Abstract
Mutations in RAS signaling pathway components cause diverse neurodevelopmental disorders, collectively called RASopathies. Previous studies have suggested that dysregulation in RAS–extracellular signal–regulated kinase (ERK) activation is restricted to distinct cell types in different RASopathies. Some cases of Noonan syndrome (NS) are associated with gain-of-function mutations in the phosphatase SHP2 (encoded by PTPN11); however, SHP2 is abundant in multiple cell types, so it is unclear which cell type(s) contribute to NS phenotypes. Here, we found that expressing the NS-associated mutant SHP2D61G in excitatory, but not inhibitory, hippocampal neurons increased ERK signaling and impaired both long-term potentiation (LTP) and spatial memory in mice, although endogenous SHP2 was expressed in both neuronal types. Transcriptomic analyses revealed that the genes encoding SHP2-interacting proteins that are critical for ERK activation, such as GAB1 and GRB2, were enriched in excitatory neurons. Accordingly, expressing a dominant-negative mutant of GAB1, which reduced its interaction with SHP2D61G, selectively in excitatory neurons, reversed SHP2D61G-mediated deficits. Moreover, ectopic expression of GAB1 and GRB2 together with SHP2D61G in inhibitory neurons resulted in ERK activation. These results demonstrate that RAS-ERK signaling networks are notably different between excitatory and inhibitory neurons, accounting for the cell type–specific pathophysiology of NS and perhaps other RASopathies.
INTRODUCTION
Dysregulation of the RAS–extracellular signal–regulated kinase (ERK) signaling pathway is associated with multiple neurodevelopmental disorders, which are collectively known as RASopathies, including Noonan syndrome (NS), neurofibromatosis, Costello syndrome, LEOPARD syndrome, cardio-facio-cutaneous syndrome, Legius syndrome, and others (1–3). Most of the mutations in RASopathies lead to hyperactivation of RAS-ERK signaling (4–7), and RASopathies share clinical symptoms such as growth delay and congenital heart defects (1, 2). RAS-ERK signaling is critically involved in synaptic plasticity, learning, and memory (8, 9). Accordingly, cognitive deficits, including learning disabilities and intellectual disability as well as synaptic plasticity impairments, are common in RASopathies (1, 2, 10, 11). Although RAS-ERK pathway is a ubiquitous signaling pathway, several studies have shown that RASopathies cause abnormalities specific to certain cell types in the brain. For example, haploinsufficiency of Nf1 enhances ERK signaling primarily in γ-aminobutyric acid (GABA)–secreting neurons (12, 13). Similarly, a mutant mouse harboring an NS-associated KRAS mutation showed enhanced ERK signaling specifically in GABAergic interneurons (14). RASopathy-associated deficits are not restricted to inhibitory neurons. For example, mutations in Syngap1 were shown to affect excitatory synaptic transmission (15). However, the determinants for these cell type–specific phenotypes in RASopathies remain unknown.
NS is relatively common among RASopathies (affecting 1 in 2500 live births), which is characterized by short stature, craniofacial problems, heart defects, and cognitive deficits (3, 6, 7, 16). SHP2 is a SRC homology 2 (SH2) domain–containing nonreceptor protein tyrosine phosphatase encoded by the PTPN11 gene (17, 18). SHP2 is required for full activation of RAS-ERK pathway in receptor tyrosine kinase and cytokine receptor signaling pathways, implying that SHP2 is a positive regulator for RAS signaling (17). Gain-of-function mutations in the PTPN11 gene, which hyperactivate RAS-ERK signaling, are responsible for the majority of NS cases (3, 6, 17). NS-associated PTPN11 mutations interrupt the interaction between the autoinhibitory N-terminal SH2 domain and the central catalytic domain, which results in the constitutive activation of SHP2 (19). SHP2 mutant also showed the increased binding affinity to its binding proteins such as GAB1 [GRB2 (growth factor receptor–bound protein 2)–associated binding protein 1], which contributes to sustained ERK activation in response to growth factors (20).
Knock-in mice expressing NS-associated SHP2 mutations show NS-like phenotypes including growth delay, heart defects, and spatial memory deficits (21–24). In addition, forebrain-specific knock-out of Ptpn11 also impairs spatial memory in mice, indicating that Ptpn11 plays an important role in memory processing (25). SHP2 is expressed in mitotically active cells in the developing brain but is restricted to neurons and activated astrocytes in the adult brain (26, 27). Shp2 expression in the adult mouse is not specific to a particular neuronal subset, and therefore, SHP2 is found in both excitatory and inhibitory neurons (25, 26, 28). Previous studies have used either mutant mice or adeno-associated virus (AAV)–mediated expression of NS-associated mutations, which cannot discriminate neuronal cell types (23, 25). Therefore, it is not clear which cell type is critically involved in the mutant SHP2–mediated deficits in synaptic plasticity and memory. In this study, we explored the cell type responsible for the synaptic plasticity and memory deficits in SHP2 mutant–associated NS.
RESULTS
Expressing SHP2D61G in hippocampal excitatory neurons impairs spatial learning and memory
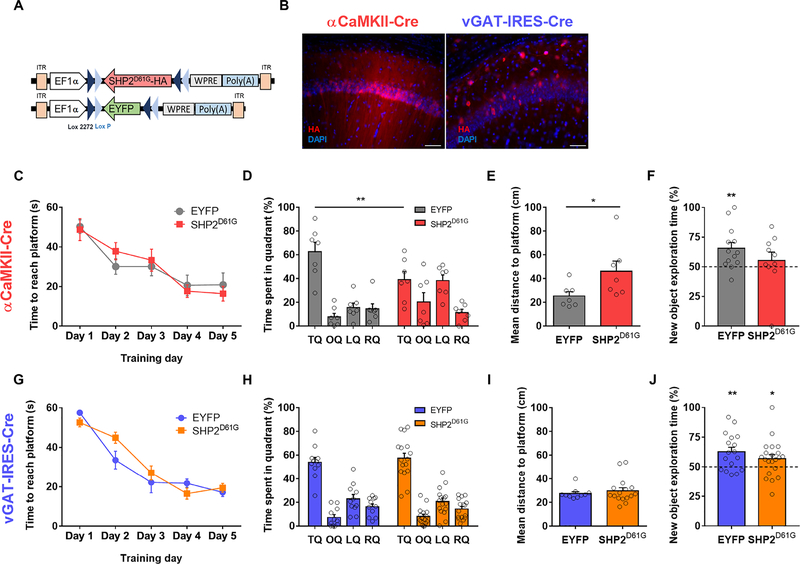
To investigate the underlying mechanism and define the cell type responsible for the deficits in synaptic plasticity and spatial memory associated with NS, we expressed the SHP2D61G mutant, which is found in a severe form of NS in a cell type–specific manner. SHP2D61G is a constitutively active gain-of-function mutant that increases the phosphatase activity of SHP2, as well as the basal activity of ERK (23, 29). We injected a Cre recombinase–dependent AAV vector expressing the SHP2D61G mutation into the dorsal hippocampus of either αCaMKII-Cre or vesicular GABA transporter (vGAT)–internal ribosomal entry site (IRES)–Cre mice (Fig. 1A). This strategy permitted selective expression of SHP2D61G in either αCaMKII+ excitatory or vGAT+ inhibitory neurons (30–32). Immunohistochemistry (IHC) analyses showed specific expression of hemagglutinin (HA)–tagged SHP2D61G in either pyramidal or nonpyramidal neurons in the hippocampal CA1 region of αCaMKII-Cre or vGAT-IRES-Cre mice, respectively (Fig. 1B).
Fig. 1. Expressing SHP2D61G in excitatory neurons impairs spatial memory.
(A) AAV constructs encoding Cre-dependent double-floxed inversed open reading frame HA-tagged αCaMKII-Cre::SHP2D61G or αCaMKII-Cre::EYFP. ITR, inverted terminal repeat sequence; WPRE, Woodchuck hepatitis virus (WHV) posttranscriptional regulatory element. (B) HA staining of SHP2D61G-expressing hippocampal slices from αCaMKII-Cre or vGAT-IRES-Cre mice. 4′,6-diamidino-2-phenylindole (DAPI) staining was used to identify nuclei. Scale bars, 100 μm. (C) Performance of αCaMKII-Cre::SHP2D61G and αCaMKII-Cre::EYFP mice in the MWM task. Data are means ± SEM from n = 7 mice per group; F1,12 = 0.01, P = 0.928 by two-way repeated measures analysis of variance (ANOVA). (D) Quadrant occupancy analysis for the probe test with mice described in (C). F3,36 = 5.459, **P < 0.01 by two-way repeated measures ANOVA with Bonferroni posttest. RQ, right quadrant; LQ, left quadrant; OQ, opposite quadrant. (E) Proximity to target platform (the average distance to the platform’s former location during the probe trial) by the mice described in (C). *P < 0.05 by unpaired t test. (F) Time exploring the relocated (new) object in OPR test by the mice described in (C), but with 13 (EYFP) and 10 (SHP2D61G) mice; **P < 0.01 and P = 0.486, respectively, compared to a hypothetical 50% (equal preference for new and old object), by one-sample t test. (G to J) As in (C) to (F) in mice overexpressing SHP2D61G in inhibitory neurons. Data are means ± SEM from n = 16 (G to I) or 21 (J) vGAT-IRES-Cre::SHP2D61G mice and n = 11 (G to I) or 17 (J) vGAT-IRES-Cre::EYFP mice. (G) F1,25 = 0.362, P = 0.553 by two-way ANOVA; (H) F3,75 = 0.257, P = 0.856 by two-way repeated measures ANOVA; (I) P = 0.523 by unpaired t test; (J) *P < 0.05 and **P < 0.01 by paired t test.
We compared the performance of αCaMKII-Cre::SHP2D61G mice and αCaMKII-Cre::enhanced yellow fluorescent protein (EYFP) control mice in the Morris water maze (MWM) task to test the effect of SHP2D61G expression in excitatory neurons on spatial learning and memory. αCaMKII-Cre::SHP2D61G and αCaMKII-Cre::EYFP mice exhibited comparable latencies to locate a hidden platform during training sessions (Fig. 1C). However, when spatial memory was assessed after 3 days of training (first probe test), αCaMKII-Cre::SHP2D61G mice spent less time in the target quadrant (TQ, where the platform was located during training sessions) than αCaMKII-Cre::EYFP mice (Fig. 1D). Moreover, αCaMKII-Cre::SHP2D61G mice searched farther from the target than control αCaMKII-Cre::EYFP mice in the first probe test (Fig. 1E), indicating that expressing SHP2D61G in αCaMKII+ neurons in the adult hippocampus is sufficient to produce spatial memory deficits. αCaMKII-Cre::SHP2D61G and αCaMKII-Cre::EYFP mice demonstrated comparable swimming speeds and similar total swimming distances in the first probe test (fig. S1, A and B), suggesting that SHP2D61G expression in hippocampal αCaMKII+ neurons does not impair motor function. αCaMKII-Cre::SHP2D61G showed comparable performance to αCaMKII-Cre::EYFP mice after 2 days of additional trainings (fig. S1, C and D), which suggests that αCaMKII-Cre::SHP2D61G mice can learn spatial memory tasks, but at a slower rate than control mice. Next, αCaMKII-Cre::SHP2D61G and αCaMKII-Cre::EYFP mice were subjected to object-place recognition (OPR) test, which is another hippocampus-dependent task (33). In the test session, 24 hours after training to test long-term memory, αCaMKII-Cre::EYFP mice showed preference for the relocated object, whereas αCaMKII-Cre::SHP2D61G mice did not (Fig. 1F). However, when the mice were tested 1 hour after training, both αCaMKII-Cre::SHP2D61G and αCaMKII-Cre::EYFP showed preference for the relocated object (fig. S2), demonstrating that short-term memory is intact in αCaMKII-Cre::SHP2D61G mice.
To examine the impact of SHP2D61G expression restricted to inhibitory neurons on learning and memory, we injected the floxed AAV vector encoding SHP2D61G or EYFP into the hippocampus of vGAT-IRES-Cre mice (Fig. 1B). vGAT-IRES-Cre::SHP2D61G mice performed comparably to control vGAT-IRES-Cre::EYFP mice during learning and probe trials (Fig. 1, G to I, and fig. S1, E and F). Moreover, both vGAT-IRES-Cre::SHP2D61G and vGAT-IRES-Cre::EYFP mice showed significant preferences for the relocated object in OPR testing (Fig. 1J). These data suggest that SHP2D61G expression in αCaMKII+, but not in vGAT+ hippocampal neurons, is critically involved in the hippocampus-dependent memory deficits associated with NS.
Expressing SHP2D61G in excitatory, but not in inhibitory neurons impairs long-term potentiation
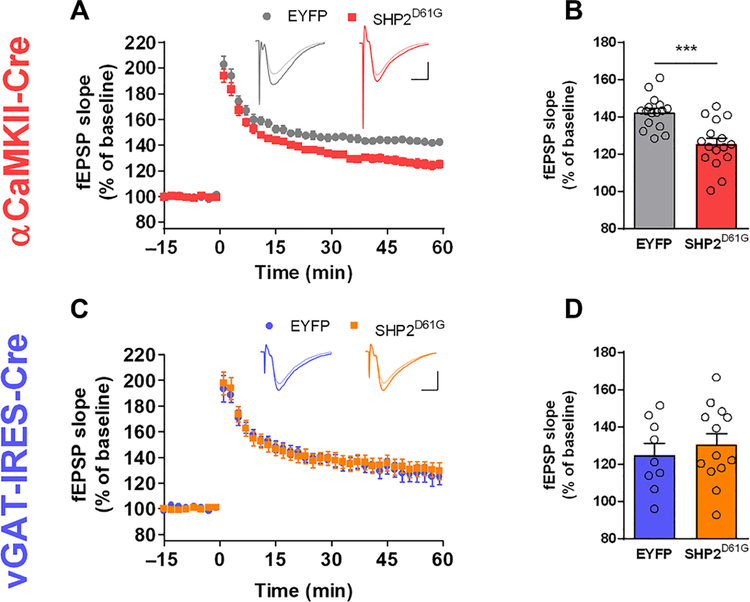
Hippocampal long-term potentiation (LTP), a cellular mechanism that is critical for spatial learning and memory, was shown to be impaired in mouse models of NS expressing SHP2D61G without cell type specificity (23). We recorded field excitatory postsynaptic potentials (fEPSPs) from hippocampal CA3-CA1 Schaffer collateral pathway to investigate the effect of cell type–specific expression of SHP2D61G on the synaptic plasticity. Expressing SHP2D61G in excitatory or inhibitory neurons did not affect basal synaptic transmission (fig. S3, A and C). Paired-pulse facilitation (PPF) ratios also did not differ between groups (fig. S3, B and D). LTP was significantly decreased in hippocampal slices from αCaMKII-Cre::SHP2D61G mice (Fig. 2, A and B). In contrast, vGAT-IRES-Cre::SHP2D61G and vGAT-IRES-Cre::EYFP mice showed similar readouts of LTP (Fig. 2, C and D). These results indicate that dysregulation of SHP2 in excitatory neurons is sufficient to impair long-term synaptic plasticity, a finding consistent with the behavioral phenotype of these mice.
Fig. 2. Expressing SHP2D61G in excitatory neurons impairs LTP.
(A) Time course of the fEPSP slope. LTP induced by theta burst stimulation (TBS; four bursts, each burst consisting of four stimuli at 100 Hz, 200-ms interburst interval) in αCaMKII-Cre::EYFP or αCaMKII-Cre::SHP2D61G slice. The fEPSP slopes were normalized to the average baseline. (B) The average fEPSP slope of 51 to 60 min after LTP induction. Average of last 10 min of LTP, αCaMKII-Cre::EYFP, n = 16 slices from 10 mice; αCaMKII-Cre::SHP2D61G, n = 16 slices from 10 mice; unpaired t test, ***P < 0.001. (C) Time course of the fEPSP slope. LTP induced by TBS in vGAT-IRES-Cre::SHP2D61G or vGAT-IRES-Cre::EYFP slices. The fEPSP slopes were normalized to the average baseline. (D) The average fEPSP slope of 51 to 60 min after LTP induction. vGAT-IRES-Cre::EYFP, n = 9 slices from seven mice; vGAT-IRES-Cre::SHP2D61G, n = 13 slices from eight mice; unpaired t test, P = 0.523.
SHP2D61G activates ERK in cell type–specific manner
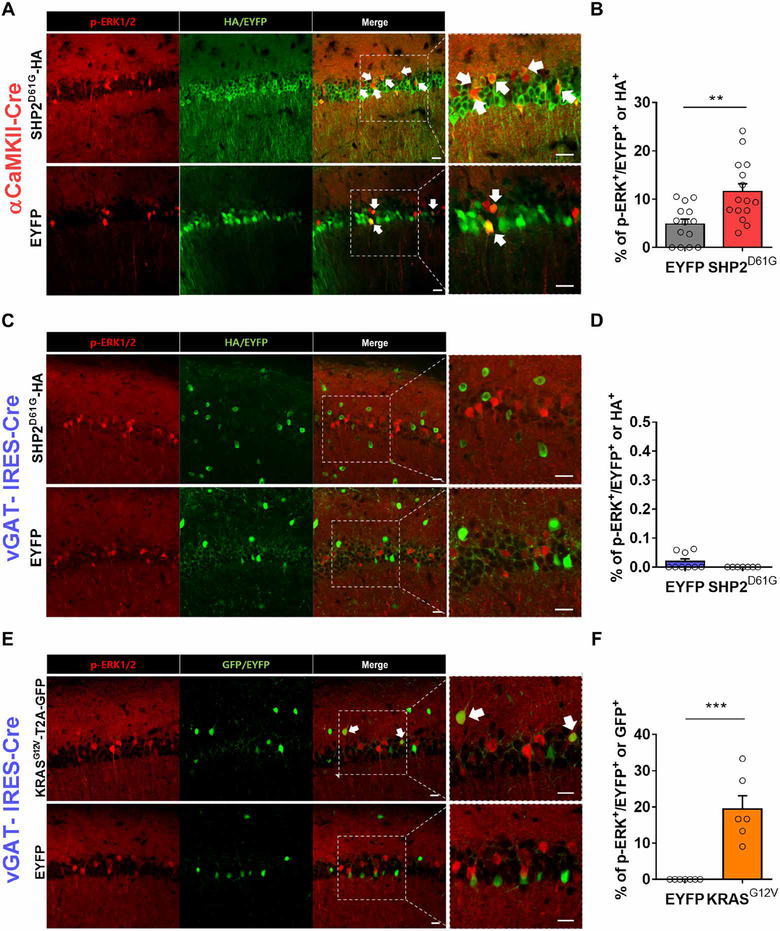
We then investigated how SHP2D61G mutation in αCaMKII+, but not in vGAT+ hippocampal neurons, selectively causes LTP and memory deficits. In a previous study, we demonstrated that SHP2D61G increases RAS-ERK signaling and subsequently impairs hippocampal LTP and learning in mice (23). Therefore, we hypothesized that SHP2D61G selectively causes ERK hyperactivation in αCaMKII+ but not vGAT+ neurons. To test this hypothesis, we examined the basal abundance of phosphorylated ERK1 and ERK2 (p-ERK1/2, or more simply p-ERK) in the hippocampal neurons expressing SHP2D61G in αCaMKII-Cre or in vGAT-IRES-Cre mice (Fig. 3, A to D). The probability of detecting p-ERK was significantly higher in SHP2D61G-expressing neurons than EYFP-expressing control neurons from αCaMKII-Cre mice (Fig. 3B). In contrast, SHP2D61G-and EYFP-expressing neurons from vGAT-IRES-Cre mice showed comparable probabilities of detecting p-ERK (Fig. 3D). In addition, we confirmed that SHP2D61G also did not increase ERK activation in another population of inhibitory neurons in parvalbumin (PV)–Cre mice (fig. S4), suggesting that SHP2D61G leads to ERK activation in αCaMKII+ but not in GABAergic inhibitory neurons such as vGAT+ and PV+ neurons. Last, to examine whether it is possible to increase ERK activation in inhibitory neurons by manipulating another ERK upstream regulator, we injected a floxed AAV vector encoding KRASG12V into the dorsal hippocampus of the vGAT-IRES-Cre mice. A previous study showed that KRASG12V increased ERK activity in interneurons in the mutant mice (14). Consistently, we found that the detection of p-ERK was significantly increased in KRASG12V-expressing neurons than in control-expressing neurons in vGAT-IRES-Cre mice, demonstrating that the finding that SHP2D61G failed to activate ERK in inhibitory neurons was not a false negative (Fig. 3, E and F). The total numbers of p-ERK1/2+ cells in the counted areas were not different between groups (fig. S5, A to C), and the numbers of cells expressing viral construct were also comparable within each cell type (fig. S5, D to F). Together, our data indicate that ERK activation is not affected by SHP2D61G expression in inhibitory neurons.
Fig. 3. SHP2D61G selectively activates RAS-ERK signaling in excitatory neurons.
(A) Representative IHC images from αCaMKII-Cre::SHP2D61G-HA and αCaMKII-Cre::EYFP mice. Slices were immunostained for p-ERK1/2 (red) and HA (green). Arrows indicate double labeling of p-ERK1/2 and SHP2D61G-HA and double labeling of p-ERK1/2 and EYFP. Higher-magnification images of boxed CA1 region are also shown (the fourth column). (B) Proportion of hippocampal neurons from the mice described in (A) that were p-ERK positive. αCaMKII-Cre::EYFP, n = 14 slices from four hippocampi; αCaMKII-Cre::SHP2D61G, n = 15 slices from four hippocampi; unpaired t test, **P < 0.01. (C and D) As described in (A) and (B) for vGAT-IRES-Cre::SHP2D61G and vGAT-IRES-Cre::EYFP hippocampi. n = 8 and 6 slices, respectively, from four hippocampi; unpaired t test, P = 0.104. (E and F) As described in (A) and (B) for vGAT-IRES-Cre::KRASG12V and vGAT-IRES-Cre::EYFP mouse hippocampal CA1 region. n = 6 and 7 slices, respectively, from four hippocampi; unpaired t test, ***P < 0.005. Scale bars, 20 μm. Data are means ± SEM.
Cell type–specific transcriptome analyses reveal distinct RAS-ERK signaling networks in excitatory and inhibitory neurons
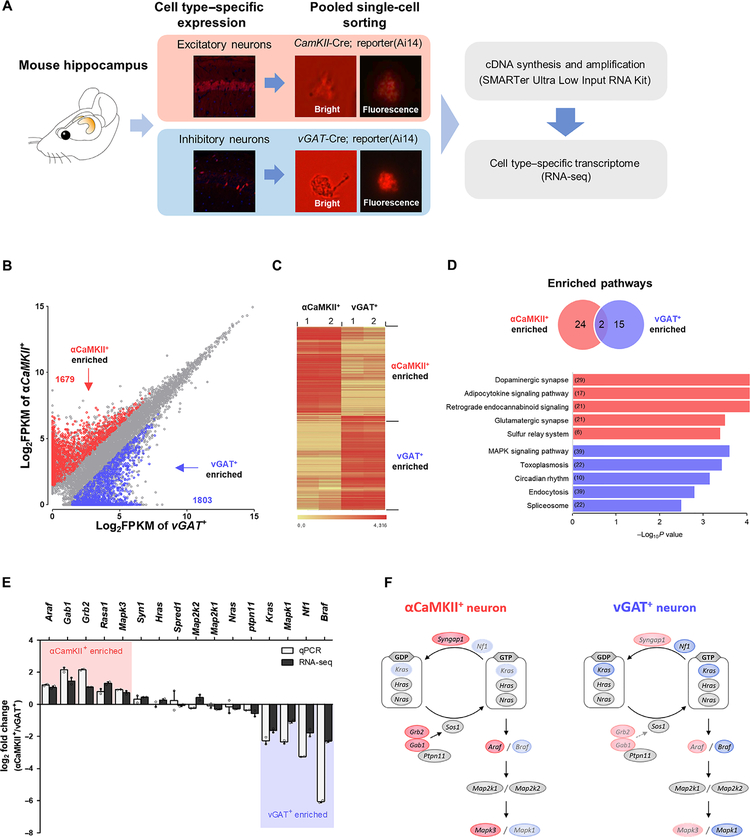
To investigate the mechanism responsible for the excitatory neuron–specific ERK activation by SHP2D61G, we examined whether RAS-ERK signaling–related genes including RASopathy-associated genes are differentially expressed between excitatory and inhibitory neurons in the mouse hippocampus. We generated cell type–specific reporter mice expressing tdTomato (Ai14 line) in excitatory neurons (αCaMKII-Cre) or in the inhibitory neurons (vGAT-IRES-Cre) in the adult hippocampus (30–32). Using fluorescent microscopy, the αCaMKII+ and vGAT+ neurons were manually collected from hippocampal lysates of the reporter mice (Fig. 4A). The quality of sorting was verified by confirming the expression pattern of well-known markers for excitatory and inhibitory neurons by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) (fig. S6A). After filtering the RNA sequencing (RNA-seq) data, 11,554 transcripts were obtained that cover 33.5% of the 34,535 Ensembl annotated transcripts [fig. S6, B and C; National Center for Biotechnology Information Gene Expression Omnibus (GEO) accession number GSE104089]. A principal components analysis revealed that biological replicates of each experimental group clustered together, confirming the high reproducibility between two libraries (fig. S6D). We identified 3482 differentially expressed genes (DEGs) (fold change > 2, P < 0.015) between two cell types (Fig. 4B and table S1). Unsupervised hierarchical clustering analysis showed a decisive shift in the neuronal cell type transcriptome in the form of genes with increased and reduced expression in excitatory and inhibitory neurons (Fig. 4C). The pathway enrichment analyses of the identified DEGs showed that glutamatergic synapse pathway and mitogen-activated protein kinase (MAPK) signaling pathway are enriched in excitatory and inhibitory neurons, respectively (Fig. 4D and table S2). Many of the RASopathy-associated genes, such as Braf, Cbl, Hras, Kras, Nf1, Rit1, and Sos2, were differentially expressed between αCaMKII+ and vGAT+ neurons (table S3). To validate the RNA-seq transcriptome data, the expression of 16 selected genes in RAS-ERK signaling were analyzed by using qRT-PCR, and we confirmed that the genes encoding key components of the RAS-ERK signaling pathway are differentially expressed between αCaMKII+ and vGAT+ neurons (Fig. 4E). These results demonstrate that RAS-ERK signaling networks are different between excitatory and inhibitory neurons (Fig. 4F).
Fig. 4. Cell type–specific transcriptome analyses reveal differential expressions of RAS signaling molecules.
(A) Workflow for cell type–specific transcriptome analysis. cDNA, complementary DNA. (B) Scatterplot illustrating genes enriched in αCaMKII+ neurons (red, 1679 transcripts) or in vGAT+ neurons (blue, 1803 transcripts) out of 11,554 transcripts. (C) Unsupervised hierarchical clustering analysis based on Pearson’s correlation of normalized fragments per kilobase million (FPKM) values shows clear segregation between two cell types. (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of 3482 DEGs. Venn diagram indicates a comparison of KEGG pathways enriched in two neuronal types. The bar graph indicates the top five KEGG pathways that are enriched in αCaMKII+ neurons (red bar) and in vGAT+ neurons (blue bar). The numbers of genes in each pathway are indicated in the bars. (E) Validation of RNA-seq data. Expression of genes were represented by log2 FC (fold change, αCaMKII/vGAT) of FPKM value and by log2 FC of ΔCt values normalized to β-actin level. The qRT-PCR and RNA-seq results represent the means of biological duplicates, which have technical triplicates. Red shading indicates enriched genes in αCaMKII+ neurons, and blue shading indicates enriched genes in vGAT+ neurons. (F) A schematic of RAS-ERK signaling in excitatory and inhibitory neurons based on the transcriptome data (genes take the place of proteins in the pathway). Genes in red are enriched in αCaMKII+ neurons, genes in blue are enriched in vGAT+ neurons, and gray represents similarly expressed genes.
SHP2D61G-GAB1 interaction mediates ERK hyperactivation
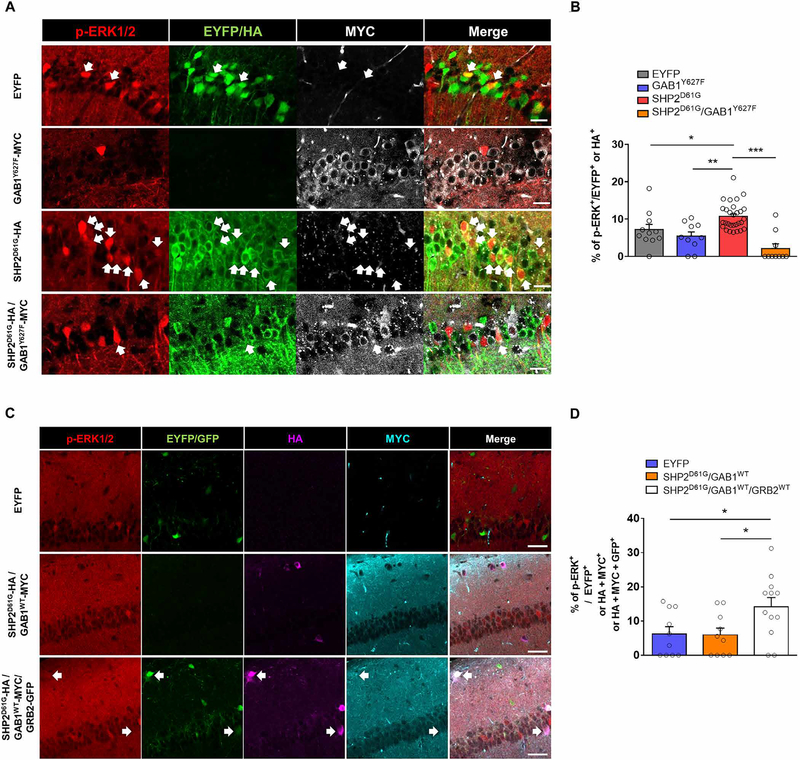
Although SHP2D61G caused ERK hyperactivation selectively in αCaMKII+ neurons, we found that Ptpn11 (encoding SHP2) was expressed at a similar abundance between these two neuronal types (Fig. 4E). We then searched for other candidate genes among αCaMKII+ neuron–enriched DEGs, which can bridge SHP2 and RAS. We found that the expression of the genes encoding two previously known SHP2-interacting proteins, GRB2, and GAB1, were significantly higher in αCaMKII+ neurons than in vGAT+ neurons (Fig. 4E). GAB1 is a member of the GAB family of docking proteins, which are critically involved in ERK activation (34, 35). Previously, a GAB1-SHP2 fusion protein was shown to hyperactivate RAS signaling, and a dominant-negative mutation of RAS was shown to block fusion protein–mediated RAS hyperactivity, indicating that the GAB1-SHP2 interaction promotes RAS-ERK signaling (36). GRB2, which also binds to RAS-activating protein son of sevenless (SOS), regulates cis interactions between the C-terminal phosphotyrosines and SH2 domain within SHP2 and subsequently regulates ERK signaling (18, 37). Because the αCaMKII+/vGAT+ ratio was higher for Gab1 than Grb2 (Fig. 4E), we decided to test whether GAB1 is a key bridging molecule between SHP2 and ERK in αCaMKII+ neurons by using a dominant-negative GAB1 mutant (GAB1Y627F) that has reduced binding affinity to SHP2 (35, 38). We also confirmed that GAB1 protein was more abundant in vGAT− neurons than in vGAT+ neurons (fig. S7). We first coexpressed SHP2D61G with GAB1Y627F in human embryonic kidney (HEK) 293T cells and confirmed that GAB1Y627F exhibited reduced interaction with SHP2D61G compared to that of wild-type GAB1 (fig. S8A). Furthermore, when coexpressed with SHP2D61G, GAB1Y627F significantly decreased ERK activation (fig. S8B). We examined whether reducing the SHP2D61G-GAB1 interaction in αCaMKII+ excitatory neurons could reverse SHP2D61G-mediated ERK activation. We confirmed that SHP2D61G expressing αCaMKII+ neurons increased the abundance of p-ERK1/2 (Fig. 5, A and B). Coexpressing GAB1Y627F significantly decreased the abundance of p-ERK1/2 in SHP2D61G-expressing αCaMKII+ neurons, whereas GAB1Y627F itself did not have a significant effect on ERK activation (Fig. 5B). Our results indicate that the interaction with GAB1 is necessary for SHP2D61G to activate ERK in αCaMKII+ excitatory neurons.
Fig. 5. Coexpressing GAB1Y627F reverses the SHP2D61G-mediated ERK hyperactivation in excitatory neurons.
(A) Immunostaining for p-ERK1/2 (red), HA (green), and MYC (white) in EYFP, GAB1Y627F-MYC, SHP2D61G-HA, and SHP2D61G-HA/GAB1Y627F-MYC expressing hippocampal slices from αCaMKII-Cre mice. Arrows indicate double labeling of p-ERK1/2 and EYFP, GAB1Y627F-MYC, SHP2D61G-HA, or SHP2D61G-HA/GAB1Y627F-MYC. Scale bars, 20 μm. (B) Proportion of p-ERK–positive neurons in each of the samples described in (A). Data are means ± SEM. EYFP, n = 11 slices from 10 hippocampi; GAB1Y627F, n = 10 slices from 10 hippocampi; SHP2D61G, n = 29 slices from 26 hippocampi, SHP2D61G/GAB1Y627F, n = 10 slices from 12 hippocampi; one-way ANOVA (P < 0.001) with Bonferroni’s multiple comparison test, *P < 0.05, **P < 0.01, and ***P < 0.005. (C and D) Representative IHC images, staining for p-ERK1/2 (red), HA (SHP2D61G; magenta), MYC (GAB1; cyan), and green fluorescent protein (GFP; GRB2, green) in hippocampal slices from vGAT-IRES-Cre::EYFP, vGAT-IRES-Cre::SHP2D61G/GAB1WT, and vGAT-IRES-Cre::SHP2D61G/GAB1WT/GRB2WT mice. Arrows indicate quadruple labeling of p-ERK1/2, SHP2D61G-HA, GAB1-MYC, and GRB2-GFP. Scale bars, 40 μm. (D) Proportion of p-ERK–positive neurons in each of the samples described in (C). Data are means ± SEM. vGAT-IRES-Cre::EYFP, n = 10 slices from four hippocampi; vGAT-IRES-Cre::SHP2D61G/GAB1WT, n = 10 slices from six hippocampi; vGAT-IRES-Cre::SHP2D61G/GAB1WT/GRB2WT, n = 12 slices from six hippocampi; one-way ANOVA, F2,29 = 4.235; P < 0.05, unpaired t test, *P < 0.05; EYFP versus SHP2D61G/GAB1WT, P = 0.916.
Then, we examined whether we can reconstitute SHP2-ERK signaling cascade in inhibitory neurons by ectopically expressing αCaMKII+ neuron–enriched genes to vGAT+ neurons. If GAB1 alone is sufficient for SHP2 to be coupled to ERK signaling network, introducing wild-type GAB1 together with SHP2D61G in vGAT+ neurons should increase p-ERK1/2 abundance in vGAT+ neurons as in αCaMKII+ neurons. However, neurons coexpressing SHP2D61G and GAB1WT did not show greater abundance of p-ERK than neurons expressing only SHP2D61G in vGAT-IRES-Cre mice (Fig. 5, C and D). Because there are multiple differentially expressed RAS-ERK pathway genes between excitatory and inhibitory neurons other than GAB1, it is highly likely that inhibitory neurons still lack other SHP2 downstream molecules required for SHP2 to activate ERK signaling (Fig. 4, E and F). We coexpressed another adaptor protein, GRB2, together with GAB1 and SHP2D61G in the hippocampus of vGAT-IRES-Cre mouse. When GRB2 and GAB1 were coexpressed in vGAT+ neurons, SHP2D61G-expressing neurons had increased abundance of p-ERK1/2 compared to control EYFP or GAB1-only expressing neurons in vGAT-IRES-Cre mice (Fig. 5, C and D). Together, our results demonstrate that SHP2D61G dysregulates the ERK signaling cascade only in excitatory neurons because inhibitory neurons lack the required adaptor proteins, such as GAB1 and GRB2.
Coexpressing GAB1Y627F in excitatory neurons reverses the SHP2D61G-mediated deficits in synaptic plasticity and spatial memory
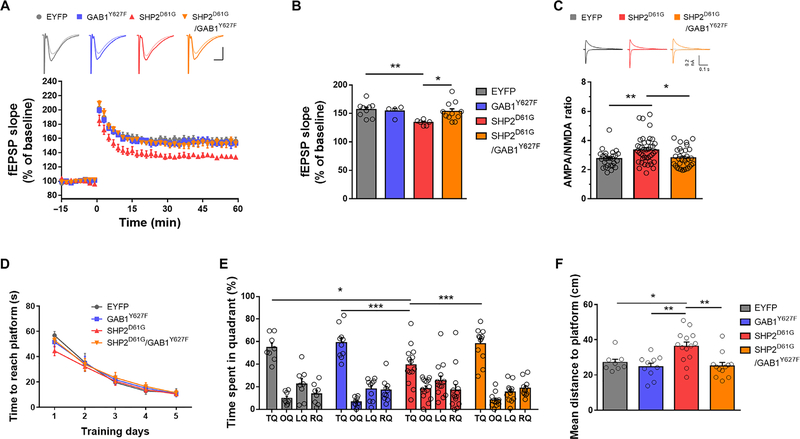
We examined whether expressing GAB1Y627F in αCaMKII+ neurons can rescue the LTP deficit in αCaMKII-Cre mice expressing SHP2D61G. We confirmed that αCaMKII-Cre::SHP2D61G mice exhibited a significant LTP deficit compared to that of control EYFP-injected αCaMKII-Cre mice (Fig. 6, A and B). Expressing GAB1Y627F in αCaMKII+ neurons rescued the LTP deficits of αCaMKII-Cre::SHP2D61G mice, whereas expression of GAB1Y627F alone did not affect LTP (Fig. 6B). This result indicates that reducing the SHP2-GAB1 interaction in excitatory neurons can rescue the LTP deficit caused by SHP2D61G expression.
Fig. 6. GAB1Y627F coexpression in excitatory neurons restores SHP2D61G-mediated LTP and memory deficits.
(A) LTP, as assessed by the time course (top) of the fEPSP slope (bottom), in hippocampal slices from αCaMKII-Cre mice expressing singly or coexpressing GAB1Y627F and SHP2D61G. Scale bars, 0.5 ms and 5 mV. Data are mean ± SEM; αCaMKII-Cre::EYFP, n = 9 slices from five mice; αCaMKII-Cre::GAB1Y627F, n = 4 slices from three mice; αCaMKII-Cre::SHP2D61G, n = 6 slices from three mice; αCaMKII-Cre::SHP2D61G/GAB1Y627F, n = 11 slices from six mice. (B) The average fEPSP slope of 51 to 60 min after LTP induction shown in (A). Two-way ANOVA with Bonferroni posttest, *P < 0.05 and **P < 0.01. (C) AMPA/NMDA ratio in SHP2D61G-expressing excitatory neurons and those coexpressing GAB1Y627F. Averages of 15 consecutive responses obtained at −70 mV (AMPA) and + 40 mV (NMDA) were used for the AMPA/NMDA ratio calculation. αCaMKII-Cre::EYFP, n = 27 cells from four mice; αCaMKII-Cre::SHP2D61G, n = 41 cells from five mice, αCaMKII-Cre::SHP2D61G/GAB1Y627F, n = 32 cells from four mice. Data are means ± SEM. One-way ANOVA with Bonferroni posttest, *P < 0.05 and **P < 0.01. (D) Latency to the platform in the MWM task during training trials. αCaMKII-Cre::EYFP, n = 8 mice; αCaMKII-Cre::GAB1Y627F, n = 10 mice; αCaMKII-Cre::SHP2D61G, n = 14 mice; and αCaMKII-Cre::SHP2D61G/GAB1Y627F, n = 12 mice. Data are means ± SEM. Two-way repeated measures ANOVA, P = 0.907. (E) Quadrant occupancy analysis for the probe test in mice expressing SHP2D61G and those coexpressing SHP2D61G and GAB1Y627F in hippocampal neurons. Data are means ± SEM. αCaMKII-Cre::EYFP, n = 8 mice; αCaMKII-Cre::GAB1Y627F, n = 10 mice; αCaMKII-Cre::SHP2D61G, n = 14 mice; αCaMKII-Cre::SHP2D61G/GAB1Y627F, n = 12 mice; two-way repeated measures ANOVA with Bonferroni posttest (time spent in TQ), *P < 0.05 and ***P < 0.005. (F) Proximity to target platform occupied by αCaMKII-Cre::GAB1Y627F mice (n = 10), αCaMKII-Cre::SHP2D61G mice (n = 14), αCaMKII-Cre::SHP2D61G/GAB1Y627F mice (n = 12), and an EYFP control group (n = 8). Data are means ± SEM. One-way ANOVA (P < 0.005) with Bonferroni’s multiple comparison test, *P < 0.05 and **P < 0.01.
We have previously reported that SHP2D61G increased both the surface expression of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit GluA1 and the AMPA/N-methyl-D-aspartate (NMDA) current ratio, which contribute to the LTP deficit in in Ptpn11D61G/− knock-in mice (23). Therefore, we examined whether SHP2D61G in excitatory neurons exploits the same cellular pathway to disrupt synaptic function by measuring AMPA/NMDA current ratio by performing whole-cell patch-clamp recordings. Consistently, we found that AMPA/NMDA ratio was increased in pyramidal neurons from αCaMKII-Cre::SHP2D61G mice compared to neurons from αCaMKII-Cre::EYFP (Fig. 6C). Moreover, coexpression of GAB1Y627F in αCaMKII+ neurons normalized the AMPA/NMDA ratio (Fig. 6C).
We tested whether reducing SHP2D61G-GAB1 interaction in αCaMKII+ excitatory neurons also improves spatial memory in SHP2D61G-expressing mice. We tested spatial memory of AAV-infused αCaMKII-Cre mice in MWM (Fig. 6, D to F). All four groups tested showed similar escape latencies during training sessions (Fig. 6D). Consistent with our LTP results, mice coexpressing SHP2D61G and GAB1Y627F in αCaMKII+ excitatory neurons spent significantly more time in the TQ than SHP2D61G-expressing mice and showed comparable performance to control groups (EYFP or GAB1Y627F alone) in the first probe test (Fig. 6E). In addition, αCaMKII-Cre::SHP2D61G/GAB1Y627F mice swam significantly closer to the platform location than αCaMKII-Cre::SHP2D61G mice during the probe test (Fig. 6F). With two additional days of training, all groups showed comparable performance in the second probe tests (fig. S9). These results demonstrate that reducing the interaction between SHP2D61G and GAB1 in αCaMKII+ excitatory neurons restores cognitive function in an NS mouse model.
DISCUSSION
Although cell type– and context-specific dysregulation of RAS signaling in RASopathies have been reported in the brain as well as in other organs, such as the heart, the underlying mechanisms of this specificity are largely not yet understood. For example, it has been shown that expressing an NS-associated SHP2 mutant only in the endocardium, but not in the myocardium or neural crest, resulted in cardiac defects (21). In the nervous system, deleting Nf1 encoding the guanosine triphosphatase activating protein neurofibromin 1 (NF1) promotes GABA release by increasing ERK-mediated phosphorylation of synapsin in the inhibitory neurons without affecting the excitatory synaptic transmission (12, 13, 39). Cell type–specific effects of RAS regulators have also been reported in glial cells (40). Induced pluripotent stem cells derived from a patient with Costello syndrome that harbored an HRASG12S mutation showed accelerated differentiation to glia, and knock-in mice expressing HRASG12S also showed dysregulation in astrocytic extracellular signaling (40). In the present study, we searched for the molecular mechanism that could account for the cell type–specific hyperactivation of RAS signaling by a SHP2 mutant found in NS by using single–cell type transcriptome analyses together with biochemical, behavioral, and electrophysiological approaches. Our transcriptome analysis provides compelling evidence supporting that the previously reported cell type–specific phenotypes are, at least in part, due to cell type–specific or cell type–enriched expression of RASopathy-associated genes. For example, we found that Nf1 is enriched in vGAT+ inhibitory neurons, which may explain why Nf1 mutant mice showed enhanced ERK activation primarily in GABAergic neurons (13). Furthermore, we found that Kras expression is also enriched in inhibitory neurons and consistently, mutant mice expressing a constitutively active KRAS mutant showed an increase in inhibitory synaptic functions (14).
We found that Ptpn11 (Shp2) expression was similar in murine αCaMKII+ neurons as in vGAT+ neurons. Nevertheless, expressing an NS-associated SHP2D61G in excitatory neurons was sufficient to increase ERK activation and to impair LTP and spatial memory in the mice, whereas expressing SHP2D61G in inhibitory neurons failed to increase ERK activation and did not affect LTP or spatial memory. In addition to vGAT+ neurons, SHP2D61G did not increase ERK activation in PV+ interneurons, either. As a control, we showed that expressing KRASG12V can increase ERK activation in vGAT+ neurons, strongly suggesting that SHP2D61G is not coupled to ERK pathway in hippocampal inhibitory neurons. These observations led us to ask whether differential expression of SHP2-interacting molecules could contribute to these cell type–specific phenotypes. Among differentially expressed RAS-ERK genes, we focused on SHP2-binding proteins such as GAB1 and GRB2. GAB1 was shown to be essential for some NS-associated SHP2 mutant–mediated ERK hyperactivation (20, 41). Previous reports have shown that NS-associated SHP2 mutants showed prolonged interaction with GAB1 compared to that of wild-type SHP2 upon the stimulation with epidermal growth factor (20, 22). Tyr627 and Tyr659 in GAB1 were reported to be important for GAB1 binding to SHP2, and mutations on these residues reduced the activation of the RAS-ERK signaling pathway (35, 38). We confirmed that GAB1Y627F suppresses ERK activation in SHP2D61G-expressing cells. We found that GAB1Y627F expression reversed SHP2D61G-mediated deficits in synaptic plasticity and memory in mice, demonstrating that the cell type–specific dysregulation of SHP2D61G-GAB1–ERK signaling in αCaMKII+ excitatory neurons causes deficits in an NS mouse model. In addition, our data suggest that SHP2 cannot be coupled to RAS-ERK pathway because of the lower expression level of adaptor proteins GAB1 and GRB2 in inhibitory neurons. Expressing GAB1 and SHP2D61G was insufficient to increase ERK signaling in vGAT+ neurons, but additional GRB2 expression together with SHP2D61G and GAB1 increased ERK activation in inhibitory neurons. These results also implicate that GRB2 functions as an effector molecule necessary for SHP2-GAB1–mediated ERK activation in excitatory neurons. SHP2 in inhibitory neurons might be involved in regulating other signaling pathways, such as phosphatidylinositol 3-kinase and Janus kinase–signal transducer and activator of transcription pathways, which remain to be explored (42, 43). In addition, we cannot exclude the possibility that SHP2D61G in inhibitory neurons caused other phenotypes in plasticity or behavior that we could not detect in this study. Although our results show that SHP2D61G does not affect RAS-ERK cascade in inhibitory neuron, it is worthy to note that the RAS-ERK cascade, which may be independent from SHP2 in inhibitory neuron, is also critically involved in synaptic plasticity and cognitive function (13, 14).
It was shown that SHP2D61G mutation induces aberrant activation of ERK signaling and dysregulates the AMPA receptor expression (23), which might contribute to the LTP deficit in excitatory neuron–specific SHP2D61G mice. Consistently, studies from the past few years showed that SHP2-ERK signaling pathway modulates the surface expression of glutamate receptors (44–47). However, SHP2 also regulates plasticity and memory through other mechanisms. For example, a study from last year showed that SHP2 phosphatase activity itself might be critical for glutamate receptor trafficking during homeostatic plasticity (48). It has been reported that a gain-of-function SHP2D61Y mutation attenuates the neuronal activity–dependent gene expression, which also can contribute to the deficits in NS (44). Other studies also suggested that SHP2 regulates NMDA receptor expression or function (24, 45). In this study, we demonstrated that reducing SHP2D61G-GAB1 interaction by expressing a dominant-negative GAB1Y627F successfully restored AMPA/NMDA ratio and rescued LTP deficit. Although the link between the excitatory neuron–specific SHP2D61G-ERK hyperactivation and LTP deficits still remains to be further investigated, our results suggest that SHP2D61G impairs LTP by affecting the excitatory synaptic function. Although distinct cell types and different genes are involved in various RASopathies, the therapeutic treatments for RASopathies have been largely focused on directly modulating the RAS-ERK pathway regardless of specific cell type or affected signaling regulators (1, 49, 50). These approaches were not always successful in treating cognitive dysfunction in patients or mutant mice (14, 51–53), and thereby, the development of an alternative effective treatment strategy is required. For example, blocking GABAergic transmission, but not ERK activation, reversed the deficits in LTP and behavior in adult mice expressing KRASG12V in neurons (14). Moreover, it has also been shown that RASopathy mutations affect more than RAS-ERK signaling in a mouse model of NS expressing SHP2D61Y mutation (44). Here, we demonstrated that selectively manipulating a cell type–enriched regulator, such as GAB1, in the affected cell type was sufficient to improve memory in an animal model of NS. Together, our study suggests that new treatments for cognitive deficits in RASopathies should selectively target the molecules in the affected cell types to gain specificity and effectiveness.
MATERIALS AND METHODS
Mice
αCaMKII-Cre mice (JAX 005359) were a gift from Y. Y. Kong at Seoul National University (SNU). vGAT-IRES-Cre (JAX 016962) and Ai14 mice (JAX 007914) were purchased from the Jackson laboratory. Mice were maintained by breeding with wild-type C57Bl/6J in the SNU Specific Pathogen Free center (LML08–404). Animals were group-housed (two to four mice per cage) on a 12-hour light/dark cycle in vivarium at Chung-Ang University (CAU) and SNU. All studies were approved by the Animal Research Committees at CAU and SNU.
Viral vectors and AAV packaging
SHP2D61G-HA was amplified by PCR using the following primers: 5′-cccgctagcgccaccatgacatcgcggagatgg-3′ and 5′-atggcgcgcctcaagcgtaatctggaacatcgtatgggtatctgaaacttttctgctgttg-3′. The sequence for the HA tag is underlined. The PCR product was digested with Nhe I and Asc I and ligated into the pAAV-EF1a-DIO-EYFP-WPRE plasmid. GAB1Y627F, GAB1WT, and GRB2-GFP were also digested with Nhe I and Asc I from the cytomegalovirus (CMV)–GAB1Y627F and pcDNA5/FRT/TO-GRB2-GFP plasmid and ligated into the pAAV-EF1a-DIO-EYFP-WPRE plasmid. CMV-GAB1Y627F was a gift from A. Yart (INSERM) (35). pcDNA5/FRT/TO-GRB2-GFP was a gift from Y. Ye (Addgene plasmid number 86873; http://n2t.net/addgene:86873; RRID: Addgene_86873) (54). AAV was prepared as previously described (55). For AAV packaging, HEK 293T cells (1.2 × 107) were plated on 150-mm culture dishes (Thermo Fisher Scientific 157150) with 15-ml D10 culture medium [Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, SH30243.01) + 10% fetal bovine serum (Thermo Fisher Scientific SH30919.03)] in an incubator at 37°C under 5% CO2 for 24 hours. Thirteen micrograms of p5E18-RxC1 plasmid and 26 μg of pAd-ΔF6 plasmid with 13 μg of pAAV plasmid (EYFP, SHP2D61G, GAB1Y627F, GAB1WT, and GRB2-GFP) were transfected into HEK 293 T cells by the CaPO4 transfection method. Cells were washed with DMEM 6 to 8 hours after transfection, and culture medium was replaced with 20-ml fresh D10 medium. After 72 hours, culture medium was harvested for AAV purification. Solutions were stacked in order (from top to bottom) in an ultracentrifuge tube (Beckman Coulter, 324214) as follows: culture medium from each dish, 6 ml of 15% iodixanol (OptiPrep; Axis-Shield, 1045) solution [1 M NaCl, 1 mM MgCl2, 2.5 mM KCl, and 25% OptiPrep in phosphate-buffered saline (PBS)], 5 ml of 25% iodixanol solution (1 mM MgCl2, 2.5 mM KCl, 0.2% phenol red, and 42% OptiPrep in PBS), 5 ml of 40% iodixanol solution (1 mM MgCl2, 2.5 mM KCl, and 67% OptiPrep in PBS), and 4 ml of 60% iodixanol solution (1 mM MgCl2, 2.5 mM KCl, and 0.2% phenol red in OptiPrep). Tubes were centrifuged at 69,000 rpm at 18°C for 1 hour using a Beckman UltimaTL-100 K ultracentrifuge and a 70Ti rotor. About 4 ml of 40% iodixanol solution was harvested from the centrifuged column using a 5-ml syringe (KOVAX ND.SY1030–005). The harvested solution was mixed with 11 ml of PBS and filtered with an Amicon ultra-15 filter tube (Millipore, UFC910024), and the filter was washed twice with 15 ml of PBS. The remaining solution was harvested, and viral particles in the solution were quantified using quantitative PCR (qPCR).
Stereotaxic viral injection
Male αCaMKII-Cre or vGAT-IRES-Cre mice (7 to 8 weeks) were anesthetized with ketamine/xylazine solution (130 and 10 mg/kg) and mounted on a stereotaxic frame. The hippocampal CA1 region was targeted using the following coordinates: anterior-posterior (AP): −1.8 mm, medial-lateral (ML): ±1.0 mm, dorsal-ventral (DV): −1.7 mm/AP: −2.5 mm, ML: ±2 mm, DV: −1.8 mm. AAV (0.5 μl of 4 × 1012 vg/ml) was injected into each point. All mice were allowed to recover for a minimum of 3 weeks before further use in experiments. Experimenters were blinded to the type of viral vector injected.
MWM test
The MWM test was performed as previously described (56). Mice were handled for 3 min at the same time of each day for seven consecutive days before testing. After handling, mice were placed into a gray opaque cylindrical tank (diameter, 140 cm; height, 100 cm) in a room with multiple spatial cues, including a water tap and a computer desk where the experimenter was seated. The tank was divided into four virtual quadrants, and a platform with a diameter of 10 cm was placed at the center of a TQ. The other three quadrants were named according to their relative position to the TQ. The tank was filled with water (20° to 22°C) until the water level was 1 cm higher than the platform. White paint was added to make the water murky. Before the first trial on training day 1, each mouse was placed onto the platform for 30 s. On training days, mice were released at the edge of the maze facing the inner wall of the tank and trained to reach the platform for 60 s. The release point was chosen at random for each trial. When mice failed to reach the platform, they were guided or placed onto the platform for 10 s and subsequently rescued from the maze. When mice successfully reached the platform and stayed on the platform more than 1 s, they were rescued from the maze after 10 s. Mice were trained with four trials per day for five consecutive days, and the interval between trials 1 and 2 or trials 3 and 4 was 1 min; between trials 2 and 3, the interval was 30 to 45 min. Probe tests were performed under the same conditions as the training trials, except the platform was absent, and mice were tracked for 1 min with a tracking program (EthoVision 3.1; Nodulus). The first probe test was performed on training day 4 before training, after which mice were trained for two more days. The second probe test was performed 24 hours after the training trials on training day 5. Experimenters were blinded to the type of viral vector injected in each mouse.
OPR test
Mice were handled for 5 min at the same time for four consecutive days and habituated in a cube-shaped acrylic box (32 cm by 32 cm by 32 cm) for 15 min for another 2 days before performing the test. One side of the box included a triangle-shaped local cue. In the training phase, mice were placed in the box containing two identical 100-ml glass bottles and were allowed to explore the objects for 10 min. Either 1 or 24 hours after training, in the test phase, mice were reexposed to the box containing the object that stayed in the same location and the other object that shifted to a new location. All locations for the objects were counterbalanced among groups, and objects were cleaned between trials. Sessions were videotaped and later analyzed manually. Experimenters were blinded to the type of injected viral vectors.
fEPSP recording
fEPSP recordings were performed as previously described (23). Mouse brains were sliced into sagittal sections (thickness, 400 μm) with a vibratome (Campden, 7000 smz-2) and incubated in artificial cerebrospinal fluid (ACSF; 120 mM NaCl, 3.5 mM KCl, 2.5 mM CaCl2, 1.3 mM MgSO4, 1.25 mM NaH2PO4, 10 mM glucose, and 26 mM NaHCO3, oxygenated with 95% O2 and 5% CO2) at room temperature for at least 30 min before the recording. Slices were transferred into a recording chamber, and fEPSPs were recorded from Schaffer collaterals in the CA3-CA1 pathway. A stimulation intensity of 30% of the maximum response was selected for these studies. Input-output ratios were presented by measuring the fEPSP slope at stimulation intensities (0 to 100 μA). The PPF ratio was analyzed over different intervals (10, 25, 50, 100, 200, and 400 ms). LTP was induced using a TBS protocol (four bursts, where each burst consisted of four pulses at 100 Hz and 200-ms interburst intervals). Data were recorded and analyzed using WinLTP software (WinLTP Ltd., Bristol, UK). Experimenters were blinded to the type of viral vector injected into each mouse.
Whole-cell patch-clamp recording
Transverse hippocampal slices (350 μm) were prepared using a vibratome (Leica, VT1200S) in ice-chilled slicing solution that contained 210 mM sucrose, 3 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 5 mM MgSO4, 10 mM D-glucose, 3 mM sodium ascorbate, and 0.5 mM CaCl2, saturated with 95% O2 and 5% CO2. The slices were transferred to an incubation chamber that contained the recording solution (ACSF: 124 mM NaCl, 3 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 2 mM MgSO4, 10 mM D-glucose, and 2 mM CaCl2, carbonated with 95% O2 and 5% CO2). Slices were allowed to recover at 32° to 34°C for 30 min and then maintained at 26° to 28°C for a minimum of 1 hour before recordings were made. Whole-cell patch-clamp recording performed at 32°C during continuous perfusion at 3 to 4 ml/min with ACSF that contained 100 μM picrotoxin (HelloBio) to prevent GABAAR transmission. CA1 pyramidal cells were visualized with infrared–differential interference contrast optics (Olympus). The whole-cell solution comprised 8 mM NaCl, 130 mM CsMeSO3, 10 mM Hepes, 0.5 mM EGTA, 4 mM Mg–adenosine triphosphate, 0.3 mM Na3–guanosine triphosphate, 5 mM QX-314, and 0.1 mM spermine. The pH was adjusted to 7.2 to 7.3 with CsOH, and osmolality was set to 285 to 290 mOsm/liter. Schaffer collateral-commissural pathway was stimulated at a constant frequency of 0.1 Hz. Borosilicate glass pipettes were used with a resistance of 4 to 6 megohms, and experiments were only accepted for analysis if series resistance values were <25 megohms and varied by 20% during the course of experiment. Signals were filtered at 10 kHz and digitized at 20 kHz using Multiclamp 700B (Molecular Devices). The peak amplitude of evoked EPSCs (pA) was monitored and analyzed using WinLTP and Clampfit. Cells were clamped at a holding potential of −70 mV to measure the peak of AMPAR-mediated synaptic transmission. NMDAR currents were measured at 50 ms after the stimulation onset. Averages of 15 consecutive responses obtained at these holding potentials were used for the AMPA/NMDA ratio calculation.
Immunohistochemical analysis
Mice were anesthetized with isoflurane (Hana Medical) and decapitated. Brains were fixed in a 4% paraformaldehyde (Sigma-Aldrich, P6148) solution in PBS for 24 hours and then transferred into a 30% sucrose (Sigma-Aldrich, S5391) solution for 48 hours. Brain samples were stored at −80°C until sectioning. Brain slices (30 to 40 μm) were cut using a cryostat and stored in a 50% glycerol PBS solution at −20°C. Brain slices were washed thrice with PBS (5 min per wash) and transferred into a blocking solution [4% goat serum (Rockland, D10400–0050), 0.2% Triton X-100 (Sigma-Aldrich, T8787) in PBS] for 1 hour at room temperature. Slices were then incubated with primary antibody [anti-HA rat immunoglobulin G (IgG), 1:50 (Roche, 11867423001); anti-MYC mouse IgG, 1:200 (Santa Cruz Biotechnology, sc-40); anti–SH-PTP2 rabbit IgG, 1:100 (Santa Cruz Biotechnology, sc-280); anti-PV mouse IgG, 1:500 (Millipore, MAB1572); or anti-somatostatin rat IgG, 1:100 (Millipore, MAB354)] in blocking solution for 48 hours at 4° to 10°C, washed thrice with PBS (5 min per wash), and incubated with the appropriate secondary antibodies [dilution range, 1:250 to 1:500; anti-rat IgG Alexa 568 conjugated (Life Technologies, A-11077), anti-rabbit IgG Alexa 488 conjugated (Life Technologies, A-11034), anti-mouse IgG Alexa 647 conjugated (Life Technologies, A-21235), anti-rat IgG Alexa 647 conjugated (Life Technologies, A-21247), or anti-mouse IgG Alexa 568 conjugated (Life Technologies, A-10037)]. Slices were then washed thrice with PBS (5 min per wash) and mounted between a slide glass and coverslip with Vectashield with DAPI (Vector Laboratories, H-1200). Slice images were acquired using a Zeiss LSM-700 confocal microscope with Zen software. Images were analyzed using ImageJ. During imaging and analysis of imaged data, experimenters were blinded to the type of viral vector injected into each mouse.
Library preparation and RNA-seq
Hippocampi were rapidly dissected from adult (8-to 9-week-old) male αCaMKII-Cre;Ai14 (tdTomato) and vGAT-IRES-Cre;Ai14 (tdTomato) mice, followed by the incubation in trypsin [0.05% in Hanks’ balanced salt solution (HBSS)] for 10 to 15 min at 37°C. Cells from both hippocampi from a mouse were pooled. Two mice were used for each group. Fluorescent cells were manually collected using a glass pipette. After washing in HBSS, collected cells were directly lysed with cell lysis buffer containing a ribonuclease inhibitor, following the manufacturer’s instruction of SMART-seq v4 (Ultra Low Input RNA Kit, Clontech Laboratories Inc.). After generation of cDNA, the construction of the library was performed by using Nextera XT DNA library prep kit (Illumina Inc.) according to the manufacturer’s instructions. High-throughput sequencing was performed as paired-end sequencing using HiSeq 2500 (Illumina Inc.). The list of qPCR primers used is listed in table S4.
Bioinformatic analyses
Transcript quantification of RNA-seq reads was performed with Cufflinks (ver. 2.1.1) by reads aligned to Ensemble v73 mouse transcriptome annotation (GRC.38.p1/Ensembl v73) using Bowtie2 (ver. 2.1.0) (57). The resulting alignments were used for assembling transcripts, estimating their abundances, and detecting differential expression of transcripts. Differentially expressed transcripts were determined on the basis of FPKM counts from unique and multiple alignments using EdgeR package in R (ver. 3.2.2). The FPKM values were processed on the basis of the quantile normalization method using Genowiz (ver. 4.0.5.6; Ocimum Biosolutions). Transcripts having greater than twofold change and P < 0.015 in any comparison were considered significantly differentially expressed and used for further analysis. Functional annotation and pathway analyses were performed by using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.ncifcrf.gov/home.jsp) and KEGG pathway, and the statistical significance level was set to a false discovery rate of < 0.25.
Statistical analysis
For MWM data, we used a two-way repeated measures ANOVA to determine whether there was a significant effect produced by the injected virus (EYFP versus SHP2D61G), with Bonferroni posttests to compare quadrant occupancies. Proximity measures between two groups were analyzed by an unpaired two-tailed t test. Learning curves were analyzed using a two-way repeated measures ANOVA. LTP data were analyzed using a repeated measures ANOVA, an unpaired two-tailed t test on averaged data collected in the last 10 min of recording, and a two-way ANOVA on averaged data collected in the last 10 min of recording, with Bonferroni posttests. For IHC data, one-way ANOVA with Newman-Keuls multiple comparison posttest was used to compare % of p-ERK1/2+ cells among viral construct–expressing cells. Robust regression and outlier removal (ROUT) test (Q = 0.5%) was used to identify outliers by using a software (GraphPad Prism 7.0) (58). Data distribution was assumed to be normal, but normality was not formally tested. All data are presented as means ± SEM.
Supplementary Material
Fig. S1. The second probe trials after extended trainings.
Fig. S2. Short-term memory test in object place recognition.
Fig. S3. Effects of expressing SHP2D61G in excitatory or inhibitory neurons on basal synaptic transmission and PPF ratio.
Fig. S4. Effect of SHP2D61G on ERK activation in PV+ neurons.
Fig. S5. The total number of p-ERK1/2+ neurons and viral vector–expressing cells were not significantly different between EYFP- and SHP2D61G-infected hippocampi.
Fig. S6. Validation of the quality of cell sorting and bioinformatic workflow.
Fig. S7. Comparison of GAB1 protein abundance in vGAT+ and vGAT− neurons in vGAT-Cre;tdTomato mice.
Fig. S8. The effect of GAB1Y627F on the interaction of SHP2D61G with GAB1 and ERK activation.
Fig. S9. The second probe trials after extended trainings in rescue experiments.
Table S1. List of 3482 DEGs.
Table S2. Functional annotation of 3482 DEGs.
Table S3. Expression profile of RASopathy-associated genes.
Table S4. Primer sequences for qRT-PCR validation.
Acknowledgments:
We thank A. Yart (INSERM, France) for sharing the GAB1 wild-type and Y627F constructs, K. Deisseroth (Stanford University, USA) for sharing the pAAV-EF1a-DIO-EYFP-WPRE plasmid, and Y. Y. Kong (SNU) for the αCaMKII-Cre mice. We would also like to thank B. G. Neel for critical discussion and S.-E. Sim and D. Han for technical help.
Funding: This work was supported by NRF-2016R1E1A1A01941939 and NRF-2017M3C7A1026959 to Y.-S.L., NRF-2016R1A4A1008035 to Y.-S.L. and J.-W.K., and the National Honor Scientist Program (NRF-2012R1A3A1050385) through a grant to B.-K.K. This study was also supported by Research Resettlement Fund for the new faculty of SNU.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq data were submitted to the GEO repository (GSE104089). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/12/571/eaau5755/DC1
REFERENCES AND NOTES
- 1.Rauen KA, The RASopathies. Annu. Rev. Genom. Hum. Genet 14, 355–369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tidyman WE, Rauen KA, The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev 19, 230–236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romano AA, Allanson JE, Dahlgren J, Gelb BD, Hall B, Pierpont ME, Roberts AE, Robinson W, Takemoto CM, Noonan JA, Noonan syndrome: Clinical features, diagnosis, and management guidelines. Pediatrics 126, 746–759 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Krab LC, Goorden SMI, Elgersma Y, Oncogenes on my mind: ERK and MTOR signaling in cognitive diseases. Trends Genet. 24, 498–510 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Stornetta RL, Zhu JJ, Ras and rap signaling in synaptic plasticity and mental disorders. Neuroscientist 17, 54–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tartaglia M, Gelb BD, Noonan syndrome and related disorders: Genetics and pathogenesis. Annu. Rev. Genomics Hum. Genet 6, 45–68 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Bentires-Alj M, Kontaridis MI, Neel BG, Stops along the RAS pathway in human genetic disease. Nat. Med 12, 283–285 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Sweatt JD, The neuronal MAP kinase cascade: A biochemical signal integration system subserving synaptic plasticity and memory. J. NeuroChem 76, 1–10 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Ye X, Carew TJ, Small G protein signaling in neuronal plasticity and memory formation: The specific role of Ras family proteins. Neuron 68, 340–361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borrie SC, Brems H, Legius E, Bagni C, Cognitive dysfunctions in intellectual disabilities: The contributions of the Ras-MAPK and PI3K-AKT-mTOR pathways. Annu. Rev. Genomics Hum. Genet 18, 115–142 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Mainberger F, Langer S, Mall V, Jung NH, Impaired synaptic plasticity in RASopathies: A mini-review. J. Neural Transm. (Vienna) 123, 1133–1138 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ, Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 415, 526–530 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, Gutmann DH, Parada LF, Mody I, Silva AJ, Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell 135, 549–560 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papale A, d’Isa R, Menna E, Cerovic M, Solari N, Hardingham N, Cambiaghi M, Cursi M, Barbacid M, Leocani L, Fasano S, Matteoli M, Brambilla R, Severe intellectual disability and enhanced gamma-aminobutyric acidergic synaptogenesis in a novel model of rare RASopathies. Biol. Psychiatry 81, 179–192 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Ozkan ED, Creson TK, Kramár EA, Rojas C, Seese RR, Babyan AH, Shi Y, Lucero R, Xu X, Noebels JL, Miller CA, Lynch G, Rumbaugh G, Reduced cognition in Syngap1 mutants is caused by isolated damage within developing forebrain excitatory neurons. Neuron 82, 1317–1333 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierpont EI, Tworog-Dube E, Roberts AE, Learning and memory in children with Noonan syndrome. Am. J. Med. Genet 161A, 2250–2257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neel BG, Gu H, Pao L, The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci 28, 284–293 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Tajan M, de Rocca Serra A, Valet P, Edouard T, Yart A, SHP2 sails from physiology to pathology. Eur. J. Med. Genet 58, 509–525 (2015). [DOI] [PubMed] [Google Scholar]
- 19.O’Reilly AM, Pluskey S, Shoelson SE, Neel BG, Activated mutants of SHP-2 preferentially induce elongation of Xenopus animal caps. Mol. Cell. Biol 20, 299–311 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fragale A, Tartaglia M, Wu J, Gelb BD, Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum. Mutat 23, 267–277 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Araki T, Chan G, Newbigging S, Morikawa L, Bronson RT, Neel BG, Noonan syndrome cardiac defects are caused by PTPN11 acting in endocardium to enhance endocardial-mesenchymal transformation. Proc. Natl. Acad. Sci. U.S.A 106, 4736–4741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, Neel BG, Mouse model of Noonan syndrome reveals cell type– And gene dosage–dependent effects of Ptpn11 mutation. Nat. Med 10, 849–857 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Lee Y-S, Ehninger D, Zhou M, Oh J-Y, Kang M, Kwak C, Ryu H-H, Butz D, Araki T, Cai Y, Balaji J, Sano Y, Nam CI, Kim HK, Kaang B-K, Burger C, Neel BG, Silva AJ, Mechanism and treatment for learning and memory deficits in mouse models of Noonan syndrome. Nat. Neurosci 17, 1736–1743 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy AD, Xiao X, Shaw JE, Sudarsana Devi SP, Katrancha SM, Bennett AM, Greer CA, Howe JR, Machida K, Koleske AJ, Noonan syndrome-associated SHP2 dephosphorylates GluN2B to regulate NMDA receptor function. Cell Rep. 24, 1523–1535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusakari S, Saitow F, Ago Y, Shibasaki K, Sato-Hashimoto M, Matsuzaki Y, Kotani T, Murata Y, Hirai H, Matsuda T, Suzuki H, Matozaki T, Ohnishi H, Shp2 in forebrain neurons regulates synaptic plasticity, locomotion, and memory formation in mice. Mol. Cell. Biol 35, 1557–1572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Servidei T, Bhide PG, Huang Z, Moskowitz MA, Harsh G, Reeves SA, The protein tyrosine phosphatase SHP-2 is expressed in glial and neuronal progenitor cells, postmitotic neurons and reactive astrocytes. Neuroscience 82, 529–543 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Reeves SA, Ueki K, Sinha B, Difiglia M, Louis DN, Regional expression and subcellular localization of the tyrosine-specific phosphatase SH-PTP2 in the adult human nervous system. Neuroscience 71, 1037–1042 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T, Matozaki T, Mizoguchi A, Kasuga M, Localization and subcellular distribution of SH-PTP2, a protein-tyrosine phosphatase with Src homology-2 domains, in rat brain. Biochem. Biophys. Res. Commun 211, 950–959 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG, Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J. Biol. Chem 280, 30984–30993 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S, Subregion- and cell type–restricted gene knockout in mouse brain. Cell 87, 1317–1326 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Vong L, Ye C, Yang Z, Choi B, Chua S Jr., Lowell BB, Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grienberger C, Milstein AD, Bittner KC, Romani S, Magee JC, Inhibitory suppression of heterogeneously tuned excitation enhances spatial coding in CA1 place cells. Nat. Neurosci 20, 417–426 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Oliveira AMM, Hawk JD, Abel T, Havekes R, Post-training reversible inactivation of the hippocampus enhances novel object recognition memory. Learn. Mem 17, 155–160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi-Tezuka M, Yoshida Y, Fukada T, Ohtani T, Yamanaka Y, Nishida K, Nakajima K, Hibi M, Hirano T, Gab1 acts as an adapter molecule linking the cytokine receptor gp130 to ERK mitogen-activated protein kinase. Mol. Cell. Biol 18, 4109–4117 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yart A, Laffargue M, Mayeux P, Chretien S, Peres C, Tonks N, Roche S, Payrastre B, Chap H, Raynal P, A critical role for phosphoinositide 3-kinase upstream of Gab1 and SHP2 in the activation of Ras and mitogen-activated protein kinases by epidermal growth factor. J. Biol. Chem 276, 8856–8864 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Cunnick JM, Meng S, Ren Y, Desponts C, Wang H-G, Djeu JY, Wu J, Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. J. Biol. Chem 277, 9498–9504 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Sun J, Lu S, Ouyang M, Lin L-J, Zhuo Y, Liu B, Chien S, Neel BG, Wang Y, Antagonism between binding site affinity and conformational dynamics tunes alternative cis-interactions within Shp2. Nat. Commun 4, 2037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunnick JM, Mei L, Doupnik CA, Wu J, Phosphotyrosines 627 and 659 of Gab1 constitute a bisphosphoryl tyrosine-based activation motif (BTAM) conferring binding and activation of SHP2. J. Biol. Chem 276, 24380–24387 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Shilyansky C, Karlsgodt KH, Cummings DM, Sidiropoulou K, Hardt M, James AS, Ehninger D, Bearden CE, Poirazi P, Jentsch JD, Cannon TD, Levine MS, Silva AJ, Neurofibromin regulates corticostriatal inhibitory networks during working memory performance. Proc. Natl. Acad. Sci. U.S.A 107, 13141–13146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krencik R, Hokanson KC, Narayan AR, Dvornik J, Rooney GE, Rauen KA, Weiss LA, Rowitch DH, Ullian EM, Dysregulation of astrocyte extracellular signaling in Costello syndrome. Sci. Transl. Med 7, 286ra66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hählen K, Hasle H, Licht JD, Gelb BD, Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet 34, 148–150 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Wu C-J, O’Rourke DM, Feng G-S, Johnson GR, Wang Q, Greene MI, The tyrosine phosphatase SHP-2 is required for mediating phosphatidylinositol 3-kinase/Akt activation by growth factors. Oncogene 20, 6018–6025 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Ernst M, Jenkins BJ, Acquiring signalling specificity from the cytokine receptor gp130. Trends Genet. 20, 23–32 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Altmüller F, Pothula S, Annamneedi A, Nakhaei-Rad S, Montenegro-Venegas C, Pina-Fernández E, Marini C, Santos M, Schanze D, Montag D, Ahmadian MR, Stork O, Zenker M, Fejtova A, Aberrant neuronal activity-induced signaling and gene expression in a mouse model of RASopathy. PLOS Genet. 13, e1006684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh J-Y, Rhee S, Silva AJ, Lee Y-S, Kim HK, Noonan syndrome-associated SHP2 mutation differentially modulates the expression of postsynaptic receptors according to developmental maturation. Neurosci. Lett 649, 41–47 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan X, Zhang B, Lu W, Peng L, Yang Q, Cao W, Lin S, Yu W, Li X, Ke Y, Li S, Yang W, Luo J, Increased Src family kinase activity disrupts excitatory synaptic transmission and impairs remote fear memory in forebrain Shp2-deficient mice. Mol. Neurobiol 54, 7235–7250 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Zhang B, du Y-L, Lu W, Yan X.-y., Yang Q, Yang W, Luo J.-h., Increased activity of Src homology 2 domain containing phosphotyrosine phosphatase 2 (Shp2) regulates activity-dependent AMPA receptor trafficking. J. Biol. Chem 291, 18856–18866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, Lu W, Src homology 2 domain–containing phosphotyrosine phosphatase 2 (Shp2) controls surface GluA1 protein in synaptic homeostasis. J. Biol. Chem 292, 15481–15488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huo X.-l., Min J.-j., Pan C.-y., Zhao C.-c., Pan L.-l., Gui F.-f., Jin L, Wang X.-t., Efficacy of lovastatin on learning and memory deficits caused by chronic intermittent hypoxiahypercapnia: Through regulation of NR2B-containing NMDA receptor-ERK pathway. PLOS ONE 9, e94278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bearden CE, Hellemann GS, Rosser T, Montojo C, Jonas R, Enrique N, Pacheco L, Hussain SA, Wu JY, Ho JS, McGough JJ, Sugar CA, Silva AJ, A randomized placebo-controlled lovastatin trial for neurobehavioral function in neurofibromatosis I. Ann. Clin. Transl. Neurol 3, 266–279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krab LC, de Goede-Bolder A, Aarsen FK, Pluijm SMF, Bouman MJ, van der Geest JN, Lequin M, Catsman CE, Arts WFM, Kushner SA, Silva AJ, de Zeeuw CI, Moll HA, Elgersma Y, Effect of simvastatin on cognitive functioning in children with neurofibromatosis type 1: A randomized controlled trial. JAMA 300, 287–294 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Vaart T, Plasschaert E, Rietman AB, Renard M, Oostenbrink R, Vogels A, de Wit M-CY, Descheemaeker M-J, Vergouwe Y, Catsman-Berrevoets CE, Legius E, Elgersma Y, Moll HA, Simvastatin for cognitive deficits and behavioural problems in patients with neurofibromatosis type 1 (NF1-SIMCODA): A randomised, placebo-controlled trial. Lancet Neurol. 12, 1076–1083 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Schreiber J, Grimbergen L-A, Overwater I, van der Vaart T, Stedehouder J, Schuhmacher AJ, Guerra C, Kushner SA, Jaarsma D, Elgersma Y, Mechanisms underlying cognitive deficits in a mouse model for Costello syndrome are distinct from other RASopathy mouse models. Sci. Rep 7, 1256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y, Anderson DE, Ye Y, The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation. Cell Discov. 2, 16040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi J-H, Yu N-K, Baek G-C, Bakes J, Seo D, Nam HJ, Baek SH, Lim C-S, Lee Y-S, Kaang B-K, Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol. Brain 7, 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S, Kim T, Lee H-R, Jang E-H, Ryu H-H, Kang M, Rah S-Y, Yoo J, Lee B, Kim J-I, Lim CS, Kim SJ, Kim U-H, Lee Y-S, Kaang B-K, Impaired learning and memory in CD38 null mutant mice. Mol. Brain 9, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL, Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Motulsky HJ, Brown RE, Detecting outliers when fitting data with nonlinear regression – A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 7, 123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The second probe trials after extended trainings.
Fig. S2. Short-term memory test in object place recognition.
Fig. S3. Effects of expressing SHP2D61G in excitatory or inhibitory neurons on basal synaptic transmission and PPF ratio.
Fig. S4. Effect of SHP2D61G on ERK activation in PV+ neurons.
Fig. S5. The total number of p-ERK1/2+ neurons and viral vector–expressing cells were not significantly different between EYFP- and SHP2D61G-infected hippocampi.
Fig. S6. Validation of the quality of cell sorting and bioinformatic workflow.
Fig. S7. Comparison of GAB1 protein abundance in vGAT+ and vGAT− neurons in vGAT-Cre;tdTomato mice.
Fig. S8. The effect of GAB1Y627F on the interaction of SHP2D61G with GAB1 and ERK activation.
Fig. S9. The second probe trials after extended trainings in rescue experiments.
Table S1. List of 3482 DEGs.
Table S2. Functional annotation of 3482 DEGs.
Table S3. Expression profile of RASopathy-associated genes.
Table S4. Primer sequences for qRT-PCR validation.