Abstract
Background:
Nontuberculous mycobacteria (NTM) species are a rising threat, especially to patients living with pulmonary comorbidities. Current point-of-care diagnostics fail to adequately identify and differentiate NTM species from M. tuberculosis (Mtb). Definitive culture- and molecular- based testing can take weeks to months and require sending samples out to specialized diagnostic labs.
Methods:
In this proof-of-concept study, we developed an assay based on PCR amplification of 16S rRNA using universal mycobacterial primers and interrogation of the amplified fragments with a panel of binary deoxyribozyme (BiDz) sensors to enable species level identification of NTM (BiDz-NTMST). Each BiDz sensor consists of two subunits of an RNA-cleaving deoxyribozyme, which form an active deoxyribozyme catalytic core only in the presence of the complimentary target sequence. The target-activated BiDz catalyzes cleavage of a reporter substrate, thus triggering either fluorescent or colorimetric (visually observed) signal depending on the substrate used. The panel included BiDz sensors for differentiation of 6 clinically relevant NTM species (M. abscessus, M. avium, M. intracellulare, M. fortuitum, M. kansasii, and M. gordonae) and Mtb.
Results:
Using the fluorescent BiDz-NTMST assay we successfully identified the species of 38 clinical isolates. In addition, a subset of strains was tested using visual BiDz sensors, providing proof-of-concept for strain typing of NTM by the naked eye.
Conclusions:
The BiDz-NTMST assay is a novel platform for rapid identification of NTM species. This method is highly specific and significantly faster than current tools and is easily adaptable for on-site diagnostic laboratories in hospitals or clinical laboratories.
Keywords: nontuberculous mycobacteria (NTM), sensors, diagnostic, deoxyribozyme
INTRODUCTION
Nontuberculous mycobacteria (NTM) comprise a group of ~150 mycobacteria species (excluding Mycobacterium tuberculosis (Mtb) and Mycobacterium leprae) which are broadly classified as either rapid (RGM) or slow (SGM) growing mycobacteria. NTM are opportunistic environmental pathogens responsible for more infections than Mtb in the United States and other industrialized countries (1). NTM are mainly associated with pulmonary infections in patients with underlying lung abnormalities or preexisting structural lung disease; i.e. cystic fibrosis (CF), chronic obstructive pulmonary disease (COPD), or bronchiectasis (2). Pulmonary NTM infections are mostly caused by Mycobacterium abscessus (Mabs), Mycobacterium kansasii (Mkan), Mycobacterium fortuitium (Mfor), Mycobacterium intracellulare (Mint), and Mycobacterium avium (Mavi) (3, 4). For example, the RGM species Mkan and Mab are the second and third most common causes of lung disease in the United States, respectively (5). The prevalence of NTM infections has been increasing worldwide and is commonly misdiagnosed as Mtb resulting in poor treatment outcomes and recurring infection (1, 6, 7). A study in Brazil reported 79% of patients diagnosed with an NTM infection had been previously treated for Mtb for up to 6 months, highlighting the need for improved diagnostic methods to accurately differentiate NTM from Mtb (8). In addition, species level identification of NTM is necessary due to differences in antimicrobial susceptibility, which warrant species-specific treatment regimens (9).
Currently, diagnosis of NTM infections is based on clinical, radiological and microbiological determination. The microbiological methods are the same techniques used for the identification of M. tuberculosis (Mtb) – culture and sputum smear microscopy (SSM). Culture based testing is a time-consuming process requiring between 4 days (RGM) and 6 weeks (SGM), and differentiation of NTM species is based on cultural characteristics (morphology, color of colony, time to colonies) (10). In addition, it lacks the specificity necessary to reliably differentiate NTM and Mtb, especially SGM, which present with similar morphology and time to colony formation. SSM cannot differentiate NTM from Mtb, often fails to detect RGM, and cannot provide information on species identification (11, 12). Currently, methods for speciation require isolation in culture and extraction of nucleic acids followed by the use of laborious molecular methods. Molecular methods for strain typing NTM rely on the differentiation of 16S rRNA or 23S rRNA by real-time PCR, line probe assays (LiPA) and sequencing of multiple loci (13–15).
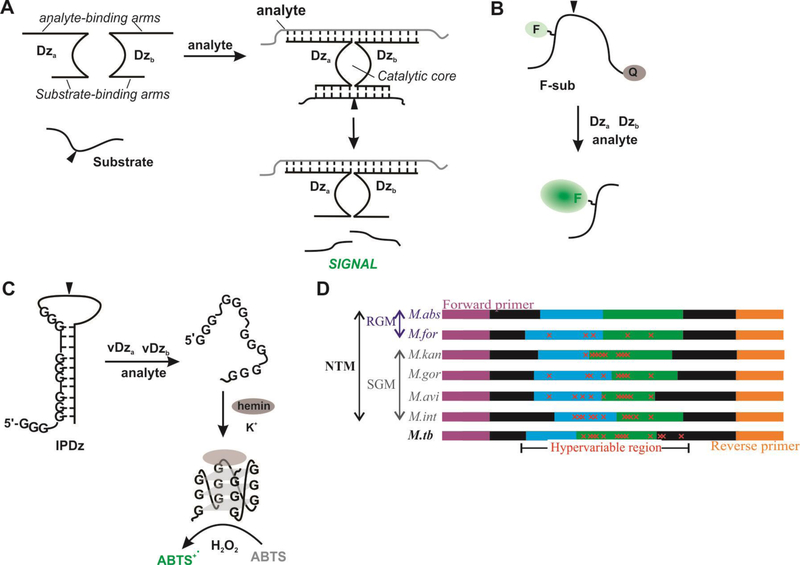
In this study, we propose the use of binary deoxyribozyme (BiDz) sensors for the detection and differentiation of NTM strains. BiDz sensors are based on the 10–23 RNA cleaving deoxyribozyme, whose catalytic core is split into two subunits, Dza and Dzb, and is re-formed only in the presence of a specific nucleic acid target (Fig 1A). The activated BiDz then cleaves a universal reporter substrate producing a signal which can either be fluorescent or colorimetric depending on the substrate used. For the dual-labeled fluorogenic substrate (F-sub) containing a fluorophore and a quencher at the opposite sides from the cleavage site, the presence of the target analyte results in cleavage of the substrate and an increase in fluorescence (Fig 1B) (16–19). Alternatively, a colorimetric substrate consisting of an inactivated peroxidase Dz (IPDz) containing a G-quadruplex sequence can be used. In the presence of the target analyte, the IPDz sequence will be cleaved releasing a G-quadruplex sequence to fold and catalyze a peroxidation reaction of an organic indicator resulting in a color change. (Fig 1C) (20). BiDz sensors offer unique advantages over traditional molecular methods. The catalytic feature of the BiDz sensors enables amplification of signal in the presence of a specific analyte resulting in lower limits of detection compared to traditional probe-based molecular detection methods (16, 21). Most importantly, careful design of BiDz sensors can enable differentiation of DNA or RNA analytes that differ by a single nucleotide (16, 21–23).
Figure 1. Schematic representation of the BiDz assay design.
A. In the presence a fully complimentary analyte, sensors assemble to reform the deoxyribozyme catalytic core, which cleaves the substrate (at site denoted by black triangle) to generate a fluorescent (B) or colorimetric (C) signal. D). Schematic diagram of 16S rRNA PCR amplicon containing hypervariable regions used for differentiation of 6 NTM species (M.abs, M.for, M.kan, M.gor, M.avi, M.int) and M.tb. Sequences are colored to indicate role in assay design: purple (forward primer), blue (Dza), green (Dzb), and orange (reverse primer). The red “x” indicates mutations contained in the hypervariable region.
A panel of BiDz sensors targeting a hypervariable region of mycobacterial 16S rRNA gene was designed to differentiate 6 NTM species (M. abscessus, M. avium M. intracellulare, M. fortuitum, M. kansasii, and M. gordonae) and Mtb (Fig 1D). The targeted fragment was PCR amplified using a universal primer pair (Table S1) followed by detection with BiDz-NTM strain typing (BiDz-NTMST) assay. We tested the BiDz-NTMST assay on 38 clinical isolates, which demonstrates the potential utility of this technology as an alternative to current molecular diagnostic tools for NTM for rapid identification of NTM species.
MATERIALS AND METHODS
Clinical Nontuberculous Mycobacterial (NTM) Isolates
NTM clinical isolates were acquired from National Jewish Health and National Reference Center for Mycobacteria, Borstel, Germany. Strains were cultured using 7H9 media supplemented with OADC. DNA was isolated from cultures grown to OD 0.6–0.8 using the standardized method (cetyltrimethylammonium bromide-NaCl (CTAB)/NaCl) and stored at −20◦C, as previously described (14). All nucleic acid stocks were then quantitated (Nanodrop) and diluted to 20 ng/uL to be used with the universal PCR protocol outlined below.
Universal PCR
A universal PCR was designed to amplify a fragment of 16S rRNA containing a hypervariable region from all targeted mycobacteria for subsequent species-specific detection BiDz sensors. A multiple sequence alignment of M. abscessus (Mabs), M. avium (Mavi), M. intracellulare (Mint), M. fortuitum (Mfor), M. kansasii (Mkan), M. gordonae (Mgor), and M. tuberculosis (Mtb) identified two highly conserved regions of 16S rRNA. Universal primers (Table S1) complementary to these conserved regions (Fig 1D, purple and orange) were designed to amplify the intervening hypervariable region for differentiation of the NTM species by BiDz sensors. The reverse primer contains a 5’ phosphate to allow for digestion of the antisense strand by lambda exonuclease. The Phusion® High-Fidelity PCR Kit (New England Biolabs) was used for these studies. PCR samples (25 μL) contained 2 units of Phusion DNA Polymerase (New England Biolabs), 2.5 μM of each primer, 200 μM of each dNTP, 1X HF buffer, and 20 ng of NTM or Mtb DNA. The DNA was then PCR amplified using the following cycling conditions: initial denaturation 98°C for 30 sec, followed by 20–30 cycles of denaturation at 98◦C for 10 sec, annealing at 65°C for 20 sec, extension at 72°C for 10 sec, and final extension at 72°C for 2 min (total time ~35 min). A no template control (NTC) containing all components except template was incubated under the PCR conditions described above. Following PCR, the samples were treated with 1–5 units of Lambda Exonuclease (New England Biolabs) and incubated at 37°C for 5–15 min followed by cooling to 12°C to yield a single-stranded product used as a target in BiDz assays. All PCR samples were analyzed using a 2% agarose gel (Data not shown). The samples were run at 120 V for 40 min followed by staining in GelRed (Biotium) for 30 min.
Fluorescent BiDz Sensor Assays
All oligonucleotides were synthesized by Integrated DNA Technologies. The fluorogenic substrate (F-sub) was synthesized and HPLC purified by TriLink BioTechnologies. All fluorescent assays contained the F-sub (200 nM), two adaptor strands (Dza and Dzb) at 15 nM each, (Table S2), and either synthetic analyte (1 nM) or 2 μL of a PCR product (after 20 cycles). Final sample volumes were 30 μL. All the components of the assay were mixed in a buffer containing 50 mM HEPES, pH 7.4, 50 mM MgCl2, 20 mM KCl, 120 mM NaCl, 0.03% Triton X-100, and 1% DMSO. Samples were placed in a 384-well plate (PerkinElmer OptiPlate), sealed (ThermalSeal 2) and incubated at 55°C for 30 mins. Fluorescence of the samples was measured on a Synergy 4 plate reader (Biotek) with a xenon flash lamp (λex=485 nm, λem=517 nm).
To test the performance of BiDz sensors in the case of a mixed NTM infection, synthetic analytes corresponding to the target 16S rRNA hypervariable region of Mabs, Mavi, or Mint (Table S2) were mixed at 1nM each (final concentration). In addition, we analyzed a mixture of Mavi and Mint analytes, with Mabs analyte omitted. Detection by individual BiDz sensors was carried out as described above. To test the full PCR + BiDz assay on mixed templates, three PCR samples were prepared: No Template Control (NTC), Mavi/Mint/Mabs (20 ng each DNA), and Mavi/Mint (20 ng each DNA). These PCR products were processed and used as analytes in BiDz-NTMST sensor assays as described above. Data from two independent times experiment, each consisting of two technical replicates (total of 4 measurements per data point) were averaged.
Colorimetric BiDz Sensor Assay
For the assay, IPDz substrate (1 μM), two adaptor strands vDza (30 nM) and vDzb (100 nM) (Table S2), and either a synthetic oligonucleotide analyte (30 nM) or 3 μL PCR product (after 30 cycles) were mixed in a buffer containing 50 mM HEPES, pH 7.4, 50 mM MgCl2, 20 mM KCl, 120 mM NaCl, 0.03% Triton X-100, and 1% DMSO. Final sample volumes were 30 μL. A “blank” sample contained no analyte with other components being the same as for other samples. The samples were incubated at 50°C for 45 min. Then, 1 mM 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS), 375 nM hemin (Sigma-Aldrich), and 1 mM hydrogen peroxide (VWR) were added to the samples, and the color change was monitored visually. After 15 min, the absorbance of the samples at 420 nm was recorded using NanoDrop OneC UV-Vis Spectrophotometer (ThermoFisher Scientific). For visual images (Fig 5B and Fig 6B), samples (60 μL) containing IPDz substrate (1000 nM), vDza (30 nM), vDzb (100 nM) and either synthetic oligonucleotide analyte (30 nM) or 6 μL PCR product in the buffer were incubated at 50°C for 45 min in microcentrifuge tubes and then transferred into a 96-well plate containing ABTS and hemin followed by addition of hydrogen peroxide. The picture was taken using a smartphone camera within 15–20 min.
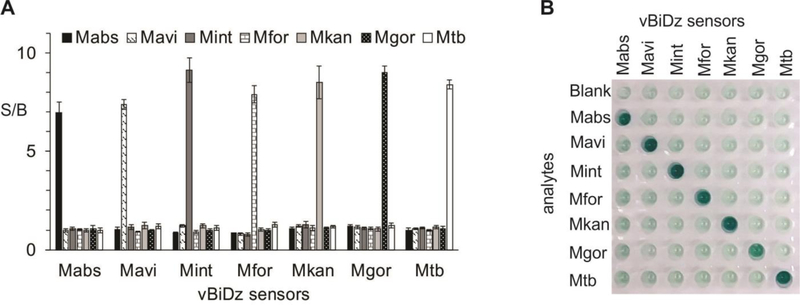
Fig 5: Validation of vBiDz sensors using synthetic oligonucleotide analytes corresponding to 6 NTM species or Mtb.
A. Absorbance at 420 nm for the samples containing each vBiDz sensor in the presence of one of the analytes (indicate in the legend). The signal-to-background ratio (S/B) was calculated by dividing the absorbance values by the absorbance of the blank (the sensor in the absence of the analyte). The data presented here is the average and standard deviation of 3 independent experiments. B. Visual image of a well plate containing samples similar to those from panel A.
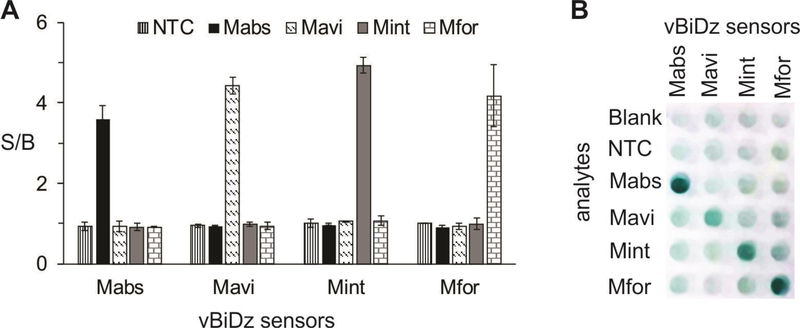
Fig 6: Results of the vBiDz-NTMST assay against NTM clinical samples containing Mavi, Mint, Mabs or Mfor.
A. Absorbance at 420 nm for the samples containing each vBiDz sensor in the presence of one of the analytes (indicate in the legend). The signal-to-background ration (S/B) was calculated by dividing the absorbance values by the absorbance of the blank (the sensor in the absence of the analyte). NTC – “no target” control for PCR amplification. The data presented here is representative of 3 independent experiments with standard deviations as error bars. B. Visual image of a well plate containing samples similar to those from panel A.
To simulate mixed infection with synthetic analytes, specific synthetic analyte (100 nM) or a mixture of all synthetic analytes for Mtb, Mabs, or Mavi (100 nM each) or all but specific analyte (100 nM each) were detected with the corresponding vBiDz sensors as described above. To test the performance of the sensors with PCR amplicons obtained when more than one mycobacterial species was present, DNA isolated from Mavi, Mint, and/or Mabs was mixed and used as a template for PCR (20 ng per 25 μL PCR reaction) under the conditions described above. As controls, individual bacterial DNA was amplified. No-target control (NTC) contained no bacterial DNA added. After PCR, the samples were treated with lambda exonuclease and used for the colorimetric assays with Mabs-, Mavi- or Mint-specific vBiDz sensor, as described above.
Limit of Detection Experiment – Visual BiDz Assay
Two-fold dilutions of total bacterial DNA were prepared and used for PCR reaction, so that the final PCR samples (25 μL) contained 0.125–4 ng DNA (corresponds to 2.5×104–8×105 genome equivalents per reaction). PCR, lambda exonuclease treatment and colorimetric assay conditions were identical to the ones described above. The limit of detection was calculated as the DNA concentration corresponding to the absorbance at 420 nm of the blank sample plus 3 standard deviation from the blank (3σ rule) using a linear trendline for the data with different DNA concentrations.
Validation by Sequencing
Clinical isolates provided by National Jewish Health were speciated by sequencing the rpoB gene. Strains provided by the National Reference Center for Mycobacteria, Borstel, Germany were whole genome sequenced. Additional validation was completed by, sequencing of the 16S rRNA PCR products for all strains tested in this study.
Statistical Analysis
The data were analyzed using Microsoft Excel. Data are presented as signal-to-background ratios (S/B). Background is defined as the signal obtained in the presence of only the substrate and sensor strands (Dza and Dzb). The average and standard deviations of 3 independent experiments are presented.
RESULTS
Validation of Fluorescent BiDz 16S rRNA Sensors
We developed a panel of BiDz sensors targeting some of the most prevalent NTM species associated with pulmonary NTM infection. BiDz sensors were designed to target a hypervariable region of 16S rRNA gene, which allows for differentiation of the following NTM species: M. abscessus (Mabs), M. avium (Mavi), M. intracellulare (Mint), M. fortuitium (Mfor), M. kansasii (Mkan), and M. gordonae (Mgor). This region can also be used to differentiate NTMs from M. tuberculosis (Mtb). We first validated the ability of the BiDz-NTMST assay to differentiate between 6 NTM species and Mtb using synthetic DNA analytes mimicking the amplified fragments of correspondent mycobacterial species (Fig 2). We used a S/B of 2 as an arbitrary threshold between negative and positive signal. We noted that only the fully complementary analytes are detected with S/B ratio >2 using each of the sensors used. S/B ratio <2 is considered insignificant.
Fig 2: Validation of BiDz sensors used in the BiDz-NTMST assay for detection of species-specific 16S rRNA sequences.
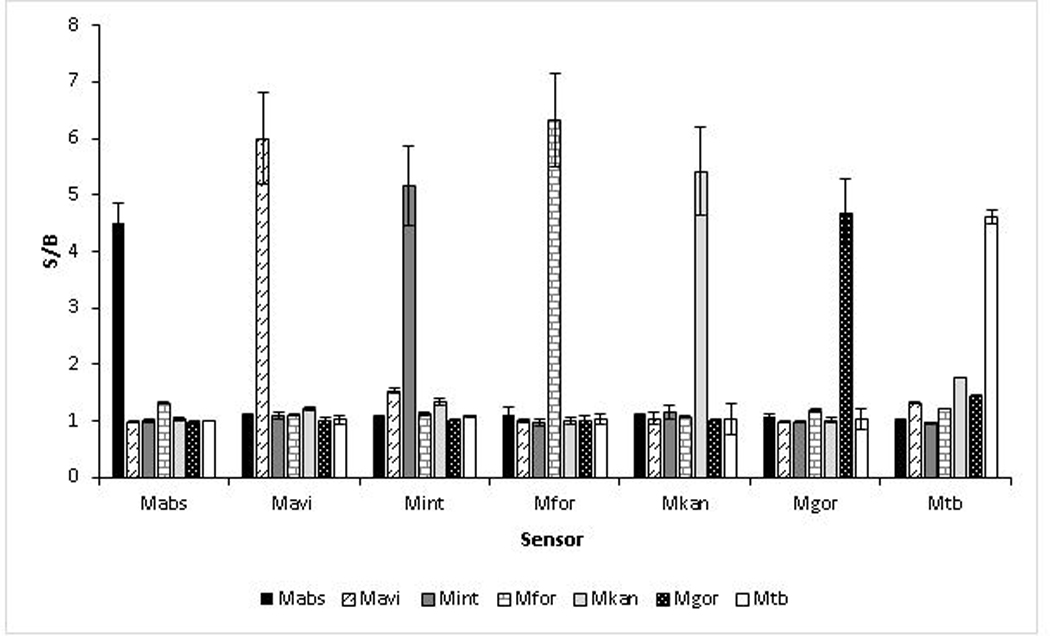
Sensors were validated using synthetic DNA analytes (1 nM) equivalent to the amplified fragments of NTM 16S rRNA gene.
Identification of species-typed clinical isolates using the BiDz-NTMST assay
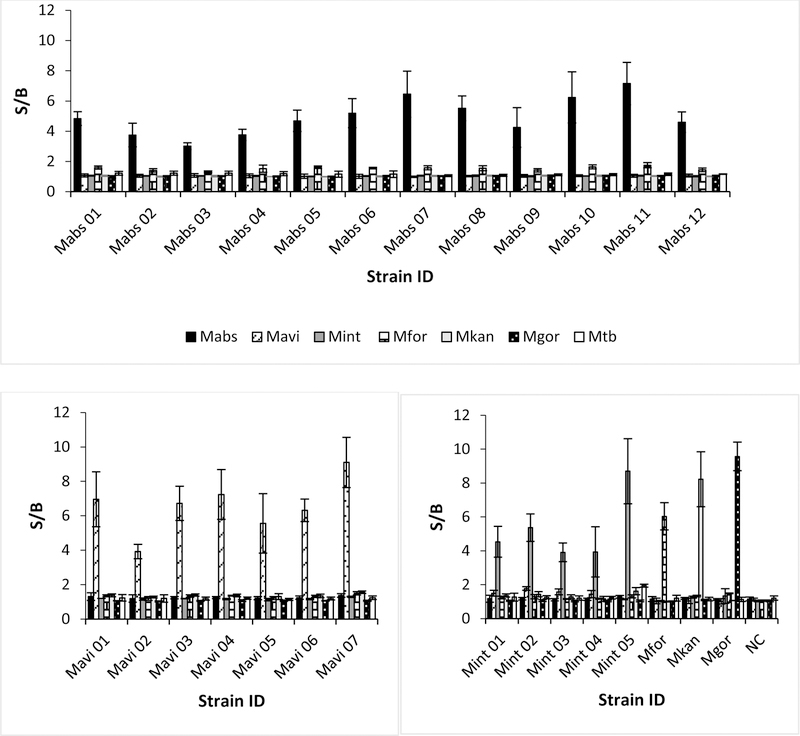
To demonstrate the utility of our BiDz-NTMST assay for NTM species typing we acquired a panel of typed NTM strains. DNA was isolated from all strains and 20 ng was used for the BiDz-NTMST assay. A fragment of 16S rRNA containing a hypervariable region was amplified by PCR with a universal primer pair followed by species identification using our panel of BiDz sensors. Fig 3 demonstrates the results of the BiDz-NTMST assay against 27 clinical isolates. We identified 12 Mabs, 7 Mavi, 5 Mint, 1 Mfor, and 1 Mgor strains. Our results were validated by comparing to the species type determined by rpoB and hsp65 sequencing (samples from National Jewish Health) or whole-genome sequence (WGS) (National Reference Center, Borstel samples). The BiDz sensor results matched the sequencing confirmation in all cases. Representative sequencing results are shown in Fig S2.
Fig 3: Speciation of NTM clinical isolates using the BiDz-NTMST assay.
Clinical isolates identified as Mabs (A), Mavi (B), or other NTM species (C). The data is average of three independent experiments with error bars as standard deviations.
Speciation of unknown clinical isolates using the BiDz-NTMST assay
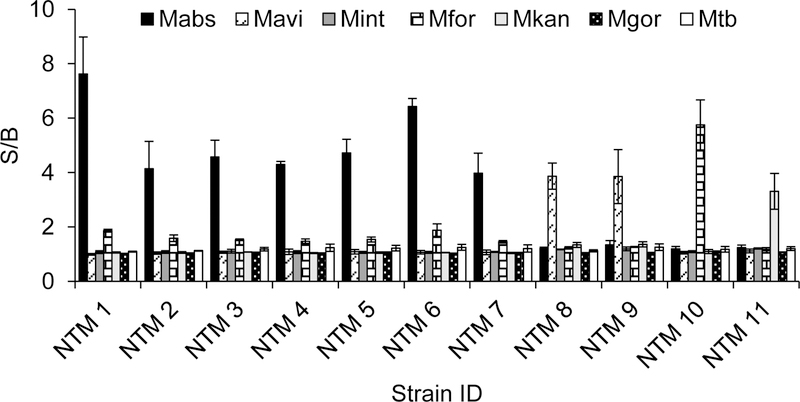
To further validate the BiDz-NTMST assay we tested 11 unknown clinical isolates provided by National Jewish Health. As a reference method of isolate speciation, we used sequencing of the PCR products tested with the BiDz-NTMST assay. As shown in Fig 4, the NTM isolates were unambiguously identified as 7 Mabs, 2 Mavi, 1 Mfor and 1 Mkan, which was confirmed by sequencing of PCR products.
Fig 4: Speciation of Unknown NTM samples using the BiDz-NTMST assay.
The data is average of three independent experiments with error bars as standard deviations.
Specific detection of NTM species in mixed infection with BiDz-NTMST assay
The occurrence of mixed infections of NTM species and Mtb or multiple distinct NTM species (i.e. Mavi plus Mabs) warrants diagnostics capable of detecting specific species even in the context of related pathogens (24–26). To test the utility of the BiDz-NTMST assay, we modeled a mixed infection by mixing synthetic analytes corresponding to three clinically prevalent NTM specifies - Mabs, Mavi, and Mint. BiDz sensors specific for each species were able to detect their target analyte to yield robust signals ~4–10 fold above background, depending on the sensor (Fig S3). In a control experiment in which one analyte was omitted (Mabs) from the mixture, only the two species present were detected (Mavi, Mint), whereas Mabs-specific sensors did not give a signal, highlighting the specificity of the sensors. A similar experiment using PCR products amplified from mixtures of chromosomal DNA (Fig S4) supported the utility of the BiDz-NTMST assay for species specific detection of mycobacteria in the context of a mixed infection.
Validation of visual BiDz sensors
In order to demonstrate versatility of BiDz design, we converted fluorescent BiDz sensors into colorimetric or visual BiDz sensors (vBiDz). For this purpose, we substituted F-sub with a stem-loop substrate containing a G-quadruplex (G4) sequence at the 5’-end of the cleavage site (IPDz reporter). In this case, cleavage of the substrate releases the G4 sequence to fold into a catalytic deoxyribozyme which binds hemin cofactor and catalyzes peroxidation of a colorless organic molecule (e.g. 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) or ABTS) into a colored oxidation product (16). The signal then can be read by the naked eye, thus eliminating the need for instrumentation for signal monitoring. Alternatively, quantitative detection could be done using any absorbance reader, for example, an inexpensive battery-operated portable spectrophotometer.
We obtained vBiDz sensors targeting the same hypervariable region of mycobacterial 16S rRNA gene as was interrogated by fluorescent sensors in BiDz-NTMST assay. In addition to changing the sequence of the substrate binding arm to be complementary to IPDz substrate, we shortened the analyte binding arms of the sensors to ensure high selectivity of the analyte recognition under the colorimetric assay conditions (50 °C instead of 55 °C used for the fluorescent assay). A panel of seven (7) vBiDz sensors was obtained – each targeting one of the NTM species (Mabs, Mavi, Mint, Mfor, Mkan, Mgor) or Mtb (Table S2). Using synthetic oligonucleotides corresponding to these bacteria it was demonstrated that the sensors produced high signal (S/B > 5) only in the presence of the complementary analyte (Fig 5A), while non-specific analytes only yielded signal at the background level. Importantly, clear differentiation of mycobacterial analytes was observed by the naked eye (Fig 5B).
We then tested the vBiDz-NTMST assay using DNA from four clinical isolates containing different NTM species – SGM Mavi and Mint and RGM Mabs and Mfor. The fragments complementary to the sensors were PCR amplified using the universal primer set that was used in fluorescent BiDz-NTMST assay, but the number of cycles was increased to ensure high concentration of the analyte. All four species were correctly identified using vBiDz sensors (Fig 6), with S/B = 3–5 for the specific NTM analytes and no signal above the background in the present of non-specific NTM or in the absence of bacterial DNA (NTC, PCR no-target control).
Specific detection of NTM species in mixed infection with vBiDz-NTMST assay
Next we sought to confirm the ability of the visual BiDz assay to accurately distinguish mycobacterial species in the case of infections involving multiple related pathogens. As shown in Fig S5 and Fig S6, vBiDz sensors were also able to specifically detect the targeted species (Mtb, Mabs, Mavi) when present in a mixture of either synthetic analytes or PCR amplicons from mixed template DNA. However, in samples where one species was omitted, no false-positive signal was generated by the analytes of the two related species.
Limit of Detection of vBiDz-NTMST assay
We have previously reported the limit of detection (LOD) of a fluorescent BiDz assay targeting Mtb as 10–15 genome equivalents (50–75 fg) (21). In this work, we determined the LOD for the IPDz-substrate based colorimetric assay by PCR amplification from 2-fold serial dilution of Mtb chromosomal DNA (0.125–4ng DNA) and subsequent detection with vBiDz sensors as described in Materials and Methods. The calculated LOD for the visual BiDz assay was 4900 genome equivalents (Fig S7). This higher detection threshold for the colorimetric assay, which is consistent with previous observations, likely stems from inherent differences in signal output and dynamic range of unquenched fluorescent substrates versus activation of ABTS substrate by the peroxidase DNAzyme.
DISCUSSION
Novel molecular diagnostic tools for the speciation of NTM are necessary to enable the initiation of the appropriate drug regimen. The lack of a quick, inexpensive, and user-friendly molecular diagnostic tool capable of strain typing NTM species near the point-of-care is an unmet clinical need. In this proof-of-concept study, we developed the BiDz-NTMST assay, which consists of two stages: (1) PCR amplification of 16S rRNA gene using a mycobacteria-specific universal primer pair; and (2) interrogation of a defined hypervariable region in the PCR-amplified DNA fragment with a panel of BiDz sensors for NTM detection and differentiation. The panel includes 7 sensors targeting one of the 6 NTM species - M. abscessus, M. avium, M. intracellulare, M. fortuitium, M. kansasii, M. gordonae - or M. tuberculosis. The sensors rely on either fluorescent or colorimetric (visual) output depending on the substrate used. The fluorescent BiDz-NTMST assay requires ~1 hr time to results with a hands-on time of ~10 min. This method is significantly faster than current tools for NTM speciation and is easily adaptable by certified diagnostic laboratories in hospitals or clinical laboratories. The colorimetric version of BiDz-NTMST assay can be completed in less than 3 hr with a hands-on time of ~15 min and is promising for point-of-care applications, especially if PCR amplification of bacterial DNA is substituted with an isothermal amplification method, such as recombinase polymerase amplification (RPA) or nucleic acid sequence based amplification (NASBA) (27, 28)
One of the major advantages of BiDz sensor technology is the straightforward design. The sensors are made of two unmodified oligonucleotides, which are inexpensive and easy to obtain custom-made from commercial vendors. The reporter substrate is more expensive than the sensor oligonucleotides but it is universal for all BiDz sensors. Therefore, the only change required to adapt the BiDz sensor technology for the detection of a new analyte is the change of the analyte-binding arm sequences for Dza and Dzb strands. Another advantage is the ability of the BiDz sensors to differentiate analytes differing by as little as one nucleotide. Here, we have demonstrated the use of BiDz sensors for discriminating between analytes which contain more than one mutation. As we have recently reported for Mtb drug susceptibility testing (21), this approach could also be applied to detection of SNPs conferring drug resistance in NTM species such as clarithromycin or aminoglycoside resistance mediated by mutations in 23S and 16S rRNA genes, respectively (29–33). In this study, we also demonstrated the applicability of the BiDz-NTMST assay for specific detection of mycobacterial species in the clinical relevant scenario of a mixed infection with multiple mycobacterial species (24–26). Due to the distinct antimicrobial regimens required for Mtb and different NTM species, accurate identification of all mycobacterial species present is of high priority. This is consistent with our previous report of a similar PCR/BiDz assay able to detect mutant DNA carrying drug resistance SNPs even when mixed with excess wild-type DNA (21).
In conclusion, we developed a new BiDz-NTMST assay for identification of multiple NTM species and Mtb. We demonstrated the use of the assay for NTM species identification in 38 clinical isolates. In all cases, the results obtained with the BiDz-NTMST assay were in agreement with strain typing provided by sequencing. The BiDz-NTMST is easily customizable. The fluorescent version of the assay can be used for a rapid laboratory-based analysis of clinical specimens requiring instrumentation already available in diagnostic or clinical labs. The colorimetric version of the assay relies on a color change as a signal detectable by the naked eye and, if combined with an isothermal amplification of bacterial DNA/RNA, can be used near the point-of-care with no expensive instrumentation required. In its current form, the assay enables identification and differentiation of 6 NTM species and Mtb. If needed, the panel of BiDz sensors can be broadened to target additional NTM strains or even other bacterial species that may be present in the same clinical specimen. The assay can be easily adapted to enable drug susceptibility testing of NTM. Our long-term vision is to incorporate this technology into a closed, cartridge or microfluidics type platform with integrated signal collection amenable to implementation in the clinical laboratory setting. Implementation of novel molecular diagnostic tools which are capable of strain typing and drug susceptibility testing can enable the rapid diagnosis and initiation of adequate antimycobacterial therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Dmitry Kolpashchikov for work in developing BiDz sensor technology and valuable input on sensor design. We acknowledge Ms. Rachael Rodger of the National Jewish Health Advanced Diagnostic Laboratory and Dr. Stefan Niemann of the National Reference Center for Mycobacteriology (Borstel, Germany) for kindly providing NTM strains and DNA. Research reported in this publication was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R21AI123876 (YG and KHR) and 1R15AI10388001A1 (KHR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- NTM
nontuberculous mycobacteria
- Mtb
Mycobacterium tuberculosis
- BiDz
binary deoxyribozyme
- RGM
rapid growing mycobacteria
- SGM
slow growing mycobacteria
- Mabs
M. abscessus
- Mkan
M. kansasii
- Mfor
M. fortuitium
- Mint
M. intracellulare
- Mavi
M. avium
- Mgor
M. gordonae
- F-sub
fluorogenic substrate
- IPDz
inactivated peroxidase deoxyribozyme
- rRNA
ribosomal RNA
- S/B
signal to background ratio
- G4
G-quadruplex
- vBiDz
visual binary deoxyribozyme
REFERENCES
- 1.Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. Prevalence of nontuberculous mycobacterial lung disease in u.S. Medicare beneficiaries. Am J Respir Crit Care Med 2012;185:881–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan ED, Iseman MD. Underlying host risk factors for nontuberculous mycobacterial lung disease. Semin Respir Crit Care Med 2013;34:110–23. [DOI] [PubMed] [Google Scholar]
- 3.Claeys TA, Robinson RT. The many lives of nontuberculous mycobacteria. J Bacteriol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koh WJ. Nontuberculous mycobacteria-overview. Microbiol Spectr 2017;5. [DOI] [PubMed] [Google Scholar]
- 5.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ats/idsa statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 2007;175:367–416. [DOI] [PubMed] [Google Scholar]
- 6.Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: A review. Clin Chest Med 2015;36:13–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shahraki AH, Heidarieh P, Bostanabad SZ, Khosravi AD, Hashemzadeh M, Khandan S, et al. “Multidrug-resistant tuberculosis” may be nontuberculous mycobacteria. Eur J Intern Med 2015;26:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Mello KG, Mello FC, Borga L, Rolla V, Duarte RS, Sampaio EP, et al. Clinical and therapeutic features of pulmonary nontuberculous mycobacterial disease, brazil, 1993–2011. Emerg Infect Dis 2013;19:393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium abscessus: A new antibiotic nightmare. J Antimicrob Chemother 2012;67:810–8. [DOI] [PubMed] [Google Scholar]
- 10.Somoskovi A, Salfinger M. Nontuberculous mycobacteria in respiratory infections: Advances in diagnosis and identification. Clin Lab Med 2014;34:271–95. [DOI] [PubMed] [Google Scholar]
- 11.Desikan P, Tiwari K, Panwalkar N, Khaliq S, Chourey M, Varathe R, et al. Public health relevance of non-tuberculous mycobacteria among afb positive sputa. Germs 2017;7:10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright PW, Wallace RJ Jr., Wright NW, Brown BA, Griffith DE Sensitivity of fluorochrome microscopy for detection of mycobacterium tuberculosis versus nontuberculous mycobacteria. J Clin Microbiol 1998;36:1046–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franco-Alvarez de Luna F, Ruiz P, Gutierrez J, Casal M. Evaluation of the genotype mycobacteria direct assay for detection of mycobacterium tuberculosis complex and four atypical mycobacterial species in clinical samples. J Clin Microbiol 2006;44:3025–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omar SV, Roth A, Ismail NA, Erasmus L, Ehlers M, Kock M, et al. Analytical performance of the roche lightcycler(r) mycobacterium detection kit for the diagnosis of clinically important mycobacterial species. PLoS One 2011;6:e24789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitarai S, Kato S, Ogata H, Aono A, Chikamatsu K, Mizuno K, et al. Comprehensive multicenter evaluation of a new line probe assay kit for identification of mycobacterium species and detection of drug-resistant mycobacterium tuberculosis. J Clin Microbiol 2012;50:884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mokany E, Bone SM, Young PE, Doan TB, Todd AV. Mnazymes, a versatile new class of nucleic acid enzymes that can function as biosensors and molecular switches. J Am Chem Soc 2010;132:1051–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolpashchikov DM. Binary probes for nucleic acid analysis. Chem Rev 2010;110:4709–23. [DOI] [PubMed] [Google Scholar]
- 18.Gerasimova YV, Peck S, Kolpashchikov DM. Enzyme-assisted binary probe for sensitive detection of rna and DNA. Chem Commun (Camb) 2010;46:8761–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolpashchikov DM. A binary deoxyribozyme for nucleic acid analysis. Chembiochem 2007;8:2039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerasimova YV, Cornett EM, Edwards E, Su X, Rohde KH, Kolpashchikov DM. Deoxyribozyme cascade for visual detection of bacterial rna. Chembiochem 2013;14:2087–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bengtson HN, Homolka S, Niemann S, Reis AJ, da Silva PE, Gerasimova YV, et al. Multiplex detection of extensively drug resistant tuberculosis using binary deoxyribozyme sensors. Biosens Bioelectron 2017;94:176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerasimova YV, Kolpashchikov DM. Nucleic acid detection using mnazymes. Chem Biol 2010;17:104–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerasimova YV, Yakovchuk P, Dedkova LM, Hecht SM, Kolpashchikov DM. Expedited quantification of mutant ribosomal rna by binary deoxyribozyme (bidz) sensors. RNA 2015;21:1834–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jun HJ, Jeon K, Um SW, Kwon OJ, Lee NY, Koh WJ. Nontuberculous mycobacteria isolated during the treatment of pulmonary tuberculosis. Respir Med 2009;103:1936–40. [DOI] [PubMed] [Google Scholar]
- 25.Kurahara Y, Tachibana K, Tsuyuguchi K, Suzuki K. Mixed pulmonary infection with three types of nontuberculous mycobacteria. Intern Med 2013;52:507–10. [DOI] [PubMed] [Google Scholar]
- 26.Shin SH, Jhun BW, Kim SY, Choe J, Jeon K, Huh HJ, et al. Nontuberculous mycobacterial lung diseases caused by mixed infection with mycobacterium avium complex and mycobacterium abscessus complex. Antimicrob Agents Chemother 2018;62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deiman B, van Aarle P, Sillekens P. Characteristics and applications of nucleic acid sequence-based amplification (nasba). Mol Biotechnol 2002;20:163–79. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Easley CJ. Isothermal DNA amplification in bioanalysis: Strategies and applications. Bioanalysis 2011;3:227–39. [DOI] [PubMed] [Google Scholar]
- 29.Maurer FP, Ruegger V, Ritter C, Bloemberg GV, Bottger EC. Acquisition of clarithromycin resistance mutations in the 23s rrna gene of mycobacterium abscessus in the presence of inducible erm(41). J Antimicrob Chemother 2012;67:2606–11. [DOI] [PubMed] [Google Scholar]
- 30.Nash KA, Brown-Elliott BA, Wallace RJ Jr., A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of mycobacterium abscessus but is absent from mycobacterium chelonae. Antimicrob Agents Chemother 2009;53:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nessar R, Reyrat JM, Murray A, Gicquel B. Genetic analysis of new 16s rrna mutations conferring aminoglycoside resistance in mycobacterium abscessus. J Antimicrob Chemother 2011;66:1719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prammananan T, Sander P, Brown BA, Frischkorn K, Onyi GO, Zhang Y, et al. A single 16s ribosomal rna substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in mycobacterium abscessus and mycobacterium chelonae. J Infect Dis 1998;177:1573–81. [DOI] [PubMed] [Google Scholar]
- 33.van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat 2012;15:149–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.