Abstract
To overcome the effect of other components of complicated biological samples on nanopore stochatic sensing, displacement chemical reaction was utilized to selectively extract the target nucleic acid from whole blood. Given its simplicity and high sensitivity for detecting nucleic acids, our developed displacement chemistry-based nanopore sensing strategy offers the potential for fieldable / point-of-care diagnostic applications.
Nanopore stochastic sensing has attracted substantial interest as a label-free technique for measuring single molecules.1–8 In addition to DNA sequencing,9–11 nanopore technology has demonstrated its potential utility as a versatile tool to explore a wide variety of applications, including disease detection,12–15 environmental monitoring,16,17 pharmaceutical screening,18,19 and homeland security and bio-defense.20,21 In spite of the past two decade’s significant advancement, it is still challenging to utilize nanopore sensors, especially protein ion channels, to analyze clinical samples with complicated matrices such as blood and serum. It has been well documented that blood and/or serum affects nanopore sensors in two ways: 1) disturbing the lipid bilayer used in the protein nanopore sensing system, which makes long time nanopore single-channel recording difficult; and 2) blocking the nanopore (including both the protein pore and solid-state nanopore) and/or inducing an increased background noise level, which leads to a smaller signal-to-noise ratio and hence a reduced sensor sensitivity.22 These two effects greatly limit the clinical applications of the nanopore sensors. To overcome these obstacles, two approaches have been attempted so far: one involves the use of highly (e.g., 100×) diluted serum samples for nanopore analysis,23 while the other employs purification kits and spin-columns to extract the target analytes and to remove the interfering blood or serum components.22,24 However, these two approaches suffer from either the significantly diminished sensor sensitivity or the tedious centrifugation separation and elution procedures, which make the nanopore analysis inappropriate for potential fieldable or point-of-care diagnostic applications. Herein, we report a displacement chemistry-based nanopore sensing method to analyze nucleic acids in complicated matrices.
In our designed displacement reaction-based detection strategy, the separation of the target nucleic acid from the sample matrix components and its sequence analysis are achieved simply by using two probes, the capture probe and the release probe. Both the probes are short single-stranded DNA molecules having sequences partially or fully complementary to the target nucleic acid. Note that the capture probe is immobilized on the magnetic beads, whose function is to capture and separate the target nucleic acid from the blood / serum matrix, while the release probe is used to pull off the nucleic acid from the magnetic beads which contains the capture probe-target nucleic acid double strand complexes. Due to the displacement reaction or competition reaction nature, it is highly important that the release probe has a much stronger hybridization affinity to the target nucleic acid than the capture probe does. As shown in Scheme 1, after addition of the capture probe to the blood or serum sample, the target nucleic acid molecules in the blood and the capture probe will form sticky end double-stranded nucleic acid molecules, and are immobilized on the magnetic beads, thus being separated from other matrix components. After removing the blood, the release probe is added. It will pull off the target nucleic acid from the magnetic beads to form release probe-nucleic acid hybridization complexes, which can then be transferred to the nanopore sensor for determining the sequence of the target nucleic acid.
Scheme 1.
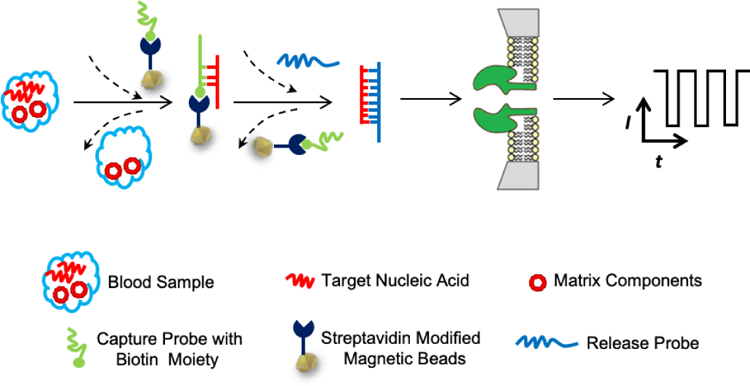
Schematic representation of the principle of displacement chemistry-based nanopore analysis of nucleic acids in complicated matrices. The separation of the target nucleic acid from the sample matrix components and its sequence analysis are achieved by using two ssDNA probes. The capture probe, which is immobilized on the magnetic beads, has sequences partially complementary to the target nucleic acid, while the release probe has a much stronger hybridization affinity to the target nucleic acid than the capture probe does. After addition of the capture probe to the blood or serum sample, the target nucleic acid molecules in the blood and the capture probe will form sticky end double-stranded nucleic acid molecules, and are immobilized on the magnetic beads, thus being separated from other matrix components. After removing the blood followed by addition of the release probe, the target nucleic acid forms more stable double-stranded nucleic acid with the release probe than with the capture probe, thus being pulled off from the magnetic beads. The produced release probe-nucleic acid hybridization complexes can then be transferred to the nanopore sensor for determining the sequence of the target nucleic acid.
For the proof of concept demonstration, a simulated blood sample was prepared by spiking a lung cancer biomarker microRNA molecule (sequence: 5’-UUAAUGCUAAUCGUGAUAGGGG-3’) into the rabbit whole blood. The capture probe used was a 20-mer biotin-modified DNA (sequence: 5’-TCACGATTAGCCCCCCCCCC-(3’-Biotin)-3’) and immobilized on the streptavidin-coated magnetic beads, while the release probe was a 30-mer DNA molecule (sequence: 5’-CCCCTATCACGATTAGCATTAAAAAAAAAA-3’), where the underlined sequences represent the nucleotide bases that can form double stranded nucleic acids with the microRNA molecule. The simulated blood sample was analyzed according to the nucleic acid detection principle as shown in Scheme 1 and using the procedure as described in the experimental section. The nanopore single-channel recording experiment was carried out using an engineered (M113F)7 α-hemolysin (αHL) protein pore at +160 mV in a pH 8.0 buffer solution containing 1 M KCl and 10 mM Tris. The +160 mV voltage was chosen based on our previous work25. Specifically, at this applied potential bias, all the event signatures, including residence time, frequency and blockage amplitude, for the unzipping of double-stranded nucleic acids in the αHL nanopore showed relatively large values, and hence, the nanopore sensor offered the best resolution without losing its stability. Compared with the wild-type αHL protein pore26, rapid unzipping of double stranded nucleic acids (a two to three order of magnitude reduction in the event residence time) could be achieved in this engineered nanopore27. As controls and also for the purpose of determining the accuracy of microRNA detection, the interactions between the αHL (M113F)7 pore and individual standards of the target microRNA, capture probe, and release probe, as well as the 1:1 ratio mixtures of capture probe and microRNA, and release probe and microRNA (all prepared in DNase, RNase free water) were also examined. As discussed previously, one important and necessary requirement for high efficient recovery of the target nucleic acid from the sample solution is that the release probe could form more stable double stranded complex with the target microRNA than the capture probe did. In our investigated system, the release probe / microRNA complex has a longer double-stranded region than the capture probe / microRNA complex (22-bp vs. 10-bp), and has a larger predicted melting temperature (83.8°C vs. 63.6°C) according to the DINA Web Server,28 thus satisfying the requirement. Note that, the different solution media (blood in the capture reaction vs. 1 M KCl in the release reaction) would affect the stability of the DNA/RNA duplex. However, since displacement chemistry reaction occurred in 1 M KCl, the stability comparison between the release probe / microRNA complex and the capture probe / microRNA duplex could be simply made in such a salt solution, which simplified the design for capture probe and release probe. To support the prediction that the release probe can pull off the target nucleic acid from the capture probe-microRNA duplex, nanopore analysis of a mixture (in 5:4:5 molar ratio) of the capture probe, microRNA, and release probe was additionally included in the investigation. Note that, the mixture solution was prepared in such a way that the capture probe and microRNA were first incubated at 95 °C for 3 minutes, then cooled down to room temperature to allow for the formation of double stranded DNA/RNA complex, followed by addition of the release probe. The results for this series of experiments were summarized in Figure 1 and Supporting Information, Figures S1 – S6.
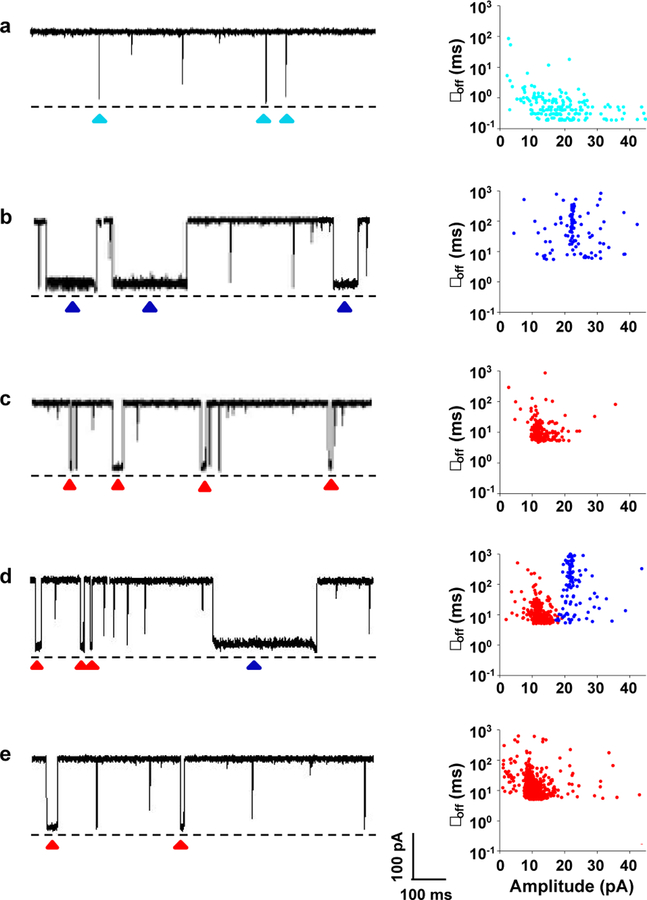
Fig. 1.
Nanopore analysis of nucleic acids. (Left) Typical single-channel recording trace segments; (Right) the corresponding scatter plots of residence time vs. residual current of triangle-marked events. (a) The target biomarker microRNA analyte; (b) hybridization mixture of capture probe and microRNA; (c) hybridization mixture of release probe and microRNA; (d) hybridization mixture of capture probe, release probe and microRNA; and (e) microRNA-containing whole blood. The experiments were performed at +160 mV in 1 M KCl solution containing 10 mM Tris-HCl at pH 8.0. The concentrations of microRNA, capture probe, and release probe used were 250 nM each except the experiment shown in Fig. 1d, where 200 nM of target microRNA was used. Dashed lines represent the levels of zero current. The characteristic events were highlighted (triangle-marked). Translocation of single-stranded capture probe and release probe molecules in the nanopore produced similar short-lived blockage events to those of the microRNA analyte. The blood sample was analyzed according to the nucleic acid detection principle as shown in Scheme 1 and using the procedure as described in the experimental section.
Our experiments showed that, the translocation of single-stranded microRNA, capture probe, or release probe molecules in the (M113F)7 α-hemolysin pore produced only one major type of short-lived blockage events with mean residence time of 0.49 ± 0.03 ms, 0.35 ± 0.02 ms, 0.20 ± 0.01 ms, respectively (Figure 1a and Supporting Information, Figure S6). In contrast, in addition to the short-lived events (Supporting Information, Figure S7), significantly longer duration events were observed when the mixture of the capture probe and microRNA or the mixture of the release probe and microRNA was present in the nanopore sensing chamber (Figures 1b and 1c), indicating the formation of DNA/RNA hybridization complexes. Since the events of the capture probe / RNA complex and the release probe / RNA complex had significantly different mean residence time (49.2 ± 1.4 ms vs. 13.8 ± 0.9 ms) and blockage amplitudes (with their event residual currents at 22.1 ± 0.7 pA, and 13.0 ± 0.1 pA, respectively), these two types of double stranded DNA/RNA molecules could be well differentiated, offering the potential for their simultaneous detection and quantification. Note that, in principle, the release probe / RNA complex should produce longer residence time events than the capture probe / RNA duplex due to its larger melting temperature, which makes the complex more stable, and more difficult to be unzipped by the nanopore. However, in our experiments, longer residence time events were observed with the presence of the capture probe / RNA complex in the nanopore, which might be attributed to the additional biotin moiety in the capture probe and/or the poly(A) tail in the release probe. Note that the phenomenon that modifying nucleotide structures or attaching new functional groups to the DNA molecule could prolong its residence time in the nanopore has been documented previously.29–32 Furthermore, utilizing a polynucleotide tail to facilitate dsDNA unzipping is well-known in the nanopore field.33,34
As to the mixture of the capture probe, microRNA, and release probe, we found that it produced two types of events, corresponding to the capture probe / microRNA, and release probe / microRNA complexes, respectively (Figure 1d). Among them, 76.2 % of the events belonged to the release probe / microRNA duplex, thus confirming that the release probe can pull off the target nucleic acid from the capture probe / microRNA complex due to the formation of more stable DNA/RNA duplex. In sharp contrast, the whole blood sample spiked with the microRNA analyte produced only one type of release probe/ RNA duplex events (Figure 1e), while the blood control sample (without the target nucleic acid) didn’t produce any current blockage events. The disappearance of the capture probe / microRNA complex events in the spiked whole blood sample was attributed to two reasons: first, most of the complexes were dissociated and the liberated microRNA molecules were hybridized with the release probe, which were present in the solution in the release probe-microRNA duplex format; and second, some of the capture probe / microRNA complex molecules remained attached to the magnetic beads, which could not be detected by the nanopore sensor due to its inability to enter the liquid phase.
To determine the efficiency of our nanopore sensing strategy, recovery of the target microRNA from the blood sample was investigated (see Supplementary Information). We found that, under our experimental condition (i.e., with the concentrations of the capture probe, target microRNA, and release probe at 250 nM, 200 nM, and 250 nM, respectively), microRNA recovery was 72.5 ± 6.9%. Note that loss of microRNA might occur during the two stages of sample preparation, i.e., utilizing the capture probe to separate microRNA from the blood matrix (stage 1), and using release probe to pull off the target microRNA from the capture probe (stage 2). Our experiments showed that 17.2 ± 2.0% of the target microRNA was lost during stage 1, suggesting the hybridization between the capture probe and microRNA was incomplete. Although use of a locked nucleic acid instead of the natural DNA as the capture probe has the potential to improve the efficiency of the hybridization reaction, another simple strategy is to raise the molar ratio between the used capture probe and target microRNA. For this purpose, we kept the concentration of capture probe constant (at 250 nM) but decreased the microRNA concentration from 200 nM to 100 nM and 50 nM, respectively. We found that, as the molar ratio of capture probe vs. microRNA increased to 5:1, 97.1 ± 1.0% of the microRNA was hybridized with the capture probe (Figure 2). Similarly, increasing the amount of release probe could also improve microRNA recovery. For example, as the molar ratio of release probe vs. microRNA increased from 5:4 to 5:2, a ~5% increase in RNA recovery was obtained. Taken together, the combined results demonstrated that our developed displacement reaction nanopore sensing approach could effectively overcome the effect of blood matrix on the protein-based nanopore sensor, offering the potential for clinical diagnostic applications.
Fig. 2.
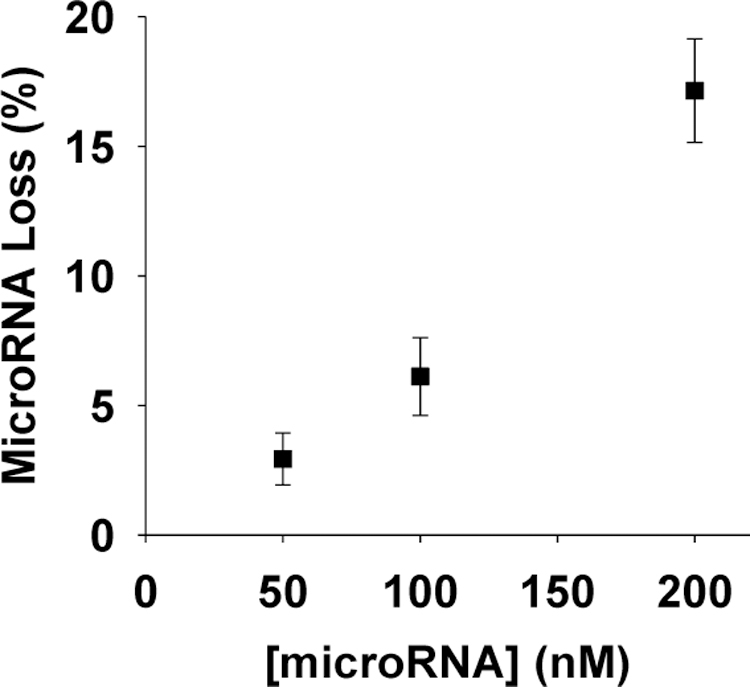
Plot of percent microRNA loss as a function of microRNA concentration. The experiments were performed at +160 mV in 1 M KCl solution containing 10 mM Tris-HCl at pH 8.0. The concentration of the capture probe was 250 nM.
Conclusions
In summary, by utilizing a capture probe and a release probe, and taking advantage of displacement chemical reaction, target nucleic acid can be selectively extracted from whole blood and accurately detected by nanopore sensor, thus overcoming the matrix effect on nanopore (especially, protein ion channel based) stochastic sensing. Compared with the triplex molecular beacon approach for detection of specific DNA30, our method is intuitive and does not involve any complicated design of DNA probes, and hence is easy to implement. Although the strategy was demonstrated by analysis of microRNA in the whole blood, it was in principle applicable to nanopore detection of nucleic acids in various complicated matrix samples (see supplementary information, Figure S8, for nanopore detection of microRNA in serum). Our method is simple and sensitive: does not involve tedious centrifugation separation and elution procedures or require sample dilution. Given the importance of nucleic acids as valuable biomarkers for various diseases, our developed displacement chemistry-based nanopore sensing strategy offers the potential for fieldable / point-of-care diagnostic applications.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (2R15GM110632–02), National Science Foundation (1708596), and Pioneer Hundred Talents Program of Chinese Academy of Sciences (to L.W.) for supporting this work.
Footnotes
† Electronic supplementary information (ESI) available: Experimental section and additional figures.
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Bayley H ; Cremer PS Nature 2001, 413, 226. [DOI] [PubMed] [Google Scholar]
- 2.Wang G; Wang L; Han Y; Zhou S ; Guan X Acc. Chem. Res 2013, 46, 2867–2877. [DOI] [PubMed] [Google Scholar]
- 3.Howorka S Nature Nanotech 2017,12, 619. [DOI] [PubMed] [Google Scholar]
- 4.Liu S-C; Li M-X; Li M; Wang Y; Ying Y-L; Wan Y-J ; Long Y-T Faraday Discussions 2018, doi: 10.1039/c8fd00023a. [DOI] [PubMed] [Google Scholar]
- 5.Misiunas K; Ermann N ; Keyser UF Nano Lett 2018, 18, 4040–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi W; Friedman AK ; Baker LA Anal. Chem 2016, 89, 157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo W; Tian Y ; Jiang L Acc. Chem. Res 2013, 46, 2834–2846. [DOI] [PubMed] [Google Scholar]
- 8.Piguet F; Ouldali H; Pastoriza-Gallego M; Manivet P; Pelta J ; Oukhaled A Nature commun 2018, 9, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laszlo AH; Derrington IM; Ross BC; Brinkerhoff H; Adey A; Nova IC; Craig JM; Langford KW; Samson JM ; Daza R Nature Biotechnol 2014, 32, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heerema SJ ; Dekker C Nature nanotechnology 2016, 11, 127. [DOI] [PubMed] [Google Scholar]
- 11.Deamer D; Akeson M ; Branton D Nature Biotechnol 2016, 34, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy E; Hokmabadi M; Dong Z; McKelvey K; Nelson EM ; Timp G Nano Lett 2018, 57, 3602–3606. [DOI] [PubMed] [Google Scholar]
- 13.Carson S; Wick ST; Carr PA; Wanunu M ; Aguilar CA ACS Nano 2015, 9, 12417–12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X; Wang L; Roozbahani GM; Zhang Y; Xiang J ; Guan X Electrophoresis 2018, doi: 10.1002/elps.201800193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L; Li T; Zhang S; Song P; Guo B; Zhao Y ; Wu HC Angew. Chem. Int. Ed 2018, doi: 10.1002/anie.201803324 [DOI] [PubMed] [Google Scholar]
- 16.Roozbahani GM; Chen X; Zhang Y; Juarez O; Li D ; Guan X Anal. Chem 2018, 90, 5938–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roozbahani GM; Chen X; Zhang Y; Xie R; Ma R; Li D; Li H ; Guan X ACS Sens 2017, 2, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper JA; Borsley S; Lusby PJ ; Cockroft SL Chem. Sci 2017, 8, 5005–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak DK; Chae H; Lee MK; Ha JH; Goyal G; Kim MJ; Kim KB ; Chi SW Angew. Chem. Int. Ed 2016, 55, 5713–5717. [DOI] [PubMed] [Google Scholar]
- 20.Guan X; Gu LQ; Cheley S; Braha O ; Bayley H ChemBioChem 2005, 6, 1875–1881. [DOI] [PubMed] [Google Scholar]
- 21.Wang L,Han Y, Zhou S, Guan X, Biosens. Bioelectron 2014, 62, 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukwikila M ; Howorka S Anal. Chem 2015, 87, 9149–9154. [DOI] [PubMed] [Google Scholar]
- 23.Wang S; Haque F; Rychahou PG; Evers BM ; Guo P ACS Nano 2013, 7, 9814–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y; Zheng D; Tan Q; Wang MX ; Gu L-Q Nature Nanotech 2011, 6, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L; Han Y; Zhou S; Wang G ; Guan X ACS Appl. Mater. Interfaces 2014, 6, 7334–7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson RP; Fleming AM; Perera RT; Burrows CJ; White HS J Am Chem Soc 2017, 139, 2750–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu A; Zhao Q; Krishantha DMM; Guan XJ Phys. Chem. Lett 2011, 2, 1372–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markham NR; Zuker M Nucleic Acids Res 2005, 33, W577–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maglia G; Restrepo MR; Mikhailova E ; Bayley H Proc. Natl. Acad. Sci. U. S. A 2008, 105, 19720–19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garaj S; Liu S; Golovchenko JA ; Branton D Proc. Natl. Acad. Sci. U. S. A 2013, 110, 12192–12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee S; Wilson J; Shim J; Shankla M; Corbin EA; Aksimentiev A; Bashir R Adv. Funct. Mater 2015, 25, 936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo B; Sheng Y; Zhou K; Liu Q; Liu L ; Wu HC Angew. Chem. Int. Ed 2018, 57, 3602–3606. [DOI] [PubMed] [Google Scholar]
- 33.Ding Y; Fleming AM; White HS ; Burrows CJ The J. Phys. Chem. B 2014, 118, 12873–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perera RT; Fleming AM; Peterson AM; Heemstra JM; Burrows CJ ; White HS Biophys. J 2016, 110, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.