Abstract
Purpose of review:
We assess the implications of recent advances in the genetics of juvenile idiopathic arthritis (JIA) for the evolving understanding of inflammatory arthritis in children.
Recent findings:
JIA exhibits prominent genetic associations with the human leukocyte antigen (HLA) region, extending perhaps surprisingly even to the hyperinflammatory systemic JIA category. Some HLA associations resemble those for adult-onset inflammatory arthritides, providing evidence for pathogenic continuity across the age spectrum. Genome-wide association studies have defined an increasing number of JIA-linked non-HLA loci, many again shared with adult-onset arthritis. Since most risk loci contain only non-coding variants, new experimental methods such as SNP-seq and innovative big-data strategies help identify responsible causative mutations, termed functional SNPs (fSNPs). Alternately, gene hunting in multiplex families implicates new genes in monogenic childhood arthritis, including the intriguing innate immune gene LACC1.
Summary:
Genetic data indicate a continuity between JIA and adult arthritis poorly reflected in current nomenclature. Advancing methodologies will help to identify new pathogenic mechanisms that inform the understanding of biologic subdivisions within JIA. Resulting insights will facilitate the application of lessons learned across the age spectrum to the treatment of arthritis in children and adults.
Keywords: juvenile idiopathic arthritis, rheumatoid arthritis, genetics, genome-wide association studies, SNP-seq, LACC1
Introduction
Juvenile idiopathic arthritis (JIA) encompasses a range of clinical phenotypes characterized by chronic inflammatory arthritis of unknown cause beginning before age 16 years [1]. Clinical and epidemiological features have been employed to categorize patients with JIA into 7 categories under the International Leagues of Associations for Rheumatology (ILAR) nomenclature. The ILAR categories represent a work in progress, and their shortcomings are now broadly appreciated. These include intricate and often counterintuitive inclusion and exclusion criteria; category-switching among patients followed over time; and growing evidence that ILAR boundaries fail to reflect underlying disease biology. More fundamentally, the age cutoff at the 16th birthday as determined without no pathophysiologic or epidemiological basis, and yet has come to represent a yawning nomenclature gap that divides pediatric and adult rheumatology [2]••. With the number of new therapies now outstripping the availability of patients for clinical studies, it is more important than ever to define biologic subgroups within childhood arthritis to identify patients most likely to benefit from mechanism-specific targeting and so that lessons learned in arthritis patients across the lifespan can be applied, without the hindrance of inaccurate nomenclature, across the whole age spectrum.
The challenge is to define patient subgroups. Recent attempts at this difficult problem have employed clinical and blood-based biomarkers, joint trajectories, and informed expert opinion [3–5]. Such studies limit their attention to pediatric patients and face the further challenge that distinct processes can yield convergent clinical phenotypes. For example, seronegative and seropositive adult rheumatoid arthritis (RA) display remarkable clinical overlap despite fundamental differences in the role of autoantibodies, complement, and synovial T cells [2]••. Phenotypic convergence is illustrated similarly in animal models of arthritis, wherein similar clinical features can emerge through remarkably different pathways [6]. Thus, while patient phenotyping is important for subgroup definition, and indeed will be required for clinical practice, complementary approaches remain essential.
Genetics as a tool to understand human polygenic disease
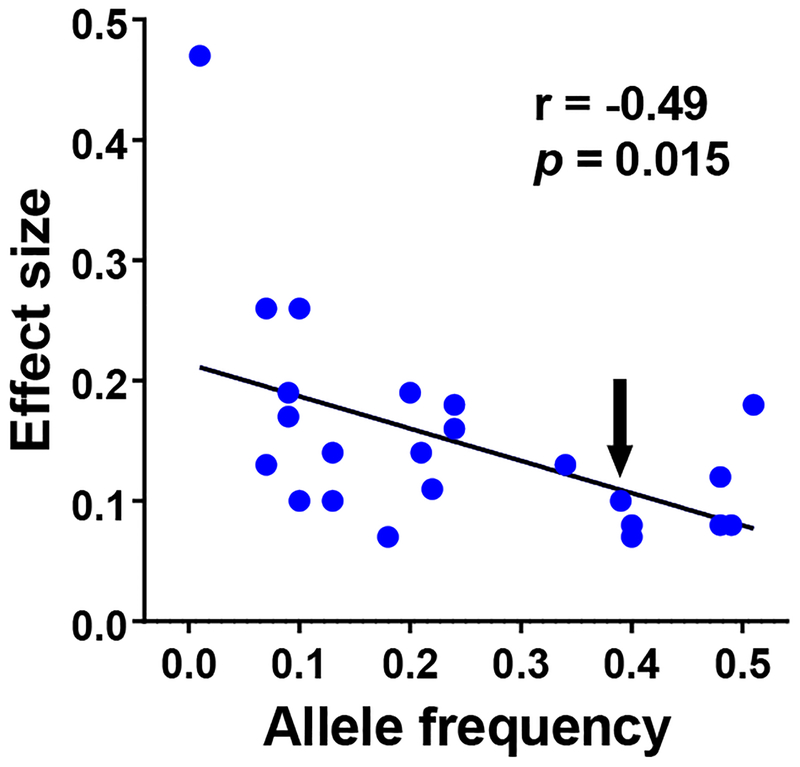
One of these approaches is genetics. Since genetic endowment precedes disease risk, genetic variants associated at a statistically-robust level with disease risk conclusively implicate related causal mechanisms. Critically, the magnitude of the associated risk bears no necessary relationship to the importance of the related gene or to its potential as a drug target. This is because observed risk reflects the impact of the variant, not of the gene itself. Genome-wide association studies (GWAS) detect risk modulation resulting from common variants, typically with a minor allele frequency (MAF) greater than 5%, whose “survival” in the population all but guarantees that their functional impact is modest. This principle was well illustrated by an early GWAS characterizing loci associated with blood lipids [7]. Figure 1 depicts the linear relationship between variant effect size and allele frequency, with the gene encoding HMG Co-A reductase – the target of blockbuster statin therapy – hidden anonymously in the middle (arrow).
Figure 1. Inverse relationship between allele frequency and effect size in GWAS data.
Depicted are 24 GWAS “hits” associated with blood lipoproteins or lipids from reference [7]. The Y axis depicts effect size as the absolute value of the β-coefficient, defined as the proportion of one standard deviation of change (corrected for age and other clinical factors) per allele copy; statistics reflect Pearson correlation. The arrow depicting a SNP with allele frequency 0.39 and effect size β (absolute value) = 0.10 is an intronic SNP in the statin target HMGCR. The outlier with allele frequency 0.01 and β (absolute value) = 0.47 is PCSK9, a newer lipid drug target.
A further challenge in the interpretation of GWAS studies is the ambiguity of each “hit.” The single nucleotide polymorphisms (SNPs) studied by GWAS tag large linkage disequilibrium (LD) blocks. Only rarely will the tagging SNP itself be causal. Unfortunately for investigators, in most cases the variant that is truly causal is only rarely evident. LD blocks often contain multiple genes, and in the large majority of cases (>85%) none of these feature any coding variant. Thus most true causal variants – we have termed these functional SNPs, or fSNPs [8]•• – reside somewhere in the vast stretches of non-coding DNA that make up 98% of the genome and act via regulation of gene expression. Since many common non-coding variants reside in LD with each tagging SNP, fSNP identification poses a challenging problem. The challenge is further compounded by the fact that some loci contain several cooperating fSNPs within the same LD block, for example in the CD40 risk locus for RA and the STAT4 risk locus for JIA [8]••.
Multiple approaches are being developed to address these challenges. The Immunochip was designed to focus genetic studies on 186 loci of immunologic interest, each characterized in greater depth by means of multiple tagging SNPs (196,524 in total) to better localize potential causal variants [9,10]. Bioinformatic strategies can help predict non-coding variants, in particular those that function by modulating the binding of DNA regulatory proteins such as transcription factors (TFs). These strategies incorporate multiple lines of evidence, including epigenetic marks and predicated and/or experimentally-determined TF binding motifs [11]. Fine mapping employs dense SNP analysis together with targeted DNA sequencing to more carefully explore loci known to contain fSNPs [12]•. Finally, tools have been developed to identify non-coding variants using high-throughput experimental strategies. These include SNP-seq, a method we developed that employs restriction enzyme protection to identify SNPs that modulate the binding TFs (discussed further below) [8]••. An alternative method is the massively parallel reporter assay (MPRA), wherein target cells are transfected simultaneously with a large number of SNP-containing DNA constructs to test which best promote expression of a reporter gene [13]. Finally, families bearing multiple affected members can be studied to identify rare but highly-potent gene variants, combining traditional Mendelian genetics with high-resolution next-generation DNA sequencing. We will discuss the impact of these methods on the evolving understanding of JIA.
HLA associations in JIA
Extensive and well-documented associations between JIA and the HLA region have been recognized for more than 40 years, differing with disease phenotype [14–16]. Principal components analysis demonstrates that age of onset, rather than oligoarticular or polyarticular presentation, is a main driver of HLA association [17]•. Adult seropositive RA risk haplotypes containing HLA-DRB1*01/*04 actually appear protective in seronegative arthritis, emerging as a risk gene only in adolescence; similarly, HLA-B27 is overrepresented only in children presenting with arthritis after early childhood [18].
Advances in serological approaches, allele-specific DNA hybridization, and most recently imputation using high-density SNP genotyping have further enhanced the resolution of these studies. Hinks, Bowes and colleagues characterized the HLA region in 5043 JIA patients by imputing SNPs, classical HLA alleles and HLA amino acids [19]••. Bivariate analysis was used to correlate these results with ILAR subcategory. They found three subtypes – persistent oligoarticular, extended oligoarticular, and seronegative polyarthritis – to be genetically indistinguishable, at least by HLA associations. Importantly, these associations overlapped markedly with those of seronegative adult RA. Correspondingly, seropositive polyarticular JIA resembled seropositive adult RA, enthesitis-related arthritis resembled adult ankylosing spondylitis, and psoriatic JIA (poorly represented in this cohort, and difficult to distinguish unambiguously in children) bore some similarity to adult psoriatic arthritis. In other words, HLA associations point to extensive continuities in arthritis across the age spectrum.
A further comparison of 214 patients with JIA-associated uveitis with 362 patients with uveitis-free JIA identified an association with serine at position 11 in HLA-DRB1 that achieved significance in girls but was less clear in boys, of whom fewer were available for study [20]. Whereas cases and controls were not fully matched on arthritis phenotype, and no correction for age of onset was employed, the specificity of this result for uveitis itself – as opposed to a uveitis-prone form of JIA with a predilection for early childhood – remains unclear. Using an in silico approach to model antigen presentation at 13 common HLA-DRB1 alleles, the authors found that alleles with Ser11 (together with 3 adjacent amino acids in nearly perfect LD) could be distinguished by peptide binding preferences, potentially helping to identify antigens involved in the development of uveitis.
HLA associations are typically regarded as strong evidence for the involvement of antigen-specific T cells in disease pathogenesis. In this context it is remarkable that the strongest GWAS signal in systemic JIA (sJIA), often considered an autoinflammatory disease, is also the HLA region. Ombrello and colleagues compared 770 sJIA patients to 6947 controls, finding that HLA-DRB1*11 and its defining amino acid glutamate 58 exhibited an odds ratio of 2.3, while only one other locus – LOC284661 on chromosome 1 – achieved genome-wide significance, with another (IL1RN, encoding the IL-1 receptor antagonist) identified via candidate gene studies [21,22]•. No evidence of shared genetic architecture was found when weighted genetic risk scores were used to compare sJIA with other JIA subtypes [23]•. Mechanisms exist whereby HLA alleles can modulate immunity without serving as antigen presenting molecules, for example through linked non-HLA genes in the locus, as targets of staphylococcal enterotoxins, and by modulating expression of gene elsewhere in the genome [24,25]. Nevertheless, since the HLA haplotype implicated in sJIA is also associated with JIA of other types, the most straightforward interpretation of this finding is that antigen-specific T cells play a role in sJIA biology, at least in some patients [17,26,27]. This conclusion is further supported by the clinical efficacy of T cell costimulatory blockade with CTLA4-Ig in sJIA [28]. Genetics thus inform, but do not yet resolve, the evolving understanding of sJIA as autoinflammatory, autoimmune, or potentially both [29,30].•
Non-HLA associations in JIA
As noted above, despite their relatively weak effect size, genetic associations outside of the HLA region are likely to provide important clues to disease pathophysiology. Prior to 2013, genetic studies in childhood arthritis (combined oligoarticular and polyarticular forms) had identified only two associated genetic loci beyond the HLA region: PTPN22 and PTPN2. The Immunochip enabled a broader study of JIA despite the relatively limited number of available patients. Hinks, Cobb and colleagues compared 2,816 oligo- and seronegative polyarticular JIA patients with 13,056 controls, identifying 14 new loci at genome-wide significance threshold (P < 5 × 10−8) and an additional 11 at a suggestive level (P < 1 × 10−6) [31]••. These 27 non-HLA genes are listed in Table 1, together with an additional 9 loci added by McIntosh and colleagues using traditional SNP microarrays encompassing regions excluded from the Immunochip (albeit none to genome-wide significance) [32]•.
Table 1. 37 genomic loci associated with oligoarticular and polyarticular JIA by GWAS.
The first 17 loci achieved genome-wide significance (< 5 × 10−8); the latter 20 approached but did not achieve this threshold, and so are considered unconfirmed. Where two sets of data are presented for a single locus, the second represents an imputed variant determined to have a level of significance greater than the measured variant. For LTBR, IL6, COG6, and PRRL5 these imputed variants achieve genome-wide significance. Genes in bold represent plausible candidates for the causative gene at that locus, because the linked allele represents a non-synonymous coding variant (TYK2, SH2B3), a truncating splice variant (ERAP2), or because dense mapping at the locus narrows the causative variant to within one gene (linkage disequilibrium r2 > 0.9; STAT4, PTPN2, IL2RA, IL2RB, ZFP36L1, COG6, PRRL5; LD data are not available for McIntosh 2017 loci). LTBR and PTH1Rc contain coding variants, but they are synonymous. The seropositive and seronegative RA columns reflect the presence of an overlapping (r2 >0.8) “hit” SNP in seropositive RA per Okada 2014 [55] (n=29,880 cases, 88.1% seropositive, 9.3% seronegative, 2.6% unknown) or Viatte 2012 [56], Viatte 2016 [57], and Wei 2017 [58] (n=3,323 cases, all CCP negative). Based on the smaller size of the seronegative cohort, some true overlap may be missed. Acknowledgements to Marc Sudman (Cincinnati Children’s Hospital Medical Center) for the data and analysis upon which this table is based.
| Locus | Chromosome | Odds ratio | Significance | Seropos RA | Seroneg RA | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Genome-wide significance | ||||||||
| 1 | HLA-DQB1 – HLA-DQA2 | 6 | 6.01 | 3.14 × 10−174 | X | X | Hinks, Cobb 2013 | |
| 2 | PTPN22 | 1 | 1.59 | 3.19 × 10−25 | X | X | Hinks, Cobb 2013 | |
| 3 | STAT4 | 2 | 1.29 | 1.28 × 10−13 | X | X | Hinks, Cobb 2013 | |
| 4 | PTPN2 | 18 | 1.31 | 1.44 × 10−12 | Hinks, Cobb 2013 | |||
| 5 | ANKRD55 | 5 | 0.78 | 4.40 × 10−11 | X | Hinks, Cobb 2013 | ||
| 0.79 | 2.73 × 10−11 | X | Hinks, Cobb 2013 | |||||
| 6 | IL2-IL21 | 4 | 0.79 | 6.24 × 10−11 | X | Hinks, Cobb 2013 | ||
| 7 | TYK2 | 19 | 0.56 | 1 × 10−10 | X | Hinks, Cobb 2013 | ||
| 8 | IL2RA | 10 | 0.72 | 8 × 10−10 | X | Hinks, Cobb 2013 | ||
| 9 | SH2B3-ATXN2 | 12 | 1.20 | 2.60 × 10−9 | X | Hinks, Cobb 2013 | ||
| 1.20 | 1.61 × 10−9 | X | Hinks, Cobb 2013 | |||||
| 10 | ERAP2-LNPEP | 5 | 1.32 | 7.50 × 10−9 | Hinks, Cobb 2013 | |||
| 1.31 | 7.37 × 10−9 | Hinks, Cobb 2013 | ||||||
| 11 | UBE2L3 | 22 | 1.24 | 6.20 × 10−9 | X | Hinks, Cobb 2013 | ||
| 12 | C5orf56-IRF1 | 5 | 0.84 | 1.02 × 10−8 | Hinks, Cobb 2013 | |||
| 0.76 | 9.73 × 10−10 | Hinks, Cobb 2013 | ||||||
| 13 | RUNX1 | 21 | 0.78 | 1.06 × 10−8 | X | Hinks, Cobb 2013 | ||
| 0.78 | 5.44 × 10−9 | X | Hinks, Cobb 2013 | |||||
| 14 | IL2RB | 22 | 0.84 | 1.55 × 10−8 | Hinks, Cobb 2013 | |||
| 15 | ATP8B2-IL6R | 1 | 1.33 | 2.75 × 10−8 | Hinks, Cobb 2013 | |||
| 1.36 | 1.26 × 10−8 | Hinks, Cobb 2013 | ||||||
| 16 | FAS | 10 | 1.18 | 2.93 × 10−8 | Hinks, Cobb 2013 | |||
| 17 | ZFP36L1 | 14 | 0.77 | 1.59 × 10−8 | Hinks, Cobb 2013 | |||
| 0.77 | 1.24 × 10−8 | Hinks, Cobb 2013 | ||||||
| Suggestive significance | ||||||||
| 18 | LTBR | 12 | 1.20 | 5.10 × 10−8 | Hinks, Cobb 2013 | |||
| 1.24 | 4.54 × 10−9 | Hinks, Cobb 2013 | ||||||
| 19 | IL6 | 7 | 1.19 | 5.80 × 10−8 | Hinks, Cobb 2013 | |||
| 1.19 | 3.36 × 10−8 | Hinks, Cobb 2013 | ||||||
| 20 | COG6 | 13 | 0.84 | 1.61 × 10−7 | X | Hinks, Cobb 2013 | ||
| 0.84 | 4.52 × 10−8 | X | Hinks, Cobb 2013 | |||||
| 21 | 13q14 | 13 | 1.18 | 1.77 × 10−7 | Hinks, Cobb 2013 | |||
| 22 | CCR1-CCR3 | 3 | 0.78 | 1.88 × 10−7 | Hinks, Cobb 2013 | |||
| 23 | PRR5L | 11 | 0.80 | 3.35 × 10−7 | Hinks, Cobb 2013 | |||
| 0.78 | 1.90 × 10−8 | Hinks, Cobb 2013 | ||||||
| 24 | PRM1-RMI2 | 16 | 0.81 | 4.46 × 10−7 | Hinks, Cobb 2013 | |||
| 0.85 | 2.40 × 10−7 | Hinks, Cobb 2013 | ||||||
| 25 | RUNX3 | 1 | 1.16 | 4.66 × 10−7 | Hinks, Cobb 2013 | |||
| 26 | TIMMDC1-CD80 | 3 | 1.20 | 6.30 × 10−7 | Hinks, Cobb 2013 | |||
| 1.22 | 3.64 × 10−7 | Hinks, Cobb 2013 | ||||||
| 27 | JAZF1 | 7 | 1.25 | 6.60 × 10−7 | X | Hinks, Cobb 2013 | ||
| 1.28 | 1.12 × 10−7 | X | Hinks, Cobb 2013 | |||||
| 28 | AFF3-LONRF2 | 2 | 1.25 | 8.83 × 10−7 | X | Hinks, Cobb 2013 | ||
| 1.24 | 8.10 × 10−7 | X | Hinks, Cobb 2013 | |||||
| 29 | JAK1 | 1 | 0.78 | 4.18 × 10−7 | McIntosh 2017 | |||
| 30 | PRR9_LOR | 1 | 1.43 | 5.12 × 10−8 | McIntosh 2017 | |||
| 31 | PTH1Rc | 3 | 1.23 | 1.87 × 10−7 | McIntosh 2017 | |||
| 32 | ILDR1_CD86 | 3 | 1.45 | 6.73 × 10−8 | McIntosh 2017 | |||
| 33 | LINC00L51 | 6 | 1.42 | 5.80 × 10−7 | McIntosh 2017 | |||
| 34 | AHI1_LINC00271 | 6 | 1.18 | 3.48 × 10−7 | McIntosh 2017 | |||
| 35 | HBP1 | 7 | 0.84 | 7.29 × 10−7 | McIntosh 2017 | |||
| 36 | WDFY4 | 10 | 1.27 | 1.79 × 10−7 | McIntosh 2017 | |||
| 37 | RNF215 | 22 | 1.19 | 3.09 × 10−7 | McIntosh 2017 | |||
Genetics as a tool to understand disease mechanisms
What do these findings tell us about the pathogenesis of JIA? Importantly, the “label” for each locus cannot be assumed to represent the gene responsible for the increment in risk. In cases where the tagging SNP correlates with a mis-sense coding variant, a causal connection is typically considered likely. But such cases are relatively uncommon – only 2 out of 17 confirmed JIA loci (Table 1) – and even these attributions remain uncertain until experimentally confirmed. For non-coding variants, the regulated gene (or genes) need not reside even within the same LD block, since modulation of expression can operate over considerable distances [25]. For some loci, regulatory effects may operate via perturbation of the densely-interconnected signaling network within the cell, without the directly-modulated gene itself participating in a key pathogenic pathway, a possibility termed the “omnigenic hypothesis” that may help explain the so-called missing heredity left over after accounting for the effects of GWAS-defined risk variants [33]•.
With these caveats, it still makes sense to consider the potential significance of the identified loci for the pathogenesis of JIA, beginning with the coding variants for which the causative gene is plausibly inferred. For JIA, as illustrated in Table 1, these are limited to TYK2 (loss of function allele protective), SH2B3 (loss of function allele associated with risk), possibly LTBR (synonymous coding variant), and the splice variant in ERAP2 (loss of function allele protective). To these we can add genes for which fine mapping localizes the fSNP to a single gene, including STAT4, PTPN2, IL2RA, IL2RB, ZFP36L1, and probably COG6 and PRR5L (i.e. where the fSNP-containing LD region r2 > 0.9 resides entirely within the gene, suggesting but not proving that the gene itself is a regulated target). The expression of these genes at the mRNA level across key hematopoietic lineages, as well as major functions, are listed in Table 2. Most are widely expressed. Broadly, most affect T cell function, supporting an important role for T cells in JIA pathogenesis, consistent with the strong HLA association, epigenetic studies that localize GWAS hits to regions of open chromatin implicated in CD4+ T cell function, and evidence for clonal skewing of T cell populations [34–37]. Expression of JIA risk genes is also abundant in innate lineages, including natural killer cells, monocyte and neutrophils, again consistent with epigenetic data available from neutrophils and concordant with the key role of neutrophils in arthritis generally [34,38].
Table 2. Expression and function in human hematopoietic cells of selected genes implicated in JIA.
Data reflect mRNA and reflect approximate relative expression as extracted manually from human gene expression data (D-Map and Garvan) of at www.ImmGen.org (version May 2019).
| CD4 T | CD8 T | B | DC | NK | Monocyte | Neutrophil | Selected functions | |
|---|---|---|---|---|---|---|---|---|
| TYK2 | X | X | XX | XX | XX | XX | XX | Signal transduction for cytokines including type 1IFN, IL-6, IL-23 |
| SH2B3 | X | X | X | XX | XX | XX | X | Negative regulator of signals from TNF and growth factors (a.k.a. LNK) |
| LTBR | XX | X | XX | XX | Receptor for lymphotoxin and LIGHT | |||
| ERAP2 | XX | XX | XX | X | XX | X | X | antigen trimming for presentation by MHC I (a.k.a. LRAP) |
| STAT4 | XXX | XXX | X | XXX | XX | Signal transduction, Th1 differentiation | ||
| PTPN2 | XX | XXX | XX | XX | XXX | XX | XX | phosphatase, negative regulator of JAKs and other signaling molecules |
| IL2RA | XX | X | X | X | X | IL-2 receptor alpha chain (a.k.a. CD25) | ||
| IL2RB | XX | XX | XXX | XX | IL-2 receptor beta chain | |||
| ZFP36L1 | XX | XX | XXX | XX | XX | XXX | XXXX | Regulate mRNA stabilty, impacting lineages incuding B cells and chondrocytes |
| PRR5L | X | XX | XX | XX | mTORC2 regulation, fibroblast migration |
New approaches to identifying non-coding variants.
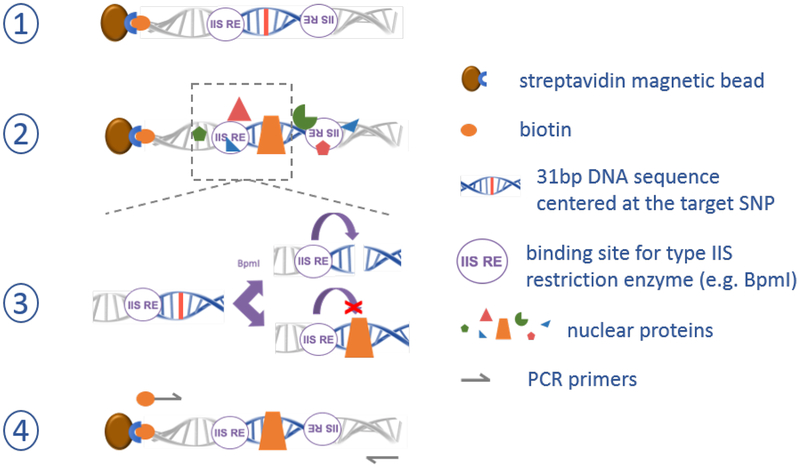
Two recent studies help to refine the analysis of JIA risk loci beyond the HLA. Our group approached the problem of finding non-coding variants using molecular biology, asking whether candidate causal SNPs could be screened experimentally to identify those that bind TFs and are therefore more likely to be causal [8]••. We developed a method termed SNP-seq, whereby all common SNPs within LD of GWAS-identified tagging SNPs (e.g. LD r2 > 0.8) are positioned within synthetic DNA constructs flanked by binding sites for the type IIS restriction enzyme BpmI (Figure 2). Unlike most restriction enzymes, type IIS enzymes cut DNA at a defined distance (for Bpm1, 15 base pairs) adjacent to their binding site, in a sequence non-specific manner. This method enables a screening approach whereby a library of constructs containing different alleles can be incubated with nuclear extract containing mixture of TFs, exposed to BpmI, regenerated by PCR, and then quantitated via next-generation sequencing (the “seq” in SNP-seq) to identify those sequences which were shielded from cleavage by a protein bound from the nuclear extract. Using this method, we screened 608 SNPs (1223 alleles) from the 27 JIA-associated loci implicated by Hinks, Bowes and colleagues to identify 148 candidate fSNPs that displayed evidence for allele-specific protein binding. Pursuing the STAT4 locus in detail, we tested each SNP, confirming that 2 of 4 SNPs (rs8179673 and rs10181656, residing 502bp apart on the same haplotype) exhibited both allele-specific protein binding and allele-dependent regulatory function in luciferase reporter constructs. To “close the loop” on these fSNPs, we employed a DNA pulldown assay termed Flanking Restriction Enhanced Pulldown (FREP), developed previously in the lab to identify TFs recognizing an fSNP of interest [39]. Knockdown of the TFs identified in this way established specific modulation of STAT4 expression, confirming regulation of this T cell regulatory gene via a pair of haplotype-linked JIA risk fSNPs.
Figure 2. SNP-seq methodology to screen non-coding DNA variants for regulatory protein binding.
Step 1) a library of synthetic DNA constructs is generated containing each allele of each candidate non-coding variant near a GWAS tagging SNP within its local 31bp context. Step 2) the library of constructs is exposed to nuclear extract from candidate cells, and non-binding proteins are washed away. Step 3) constructs are incubated with a type IIS restriction factor (here BpmI); constructs in which a protein binds the candidate SNP are shielded from cleavage. Step 4) PCR using one biotinylated and one non-biotinylated PCR primer are used to regenerate the library for another round of screening, typically repeated 5–10 times to enable sufficient amplification, followed by next-generation sequencing to identify surviving sequences as candidate fSNPs.
Harley and colleagues used new computational methods developed by the Weirauch lab to show that particular transcription factors (TFs) occupy multiple loci associated with individual complex genetic disorders [40]••. Interestingly it was noted that that about half of gene loci for systemic lupus erythematosus (SLE) and multiple sclerosis are occupied by the Epstein-Barr virus (EBV) enhancer protein EBNA2, along with co-clustering human TFs. EBNA2 interacts with multiple human TFs, reprogramming cells to promote viral survival, suggesting a mechanism by which EBV infection could distort immune responses to favor development of SLE. Intriguingly, a similar EBNA2 signature was found in other autoimmune diseases, including JIA, but not for unrelated diseases such as breast or prostate cancer. This finding raises the possibility that many loci are associated genetically with JIA because they are susceptible to aberrant regulation by EBV, or potentially by other viruses, such that infection with this nearly ubiquitous pathogen could help trigger JIA in a genetically susceptible host. The remarkable corollary is that development of a successful strategy to vaccinate against EBV – still a distant goal – could markedly reduce the incidence of JIA and other autoimmune diseases. This earthshattering possibility remains to be tested using other methods.
Subdivisions within arthritis as defined by non-HLA loci.
Just as HLA associations have been employed to explore the similarities between forms of arthritis, non-HLA loci can serve a similar purpose. McIntosh and colleagues evaluated the overlap between JIA “hits” and those from adult RA, principally from seropositive patients. A large meta-analysis of 30,000 RA patients and 74,000 controls had defined 101 genetic loci associated with either increased or decreased risk of RA [32]. McIntosh sought overlap between these loci and those associated with oligo- and seronegative polyarticular JIA. Correcting for the different tagging SNPs used in these studies, and the more limited power afforded by the smaller JIA cohort, they estimated that if these diseases were identical, then overlap should emerge in 25.9 loci (95% confidence interval 16.5–32.7). In fact, only 9 were found among non-HLA GWAS-wide hits (Table 1). This finding illustrates that seronegative forms of JIA – like seronegative forms of adult RA – bear only modest genetic similarity to adult seropositive RA.
Seronegative JIA is even more distant from sJIA. Ombrello and colleagues developed a genetic risk score from their sJIA GWAS study and showed essentially no overlap at all with seronegative JIA, confirming that the marked clinical division between these subgroups corresponds to an equally stark genetic division [23]•.
By contrast, the similarity between seropositive forms extends across the age spectrum. Prahalad and colleagues demonstrated that children with RF+ JIA shared both HLA and non-HLA associations with RF+ adult RA [41,42]•. While far more limited data are available for seronegative adult RA, some associated loci confer risk for both seronegative RA and seronegative JIA, but not seropositive RA (Table 1) [2]••. Thus, even beyond the HLA, genetic signatures provide strong evidence for the continuity of arthritis across the age spectrum.
Rare variants that present as childhood-onset arthritis
While most childhood arthritis likely originates via a complex interplay between multiple predisposing genes, a subset of cases may arise through highly-penetrant single-gene defects. For example, Blau syndrome is an autosomal dominant autoinflammatory disease arising through activating mutations affecting the intracellular sensor NOD2. Patients with Blau syndrome often present in childhood with arthritis affecting the hands and wrists, sometimes together with uveitis, and so can be easily mistaken for JIA, especially when the characteristic granulomatous skin rash remains absent. Severe erosive polyarthritis has recently been described from a spontaneous gain-of-function mutation in MYD88, encoding a signal transducer downstream of IL-1 and Toll-like receptor signaling [43]•. A small number of such familial cases admixed with “garden-variety” JIA could markedly skew the apparent recurrence risk of JIA in siblings, λs, estimated currently at 11.6 [44].
A gene recently implicated in “familial JIA” is LACC1 (laccase domain containing 1). Positional techniques identified homozygous LACC1 coding variants in members of several consanguineous Saudi Arabian families presenting with childhood-onset febrile arthritis resembling sJIA [45]••. Other forms of familial childhood-onset arthritis, including highly-inflammatory polyarthritis (without fever) and spondyloarthritis, have since been identified in other patients bearing LACC1 mutations, though a screen of 23 systemic JIA and 44 polyarticular JIA patients from monoplex families failed to identify pathogenic LACC1 mutations, suggesting that this gene may not drive ‘garden-variety’ JIA [46–48]. Murine studies show that LACC1 encodes a regulator of fatty acid metabolism, termed FAMIN (fatty acid metabolism-immunity nexus) by the authors, expressed principally in myeloid cells (macrophages, dendritic cells, and neutrophils) but also in B cells [49,50]•. The human expression profile is likely similar, although protein was rarely observed in B cells [51]. LACC1 deficiency impairs the energy reserves of macrophages and neutrophils, translating into a reduced production of reactive oxygen species that may contribute to the association of LACC1 mutations with Crohn’s disease and leprosy [52,53]. LACC1-deficient mice demonstrate exaggerated inflammatory responses to immune stimuli, potentially reflecting aberrant function of energetically-deprived macrophages [49]. Under inflammatory conditions, mice lacking LACC1 also generate more TNF and IL-17, with enhanced Th17 cells development and exacerbated experimental arthritis and colitis, though inflammatory disease did not occur spontaneously [50]. By contrast, in human carriers of a LACC1 mutation associated with Crohn’s disease, production of IL-1β and other cytokines was actually reduced in response to innate immune stimulants such as NOD2 ligands, implicating this protein in the regulation of related intracellular pathogen detection and thereby potentially in pathogen clearance [54]. Together, these studies implicate LACC1 as an interesting new “node” in the pathogenesis of inflammatory diseases including arthritis, though its pathophysiological role remains to be clarified.
Summary and implications
Technical and bioinformatic advances have yielded important new insights into the genetics of JIA. At a most basic level, studies of both HLA and non-HLA associations show that the “hard stop” between JIA and adult arthritis at age 16 fails to correspond to a biologic dividing line. Rather, inflammatory arthritis is likely to represent a continuum across the age spectrum. This general claim does not preclude the existence of specific forms of arthritis restricted to one age group or another. In particular, oligoarticular arthritis accompanied by chronic anterior uveitis is common in early childhood and essentially nonexistent as a de novo phenotype in adults, whereas seropositive arthritis is rare in young children and HLA-B27+ axial spondyloarthritis tends to appear first in the teenage years. Whether residual differences between pediatric and adult arthritis reflect true pathological differences, or instead differences in “substrate” – such as immune function, environmental exposures (including microbiome), and tissues that are growing or senescing – remains to be determined. Genetics will help to guide this determination and should be considered a key “litmus test” for proposed revisions in arthritis nomenclature.
We remain distant from the ultimate goal of personalized medicine in arthritis care, which is to define which therapy will provide each patient with the most benefit at the lowest risk. We remain even further from the prospect of specifically reprogramming immunity to achieve a cure, although the EBNA2 association with JIA presents the tantalizing prospect of prevention. Study of the loci implicated in JIA as well as adult arthritis over coming years should provide further insights onto genes, pathways and cells critical for disease pathogenesis, and thereby new roads forward in the understanding and management of arthritis across the age spectrum.
KEY POINTS.
Juvenile idiopathic arthritis represents a heterogenous group of clinical phenotypes with divergent genetic signatures.
Some forms of pediatric and adult inflammatory arthritis exhibit overlapping genetic associations, strongly suggesting pathophysiological continuity across the age spectrum.
Genetic findings represent important clues to key disease pathways, but challenges in defining causal variants and the mechanisms they engage limit the degree of insight gained so far into disease processes.
Experimental methods such as SNP-seq, as well as advanced bioinformatic methods, hold the potential for illuminating new mechanisms in juvenile idiopathic arthritis as well as potential new avenues to therapy.
Acknowledgments
Funding for this work is provided by NIH grants R01 AR073201, R01 AR065538 and P30 AR070253; the Fundación Bechara; and the Arbuckle Family Fund for Arthritis Research (to PAN); a P30 AR070253 Joint Biology Consortium Microgrant (to MM-B); and NIH grant P30 AR070549 to SDT.
The authors report no conflicts of interest directly relevant to this work. PAN is the recipient of investigator-initiated research grants from Novartis, Pfizer and Bristol-Myers Squibb; consulting fees from Sobi and AbbVie; salary support from the Childhood Arthritis & Rheumatology Research Alliance; and royalties from UpToDate, Inc. and the American Academy of Pediatrics. MM-B and SDT report no disclosures.
REFERENCES AND RECOMMENDED READING
Papers of particular interest have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, He X, Maldonado-Cocco J, Orozco-Alcala J, Prieur AM, et al. : International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol 2004, 31:390–392. [PubMed] [Google Scholar]
- 2.Nigrovic PA, Raychaudhuri S, Thompson SD: Genetics and the classification of arthritis in adults and children. Arthritis Rheumatol 2018, 70:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eng SW, Duong TT, Rosenberg AM, Morris Q, Yeung RS: The biologic basis of clinical heterogeneity in juvenile idiopathic arthritis. Arthritis Rheumatol 2014, 66:3463–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eng SWM, Aeschlimann FA, van Veenendaal M, Berard RA, Rosenberg AM, Morris Q, Yeung RSM, Re A-ORC: Patterns of joint involvement in juvenile idiopathic arthritis and prediction of disease course: A prospective study with multilayer non-negative matrix factorization. PLoS Med 2019, 16:e1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martini A, Ravelli A, Avcin T, Beresford MW, Burgos-Vargas R, Cuttica R, Ilowite NT, Khubchandani R, Laxer RM, Lovell DJ, et al. : Toward New Classification Criteria for Juvenile Idiopathic Arthritis: First Steps, Pediatric Rheumatology International Trials Organization International Consensus. J Rheumatol 2019, 46:190–197. [DOI] [PubMed] [Google Scholar]
- 6.Chang MH, Nigrovic PA: Antibody-dependent and -independent mechanisms of inflammatory arthritis. JCI Insight 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. : Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet 2008, 40:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G, Martinez-Bonet M, Wu D, Yang Y, Cui J, Nguyen HN, Cunin P, Levescot A, Bai M, Westra HJ, et al. : High-throughput identification of noncoding functional SNPs via type IIS enzyme restriction. Nat Genet 2018, 50:1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, Bakker SF, Bardella MT, Bhaw-Rosun L, Castillejo G, et al. : Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet 2011, 43:1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortes A, Brown MA: Promise and pitfalls of the Immunochip. Arthritis Res Ther 2011, 13:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT: The Human Transcription Factors. Cell 2018, 175:598–599. [DOI] [PubMed] [Google Scholar]
- 12.Westra HJ, Martinez-Bonet M, Onengut-Gumuscu S, Lee A, Luo Y, Teslovich N, Worthington J, Martin J, Huizinga T, Klareskog L, et al. : Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat Genet 2018, 50:1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue F, Ahituv N: Decoding enhancers using massively parallel reporter assays. Genomics 2015, 106:159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rachelefsky GS, Terasaki PI, Katz R, Stiehm ER: Increased prevalence of W27 in juvenile rheumatoid arthritis. N Engl J Med 1974, 290:892–893. [DOI] [PubMed] [Google Scholar]
- 15.Glass D, Litvin D, Wallace K, Chylack L, Garovoy M, Carpenter CB, Schur PH: Early-onset pauciarticular juvenile rheumatoid arthritis associated with human leukocyte antigen-DRw5, iritis, and antinuclear antibody. J Clin Invest 1980, 66:426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forre O, Dobloug JH, Hoyeraal HM, Thorsby E: HLA antigens in juvenile arthritis. Genetic basis for the different subtypes. Arthritis Rheum 1983, 26:35–38. [DOI] [PubMed] [Google Scholar]
- 17.Hollenbach JA, Thompson SD, Bugawan TL, Ryan M, Sudman M, Marion M, Langefeld CD, Thomson G, Erlich HA, Glass DN: Juvenile idiopathic arthritis and HLA class I and class II interactions and age-at-onset effects. Arthritis Rheum 2010, 62:1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray KJ, Moroldo MB, Donnelly P, Prahalad S, Passo MH, Giannini EH, Glass DN: Age-specific effects of juvenile rheumatoid arthritis-associated HLA alleles. Arthritis Rheum 1999, 42:1843–1853. [DOI] [PubMed] [Google Scholar]
- 19.Hinks A, Bowes J, Cobb J, Ainsworth HC, Marion MC, Comeau ME, Sudman M, Han B, Juvenile Arthritis Consortium for I, Becker ML, et al. : Fine-mapping the MHC locus in juvenile idiopathic arthritis (JIA) reveals genetic heterogeneity corresponding to distinct adult inflammatory arthritic diseases. Ann Rheum Dis 2017, 76:765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haasnoot AJW, Schilham MW, Kamphuis S, Hissink Muller PCE, Heiligenhaus A, Foell D, Minden K, Ophoff RA, Radstake T, Den Hollander AI, et al. : Identification of an Amino Acid Motif in HLA-DRbeta1 That Distinguishes Uveitis in Patients With Juvenile Idiopathic Arthritis. Arthritis Rheumatol 2018, 70:1155–1165. [DOI] [PubMed] [Google Scholar]
- 21.Ombrello MJ, Remmers EF, Tachmazidou I, Grom A, Foell D, Haas JP, Martini A, Gattorno M, Ozen S, Prahalad S, et al. : HLA-DRB1*11 and variants of the MHC class II locus are strong risk factors for systemic juvenile idiopathic arthritis. Proc Natl Acad Sci U S A 2015, 112:15970–15975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arthur VL, Shuldiner E, Remmers EF, Hinks A, Grom AA, Foell D, Martini A, Gattorno M, Ozen S, Prahalad S, et al. : IL1RN Variation Influences Both Disease Susceptibility and Response to Recombinant Human Interleukin-1 Receptor Antagonist Therapy in Systemic Juvenile Idiopathic Arthritis. Arthritis Rheumatol 2018, 70:1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ombrello MJ, Arthur VL, Remmers EF, Hinks A, Tachmazidou I, Grom AA, Foell D, Martini A, Gattorno M, Ozen S, et al. : Genetic architecture distinguishes systemic juvenile idiopathic arthritis from other forms of juvenile idiopathic arthritis: clinical and therapeutic implications. Ann Rheum Dis 2017, 76:906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scholl PR, Diez A, Karr R, Sekaly RP, Trowsdale J, Geha RS: Effect of isotypes and allelic polymorphism on the binding of staphylococcal exotoxins to MHC class II molecules. J Immunol 1990, 144:226–230. [PubMed] [Google Scholar]
- 25.Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, Jostins L, Plant K, Andrews R, McGee C, et al. : Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 2014, 343:1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson W, Barrett JH, Donn R, Pepper L, Kennedy LJ, Ollier WE, Silman AJ, Woo P, Southwood T: Juvenile idiopathic arthritis classified by the ILAR criteria: HLA associations in UK patients. Rheumatology (Oxford) 2002, 41:1183–1189. [DOI] [PubMed] [Google Scholar]
- 27.Angeles-Han ST, McCracken C, Yeh S, Jang SR, Jenkins K, Cope S, Bohnsack J, Hersh A, Thompson SD, Prahalad S: HLA Associations in a Cohort of Children With Juvenile Idiopathic Arthritis With and Without Uveitis. Invest Ophthalmol Vis Sci 2015, 56:6043–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruperto N, Lovell DJ, Quartier P, Paz E, Rubio-Perez N, Silva CA, Abud-Mendoza C, Burgos-Vargas R, Gerloni V, Melo-Gomes JA, et al. : Abatacept in children with juvenile idiopathic arthritis: a randomised, double-blind, placebo-controlled withdrawal trial. Lancet 2008, 372:383–391. [DOI] [PubMed] [Google Scholar]
- 29.Nigrovic PA: Review: is there a window of opportunity for treatment of systemic juvenile idiopathic arthritis? Arthritis Rheumatol 2014, 66:1405–1413. [DOI] [PubMed] [Google Scholar]
- 30.Nigrovic PA: Autoinflammation and autoimmunity in systemic juvenile idiopathic arthritis. Proc Natl Acad Sci U S A 2015, 112:15785–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, Martin P, Comeau ME, Sajuthi S, Andrews R, et al. : Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet 2013, 45:664–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIntosh LA, Marion MC, Sudman M, Comeau ME, Becker ML, Bohnsack JF, Fingerlin TE, Griffin TA, Haas JP, Lovell DJ, et al. : Genome-wide association meta-analysis reveals novel juvenile idiopathic arthritis susceptibility loci. Arthritis Rheumatol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle EA, Li YI, Pritchard JK: An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang K, Zhu L, Buck MJ, Chen Y, Carrier B, Liu T, Jarvis JN: Disease-Associated Single-Nucleotide Polymorphisms From Noncoding Regions in Juvenile Idiopathic Arthritis Are Located Within or Adjacent to Functional Genomic Elements of Human Neutrophils and CD4+ T Cells. Arthritis Rheumatol 2015, 67:1966–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spreafico R, Rossetti M, Whitaker JW, Wang W, Lovell DJ, Albani S: Epipolymorphisms associated with the clinical outcome of autoimmune arthritis affect CD4+ T cell activation pathways. Proc Natl Acad Sci U S A 2016, 113:13845–13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson LA, Volpi S, Frugoni F, Janssen E, Kim S, Sundel RP, Dedeoglu F, Lo MS, Hazen MM, Beth Son M, et al. : Next-Generation Sequencing Reveals Restriction and Clonotypic Expansion of Treg Cells in Juvenile Idiopathic Arthritis. Arthritis Rheumatol 2016, 68:1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spreafico R, Rossetti M, van Loosdregt J, Wallace CA, Massa M, Magni-Manzoni S, Gattorno M, Martini A, Lovell DJ, Albani S: A circulating reservoir of pathogenic-like CD4+ T cells shares a genetic and phenotypic signature with the inflamed synovial micro-environment. Ann Rheum Dis 2016, 75:459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grieshaber-Bouyer R, Nigrovic PA: Neutrophil heterogeneity as therapeutic opportunity in immune-mediated disease. Front Immunol 2019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, Cunin P, Wu D, Diogo D, Yang Y, Okada Y, Plenge RM, Nigrovic PA: The Rheumatoid Arthritis Risk Variant CCR6DNP Regulates CCR6 via PARP-1. PLoS Genet 2016, 12:e1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harley JB, Chen X, Pujato M, Miller D, Maddox A, Forney C, Magnusen AF, Lynch A, Chetal K, Yukawa M, et al. : Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet 2018, 50:699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prahalad S, Thompson SD, Conneely KN, Jiang Y, Leong T, Prozonic J, Brown MR, Ponder LA, Angeles-Han ST, Vogler LB, et al. : Hierarchy of risk of childhood-onset rheumatoid arthritis conferred by HLA-DRB1 alleles encoding the shared epitope. Arthritis Rheum 2012, 64:925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinks A, Marion MC, Cobb J, Comeau ME, Sudman M, Ainsworth HC, Bowes J, Juvenile Idiopathic Arthritis Consortium for I, Becker ML, Bohnsack JF, et al. : Brief Report: The Genetic Profile of Rheumatoid Factor-Positive Polyarticular Juvenile Idiopathic Arthritis Resembles That of Adult Rheumatoid Arthritis. Arthritis Rheumatol 2018, 70:957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikora KA, Bennett JR, Vyncke L, Deng Z, Tsai WL, Pauwels E, Layh-Schmitt G, Brundidge A, Navid F, Zaal KJM, et al. : Germline gain-of-function myeloid differentiation primary response gene-88 (MYD88) mutation in a child with severe arthritis. J Allergy Clin Immunol 2018, 141:1943–1947 e1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prahalad S, Zeft AS, Pimentel R, Clifford B, McNally B, Mineau GP, Jorde LB, Bohnsack JF: Quantification of the familial contribution to juvenile idiopathic arthritis. Arthritis Rheum 2010, 62:2525–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakil SM, Monies DM, Abouelhoda M, Al-Tassan N, Al-Dusery H, Naim EA, Al-Younes B, Shinwari J, Al-Mohanna FA, Meyer BF, et al. : Association of a mutation in LACC1 with a monogenic form of systemic juvenile idiopathic arthritis. Arthritis Rheumatol 2015, 67:288–295. [DOI] [PubMed] [Google Scholar]
- 46.Kallinich T, Thorwarth A, von Stuckrad SL, Rosen-Wolff A, Luksch H, Hundsdoerfer P, Minden K, Krawitz P: Juvenile arthritis caused by a novel FAMIN (LACC1) mutation in two children with systemic and extended oligoarticular course. Pediatr Rheumatol Online J 2016, 14:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karacan I, Ugurlu S, Sahin S, Everest E, Kasapcopur O, Tolun A, Ozdogan H, Turanli ET: LACC1 Gene Defects in Familial Form of Juvenile Arthritis. J Rheumatol 2018, 45:726–728. [DOI] [PubMed] [Google Scholar]
- 48.Rabionet R, Remesal A, Mensa-Vilaro A, Murias S, Alcobendas R, Gonzalez-Roca E, Ruiz-Ortiz E, Anton J, Iglesias E, Modesto C, et al. : Biallelic loss-of-function LACC1/FAMIN Mutations Presenting as Rheumatoid Factor-Negative Polyarticular Juvenile Idiopathic Arthritis. Sci Rep 2019, 9:4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cader MZ, Boroviak K, Zhang Q, Assadi G, Kempster SL, Sewell GW, Saveljeva S, Ashcroft JW, Clare S, Mukhopadhyay S, et al. : C13orf31 (FAMIN) is a central regulator of immunometabolic function. Nat Immunol 2016, 17:1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skon-Hegg C, Zhang J, Wu X, Sagolla M, Ota N, Wuster A, Tom J, Doran E, Ramamoorthi N, Caplazi P, et al. : LACC1 Regulates TNF and IL-17 in Mouse Models of Arthritis and Inflammation. J Immunol 2019, 202:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Assadi G, Vesterlund L, Bonfiglio F, Mazzurana L, Cordeddu L, Schepis D, Mjosberg J, Ruhrmann S, Fabbri A, Vukojevic V, et al. : Functional Analyses of the Crohn’s Disease Risk Gene LACC1. PLoS One 2016, 11:e0168276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. : Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Irwanto A, Fu X, Yu G, Yu Y, Sun Y, Wang C, Wang Z, Okada Y, Low H, et al. : Discovery of six new susceptibility loci and analysis of pleiotropic effects in leprosy. Nat Genet 2015, 47:267–271. [DOI] [PubMed] [Google Scholar]
- 54.Lahiri A, Hedl M, Yan J, Abraham C: Human LACC1 increases innate receptor-induced responses and a LACC1 disease-risk variant modulates these outcomes. Nat Commun 2017, 8:15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, et al. : Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viatte S, Plant D, Bowes J, Lunt M, Eyre S, Barton A, Worthington J: Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann Rheum Dis 2012, 71:1984–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viatte S, Massey J, Bowes J, Duffus K, arc OC, Eyre S, Barton A, Worthington J: Replication of Associations of Genetic Loci Outside the HLA Region With Susceptibility to Anti-Cyclic Citrullinated Peptide-Negative Rheumatoid Arthritis. Arthritis Rheumatol 2016, 68:1603–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei WH, Viatte S, Merriman TR, Barton A, Worthington J: Genotypic variability based association identifies novel non-additive loci DHCR7 and IRF4 in sero-negative rheumatoid arthritis. Sci Rep 2017, 7:5261. [DOI] [PMC free article] [PubMed] [Google Scholar]