Abstract
The GGGGCC repeat expansion in C9ORF72 is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). However, it is not known which dysregulated molecular pathways are primarily responsible for disease initiation or progression. We established an inducible mouse model of poly(GR) toxicity in which (GR)80 gradually accumulates in cortical excitatory neurons. Low-level poly(GR) expression induced FTD/ALS-associated synaptic dysfunction and behavioral abnormalities, as well as age-dependent neuronal cell loss, microgliosis and DNA damage, probably caused in part by early defects in mitochondrial function. Poly(GR) bound preferentially to the mitochondrial complex V component ATP5A1 and enhanced its ubiquitination and degradation, consistent with reduced ATP5A1 protein level in both (GR)80 mouse neurons and patient brains. Moreover, inducing ectopic Atp5a1 expression in poly(GR)-expressing neurons or reducing poly(GR) level in adult mice after disease onset rescued poly(GR)-induced neurotoxicity. Thus, poly(GR)-induced mitochondrial defects are a major driver of disease initiation in C9ORF72-related ALS/FTD.
FTD, the second most common form of presenile dementia, overlaps clinically, pathologically and genetically with the motor neuron disease ALS1,2. Many genetic mutations cause both ALS and FTD; the most prevalent is a GGGGCC (G4C2) repeat expansion in the non-coding region of C9ORF72 (refs. 3,4). Studies in cellular and animal models have uncovered several downstream molecular pathways that are dysregulated in C9ORF72-related ALS/FTD, including the autophagy, nucleocytoplasmic transport, premessenger RNA splicing, stress granule dynamics and DNA damage repair pathways5. However, it is unclear which molecular events initiate disease.
One of the major unusual features of C9ORF72-related ALS/ FTD is the accumulation of proteins containing dipeptide repeats (DPRs) and the formation of DPR aggregates in patient neurons, mostly in the cytoplasm6–15. These endogenous abnormal proteins, such as poly(GA), poly(GR), poly(GP), poly(PR) and poly(PA), can be synthesized from either sense or antisense transcripts containing expanded hexanucleotide repeats, potentially through different translation mechanisms in human patient cells, including repeat-associated non-AUG (RAN) translation16,17. Multiple lines of evidence suggest that arginine-containing DPR proteins are highly toxic when ectopically expressed in experimental systems. In Drosophila, for instance, poly(GR) and poly(PR) expression induces the death of neuronal and non-neuronal cells18–20. In mammalian cells, poly(GR) and poly(PR) expression causes nucleolar stress19–22, increases DNA damage23,24 and blocks the nuclear pore through interactions with proteins containing FG domains25. However, it is not known whether low-level expression of arginine-containing DPR proteins in mice induces ALS/FTD-relevant phenotypes and which cellular defect occurs first during disease onset.
The exact pathogenic roles of individual DPR proteins in patient brains are unclear, in part because different DPR proteins are present in both aggregated and diffusible forms10–13. Two recent studies showed that poly(GR) is associated with neurodegeneration in the brains of patients with C9ORF72-related ALS/FTD14,15, suggesting a central role for poly(GR) in disease pathogenesis. To help understand how poly(GR) initiates neurodegenerative processes in vivo, we established a mouse model in which (GR)80 is expressed in the brain at a substantially lower level than in C9ORF72-related ALS/FTD brain tissues, under the control of an inducible promoter. Using this model, we discovered that mitochondrial defects are a major driver of disease initiation in C9ORF72-related ALS/FTD.
Results
Establishment of a mouse model of C9ORF72-ALS/FTD with inducible low level of poly(GR) expression.
To determine whether low-level poly(GR) expression can cause ALS/FTD disease-associated phenotypes in mice, we took advantage of the tetracyclinecontrolled inducible expression system to express (GR)80 postnatally in excitatory neurons of all layers of the cortex (Fig. 1a). To this end, we used the same method and transgenic vector as we used before to express FTD3-associated mutant charged multivesicular body protein 2B (CHMP2B) protein at a level substantially lower than endogenous mouse Chmp2B (ref. 26). Eighty copies of GR are probably a disease-relevant size, because even a few dozen G4C2 repeats can cause disease in humans and mice27,28. We engineered a transgenic vector containing the tetracycline response element, an ATG codon and a sequence encoding the FLAG epitope tag followed by a (GGXCGX)80 sequence encoding (GR)80 (Fig. 1a). Transgenic mice were generated, and three founder lines were crossed with calcium/calmodulin-dependent protein kinase II alpha-tetracycline transactivator (CamKII-tTA) mice for further expression analysis. At 2 months of age, the (GR)80 mRNA expression levels in the cortex of CamKII-tTA;(GR)80 mice were about 9 times higher in line 8 than in line 16, and 4 times higher in line 16 than in line 18 (Fig. 1b). To avoid the overexpression problem often encountered with transgenic mouse models of neurodegenerative diseases, the intermediate expresser line 16 was used for all experiments.
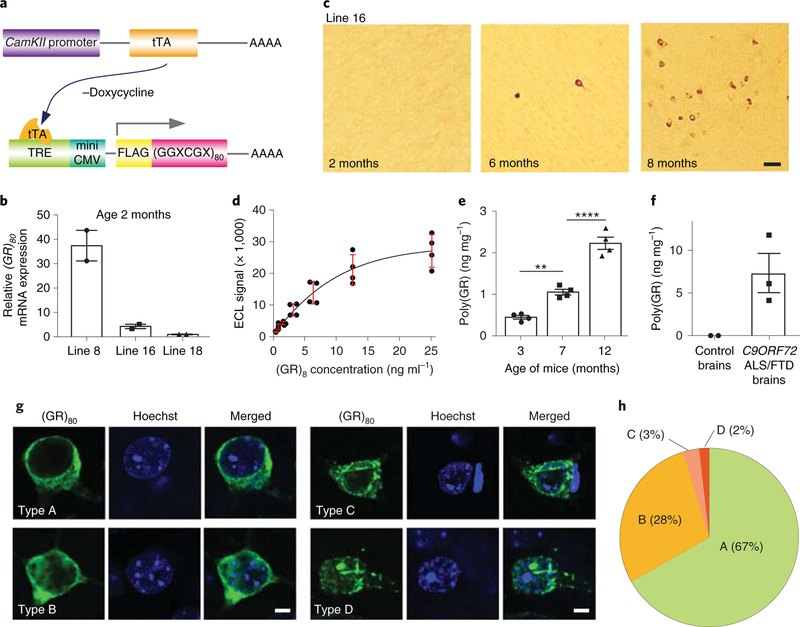
Fig. 1 |. Age-dependent accumulation of low level of poly(GR) in CamKII;(GR)80 mice.
a, Schematic of CamKII-tTA and (GR)80 constructs. b, Relative expression levels of (GR)80 mRNA in the frontal cortex of different CamKII;(GR)80 mouse lines at 2 months of age. Line 8 = 37.4±6.27, line 16 = 4.27±0.89, line 18 = 1.00±0, n = 2 mice per line. c, Age-dependent accumulation of poly(GR) in frontal cortex neurons of line 16 CamKII;(GR)80 mice (from 3 independently repeated experiments). Scale bar, 50 μm. d, Standard curve for the mesoscale discovery immunoassay with a synthetic (GR)8 peptide. Values are mean±s.d. of two independent experiments. e, Poly(GR) levels in frontal cortex tissues of CamKII;(GR)80 mice were measured by immunoassay; 3 months = 0.44±0.04, 7 months = 1.05±0.06, 12 months = 2.22±0.64 (n = 4 mice). Values are mean±s.e.m., F(2,9) = 86.57, **P = 0.0044, ****P= 0.0001 by one-way ANOVA with Tukey’s post hoc analysis for multiple comparisons. f, Poly(GR) levels in frontal cortex tissue of 2 control brains and 3 C9ORF72-related ALS/FTD brains measured by immunoassay. Control brain sample values were subtracted. Control brains = 0± 0, C9ORF72-related ALS/FTD brains = 7.34± 2.29. g, Neuronal types with different poly(GR) expression patterns in CamKII;(GR)80 mice at 8 months of age (repeated 3 times independently with similar results). Scale bar, 5 μm. h, Pie chart showing percentage of neuronal types A-D. Over 150 cells expressing poly(GR) were quantified. ECL, electrochemiluminescence.
At 2 months of age, cortical expression of (GR)80 protein in line 16 CamKII-tTA;(GR)80 mice was undetectable (Fig. 1c), even though CamKII-tTA drives transgene expression postnatally. By 6 months of age, (GR)80 was still undetectable with a fluorescein-conjugated secondary antibody; however, 3,3’-diaminobenzidine (DAB) staining after amplification of the signal with a biotinylated secondary antibody detected (GR)80 expression in the soma and dendrites of a few neurons, mostly in the frontal cortex (Fig. 1c). By 8 months of age, the number of (GR)80-positive neurons had increased (Fig. 1c). Poly(GR) accumulated mostly in the frontal cortex; this finding was confirmed in 8-month old mice of high-expresser line 8 (Supplementary Fig. 1a, b).
The difficulty of detecting (GR)80 expression in young mice of line 16 raises the possibility that it is expressed at a relatively low level. Therefore, we generated a polyclonal antibody specific to poly(GR) with no reactivity to poly(GA) and poly(GP) (Supplementary Fig. 1c). Using this antibody, we established a sensitive enzyme-linked immunosorbent assay (ELISA) that can detect poly(GR) in the range of a few nanograms per milligram (Fig. 1d). This assay confirmed the age-dependent accumulation of poly(GR) in CamKII-tTA;(GR)80 mice (Fig. 1e). More importantly, the poly(GR) level in the frontal cortex of 3–7-month-old CamKII-tTA;(GR)80 mice was only about 5–15% of that in postmortem frontal cortex tissues of patients with C9ORF72-related ALS/FTD (Fig. 1f). As in C9ORF72 induced pluripotent stem cell (iPSC)-derived neurons23 and in CamKII-tTA;(GR)80 mice (Fig. 1e), it is highly likely that poly(GR) also accumulates in an age-dependent manner in patient brains. Nonetheless, the poly(GR) level in mice is regarded as low and well within the disease-relevant range.
Similar to endogenous poly(GR) in patient brains12–14, poly(GR) in CamKII-tTA;(GR)80 mice is mostly found in the cytoplasm of neurons. Detailed analysis revealed that poly(GR)-expressing neurons can be grouped into four classes: neurons with poly(GR) diffusely distributed in the cytosol (type A), neurons with cytosolic and nuclear diffuse form of poly(GR) (type B), neurons with cytosolic aggregated form of poly(GR) (type C) and neurons with cytosolic and nuclear aggregates of poly(GR) (type D) (Fig. 1g). The nuclear localization of some poly(GR) aggregates in type B and type D neurons was further confirmed by analysis of confocal z-stack section (Supplementary Fig. 1d). Interestingly, in about 95% of neurons, poly(GR) did not form large aggregates (Fig. 1h), raising the possibility that diffusible poly(GR) can be neurotoxic in vivo.
CamKII;(GR)80 mice show age-dependent ALS/FTD-like social behavioral deficits and impaired synaptic function.
C9ORF72-related FTD often manifests as the behavioral variant FTD1. To determine whether low-level poly(GR) expression in excitatory neurons of CamKII;(GR)80 mice causes ALS/FTD-like social behavioral deficits, we used the three-chamber social interaction test29. The test mouse is placed in the middle chamber and allowed to explore freely for 10 min (habituation). Then, a stranger mouse is placed in a wire cage in the left chamber and a novel wooden object is placed in the right chamber (Fig. 2a). Both CamKII mice and CamKII;(GR)80 mice of different ages spent equally greater amounts of time in the left chamber than in the two other chambers, and, in the left chamber, both CamKII mice and CamKII;(GR)80 mice spent equal amounts of time in the left chamber (Fig. 2b,c and Supplementary Fig. 2a), suggesting a normal ability to recognize a conspecific subject. However, 6- and 9-month-old, but not 3-month-old, CamKII;(GR)80 mice spent less time interacting with the stranger mouse in the left chamber, even though they stayed there as long as control mice (Fig. 2b,c and Supplementary Fig. 2a), indicating an age-dependent deficit in social behavior, as seen in patients with FTD1. Similarly, 6- and 9-month-old, but not 3-month-old, CamKII;(GR)80 mice exhibited an age-dependent increase in anxiety in the elevated plus maze test (Fig. 2d and Supplementary Fig. 2b–d). These behavioral deficits most probably do not reflect a general deterioration in health, since CamKII;(GR)80 mice at these ages weighed the same as controls (Supplementary Fig. 3a). Moreover, the total distance traveled, the average speed and the frequency of passing through the center chamber were indistinguishable between CamKII;(GR)80 and CamKII mice at 3, 6 and 9 months of age in the open-field test (Supplementary Fig. 3b–d). In contrast, working memory of CamKII;(GR)80 mice in the T-maze test remained intact (Supplementary Fig. 4). These results indicate a specific deficit in social behavior, which is probably caused by synaptic defects in the neuronal circuit involving the prefrontal cortex, where poly(GR) accumulates. Indeed, at 6 months of age, CamKII;(GR)80 mice had significantly reduced cortical expression of the postsynaptic scaffolding protein discs large MAGUK scaffold protein 4 (Dlg4), also known as PSD-95 (Fig. 2e), suggesting a defect in synaptic structure or function.
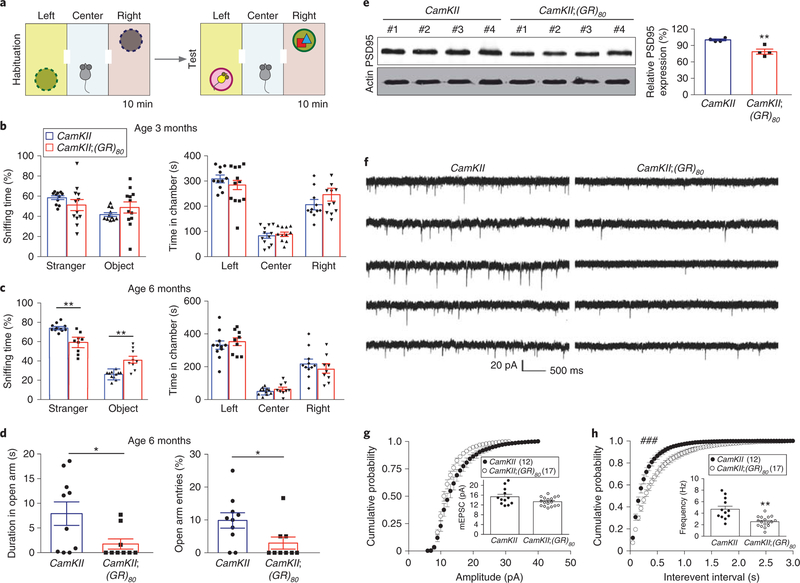
Fig. 2 |. CamKII;(GR)80 mice have age-dependent behavioral deficits and synaptic impairment.
a, Schematic of the three-chamber social interaction test. CamKII and CamKII;(GR)80 mice were allowed to habituate alone for 10 min in the 3-chambered box with empty wire cages. A stranger mouse was then placed under the wire cage in the left chamber, and a novel object was placed in the right chamber. b,c, Quantification of the 3-chamber social interaction test for CamKII and CamKII;(GR)80 mice at 3 months (b) and 6months (c) of age. In b, CamKIIS + stranger = 58.47±1.86 (n = 11 mice), CamKII;(GR)80 + stranger = 48.79 ±5.46 (n = 12 mice), CamKII + object = 41.53 ±1.86 (n = 11 mice), CamKII;(GR)80 + object = 51.21 ± 5.46 (n = 12 mice), CamKII left = 307.07±12.04 (n = 11 mice), CamKII;(GR)80 left = 269.28± 22.34 (n = 12 mice), CamKII center = 84.58±10.47 (n = 11 mice), CamKII; (GR)80 center = 87.62 ± 8.09 (n = 12 mice), CamKII right = 205.14± 16.07 (n = 11 mice), CamKII;(GR)80 right = 245.77 ±25.38 (n = 12 mice). Values are mean± s.e.m.; F(1,21) = 9.377, P = 0.1209 for CamKII + stranger versus CamKII;(GR)80 + stranger; F(1,21) = 9.377, P = 0.1209 for CamKII + object versus CamKII;(GR)80 + object; F(1,21) = 3.450, P = 0.1653 for CamKII left versus CamKII;(GR)80 left; F(1,21) = 0.22, P = 0.8251 for CamKII center versus CamKII;(GR)80 center; F(1,21) = 2.490, P = 0.2045 for CamKII right versus CamKII;(GR)80 right by two-sided Student’s t-test. In c, CamKII + stranger = 73.96±1.71 (n = 11 mice), CamKII;(GR)80 + stranger = 59.06±4.78 (n = 9 mice), CamKII + object = 26.04±1.71 (n = 11 mice), CamKII;(GR)80 + object = 43.46 ±4.78 (n = 9 mice), CamKII left = 331.6± 25.60 (n = 11 mice), CamKII;(GR)80 left = 352.66± 24.09 (n = 9 mice), CamKII center = 49.09 ±6.36 (n = 11 mice), CamKII;(GR)80 center = 62.28 ±12.26 (n = 9 mice), CamKII right = 216.8± 26.69 (n = 11 mice), CamKII;(GR)80 right = 185±27.24 (n = 9 mice). Values are mean±s.e.m.; F(1,18) = 6.570, P = 0.0019 for CamKII + stranger versus CamKII;(GR)80 + stranger; F(1,18) = 6.570, P = 0.0019 for CamKII + object versus CamKII;(GR)80 + object; F(1,18) = 1.381, P = 0.5635 for CamKII left versus CamKII;(GR)80 left; F(1,18) = 3.040, P= 0.3268 for CamKII center versus CamKII;(GR)80 center; F(1,18) = 1.17, P= 0.4187 CamKII right versus CamKII;(GR)80 right by twosided Student’s t-test. d, Time spent (left graph) and percentage of entries (right graph) in the open arm of the elevated plus maze by CamKII and CamKII;(GR)80 mice at 6months of age. Each dot represents one mouse. Left: CamKII = 7.91 ±2.38 (n = 10 mice), CamKII;(GR)80 = 1.75±1.03 (n = 9 mice), F(1,17) = 5.980, *P= 0.0358. Right: CamKII = 9.86±2.34 (n = 10 mice), CamKII;(GR)80 = 2.96±1.86 (n = 9 mice), F(1,17) = 1.76, *P=0.0362. Values are mean ± s.e.m., by two-sided Student’s t-test. e, Western blot analysis of PSD-95 expression level in the cortex of CamKII and CamKII;(GR)80 mice at 6 months of age and densitometric quantification of the western blot in e. CamKII = 100.2 ±1.62, CamKII;(GR)80 = 78.66± 4.65, values are mean ± s.e.m.; F(1,6) = 8.290, **P = 0.0047 by two-sided Student’s t-test. f, mEPSCs in layer V pyramidal neurons of prefrontal cortex. Representative traces recorded at −70 mV are selected from 12 neurons in 4 CamKII mice and 17 neurons in 4 CamKII;(GR)80 mice. g,h, Cumulative probabilities of amplitude (g) and interevent interval (h) distributions of mEPSCs. In g, CamKII = 14.38±0.91 (n = 12 cells from 4 mice), CamKII;(GR)80 = 12.50±0.40 (n = 17 cells in 4 mice), values are mean±s.e.m.; D = 0.225, P= 0.2634, by two-tailed Kolmogorv-Smirnov test. In h, CamKII = 4.696±0.529 (n = 12 cells in 4 mice), CamKII;(GR)80 = 2.528±0.216 (n = 17 cells in 4 mice). Values are mean±s.e.m. D = 0.3667, ###P = 0.0006 by two-tailed Kolmogorv-Smirnov test; t = 3.79, **P = 0.0020 by two-sided Student’s t-test.
To investigate the effects of poly(GR) on synaptic transmission in prefrontal cortex neurons in CamKII;(GR)80 mice, we analyzed AMPA receptor-mediated miniature excitatory postsynaptic currents (mEPSCs) in layer V pyramidal neurons, the major cortical output relay that exerts top-down control of subcortical brain centers and behavior30. Whole-cell patch-clamp recordings were made from acute medial prefrontal cortex slices derived from 4.5-month-old CamKII;(GR)80 and CamKII mice (Fig. 2f). mEPSCs were recorded at −70 mV in the presence of tetrodotoxin (1 μM) and picrotoxin (100 μM) to block voltage-gated Na+ currents and GABAA receptor-mediated inhibitory responses, respectively. The amplitude of mEPSCs did not differ in CamKII;(GR)80 and CamKII mice (Fig. 2g), suggesting that postsynaptic AMPA receptor activity is largely unaltered in the prefrontal cortex of mutant mice. In contrast, the frequency of mEPSCs was significantly reduced in CamKII;(GR)80 mice (Fig. 2h), indicating a loss of functional synapses, a decrease in the probability of presynaptic glutamate release or both. These findings show that poly(GR) impairs synaptic connection and efficacy in the prefrontal cortex in vivo.
CamKII;(GR)80 mice show other ALS/FTD-related cellular phenotypes.
In addition to behavioral and synaptic deficits, low-level poly(GR) expression alone can induce other ALS/FTD-relevant cellular phenotypes in vivo. At 6 months but not 3 months of age, neurons containing cleaved caspase 3 were very rare but more abundant in CamKII;(GR)80 mice than in controls (only 20 cells or so per cortical section) (Fig. 3a,b), and more abundant in the frontal cortex than in the parietal and occipital cortex of CamKII;(GR)80 mice (Fig. 3c), indicating a strong correlation between poly(GR) accumulation and neuronal cell death in this cortical region. Those caspase 3-positive neurons were mostly type C and type D poly(GR)-expressing neurons (Supplementary Fig. 5), indicating that aggregated poly(GR) is associated with dying neurons; however, the vast majority of neurons are probably functionally compromised by diffusible poly(GR), accounting for the behavioral defects. Accordingly, there was an age-dependent loss of neuronal cells, as shown by the quantification of RNA-binding protein fox-1 homolog 3 (Rbfox3, also known as NeuN)-positive neurons in the frontal cortex of 3-, 6- and 9-month-old CamKII;(GR)80 mice (Fig. 3d). These mice also had an age-dependent increase in microglia, as judged from the higher number of Ibal-positive microglia and the greater branching of their processes than in controls (Fig. 3e,f). Some microglial cells seem to be activated, as they expressed the marker Clec7a and had round, IbaI-positive cell bodies (Fig. 3g). Astrogliosis seemed to be elevated as well, as shown by increased glial fibrillary acidic protein (Gfap) expression (Supplementary Fig. 6). However, Rangap1 mislocalization, TAR DNA binding protein 43 (TDP-43) aggregation and altered Sequestosome 1 (Sqstm1), also known as p62, levels were not found in the cortex of >6-month-old CamKII;(GR)80 mice, suggesting that these molecular defects are not associated with the initiation of poly(GR)-induced disease phenotypes (Supplementary Fig. 7).
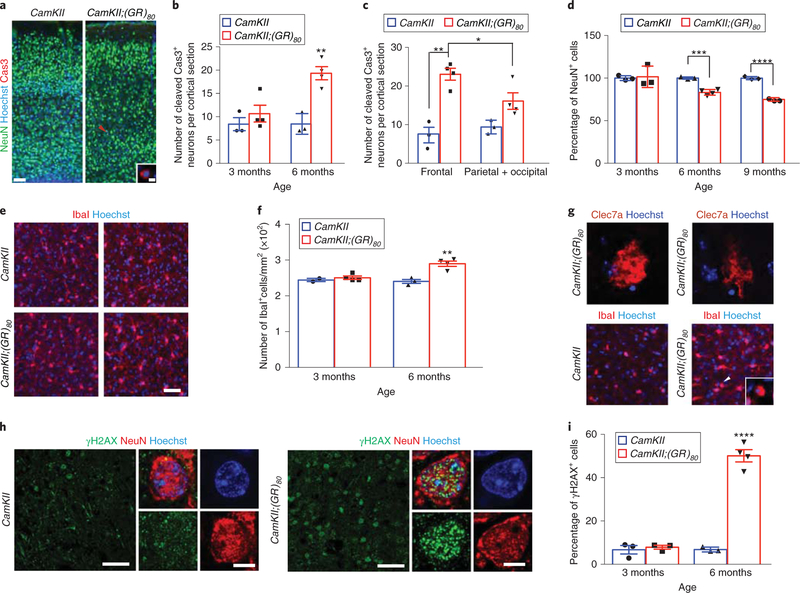
Fig. 3 |. Pathological features in the cortex of CamKII;(GR)80 mice.
a, Double-immunostaining for NeuN and cleaved caspase 3 in the cortex of 6-month-old mice (repeated 3 times independently with similar results). Arrowhead indicates the cell shown in the inset. Scale bars, 100 μm (5μm in the inset). Cas3, cleaved caspase 3. b, Quantification of the number of cleaved caspase 3-positive cells per cortical section. CamKII: 3 months = 8.40±1.40 (n = 3 mice), CamKII;(GR)80: 3 months = 10.67±1.81 (n = 4 mice), CamKII: 6 months = 8.46±1.28 (n = 3 mice), CamKII;(GR)80: 6 months = 19.33 ±1.41 (n = 4 mice). Values are mean±s.e.m. F(1,5) = 2.217, P = 0.3949 for CamKII: 3 months versus CamKII;(GR)80: 3 months; F(1,5) = 1.630, **P = 0.0028 by two-sided Student’s t-test. c, Quantification of the number of cleaved caspase 3-positive cells per section in the frontal cortex versus other cortical regions of 6-month-old mice. CamKII: frontal = 7.00±2.02 (n = 3 mice), CamKII;(GR)80: frontal = 23.04±1.53 (n = 4 mice), CamKII: parietal + occipital = 9.39±1.03 (n = 3 mice), CamKII;(GR)80: parietal + occipital = 16.10±2.15 (n = 4 mice). Values are mean±s.e.m. F(1,6) = 1.950, *P = 0.039, F(1,5) = 1.300, **P = 0.0013 by two-sided Student’s t-test. d, Quantification of the number of NeuN+ cells in frontal cortex region. CamKII: 3 months = 100±1.65 (n = 3 mice), CamKII;(GR)80: 3 months = 101.5±7.25 (n = 3 mice), CamKII: 6 months = 100±0.78 (n = 3 mice), CamKII;(GR)80: 6 months = 83.23±1.67 (n = 4 mice), CamKII: 9 months = 100±1.02 (n = 3 mice), CamKII;(GR)80: 9 months = 74.95±1.06 (n = 4 mice). Values are mean±s.e.m. F(1,4) = 19.25, P = 0.8470 for 3-month-old CamKII versus 3-month-old CamKII;(GR)80, F(1,5) = 6.100, ***P = 0.0005, F(1,5) = 1.441, ****P< 0.0001 by two-sided Student’s t-test. e, Immunostaining for the microglia-specific marker Ibal in the cortex of CamKII and CamKII;(GR)80 mice at 3 and 6 months of age (repeated 3 times independently with similar results). Scale bar, 50μm. f, Quantification of lbal+ cells in the cortex of CamKII (n = 3) and CamKII;(GR)80 (n = 4) mice at 3 and 6 months of age. CamKII: 3months = 244.3 ±4.34 (n = 2 mice), CamKII;(GR)80: 3 months = 249.6±4.79 (n = 4 mice), CamKII: 6 months = 240 ±5.50 (n = 3 mice), CamKII;(GR)80: 6 months = 289.9 ±7.3 (n = 4 mice). Values are mean±s.e.m. F(1,5) = 2.35, **P= 0.0039 by two-sided Student’s t-test. g, Activated Clec7a+ microglial cells (top; scale bar, 5μm) and round lbal+ microglial cells (indicated by the arrowhead and enlarged in the white square in lower panels) in the frontal cortex of 9-month-old CamKII;(GR)80 mice (repeated independently 3 times with similar results). Scale bar, 25μm. h, Double-immunostaining for γH2AX and NeuN in the cortex of 6-month-old CamKII and 4 CamKII;(GR)80 mice (repeated 3 times independently with similar results). Scale bars, 50 μm for large images and 5μm for small images. Note: γH2AX antibody gives rise to some background signal. i, Quantification of γH2AX+ and NeuN+ cells in the cortex. CamKII: 3 months = 6.74±1.94 (n = 3 mice), CamKII;(GR)80: 3 months = 7.86±0.90 (n = 3 mice), CamKII: 6 months = 6.83±0.62 (n = 3 mice), CamKII;(GR)80: 6 months = 50.1 ±2.82 (n = 4 mice). Values are mean±s.e.m. F(1,4) = 4.68, P = 0.6270 for 3-month old CamKII: versus 3-month-old CamKII;(GR)80:, F(1,5) = 27.48, ****P< 0.0001 by two-sided Student’s t-test.
Poly(GR) compromises mitochondrial morphology and function in CamKII;(GR)80 mice.
Previously, we reported that neurons derived from C9ORF72 iPSCs have an age-dependent increase in DNA damage and that ectopic expression of poly(GR) increases DNA damage in control iPSC-derived neurons23. Consistent with these findings, cortical neurons with elevated DNA damage were abundant in 6-month-old but not 3-month-old CamKII;(GR)80 mice, as shown by immunostaining for histone H2AX (γH2AX), a marker of double-stranded DNA damage31 (Fig. 3h,i). All neurons with increased DNA damage expressed poly(GR) (Supplementary Fig. 8), suggesting a cell-autonomous effect of poly(GR) toxicity.
To study molecular defects that lead to increased DNA damage in CamKII;(GR)80 mice, we examined the morphology and function of mitochondria, a major source of reactive oxygen species that damage DNA31. Strikingly, electron microscopy analysis revealed that disruption of the inner mitochondrial membrane and loss of cristae were increased in the prefrontal cortex of CamKII;(GR)80 mice at 3 months of age (Fig. 4a,b). The defect was more pronounced in 6-month-old CamKII;(GR)80 mice, indicating an age-dependent accumulation of mitochondrial damage. Because some cellular defects and behavioral deficits were not obvious in 3-month-old CamKII;(GR)80 mice, our finding raises the interesting possibility that mitochondrial defects are an early pathogenic event in this mouse model of C9ORF72-ALS/FTD. We performed additional assays to measure mitochondrial length of primary cortical neurons isolated from CamKII and CamKII;(GR)80 mice at 14 d in vitro (Fig. 4c). Mitochondria in CamKII;(GR)80 neurons seemed to be shorter (Fig. 4c); indeed, detailed quantification indicated that mitochondria less than 1.0-μm long were more numerous in CamKII;(GR)80 neurons than in CamKII neurons (Fig. 4d). Moreover, the average mitochondrial length in neurites was shorter in many CamKII;(GR)80 neurons than in CamKII neurons (Fig. 4e). Moreover, mitochondria in cultured CamKII;(GR)80 neurons were less motile than those in control neurons (Supplementary Fig. 9). Interestingly, in accordance with mitochondrial fragmentation, the level of the mitochondrial fission protein dynamin 1-like (Dnm1l), also known as DRP1, was increased and that of the mitochondrial fusion protein optic atrophy 1 (OPA1) was decreased in the cortex of 9-month-old CamKII;(GR)80 mice (Supplementary Fig. 10), further indicating a defect in mitochondrial dynamics. Next, we sought to test whether these morphology changes would have an effect on the main mitochondrial function, the production of ATP through oxidative phosphorylation mediated by respiratory chain complexes. We found decreased activities of mitochondrial complexes I and V in the frontal cortex of 6-month-old mice (Fig. 4f).
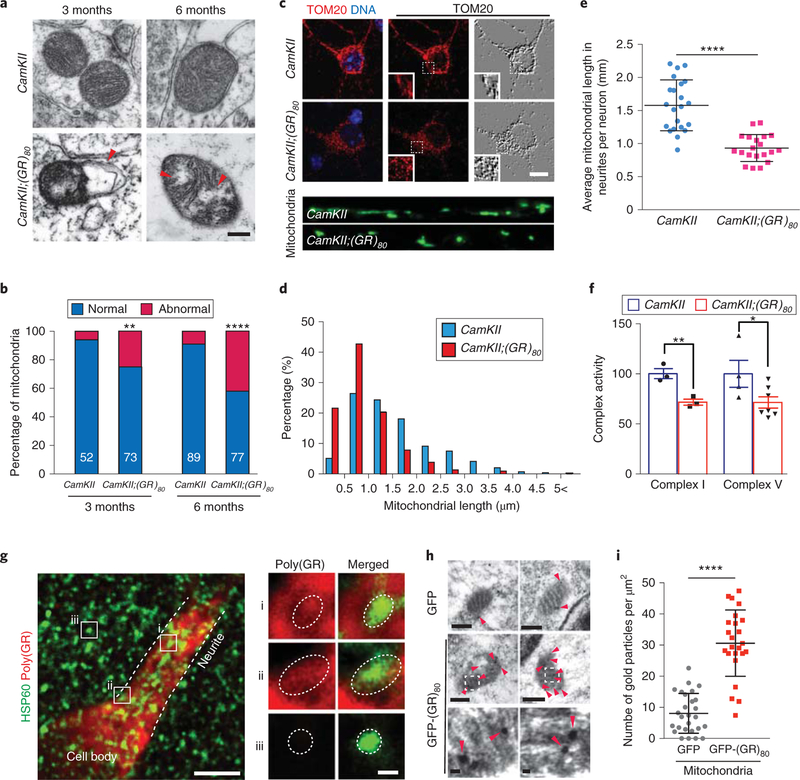
Fig. 4 |. Poly(GR) induces mitochondrial dysfunction in the cortex of CamKII;(GR)80 mice.
a, Mitochondrial morphology in prefrontal cortex of 3- and 6-month-old CamKII and CamKII;(GR)80 mice analyzed by transmission electron microscopy in 3 independent experiments with similar results. Red arrowhead indicates disruption of the mitochondrial inner membrane. Scale bar, 200 nm. b, Quantification of the percentages of normal and abnormal mitochondria in the prefrontal cortex of CamKII and CamKII;(GR)80 mice at 3 and 6 months of age. The numbers of analyzed mitochondria are indicated in each column. Values are mean±s.d. **P = 0.0067, ****P<0.0001 by two-tailed chi-squared test for categorical data. c, Representative images of mitochondria in poly(GR)-expressing mouse primary cortical neurons at 14d in vitro (DIV14) from 3 independent experiments. Scale bar, 10 μm. d, The distribution of mitochondrial length in 22 control and 20 poly(GR)-expressing mouse primary cortical neurons at DIV14. e, Average mitochondrial length in mouse primary cortical neurons at DIV14. Each dot represents 1 neuron and in total 323 mitochondria in CamKII neurons and 275 mitochondria in CamKII;(GR)80 neurons were measured. CamKII = 1.58±0.08 (n = 22 neurons), CamKII;(GR)80 = 0.93±0.045 (n = 20 neurons). Values are mean± s.d. F(1,40) = 3.57, ****P< 0.0001 by two-sided Student’s t-test. f, The activities of mitochondrial complexes I and V in the frontal cortex of 6-month-old CamKII and CamKII;(GR)80 mice. CamKII: complex I =100.2 ±4.98 (n = 3 mice), CamKII;(GR)80: complex I =71.67±3.00 (n = 3 mice), CamKII: complex V = 100±13.45 (n = 4 mice), CamKII;(GR)80: complex V = 71.45±5.64 (n = 7 mice). Values are mean±s.e.m. F(1,9) = 3.250, *P = 0.0464; F(1,4) = 2.750, **P= 0.008 by two-sided Student’s t-test. g, Double immunostaining for hsp60, a mitochondrial marker, and for poly(GR) in type A poly(GR)-expressing neurons of 9-month-old CamKII;(GR)80 mice (repeated 5 times independently with similar results). Scale bar, 5μm. Enlarged squares i, ii and iii are shown on the right. Dotted circles indicate the mitochondrial location. Scale bar, 0.5 μm. h, Representative images obtained by immuno-electron microscopy using gold particles to label poly(GR) in HEK293 cells overexpressing GFP and GFP-(GR)80 (repeated 3 times independently with similar results). Red arrowheads indicate immunogold-labeled poly(GR). Scale bars, top and middle images, 200 nm; bottom images, 20 nm. i, Number of gold particles in mitochondria from each HEK293 cell overexpressing GFP and GFP-(GR)80: GFP = 8.08±1.23 (n = 25 cells), GFP-(GR)80 = 30.61 ±2.17 (n = 24 cells). Values are mean±s.d. F(1,49) = 2.770, ****P<0.0001 by two-sided Student’s t-test.
To investigate how poly(GR) affects mitochondrial function, we first analyzed the subcellular location of poly(GR) in mouse brain neurons at high resolution. In type A poly(GR)-expressing neurons of 9-month-old CamKII;(GR)80 mice, poly(GR) was diffusely expressed in the soma and proximal dendrites, including inside mitochondria (Fig. 4g), although the intensity of the poly(GR) signal was generally weaker inside than outside of mitochondria (Fig. 4g). The specificity of the poly(GR) signal was demonstrated by its absence in mitochondria in cells that did not express poly(GR) (Fig. 4g). To further confirm the presence of poly(GR) in mitochondria, we examined type C neurons of 9-month-old CamKII;(GR)80 mice with a high level of poly(GR) expression (line 8), where poly(GR) formed droplets or small aggregates in the soma and proximal dendrites (Supplementary Fig. 11). Most poly(GR) aggregates did not overlap with mitochondria; however, mitochondria often appeared to be in close contact with poly(GR) droplets and diffusible poly(GR) was clearly present in mitochondria (Supplementary Fig. 11a′′, b′). The poly(GR) signal was specific, as no signal was observed in mitochondria that did not express poly(GR) (Supplementary Fig. 11c′). This notion is further supported by immuno-electron microscopy analysis of HEK293 cells expressing green fluorescent protein (GFP)-(GR)80. Judging from the number of gold particles coated with poly(GR)-specific antibody, poly(GR) was indeed present in mitochondria (Fig. 4h,i), in contrast to a recent report that poly(GR) is not localized in mitochondria32. Thus, nonaggregated or diffusible forms of poly(GR) may directly affect mitochondrial morphology and function.
Poly(GR) interacts with and decreases expression of Atp5a1 in CamKII;(GR)80 mice.
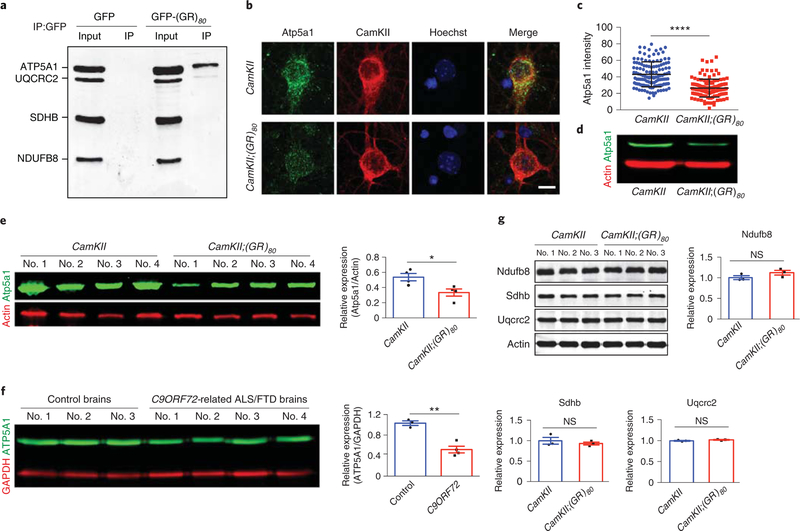
To investigate the molecular mechanism of poly(GR)-induced mitochondrial dysfunction, we did coimmunoprecipitation experiments to pull down poly(GR) in transfected HeK293 cells and then used an OXPHOS antibody cocktail to identify mitochondrial respiratory chain complex proteins that may interact with poly(GR). We found that poly(GR) preferentially binds to ATP synthase F1 subunit alpha (ATP5FA1), also known as ATP5A1, a subunit of mitochondrial respiratory chain complex V (Fig. 5a). This interaction was confirmed by reverse immunoprecipitation experiments in which an ATP5A1 antibody was used to pull down poly(GR) (Supplementary Fig. 12a). To determine how the binding between poly(GR) and ATP5A1 affects mitochondrial function, we found that the endogenous expression level of Atp5a1 was decreased in poly(GR)-expressing primary cortical neurons cultured from CamKJI;(GR)80 mice, as shown by immunostaining and western blot analyses (Fig. 5b–d), as well as in poly(GR)-expressing neurons in the CamKII;(GR)80 mouse brain (Supplementary Fig. 12b). More importantly, the Atp5a1 protein level was also reduced in the cortex of 6-month-old CamKII;(GR)80 mice (Fig. 5e) and in the frontal cortex of 4 C9ORF72-related ALS/FTD patients compared with 3 control brains (Fig. 5f). This decrease is not due to transcriptional regulation since the Atp5a1 mRNA level was not affected in CamKII;(GR)80 mice at 6 months of age (Supplementary Fig. 12c). Moreover, protein levels of some other mitochondrial proteins such as NADH:ubiquinone oxidoreductase subunit B8 (Ndufb8); succinate dehydrogenase complex iron sulfur subunit B (Sdhb); ubiquinol cytochrome c reductase core protein 2 (Uqcrc2); Hspd1 heat shock protein 1 (Hspd1, also known as HSP60); and ATP synthase, H+-transporting, mitochondrial F0 complex, subunit D (Atp5h), were not affected in the cortex of CamKII;(GR)80 mice or C9ORF72-related ALS/FTD brains (Fig. 5g and Supplementary Fig. 13). Together, these results suggest that poly(GR) specifically decreases Atp5a1 levels in CamKJI;(GR)80 mice and that this interaction may be partly responsible for compromised mitochondrial morphology and function.
Fig. 5 |. Poly(GR) binds to ATP5A1 and decreases Atp5a1 expression level in the cortex of CamKII;(GR)S0 mice.
a, Proteins that coimmunoprecipitated with anti-GFP antibody in 4 independently repeated experiments were analyzed by western blots with an OXPHOS cocktail of antibodies, which showed that poly(GR) preferentially binds to the mitochondrial complex V protein ATP5A1 in HEK293 cells. b, Immunostaining analysis of Atp5a1 protein levels in the DIV14 primary cortical neurons (repeated 3 times independently with similar results). Scale bar, 10 μm. c, Atp5a1 intensity in primary neurons in b. Each dot represents one neuron. CamKII = 43.07±1.35 (n = 126 neurons), CamKII;(GR)80 = 26.39±0.93 (n = 135 neurons). Values are mean±s.d. F(1, 259) = 1.960, ****P< 0.0001 by two-sided Student’s t-test. d, The representative western blot analysis (of 4 independently performed experiments) of Atp5a1 level in the DIV14 primary cortical neurons. e, Western blot analysis and relative Atp5a1 expression in the frontal cortex of 6-month-old CamKII and CamKII;(GR)80 mice. CamKII = 0.54±0.05 (n = 4 mice), CamKII;(GR)80 = 0.33 ± 0.05 (n = 4 mice). Values are mean± s.e.m. F(1,6) = 1.042, *P = 0.0240 by two-sided Student’s t-test. f, Western blot analysis and ATP5A1 levels in the frontal cortex of C9ORF72-related ALS/FTD and control brains. Control brains = 1.03±0.04 (n = 3 brains), C9ORF72-related ALS/FTD brains = 0.52±0.07 (from n = 4 patients). Values are mean±s.e.m. F(1,5) = 2.810, **P = 0.0018 by two-sided Student’s t-test. g, Western blot analysis and quantification of Ndufb8 for complex I, Sdhb for complex II and Uqcrc2 for complex III in the frontal cortex of CamKII and CamKII;(GR)80 mice at 9 months of age. CamKII: Ndufb8 = 1.00 ±0.05 (n = 3 mice), CamKII;(GR)80: Ndufb8 = 1.12±0.06 (n = 3 mice), CamKII: Sdhb = 1.00±0.08 (n = 3 mice), CamKII;(GR)80: Sdhb = 0.93±0.03 (n = 3 mice), CamKII: Uqcrc2 = 1.00±0.01 (n = 3 mice), CamKII;(GR)80: Uqcrc2 = 1.02±0.01 (n = 3 mice). Values are mean±s.e.m. F(1,4) = 1.440, P = 0.1799 for Ndufb8; F(1,4) = 5.770, P=0.5024 for Sdhb; F(1,4) = 1.470, P= 0.1454 for Uqcrc2 graph by two-sided Student’s t-test. IP, immunoprecipitation; NS, not significant.
Poly(GR) increases ATP5A1 ubiquitination and degradation via the proteasome pathway and ectopic expression of Atp5a1 rescues neurodegeneration of CamKII;(GR)80 neurons.
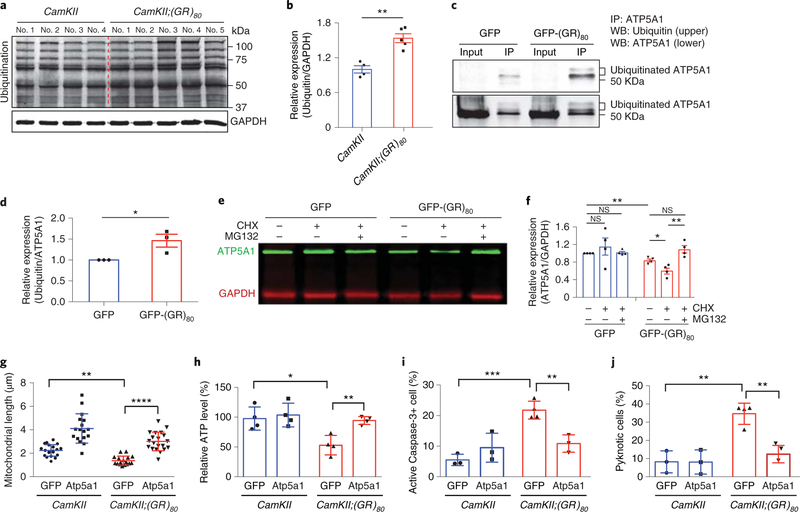
To understand the molecular mechanism by which poly(GR) decreases the Atp5a1 level in CamKII;(GR)80 mice, we did a series of experiments. First, we found that ubiquitination was increased in the cortex of CamKII;(GR)80 mice at 6 months of age (Fig. 6a,b), suggesting that some proteins have increased ubiquitination and/or some ubiquitinated proteins accumulate. Next, to examine the ubiquitination status of ATP5A1, we immunoprecipitated endogenous ATP5A1 and then used anti-ubiquitin and anti-ATP5A1 antibodies for western blot analysis, and, under this experimental condition, ATP5A1 with higher molecular weight was greatly enriched in the immunoprecipitated fraction, which is probably due to increased accessibility of ATP5A1 epitopes to this antibody on these isoforms (Fig. 6c). We found that ubiquitination of ATP5A1 was increased in (GR)80-transfected HEK293 cells compared with GFP-transfected cells (Fig. 6c,d). Finally, to examine whether the increased ubiquitination of ATP5A1 could in part explain its reduced levels in the presence of (GR)80, we treated GFP- or (GR)80-transfected HEK293 cells with cycloheximide (CHX) for 30 h to block protein synthesis and then examined degradation of ATP5A1. In GFP-transfected cells, the ATP5A1 level was not significantly affected by treatment with CHX or the proteasome inhibitor MG132 with this time window, suggesting a relatively long half-life (Fig. 6e,f) consistent with an earlier study on ATP5A1 protein stability33, although some other mitochondrial proteins are more susceptible to degradation through the ubiquitin-proteasome pathway33,34. Interestingly, in cells expressing (GR)80 and treated with CHX, ATP5A1 degraded faster, which was probably through the proteasome pathway because concurrent treatment with MG132 prevented its degradation (Fig. 6e,f). Thus, poly(GR) increases ubiquitination of ATP5A1 and its degradation through the proteasome pathway.
Fig. 6 |. Poly(GR) increases ubiquitination and degradation of ATP5A1, and Atp5a1 overexpression rescues neuronal survival of CamKII;(GR)80 neurons.
a, Western blot analysis of protein ubiquitination in the frontal cortex of CamKII mice (n = 4) and CamKII;(GR)80 mice (n = 5) at 6 months of age. b, Quantification of the western blot in a. CamKII = 1.00±0.06 (n = 4 mice), CamKII;(GR)g0 = 1.54±0.08 (n = 5 mice). Values are mean±s.e.m. F(1,7) = 1.833, **P= 0.0012 by two-sided Student’s t-test. c, Immunoprecipitated ATP5A1 from HEK293 cells expressing GFP and GFP-(GR)80 was analyzed by western blot with anti-ubiquitin (top) and anti-ATP5A1 (bottom) antibodies. This experiment was repeated independently 3 times. d, Quantification of 3 independent experiments, as in c. GFP = 1.00±0, GFP-(GR)80 = 1.46±0.15, from n = 3 experiments. Values are mean±s.e.m. t = 3.01, *P = 0.0396 by two-sided Student’s t-test. e, ATP5A1 degradation in HEK293 cells expressing GFP and GFP-(GR)80 as revealed by treatment with CHX with or without MG132 for 30h (repeated independently 3 times). f, Quantification of 4 independent experiments, as in e. GFP = 1.00 ±0, GFP + CHX = 1.15±0.19, GFP + CHX + MG132 = 1.02±0.03, GFP-(GR)80 = 0.84±0.03, GFP-(GR)80 + CHX = 0.61 ±0.07, GFP-(GR)80 + CHX + MG132 = 1.09 ±0.08 from n = 4 experiments. Values are mean±s.e.m. F(1,6) = 454,589.000, P = 0.4647 for GFP versus GFP + CHX; F(1,6) = 9,745.00, P = 0.5383 for GFP versus GFP + CHX + MG132; F(1,6) = 14,602, **P = 0.0039 for GFP versus GFP-(GR)80; F(1,6) = 3.684, *P = 0.0213 for GFP-(GR)80 versus GFP-(GR)80 + CHX; F(1,6) = 2.724, **P = 0.0082 for GFP-(GR)80 + CHX versus GFP-(GR)80 + CHX + MG132; F(1,6) = 10.03, P= 0.0604 for GFP-(GR)80 versus GFP-(GR)80 + CHX + MG132 by two-sided Student’s t-test. g, Length of mitochondria in GFP-expressing and GFP-Atp5a1-overexpressing DIV14 primary cortical neurons from CamKII and CamKII;(GR)80 mice. Each data point is the average of 20 mitochondria in neurites of 1 neuron. CamKII GFP = 2.24±0.49 (n = 20 neurons), CamKII Atp5a1 =4.11 ±1.25 (n = 17 neurons), CamKII;(GR)80 GFP = 1.38± 0.38 (n = 17 neurons), CamKII;(GR)80 Atp5a1 = 3.01 ±0.79 (n = 20 neurons). Values are mean±s.d. F(3,70) = 37.27, **P = 0.0024 for CamKII GFP versus CamKII;(GR)80 GFP; ****P<0.0001 for CamKII;(GR)80 GFP versus CamKII;(GR)80 Atp5a1 by one-way ANOVA with Tukey’s post hoc analysis for multiple comparisons. h, ATP production in GFP-expressing and GFP-Atp5a1-overexpressing primary cortical neurons at DIV14. CamKII GFP = 97.60 ±19.48, CamKII Atp5a1 =103.61 ±19.87, CamKII;(GR)80 GFP = 52.99±16.37, CamKII;(GR)80 Atp5a1 = 94.39±6.58 from 4 independent primary cultures. Values are mean±s.d. F(3,12) = 7.860, *P = 0.0111 for CamKII GFP versus CamKII;(GR)80 GFP; *P = 0.0180 for CamKII;(GR)80 GFP versus CamKII;(GR)80 Atp5a1 by one-way ANOVA with Tukey’s post hoc analysis for multiple comparisons. i,j, Number of cleaved caspase 3-positive cells (i) and pyknotic cells (j) in GFP-expressing and GFP-Atp5a1-expressing DIV14 primary cortical neurons. Values are mean±s.d., analyzed by one-way ANOVA followed by Tukey’s post hoc test. In i, CamKII GFP = 5.51 ±1.85, CamKII Atp5a1 = 9.52 ±4.74, CamKII;(GR)80 GFP = 21.77± 2.91, CamKII;(GR)80 Atp5a1 =10.85±2.85 from n = 3 independent primary cultures. F(3,9) = 16.85, ***P = 0.0005, **P = 0.0073. In j, CamKII GFP = 8.22 ±6.06, CamKII Atp5a1 = 8.12± 6.65, CamKII;(GR)80 GFP = 34.64±5.82, CamKII;(GR)80 Atp5a1 = 12.42 ±4.79 from n = 3 independent primary cultures. F(3,9) = 17.22, **P = 0.0011 for CamKII:GFP versus CamKII;(GR)80 GFP; **P = 0.0035 for CamKII;(GR)80 GFP versus CamKII;(GR)80 Atp5a1. WB, western blot.
To determine whether an increase in ectopic Atp5a1 protects from poly(GR) toxicity, we overexpressed Atp5a1 using lentiviral transduction in primary cortical neurons isolated from CamKII;(GR)80 mice. Atp5a1 overexpression decreased mitochondrial fragmentation in poly(GR)-expressing cortical neurons and restored ATP levels (Fig. 6g,h). Moreover, Atp5a1 expression reduced the number of cleaved caspase 3-positive and pyknotic cells in poly(GR)-expressing primary cortical neurons (Fig. 6i,j). These findings suggest that the direct interaction between poly(GR) and Atp5a1 results in its degradation via the ubiquitin-proteasome pathway and that restoring levels of Atp5a1 attenuates poly(GR) toxicity, resulting in increased ATP levels and neuronal survival.
Reducing poly(GR) production in adult mice rescues poly(GR)-induced neurodegeneration.
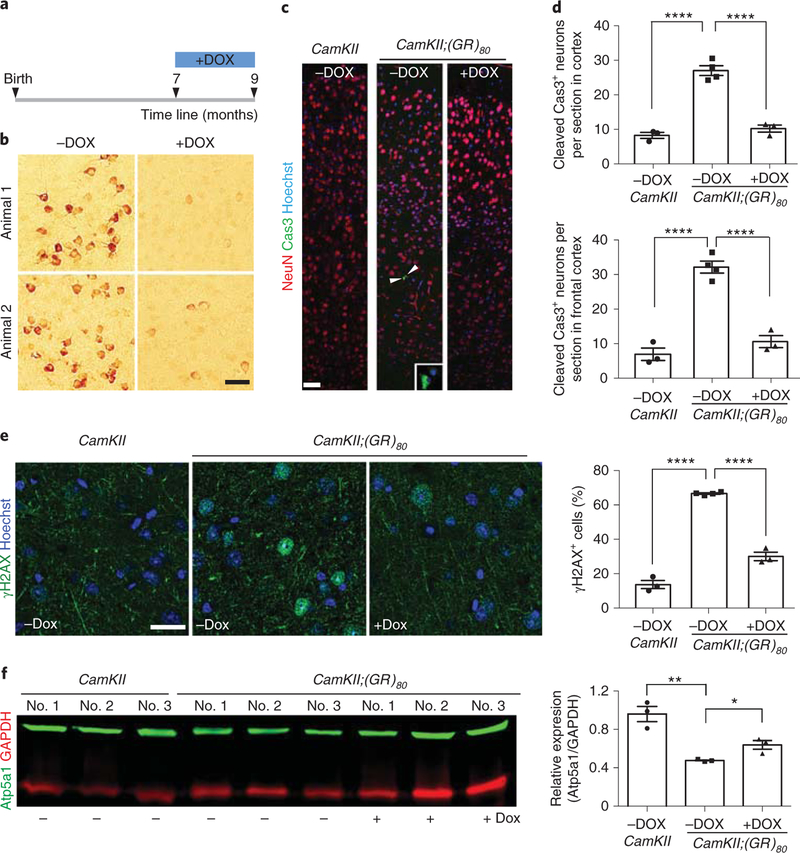
Next, we determined whether reducing the poly(GR) level alters the behavioral and cellular phenotypes in CamKII;(GR)80 mice. Feeding mice with chow containing doxycycline beginning at 1 month of age to shut down expression of (GR)80 prevented social behavioral deficits and reduced anxiety level at 6 months of age (Supplementary Fig. 14). To further test whether disease phenotypes can be at least partially reversed after disease onset, we turned off (GR)80 expression in CamKII;(GR)80 mice beginning at 7 months of age (Fig. 7a), when disease-relevant behavioral and cellular phenotypes are already present. After 2 months of doxycycline treatment, the (GR)80 level in the cortex greatly reduced in the 9-month-old mice (Fig. 7b). As anticipated, the number of cleaved caspase 3-positive neurons in the cortex, especially the frontal cortex, was further elevated in 9-month-old (Fig. 7d) than 6-month-old (Fig. 3b) CamKJI;(GR)80 mice. Similarly, some other disease phenotypes such as DNA damage (Fig. 7e versus Fig. 3i) and microgliosis (Supplementary Fig. 15b versus Fig. 3f) were also worsened in an age-dependent manner. Remarkably, these phenotypes were reduced in CamKII;(GR)80 mice fed doxycycline for 2 months (Fig. 7c–e and Supplementary Fig. 15a,b). Also reversed were the increased levels of astrogliosis induced by poly(GR) in vivo (Supplementary Fig. 15c,d). Finally, the decrease in Atp5a1 level in the cortex of CamKII;(GR)80 mice was partially reversed after the poly(GR) expression level was greatly reduced (Fig. 7f). Thus, therapeutic strategies to lower poly(GR) levels can reverse disease phenotypes even after disease onset.
Fig. 7 |. Reducing (GR)80 expression in adult mice rescues pre-existing pathological features of CamKII;(GR)80 mice.
a, Schematic of doxycycline (DOX) treatment of CamKII;(GR)80 mice from 7–9 months of age. b, Poly(GR) expression in the frontal cortex of 9-month-old CamKII;(GR)80 mice with and without doxycycline treatment (repeated 3 times independently with similar results). Scale bar, 50 μm. c, Immunostaining for NeuN and cleaved caspase 3 in the cortex of 9-month-old CamKII or CamKII;(GR)80 mice with or without doxycycline treatment (repeated 3 times independently with similar results). Arrowheads indicate cells positive for cleaved caspase 3. Scale bar, 50 μm. d, Average number of cleaved caspase 3-positive (Cas3+) neurons per section in the cortex (top) or frontal cortex only (bottom) of 9-month-old CamKII and CamKII;(GR)80 mice. All values are mean± s.e.m. and analyzed by one-way ANOVA with Tukey’s post hoc analysis for multiple comparisons. Top: CamKII -DOX = 8.26±0.87 (n = 3 mice), CamKII;(GR)80 -DOX = 27.00±1.42 (n = 4 mice), CamKII;(GR)80 +DOX = 10.21 ±1.03 (n = 3 mice). F(2,8) = 75.96, ****P< 0.0001. Bottom: CamKII-DOX = 6.92 ±1.81 (n = 3 mice), CamKII;(GR)80 -DOX = 32.14± 1.73 (n = 4 mice), CamKII;(GR)80 +DOX = 10.58±1.73 (n = 3 mice). F(2,8) = 63.39, ****P< 0.0001. e, Immunostaining and quantification of γH2AX+ and NeuN+ cells in the cortex of 9-month-old CamKII and CamKII;(GR)80 mice. CamKII -DOX = 13.66± 2.39 (n = 3 mice), CamKII;(GR)80 -DOX = 66.61 ±0.43 (n = 4 mice), CamKII;(GR)80 +DOX = 30.03±2.46 (n = 3 mice). Values are mean± s.e.m. F(2,8) = 250.800, ****P< 0.0001 by one-way ANOVA with Tukey’s post hoc analysis for multiple comparisons. Scale bar, 25 μm. f, Atp5a1 protein level in the cortex of 9-month-old mice. CamKII -DOX = 0.96± 0.08, CamKII;(GR)80 -DOX = 0.48± 0.01, CamKII;(GR)80 +DOX = 0.64± 0.05 from n = 3 mice. Values are mean ± s.e.m. F(2,6) = 21.69, **P = 0.0016, *P = 0.0120 by two-sided Student’s t-test.
Discussion
In this study, we established a mouse model of C9ORF72-related ALS/FTD in which poly(GR) is expressed in the brain in a spatially and temporally controlled manner. We found that the poly(GR) level in the frontal cortex is about 85–95% lower in these mice at 3–7 months of age than in patients with C9ORF72-related ALS/FTD and that poly(GR) accumulates in the soma and dendrites of neurons in an age-dependent manner. Poly(GR) expression induced ALS/FTD-associated synaptic dysfunction and social behavioral deficits as well as neuronal cell loss, microgliosis and DNA damage. The increased DNA damage is probably a consequence of compromised mitochondrial morphology and function, which may result in increased oxidative stress that in turn causes DNA damage such as in C9ORF72 iPSC-derived motor neurons23. More importantly, poly(GR) preferentially bound to Atp5a1, a subunit of mitochondrial complex V, whose expression in the frontal cortex of CamKII;(GR)S0 mice and C9ORF72-related ALS/FTD brains was decreased probably in part through enhanced degradation via the ubiquitin-proteasome pathway—a molecular mechanism for poly(GR) toxicity. Moreover, several disease-relevant cellular phenotypes were reversed by ectopic expression of Atp5a1 in cultured neurons or by reducing the poly(GR) level in adult mice. Thus, it may be possible to develop therapies to decrease poly(GR) level and halt progression of C9ORF72-related ALS/FTD even after the onset of disease.
A key molecular event downstream of C9ORF72 repeat expansion is the production of DPR proteins, which are mostly localized in the cytoplasm6–15. The composition of DPR proteins in C9ORF72-related ALS/FTD neurons may be much more complex than initially thought and may depend on which translation mechanisms are in play17. Thus, mouse models of individual DPR protein toxicity are highly informative. Studies in patient brain tissues indicate that only poly(GR) is significantly more abundant in clinically related brain areas than in unrelated areas14,15, suggesting that poly(GR) has a central role in disease pathogenesis. A major feature of our mouse model is that poly(GR) is expressed at a substantially lower level than in C9ORF72-related ALS/FTD brain tissues. Thus, a relatively low level of diffusible poly(GR) without aggregate formation can initiate disease in vivo.
Synaptic dysfunction in the prefrontal cortex probably underlies the social behavioral deficits in our mouse model, as this brain region is critically important for social cognition30. Interestingly, poly(GR) accumulated preferentially in the soma and dendrites of frontal cortex neurons, especially those in deep layers (Fig. 1). This accumulation may result from a compromised degradation and might help explain the selective vulnerability of these neurons in patients with FTD. The observed synaptic dysfunction in CamKII;(GR)80 mice can be caused at least in part by mitochondrial dysfunction as in other experimental systems35–37. Remarkably, morphological defects in mitochondria were present in CamKII;(GR)80 mice at 3 months of age, before social behavioral and other cellular defects become apparent, suggesting that mitochondrial dysfunction is an early pathogenic event. Other cellular phenotypes, such as increased proliferation of microglia, could result from synaptic loss caused by cell-autonomous toxicity of poly(GR) in neurons, as microglia have a key role in the formation and maintenance of functional synapses38. Interestingly, at 6 months of age, when poly(GR)-induced disease phenotypes are present in CamKII;(GR)80 mice, TDP-43 pathology seems to be absent (Supplementary Fig. 7). Thus, poly(GR) itself may initiate disease, and the TDP-43 pathology in autopsy tissues of patients may be a late event in disease progression. Indeed, a case of C9ORF72-related FTD with RNA foci and DPR protein accumulation but without TDP-43 aggregation 13 y before autopsy has been reported13, and overexpression of poly(GA) also induces neurodegeneration in mice without obvious TDP-43 mislocalization and inclusion formation39,40.
At the mechanistic level, one important insight comes from the finding that poly(GR) preferentially binds to ATP5A1, a stable protein often used as a marker for mitochondrial complex V. Our immunostaining and immuno-electron microscopy analyses revealed that poly(GR) is present within mitochondria. Poly(GR) increased the ubiquitylation of ATP5A1 and its degradation via the ubiquitin- proteasome pathway. Indeed, expression of Atp5a1, but not some other mitochondrial proteins, was reduced in CamKII;(GR)80 mice and in brain tissues of patients with C9ORF72-related ALS/FTD. It will be interesting to determine at the molecular level how a low level of poly(GR) increases Atp5a1 ubiquitination, and what other mechanisms contribute to selective reduction in the Atp5a1 protein level in CamKII;(GR)80 mice. The biochemical interaction between poly(GR) and Atp5a1 seems to be a key pathogenic mechanism during disease initiation, since ectopic expression of Atp5a1 in primary neurons isolated from CamKII;(GR)80 mice rescued poly(GR)-induced mitochondrial defects and neuronal cell death (Fig. 6).
Finally, our finding that some aspects of poly(GR)-induced toxicity can be reversed even after disease onset is encouraging because it suggests that diffusible poly(GR) induces functional deficits long before neuronal cell death. Thus, therapeutic approaches to lower the poly(GR) level may be beneficial for C9ORF72-related ALS/ FTD even after the onset of clinical symptoms. Our poly(GR) mouse model will be useful to test these therapeutic approaches and to examine the contribution of genetic modifiers of poly(GR) toxicity, identified in other disease models.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41593-019-0397-0.
Methods
Mice and genotyping.
All procedures involving mice were approved by The University of Massachusetts Medical School Institutional Animal Care and Use Committee. A DNA sequence encoding the Flag tag followed by 80 copies of the dipeptide GR (sequence (GGXCGX)80, FLAG-(GR)80) was cloned into the pTRE-Tight-BI-ZsGreen1 mammalian expression vector (Clontech) with the restriction enzymes NotI and SalI. Before microinjection into mouse oocytes, the vector was linearized with BbsI, which cuts the vector twice in the region encoding ZsGreen1 and removes a 447-base pair fragment to prevent expression of the ZSGreen 1 gene. The F1 litters were analyzed by PCR for transgene insertion with the following set of primers: forward 5′-GATTACAAGGACGACGACGA-3′ and reverse 5′-GACAAGTTATCAACGTCCAC-3′. Six germline-transmitting founders were obtained and backcrossed to wild-type C57BL/6 mice to maintain hemizygous lines. To express the transgene in the forebrain, the (GR)80 transgenic male mice were crossed with CamKII-tTA female mice (Jackson Laboratory, 007004). Equal or near equal numbers of male and female double-transgenic offspring were used in all experiments. Except unexpected accidental death, all animals and data points are used in the analyses. Mice were randomly assigned to the various experimental groups and selected for behavioral experiments.
Human brain tissues.
Human brain tissue was obtained from the Neurodegenerative Diseases Brain Bank at the University of California, San Francisco. Authorization for autopsy was provided by all participants or their next-of-kin in accordance with the Declaration of Helsinki. All procedures were approved by the University of California, San Francisco Committee on Human Research. Neuropathological diagnoses are summarized in Supplementary Table 1.
Brains were selected on the basis of neuropathological diagnosis and genetic analyses. Fresh-frozen blocks of the superior frontal gyrus were obtained from three individuals with frontotemporal lobar degeneration with transactive response DNA-binding protein 43KDa proteinopathy (FTLD-TDP) type B carrying a C9ORF72 repeat expansion and from three control participants. Control tissue was obtained from individuals without dementia who had minimal age-related neurodegenerative changes. Further details are provided in Supplementary Table 1.
Immunohistochemistry.
Mice were anesthetized and perfused through a transthoracic cardiac puncture with prechilled 0.9% saline and then 4% paraformaldehyde (PFA). The brains were dissected and postfixed in 4% PFA at 4 °C for 2 d. After fixation, the brains were washed in PBS, allowed to sink in 30% sucrose solution at 4 °C, cut into 40-p.m sections with a cryostat and stored in 50% glycerol solution at –20 °C.
For immunofluorescence studies, the brain tissues were washed in PBS and incubated in 3% BSA with 0.2% Triton X-100 (blocking solution) for 30 min at room temperature. The tissues were then incubated with primary antibodies in PBS containing 3% BSA and 0.2% Triton X-100 overnight on a shaker at room temperature. The primary antibodies were for NeuN (ABN78 and MAB377, Millipore; 1:1,000), cleaved caspase 3 (9661S, Cell Signaling; 1:500; 347683, United States Biological Life Science; 1:200), Ibal (019–19741, Wako; 1:1,000), glial fibrillary acidic protein (GFAP) (Z0334, Dako; 1:1,000), γH2AX (AB10022, Millipore; 1:500), HSP60 (MAB1800, R&D Systems; 1:500), Atp5a1 (14748, Abcam; 1:500), CamKII (611293, BD Biosciences; 1:500), Ran GAP1 (SC-25630, Santa Cruz Biotechnology; 1:500), CLE7A (217331, Abcam; 1:500) and poly-GR (ABN1361, Millipore; 1:2,000). Each antibody had been validated by the manufacturers. The brain tissues were then incubated with secondary antibodies conjugated with Alexa 488 and 586 (Invitrogen), washed in PBS and mounted in CC mounting medium (C9368, Sigma).
For DAB staining, the brain tissues were washed in PBS and pretreated with methanol containing H2O2 to eliminate endogenous peroxidase. The tissues were then washed in PBS and incubated first in blocking solution for 30 min at room temperature and then with the poly-GR (ABN1361, Millipore; 1:2,000) or TDP-43 (12892–1-AP, Proteintech; 1:1,000) primary antibodies overnight on a shaker at room temperature. The tissues were washed with PBS, incubated with biotinylated secondary antibody (Vector), treated with avidin-biotin complex/horseradish peroxidase reagent solution (PK-7100, Vector) for 30 min, developed with a DAB substrate kit (SK-4105, Vector), stained with hematoxylin solution and mounted in Permount solution (SP15–100, Fisher).
Behavioral tests.
Mice were singly housed in home cages and kept under specific-pathogen-free conditions in an animal facility with a 12-h light/12-h dark cycle (light off at 7:00). All behavioral tests were done on 3-, 6- and 9-month-old mice between 9:00 and 17:00. To avoid effects of stress, behavioral tests were done at 2–3-d intervals. Before testing, mice were placed in the operating room to habituate for 20–30 min.
Open-field.
General exploratory locomotion was assessed with the open-field test. The middle chamber (20 × 20 cm2) and border area (10 cm from the wall) of a white polypropylene box (40 × 40 × 40 cm3) were recorded with an EthoVision system (Noldus) program to evaluate the number of entries and time spent. The experimental mice were placed in the center of the chamber and allowed to explore freely for 10 min. Their behavior was recorded and analyzed automatically by the EthoVision system.
Elevated plus maze.
The elevated plus maze, made of black polypropylene and elevated about 1 m above the ground, consists of 2 opposite closed arms (each 6 cm wide and 40 cm long) enclosed by 20-cm-high walls, 2 opposite open arms (each 6 × 40 cm2) and a central square (6 × 6 cm2). The open arms have a 0.5-cm-high railing to prevent falls. To assess anxiety, the mouse was placed in the central square facing an open arm and allowed to explore freely for 5 min. The number of entries into the open and closed arms and the time spent in each arm were analyzed with MED-PC to Excel (Med Associates).
Sociability.
The sociability task apparatus has 3 chambers, each 40 cm long and 30 cm wide and separated by walls with a removable door (3 × 5 cm2) between each chamber. For habituation, mice were placed in the center chamber and allowed to explore all chambers freely. After 10 min, a stranger mouse (randomly selected from a group of mice of the same sex and age as the experimental mice) and a novel wooden object (of the size and weight of a mouse) were placed under wire cages in the flanking chambers. Social ability was evaluated with the EthoVision system by measuring how long the test mouse spent sniffing the stranger mouse and the wooden object and the total time spent in each chamber.
T-maze.
The T-maze test apparatus has 3 arms (30 × 10 cm2) and is elevated 30 cm above the ground. During the first 24 h, mice were fed with small pieces of chow (1 g). On the next day, each mouse was kept for 20 min in the testing room to calm down and was then forced to run from the start arm to the left arm where food was placed as reward, while the door to the right arm was blocked. After 10 s, the mouse was allowed to enter either the left or right arm. If the mouse chose the left arm where food was placed, the event was counted as a success. This test was repeated 10 times for each mouse; 9–10 mice of each genotype were tested.
Electrophysiology.
CamKII and CamKII;(GR)80 mice (4.5 months of age) were killed. Their brains were rapidly removed and placed in saturated (95% O2/5% CO2) ice-cold artificial cerebrospinal fluid (ACSF) containing 126 mM NaCl, 2.5 mM KCl, 2.5 mM CaCl2, 1.2 mM MgCl2, 25 mM NaHCO3, 1.2 mM NaH2PO4 and 11 mM D-glucose. Coronal slices (300 μm) containing the medial prefrontal cortex (1.54–2.8 mm anterior to the bregma) were cut with a Leica VT1200 vibratome. Slices were collected into a tissue incubator (Harvard Apparatus), incubated in ACSF for at least 1 h at room temperature (21–23 °C), transferred to a recording chamber and continuously perfused with oxygenated ACSF by a gravity-driven perfusion system. Recordings were made at 32 °C; temperature was maintained with a temperature controller (Warner Instruments).
Whole-cell voltage-clamp recordings of individual layer V pyramidal neurons in the anterior cingulated cortex, prelimbic cortex or both were done with an Axopatch 200B amplifier (Molecular Devices). Pyramidal neurons were identified morphologically by infrared differential interference contrast microscopy. Recording pipettes (4.5–5.5 MΩ) were filled with solution containing 142 mM Cs-gluconate, 8 mM NaCl, 10 mM HEPES, 0.4 mM EGTA, 2.5 mM QX-314 (N-(2,6-dimethylphenylcarbamoylmethyl) triethylammonium bromide), 2 mM Mg-ATP and 0.25 mM GTP-Tris, pH 7.25. The superfusion solution contained 1 μM tetrodotoxin (Sigma) (to block voltage-gated Na+ channels) and 100 μM picrotoxin (to block GABAA receptor-mediated inhibitory synaptic responses). Series resistance was monitored throughout the recordings, and data were discarded if the resistance changed by more than 15%. All data were digitized and collected with Digidata 1322A and pClamp software (version 9.2; Molecular Devices). Signals were filtered at 2 kHz and analyzed with Mini Analysis 6 (Synaptosoft).
Western blot analysis.
Mice of each genotype were anesthetized, and the cortex was dissected, quickly frozen at –80 °C, homogenized and sonicated in RIPA buffer with proteinase and phosphatase inhibitors (Thermo Fisher, catalog no. 89900). The protein extract was centrifuged to remove tissue debris, boiled for 15 min, subjected to 10% SDS-PAGE and transferred to a western blot membrane. The membrane was incubated with primary antibodies against PSD-95 (MA1–045, Thermo Fisher; 1:1,000), GFAP (Z0334, Dako; 1:2,000), ATP5A1 (14748, Abcam; 1:1,000), ATP5h (173006, Abcam; 1:1,000), OXPHOS (ab110413, Abcam; 1:1,000), DLP1 (61112, BD; 1:1,000), OPA1 (80471, Cell Signaling; 1:1,000), ubiquitinylated conjugation (PW8810, Enzolifescience; 1:1,000), p62 (ab91526, Abcam; 1:1,000), β-tubulin (T3592–100UG, Sigma; 1:1,000), GAPDH (10494–1-AP, Proteintech; 1:1,000) and actin (A2228, Sigma-Aldrich; 4967, Cell Signaling; 1:1,000) overnight at 4 °C and then with IRDye secondary antibodies (LI-COR Bioscience) for 1 h at room temperature and analyzed with a LI-COR Odyssey image system.
Western blot analysis of human brain samples.
Human frontal cortex tissues from 3 control brains and brains of 4 patients with behavioral variant FTD with motor neuron disease (FTD-MND) were lysed in RIPA buffer (Thermo Scientific) containing protease and phosphatase inhibitors. Total protein (30 μg) from each sample was analyzed by western blot (as described above) with primary antibodies against ATP5A1, ATP5H, HSP60, GAPDH and actin.
Immunoprecipitation of poly(GR), ATP5A1 and ubiquitin.
For coimmunoprecipitation experiments, HEK293 cells were cultured in DMEM medium with 10% fetal bovine serum (Life Technologies). At 70% confluency, cells were transfected with EGFP-C1 empty vector or EGFP-(GR)80 plasmid using Lipofectamine 2000 (Life Technologies). After 48 h, transfected cells were collected and homogenized in lysis buffer (50 mM Tris-HCl, pH 7.5, 125 mM NaCl, 5% glycerol, 0.5% NP40, 1.5 mM MgCl2, 25 mM NaF, 0.2 mM dithiothreitol) and protease and phosphatase inhibitors. For ATP5A1 and ubiquitin immunoprecipitation experiments, we used the Pierce direct IP/ColP kit (Thermo Scientific) by incubating beads with the respective antibodies: mouse anti-ATP5A1 (10 μg, Abcam, ab14748) and mouse anti-ubiquitin (10 μg Enzo life Sciences, PW8810–0100). Then, beads were incubated with lysates of cells expressing EGFP-C1 or EGFP-(GR)80 and washed 3 times for 5 min each. The beads were resuspended in gel-loading buffer and boiled for 10 min. The immunoprecipitation samples were analyzed by western blot (as described above) with the following primary antibodies: anti-total OXPHOS antibody cocktail (Abcam, ab110413; 1:1,000), rabbit polyclonal anti-(GR)8 antibody (custom made by Covance, 1:100) and mouse anti-ATP5A1 (Abcam, ab14748; 1:1,000).
Cell counting.
After NeuN, Ibal and Histone H2AX immunostaining, fluorescence images of the cortex of CamKII and CamKII;(GR)80 mice were examined by confocal microscopy (Leica). NeuN+, IbaI+ and Histone H2AX+ cells were counted with the ImageJ cell-counter plug-in. Cells positive for cleaved caspase 3 were counted on every 12th section throughout the cortex.
Quantitative PCR with reverse transcription.
The prefrontal cortex was dissected from 2-month-old mice and quickly frozen at −80 °C. RNA was extracted with an RNeasy kit (Qiagen), and complementary DNA was synthesized with random hexamers and TaqMan reverse transcriptase (Applied Biosystems). For quantitative PCR, primers specific for Flag-(GR)80 are: forward primer 5′-GGCGCTTTCTCATAG CTCAC-3′ and reverse primer 5′-GCCTACATACCTCGCTCTGC-3′. Primers for Atp5a1 are: forward primer 5′-GCCCTCGGTAATGCTATTGA-3′ and reverse primer 5’-GCAATCGATG TTTTCCCAGT-3′. For quantitative PCR, these primers were used in reactions with SYBR Green PCR master mix (Applied Biosystems). Gapdh-specific primers were used as an internal control: forward primer 5′-AACTTTGGCATTGTGGAAGG-3′ and reverse primer 5′-ACACATTGGG GGTAGGAACA-3′.
Transmission electron microscopy.
Frontal cortex tissues from CamKII and CamKII;(GR)go mice were cut into ~1-mm3 pieces, fixed in glutaraldehyde-containing phosphate buffer, pH 7.2, at room temperature and postfixed in 1% OsO4 solution for 1 h at 4 °C. The specimens were then dehydrated, embedded in araldite, cut into semi-thin and thin sections, stained with uranyl acetate and lead citrate, examined with a Philips CM10 electron microscope and photographed with a Gatan TEM CCD camera.
Immuno-gold electron microscopy.
HEK293 cells expressing EGFP-C1 and EGFP-(GR)80 plasmids were fixed with 4% PFA (v/v) in 0.75 M sodium phosphate buffer (pH 7.2) solution for 1 h, pelleted and dehydrated with ethanol, embedded in LR white resin (Sigma-Aldrich) and cut into ultrathin sections (80 nm) in gold support grids. The grids were incubated first in blocking solution (10% normal goat serum in PBS) for 1 h and then with rabbit polyclonal anti-(GR)8 antibody (Covance; 1:100) overnight at room temperature in blocking solution. The grids were washed 5 times with PBS 1× and incubated with 1 μg/μl rabbit anti-IgG (1:20) conjugated with 12-nm gold particles in blocking buffer. The samples were examined on a Philips CM 10 transmission electron microscope using 80 kV accelerating voltage. Images were captured with a Gatan TEM CCD camera.
Lentiviral infection and ATP assay.
Cortical neurons from CamKII and CamKII;(GR)80 neurons were cultured for 9 d and then transduced with the supernatants of lentiviral particles pseudotyped with VSGV envelope expressing GFP or Atp5a1 for an additional 5 d. Then, cells were fixed with 4% PFA for immunostaining experiments or collected for ATP measurement. Primary cortical neurons were collected on day 14 in vitro and homogenized in ATP assay buffer (ATP assay kit, catalog no. ab83355, Abcam), and the protein content was measured. Samples in each group were deproteinized with Deproteinizing Sample Kit-TCA (catalog no. ab204708, Abcam). The ATP level of each group was then determined as recommended by the manufacturer of the ATP assay kit.
Proteasome and protein synthesis inhibitor treatment.
For proteasome and protein synthesis inhibitor experiments, HEK293 cells transfected with EGFP-C1 or EGFP-(GR)80 plasmids were cultured for 16 h, then treated with 10 μM proteasome (MG123, Sigma-Aldrich) and 50 μg ml−1 protein synthesis inhibitor (Cycloheximide, Sigma-Aldrich) for 30 h. Cell were then collected for western blot analysis.
Confocal and fluorescence microscopy.
Confocal images from mouse tissue and primary cortical neurons were acquired with a Leica TCS SP5 II laser scanning confocal microscope, and each color channel (blue, green and red) image was processed with Leica LAS AF software. For the measurement of mitochondrial movement, at 12 d in vitro, primary cortical neurons from CamKII and CamKII;(GR)80 mice were transfected with DsRed-Mito CalPhos kit (631312, Clontech). After 2 d, mitochondrial movement in neurites was recorded every 2 s for 5 min and images were acquired with a Nikon Eclipse TiE wide-field microscope equipped with a cooled CMOS camera (Andor Zyla) and a temperature- and CO2-controlled environmental chamber; images were processed with NIS elements software and analyzed with the Manual tracking plug-in of ImageJ.
Immunoassay analysis of poly(GR).
To prepare human cortex tissue homogenates, approximately 30 mg frontal cortex was homogenized in 10% (w/v) ice-cold RIPA buffer containing 25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% sodium deoxycholate, 1% Nonidet P-40, 1% sodium dodecyl sulfate and 2× protease and phosphatase inhibitors and sonicated at a 20% pulse rate for 15–20 s on ice. Lysates were centrifuged at 100,000g for 30 min at 4 °C. The supernatant was collected into a fresh tube. The pellet was resuspended in RIPA buffer, resonicated and recentrifuged, and the concentration of RIPA-soluble protein was determined by BCA assay (Pierce). Approximately 30 mg frozen cortex tissue from GR80 and CAMK mice of different ages was homogenized in 10% (w/v) ice-cold lysis buffer containing 150 mM NaCl, 25 mM Tris, pH 7.5, 5 mM EDTA, 1% Triton X-100, 1% SDS and 2× protease and phosphatase inhibitors. The homogenate was sonicated at a 20% pulse rate for 15–20 s on ice. Lysates were centrifuged at 16,000g for 15–20 min at 4 °C. The supernatant was collected into a fresh tube, and the protein concentration was determined by bicinchoninic acid assay.
A poly(GR) sandwich immunoassay was established with a newly generated affinity-purified polyclonal rabbit anti-(GR)8 antibody (custom made by Covance). The experiment validating this antibody is presented in Supplementary Fig. 1c. For use as a capture antibody, this antibody (1 μg μl−1) was coated on a 96-well single spot plate (Meso Scale Discovery) overnight at 4 °C. The next day, after blocking and washing of the plate, tissue lysates were diluted to the same concentration (human brain lysate 1 μg μl−1, mouse brain lysate 1.7 μg μl−1) with lysis buffer or Tris-buffered saline and tested in duplicate wells. GOLD SULFO-tagged anti-(GR)8 antibody (0.5 μg μl−1; Meso Scale Discovery) was used as a detection antibody. Serial dilutions of the recombinant (GR)8 peptide in 1% BSA + Tris-buffered saline with Tween 20 (BSA-TBST) were used to prepare the standard curve. A QuickPlex SQ120 instrument (Meso Scale Discovery) was used to acquire response signal values of emitted light on electrochemical stimulation of the assay plate. A four-parameter logistic regression curve program (GraphPad Prism) was used to fit the values obtained with the recombinant (GR)8 peptide calibrators, and unknown concentrations from test samples were interpolated and expressed as nanograms per milligram. Specificity was confirmed with a peptide cross-reactivity assay using (GP)8 and (GA)8 synthetic peptides (Covance) at a concentration of 100 ng ml-1. For mouse tissue, values from control mice of different ages were subtracted from the corresponding test samples to correct for background.
Doxycycline treatment.
To suppress (GR)80 expression, mice in each group were fed a diet containing doxycycline (200 mg kg−1, Bioserv) from 7 to 9 months or from 1 to 6 months of age.
Statistical analysis.
For statistical analyses, we used unpaired or two-sided t-tests, one-way analysis of variance (ANOVA) followed by Tukey post hoc comparison or two-way ANOVA followed by Bonferroni correction for multiple comparisons; the chi-squared test was used for categorical data. Values are presented as mean ± s.e.m. or mean ± s.d.; P < 0.05 was considered statistically significant. The exact value of P is not provided by the software when it is less than 0.0001. Data distribution was assumed to be normal but this was not formally tested. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications, such as minimal number of mice needed for behavioral experiments (for example, ref. 26).
Data collection.
Data collection and analysis were not performed blind to the conditions of the experiments, except for two independent experiments in Fig. 1e.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
We thank A. Tapper for sharing mouse behavioral equipment. This work was supported by the National Institutes of Health (grant no. R01NS093097 to W.-D.Y. and F.-B.G., grant nos. R37NS057553 and R01NS101986 to F.-B.G., and grant nos. P01AG019724 and P50AG023501 to W.W.S.) and by grants from the Packard Center for ALS Research, the Target ALS Foundation and the Muscular Dystrophy Association (F.-B.G.), Frick Foundation for ALS Research, the ALS Association and Angel Fund (S.A.), the Alzheimer’s Association (grant no. 2016-NIRG-396129 to S.A. and grant no. 2018- AARFD-592264 to R.L.-G.), the Tau Consortium and the Bluefield Project to Cure Frontotemporal Dementia (W.W.S.). F.-B.G. thanks the Cellucci family for their inspiration.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41593-019-0397-0.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Code availability
In this study, we used only publicly available software for data analysis such as ImageJ. Please send a request to the corresponding author for further details.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Olney NT, Spina S. & Miller BL Frontotemporal dementia. Neurol. Clin 35, 339–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardiman O. et al. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 3, 17085 (2017). [DOI] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton AE et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao FB, Almeida S. & Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J 36, 2931–2950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori K. et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 1335–1338 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Ash PE et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zu T. et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl Acad. Sci. USA 110, E4968–E4977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackenzie IR et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 126, 859–879 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Davidson YS et al. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol. Commun 2, 70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackenzie IR et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130, 845–861 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Schludi MH et al. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol 130, 537–555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vatsavayai SC et al. Timing and significance of pathological features in C9orf72 expansion-associated frontotemporal dementia. Brain 139, 3202–3216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saberi S. et al. Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol 135, 459–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakae N. et al. Poly-GR dipeptide repeat polymers correlate with neurodegeneration and clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathol. Commun 6, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleary JD & Ranum LP New developments in RAN translation: insights from multiple diseases. Curr. Opin. Genet. Dev 44, 125–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao FB, Richter JD & Cleveland DW Rethinking unconventional translation in neurodegeneration. Cell 171, 994–1000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizielinska S. et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen X. et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang D. et al. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol 130, 525–535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon I. et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao Z. et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet 24, 2426–2441 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Gonzalez R. et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron 92, 383–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farg MA, Konopka A, Soo KY, Ito D. & Atkin JD The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum. Mol. Genet 26, 2882–2896 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Shi KY et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl Acad. Sci. USA 114, E1111–E1117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gascon E. et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat. Med 20, 1444–1451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sareen D. et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med 5, 208ra149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chew J. et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348, 1151–1154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moy SS et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav 3, 287–302 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Forbes CE & Grafman J. The role of the human prefrontal cortex in social cognition and moral judgment. Annu. Rev. Neurosci 33, 299–324 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Madabhushi R, Pan L. & Tsai LH DNA damage and its links to neurodegeneration. Neuron 83, 266–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YJ et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med 24, 1136–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavie J. et al. Ubiquitin-dependent degradation of mitochondrial proteins regulates energy metabolism. Cell Rep 23, 2852–2863 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Franz A, Kevei E. & Hoppe T. Double-edged alliance: mitochondrial surveillance by the UPS and autophagy. Curr. Opin. Cell Biol 37, 18–27 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Li Z, Okamoto K, Hayashi Y. & Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119, 873–887 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Pathak D. et al. The role of mitochondrially derived ATP in synaptic vesicle recycling. J. Biol. Chem 290, 22325–22336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebrahimi-Fakhari D. et al. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep 17, 1053–1070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong S, Dissing-Olesen L. & Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr. Opin. Neurobiol 36, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schludi MH et al. Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol 134, 241–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang YJ et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci 19, 668–677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.