Abstract
Several pathways exist to bypass DNA damage during replication. One such pathway is template switching. The Rad5 protein plays two important roles in template switching: it is an E3 ubiquitin ligase that catalyzes PCNA poly-ubiquitylation and it is a helicase that converts replication forks to chicken foot structures. To understand the structure, conformational flexibility, and mechanism of Rad5, we used a full-ensemble hybrid method combining Langevin dynamics simulations and small-angle X-ray scattering. From these studies, we generated the first experimentally validated, high-resolution structural model of Rad5. We found that Rad5 is more compact and less extended than is suggested by its large amount of predicted intrinsic disorder. Thus, Rad5 likely has a novel intra-molecular interaction that limits the range of conformational space it can sample. We provide evidence for a novel interaction between the HIRAN and the helicase domains of Rad5, and we discuss the biological and mechanistic implications of this.
1. Introduction
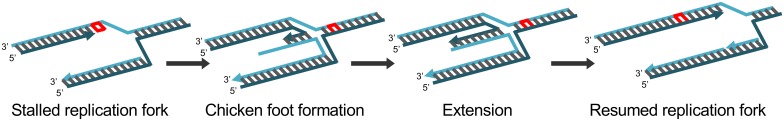
DNA damage in the template strand blocks DNA replication by classical DNA polymerases. Cells, consequently, have evolved several pathways to bypass DNA damage during DNA replication. One pathway is translesion synthesis, which is initiated by the mono-ubiquitylation of replication accessory factor PCNA (proliferating cell nuclear antigen) [1–6]. During translesion synthesis, the stalled classical DNA polymerase is replaced by one or more non-classical DNA polymerases that catalyze DNA synthesis using the damaged strand as the template [7–16]. Typically, this is an error-prone process. Another pathway for damage bypass is template switching, which is initiated by the poly-ubiquitylation of PCNA [1–4]. During template switching, the stalled replication fork is converted to a chicken foot intermediate (Fig 1) [17,18]. Damage bypass is then accomplished by DNA synthesis using the newly synthesized sister strand as the template. Typically, this is an error-free process. In yeast, the Rad5 protein plays two critical roles in template switching. First, it binds Ubc13 (an E2 ubiquitin-conjugating enzyme) and Mms2 (a ubiquitin-conjugating enzyme variant) and acts as an E3 ubiquitin ligase to catalyze the poly-ubiquitylation of PCNA [1,2]. Second, it acts as a helicase to catalyze the conversion of the stalled replication fork to the chicken foot intermediate [17,18].
Fig 1. Illustration of replication fork remodeling in template switching.
A stalled replication fork is shown with damage on the leading strand. Rad5 rearranges the DNA such that the stalled primer strand base pairs with the newly synthesized primer strand from the lagging strand. This results in a chicken foot intermediate. In this configuration, classical DNA polymerases can extend the stalled primer strand resulting in error-free bypass of DNA damage. Once replication has progressed beyond the site of damage, the replication fork can be reestablished and conventional DNA replication can resume.
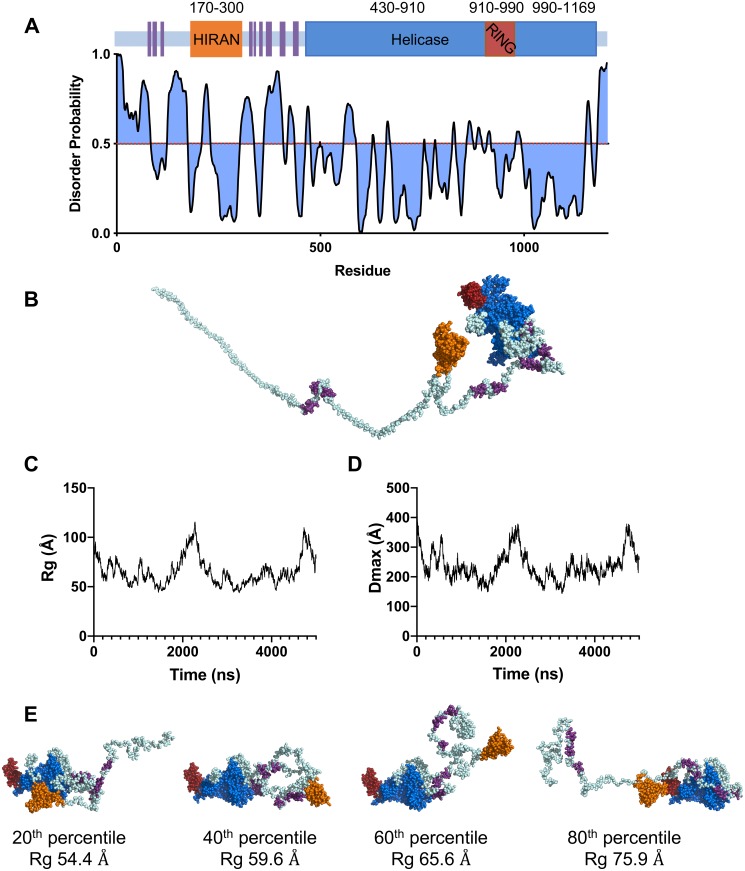
Despite the critical role that Rad5 plays in DNA damage bypass, very little is known about the structure or mechanism of Rad5 or its two human homologs HLTF (helicase-like transcription factor) and SHPRH (SNF2 histone linker PHD RING helicase) [19]. Rad5 is comprised of 1169 amino acid residues that form three folded domains: a HIRAN (HIP116 Rad5p N-terminal) domain (approximately residues 170 to 300), a Swi/Snf superfamily 2 helicase domain (approximately residues 430 to 910 and 990 to 1169), and a RING (really interesting new gene) domain (approximately residues 910 to 990). The HIRAN domain, which binds free 3′ ends of DNA, and the helicase domain are both required for catalyzing fork reversal [18,20]. The RING domain and the helicase domain are required for poly-ubiquitylation of PCNA [20,21]. Only the structure of the HIRAN domain of human HLTF has been determined [18]. Protein disorder predictions indicate that the regions outside of these three domains are intrinsically disordered (Fig 2). These putative disordered regions include an N-terminal tail region (approximately residues 1 to 170) and a region between the HIRAN domain and the helicase domain (approximately residues 300 to 430). These disordered regions are likely to have high conformational flexibility.
Fig 2. Initial LD simulations of Rad5.
(A) Disorder probability prediction plot obtained using the PrDOS server [22,23] and an illustration of the structured and disordered regions of Rad5. Structured regions include the helicase domain (blue), RING finger domain (red), HIRAN domain (orange) and nine, putative α-helices (purple). Disordered regions are shown in light blue. (B) The starting model of Rad5 used for initial LD simulations. (C) Rg plotted over time for one 5 μs initial LD simulation. (D) Dmax plotted over time for one 5 μs initial LD simulation. (E) Individual structures of Rad5 from the initial LD simulations representing the 20th, 40th, 60th, and 80th percentile of Rg values.
Obtaining high-resolution structural models of proteins with high conformational flexibility is one of the most difficult challenges in contemporary structural biology. Recently, our group has successfully employed a full-ensemble hybrid method that combines Langevin dynamics (LD) simulations and small-angle X-ray scattering (SAXS) to generate experimentally validated, high-resolution structural models of several proteins with high conformational flexibility [24–26]. These included one protein with a disordered C-terminal tail region, non-classical DNA polymerase eta (pol η), as well as two proteins with short tether regions: ubiquitin-modified PCNA and SUMO (small ubiquitin-like modifier)-modified PCNA. In these cases, the LD simulations were used to generate large ensembles of several thousand structures, and these full ensembles were compared to experimental SAXS data. This was all done without resorting to curve fitting in order to avoid over fitting the experimental data.
In order to better understand the structure, conformational flexibility, and mechanism of Rad5, we have utilized this full-ensemble hybrid method. Initial LD simulations of Rad5 resulted in an ensemble in which the predicted radius of gyration (Rg) and maximal distance (Dmax) values were significantly greater than those obtained from the experimental SAXS data. To obtain better agreement with the experimental data, we postulated a novel interaction between the HIRAN domain and the helicase domain. Including this interaction in the simulations resulted in an ensemble in which the predicted Rg and Dmax values closely matched the experimental values. Thus, we have generated an experimentally validated structural model of Rad5 that has high conformational flexibility, yet retains a large, folded core comprised of the HIRAN, helicase, and RING domains.
2. Materials and methods
2.1. Protein expression and purification
Rad5 was codon-optimized for bacterial expression and cloned into pET11a with an N-terminal 6xHis tag and a C-terminal Twin-Strep tag, resulting in plasmid pKW746. Rad5 was over-expressed in BL21 Star (DE3) cells by induction at an OD600 of 0.6 with 1 mM IPTG for 16 hours at 16°C. Cells were lysed at 4°C using an EmulsiFlex (Avestin) in the presence of 1 mM PMSF, Complete, EDTA-free Protease Inhibitor Cocktail (Roche), and DNase. The crude extracts were clarified by ultracentrifugation. The proteins were purified using a Strep-Tactin XT resin (IBA) in 100 mM Tris pH 8.0, 150 mM KCl, 5% glycerol, and 1 mM DTT. Rad5 was eluted with 50 mM biotin and further purified using a HiLoad Superdex 200 size-exclusion column (GE Healthcare).
2.2. Small-angle X-ray scattering
SAXS data were collected at the BioCAT beamline 18-ID at the Advanced Photon Source. In-line size exclusion chromatography was performed using a Superdex 200 Increase 10/300 GL column, which was run at 0.7 ml/min by an AKTA Pure FPLC instrument (GE Healthcare Life Sciences). The eluate was passed through the UV monitor and through a 100 μL quartz flow cell and exposed to the X-ray beam every 2 s with 0.5 s exposures. Data were collected at room temperature using a wavelength equal to 1.033 Å, a Pilatus3 1M detector (Dectris), and a sample-to-detector distance equal to 3.5 m. Buffer subtraction was performed using BioXTAS RAW [27]. PRIMUS and AUTORG were used to calculate Rg values [28,29], and GNOM was used to generate Dmax values and pair-wise distribution plots [29,30].
2.3. Construction of starting models for simulations
The starting model of Rad5 was built using homology models of the HIRAN domain (SWISS-MODEL, based on 4XZG.pdb) [18], RING domain (SWISS-MODEL, based on 4R8P.pdb) [31], and helicase domain (Phyre2, based on 1Z3I.pbd, 6GEJ.pbd, 3MWY.pbd, 5O9G.pbd, 6FML.pbd, and 6G7E.pbd) [32–37]. Nine putative α-helices within the intrinsically disordered regions of Rad5 were also identified and modeled using Phyre2 [38]. For our refined simulations, the ClusPro and ZDOCK docking servers were used to determine reasonable orientations of the HIRAN domain docked to the helicase domain [39,40]. Intrinsically disordered loops were built in PyMol or generated as unstructured regions in the Phyre2 models. The positions of these disordered regions were adjusted to accommodate different HIRAN domain positions using PyMol. Models were coarse-grained such that each amino acid was replaced by one to four pseudoatoms depending on the size, shape, and charge of the residue as described previously [41].
2.4. Langevin dynamics simulations
All simulations were carried out using the simulation code uiowa_BD [24]. The partial charges of the ionizable groups, the hydrodynamic radii of the pseudo-atoms, and the energy function used in the simulations were as described previously [24]. A time step of 125 fs was used, and snapshots (PDB files) were recorded after every ns of simulation time for a total of 5 μs of simulation time. This yielded an ensemble of 5,000 individual structures per simulation.
2.5. Comparisons of the SAXS data and the simulations
Each simulation produced an ensemble of 5,000 structures that were output as sequential PDB files. Predicted scattering curves for each of the individual structures were generated using CRYSOL [42]. The predicted scattering curve for the full ensembles were obtained by averaging the scattering curves of the individual structures constituting each of the ensembles. χ2 values were determined by comparing the predicted scattering curves to the experimental scattering curves as described [24]. The predicted P(r) plots for the full ensembles were obtained by summing all of the inter-atomic distances in all of the individual structures constituting each of the ensembles and generating histograms in GraphPad Prism. The cut off for determining Dmax was set to include 99% of the inter-atomic distances.
2.6. Chemical crosslinking reactions
A 100-μl reaction containing 50 μM of full length Rad5 was treated with BS2G and BS3 crosslinkers (ProteoChem) that were freshly dissolved in sodium phosphate and added to a final concentration of 1 mM. The crosslinking reactions and a sodium phosphate negative control were incubated at room temperature for 1 hour. To quench the reactions, Tris buffer was added to a final concentration of 60 mM and the samples were maintained at room temperature for 15 minutes. The excess crosslinker was removed via size exclusion chromatography. Samples were then precipitated with 15% trichloroacetic acid, washed with acetone, and re-suspended in a urea solution. Samples were then reduced with dithiothreitol and alkylated with iodoacetamide.
2.7. Mass spectrometry and analysis
Samples were digested in solution with trypsin/Lys-C. After digestion, the solution was dried and reconstituted to 1μg/μl in 5% acetonitrile/water (0.1% formic acid). The peptides were then separated by liquid chromatography (Thermo Scientific EASY nLC-1200 coupled to a Thermo Scientific Nanospray FlexIon source) using a pulled glass emitter 75um X 20 cm (Agilent capillary, part#160-2644-5), with the tip packed with Agilent SB-C18 Zorbax 5um packing material (part #820966–922) and the remaining emitter packed with nanoLCMS Solutions UChrom C18 3um packing material (part #80002) and analyzed by MS/MS on a Thermo Scientific Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer.
The raw data were analyzed using Thermo Scientific’s Proteome Discoverer Software. The data was searched using Mascot and Sequest HT against the Rad5 sequence [43,44]. Identification of crosslinks was carried out using Byonic (Protein Metrics) [45]. The following parameters were used for searching the Thermo raw files in Byonic: Cleavage residues were set to RK and digest cutter was set to C-terminal cutter. Peptide termini were set to Fully Specific and maximum number of missed cleavages was set to 3. Fragmentation type was set to QTOF/HCD with a precursor tolerance of 100 ppm, and a fragment tolerance of 0.02 Da. Precursor isotope off by x was set to off by one or two. Protein FDR was set to 1% FDR. Crosslinks were enabled as appropriate for each sample. The crosslinking candidates were filtered by Xlink score. Candidates with positive Xlink scores, Byonic scores above 200, and PEP2D scores below .001 were selected as likely crosslinking sites. Candidates containing consecutive peptides were not considered in analysis.
2.8. Accession numbers
The SAXS data, all of the individual snapshots from the triplicate LD simulations, and a description of all relevant methods have been deposited in the SASBDB under the accession code SASDG25.
3. Results
3.1. Initial Langevin dynamics simulations of Rad5
We carried out LD simulations of Rad5 to better understand its conformational flexibility. Because there are currently no high-resolution structures of Rad5, we used homology modeling to build the starting model for the simulations (Fig 2). We used SWISS-MODEL [46] to generate a homology model of the Rad5 HIRAN domain that was based on the X-ray crystal structure of the HIRAN domain of HLTF (4XZG.pdb) [18] as well as a homology model of the Rad5 RING domain that was based on the structure of the RING domain of PRC1 (4R8P.pdb) [31]. We used Phyre2 [38] to generate a homology model of the Rad5 helicase domain that was based on the structures of the helicase domains of Rad54 (1Z3I.pdb) [32], SWR1 (6GEJ.pdb) [33], Chd1 (3MWY.pdb and 5O9G.pdb) [34,35], INO80 (6FML.pdb) [36], and Mot1 (6G7E.pdb) [37]. Phyre2 was also used to identify nine putative α-helices within the intrinsically disordered regions of Rad5. These putative helices were included in the starting model because our previous studies of non-classical polymerase pol η showed that the inclusion of such putative α-helices substantially improves the agreement between simulations and experimental X-ray scattering data [24]. The starting model also contained an N-terminal 6xHis tag and a C-terminal Twin-Strep tag.
Simulating Rad5 at full atomic resolution is not currently feasible. Thus, the starting model was coarse-grained such that, depending on the size, shape, and charge of the residue in question, each amino acid residue was replaced by one to four pseudo-atoms [41]. The LD simulations were carried out in duplicate using uiowa_BD as previously described [24]. Briefly, the positions of each pseudo-atom were calculated using 125 fs time steps and snapshots (PDB files) were recorded every ns for 5 μs of simulation time. These simulations each generated ensembles containing 5,000 individual structures. To ensure sufficient sampling of conformational space, the Rg and Dmax values for the individual structures in the ensembles were graphed as a function of time (Fig 2). We used CRYSOL to generate scattering curves for all of the individual structures [42], and these were averaged to obtain theoretical scattering curves for the full ensembles (see below). The Rg and Dmax values for the full ensembles for this starting model are 62.9 Å and 229 Å, respectively (Table 1).
Table 1. SAXS and simulation parameters.
| Rg (Å) | Dmax (Å) | χ2 | |
|---|---|---|---|
| Experimental SAXS data | 47.0 | 178 | N/A |
| Initial simulations | 62.9 | 229 | 3.73 |
| Refined simulations (ClusPro) | 48.5 | 182 | 1.74 |
| Refined simulations (ZDOCK) | 49.6 | 192 | 1.21 |
To visualize the structures that constitute the full ensemble, we ordered the 5,000 individual structures in order of increasing Rg. The individual structures corresponding to the 20th, 40th, 60th, and 80th percentiles are shown in Fig 2. Overall, these simulations show that Rad5 possesses a high degree of conformational flexibility. Its flexible N-terminal tail region, which includes the HIRAN domain, samples a wide range of conformations. Interestingly, the HIRAN and helicase domains are in contact or in close proximity with each other (within 6 Å) in approximately 15% of the individual structures in the ensemble. These periods of contact or close proximity persist from as short as 2 ns to as long as 200 ns.
3.2. Small-angle X-ray scattering of Rad5
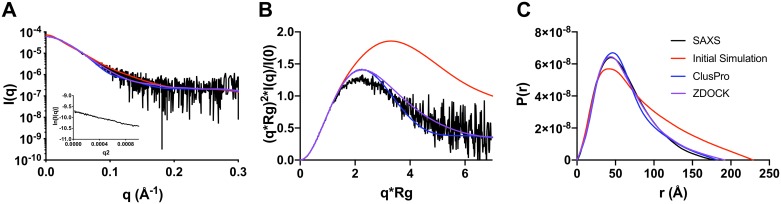
Rad5 containing an N-terminal 6xHis tag and a C-terminal Twin-Strep tag was overexpressed in E. coli and purified using a Strep-Tactin XT affinity column and Superdex 200 size exclusion column. To experimentally validate the LD simulations, we carried out size exclusion chromatography (SEC)-SAXS (S1 Table and S1 Fig). Buffer subtraction was performed using BioXTAS RAW [27] to obtain an experimental scattering curve (Fig 3). We compared this experimental scattering curve to the theoretical scattering curve obtained from the initial LD simulations using the equation described previously [42], and found that the agreement was rather poor with a χ2 equal to 3.73. We used PRIMUS and AUTORG to carry out Guinier analysis and to obtain an experimental Rg [28,29]. The linear Guinier plot (Fig 3) shows that the protein is free from aggregation, and the experimental Rg is equal to 47.0 Å (Table 1). This experimental value is considerably less than the theoretical Rg obtained from the initial simulations (62.9 Å).
Fig 3. SEC-SAXS analysis of Rad5.
(A) Experimental SAXS scattering curve (black) overlaid with theoretical scattering curves generated from full ensemble simulations of the initial simulations of Rad5 (red), as well as two refined simulations utilizing ClusPro (blue) or ZDOCK (purple). (B) Dimensionless Kratky plots from the experimental data (black), the initial simulation (red) and the two refined simulations (blue and purple). (C) Pairwise distribution plots from the experimental data (black), the initial simulation (red) and the refined simulations (blue and purple).
The experimental, dimensionless Kratky plot (Fig 3) suggests that Rad5 behaves as a partially disordered protein in solution. The theoretical, dimensionless Kratky plot derived from the initial simulations suggests a greater degree of disorder and conformational flexibility than is observed with the experimental data. Furthermore, the experimental pairwise distribution plot (Fig 3) generated using GNOM [29,30] yielded a Dmax equal to 178 Å (Table 1). This experimental value is considerably less than the theoretical Dmax obtained from the initial simulations (229 Å). Moreover, the theoretical, pairwise distribution plot shows a greater degree of extension than is observed with the experimental data. Overall, these results imply that the initial simulations over-estimated the degree of disorder and conformational flexibility in Rad5.
3.3. Refined Langevin dynamics simulations of Rad5
To bring the LD simulations of Rad5 into better agreement with the results from the SAXS experiments, we repeated the LD simulations with adjustments made to the starting model. We built two new starting models, both of which had an interaction between the HIRAN domain and the helicase domain. This hypothetical intra-molecular interaction was chosen for three reasons. First, the SAXS results showed that Rad5 is less extended and more compact than the initial LD simulations suggested. Second, we extracted the 58 best-fit structures from the initial ensemble (those with χ2 values less than 1.22), and all of them featured the HIRAN and helicase domains in contact or in close proximity. Third, 15% of the individual structures in the initial ensemble had these domains in contact or close proximity.
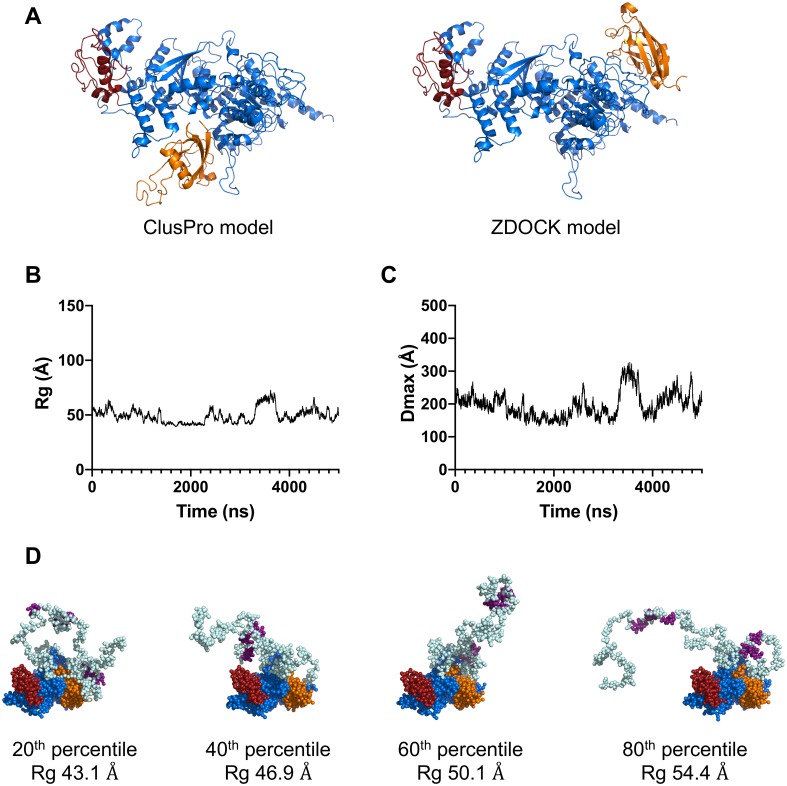
To build these new starting models, we used two docking servers (ClusPro and ZDOCK) to generate two different docked poses of the HIRAN domain on the helicase domain (Fig 4) [39,40]. Distance restraints were not imposed with ClusPro in order to ensure that the placement of the HIRAN domain on the helicase domain was not biased by the initial LD simulations. By contrast, distance restraints were imposed with ZDOCK to ensure that the HIRAN domain was placed on the helicase domain in the most common position observed in the initial LD simulations. We used both approaches in order to determine whether the position of the HIRAN domain on the helicase domain would significantly affect the Rg and Dmax values as well as the quality of the fit.
Fig 4. Refined LD simulations of Rad5.
(A) Refined starting models built using the ClusPro server (left) or the ZDOCK server (right). The helicase domain is shown in blue, the HIRAN domain is shown in orange, and the RING finger domain is shown in red. The disordered regions have been removed for clarity. (B) Rg plotted over time for one 5 μs refined ClusPro simulation of Rad5. (C) Dmax plotted over time for one 5 μs refined ClusPro simulation of Rad5. (D) Individual structures of Rad5 from the refined ClusPro simulation representing the 20th, 40th, 60th, and 80th percentile of Rg values.
As with the initial LD simulations, these two new starting models were coarse-grained such that each amino acid residue was replaced by one to four pseudo-atoms. The two sets of refined LD simulations were carried out in triplicate using 125 fs time steps. Snapshots were recorded every ns for 5 μs of simulation time, such that each simulation generated an ensemble containing 5,000 individual structures. The Rg and Dmax values for the individual structures in the ClusPro-derived ensemble were graphed as a function of time (Fig 4). The corresponding graphs for the ZDOCK-derived ensemble are shown in S2 Fig. We generated scattering curves for all of the individual structures in a given ensemble, and these were averaged to obtain theoretical scattering curves for the full ensemble. The Rg and Dmax values for the ClusPro-derived full ensemble are 48.5 Å and 182 Å, respectively (Table 1). The Rg and Dmax values for the ZDOCK-derived full ensemble are 49.6 Å and 192 Å, respectively (Table 1). The values from these refined Rad5 LD simulations are in much better agreement with the experimentally derived values than are the values from the initial LD simulations.
We compared the theoretical scattering curves for both the ClusPro-derived ensemble and the ZDOCK-derived ensemble with the experimental scattering curve (Fig 3). We found that the agreement using both starting models was significantly improved compared to the initial simulation. The ClusPro-derived ensemble had a χ2 equal to 1.74 and the ZDOCK-derived ensemble had a χ2 equal to 1.21. The lower χ2 value with the ZDOCK-derived ensemble reflects a better fit of the theoretical and experimental scattering curves in the intermediate q region. Overall, the Rg and Dmax values for the ClusPro-derived ensemble agree with the experimental values marginally better than the ZDOCK-derived ensemble values do, while the scattering curve for the ZDOCK-derived ensemble agrees with the experimental curve marginally better than the ClusPro-derived ensemble does. Given these results and the resolution limitations of SAXS, it is extremely difficult to predict the precise position and orientation of the HIRAN domain on the helicase domain. Nevertheless, these results provide strong support for an intra-molecular interaction between the HIRAN and helicase domains of Rad5.
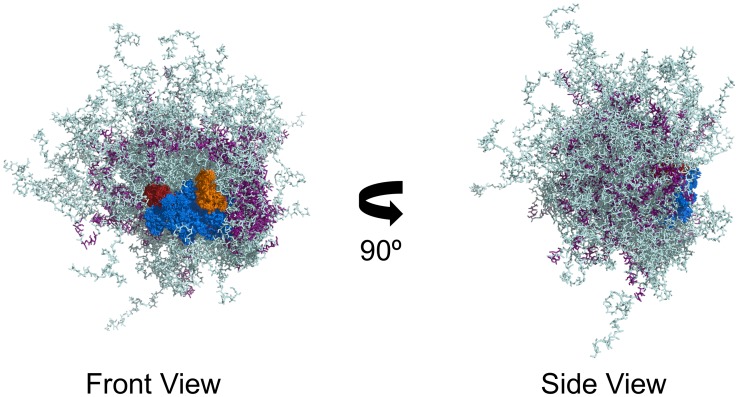
We also generated theoretical dimensionless Kratky plots and pairwise distribution plots for both the ClusPro-derived ensemble and the ZDOCK-derived ensemble and compared these with the corresponding experimental plots (Fig 3). The plots from both of these refined Rad5 LD simulations agree better with the experimental plots than do the plots from the initial Rad5 LD simulations. Finally, to visualize the structures that constitute the full, refined ensembles, we ordered the 5,000 individual structures in order of increasing Rg. The individual structures from the ClusPro-derived ensemble corresponding to the 20th, 40th, 60th, and 80th percentiles are shown in Fig 4. The corresponding structures for the ZDOCK-derived ensemble are shown in S2 Fig. Overall, these simulations show that, while the HIRAN domain is bound to the helicase domain, Rad5 still possesses a high degree of conformational flexibility. This flexibility is mainly in the N-terminal tail region (approximately residues 1 to 170) and the region between the HIRAN domain and the helicase domain (approximately residues 300 to 430) (Fig 5).
Fig 5. Conformational flexibility of Rad5.
Overlay of 100 random individual structures from the refined ClusPro simulation. The helicase domain is shown in blue, the RING finger domain is shown in red, the HIRAN domain is shown in orange, the nine, putative α-helices are shown in purple, and the disordered regions are shown in light blue. Rad5 can sample a wide range of conformational space, mostly due to the intrinsically disordered N-terminal tail and the intrinsically disordered region between the HIRAN and helicase domains.
3.4. Crosslinking the Rad5 HIRAN and helicase domains
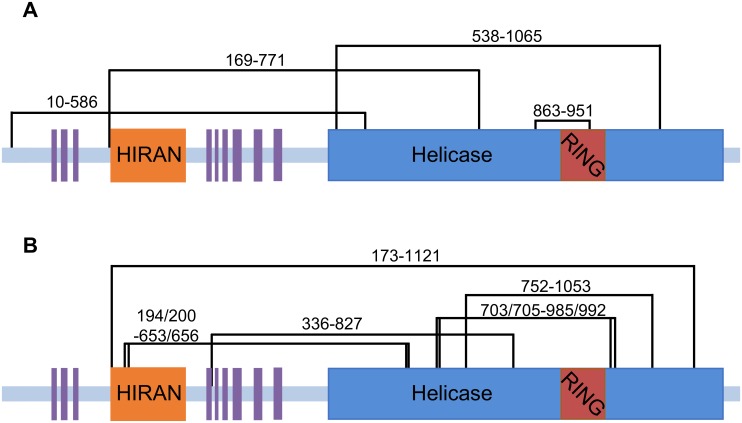
To provide independent, experimental evidence that the Rad5 HIRAN and helicase domains interact, we carried out crosslinking followed by peptide analysis by mass spectrometry (S2 Table and S3 Fig). The Rad5 protein was treated with homobifunctional crosslinking agents BS2G or BS3. The crosslinked protein was precipitated, washed, re-suspended, and digested with trypsin and Lys-C. The desalted peptides were subjected to LC-MS/MS. From the BS2G experiment, one crosslink was identified between Lys-169 of the HIRAN domain and Lys-771 of the helicase domain (Fig 6). From the BS3 experiment, two peptide pairs containing a total of three crosslinks were identified. Lys-173 of the HIRAN domain was crosslinked to Lys-1121 of the helicase domain, and Lys-194 and Lys-200 of the HIRAN domain were crosslinked to Lys-653 and Lys-656 of the helicase domain (Fig 6). Most of the remaining crosslinks from both experiments were between residues within the helicase domain.
Fig 6. Crosslinking the Rad5 HIRAN and helicase domains.
(A) Black lines represent the most statistically significant crosslinks in full length Rad5 using the BS2G crosslinker. Shown are the helicase domain (blue), the RING finger domain (red), the HIRAN domain (orange) and the nine, putative α-helices (purple). Disordered regions are shown in light blue. (B) Black lines represent the most statistically significant crosslinks in full length Rad5 using the BS3 crosslinker. The structured and disordered regions of Rad5 are depicted as described above.
From our crosslinking experiments, three peptide pairs containing four individual crosslinks between the HIRAN and helicase domains were identified. While it is technically possible that these represent inter-molecular crosslinks between two Rad5 molecules, this is unlikely because multi-angle light scattering (MALS) measurements show Rad5 to be a monomer in solution (S4 Fig). The locations of two of these crosslinking sites on the helicase domain, Lys-771 from the BS2G experiment and Lys-1121 from the BS3 experiment, are in close proximity to the docking position generated by ZDOCK that was used in our refined LD simulations. The locations of the other two crosslinking sites, Lys-653 and Lys-656, were identified in a single peptide pair from the BS3 experiment and are in proximity to the position generated by ClusPro that was used in our refined simulations.
4. Discussion
The bypass of DNA damage during DNA replication is critical to reducing the frequency of mutagenesis and avoiding genome instability. Rad5 plays two important roles in DNA damage bypass. It functions as an E3 ubiquitin ligase to catalyze the poly-ubiquitylation of PCNA, and it functions as a fork-remodeling helicase to catalyze the conversion of stalled replication forks to chicken foot structures. To begin to understand the structure and conformational flexibility of Rad5, we have employed a full ensemble hybrid method that combines molecular simulations with SAXS [26].
Full ensemble hybrid methods have substantial advantages over the more widely used minimal ensemble hybrid methods [26]. Traditionally, molecular simulations are used to generate a large ensemble of structures. These structures are used in a minimal ensemble search to obtain the fewest number of structures that best fit the experimental SAXS data. These minimal ensembles are highly unrealistic in that they represent the conformational flexibility of a protein by typically only two to four individual structures. By contrast, full ensemble methods are far more realistic in that they represent the conformational flexibility of a protein by thousands of individual structures, each related to one other by a series of time steps in a molecular simulation. In full ensemble methods, the simulations are generally not used to fit the experimental SAXS data [26]. Instead the experimental SAXS data is used simply to validate the simulations.
The present study of the conformational flexibility of Rad5 extends beyond prior full ensemble hybrid studies. This is because there are no experimentally determined structures of any region of Rad5 upon which to build starting models for the LD simulations. For this reason, we had to rely on disorder predictions and on homology modeling. Despite these obvious limitations, we still achieved the same remarkable agreement between the experimental SAXS data and the LD simulations as in prior studies with other proteins [24,25,47]. This remarkable agreement is a testament to the strength of the full ensemble hybrid approach, the accuracy of the LD simulations, the reliability of protein disorder prediction methods, and the accuracy of homology modeling methods.
Our initial LD simulations of Rad5 placed few constraints on the structure of Rad5. It enforced the structure of the three folded domains as well as the putative α-helices within the disordered regions. Interestingly, by comparing the theoretical Rg and Dmax values derived from these simulations with the experimental Rg and Dmax values obtained from the SAXS data, we found that Rad5 is actually more compact and less extended than indicated by the initial simulations. This suggested that Rad5 has an intra-molecular interaction that limits the range of conformational space it can sample to less extended states.
Given these considerations, we carried out refined LD simulations of Rad5 by placing one additional constraint on the starting model: an interaction between the HIRAN and helicase domains. While other intra-molecular interactions would also limit the range of conformational space to less extended states, we chose this particular one because it was the simplest given our knowledge of Rad5 structure. We positioned the HIRAN domain in two different locations on the helicase domain (one derived from ClusPro and the other from ZDOCK) to see if we could discriminate between these two models. We achieved excellent agreement between both of these refined LD simulations of Rad5 and the experimental SAXS data. We were not, however, able to discriminate between the two models. This is probably due to the resolution limitations of the experimental SAXS data. Thus, while one can conclude that the HIRAN and helicase domains likely interact with each other, we must await a high-resolution structure of Rad5 to understand the precise nature of this interaction.
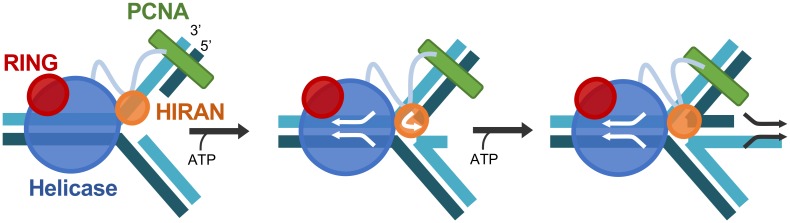
Further support for an intra-molecular interaction between the HIRAN and helicase domains comes from the crosslinking/mass spectrometry data, which shows direct interactions between the HIRAN and helicase domains. This has several important biological and mechanistic implications. First, the Rad5 helicase domain is a Swi/Snf superfamily 2 helicase. These helicase domains are believed to bind to and translocate along double-stranded DNA [48]. One key feature of superfamily 2 helicases is the presence of accessory domains that are associated with the helicase domain that gives each helicase in this superfamily its distinct function [48]. The association between the HIRAN and helicase domains of Rad5 suggests that the HIRAN domain may be the critical accessory domain needed for fork re-modeling. Support for this comes from the fact that the HIRAN domain binds the 3′ end of the DNA, presumably at the end of the primer strand on the leading strand of the replication fork [18]. It is possible that the HIRAN domain binds the primer terminus, while the helicase core domain translocates along the double-stranded DNA directly in front of the replication fork. This would result in the unwinding and peeling back of the primer stands from both the leading and lagging strands and would result in the formation of the chicken foot intermediate and regression of the replication fork (Fig 7). It should be pointed out, however, that this model is speculative as it is based full ensemble studies of Rad5 performed in the absence of the DNA substrate.
Fig 7. Model of Rad5-catalyzed fork re-modeling.
Illustration of a model of Rad5 converting a stalled replication fork to a chicken foot intermediate. Here, a replication fork that has stalled because of damage on the leading strand is shown with Rad5 bound to PCNA (green) via the Rad5 N-terminal tail. The RING domain is red, the helicase domain is blue, and the HIRAN domain is orange. ATP hydrolysis facilitates translocation of double-stranded DNA, which leads to unwinding of the replication fork. Arrows indicate the direction in which Rad5 moves the DNA strands. The HIRAN domain binds the 3’ end of the newly synthesized leading strand and redirects its pair with the newly synthesized lagging strand to form the chicken foot structure.
Second, Rad5 contains a PIP (PCNA-interacting protein)-like motif, which binds non-classical polymerase Rev1, a common protein present at stalled replication forks, and which likely binds PCNA [49]. This PIP-like motif (residues 9 to 17) is located at the beginning of the N-terminal disordered region of Rad5 [50]. As we have shown, this region has high conformational flexibility (Fig 5). Thus, Rad5 would be able to maintain protein-protein interactions with Rev1 or with PCNA without having significant constraints placed on the position and orientation of its catalytic core. For example, this N-terminal region could act as a tether providing the necessary flexibility to allow Rad5 first to participate in the poly-ubiquitylation of PCNA and then to move to the fork junction to participate in fork re-modeling without having to dissociate from Rev1, PCNA, or any other binding partner. In this way, the conformational flexibility of Rad5 may be critical to regulating and coordinating the two activities of this protein in template switching.
Supporting information
(PDF)
(PDF)
The integrated intensity is shown in blue. The Rg values for frames used in further analysis shown in black.
(PDF)
(A) Rg is plotted as a function of time for a 5 μs simulation. (B) Dmax is plotted as a function of time for a 5 μs simulation. (C) Individual structures from the resulting ensemble arranged in order of increasing Rg values.
(PDF)
The crosslinks in proximity to the ZDOCK position, which is shown in light pink, are indicated in dark pink. Crosslinks in proximity to the ClusPro position, which is shown in light green, are indicated in dark green.
(PDF)
The A280 is shown in black. The molar mass for peak 1 shown in red. The calculated molecular weight is 140 kDa, which is in close agreement with the expected molecular weight of the Rad5 monomer, which is 134 kDa.
(PDF)
Acknowledgments
We thank Adrian Elcock, Lokesh Gakhar, Christine Kondratick, Brittany Ripley, Devin Reusch, Justin Ling, and Maria Spies for discussions. We thank Srinivas Chakravarthy and Zhen Xu for assistance with SAXS data collection. We thank Joel Nott for assistance with the mass spectrometry data collection. The project described was supported by award number GM081433 to M.T.W. from the National Institute of General Medical Sciences (NIGMS) and award number F30ES028078 to M.S.G. from the National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH). We acknowledge the use of resources at the Carver College of Medicine’s Protein Crystallography Facility at the University of Iowa. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This project was supported by grant 9 P41 GM103622 from NIGMS of the NIH. Use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS. This research also used resources of the Protein Facility of the Iowa State University Office of Biotechnology. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS, NIEHS, or NIH.
Abbreviations
- Dmax
maximal distance
- HIRAN
HIP116 Rad5p N-terminal
- HLTF
helicase-like transcription factor
- LD
Langevin dynamics
- MALS
multi-angle light scattering
- MD
molecular dynamics
- PCNA
proliferating cell nuclear antigen
- PIP
PCNA-interacting protein
- pol η
DNA polymerase eta
- Rg
radius of gyration
- RING
really interesting new gene
- SAXS
small-angle X-ray scattering
- SEC
size exclusion chromatography
- SHPRH
SNF2 histone linker PHD RING helicase
- SUMO
small ubiquitin-like modifier
Data Availability
The minimal data set - which includes (a) the experimental SAXS data, (b) the experimental pair-wise distribution plot, (c) the fits of the SAXS data and pair-wise distribution plot to the ClusPro-derived model and the ZDOCK-derived model, (d) all three triplicate sets of snapshots from both the ClusPro-derived model (a total of 15,000 pdb files) and the ZDOCK-derived model (a total of 15,000 pdb files), and (e) a description of all relevant methods - was deposited in the Small Angle Scattering Biological Data Back (SASBDB) under to accession code SASDG25.
Funding Statement
The project described was supported by award number GM081433 to M.T.W. from the National Institute of General Medical Sciences (NIGMS) and award number F30ES028078 to M.S.G. from the National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This project was supported by grant 9 P41 GM103622 from NIGMS of the NIH. Use of the Pilatus 3 1M detector was provided by grant 1S10OD018090-01 from NIGMS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., and Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- 2.Stelter P., and Ulrich H. D. (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- 3.Bergink S., and Jentsch S. (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 10.1038/nature07963 [DOI] [PubMed] [Google Scholar]
- 4.Ulrich H. D., and Walden H. (2010) Ubiquitin signalling in DNA replication and repair. Nature reviews. Molecular cell biology 11, 479–489 10.1038/nrm2921 [DOI] [PubMed] [Google Scholar]
- 5.Dieckman L. M., Freudenthal B. D., and Washington M. T. (2012) PCNA structure and function: insights from structures of PCNA complexes and post-translationally modified PCNA. Sub-cellular biochemistry 62, 281–299 10.1007/978-94-007-4572-8_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehm E. M., Gildenberg M. S., and Washington M. T. (2016) The Many Roles of PCNA in Eukaryotic DNA Replication. The Enzymes 39, 231–254 10.1016/bs.enz.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prakash S., and Prakash L. (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes & development 16, 1872–1883 [DOI] [PubMed] [Google Scholar]
- 8.Prakash S., Johnson R. E., and Prakash L. (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annual review of biochemistry 74, 317–353 10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- 9.Lehmann A. R. (2005) Replication of damaged DNA by translesion synthesis in human cells. FEBS letters 579, 873–876 10.1016/j.febslet.2004.11.029 [DOI] [PubMed] [Google Scholar]
- 10.Lehmann A. R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing, et al. (2007) Translesion synthesis: Y-family polymerases and the polymerase switch. DNA repair 6, 891–899 10.1016/j.dnarep.2007.02.003 [DOI] [PubMed] [Google Scholar]
- 11.Guo C., Kosarek-Stancel J. N., Tang T. S., and Friedberg E. C. (2009) Y-family DNA polymerases in mammalian cells. Cellular and molecular life sciences: CMLS 66, 2363–2381 10.1007/s00018-009-0024-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waters L. S., Minesinger B. K., Wiltrout M. E., D’Souza S., Woodruff R. V., and Walker G. C. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiology and molecular biology reviews: MMBR 73, 134–154 10.1128/MMBR.00034-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Washington M. T., Carlson K. D., Freudenthal B. D., and Pryor J. M. (2010) Variations on a theme: eukaryotic Y-family DNA polymerases. Biochimica et biophysica acta 1804, 1113–1123 10.1016/j.bbapap.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sale J. E., Lehmann A. R., and Woodgate R. (2012) Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nature reviews. Molecular cell biology 13, 141–152 10.1038/nrm3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pryor J. M., Dieckman L. M., Boehm E. M., and Washington M. T. (2014) Eukaryotic Y-Family Polymerases: A Biochemical and Structural Perspective. Nucleic Acids Mol Bi 30, 85–108 [Google Scholar]
- 16.Powers K. T., and Washington M. T. (2018) Eukaryotic translesion synthesis: Choosing the right tool for the job. DNA repair 71, 127–134 10.1016/j.dnarep.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., and Haracska L. (2007) Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Molecular cell 28, 167–175 10.1016/j.molcel.2007.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kile A. C., Chavez D. A., Bacal J., Eldirany S., Korzhnev D. M., Bezsonova I., et al. (2015) HLTF’s Ancient HIRAN Domain Binds 3' DNA Ends to Drive Replication Fork Reversal. Molecular cell 58, 1090–1100 10.1016/j.molcel.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unk I., Hajdu I., Blastyak A., and Haracska L. (2010) Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA repair 9, 257–267 10.1016/j.dnarep.2009.12.013 [DOI] [PubMed] [Google Scholar]
- 20.Gangavarapu V., Haracska L., Unk I., Johnson R. E., Prakash S., and Prakash L. (2006) Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Molecular and cellular biology 26, 7783–7790 10.1128/MCB.01260-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ball L. G., Xu X., Blackwell S., Hanna M. D., Lambrecht A. D., and Xiao W. (2014) The Rad5 helicase activity is dispensable for error-free DNA post-replication repair. DNA repair 16, 74–83 10.1016/j.dnarep.2014.02.016 [DOI] [PubMed] [Google Scholar]
- 22.Ishida T., and Kinoshita K. (2007) PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic acids research 35, W460–464 10.1093/nar/gkm363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishida T., and Kinoshita K. (2008) Prediction of disordered regions in proteins based on the meta approach. Bioinformatics 24, 1344–1348 10.1093/bioinformatics/btn195 [DOI] [PubMed] [Google Scholar]
- 24.Powers K. T., Elcock A. H., and Washington M. T. (2018) The C-terminal region of translesion synthesis DNA polymerase eta is partially unstructured and has high conformational flexibility. Nucleic acids research 46, 2107–2120 10.1093/nar/gky031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powers K. T., Lavering E. D., and Washington M. T. (2018) Conformational Flexibility of Ubiquitin-Modified and SUMO-Modified PCNA Shown by Full-Ensemble Hybrid Methods. Journal of molecular biology 430, 5294–5303 10.1016/j.jmb.2018.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers K. T., Gildenberg M. S., and Washington M. T. (2019) Modeling Conformationally Flexible Proteins With X-ray Scattering and Molecular Simulations. Comput Struct Biotechnol J 17, 570–578 10.1016/j.csbj.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen S. S., Toft K. N., Snakenborg D., Jeppesen M. G., Jacobsen J. K., Vestergaard B., et al. (2009) BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Crystallogr 42, 959–964 [Google Scholar]
- 28.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr 36, 1277–1282 [Google Scholar]
- 29.Petoukhov M. V., Konarev P. V., Kikhney A. G., and Svergun D. I. (2007) ATSAS 2.1—towards automated and web-supported small-angle scattering data analysis. J Appl Crystallogr 40, S223–S228 [Google Scholar]
- 30.Svergun D. I. (1992) Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Crystallogr 25, 495–503 [Google Scholar]
- 31.McGinty R. K., Henrici R. C., and Tan S. (2014) Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 10.1038/nature13890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoma N. H., Czyzewski B. K., Alexeev A. A., Mazin A. V., Kowalczykowski S. C., and Pavletich N. P. (2005) Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature structural & molecular biology 12, 350–356 [DOI] [PubMed] [Google Scholar]
- 33.Willhoft O., Ghoneim M., Lin C. L., Chua E. Y. D., Wilkinson M., Chaban Y., et al. (2018) Structure and dynamics of the yeast SWR1-nucleosome complex. Science 362 [DOI] [PubMed] [Google Scholar]
- 34.Hauk G., McKnight J. N., Nodelman I. M., and Bowman G. D. (2010) The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Molecular cell 39, 711–723 10.1016/j.molcel.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farnung L., Vos S. M., Wigge C., and Cramer P. (2017) Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 10.1038/nature24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eustermann S., Schall K., Kostrewa D., Lakomek K., Strauss M., Moldt M., et al. (2018) Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390 10.1038/s41586-018-0029-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butryn A., Woike S., Shetty S. J., Auble D. T., and Hopfner K. P. (2018) Crystal structure of the full Swi2/Snf2 remodeler Mot1 in the resting state. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozakov D., Hall D. R., Xia B., Porter K. A., Padhorny D., Yueh C., et al. (2017) The ClusPro web server for protein-protein docking. Nat Protoc 12, 255–278 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierce B. G., Wiehe K., Hwang H., Kim B. H., Vreven T., and Weng Z. (2014) ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 10.1093/bioinformatics/btu097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frembgen-Kesner T., and Elcock A. H. (2009) Striking Effects of Hydrodynamic Interactions on the Simulated Diffusion and Folding of Proteins. Journal of chemical theory and computation 5, 242–256 10.1021/ct800499p [DOI] [PubMed] [Google Scholar]
- 42.Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL—A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28, 768–773 [Google Scholar]
- 43.Eng J. K., McCormack A. L., and Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5, 976–989 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- 44.Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 45.Bern M., Kil Y. J., and Becker C. (2012) Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics Chapter 13, Unit13 20 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research 42, W252–258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brosey C. A., Yan C., Tsutakawa S. E., Heller W. T., Rambo R. P., Tainer J. A., et al. (2013) A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic acids research 41, 2313–2327 10.1093/nar/gks1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beyer D. C., Ghoneim M. K., and Spies M. (2013) Structure and Mechanisms of SF2 DNA Helicases. Adv Exp Med Biol 767, 47–73 10.1007/978-1-4614-5037-5_3 [DOI] [PubMed] [Google Scholar]
- 49.Xu X., Lin A., Zhou C., Blackwell S. R., Zhang Y., Wang Z., et al. (2016) Involvement of budding yeast Rad5 in translesion DNA synthesis through physical interaction with Rev1. Nucleic acids research [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moldovan G. L., Pfander B., and Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 10.1016/j.cell.2007.05.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
The integrated intensity is shown in blue. The Rg values for frames used in further analysis shown in black.
(PDF)
(A) Rg is plotted as a function of time for a 5 μs simulation. (B) Dmax is plotted as a function of time for a 5 μs simulation. (C) Individual structures from the resulting ensemble arranged in order of increasing Rg values.
(PDF)
The crosslinks in proximity to the ZDOCK position, which is shown in light pink, are indicated in dark pink. Crosslinks in proximity to the ClusPro position, which is shown in light green, are indicated in dark green.
(PDF)
The A280 is shown in black. The molar mass for peak 1 shown in red. The calculated molecular weight is 140 kDa, which is in close agreement with the expected molecular weight of the Rad5 monomer, which is 134 kDa.
(PDF)
Data Availability Statement
The minimal data set - which includes (a) the experimental SAXS data, (b) the experimental pair-wise distribution plot, (c) the fits of the SAXS data and pair-wise distribution plot to the ClusPro-derived model and the ZDOCK-derived model, (d) all three triplicate sets of snapshots from both the ClusPro-derived model (a total of 15,000 pdb files) and the ZDOCK-derived model (a total of 15,000 pdb files), and (e) a description of all relevant methods - was deposited in the Small Angle Scattering Biological Data Back (SASBDB) under to accession code SASDG25.