Abstract
Introduction:
Microtia is a congenital malformation of the external and middle ear caused by the abnormal development of the first and second zygomatic arch and the first sulcus. There is currently no consensus concerning the pathogenesis and etiology of microtia; genetic and environmental factors may play a role. Gene-based studies have focused on finding the genes that cause microtia and on gene function defects. However, no clear pathogenic genes have so far been identified. Microtia is multifactorial; gene function defects cannot completely explain its pathogenesis. In recent years, the epigenetic aspects of microtia have begun to receive attention.
Conclusions:
Analysis of the existing data suggests that certain key genes and pathways may be the underlying cause of congenital microtia. However, further exploration is needed.
Keywords: epigenetics, heredity, microtia, pathogenesis
1. Introduction
Microtia is a congenital malformation of the external and middle ear caused by the abnormal development of the first and second arch, and the first sulcus in the embryonic stage; it may be accompanied by hearing loss, and also mandibular and facial soft tissue dysplasia.[1,2] It can be unilateral or bilateral, and the degree of auricular deformity can range from mild structural abnormalities to complete absence of the ear. Epidemiological prevalence surveys carried out both in China and abroad indicate that the prevalence of microtia varies depending on region,[3,4] race,[5,6] sex, and even altitude.[5] Risk factor analysis suggests that parental status and pregnancy issues, such as age and education level of the mother,[7] diabetes of the mother, early pregnancy infection, multiple births, taking medicine,[8] smoking, or drinking during pregnancy,[9] have a major impact on the incidence rate. Insufficient folic acid intake during early pregnancy can also increase the risk of microtia.[10]
Thus, microtia is a multifactorial disease in which environmental and genetic factors, and also interactions between the 2, may be involved. Researchers have tried to identify the genetic factors in the development of microtia and their mechanism of action; however, no clear disease-causing genes have been found, and there is no consensus on the mechanism of action of microtia.
Over the past decade, scientists have grappled with the implications of epigenetics: genetic information is never solely determined by genes.[11] Just as most cells in a multicellular organism carry the same genetic material, the morphology and function of different types of cells vary widely. This heterogeneity is caused by different gene expression patterns in various cells. Epigenetics regulates these different gene expression patterns to determine cell fate.[12] As early as 2009,[11] a group of researchers from the Eidgenössische Technische Hochschule in Zürich led by Professor Renato Paro conducted cross-cultivation of fruit flies for 6 generations, observing the effect of temperature changes on their eye color. The results showed that temperature change can turn the eyes of this species of fly from white to red. After several generations of cultivation, the offspring still had red eyes. However, the genetic DNA sequences responsible for eye color in the white-eye parents and the red-eye progeny remained the same. These results contradicted the predictions of traditional genetic theory. The phenotype of some organisms may change without changing the corresponding gene base sequence.[13,14] This is the so-called epigenetic effect, by which the genome can be modified without affecting the DNA sequence. This change can not only affect the development of an individual, but also be inherited. This discovery challenged traditional genetic concepts. In February, 2010, an article in Nature magazine[15] pointed out that while genetic sequences contain genetic information, modifications to those sequences also contain genetic information. Since then, the emerging discipline of epigenetics has become an important branch and research hotspot of modern genetics. Meanwhile, large-scale genome sequencing and functional studies have created a major problem for scientists: in a nutshell, the intra and interspecies genomic sequences are too similar to explain the diversity of life. Epigenetics, the chemical modification of DNA or its related proteins, changes in gene expression caused by noncoding RNA, can shed light on how these similar genetic codes are expressed specifically in different cells, different environmental conditions, and different periods.[15] Unfortunately, microtia has largely been ignored by the intense research activity in the field of epigenetics over the past decade; very few articles have been published directly relating to microtia.
Environmental factors include not only the environment in which we live, but also the intrauterine environment in which fetal embryos develop, which is critical for fetal growth. Many research studies have reported that changes of the epigenetic pattern in the intrauterine environment impact the incidence of birth defects such as fetal growth restriction,[16–19] neural tube defects,[20–22] cleft lip and palate,[23–28] craniosynostosis,[9] hypospadias,[9,29–31] and congenital heart disease[9,32–34] at a much higher rate than that of control groups; however, when the DNA of people with birth defects is compared with that of normal people, it is difficult to identify with certainty a pathogenic gene or genes.
In view of this, we speculate that there may be an epigenetic dimension to the occurrence of microtia. Epigenetics mainly includes DNA modification, histone modification, chromatin remodeling, and noncoding RNA.[35] Based on epigenetic studies of microtia to date, this article reviews the effects of DNA methylation and noncoding RNA including micro ribonucleic acid (miRNA) and long non-coding ribonucleic acid (lncRNA) on microtia.
2. Effect of epigenetic modification of DNA on microtia
DNA methylation is a common epigenetic way of regulating gene expression in eukaryotic cells and is the major epigenetic modification of genomic DNA.[36] DNA methylation not only affects the process of gene expression, it can be inherited and persist with mitosis and meiosis in cells.[37] The study found that 9.5-day-old fetal rat craniofacial tissue showed positive expression of DNA methyltransferase,[26] suggesting that DNA methylation is regulated in the early phase of the embryo. The Sox4 gene is reportedly an important regulatory gene for the differentiation and development of neural crest cells during embryonic development, and also a strong candidate gene for cleft lip and palate.[27] Its methylation abnormality can affect a series of development-related signal transductions and gene transcriptions, and is involved in the development of multiple organ malformations including cleft lip and palate.[28]
In 2011, a systematic review of 173,687 cases of deformed children and 11.7 million controls showed that smoking during pregnancy was associated with multiple neonatal defects including microtia.[9] The study also found that adverse environmental factors such as tobacco and alcohol can cause craniofacial morphological abnormalities[38–40] by inducing abnormal methylation of related susceptibility genes during craniofacial morphogenesis. Methionine, VitB12, and folic acid in the diet are donors of the DNA methylated methyl group.[41–43] If there is a lack of folic acid, methionine, VitB12, or selenium in the diet, the methylation status of the gene will change,[44,45] resulting in neural tube defects,[46–48] ataxia,[48] cleft lip and palate,[25] congenital deafness,[48] microtia,[48] and so on. Environmental pollution, such as occupational chemicals, fossil fuel emissions, water pollution, and smoking, releases harmful substances such as arsenic and polycyclic aromatic hydrocarbons (benzoxene), which can increase genetic instability and alter cellular material metabolism.[27,49]
3. Effect of noncoding RNA regulation on microtia
The human gene coding sequence only accounts for 1.2% of the genome, whereas more than 98% of the genome is transcribed to generate noncoding RNA.[50–52] Of this 98%, 50% of miRNA is located in the chromosomal region which is susceptible to change, and the post-transcriptional expression level of the gene can be regulated by gene silencing.[53] Many developmental genes that play an important role during cranial morphogenesis are regulated by miRNA.[54,55] In zebrafish, miR-140 has been shown to negatively regulate platelet-derived growth factor alpha receptors, and exogenously injected excess miRNA-140 can cause extensive craniofacial developmental defects including cleft lip and palate, whereas miRNA-140 function loss interferes with the normal localization of neural crest cells and the normal occurrence of sputum organ morphology.[23] In a 2013 literature report,[56] miRNA microarray analysis of 9 microtia and 3 normal ear tissue samples revealed that 11 miRNAs were differentially expressed. Among them, 6 were up-regulated (miR-16, miR-140–3p, miR-126, miR-185, miR-451, and miR-486-5p) and 5 were down-regulated (respectively, miR-203, miR-205, miR-200c, miR-708, and miR-1308). Subsequently, 58 microtia and 16 normal ear tissues were subjected to amplified verification. It was found that miR-451 and miR-486-5p were up-regulated in microtia, and the expression of miR-200c was down-regulated (P < .05). These differentially expressed miRNAs may play an important role in the development and progression of microtia.
Compared with miRNAs, lncRNA sequences are longer, larger than 200 nt, and have a more complex spatial structure, so the information content is more abundant. LncRNAs make up a significant proportion of total noncoding RNA. Abnormally expressed lncRNAs participate in a variety of pathogenic genes through transcription or post-transcriptional regulation, or exert biological functions on their regulated expression networks, which have different degrees of effect on the occurrence and development of diseases.[57] Many studies have confirmed that lncRNAs can silence or activate the expression of some genes through direct interaction with chromosomes, especially in the embryonic development stage. LncRNAs participate in the expression silencing of the heritable alleles and maintain the epigenetic traits, which is essential for the normal development and cell differentiation of multicellular organisms. Ozturk et al[58] used RNA sequencing to identify cleft palate causative genes in mice in a knockout model. Gao et al[59] found that IncRNA H19 may participate in the occurrence of mouse cleft palate by interacting with insulin growth factor. Babajko et al[60] found that Msx1 antisense RNA regulated the expression of Msx1 gene; when the balance between the 2 was disturbed, tooth hypoplasia and bone loss resulted. LncRNA not only affects the development of cleft palate and jaw bone, the growth and development of the nervous system also depends on the accurate expression of lncRNA in time and spatial specificity. The brain is rich in lncRNA expression and plays an important role in regulating neural cell differentiation, synaptic plasticity, and brain development.[61] Guttman et al[62] analyzed more than 1000 intergenic lncRNAs by analyzing mouse neuronal chromatin tags. These lncRNAs are associated with oligodendrocyte myelination, GABAergic neuron differentiation, cAMP-response element binding protein (CREB)-mediated transcriptional regulation, mouse hippocampus development, brain aging, G-protein-coupled receptors, and calcineurin-dependent signal transduction pathways. In 2017, through the detection of lncRNA microarray in autologous residual ear cartilage and normal lateral ear cartilage in 3 microtia patients, a total of 180 differentially expressed lncRNAs were found, of which 74 were up-regulated and 106 were down-regulated.[63] These differential changes in noncoding RNA and normal tissues may provide biological information for the etiology and pathogenesis of microtia.
4. Identification of key candidate genes by bioinformatical analysis
Eleven miRNAs reported in a study by Li et al[56] were analyzed and their target genes predicted by searching 5 databases: MicroCosm, miRanda, miRDB, TargetScan, and PicTar. All analyses were based on previous published studies, thus no ethical approval and patient consent were required. The only miRNA in this group of 11 that contained experimentally verified target genes was hsa-miR-140-3p. A total of 94 predicted target genes was identified (Fig. 1). Gene ontology (GO) analysis results (Fig. 2) show that genes in the biological process (BP) group were principally enriched by cellular process, single-organism process, biological regulation, and regulation of BP. In the cellular component (CC) group, genes were mainly enriched in cell, cell part, organelle part, and membrane. In the molecular function (MF) group, genes were mainly enriched in binding, catalytic activity, MF regulator, and signal transducer activity. The ranking of enriched GO terms in Fig. 3 shows that most genes were significantly enriched in protein kinase binding, kinase binding, cellular response to growth factor stimulus, and axon.
Figure 1.
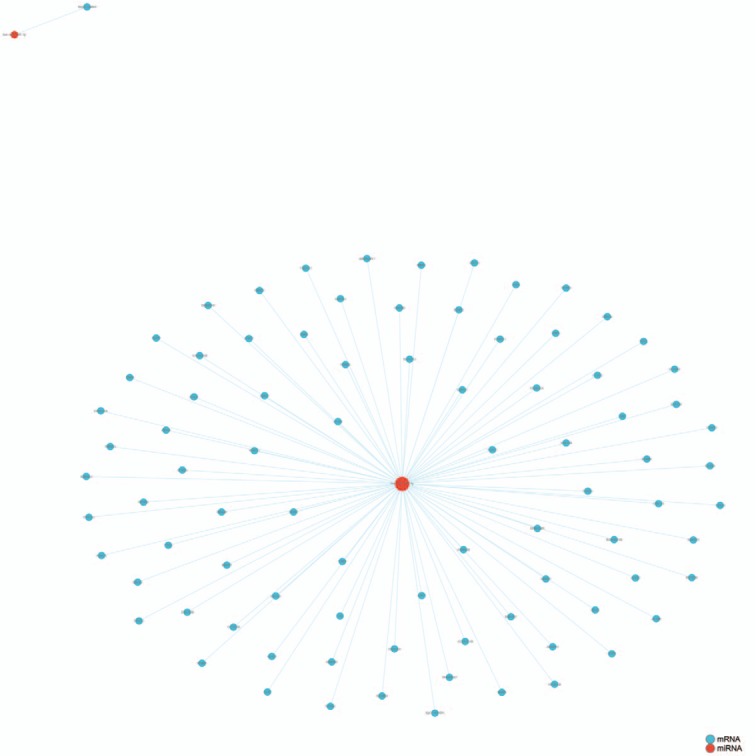
Experimentally verified target genes of hsa-miR-140-3p identified in microtia.
Figure 2.
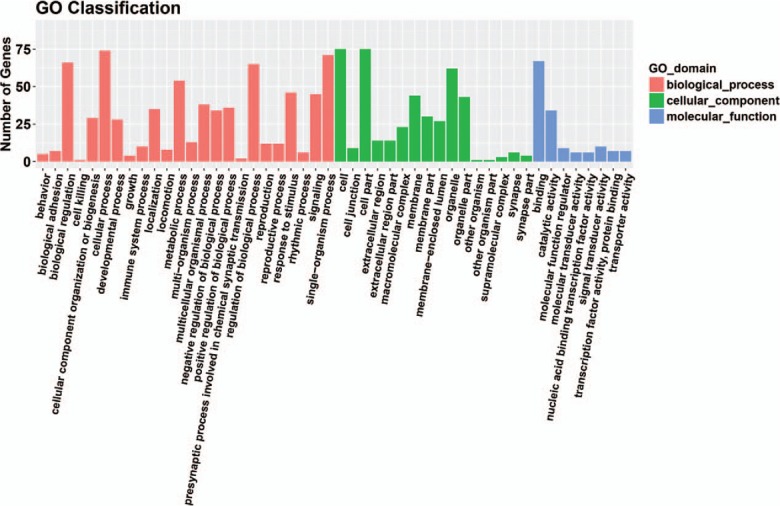
Gene ontology analysis results in three groups: biological process group, cellular component group and molecular function group.
Figure 3.
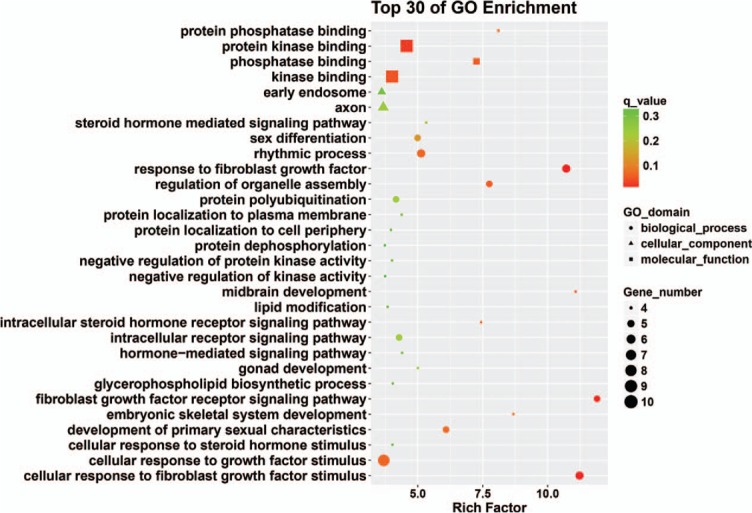
Top 30 enriched GO terms. GO = gene ontology.
The kyoto encyclopedia of genes and genomes classification results showed that genes were mainly enriched in cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems (Fig. 4). The top 30 signaling pathway results showed that genes were mainly enriched in cell cycle, pyrimidine metabolism, and RNA polymerase (Fig. 5).
Figure 4.
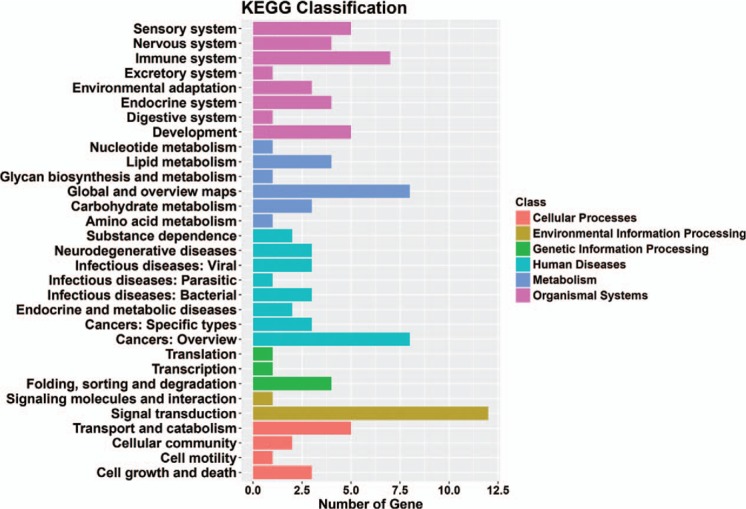
Significant enriched KEGG terms in different classes of target genes in microtia. KEGG = kyoto encyclopedia of genes and genomes.
Figure 5.
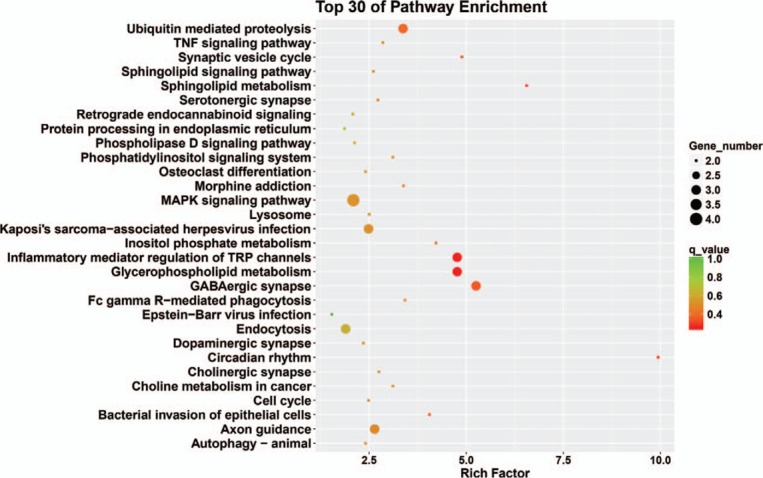
Top 30 enriched pathway terms of target genes in microtia.
Through integrated bioinformatical analysis, we identified candidate genes for microtia, and also key pathways. These findings improve our understanding of the etiology and occurrence of microtia, and also microtia molecular events. The candidate genes and pathways can therefore be used as therapeutic targets.
5. Outlook
The latest research in the field of epigenetics sheds new light on a range of vital processes, challenging “genetic determinism.” A deeper understanding of microtia epigenetics will not only shed light on the occurrence of this defect, but also facilitate more accurate assessment and evaluation of pregnant women at risk of giving birth to babies with microtia. This will lead to better prevention and better treatment of microtia.
6. Compliance with ethical standards
-
(1)
No financial support or benefits have been received by any co-author, by any member of our immediate family or any individual or entity with whom or with which we have a relationship from any commercial source which is related directly or indirectly to the scientific work which is reported on in the article. We have no financial interest or commercial association with any of the subject matter or products mentioned in our manuscript.
-
(2)
This article does not contain any studies with human participants or animals performed by any of the authors.
-
(3)
For this type of study formal consent is not required.
Author contributions
Supervision: Ruhong Zhang.
Writing – review & editing: Xia Chen.
Footnotes
Abbreviations: BP = biological process, CC = cellular component, CREB = cAMP-response element binding protein, GO = gene ontology, KEGG = kyoto encyclopedia of genes and genomes, lncRNA = long non-coding ribonucleic acid, MF = molecular function, miRNA = micro ribonucleic acid.
How to cite this article: Chen X, Zhang R. Microtia epigenetics. Medicine. 2019;98:41(e17468).
The authors have no conflicts of interest to disclose.
References
- [1].Piceci F, Morlino S, Castori M, et al. Identification of a second HOXA2 nonsense mutation in a family with autosomal dominant non-syndromic microtia and distinctive ear morphology. Clin Genet 2017;91:774–9. [DOI] [PubMed] [Google Scholar]
- [2].Chen X, Zhang R, Zhang Q, et al. Microtia patients: auricular chondrocyte ECM is promoted by CGF through IGF-1 activation of the IGF-1R/PI3K/AKT pathway. J Cell Physiol 2019;234:21817–24. [DOI] [PubMed] [Google Scholar]
- [3].Suutarla S, Rautio J, Ritvanen A, et al. Microtia in Finland: Comparison of characteristics in different populations. Int J Pediatr Otorhinolaryngol 2007;71:1211–7. [DOI] [PubMed] [Google Scholar]
- [4].Forrester MB, Merz RD. Descriptive epidemiology of anotia and microtia, Hawaii, 1986–2002. Congenit Anom (Kyoto) 2005;45:119–24. [DOI] [PubMed] [Google Scholar]
- [5].Gonzalez-Andrade F, Lopez-Pulles R, Espin VH, et al. High altitude and microtia in Ecuadorian patients. J Neonatal Perinatal Med 2010;3:109–16. [Google Scholar]
- [6].Canfield MA, Langlois PH, Nguyen LM, et al. Epidemiologic features and clinical subgroups of anotia/microtia in Texas. Birth Defects Res A Clin Mol Teratol 2009;85:905–13. [DOI] [PubMed] [Google Scholar]
- [7].Shaw GM, Carmichael SL, Kaidarova Z, et al. Epidemiologic characteristics of anotia and microtia in California, 1989–1997. Birth Defects Res A Clin Mol Teratol 2004;70:472–5. [DOI] [PubMed] [Google Scholar]
- [8].Klieger-Grossmann C, Chitayat D, Lavign S, et al. Prenatal exposure to mycophenolate mofetil: an updated estimate. J Obstet Gynaecol Can 2010;32:794–7. [DOI] [PubMed] [Google Scholar]
- [9].Hackshaw A, Rodeck C, Boniface S. Maternal smoking in pregnancy and birth defects: a systematic review based on 173,687 malformed cases and 11.7 million controls. Hum Reprod Update 2011;17:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ma C, Carmichael SL, Scheuerle AE, et al. Association of microtia with maternal obesity and periconceptional folic acid use. Am J Med Genet 2010;152A:2756–61. [DOI] [PubMed] [Google Scholar]
- [11].Renato Paro. Epigenetics: DNA isn’t everything; April 13, 2009. Available at: https://www.sciencedaily.com/releases/2009/04/090412081315.htm. [Google Scholar]
- [12].Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet 2019;42920:109–27. [DOI] [PubMed] [Google Scholar]
- [13].Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol 2018;19:774–90. [DOI] [PubMed] [Google Scholar]
- [14].Tucci V, Isles AR, Kelsey G, et al. Genomic imprinting and physiological processes in mammals. Cell 2019;176:952–65. [DOI] [PubMed] [Google Scholar]
- [15].Time for the epigenome. Nature 2010;463:587. [DOI] [PubMed] [Google Scholar]
- [16].Monteagudo-Sánchez A, Sánchez-Delgado M, Mora JRH, et al. Differences in expression rather than methylation at placenta-specific imprinted loci is associated with intrauterine growth restriction. Clin Epigenetics 2019;11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Malnou EC, Umlauf D, Mouysset M, et al. Imprinted microRNA gene clusters in the evolution, development, and functions of mammalian placenta. Front Genet 2019;9:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ding YX, Cui H. Integrated analysis of genome-wide DNA methylation and gene expression data provide a regulatory network in intrauterine growth restriction. Life Sci 2017;179:60–5. [DOI] [PubMed] [Google Scholar]
- [19].Burris HH, Baccarelli AA. Air pollution and in utero programming of poor fetal growth. Reprod Sci 2017;9:213–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin Y, Yu J, Wu J, et al. Abnormal level of CUL4B-mediated histone H2A ubiquitination causes disruptive HOX gene expression. Epigenetics Chromatin 2019;12:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li H, Niswander L. Does DNA methylation provide a link between folate and neural tube closure? Epigenomics 2018;10:1263–5. [DOI] [PubMed] [Google Scholar]
- [22].Price EM, Peñaherrera MS, Portales-Casamar E, et al. Profiling placental and fetal DNA methylation in human neural tube defects. Epigenetics Chromatin 2016;9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Eberhart JK, He X, Swartz ME, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet 2008;40:290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gonseth S, Shaw GM, Roy R, et al. Epigenomic profiling of newborns with isolated orofacial clefts reveals widespread DNA methylation changes and implicates metastable epiallele regions in disease risk. Epigenetics 2019;14:198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wehby GL, Murray JC. Folic acid and orofacial clefts: a review of the evidence. Oral Dis 2010;16:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Trasler JM, Trasler DG, Bestor TH, et al. DNA methyltransferase in normal and Dnmtn/Dnmtn mouse embryos. Dev Dyn 1996;206:239–47. [DOI] [PubMed] [Google Scholar]
- [27].Juriloff DM, Harris MJ. Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol 2008;82:63–77. [DOI] [PubMed] [Google Scholar]
- [28].Clark SJ, Harrison J, Paul CL, et al. High sensitivity mapping of methylated cytosines. Nucl Acids Res 1994;22:2990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bouty A, Ayers KL, Pask A, et al. The genetic and environmental factors underlying hypospadias. Eur J Pediatr 2015;9:239–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr 2009;21:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tournaire M, Devouche E, Epelboin S, et al. Birth defects in children of men exposed in utero to diethylstilbestrol (DES). Therapie 2018;73:399–407. [DOI] [PubMed] [Google Scholar]
- [32].Cho E, Mysliwiec MR, Carlson CD, et al. Cardiac-specific developmental and epigenetic functions of Jarid2 during embryonic development. J Biol Chem 2018;293:11659–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lyu G, Zhang C, Ling T, et al. Genome and epigenome analysis of monozygotic twins discordant for congenital heart disease. BMC Genomics 2018;19:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mao F, Liu Q, Zhao X, et al. EpiDenovo: a platform for linking regulatory de novo mutations to developmental epigenetics and diseases. Nucleic Acids Res 2018;46(D1):D92–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rivera CM, Ren B. Mapping human epigenomes. Cell 2013;155:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Park C, Rosenblat J, Brietzke E, et al. Stress, epigenetics and depression: a systematic review. Neurosci Biobehav Rev 2019;doi: 10.1016/j.neubiorev.2019.04.010. [DOI] [PubMed] [Google Scholar]
- [37].Bajrami E, Spiroski M. Genomic imprinting. Open Access Maced J Med Sci 2016;4:181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Joubert BR, Felix JF, Yousefi P, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 2016;98:680–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Grewal J, Carmichael SL, Ma C, et al. Maternal periconceptional smoking and alcohol consumption and risk for select congenital anomalies. Birth Defects Res A Clin Mol Teratol 2008;82:519–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet 2010;71:3–9. [DOI] [PubMed] [Google Scholar]
- [41].McKay JA, Mathers JC. Diet induced epigenetic changes and their implications for health. Acta Physiol (Oxf) 2011;202:103–18. [DOI] [PubMed] [Google Scholar]
- [42].Santos-Guzmán J, Arnhold T, Nau H, et al. Antagonism of hypervitaminosis A-induced anterior neural tube closure defects with a methyl-donor deficiency in murine whole-embryo culture. J Nutr 2003;133:3561–70. [DOI] [PubMed] [Google Scholar]
- [43].Imbard A, Benoist JF, Blom HJ. Neural tube defects, folic acid and methylation. Int J Environ Res Public Health 2013;10:4352–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McKay JA, Groom A, Potter C, et al. Genetic and non-genetic influences during pregnancy on infant global and site specific DNA methylation: role for folate gene variants and vitamin B12. PLoS One 2012;7:e33290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Padmanabhan N, Jia D, Geary-Joo C, et al. Mutation in folate metabolism causes epigenetic instability and transgenerational effects on development. Cell 2013;155:81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jia Xiao-hui, Lu Su-qin, Bai Yun, et al. Correlation study of biochemical metabolic indexes of pregnant women and fetal neural tube defects. China Womens Childrens Health Res 2014;25:69–71. [Google Scholar]
- [47].Ray JG, Wyatt PR, Thompson MD, et al. Vitamin B12 and the risk of neural tube defects in a folic-acid-fortified population. Epidemiology 2007;18:362–6. [DOI] [PubMed] [Google Scholar]
- [48].Dill P, Schneider J, Weber P, et al. Pyridoxal phosphate-responsive seizures in a patient with cerebral folate deficiency (CFD) and congenital deafness with labyrinthine aplasia, microtia and microdontia (LAMM). Mol Genet Metab 2011;104:362–8. [DOI] [PubMed] [Google Scholar]
- [49].Schwartz JL. Environmental mutagenesis and radiation biology: the legacy of William Morgan. Mutat Res 2017;806:63. [DOI] [PubMed] [Google Scholar]
- [50].Birney E, Stamatoyannopoulos JA, Dutta A. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol 2017;1008:1–46. [DOI] [PubMed] [Google Scholar]
- [52].Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet 2010;19(R2):R162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol 2019;51:11–7. [DOI] [PubMed] [Google Scholar]
- [54].Powder KE, Ku YC, Brugmann SA, et al. A cross-species analysis of microRNAs in the developing avian face. PLoS One 2012;7:1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu Y, Huang T, Zhao X, et al. MicroRNAs modulate the Wnt signaling pathway through targeting its inhibitors. Biochem Biophys Res Commun 2011;408:259–64. [DOI] [PubMed] [Google Scholar]
- [56].Li C, Hao S, Wang H, et al. MicroRNA expression profiling and target genes study in congenital microtia. Int J Pediatr Otorhinolaryngol 2013;77:483–7. [DOI] [PubMed] [Google Scholar]
- [57].Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018;172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ozturk F, Li Y, Zhu X, et al. Systematic analysis of palatal transcriptome to identify cleft palate genes within TGFβ3 - knockout mice alleles: RNA -Seq analysis of TGFβ3 mice. BMC Genomics 2013;14:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gao L, Yin J, Wu W. Long non -coding RNA H19 -mediated mouse cleft palate induced by 2,3,7,8 -tetrachlorodibenzo -p - dioxin. Exp Ther Med 2016;11:2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Babajko S, Méary F, Petit S, et al. Transcriptional regulation of MSX1 natural antisense transcript. Cells Tissues Organs 2011;194:151–5. [DOI] [PubMed] [Google Scholar]
- [61].Sasaki YT, Sano M, Ideue T, et al. Identification and characterization of human non-coding RNAs with tissue-specific expression. Biochem Biophys Res Commun 2007;357:991–6. [DOI] [PubMed] [Google Scholar]
- [62].Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non -coding RNAs in mammals. Nature 2009;458:223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang L, Lin L, Song YP, et al. Differential expression of long noncoding RNAs in congenital microtia. Gene Expr Patterns 2017;25–6. [DOI] [PubMed] [Google Scholar]


