Abstract
A disintegrin and metalloproteinase 17 (ADAM17) has become a novel biomarker and potential therapeutic target for the early detection and treatment of human cancers. In this work, by covalently attaching fluorescent labeled ADAM17 substrate peptide (Pep-FAM) molecules to carboxylated graphene oxide (cGO), and monitoring the cleavage of the peptide substrate by ADAM17, we developed a cGO-Pep-FAM fluorescent sensor for the rapid, sensitive and accurate detection of ADAM17. The sensor was highly sensitive with a detection limit of 17.5 picomolar. Furthermore, the sensor was selective: structure similar proteases such as ADAM9 and MMP-9 would not interfere with ADAM17 detection. In addition, simulated serum samples were successfully analyzed. Our developed cGO-Pep-FAM sensing strategy should find useful applications in disease diagnosis and drug screening.
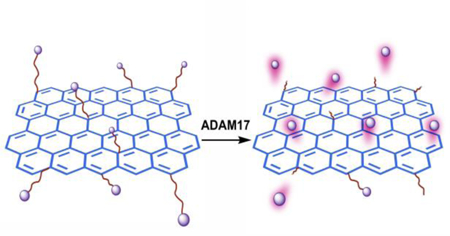
Graphical Abstract
The cleavage of the peptide substrate by ADAM17 releases a dye-labelled short peptide fragment into the solution, thus producing fluorescence
1. Introduction
ADAMs (short for a disintegrin and metalloproteinase) play important roles in cell surface remodeling, ectodomain shedding, regulation of growth factor availability, and mediating cell-cell and cell-matrix interaction in both normal development and pathological states such as Alzheimer’s diseases, arthritis, cancer, and cardiac hypertrophy.1 As a member of ADAM family, a disintegrin and metalloproteinase 17 (ADAM17) is also known as a tumor necrosis factor-α-converting enzyme.2 An increased expression of ADAM17 has been found in various inflammatory diseases such as rheumatoid arthritis (RA), Crohn’s disease, pulmonary inflammation, endotoxin shock, multiple sclerosis and myocardial infarction.1–3 Recent studies showed that ADAM17 was highly expressed in many human tumors and can promote tumor invasion and metastasis.4 For example, significantly enhanced expression of ADAM17 was found in hepatocellular carcinoma than non-cancerous liver tissues.5 It was also demonstrated that ADAM17 was involved in the progression of breast, ovarian, and colorectal cancers.6–8 Therefore, ADAM17 has become a novel biomarker and potential therapeutic target for the early detection and treatment of human cancers.9 Thus far, two major approaches have been used for ADAM17 detection. One is enzyme-linked immunosorbent assay (ELISA), while the other is western blotting. These two methods, although sensitive, are time-consuming, which contain complicated steps such as multi-steps of washing / immunoblotting and incubation, and usually take hours or even days to provide results.10, 11 Furthermore, another limitation of them is that they only detect protease abundance but could not measure protease activity.12, 13 However, proteases are often tightly regulated on a post-translational level leading to a potentially significant divergence of abundance and activity. Hence, it is important to develop a simple, rapid and sensitive method for detection of the activity of ADAM17.
In our previous study, we have successfully developed a graphene oxide (GO) based fluorescence resonance energy transfer (FRET) biosensing platform for the detection of HIV-1 protease activity.14 Due to the unique heterogeneous structure (coexistence of π state sp2 carbon clusters and σ state sp3 C-O matrix),15 GO not only provides a broad absorbance from 200 nm to 800 nm, but also supplies a mass of chemical binding sites for further modification.16 Note that, one requirement for the development of highly sensitive FRET sensors is the delicate matching of fluorophore and quencher (i.e., the overlap of the emission spectrum of the fluorophore and the absorption spectrum of the quencher). The capability of a quencher to cover a broad range of absorbance spectra has the advantage of constructing a fluorescent sensor to detect multiple analytes in one sample without employing multiple different quenchers.17 Furthermore, both theoretical calculation and experiments have demonstrated that GO was an efficient quencher for various fluorophores with the quenching distance reaching as far as 30 nm.18 Compared with other fluorescent quenchers such as quantum dots, which typically allow less than 10 nm distance between donor and acceptor, such a long quenching distance offers unique opportunities to detect large biomolecules and study biomolecule-biomolecule (e.g., protein-antibody) interactions.19–21 In this work, by attaching a dye labeled ADAM17 substrate peptide to the GO surface, and monitoring the cleavage of the substrate by ADAM17 (Scheme 1), we accomplished quantitative ADAM17 detection with good sensitivity, great simplicity and high reproducibility.
Scheme 1.
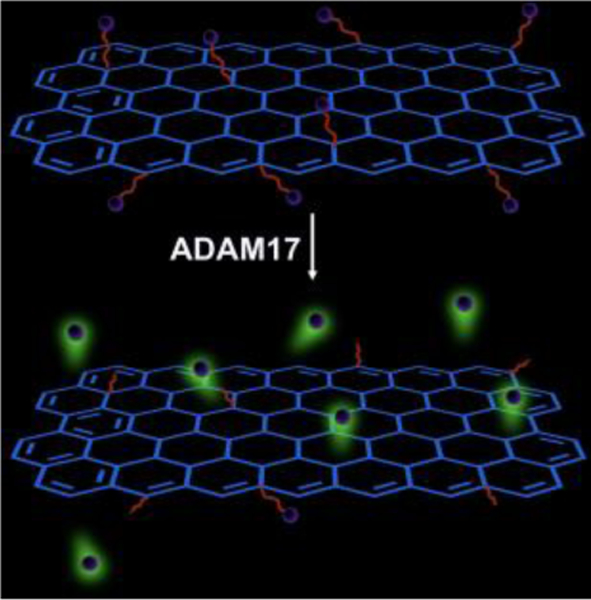
The principle of detecting ADAM17. The cleavage of the peptide substrate by ADAM17 releases a dye-labeled short peptide fragment into the solution, thus producing fluorescence.
2. Experimental Section
2.1. Chemicals and Reagents.
ADAM9 and ADAM17 were purchased from R&D Systems (Minneapolis, MN), while the 5-FAM labeled ADAM17 protease substrate peptide (Pep-FAM) with a sequence of NH2-CALNNLAQAVRSSSARK(5-FAM) (95.22% pure) was ordered from WatsonBio Sciences (Houston, TX). Human serum and all the other chemicals, including graphene oxide and MMP-9, were obtained from Sigma-Aldrich (St. Louis, MO). ADAM17 and its substrate peptide were dissolved in HPLC-grade water (ChromAR, Mallinckrodt Baker). The stock solution of ADAM17 was prepared at 200 μg mL−1 and kept at −80 °C before and after immediate use, while that of the peptide substrate was prepared at a concentration of 1 mM and stored at −20 °C before and after use. Two buffer solutions were used in this study: (1) MES buffer (containing 100 mM 2-(N-morpholino)-ethanesulfonic acid, pH 6.0); and (2) Tris buffer, which was consisted of 50 mM Tris, 15 mM NaCl, and 5 μM ZnCl2 with the solution pH adjusted to 7.5 using HCl.
2.2. Instruments.
Fluorescence spectra were obtained at λex/em = 492/515 nm by using a luminescence spectrophotometer (LS50B, PerkinElmer, Waltham, MA, USA). Infrared (IR) spectra were recorded from 1000 cm−1 to 4000 cm−1 with an infrared spectrophotometer (NEXUS 470 FT-IR, Thermo Nicolet, Waltham, MA, USA) equipped with pressed KBr pellets. UV-vis absorption spectra (from 200 nm to 800 nm) were collected using a UV-Vis-NIR spectrophotometer (Varian Cary 500 Scan, Agilent, Santa Clara, CA, USA). Raman experiments were performed in the range of 2000 cm−1 −1000 cm−1 using a confocal Raman spectrometer with a 514 nm laser (25 mW) as the excitation source and collected with 30-s exposure through 5 times accumulation under x20 objective lens (Renishaw, United Kingdom).
2.3. Synthesis of cGO-Pep-FAM.
Production of cGO-Pep-FAM sensor was carried out using GO as starting material and based on two step reactions. The first step was involved with a well-documented technique, i.e., use of chloroacetic acid to transform unreactive hydroxyls of the GO surface into carboxylate groups under strongly basic conditions 22, while the second step took advantage of carbodiimide coupling reaction with EDC and sulfo-NHS. Briefly, 4 mg mL−1 GO suspension was mixed with 4.8 g NaOH and 4 g ClCH2COOH, and sonicated until the hydroxyl groups of GO were converted to carboxyl groups. The resultant solution was neutralized by HCl (1 M), and purified by repeated rinsing with water until carboxylated GO (cGO) could be well dispersed in water. Then, cGO (1 mg mL−1, 50 μL) was dispersed in 1.0 mL of MES buffer (100 mM, pH 5.2). 20 μL of 200 mM 1-ethyl-3-(−3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and 100 μL of 200 mM N-hydroxysulfosuccinimide (Sulfo-NHS) were added to the cGO suspension and sonicated for 30 min under an ice-water bath. The resulting mixture was centrifuged at 12,000 rpm for 10 min, and the supernatant was discarded. After washing the precipitate for three more times to remove excess EDC and Sulfo-NHS, it was dispersed in 1.0 mL of MES buffer and adjusted to pH 7.4 with NaOH, followed by addition of 50 μL peptide substrate (1 mM). The mixture was stirred at room temperature for 2 hours in darkness. The product was purified by repeated centrifugation and rinsing with distilled water three times to remove the unreacted peptide, and then rinsing with 20 μL bull serum albumin (BSA, 20 mg mL−1) three times to remove non-specifically absorbed peptide on the GO surface. The final product was dispersed in 1 mL water and stored in a refrigerator at 4 °C. Note that the molar ratio of the peptide substrate Pep-FAM to cGO used to prepare the cGO-Pep-FAM sensor as described above was determined from the fluorescence quenching experiment. Under this experimental condition, fluorescence quenching efficiency was relatively high (90%), while the amount of cGO consumed was relatively small (Supplementary Material, Fig. S1).
3. Results and discussion
3.1. Infrared, Raman, UV-vis Spectroscopy and Fluorescence Spectra.
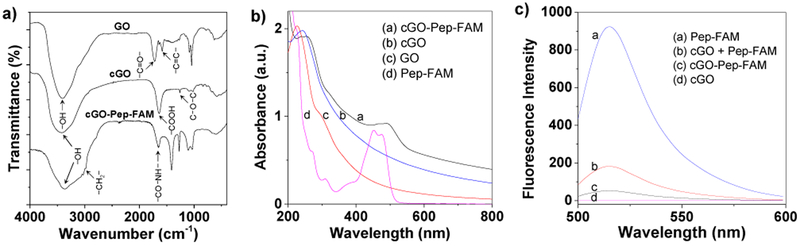
The chemical structures of GO, cGO and cGO-Pep-FAM samples were confirmed by infrared spectroscopy (FT-IR). As shown in Fig. 1a and Supplementary Material, Fig. S2, characteristic absorption peaks appeared at 3400 cm−1 (stretching vibration of OH), 1722 cm−1 (stretching vibration of C=O), and 1580 cm−1 (stretching vibration of C=C), revealing the presence of OH, C=O and C=C functional groups on GO. After activation of GO with chloroacetic acid, in addition to the stretching vibration of OH at 3400 cm−1, cGO showed a stronger absorption band at 1634 cm−1 (stretching vibration of COOH) and a small peak at 1258 cm−1 (stretching vibration of C-O-C). The combined results indicated the formation of COCOOH moieties. After conjugation of the peptide with cGO, a strong characteristic band appeared at 1655 cm−1 (stretching vibration of CO-NH) with an increased small peak near 2900 cm−1 (stretching vibration of CH2), which indicated the successful formation of covalent bond between cGO and the peptide molecule. The structure of cGO was also characterized by Raman spectroscopy. As depicted in Supplementary Material, Figure S3, cGO showed a strong Raman shift at 1344 cm−1 and 1587 cm−1, corresponding to the D- and G-bands, respectively. It should be noted that the high intensity ratio of D band and G band indicated a more amorphous carbon structure.23 The significant difference between the structures of GO, cGO and cGO-Pep-FAM was also supported by our UV-vis experiments. For example, as shown in Fig. 1b, the cGO-Pep-FAM sample (50 μg mL−1) had much larger absorbance values in the visible region than cGO (50 μg mL−1), and showed a small peak at 492 nm. This peak was located at a similar position to that of 5-FAM labeled peptide (Pep-FAM), which was not observed with the GO and cGO samples. Moreover, the huge difference between the Raman spectra of cGO and cGO-Pep-FAM provided further evidence for the difference in their structures. In addition, fluorescence of the free substrate peptide Pep-FAM, cGO, mixture of cGO and Pep-FAM, and covalently-conjugated cGO-Pep-FAM were recorded at λex/em = 492/515 nm. As shown in Fig. 1c, these four samples produced significantly different fluorescence intensities, again indicating the quite different structures among them.
Figure 1.
Characterization of cGO-Pep-FAM sensor. (a) FT-IR spectra of GO, cGO and cGO-Pep-FAM; (b) UV-Vis absorbance spectra of GO, Pep-FAM, cGO, and cGO-Pep-FAM; (c) fluorescence spectra of Pep-FAM, cGO, cGO / Pep-FAM mixture, and covalently-conjugated cGO-Pep-FAM. With the exception of Pep-FAM (50 μM), the concentrations of GO, cGO, and cGO-Pep-FAM used in Fig. 1b were 50 μg mL−1 each, while those of Pep-FAM, cGO, and cGO-Pep-FAM used in Fig. 1c were 5 μM, 1 μg mL−1, 1 μg mL−1, respectively. An enlarged view of the FTIR spectra in the region from 1500 cm−1 to 2000 cm−1 was displaced in Supplementary Material, Fig. S2.
3.2. Effect of Solution pH on the cGO-Pep-FAM Sensor.
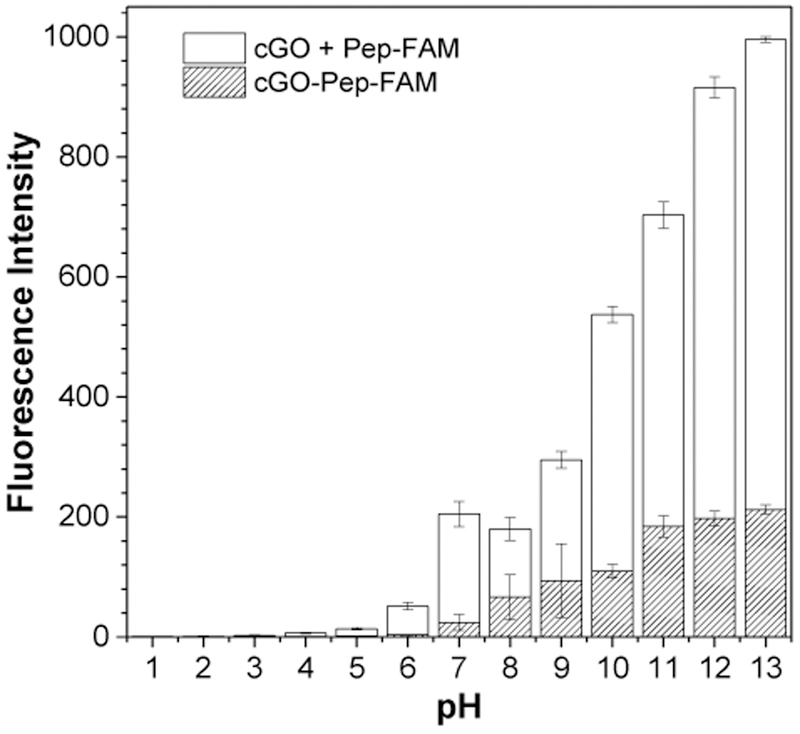
Although covalently conjugated GO sensors are more resistant to various experimental conditions than non-specific adsorption-based graphene sensors, they are still slightly affected by solution pH since fluorescein has four species (i.e., cation, neutral, monoanion, and dianion) with the dominant component changing with the pH of the solution.24 To find an appropriate pH for sensitive detection of ADAM17, the fluorescence intensities of the 0.5 μg mL−1 covalently-conjugated cGO-Pep-FAM sensor were investigated at a series of pH values ranging from 1 to 13. As a control, a mixture of cGO (0.5 μg mL−1) and peptide (0.5 μM) was also prepared. Our experimental results (Fig. 2) showed that, with an increase in the solution pH, the background fluorescence intensity of cGO-Pep-FAM gradually increased (but didn’t change much until pH 10), while that of the mixture of GO and Pep-FAM increased drastically. Hence, the effect of the solution pH on cGo-Pep-FAM was much less significant than that on cGO / Pep-FAM mixture due to the covalent conjugation. Furthermore, cGO-Pep-FAM biosensor had smaller background fluorescence intensity values than the non-specific adsorption-based cGO / Pep-FAM mixture at all the pH values investigated, and hence it is more suitable for use as sensing element for ADAM17 detection. Due to the low background noise and high signal to noise ratio of the cGO-Pep-FAM sensor at pH 7.5 as well as the optimum enzyme activity25 of ADAM17 under this condition, a buffer solution with a pH of 7.5 was used in the remaining experiments.
Figure 2.
Effect of solution pH on fluorescence intensities of cGO-Pep-FAM and mixture of cGO and Pep-FAM. The concentrations of cGO-Pep-FAM, cGO, and Pep-FAM used were 0.5 μg mL−1, 0.5 μg mL−1, and 0.5 μM, respectively. Each data point represents the average from three replicate analyses ± one standard deviation.
3.3. Effect of Incubation Time on ADAM17 Detection.
To optimize the experimental condition for ADAM17 detection, the effect of reaction time on ADAM17 cleavage of the peptide substrate was investigated. For this purpose, a mixture sample, which contained 0.5 μg mL−1 cGO-Pep-FAM, 100 ng mL−1 ADAM17 and Tris buffer, was prepared. Then, the mixture was incubated at 37 °C for a period of time ranging from 0 to 120 min, followed by fluorescence measurement. The experimental results were summarized in Fig. S4 (Supplementary Material). We could see that the fluorescence intensity of the mixture sample increased with an increase in the reaction time until 60 minutes, after which the fluorescence signal began to saturate. To achieve rapid detection of ADAM17, 60 min was chosen as the optimum reaction time and used in all the subsequent experiments.
3.4. Dose Response Curve for ADAM17.
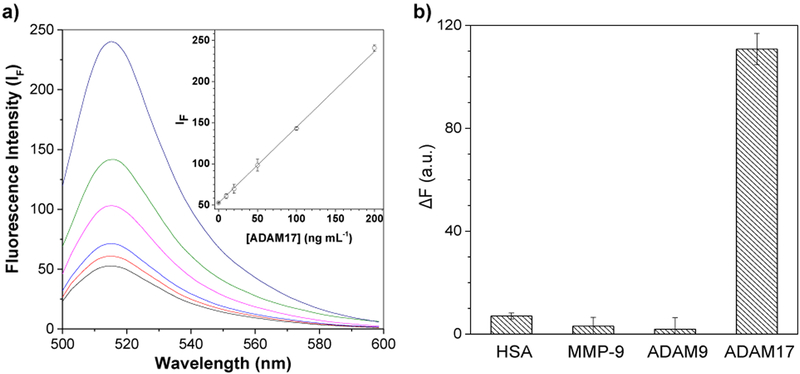
Utilizing the current physical conditions (i.e., pH 7.5, 60 min reaction time, and 37 °C incubation temperature), dose response curve for ADAM17 detection was constructed by monitoring the interaction between the cGO-Pep-FAM solution (0.5 μg mL−1) and ADAM17 at various concentrations, ranging from 5 to 200 ng mL−1. The fluorescence intensity of each mixture sample was collected in the range of 500 nm to 600 nm (excited at 492 nm). Our experimental results (Fig. 3a) showed that the fluorescence intensity of the mixture was linearly correlated with ADAM17 concentration, indicating that the peptide substrate (NH2-CALNNLAQAVRSSSARK(5-FAM)) attached to the GO surface was being cleaved by ADAM17. In addition to a wide dynamic range (linear regression with the ADAM17 concentration ranging from 5 to 200 ng mL−1, R2 = 0.9998), this ADAM17 sensor also showed a detection limit (defined as the ADAM17 concentration corresponding to three times the standard deviation of blank signal) of 0.91 ng mL−1, which is equivalent to 17.5 pM. Such a detection limit is better than that (2 ng mL−1) obtained with ELISA26, and more than good enough for analyzing ADAM17 in clinical samples (note that the mean serum level of ADAM 17 in patients with colorectal cancer is 2.09 ng mL−1 27, 28).
Figure 3.
cGO-Pep-FAM detection of ADAM17. (a) Fluorescence spectra of cGO-Pep-FAM in the presence of ADAM17 at various concentrations; and (b) plot of fluorescence intensity versus analyte species, showing the selectivity of the cGO-Pep-FAM sensor. The inset in Fig. 3a displays the dose-response curve for ADAM17. The fluorescence intensity values in Fig. 3b were background corrected, which were obtained by subtracting the blank fluorescence intensity from that of the analyte species. Experiments were performed by incubating cGO-Pep-FAM suspension solutions (0.5 μg mL−1, Tris buffer, pH 7.5) with ADAM17 or other species for 60 min at 37 °C, followed by measuring their fluorescence intensities with λex/em = 492/515 nm at room temperature. The concentrations of MMP-9, ADAM9, ADAM17, and HSA used in Fig. 3b were 100 ng mL−1, 100 ng mL−1, 100 ng mL−1, and 10 μg mL−1, respectively. Each data point represents the average from three replicate analyses ± one standard deviation.
3.5. Selectivity and Specificity of the cGO-Pep-FAM Sensor.
Two structure similar proteases, including ADAM 9 and matrix metalloproteinases 9 (MMP-9), were selected as potential interfering species to examine the cross-reactivity of the cGO-Pep-FAM sensor. Like ADAM17, ADAM9 and MMP-9 are also important cancer biomarkers.29, 30 For example, serum and tissue levels of MMP-9 are significantly higher in patients with pancreatic ductal adenocarcinoma than in patients with chronic pancreatitis and healthy controls,31 while plasma and/or tumor tissues from patients with pancreatic ductal adenocarcinoma have significantly elevated levels of ADAM9,32 which may predict shortened survival of patients.32,33 Furthermore, human serum albumin (HSA, an abundant serum protein) was tested to investigate the matrix effect. The experimental results were summarized in Fig. 3b. We could see that all these three samples produced significantly smaller fluorescence signals than ADAM17, thus suggesting the high selectivity and specificity of our cGO-Pep-FAM sensor.
3.6. Simulated Serum Sample Analysis.
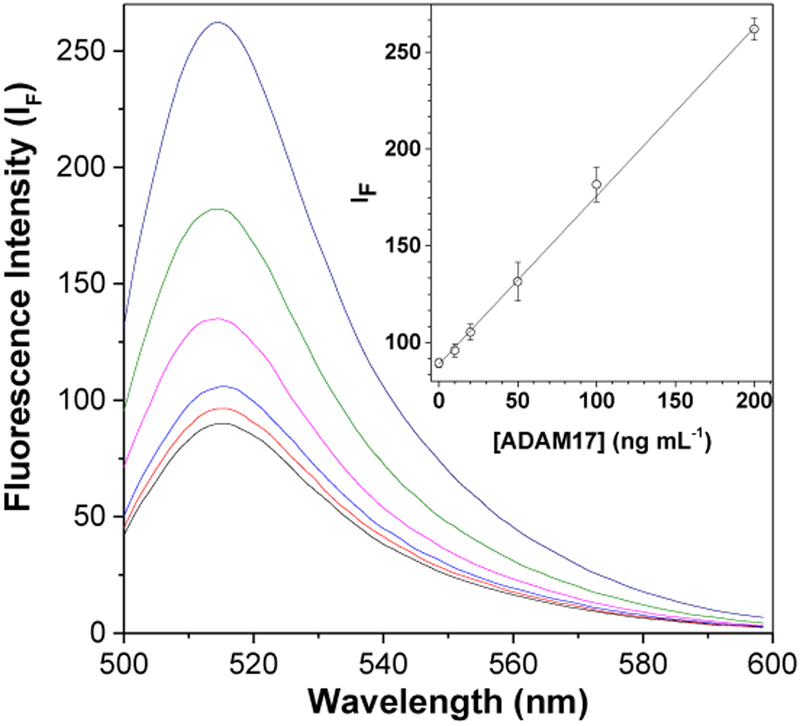
Previous studies have shown that some molecules, especially proteins, could competitively bind to the GO surface, thus affecting the fluorescence signal 14. In the previous selectivity section, we found that low concentrations of HSA would not significantly interfere with ADAM17 detection by our cGO-Pep-FAM sensor. In order to accurately detect ADAM17 in serum, which contains large amounts of HSA, IgG, hemoglobin and fatty acids, and also has trace amounts of short DNA/RNA, small peptides, and other molecules, the effect of the concentration of HSA on the cGO-Pep-FAM sensor was further investigated. Our experimental results (Supplementary Material, Fig. S5) showed that, with an increase in the concentration of added HSA, the background fluorescence intensity of the cGO-Pep-FAM suspension increased slightly until the concentration of HSA reached 200 μg mL−1, after which the fluorescence signal began to saturate. To address the serum matrix effect, a modified dose response curve (Fig. 4) was constructed, where the interactions between the cGO-Pep-FAM solution (0.5 μg mL−1) and ADAM17 at various concentrations were held and monitored in the presence of 400 μg mL−1 HSA. Then, 4 simulated serum samples, which were prepared by spiking 20 ng mL−1 to 200 ng mL−1 ADAM17 into 20 μL human serum, were analyzed by the cGO-Pep-FAM sensor. Our experimental results (Table 1) showed that these ADAM17-spiked serum samples could successfully be quantitated, supporting the feasibility of our developed cGO-Pep-FAM sensor for potential clinical applications.
Figure 4.
Effect of HSA on the fluorescence spectra of cGO-Pep-FAM in the presence of ADAM17 at various concentrations. The inset displays the plot of fluorescence intensity versus ADAM17 concentration. Experiments were performed by incubating the mixtures of cGO-Pep-FAM suspension (0.5 μg mL−1, Tris buffer, pH 7.5), HSA (400 μg mL−1), and ADAM17 at concentrations ranging from 5 ng mL−1 to 200 ng mL−1 at 37 °C, followed by measuring their fluorescence intensity with λex/em = 492/515 nm at room temperature. Each data point represents the average from three replicate analyses ± one standard deviation.
Table 1.
Recovery of ADAM17 from serum by use of the cGo-Pep-FAM sensor. Each value represents the mean of three replicate analyses ± one standard deviation.
| Sample number |
Theoretical value (ng mL−1) |
Experimental value ± SD (ng mL−1) |
|---|---|---|
| 1 | 20 | 19.0 ± 4.8 |
| 2 | 50 | 49.0 ± 6.5 |
| 3 | 100 | 106.8 ± 7.1 |
| 4 | 200 | 199.1 ± 6.6 |
4. Conclusions
In summary, by covalently attaching fluorescently labeled ADAM17 substrate peptide to the GO surface, and based on enzyme-substrate cleavage reaction, we successfully developed a cGO-Pep-FAM fluorescent biosensor for rapid and accurate detection of ADAM17. The sensor was highly sensitive with a detection limit of 17.5 picomolar. Furthermore, the sensor was selective: structure similar proteases such as ADAM 9 and MMP-9 would not interfere with ADAM17 detection. In addition, simulated serum samples were accurately analyzed. It could be visualized that, with the same ADAM17 detection strategy, highly sensitive and selective sensors for a variety of other proteases could be developed by changing the peptide substrates. These GO-based fluorescent sensors may find useful applications in many fields such as diagnosis of protease-related diseases and high-throughput screening of drug candidates.
Supplementary Material
Acknowledgments
This work was financially supported by the National Institutes of Health (2R15GM110632-02) and National Science Foundation (1708596).
Footnotes
Electronic Supplementary Information (ESI) available: [Additional figures, including effect of cGO concentration on fluorescence intensity and quenching efficiency of peptide Pep-FAM, raman spectra of cGO and cGO-Pep-FAM, effect of reaction time on cGO-Pep-FAM detection of ADAM17, and effect of HSA on the background fluorescence intensity of the cGO-Pep-FAM sensor]. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Saftig P and Reiss K, Eur. J. Cell Biol, 2011, 90, 527–535. [DOI] [PubMed] [Google Scholar]
- 2.Le Gall SM, Bobé P, Reiss K, Horiuchi K, Niu X-D, Lundell D, Gibb DR, Conrad D, Saftig P and Blobel CP, Mol. Biol. Cell, 2009, 20, 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satoh M, Ishikawa Y, Itoh T, Minami Y, Takahashi Y and Nakamura M, European Journal of Clinical Investigation, 2008, 38, 97–105. [DOI] [PubMed] [Google Scholar]
- 4.H. B. Liu, Y. Zhu, Q. C. Yang, Y. Shen, X. J. Zhang and H. Chen, 2015, 14, 4391–4398. [DOI] [PubMed]
- 5.Ding X, Yang L-Y, Huang G-W, Wang W and Lu W-Q, World Journal of Gastroenterology : WJG, 2004, 10, 2735–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGowan PM, Ryan BM, Hill ADK, McDermott E, Higgins N and Duffy MJ, Clinical Cancer Research, 2007, 13, 2335. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Miyamoto S, Suzuki SO, Oki E, Yagi H, Sonoda K, Yamazaki A, Mizushima H, Maehara Y, Mekada E and Nakano H, Clinical Cancer Research, 2005, 11, 4783. [DOI] [PubMed] [Google Scholar]
- 8.Merchant NB, Voskresensky I, Rogers CM, LaFleur B, Dempsey PJ, Graves-Deal R, Revetta F, Foutch AC, Rothenberg ML, Washington MK and Coffey RJ, Clinical Cancer Research, 2008, 14, 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy R, Yang J and Moses MA, Journal of Clinical Oncology, 2009, 27, 5287–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saraheimo S, Hepojoki J, Nurmi V, Lahtinen A, Hemmilä I, Vaheri A, Vapalahti O and Hedman K, PLoS One, 2013, 8, e62739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass JJ, Wilkinson DJ, Rankin D, Phillips BE, Szewczyk NJ, Smith K and Atherton PJ, Scandinavian Journal of Medicine & Science in Sports, 2016, 27, 4–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulli B, Ali M, Forghani R, Schob S, Hsieh KLC, Wojtkiewicz G, Linnoila JJ and Chen JW, PLoS One, 2013, 8, e67976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou S, Wang L, Chen X and Guan X, ACS Sensors, 2016, 1, 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Chen X, Roozbahani GM and Guan X, Anal. Bioanal. Chem, 2018, 410, 6177–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreyer DR, Park S, Bielawski CW and Ruoff RS, Chem. Soc. Rev, 2010, 39, 228–240. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Cote LJ, Kim F, Yuan W, Shull KR and Huang J, J. Am. Chem. Soc, 2010, 132, 8180–8186. [DOI] [PubMed] [Google Scholar]
- 17.Crisalli P and Kool ET, Bioconjugate Chem, 2011, 22, 2345–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swathi RS and Sebastian KL, The Journal of Chemical Physics, 2009, 130, 086101. [DOI] [PubMed] [Google Scholar]
- 19.Tian F, Lyu J, Shi J and Yang M, Biosensors and Bioelectronics, 2017, 89, 123–135. [DOI] [PubMed] [Google Scholar]
- 20.Mayilo S, Kloster MA, Wunderlich M, Lutich A, Klar TA, Nichtl A, Kürzinger K, Stefani FD and Feldmann J, Nano Lett, 2009, 9, 4558–4563. [DOI] [PubMed] [Google Scholar]
- 21.Bojarski P, Kulak L, Walczewska-Szewc K, Synak A, Marzullo VM, Luini A and D’Auria S, The Journal of Physical Chemistry B, 2011, 115, 10120–10125. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Liu Z, Welsher K, Robinson JT, Goodwin A, Zaric S and Dai H, Nano research, 2008, 1, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucchese MM, Stavale F, Ferreira EHM, Vilani C, Moutinho MVO, Capaz RB, Achete CA and Jorio A, Carbon, 2010, 48, 1592–1597. [Google Scholar]
- 24.Ferrari L, Rovati L, Fabbri P and Pilati F, Sensors (Basel, Switzerland), 2013, 13, 484–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan MJ, Seaton T, Mitchell J, Howe A, Blackburn K, Burkhart W, Moyer M, Patel I, Waitt GM, Becherer JD, Moss ML and Milla ME, Biochemistry, 2002, 41, 9462–9469. [DOI] [PubMed] [Google Scholar]
- 26.Trad A, Hedemann N, Shomali M, Pawlak V, Grötzinger J and Lorenzen I, J. Immunol Methods, 2011, 371, 91–96. [DOI] [PubMed] [Google Scholar]
- 27.K. Walkiewicz, E. Nowakowska-Zajdel, J. Strzelczyk, S. Dzięgielewska-Gęsiak and M. Muc-Wierzgoń, 2017, 31, 929–934. [PubMed]
- 28.Paweł K, Katarzyna W, Joanna S, Sylwia D-G, Dariusz W, Angelika C, Malgorzata M-W and Ewa Nowakowska Z, Journal of Gastrointestinal & Digestive System, 2016, 8. [Google Scholar]
- 29.Merdad A, Karim S, Schulten H-J, Dallol A, Buhmeida A, Al-Thubaity F, Gari MA, Chaudhary AG, Abuzenadah AM and Al-Qahtani MH, Anticancer Res, 2014, 34, 1355–1366. [PubMed] [Google Scholar]
- 30.Zhao S, Ma W, Zhang M, Tang D, Shi Q, Xu S, Zhang X, Liu Y, Song Y, Liu L and Zhang Q, Medical Oncology, 2012, 30, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian M, Cui Y-Z, Song G-H, Zong M-J, Zhou X-Y, Chen Y and Han J-X, BMC Cancer, 2008, 8, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grützmann R, Lüttges J, Sipos B, Ammerpohl O, Dobrowolski F, Alldinger I, Kersting S, Ockert D, Koch R, Kalthoff H, Schackert HK, Saeger HD, Klöppel G and Pilarsky C, Br. J. Cancer, 2004, 90, 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones LE, Humphreys MJ, Campbell F, Neoptolemos JP and Boyd MT, Clinical Cancer Research, 2004, 10, 2832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.