Abstract
Among thousands of non-protein-coding RNAs which have been found in humans, a significant group represents snoRNA molecules that guide other types of RNAs to specific chemical modifications, cleavages, or proper folding. Yet, hundreds of mammalian snoRNAs have unknown function and are referred to as “orphan” molecules. In 2006, for the first time, it was shown that a particular orphan snoRNA (HBII-52) plays an important role in the regulation of alternative splicing of the serotonin receptor gene in humans and other mammals. In order to facilitate the investigation of possible involvement of snoRNAs in the regulation of pre-mRNA processing, we developed a new computational web resource, snoTARGET, which searches for possible guiding sites for snoRNAs among the entire set of human and rodent exonic and intronic sequences. Application of snoTARGET for finding possible guiding sites for a number of human and rodent orphan C/D-box snoRNAs showed that another subgroup of these molecules (HBII-85) have statistically elevated guiding preferences toward exons compared to introns. Moreover, these energetically favorable putative targets of HBII-85 snoRNAs are non-randomly associated with genes producing alternatively spliced mRNA isoforms. The snoTARGET resource is freely available at http://hsc.utoledo.edu/depts/bioinfo/snotarget.html.
Keywords: Genome, Mammals, Splicing, Exon, Intron, Non-coding RNA
1. Introduction
Pre-mRNA splicing in mammals and other multicellular eukaryotes is a very intricate process, involving hundreds of proteins as well as U1–U12 small nuclear RNA molecules, which self-organize into spliceosomes — particles comparable in size and complexity to ribosomes. In complex organisms with large genomes, spliceosomes must distinguish true splicing exon/intron junctions from many similar motifs, which are randomly dispersed along introns and are known as pseudoexons (Sun and Chasin, 2000). Moreover, the pattern of splicing often varies between different cell types and under different conditions and stages of development, producing alternative mRNA isoforms from the same gene (see reviews Wu et al., 2004; Matlin et al., 2005). In mammals, alternative splicing occurs in every second gene and increases the proteome diversity by a factor of three (Lee and Wang, 2005). How has this pre-mRNA processing achieved such intricacy and perfection? As early as 1979, it was proposed that non-coding RNA could play an important role in the determination of splicing sites (Murray and Holliday, 1979). However, the first experimentally confirmed description of non-coding RNA (besides U1–U12 snRNAs) involvement in splicing was published in 2006 (Kishore and Stamm, 2006). The authors demonstrated that an antisense element (ASE) of a human HBII-52 C/D-box snoRNA specifically pairs to a target site within alternative exon 5b of HTR2C — the serotonin receptor 2C gene (snoRNA molecules have been reviewed in Fatica and Tollervey, 2003, Bachellerie et al., 2002, Kiss 2002). This snoRNA interaction occurs over the exonic splicing silencer motif of pre-mRNA, making it dysfunctional. The masking of this splicing silencer promotes the inclusion of exon 5b into mature HTR2C mRNA. In addition to the regulation of alternative splicing, HBII-52 snoRNAs might be involved in HTR2C mRNA editing (Vitali et al., 2005). This interesting and functionally important group of HBII-52 snoRNAs is located in the imprinted region of chromosome 15q11–q13, which expresses a complex transcription unit known as IC–SNURF-SNRPN (Runte et al., 2001). Several strong yet indirect evidences testify that IC–SNURF-SNRPN is built from at least 148 exons that code for two proteins, SNURF and SmN spliceosomal protein (Runte et al., 2001; Gray et al., 1999; Ozcelik et al., 1992), as well as a segment of antisense non-coding RNA, which is probably involved in the development of imprinted properties of another gene (UBE3A) from the same locus (Runte et al., 2001). At the same time, introns from IC–SNURF-SNRPN transcription unit harbor a number of C/D-box snoRNAs, including 47 copies ofHBII-52, 27 copies ofHBII-85, 2 copies of HBII-438, and a single copy of HBII-13, HBII-436, and HBII-437. These snoRNAs have been designated as “orphan” since they do not have canonical guiding targets for chemical modification of rRNAs or snRNAs (Bachellerie et al., 2002). The deletion or dysfunction of the IC–SNURF-SNRPN locus causes Prader–Willi syndrome (PWS) — a clinically and genetically heterogeneous imprinting disorder with complex manifestations which has separate clinical phases (Bittel and Butler, 2005). PWS is due to genomic imprinting caused by a deficiency in expression of paternal 15q11–q13 genes through a paternal deletion or maternal 15 disomy (UPD). There are at least two subtypes for deletion (Types I and II) and phenotypic differences exist among individuals with UPD, either Type I or Type II deletions (Bittel and Butler, 2005; Butler et al., 2004). Presumably, the lack of snoRNA expression is responsible for some of PWS’s symptoms (Ding et al., 2005; Gallagher et al., 2002; Runte et al., 2005). A majority of snoRNAs within IC–SNURF-SNRPN are evolutionarily conserved in mammals (they are present in human, mouse, rat, and dog, yet in different copy numbers (Kishore and Stamm, 2006). In addition to IC–SNURF-SNRPN, dozens of other evolutionarily conserved orphan snoRNAs have been found in different locations throughout the human genome, as reviewed in (Bachellerie et al., 2002) or have recently been found by the new snoSeeker software (Yang et al., 2006). It was demonstrated that snoRNA can play different roles in the maturation of RNAs (Gerbi 1995; Brown et al., 2001; Barneche et al., 2001; Omer et al., 2000; Gaspin et al., 2000; Jady et al., 2004). Finally, thousands of snoRNA-like sequences were revealed inside human and rodent introns of protein-coding genes by computational approaches (Fedorov et al., 2005; Washietl et al., 2005; Weber, 2006; Yang et al., 2006; Luo and Li, 2007). These snoRNA-like sequences possess all features characteristic of real snoRNAs, hence, a number of them likely represent genuine and properly processed molecules. Therefore, it is plausible that, in addition to a sole example of HBII-52 snoRNA involvement in alternative splicing regulation, other orphan snoRNAs could have similar functions related to splicing.
In order to facilitate the investigation of possible involvement of orphan snoRNAs in regulation of splicing, we developed a computer program for broad usage, snoTARGET, which finds targets for snoRNAs within human and rodent protein-coding genes. snoTARGET operates from the Internet and is easy to use. While there are other applications for searching miRNA targets, including RNAhybrid (Krüger and Rehmsmeier, 2006), TargetScanS (Lewis, et al., 2005), TargetBoost (SaeTrom, et al., 2005), MicroTar (Thadani and Tammi, 2006), and NBmiRTar (Yousef, et al., 2007), these applications are specific to miRNA targets within mRNAs, and cannot be applied for studies involving splice site locations inside exons and introns. snoTARGET finds snoRNA targets within the entire set of exons and introns and calculates the distance from the target to the nearest splicing junctions, as well as the stringency of this RNA–RNA interaction. There are multiple choices for sorting the discovered snoRNA targets. Using our software, we examined in detail possible guiding sites for the C/D-box snoRNAs from the 15q11–q13 locus. It appears that the HBII-85 group of these molecules has elevated guiding specificity for exons compared to introns. Moreover, their guiding sites have strong and statistically significant association with alternatively spliced genes.
2. Materials and methods
2.1. snoTARGET software
We generated the first release of a computer program, snoTARGET, freely available on-line (http://hsc.utoledo.edu/depts/bioinfo/snotarget.html), which finds targets for snoRNA ASE within exon/intron sequences of human, mouse, and rat genes. As input, snoTARGET takes the ASE sequence and the number of allowed non-Watson–Crick G–U pairs for the ASE-target match. It is also possible to allow a single mismatch in the ASE at positions 2 and > 11, according to Chen et al. (2007). In addition, preferences for how the ASE targets are sorted must be specified, although defaults are present. A simple search will take about 40 s, while a more complicated sequence can take over a minute. When completed, it returns the total number of targets within exons and introns of the examined genome. For each target, it computes 1) the energy of interaction between the ASE and the target; 2) the consecutive number of the exon or intron containing the target and the exact position of the target in the gene; and 3) the distance from the target to the nearest splicing junction upstream or downstream of its position. An example of the output of the snoTARGET is shown in Fig. 1. The search is performed in parallel fashion in order to maximize computational efficiency. This is done using Sun Microsystem’s Grid Engine application (http://gridengine.sunsource.net/), which is used for distributing computational processes within computer clusters. A Perl script takes the web input and initiates the parallel operation by submitting Sun Grid Engine processes to twenty-five CPUs in our local computer cluster. Each process performs a search of the query sequence over a portion of the Exon–Intron Database (see below). The results of the 25 searches are then collated into the final output, which is an ASCII text file. The Perl script for snoTARGET, designed for a single CPU, can be viewed/downloaded from our web page (http://hsc.utoledo.edu/depts/bioinfo/snotarget.html). The algorithm for snoTARGET can be viewed by looking at this code, and the search can be modified as needed for specialized analyses. The algorithm utilizes Perl regular expressions to find the targets. Minimum free energy (MFE) is calculated by invoking the RNAcofold program from the Vienna RNA package, version 1.6.1 (Hofacker 2003), running with default parameters.
Fig. 1.
Example of the snoTARGET output for a single HBII-85-12 ASE target. The second line shows the position of the 5′-end of the target inside the gene; the consecutive number of exon or intron containing the target; and the distance from the 5′-end of the target to the nearest splicing junction upstream and downstream of the target, respectively. In case the target is within the first/last exon, the distance would be to the end of the gene. The third line shows the number of G–U pairs between the ASE and the target. The fourth line shows the calculated minimum free energy between the ASE and the target (kcal/mol). The fifth line presents the target sequence itself (in the middle) and also the 30 nucleotides adjacent to it from the 5′-end and from the 3′-end, respectively. Exons are shown in uppercase while the introns in lowercase. In this example, intron #24 ends in the middle of the 5′-adjacent sequence. Finally, at the bottom, the program prints the exon/intron information of the gene containing the target, which is the entire informational line of the gene from the Exon–Intron Database (Saxonov et al., 2000).
2.2. Datasets
snoTARGET uses the Exon–Intron Database (EID) as a source of mammalian exon and intron sequences (Saxonov et al., 2000). The snoTARGET algorithm takes advantage of the specialized database indexing scheme utilized within the EID, and uses exon/intron location information contained within the FASTA-comment line, and therefore is not appropriate to search against an arbitrary flat-file database. The latest updates and statistics on EID can be found in (Shepelev and Fedorov, 2006). The supplementary files showing all orphan C/D-box snoRNAs described in this paper can be viewed from our snoTARGET web page (http://hsc.utoledo.edu/depts/bioinfo/snotarget.html; file 15q11q13snoRNA.doc).
3. Results
3.1. Characterization of targets for human HBII-52 snoRNAs
HBII-52 snoRNAs are a group of 47 genes with high sequence similarity, which are produced from the same IC–SNURF-SNRPN transcriptional unit (Runte et al., 2001). Twenty-nine of HBII-52 snoRNAs have identical sequences of the second antisense element (ASE-2) located upstream of the D-box and designated as type-I (see supplementary file at the project Web site). We obtained these snoRNAs from the snoRNA-LBME Web site (Lestrade and Weber, 2006). Application of snoTARGET to the entire set of human genes confirmed the previous notion (Kishore and Stamm, 2006; Vitali et al., 2005; Cavaille et al., 2000) that an 18-nt-long ASE-2 of HBII-52 type-I snoRNA has a unique target in the HTR2C gene. This ASE perfectly matches a target in the alternative 5b exon of HTR2C and has a considerable energy of interaction between the guiding sequence and its target (minimum free energy (mfe)=−27.6 kcal/mol). No other targets of similar strength have been found among all human exons and introns. Even after shortening the length of the ASE by three nucleotides (two from the 5′-end and one from the 3′-end), the HTR2C target remains unique amidst the whole set of human exons and introns. In addition, the remarkable specificity of this particular snoRNA was also observed for mouse and rat homologs (MBII-52 and RBII-52, respectively). Because there is little overlap in the brain between the cell types where HBII-52 snoRNA and HTR2C mRNA are expressed (Vitali et al., 2005), evolution presumably resolved this problem by increasing the number of HBII-52 snoRNAs in the IC–SNURF-SNRPN transcriptional unit to 47 copies. Due to constant mutational flow, it is impossible to maintain identical sequences in all multiple copies. At present, 29 copies of type-I HBII-52 snoRNAs seem functional in humans while the remaining copies contain at least one nucleotide change in their ASE-2. These modified HBII-52 sequences could be divided into three types (II, III, and IV) based on their similarity (see supplementary file at the project web page). The majority of types II–IV HBII-52 representatives have ASE-2 sequences that differ by only 1–3 nucleotides from their counterpart in type-I sequences. Application of snoTARGET showed that these mutant HBII-52 snoRNAs from types II–IV do not exhibit any trace of exon-specificity that is significant for type-I HBII-52. The search for targets for types II–IV of HBII-52 snoRNAs resulted in a total of 122 matches within intronic sequences, and only 4 within exons, excluding HTR2C exon 5b (parameters: ASE length 15 nt; one G–U pair allowed). This corresponds well with random matches, since the total length of human introns is 37 times longer than the total length of exons.
3.2. Characterization of targets for the 27 human HBII-85 C/D-box snoRNAs
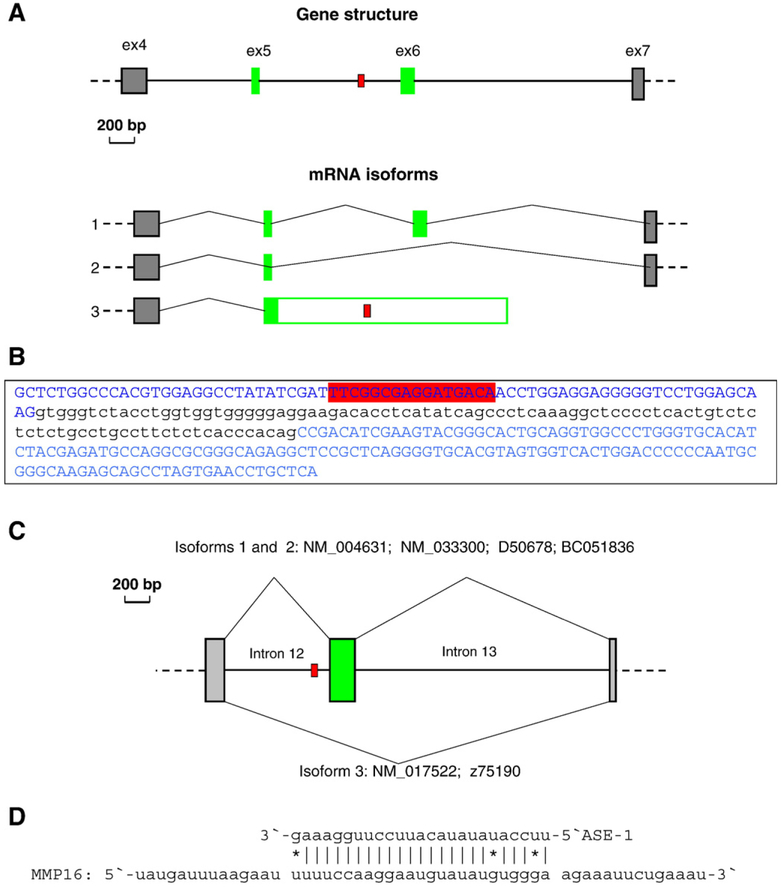
We applied snoTARGET for investigation of possible guiding sites for another group of 27 human HBII-85 snoRNAs, located in the same IC–SNURF-SNRPN transcription unit as the HBII-52 snoRNAs (Runte et al., 2001). These sequences were also obtained from the snoRNA-LBME Web site (Lestrade and Weber, 2006). Because the C′-box of all HBII-85 snoRNA (TGAGTG) differs considerably from its consensus (TGATGA) and the sequence between the D′- and C′-boxes (5′-ACAAAA-3′) does not have the ability to form stem-loop structure, the first antisense elements (ASE-1) located upstream of the D′-box could be non-functional. Therefore, we assumed that ASE-2 sequences located upstream of the D-box of HBII-85 molecules are the prime candidates for true guiding sequences. Using snoTARGET, we examined possible targets for 27 ASE-2 sequences from the entire group of HBII-85 snoRNAs. By default, we allowed one G–U mismatch and began our search with a relatively long (17 nt) ASE, whose 3′-end is located one nucleotide in front of the D-box. These stringent parameters usually produce few search results, if any. When zero targets had been found, we gradually reduced the length of the ASE until several targets came into view. It appeared that fourteen of these snoRNAs have significantly elevated specificity of their guidance toward exons compared to intronic sequences (results are presented in Table 1). Among them, eight HBII-85 snoRNAs (#1, 2, 3, 5, 6, 7, 19, and 22) have unique, strong targets exclusively within exons, while exhibiting no targets among the entire set of introns. Overall, for the targets shown in Table 1, eleven occur within alternatively splicing genes, according to the annotations in GenBank Build 35.1, and eleven targets occur within genes without annotated alternatively spliced isoforms. Since only 12% of the genes in this release of GenBank have annotated alternative splicing (Shepelev and Fedorov, 2006), this association of targets with alternatively spliced genes is statistically significant according to the binomial distribution (P-value<1×10−6). Even the most generous estimations of target association with alternatively spliced genes, which take into account gene lengths and other precautions, still give the P-value of this association less than 0.001. Two of the described targets for the HBII-85-13 and HBII-85-19 snoRNAs are inside the DRF1 and GTPBP3 genes, respectively, and are in close proximity to alternative splice junctions (see Fig. 2A,B). Moreover, among the thirteen HBII-85 snoRNAs (#4, 10, 11, 14, 16, 17, 18, 20, 21, 24, 25, 26, 27) that do not exhibit elevated specificity toward exons, and thus are absent in the Table 1, some targets are located very close to alternatively spliced junctions. One example is illustrated in Fig. 2C for the HBII-85-24 and HBII-85-25 snoRNAs, having their targets within intron #12 of the LRP8 gene (EID identifier 715A; ASE — GTGCCACTTCTGTGAg; target — ttcacagaagtggcat; mfe=−26.7 kcal/mol).
Table 1.
Exon-specific targets for HBII-85 C/D-box snoRNA genes
| SnoRNA | Common name of gene containing ASE target (protein RefSeq ID) |
The EID gene identifier; (exon containing target) |
Exon specificity | ASE sequence (5′->3′) |
Target sequence (5′->3′) |
mfe (kcal/mol) |
|---|---|---|---|---|---|---|
| HBII-85-1,2,6 | BAT1 (NP_004631.1) | 6563A; (ex #10) | +++ | CGUCAUUCUCAUCGGA | UCCGAUGAGAAUGAUG | −25.3 |
| HBII-85-1,2,6 | CDH23 (NP_071407.2) | 10252A; (ex #45) | +++ | GUCAUUCUCAUCGGAa | uUCUGAUGAGAAUGAC | −23.5 |
| HBII-85-3,8,9 | RASAL2 (NP_733793.1) | 1626A; (ex #8) | +− | GUUCUCAUCGGA | UCCGAUGAGAAC | −19.1 |
| HBII-85-3,8,9 | SH2BP1a (NP_055448.1) | 10875; (ex #25) | +− | GUUCUCAUCGGAa | uUCCGAUGAGAAC | −20.1 |
| HBII-85-3,8,9 | RGL3 (XP_290867) | 17313; (ex #9) | +− | GUUCUCAUCGGA | UCCGAUGAGAAC | −19.1 |
| HBII-85-3,8,9 | BAT1 (NP_004631.1) | 6563A; (ex #10) | + | GUCGUUCUCAUCGGA | UCCGAUGAGAAUGAU | −23 |
| HBII-85-5,7 | C8A (NP_000553.1) | 749; (ex #4) | +++ | GUCGUUCUCAUCAGA | UCUGAUGAGGACGAC | −24.1 |
| HBII-85-5,7 | CDH23 (NP_071407.2) | 10252A (ex #45) | +++ | GUCGUUCUCAUCAGAa | uUCUGAUGAGAAUGAC | −23.8 |
| HBII-85-12 | NCKAP1 (NP_038464.1) | 3161A; (ex #25) | +− | UGUCAUCCUCUUCAA | UUGAAGAGGAUGACA | −23.6 |
| HBII-85-12 | BAP1 (NP_004647.1) | 3929; (ex #13) | +− | CUGUCAUCCUCUUCAAa | uUUGGAGAGGAUGACAG | −26.5 |
| HBII-85-12 | NANS (NP_061819.2) | 9540; (ex #6) | +− | UGUCAUCCUCUUCAA | UUGAAGAGGAUGACA | −23.6 |
| HBII-85-13 | PPP1R8 (NP_054829.2) | 414A; (ex #5) | −+ | CAUCAUCCUCAUUGAa | uUCAGUGAGGAUGAUG | −24 |
| HBII-85-13 | RNF137b (NP_060543.5) | 10800; (ex #7) | −+ | CAUCAUCCUCAUUGA | UCAGUGAGGAUGAUG | −23 |
| HBII-85-13 | PITPNM2 (NP_065896.1) | 12794; (ex #5) | −+ | CAUCAUCCUCAUUGAa | uUCAAUGAGGAUGGUG | −23.4 |
| HBII-85-13 | DRF1c (NP_079380.1) | 16188B; (ex #5) | −+ | ACCAUCAUCCUCAUUGA | UCAGUGAGGAUGAUGGU | −28 |
| HBII-85-15 | LOC389550d (XP_371949.3) | 8076; (ex #1) | ++ | UCAUCCUCGUCGA | UCGACGAGGAUGA | −22.5 |
| HBII-85-15 | ERCC1 (NP_001974.1) | 17928A; (ex #1) | ++ | UCAUCCUCGUCGA | UCGACGAGGAUGA | −22.5 |
| HBII-85-15 | PPIL2 (NP_680481.1) | 19153A; (ex #12) | + | CAUCCUCGUCGA | UCGACGAGGAUG | −20.6 |
| HBII-85-19 | GTPBP3 (NP_598399.1) | 17468A; (ex #5) | +++ | UGUCAUCCUCGUCGAa | uUCGGCGAGGAUGACA | −27.3 |
| HBII-85-22 | NY-REN-41e (NP_542385.1) | 10957; (ex #1) | +++++ | UACCGUCGUCCUCGUCAAa | uUUGACGAGGACGACGGUG | −34.2 |
| HBII-85-22 | ANKRD11 (NP_037407.3) | 15508; (ex #9) | +++ | CGUCGUCCUCGUCAAa | uUUGAUGAGGACGACG | −26.4 |
| HBII-85-23 | NTRK2 (NP_006171.2) | 9438; (ex #3) | −+ | CGUCAUCUUCGUUGA | UCAACGAAGAUGAUG | −21.6 |
| HBII-52-type-I | HTR2C (NP_000859.1) | 20027; (ex #5) | +++++ | AUGCUCAAUAGGAUUACg | cGUAAUCCUAUUGAGCAU | −27.6 |
Column three shows the EID identifier for the gene containing the target, which is the necessary reference for obtaining the gene sequence from the Exon/Intron Database (Saxonov et al., 2000). Exon specificity (column 4) demonstrates the preference of an ASE toward guiding exons compared to introns: five pluses ‘+++++’ means super-specificity of the ASE toward exons (even when a 2–3 nt shorter ASE does not have targets among all human introns); ‘+++’ means that the ASE matches only exons and no introns; ‘+’ means that the ASE has approximately equal number of targets within exons and introns; ‘+−’ means that the ASE has 2–4 times more targets within introns than in exons; and ‘−+’ means that the ASE has 5–7 times more targets within introns than in exons. G–U non-Watson–Crick pairs between the ASE and its target in columns 5 and 6, respectively, are underlined and shown in red. If the match between the ASE and its target extends to the closest nucleotide adjacent to the D-box, then this nucleotide is shown in lowercase at the 3′-end of the ASE sequence.
Another common name for this gene is CTR9.
Another common name for this gene is TRIM68.
Another common name for this gene is DBF4B.
Another common name for this gene is LMOD2.
Another common name for this gene is CCDC34.
Fig. 2.
A. Location of the ASE target relative to the alternative splice junctions in the DRF1 gene. Homo sapiens DBF4 yeast homolog B gene (GenBank mRNA identifier NM_025104). The target is present in the 3′-terminal alternative exon of isoform #3 (mRNAs NM_025104 and AK023149); while it is within alternative introns of isoforms #1 (AF448801 and BC016158) and isoform #2 (BC033660). Exons are shown as boxes, whose colored regions represent coding segments and the white region in isoform #3 represents a 3′-untranslated region. Alternative exons are shown as green boxes. The location of the target is shown by a small, red box. Exons are numbered according to isoform 16188A in the Exon–Intron Database (Saxonov et al., 2000). B. Location of the ASE target relative to the alternative splice junctions in the GTPBP3. The GTP binding protein 3 gene (UniGene identifier Hs.334885). Exon #5 (73 nt) and Exon #6 (144 nt) are shown in uppercase and are blue; retained intron #5 is shown in lowercase and is black. This intron is present in isoform IV (NM_133644) and is excluded from other isoforms (e.g. NM_032620). The target site within Exon 5 is marked in red. C. Location of the ASE target relative to the alternative splice junctions in the LRP8 gene. The low-density lipoprotein receptor-related protein 8 precursor, apolipoprotein E receptor 2 (UniGene identifier Hs.576154). The target is present inside the 854-nt-long intron 12 of the LRP8 gene. On the diagram, the constitutive exons are shown as shadowed grey boxes and the alternatively skipped exon as a green box. The location of the target is shown by a small, red box. Introns are numbered according to isoform 717A in the Exon–Intron Database (Saxonov et al., 2000). D. Scheme of the 25-nt-long ASE-1 of HBII-85-13, -14, and -16-19 snoRNAs and its target within intron 4 of the matrix metalloproteinase 16 gene (MMP16). Asterisks show U–G base-pairing. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The energy of interaction between the majority of ASE and their targets presented in Table 1 are slightly less than the energy (mfe=−27.6 kcal/mol) for the HBII-52 type-I snoRNA and its target in the serotonin receptor 2C gene (see previous paragraph). However, two HBII-85 targets, one within the DRF1 gene and another within the NY-REN-41 gene, have stronger interactions with corresponding snoRNAs (HBII-85-13 (mfe=−28.0 kcal/mol) and HBII-85-22 (mfe=−34.2 kcal/mol), respectively) than the canonical example of HBII-52-type-I with its HTR2C target.
Examination of targets for HBII-85 snoRNA homologs in mouse (MBII-85) and rat (RBII-85) did not reveal any single evolutionarily conserved exonic target in the same gene for human and rodents, among those genes listed in Table 1.
In order to evaluate whether or not ASE-1 sequences of HBII-85 are functional, we also examined their possible targets. Because all of these ASE-1 are AT-rich (72% on average), not surprisingly the vast majority of their putative targets detected by snoTARGET are inside introns. Some of these putative targets for ASE-1 show significant nonrandomness in targeting non-coding parts of the genes. For example, analysis of ASE-1 of the group of six HBII-85-13,-14, and -16-19 snoRNAs shows a very strong matching site (mfe −35.9 kcal/mol). This 25-nt-long target locates within 48,807 bp of intron 4 of the matrix metalloproteinase 16 (MMP16) gene (Fig. 2D). This target is significantly removed from the nearest splicing junctions (2748 bp and 46,059 bp, respectively, to the 5′- and 3′-ends). It exhibits statistically significant non-randomness of matching with the described ASE-1 because the second strongest target only has a 17-nt length, with one G–U pair, and a mfe value of −20.40 kcal/mol. Therefore, the probability of finding such a 25-nucleotide-long match shown in Fig. 2D by chance in the entire human genome is less than 0.001. The results obtained by snoTARGET for ASE-1 of HBII-85 are presented in our supplementary files at the snoTARGET Web site (see Implementation section).
3.3. Characterization of targets for other groups of orphan C/D-box snoRNAs
The results obtained by snoTARGET for 14qI, 14qII, HBII13, HBII437, and HBII438 orphan C/D-box snoRNAs from 15q11–q13 are also presented in our supplementary files. These sequences were obtained from the snoRNA-LBME Web site as well (Lestrade and Weber, 2006). Most of the ASE of these snoRNAs are AT-rich, which is most likely the reason why a majority of their putative targets are located inside introns. No significant preference toward exons has been found for these groups of snoRNAs.
4. Discussion
4.1. Involvement of orphan snoRNAs in the regulation of alternative splicing
The pool of characterized mammalian orphan snoRNAs is expanding rapidly. There are about a hundred well-known orphan snoRNA genes in humans (Bachellerie et al., 2002). A few months ago, a new computational algorithm, snoSeeker, revealed 26 novel evolutionarily conserved (between human and rodents) snoRNA genes (Yang et al., 2006). In addition, there are thousands of snoRNA-like sequences inside mammalian introns (Fedorov et al., 2005). A vast majority of them are species-specific. Their location inside introns provides an opportunity to be co-transcribed with the harboring gene. Because snoRNA-like sequences have all the features characteristic of real snoRNAs, there is a good chance that they also undergo post-splicing processing and maturate into functional molecules. Thus, it is plausible that the involvement of snoRNA molecules in the regulation of gene expression is much broader than previously thought. We designed our snoTARGET program as a useful public tool for the investigation of orphan snoRNAs. A single CPU, command-line version of this program is available for download for individual customization. In addition, this program will be updated regularly and eventually will cover many more species and will search for possible targets in a broad range of non-protein-coding genes. For diversification of snoTARGET for different purposes, it has a complex sorting system for output results. Particularly, for those who are interested in snoRNAs guiding to exons or involvement in regulation of splicing, it is possible to sort the data by exons versus introns and/or by the distance to the nearest splicing junction.
Implementation of snoTARGET for the examination of possible targets for a group of 27 human orphan HBII-85 snoRNAs showed that half of them have significantly elevated guiding specificity toward exons compared to introns. Moreover, these exonic targets for HBII-85 snoRNAs are non-randomly associated with alternatively spliced genes (P-value<0.001). What is the reason for non-random guiding specificity in dozens of orphan snoRNAs toward exons rather than the much longer intronic sequences? Was this specificity created by a stochastic mutational process during millions of years, or have some molecular processes facilitated and sped-up this snoRNA-target recognition during evolution? Presumably, the answer to these questions to some extent lies in the difference in the GC-content of exons (53% GC for human) versus introns (42% GC). It is especially important to consider the frequency of the rarest CG-dinucleotide in the mammalian genome (a methylation and mutation-prone site), which is three times more abundant in coding versus non-coding sequences of mammals. Among the 23 ASE sequences listed in Table 1, there are 27 CG-dinucleotides. Taking into account that the average length of an ASE is 15 nt, the concentration of CG-dinucleotides is well above the average expectation (1/96 nts inside human introns, and 1/31 nts inside exons). Therefore, the guiding specificity of many orphan snoRNAs to exonic sequences should be at least in part due to GC-content and harboring of CG-dinucleotides. When we studied AT-rich ASE of orphan snoRNAs (ASE-1 of HBII-85, and ASEs of 14qI, 14qII, HBII-13 and HBII-438), we found that the majority of the possible targets are located inside introns.
Currently, we are studying a possible involvement of HBII-85 snoRNAs in the regulation of alternative splicing for several genes, listed in Table 1. To validate our results, we are evaluating posttranscriptional modifications of the predicted target genes by RT-PCR in subjects without functional snoRNAs, as in the case of patients with PWS. One case of splicing abnormality in the snoRNA-targeted gene in a PWS patient lacking the HBII-85 locus has already been found (paper by Talebizadeh, Theodoro, Fedorov, and Butler, in preparation).
4.2. Algorithms for snoRNA-target searching
Searching for guiding sites of orphan snoRNAs is ambiguous since nobody knows how long the base-pairing between a given ASE and its target is or how many mismatches are allowed between them. So far, only a sole example of a HBII-52 snoRNA that guides to the protein-coding gene of serotonin receptor 2C (Kishore and Stamm, 2006) is known. This snoRNA–pre-mRNA pairing has perfect Watson–Crick complementarity in the 18-nucleotide-long match, which is absolutely conserved for human, rodents, and dog. On the other hand, those snoRNAs that are known in hundreds of genes and guide to rRNA and snRNA chemical modifications, could have up to three non-Watson–Crick U–G pairs between the ASEs and their targets (Huttenhofer et al., 2004). In rare cases mismatches between ASE and their targets could be observed (Chen et al., 2007). The length of an ASE is also variable, from 9 to 20 nucleotides (Huttenhofer et al., 2004). Therefore, snoTARGET has an option to choose the maximal number of allowed G–U mismatches (0, 1, 2, 3, etc.). By default, we recommend to start with 1 allowed G–U pair and not to include the first nucleotide upstream of the D- or D′-boxes in the ASE sequence, since frequently it is not involved in the pairing of an ASE with its guiding site (Huttenhofer et al., 2004; Chen et al., 2007). These particular parameters resulted in a preference of many HBII-85 snoRNAs toward exons. Increasing the number of allowed G–U pairs up to 3 considerably increases the number of possible targets and diminishes the exon/intron preferences toward random matches (about one exonic target per 40 intronic, now in accordance with the exon/intron length proportion (1/40) in mammals). However, for this latter set of less stringent parameters, the advanced sorting of the targets by energy of ASE-target interaction, and exons versus introns, could be helpful in resolving the real targets as well as determining their specificity. Finally, for the interpretation of the obtained results that have too many intron targets, it is important to compare the expression profile of the targeted gene and the orphan snoRNAs. If they express in different tissues, then the target has no biological meaning.
Acknowledgements
We are grateful to Jason Bechtel from the Bioinformatics Program at the University of Toledo for his help in optimizing cluster parameters and management of our software. This project is supported by NSF Career grant MCB-0643542 “Investigation of intron cellular roles”.
Abbreviations:
- ASE
antisense element
- PWS
Prader–Willi syndrome
References
- Bachellerie JP, Cavaille J, Huttenhofer A, 2002. The expanding snoRNA world. Biochimie 84, 775–790. [DOI] [PubMed] [Google Scholar]
- Barneche F, Gaspin C, Guyot R, Echeverria M, 2001. Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2¢-O-methylation sites. J. Mol. Biol 311, 57–73. [DOI] [PubMed] [Google Scholar]
- Bittel DC, Butler MG, 2005. PWS: clinical genetics and molecular biology. Expert Rev. Mol. Med 7, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Clark GP, Leader DJ, Simpson CG, Lowe T, 2001. Multiple snoRNA gene clusters from Arabidopsis. RNA 7, 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Bittel DC, Kibiryeva N, Talebizadeh Z, Thompson T, 2004. Behavioral differences among subjects with Prader–Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics 113, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J, Buiting K, Kiefmann M, et al. , 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Perasso R, Qu LH, Amar L, 2007. Exploration of pairing constraints identifies a 9 base-pair core within box C/D snoRNA–rRNA duplexes. J. Mol. Biol 369, 771–783. [DOI] [PubMed] [Google Scholar]
- Ding F, Prints Y, Dhar MS, et al. , 2005. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader–Willi syndrome mouse models. Mamm. Genome 16, 424–431. [DOI] [PubMed] [Google Scholar]
- Fatica A, Tollervey D, 2003. Insights into the structure and function of a guide RNP. Nat. Struct. Biol 10, 237–239. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Stombaugh J, Harr MW, Yu S, Nasalean L, Shepelev V, 2005. Computer identification of snoRNA genes using a Mammalian Orthologous Intron Database. Nucl. Acids Res 33, 4578–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RC, Pils B, Albalwi M, Francke U, 2002. Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader–Willi syndrome. Am. J. Hum. Genet 71, 669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspin C, Cavaille J, Erauso G, Bachellerie JP, 2000. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol 297, 895–906. [DOI] [PubMed] [Google Scholar]
- Gerbi SA, 1995. Small nucleolar RNA. Biochem. Cell. Biol 73, 845–858. [DOI] [PubMed] [Google Scholar]
- Gray TA, Saitoh S, Nicholls RD, 1999. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA 96, 5616–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, 2003. Vienna RNA secondary structure server. Nucl. Acids Res 31, 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A, Cavaille J, Bachellerie JP, 2004. Experimental RNomics. A global approach to identifying small nuclear RNAs and their targets in different model organisms. Methods Mol. Biol 265, 409–428. [DOI] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T, 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol 164, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Stamm S, 2006. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311, 230–232. [DOI] [PubMed] [Google Scholar]
- Kiss T, 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148. [DOI] [PubMed] [Google Scholar]
- Krüger J, Rehmsmeier M, 2006. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucl. Acids Res 34, W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ, 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucl. Acids Res 34, 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Wang Q, 2005. Bioinformatics analysis of alternative splicing. Brief Bioinform 6, 23–33. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP, 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15. [DOI] [PubMed] [Google Scholar]
- Luo Y, Li S, 2007. Genome-wide analyses of retrogenes derived from the human box H/ACA snoRNAs. Nucl. Acids Res 35, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW, 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol 6, 386–398. [DOI] [PubMed] [Google Scholar]
- Murray V, Holliday R, 1979. A mechanism for RNA–RNA splicing and a model for the control of gene expression. Genet. Res 34, 173–188. [DOI] [PubMed] [Google Scholar]
- Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP, 2000. Homologs of small nucleolar RNAs in Archaea. Science 288, 517–522. [DOI] [PubMed] [Google Scholar]
- Ozcelik T, Leff S, Robinson W, et al. , 1992. Small nuclear ribonucleoprotein polypeptide N (SNRPN), an expressed gene in the Prader–Willi syndrome critical region. Nat. Genet 2, 265–269. [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K, 2001. The IC–SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet 10, 2687–2700. [DOI] [PubMed] [Google Scholar]
- Runte M, Varon R, Horn D, Horsthemke B, Buiting K, 2005. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader–Willi syndrome. Hum Genet. 116, 228–230. [DOI] [PubMed] [Google Scholar]
- SaeTrom OLA, Snove OJ, SaeTrom PAL, 2005. Weighted sequence motifs as an improved seeding step in microRNA target prediction algorithms. RNA 11, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Daizadeh I, Fedorov A, Gilbert W, 2000. EID: the Exon–Intron Database: an exhaustive database of protein-containing genes. Nucl. Acids Res 28, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelev V, Fedorov A, 2006. Advances in the Exon–Intron Database (EID). Brief Bioinformatics 7, 178–185. [DOI] [PubMed] [Google Scholar]
- Sun H, Chasin LA, 2000. Multiple splicing defects in an intronic false exon. Mol. Cell Biol 20, 6414–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Tammi M, 2006. MicroTar: predicting microRNA targest from RNA duplexes. BMC Bioinformatics, 7, S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, Basyuk E, Le Meur E, et al. , 2005. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol 169, 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Hüttenhofer A, Stadler PF, 2005. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nature Biotech. 23, 1383–1390. [DOI] [PubMed] [Google Scholar]
- Weber MJ, 2006. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2, e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Havlioglu N, Yuan L, 2004. In: Meyers RA (Ed.), 2nd edition. Alternatively spliced genes In encyclopedia of molecular cell biology and molecular medicine, Volume 1 Wiley-VCH, pp. 125–177. [Google Scholar]
- Yang JH, Zhang XC, Huang ZP, et al. , 2006. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 34, 5112–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M, Jung S, Kossenkov AV, Showe LC, Showe MK, 2007. Naïve Bayes for MicroRNA Target Predictions Machine Learning for MicroRNA Targets. Bioinformatics. Advance Access. October 8. [DOI] [PubMed] [Google Scholar]