Google Scholar]
- Krammer, Baumann & Henrich (2006).Krammer R, Baumann K-H, Henrich R. Middle to late Miocene fluctuations in the incipient Benguela upwelling system revealed by calcareous nannofossil assemblages (ODP Site 1085A) Palaeogeography, Palaeoclimatology, Palaeoecology. 2006;230(3–4):319–334. doi: 10.1016/j.palaeo.2005.07.022. [DOI] [Google Scholar]
- Kundu & Faulkes (2004).Kundu S, Faulkes CG. Patterns of MHC selection in African mole–rats, family Bathyergidae: the effects of sociality and habitat. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2004;271(1536):273–278. doi: 10.1098/rspb.2003.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey, Patton & Cameron (2000).Lacey E, Patton JL, Cameron GN. Life underground: the biology of subterranean rodents. Chicago: The University of Chicago Press; 2000. [Google Scholar]
- Lavocat (1973).Lavocat R. Les rongeurs du Miocene d’Afrique Orientale. I. Miocene inferieur. Memoires et Travaux de L’ephe, Institut de Montpellier. 1973;1:1–284. [Google Scholar]
- Lavocat (1978).Lavocat R. Rodentia and lagomorpha. In: Maglio VJ, Cooke HBS, editors. Evolution of African Mammals. Cambridge: Harvard University Press; 1978. pp. 69–89. [Google Scholar]
- Li et al. (2015).Li K, Hong W, Jiao H, Wang G, Rodriguez KA, Buffenstein R, Zhao Y, Nevo E, Zhao H. Sympatric speciation revealed by genome-wide divergence in the blind mole rat Spalax. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(38):11905–11910. doi: 10.1073/pnas.1514896112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2016).Li K, Wang L, Knisbacher BA, Xu Q, Levanon EY, Wang H, Frenkel-Morgenstern M, Tagore S, Fang X, Bazak L, Buchumenski I, Zhao Y, Lövy M, Li X, Han L, Frenkel Z, Beiles A, Cao YB, Wang ZL, Nevo E. Transcriptome, genetic editing, and microRNA divergence substantiate sympatric speciation of blind mole rat, Spalax. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(27):7584–7589. doi: 10.1073/pnas.1607497113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado & Rozas (2009).Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lövy et al. (2015).Lövy M, Šklíba J, Hrouzková E, Dvořáková V, Nevo E, Šumbera R. Habitat and burrow system characteristics of the blind mole rat Spalax galili in an area of supposed sympatric speciation. PLOS ONE. 2015;10(7):e0133157. doi: 10.1371/journal.pone.0133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor (2015).Macgregor D. History of the development of the East African Rift System: a series of interpreted maps through time. Journal of African Earth Sciences. 2015;101:232–252. doi: 10.1016/j.jafrearsci.2014.09.016. [DOI] [Google Scholar]
- Macholan et al. (1998).Macholan M, Scharff A, Burda H, Zima J, Grutjen O. The karyotype and taxonomic status of Cryptomys amatus (Wroughton, 1907) from Zambia (Rodentia, Bathyergidae) International Journal of Mammalian Biology. 1998;63:186–190. [Google Scholar]
- Maud & Partridge (1987).Maud RR, Partridge TC. Regional geomorphic evidence for climatic change in southern Africa since the Mesozoic. Palaeoecology of Africa. 1987;18:337–348. [Google Scholar]
- Nevo (1985).Nevo E. Speciation in action and adaptation in subterranean mole rats: patterns and theory. Italian Journal of Zoology. 1985;52:65–95. [Google Scholar]
- Nevo (1999).Nevo E. Mosaic evolution of subterranean mammals: Regression, progression and global convergence. Oxford: Oxford University Press; 1999. [Google Scholar]
- Nevo & Bar-El (1976).Nevo E, Bar-El H. Hybridization and speciation in fossorial mole rats. Evolution. 1976;30(4):831–840. doi: 10.1111/j.1558-5646.1976.tb00964.x. [DOI] [PubMed] [Google Scholar]
- Nevo et al. (1994).Nevo E, Filippucci MG, Redi C, Korol A, Beiles A. Chromosomal speciation and adaptive radiation of mole rats in Asia Minor correlated with increased ecological stress. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(17):8160–8164. doi: 10.1073/pnas.91.17.8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo et al. (2000).Nevo E, Ivanitskaya E, Filippucci MG, Beiles A. Speciation and adaptive radiation of subterranean mole rats, Spalax ehrenbergi superspecies, in Jordan. Biological Journal of the Linnean Society. 2000;69(2):263–281. doi: 10.1111/j.1095-8312.2000.tb01202.x. [DOI] [Google Scholar]
- Nevo et al. (1974).Nevo E, Kim YJ, Shaw CR, Thaeler CS., Jr Genetic variation, selection and speciation in Thomomys talpoides pocket gophers. Evolution. 1974;28(1):1–23. doi: 10.1111/j.1558-5646.1974.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Nevo & Reig (1990).Nevo E, Reig OA. Evolution of subterranean mammals at the organismal and molecular levels. In: Nowak RM, editor. Walker’s Mammals of the World. Vol. 2. Baltimore and London: The Johns Hopkins University Press; 1990. pp. 1635–1644. [Google Scholar]
- Ocampo-Peñuela et al. (2016).Ocampo-Peñuela N, Jenkins CN, Vijay V, Li BV, Pimm SL. Incorporating explicit geospatial data shows more species at risk of extinction than the current Red List. Science Advances. 2016;2(11):e1601367. doi: 10.1126/sciadv.1601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo et al. (2008).Opazo JC, Bugueño MP, Carter MJ, Palma RR, Bozinovic F. Phylogeography of the subterranean rodent Spalacopus cyanus (Caviomorpha, Octodontidae) Journal of Mammalogy. 2008;89(4):837–844. doi: 10.1644/07-MAMM-A-068.1. [DOI] [Google Scholar]
- Papadopouloua & Knowles (2016).Papadopouloua A, Knowles LL. Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(29):8018–8024. doi: 10.1073/pnas.1601069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge & Maud (1987).Partridge TC, Maud RR. Geomorphic evolution of southern Africa since the Mesozoic. South African Journal of Geology. 1987;90:179–208. [Google Scholar]
- Partridge & Maud (2000).Partridge TC, Maud RR. Macro-scale geomorphic evolution of southern Africa. In: Partridge TC, Maud RR, editors. The Cenozoic of Southern Africa. New York: Oxford University Press; 2000. pp. 3–18. [Google Scholar]
- Patton & Yang (1977).Patton JL, Yang SY. Genetic variation in Thomomys bottae pocket gophers: Macrogeographic patterns. Evolution. 1977;31(4):697–720. doi: 10.1111/j.1558-5646.1977.tb01064.x. [DOI] [PubMed] [Google Scholar]
- Pimm et al. (2014).Pimm S, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO. The biodiversity of species and their rates of extinction, distribution, and protection. Science. 2014;344(6187):1246752. doi: 10.1126/science.1246752. [DOI] [PubMed] [Google Scholar]
- Rambaut & Drummond (2003).Rambaut A, Drummond AJ. Tracer [computer program] 2003. http://evolve.zoo.ox.ac.uk/software/ [10 October 2013]. http://evolve.zoo.ox.ac.uk/software/
- Rogers (1987).Rogers J. The evolution of the continental terrace between St Helena Bay and Lambert’s Bay. In: Parkington J, Hall M, editors. Papers in the Prehistory of the Western Cape, South Africa. Vol. 332. Oxford: BAR International Series; 1987. pp. 35–45. [Google Scholar]
- Ronquist et al. (2012).Ronquist F, Teslenko M, Van der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi et al. (2018).Salvi D, Perera A, Sampaio FL, Carranza S, Harris DJ. Underground cryptic speciation within the Maghreb: Multilocus phylogeography sheds light on the diversification of the checkerboard worm lizard Trogonophis wiegmanni. Molecular Phylogenetics and Evolution. 2018;120:118–128. doi: 10.1016/j.ympev.2017.11.013. [DOI] [PubMed] [Google Scholar]
- Savić, Ćirović & Bugarski-Stanojević (2017).Savić I, Ćirović D, Bugarski-Stanojević V. Exceptional chromosomal evolution and cryptic speciation of blind mole rats Nannospalax leucodon (Spalacinae, Rodentia) from south-eastern Europe. Genes. 2017;8:1–22. doi: 10.3390/genes8110292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff, Macholan & Burda (2001).Scharff A, Macholan M, Burda H. A new karyotype of Heliophobius argenteocinereus (Bathyergidae, Rodentia) from Zambia with field notes on the species. Zeitschrift für Säugetierkunde. 2001;66:376–378. [Google Scholar]
- Shackleton & Kennett (1975a).Shackleton NJ, Kennett JP. Late Cenozoic oxygen and carbon isotopic changes at DSDP site 284: implications for glacial history of the Northern Hemisphere and Antarctica. In: Kennett JP, Houtz RE, Andrews PB, Edwards AR, Gostin VA, Hajós M, Hampton MA, Jenkins DG, Margolis SV, Ovenshine AT, Perch-Nielsen K, editors. Initial Reports of the Deep Sea Drilling Project. Vol. 29. Washington, D.C.: U.S. Government Printing Office; 1975a. pp. 801–807. [Google Scholar]
- Shackleton & Kennett (1975b).Shackleton NJ, Kennett JP. Paleotemperature history of the Cenozoic and the initiation of Antarctic glaciation: oxygen and carbon isotope analyses in DSDP sites 277, 279 and 281. In: Kennett JP, Houtz RE, Andrews PB, Edwards AR, Gostin VA, Hajós M, Hampton MA, Jenkins DG, Margolis SV, Ovenshine AT, Perch-Nielsen K, editors. Initial Reports of the Deep Sea Drilling Project. Vol. 29. Washington D.C.: U.S. Government Printing Office; 1975b. pp. 801–807. [Google Scholar]
- Sherman, Jarvis & Alexander (1991).Sherman PW, Jarvis JUM, Alexander RD. The biology of the naked mole-rat. Princeton: Princeton University Press; 1991. [Google Scholar]
- Siesser (1980).Siesser WG. Late Miocene origin of the Benguela upwelling system off Northern Namibia. Science. 1980;208(4441):283–285. doi: 10.1126/science.208.4441.283. [DOI] [PubMed] [Google Scholar]
- Siesser & Dingle (1981).Siesser WG, Dingle RV. Tertiary sea-level movements around Southern Africa. Journal of Geology. 1981;89(4):523–536. doi: 10.1086/628618. [DOI] [Google Scholar]
- Stamakis (2014).Stamakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2010).Tang L-Z, Wang L-Y, Cai Z-Y, Zhang T-Z, Ci H-X, Lin G-H, Su J-P, Liu J-Q. Allopatric divergence and phylogeographic structure of the plateau zokor (Eospalax baileyi), a fossorial rodent endemic to the Qinghai-Tibetan Plateau. Journal of Biogeography. 2010;37(4):657–668. doi: 10.1111/j.1365-2699.2009.02232.x. [DOI] [Google Scholar]
- Thomas & Shaw (1988).Thomas DSG, Shaw PA. Late Cainozoic drainage evolution in the Zambezi basin: geomorphological evidence from the Kalahari rim. Journal of African Earth Sciences (and the Middle East) 1988;7(4):611–618. doi: 10.1016/0899-5362(88)90111-X. [DOI] [Google Scholar]
- Tyson & Partridge (2000).Tyson PD, Partridge TC. The evolution of Cenozoic climates. In: Partridge TC, Maud RR, editors. The Cenozoic of Southern Africa. New York: Oxford University Press; 2000. pp. 371–387. [Google Scholar]
- Van Couvering & Van Couvering (1976).Van Couvering JAH, Van Couvering JA. Early Miocene mammal fossils from East Africa: aspects of geology, faunistics and paleontology. In: Issac GL, McCown ER, editors. Human Origins: Louis Leakey and the East African Evidence. San Francisco: W.A. Benjamin Reading; 1976. pp. 155–207. [Google Scholar]
- Van Daele et al. (2013).Van Daele PAAG, Blondé P, Stjernstedt R, Adriaens D. A new species of African Mole-rat (Fukomys, Bathyergidae, Rodentia) from the Zaire-Zambezi Watershed. Zootaxa. 2013;3636(1):171–189. doi: 10.11646/zootaxa.3636.1.7. [DOI] [PubMed] [Google Scholar]
- Van Daele et al. (2004).Van Daele PAAG, Dammann P, Meier JL, Kawalika M, Van De Woestijne C, Burda H. Chromosomal diversity in mole-rats of the genus Cryptomys (Rodentia: Bathyergidae) from the Zambezian region: with descriptions of new karyotypes. Journal of Zoology (London) 2004;264(3):317–326. doi: 10.1017/S0952836904005825. [DOI] [Google Scholar]
- Van Daele et al. (2007a).Van Daele PAAG, Faulkes CG, Verheyen E, Adrians D. African mole-rats (Bathyergidae): a complex radiation in Afrotropical soils. In: Begall S, Burda H, Schleich CE, editors. Subterranean Rodents: News from Underground. Heidelberg: Springer-Verlag; 2007a. pp. 357–373. [Google Scholar]
- Van Daele et al. (2007b).Van Daele PAAG, Verheyen E, Brunain M, Adriaens D. Cytochrome b sequence analysis reveals differential molecular evolution in African mole-rats of the chromosomally hyperdiverse genus Fukomys (Bathyergidae, Rodentia) from the Zambezian region. Molecular Phylogenetics and Evolution. 2007b;45(1):142–157. doi: 10.1016/j.ympev.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Van Zinderen Bakker (1967).Van Zinderen Bakker EM. The ‘Arid Corridor’ between southwest Africa and the Horn of Africa. In: Van Zinderen Bakker EM, editor. Paleoecology of Africa. Vol. 2. Amsterdam: A.A. Balkema; 1967. pp. 76–79. [Google Scholar]
- Van Zinderen Bakker & Mercer (1986).Van Zinderen Bakker EM, Mercer JH. Major late Cainozoic climatic events and palaeoenvironmental changes in Africa viewed in a world wide context. Palaeogeography, Palaeoclimatology, Palaeoecology. 1986;56(3–4):217–235. doi: 10.1016/0031-0182(86)90095-7. [DOI] [Google Scholar]
- Visser, Bennett & Jansen van Vuuren (2014).Visser JH, Bennett NC, Jansen van Vuuren B. Local and regional scale genetic variation in the Cape dune mole-rat, Bathyergus suillus. PLOS ONE. 2014;9(9):e107226. doi: 10.1371/journal.pone.0107226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, Bennett & Jansen van Vuuren (2018).Visser JH, Bennett NC, Jansen van Vuuren B. Spatial genetic diversity in the Cape mole-rat, Georychus capensis: extreme isolation of populations in a subterranean environment. PLOS ONE. 2018;13(3):e0194165. doi: 10.1371/journal.pone.0194165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, Bennett & Jansen van Vuuren (2019).Visser JH, Bennett NC, Jansen van Vuuren B. Evolutionary and ecological patterns within the South African Bathyergidae: Implications for taxonomy. Molecular Phylogenetics and Evolution. 2019;130:181–197. doi: 10.1016/j.ympev.2018.10.017. [DOI] [PubMed] [Google Scholar]
- Walton, Nedbal & Honeycutt (2000).Walton AH, Nedbal MA, Honeycutt RL. Evidence from Intron 1 of the nuclear transthyretin (prealbumin) gene for the phylogeny of African mole-rats (Bathyergidae) Molecular Phylogenetics and Evolution. 2000;16(3):467–474. doi: 10.1006/mpev.2000.0808. [DOI] [PubMed] [Google Scholar]
- Zachos et al. (2001).Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292(5517):686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- Zhao et al. (2016).Zhao Y, Tang J-W, Yang Z, Cao Y-B, Ren J-L, Ben-Abu Y, Li K, Chen X-Q, Du J-Z, Nevo E. Adaptive methylation regulation of p53 pathway in sympatric speciation of blind mole rats, Spalax. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(8):2146–2151. doi: 10.1073/pnas.1522658112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogeny of the Bathyergidae constructed through Bayesian analysis (in BEAST) and based on the cytochrome b sequence data for all the available sample haplotypes listed in Table S1. The various mole-rat species included and identified within each genus are indicated. Values above nodes represent posterior probability values derived from the Bayesian analysis in BEAST and MrBayes respectively, while values below nodes indicate bootstrap values derived after Maximum Likelihood analysis in RAxML.
Accession numbers of the cytochrome b haplotypes used in this study, also indicating the species (as designated on the public database), reference to the original study in which the sequence was generated, the species designation in the original study, the species designation in the current study and geographic information and coordinates of the sampling locality from where the species originates.
Pairwise estimates of uncorrected sequence divergence among the six bathyergid genera.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Heliophobius.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Georychus.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Bathyergus.
Pairwise estimates of uncorrected sequence divergence among the various species (subspecies) included and identified within the genus Cryptomys.
Pairwise estimates of uncorrected sequence divergence among the various species included and identified within the genus Fukomys.
Data Availability Statement
The following information was supplied regarding data availability:
The data used in this study are available from public databases (EMBL and Genbank). Their accession numbers are available in the Supplemental File.