Abstract
Encoded in mammalian cells by 33 genes, the transforming growth factor-β (TGF-β) family of secreted, homodimeric and heterodimeric proteins controls cell differentiation of most, if not all, lineages and many aspects of cell and tissue physiology in multicellular eukaryotes. Deregulation of TGF-β family signaling leads to developmental anomalies and disease, and enhanced TGF-β signaling contributes to cancer and fibrosis. Here, we review the fundamentals of the signaling mechanisms that are initiated upon TGF-β ligand binding to its cell-surface receptors, and the dependence of the signaling responses on input from and cooperation with other signaling pathways. We discuss how cells exquisitely control the functional presentation and activation of heteromeric receptor complexes of transmembrane dual-specificity kinases and, thus, define their context-dependent responsiveness to ligands. We also introduce the mechanisms through which proteins called Smads act as intracellular effectors of ligand-induced gene expression responses, and that the specificity and impressive versatility of Smad signaling depend on crosstalk from other pathways. Finally, we discuss how non-Smad signaling mechanisms, initiated by distinct ligand-activated receptor complexes, complement Smad signaling and thus contribute to the cell responses.
INTRODUCTION
Cells communicate through membrane-associated proteins or secreted molecules that define their proliferation, differentiation, metabolism and behavior. Secreted hormones, cytokines, chemokines and growth factors interact with cell surface receptors over long distances or in close proximity. The latter involves paracrine communication between neighboring cell populations, or autocrine responses within the same cell population that produces the secreted factors. Many secreted proteins have been named growth factors, since cell proliferation was historically easy to score, yet proliferation is closely linked to cell differentiation and metabolism, and gene expression that control cell phenotype and behavior. Most growth factors are grouped in structurally related families that reflect evolutionary diversification from ancestral genes.
Among the secreted growth factors and cytokines, the transforming growth factor-β (TGF-β) family receives a lot of attention because of its many functions at the cellular level and in development, and roles in many diseases including cancer (1). TGF-β family proteins are secreted and function as homo- or heterodimers, and recognized by a characteristic spacing of seven cysteines in the mature, fully processed polypeptide (2). Among these, the TGF-β1 homodimer is seen as prototype, since it was the first TGF-β family protein to be biochemically characterized and defined by complementary DNA (cDNA) cloning (3, 4). It was also the first one to be readily available as a purified protein for experimental use (3, 4), and, thus, has been most extensively studied. Many concepts about the modes of actions of the other TGF-β family proteins have been inferred by analogy with TGF-β; in how far this is fully justified should be further explored.
Many TGF-β family proteins stimulate cell proliferation, albeit often modestly compared to other growth factors, and depending on the cell type and environment. TGF-β, however, strongly inhibits the proliferation of various cell types, including epithelial, endothelial, hematopoietic and immune cells (5). Most prominent are the many effects of TGF-β family proteins on cell differentiation, as they control differentiation of all cell lineages at multiple steps in development (1, 2). Besides cell proliferation and differentiation, they exert many additional cell functions; they promote or protect against cell death, promote extracellular matrix (ECM) protein expression, cell motility and invasion, and control cell metabolism (1, 2). Yet, in spite of a wealth of information, the developmental and physiological functions of most TGF-β family proteins remain poorly defined, with most attention focused on only few TGF-β proteins. TGF-β family proteins are also well studied because of their roles in disease, with most attention given to the roles of TGF-β1 in cancers, primarily carcinomas (6–8), and fibrosis (9), and the deregulated activities of TGF-β and related Bone Morphogenetic Proteins (BMPs) in connective tissue diseases (10). The activities and functions of TGF-β family proteins at the cellular level, in development and disease, have been extensively reviewed (1).
Consistent with its roles in cell-cell communication and cell differentiation, the signaling system activated by the TGF-β family emerged during evolution with the appearance of multicellular metazoa. The simplest extant metazoan consisting of two cell layers, Trichoplax adhaerens, has a minimal yet complete TGF-β family signaling system with recognizable receptors and Smads, the intracellular signaling effectors (11). The receptor-Smad system duplicated and diversified with the emergence of chordates to give rise to the vertebrate receptor-Smad signaling network (11). Plants have a heteromeric receptor system that structurally resembles the heteromeric TGF-β receptor system, yet is activated by brassinosteroids as ligands (12). No Smad genes have been found in unicellular organisms or plants.
LIGAND DIVERSITY
The mouse and human genomes have 33 functional genes encoding TGF-β family polypeptides (2). Each of these consists of a signal peptide, required for secretion, a long pro-polypeptide, and the mature polypeptide that, as a dimer, binds and activates the receptors (13) (Fig. 1A). Besides three TGF-β and five inhibin (activin) polypeptides, the TGF-β family sequences comprise many that are named BMPs or “growth and differentiation factors” (GDFs) (2). The nomenclature is confusing; many proteins received several names that do not reflect their natural functions (2).
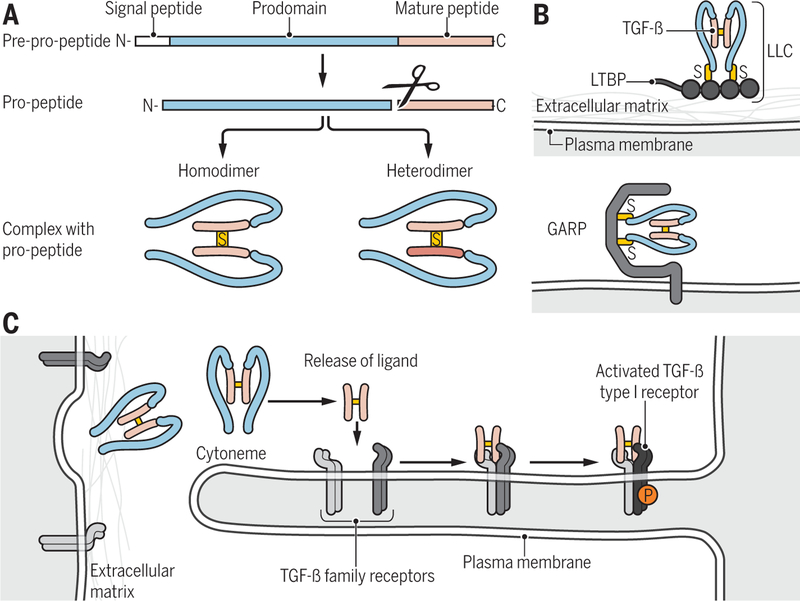
Figure 1. Ligand processing and presentation.
(A) TGF-β family proteins are synthesized as precursor molecules consisting of a signal peptide, a prodomain (termed latency-associated peptide, LAP, for TGF-β), and the mature polypeptide. After signal peptide removal, the precursor is further processed by proteolytic cleavage at basic residues, thus separating the prodomain from the mature polypeptide, which remain non-covalently associated. Concomitant, disulfide-linked dimerization of the mature polypeptides into mature homo- and heterodimeric proteins is shown. (B) Latent TGF-β complex can associate through disulfide bonding with LTBP into a large latent complex (LLC) that in turn associates with the extracellular matrix (top), or with the plasma membrane-associated GARP (bottom). (C) Cytoneme-associated activation of TGF-β family signaling. Long cytonemes extend from the cell body, and present TGF-β family receptors to ligand complexes. Binding of ligand to the receptors results in activation of the type I receptors (light to dark blue).
Credit: Veronica Falconieri Hays/Science Signaling
Four structurally related, dimeric secreted proteins, named glial-derived neurotrophic factor (GDNF), neurturin, artemin and persephin, share key structural similarities with the mature TGF-β family ligands, but are, based on sequence similarities, more distant from the TGF-β family (14, 15). These neurotrophins signal through a small family of receptors that is distinct from TGF-β family receptors; that is through cell surface-linked GDNF family receptor α (GFRα) proteins in combination with the Ret1 transmembrane tyrosine kinase (14, 15). They are therefore no longer seen as members of the TGF-β family. Growth/differentiation factor 15 (GDF15), which is structurally seen as a bona fide member of the TGF-β family (2), does not act through TGF-β family receptors either. Rather, it binds to GFRα-like (GFRAL), a transmembrane receptor that is related to the GFRα receptors and also signals through association with Ret1, thus activating signaling pathways commonly associated with receptor tyrosine kinases (RTKs) (16, 17). Consequently, GDF15 has signaling properties that link it to the GDNF family of neurotrophins. The dimeric nerve growth factor (NGF) and platelet-derived growth factor (PDGF) also show similarities with TGF-β in their three-dimensional topologies, yet have no apparent sequence similarities, suggesting an ancient structural conservation and phylogenic relationship among these growth factor families (18). Thus, TGF-β family proteins, GDNF-related neurotrophins, NGF and PDGF proteins combine into a superfamily (18).
The mature TGF-β family polypeptides are recognized by a conserved cysteine pattern and sequence features that define their structural folding and enable their dimerization (13). Basic residues that are predicted cleavage sites of furin proteases separate the pro-segments from the conserved mature polypeptide (13) (Fig. 1A). Cleavage by furins occurs generally but not always in the endoplasmic reticulum, and may be constitutive for the TGF-βs, yet regulated for some TGF-β family proteins (19). Such regulation is apparent with Gbb, the Drosophila homolog of BMP-5, −6 and −7, and illustrates an underappreciated scenario of differential proteolytic processing of the precursor as basis for tissue-specific activities (20–22). The pro-segments act as chaperones required for folding and secretion of TGF-β family dimers (19, 23).
Among the three TGF-βs, TGF-β1 is abundant in platelets, which allowed its initial purification and the discovery that it is a disulfide-linked homodimer (3, 24). Biochemical studies showed that inhibin βA and βB naturally exist as disulfide-linked homodimers and heterodimers, both designated as activins, whereas inhibin α and inhibin β combine in a heterodimer, named inhibin (13, 25–27). Whether the other TGF-β family proteins naturally occur as homo- or heterodimers has largely remained uncharacterized. The commercial availability of BMPs and GDFs as homodimers and the preferred concept of homodimerization explain why most studies define their activities as homodimers, and the assumption that they physiologically act as homodimers. Increasing evidence, however, much of it from developmental genetics, indicates or suggests that TGF-β family proteins often act as heterodimers with activities that are stronger or distinct from the homodimers, as illustrated with the BMP-2/−7 (28) and Vg1/Nodal (29, 30) heterodimers (Fig. 1A), a new concept that requires increased appreciation.
Three-dimensional structures have been elucidated for ten fully mature and active TGF-β family protein homodimers, including all three TGF-βs and several BMPs (13). They show symmetric conformations with a single disulfide between the two monomers and overall structural similarities, including a cystine knot (13, 31, 32). Some have a “closed” configuration of the two monomers, and others show an open conformation with little contact between the two monomers except for the disulfide (13). With TGF-β3 able to acquire both conformations, conformational transitions may provide inherent flexibility in the association of a TGF-β family ligand with inhibitor proteins or receptor ectodomains (33, 34). BMP-9 exists as disulfide-bonded dimers as well as stable dimers without a disulfide bond, and both forms are both able to bind and activate the receptors (35). Redox-dependent interconversion between both dimers controls their balance, with the non-linked dimer showing increased susceptibility to proteolytic degradation (35). Whether similar transitions control the properties of other TGF-β family ligands remains to be seen. A few TGF-β family polypeptides lack the cysteine that mediates dimerization, suggesting other means to dimerize, or act as competitive antagonists (13). Much has to be learnt about the nature of the physiological ligands, and structures for asymmetric heterodimers need to be defined.
Mature TGF-β family dimers were shown or are thought to be secreted in a complex, in which the large prosegments remain associated with the disulfide-linked mature protein (19). For all three TGF-βs, this association confers latency, meaning that it prevents binding of active TGF-β to its receptors. This complex is most often associated, through a disulfide linkage, with a latent TGF-β binding protein (LTΒP) that directs its deposition in the ECM (19) (Fig. 1B). Instead of association with an LTBP, the latent TGF-β complex has also been found to be disulfide-linked to a membrane-associated protein named GARP (glycoprotein-A repetitions predominant), also known as LRRC32 (leucine-rich repeat containing 32) (19, 36–39), or the closely related LRRC33 protein (40) (Fig. 1B). The secretion and deposition of TGF-βs in large latent complexes necessitates activation mechanisms to locally release active TGF-β that can bind its receptors on target cells. When expressed in transfected cells, myostatin, also known as GDF8, and GDF11 are also secreted as latent complexes, in which the pro-polypepide association prevents mature ligand from binding and activating the receptors (19, 41). These findings suggest that the ability of the propeptide to confer latency may extend to additional TGF-β proteins. In contrast, activins and several BMPs are not secreted as latent complexes, even though they associate with their pro-segments. The structural basis for how the pro-peptide confers latency to TGF-β1, and the BMP-9 pro-polypeptide does not confer latency to BMP-9, has been addressed (42, 43).
LIGAND PRESENTATION TO THE RECEPTORS
While some TGF-β family proteins may freely diffuse, the complexes of mature ligands with their pro-segments are most often locally retained in the ECM. With only a few TGF-β family proteins studied, ECMs, basement membranes, and cartilage and bone matrices, act as reservoirs from which the active proteins can be released. The secreted complexes are targeted to the ECM through association of either the mature proteins or their pro-polypeptides with select ECM proteins (19, 44). Among the many ECM proteins, collagens bind and sequester TGF-βs, BMPs and activins, whereas fibulin glycoproteins selectively bind BMPs and TGF-β, and fibronectin and thrombospondin bind TGF-β (10, 19). Fibrillins interact with the pro-polypeptides of several BMPs, and are thought to retain secreted BMP complexes (10, 19, 45). These associations also control TGF-β deposition and activation, given that LTΒP1, for example, with its structural similarity to fibrillins, directs the latent TGF-β1 complexes to interact with fibrillins, and thus prevents or attenuates the activation of latent TGF-β (10). Mutations in the fibrillin-1 gene lead to increased TGF-β activation, and Marfan syndrome in humans (10, 46). Proteolytic degradation of ECM proteins, during physiological remodeling or in response to injury or cell invasion, is thought to release the ligands or their complexes, enabling their activation and ligand-induced cell responses. Thus, depending on the protein association and the secreted ligand complex, ECM proteins not only sequester TGF-β family proteins, but also facilitate or direct their activities (47).
For those ligands that are secreted as latent complexes, specialized mechanisms control the release of the active ligand in response to stimuli. The latent complexes of the three TGF-βs, myostatin/GDF8 and GDF11 require activation mechanisms to release the mature ligand; however, only the activation of the TGF-β1 complex has been well characterized (19). The diverse modes of latent TGF-β1 activation strongly suggest cell type- or tissue-selective mechanisms that may depend on the signaling context (19). In various contexts, interaction of the pro-segment with integrins—notably the integrins αvβ1, αvβ6, and αvβ8—enables stress-induced, integrin-mediated changes in conformation of the latent TGF-β complex that result in the release of active TGF-β1 (9, 13, 19, 42, 48). Various proteases also confer activation of latent TGF-β1 (19). Together, the many studies strongly suggest that physiological activation of latent TGF-β1 requires combined activities of integrins and proteases (19, 49). The ECM proteins thrombospondin and fibronectin, as well as fibrillin-1, also direct TGF-β activation, but their roles are less defined (19). Latent TGF-β2 and -β3 activation are expected to require distinct mechanisms and regulation, because of sequence divergence among the three pro-polypeptides (13, 19). No evidence exists for integrin-mediated activation of latent myostatin and GDF11 complexes. Rather, the release of myostatin and GDF11 from latency depends on cleavage of the pro-domain by BMP1 and mTolloid metalloproteases (50, 51). With increasing appreciation that differential splicing controls the nature and activities of the secreted TGF-β family proteins and cell responses, one may speculate that such regulation, for example that resulting in an additional 28 amino acids in the TGF-β2 pro-segment (52), might control the association of ligand complexes with ECM proteins and their activation.
The localized activities of TGF-β family proteins are further controlled and restricted by a repertoire of proteins that selectively bind active TGF-β family proteins to thus prevent their binding to the receptors (44). Most known inhibitors inactivate BMPs, whereas few inactivate activins or TGF-βs. Others, however, can act as agonists that potentiate ligand binding to the receptor and enhance ligand-induced cell signaling (44). Among the secreted BMP-binding inhibitors, noggin, chordin and Twisted Gastrulation (Tsg) have been extensively studied (44). Tsg can either promote or inhibit BMP activities, and associate with chordin and BMP to prevent BMP receptor activation (44, 53–56). The most studied activin inhibitor is follistatin, the binding of which prevents activin binding to the receptor (25, 44, 57). Other proteins form stable complexes with TGF-β, thus preventing TGF-β receptor activation. For example, decorin and biglycan, which are localized in the ECM, and α2 macroglobulin, which is abundant in plasma, have high affinity for TGF-β and enable its sequestration (44). The regulation of ligand inhibition by soluble or ECM-associated proteins is complex, involving a plethora of proteins with tissue-dependent regulation.
The exquisite regulation of ligand availability and receptor binding through protein associations, at the cell surface or in the ECM, which is here only touched upon, ensures that most TGF-β family proteins act locally in a highly controlled fashion, rather than by free diffusion. Inadvertent escape from the site of activation is likely to result in sequestration to prevent unwanted activities in unintended cells. Accordingly, tissue-specific activities of TGF-β family proteins and morphogen activity gradients of TGF-β family proteins must invoke scenarios beyond mere ligand diffusion that take into account the localized control. Besides diffusion-based models (58), substantial evidence has emerged that ligands and receptors are presented to each other over distances that span many cell diameters, through thin, filipodia-like membrane protrusions, named cytonemes (Fig. 1C). These have been seen in Drosophila and vertebrate tissues, as well as tumors. Cytoneme-mediated ligand presentation can explain the highly localized ligand-receptor activities, as well as gradients of morphogen signaling (59, 60). Considering the narrow diameter of the cytoneme and the large surface of the target cell, cytoneme-mediated receptor activation and signaling are expected to initiate in synapse-like structures at sites of contact that could function as highly localized signaling centers (60). As cytonemes are selective for ligands or receptors (59), they could confer differential signaling in distinct and different subcellular signaling centers. In contrast, signaling mechanisms by TGF-β family proteins or any soluble ligands are studied in cell culture, in which soluble ligand is made available to all cell surface receptors, not taking into account localized activation at membrane microdomains or selective signaling centers.
TGF-β FAMILY RECEPTORS
Mature, dimeric TGF-β family ligands bind and signal through cell surface receptor complexes that combine two “type II” and two “type I” receptors (61) (Fig. 2), initially designated as such based on the mobilities in gel of the 125I-TGF-β-crosslinked proteins (3, 62). Both receptor types are transmembrane kinases and share structural similarities; they have a glycosylated, disulfide-rich ectodomain of about 100 amino acids, a transmembrane region, a short juxtamembrane sequence and a cytoplasmic kinase with its 11 subdomains organized in an N- and a C-lobe (61, 63). The juxtamembrane sequences of type I receptors have a short Gly-Ser-rich sequence, named the GS domain, that is phosphorylated by the type II receptor kinase in response to ligand binding (61, 63, 64). The mammalian genome encodes five type II and seven type I receptors, and different ectodomain combinations enable selective or specific binding of TGF-β family ligands, and ligand-induced activation of signaling (61).
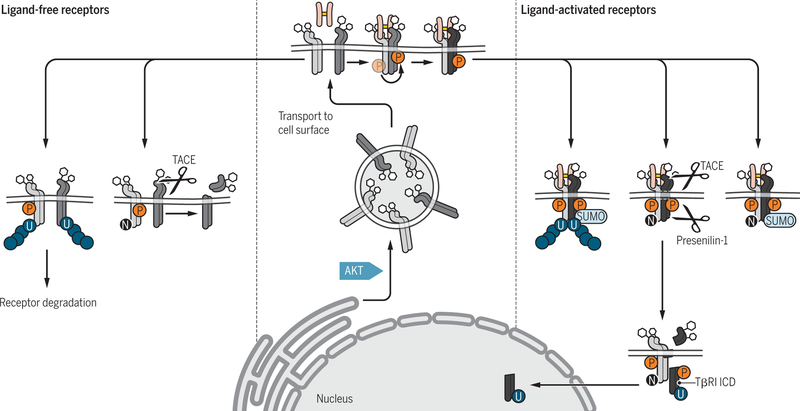
Figure 2. Posttranslational and functional modifications of the glycosylated TGF-β receptors TβRII and TβRI before or after ligand-induced activation.
AKT activation in response to insulin or other stimuli drives TGF-β receptor transport from intracellular compartments to the cell surface (center). Prior to activation (left, light blue), the plasma membrane-associated TβRI (blue) can undergo TACE-mediated ectodomain cleavage, thus preventing ligand-induced activation of signaling, or polyubiquitylation that leads to receptor degradation. Ligand-induced activation of the TβRI receptor (right, dark blue) results in phosphorylation (P) of its GS domain by TβRII (green), and phosphorylation of TβRII and TβRI lead to TβRII neddylation (N), TβRI sumoylation (SUMO), and TβRII and TβRI ubiquitylation (U). Activation of the TβRI receptor may also result in proteolytic release of its intracellular cytoplasmic domain (ICD) by presenilin-1 after TACE-mediated ectodomain cleavage, and then nuclear translocation of the ICD.
Credit: Veronica Falconieri Hays/Science Signaling
Based on sequence characteristics, the type I and type II receptors are predicted dual-specificity kinases, able to phosphorylate on serine and threonine as well as tyrosine, and are phylogenetically clustered, proximal to other dual-specificity kinases such as MEK kinases (65, 66). Because the type I receptors activate Smads through phosphorylation at two C-terminal serines, and Smads are seen as the major signaling effectors of TGF-β proteins, the TGF-β family receptors have often been referred to as Ser/Thr kinases (61). However, their ability to phosphorylate on Tyr is illustrated by the autophosphorylation on tyrosine of the type II TGF-β receptor, TβRII (67), and ShcA phosphorylation on serine and tyrosine by the type I TGF-β receptor, TβRI (also known as ALK-5), and tyrosine phosphorylation then activating mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK)-kinase (MEK)–ERK (also collectively referred to as ERK/MAPK) pathway in response to TGF-β (68). Similarly to other dual-specificity kinases (69), TGF-β family receptors have a lower ability to phosphorylate on tyrosine than on serine and threonine (61, 67, 68).
In the absence of ligand, the type I and type II receptors at the plasma membrane exist as monomers, homodimers, as well as heterodimers, strongly suggesting inherent, non-random affinities of both receptor types, and a need for protein associations that prevent signaling activation in the absence of ligand (70–72). Ligand binding promotes the formation and stabilization of the heteromeric receptor complexes, with the dimeric ligand interfacing with the four receptor ectodomains (61, 63, 70). TGF-β1 and TGF-β3 bind with high affinity to TβRII receptor dimers without the need for TβRI receptors, whereas TβRI receptors have only low affinity for TGF-β and require TβRII for ligand binding (13). This difference in ligand affinities suggests an ordered sequential assembly of the TGF-β dimer with the four receptors that is initiated by TGF-β binding to TβRII (70). In contrast, TGF-β2 has low affinity for both the TβRII or TβRI receptors in the absence of TβRI or TβRII, respectively, suggesting a random sequential assembly process with TGF-β2 initially binding to either the TβRI or TβRII receptors in preformed receptor complexes (70). Similarly to TGF-β1 and TGF-β3, activins bind with high affinity to their type II receptors, prior to binding to type I receptors in heteromeric receptor complexes (61, 70). In contrast, BMPs have generally higher affinities for their type I receptors than for the type II receptors, and BMP binding to preformed complexes ensures high affinity binding (61, 70).
The three TGF-βs bind and activate signaling through heteromeric complexes of the type II receptor TβRII with the type I receptor TβRI, but have also been shown to act through the type I receptors ALK-1 (also known as ActRL1 or ACVRL1) and ALK-2 (also known as ActRIA or ACVR1). Activins signal by binding to the type II receptors ActRII or ActRIIB in combination with the type I receptors ActRIB (also known as ALK-4) or ActRIC (also known as ALK-7). BMPs and GDFs act primarily through the type II receptors ActRII, ActRIIB, or BMPRII in partnership with the type I receptors ALK-2, BMPRIA, or BMPRIB (also known as ActRIA, ALK-3, and ALK-6, respectively) (61). The diversity of ligand binding to functional combinations of type II and type I receptors has been summarized elsewhere (61), and structural studies of ligand-ectodomain interactions have provided insights into the nature and specificity of ligand binding (13).
TGF-β family ligands bind to the heteromeric receptor complexes with very high affinity, mostly with KD values around 10−11, which contrasts with the lower affinities of growth factor ligands for RTKs (KD values around 10−9) (73, 74). Receptor abundance at the plasma membrane is much lower, often 10-fold lower, than that of RTKs, ranging around <1000–5000 per cell. The low abundance of cell surface receptors and very high affinity ligand binding allow the cells to exquisitely regulate their ligand sensitivity in response to cell-intrinsic or extracellular stimuli, as will be discussed. Such regulation complements the control of ligand availability by secreted proteins that often prevent ligand-induced receptor activation, as discussed above, and by membrane-associated proteins (discussed below).
TGF-β FAMILY CORECEPTORS
In addition to extensive control of TGF-β ligand presentation through association with secreted proteins, cells control ligand binding to the receptors and, thus, activation of signaling, by expressing transmembrane or membrane-anchored co-receptors that selectively aid in ligand binding to the receptors. Some co-receptors enhance ligand binding, while others help specify ligand binding, and their secreted ectodomains may sequester ligands (13, 61, 75) (Fig. 3). The best characterized TGF-β family coreceptors are the structurally related betaglycan, first described as TGF-β type III receptor, and endoglin (13, 61, 75–77). Betaglycan is a single-pass transmembrane proteoglycan with chondroitin and heparin sulfate side chains in its extracellular domain that binds all three TGF-βs, yet with highest affinity for TGF-β2 (78). This high affinity may be particularly relevant for TGF-β2, which binds TβRII and TβRI with lower affinities than TGF-β1 and TGF-β3. Betaglycan also binds TβRI and TβRII independently, and stabilizes the TβRI-TβRII association, which may help TGF-β2 binding to preformed TβRI-TβRII complexes. Consequently, cell surface betaglycan facilitates and enhances ligand-induced TGF-β signaling (13, 61, 75, 76). Conversely, however, betaglycan can be cleaved to release its ectodomain that then functions to sequester TGF-β, thus dampening TGF-β responses (78, 79). The expression and ectodomain shedding of betaglycan are major determinants of TGF-β responsiveness. Betaglycan also binds other TGF-β family proteins, notably BMP-2 and −4, GDF5 and inhibins, and therefore may play a broader role in promoting TGF-β family signaling (75, 76). Additionally, it binds basic fibroblast growth factor (bFGF or FGF2) through separate domains, enabling betaglycan to control FGF-induced RTK signaling (80), and raising the possibility for coordinated regulation of distinct pathways by betaglycan.
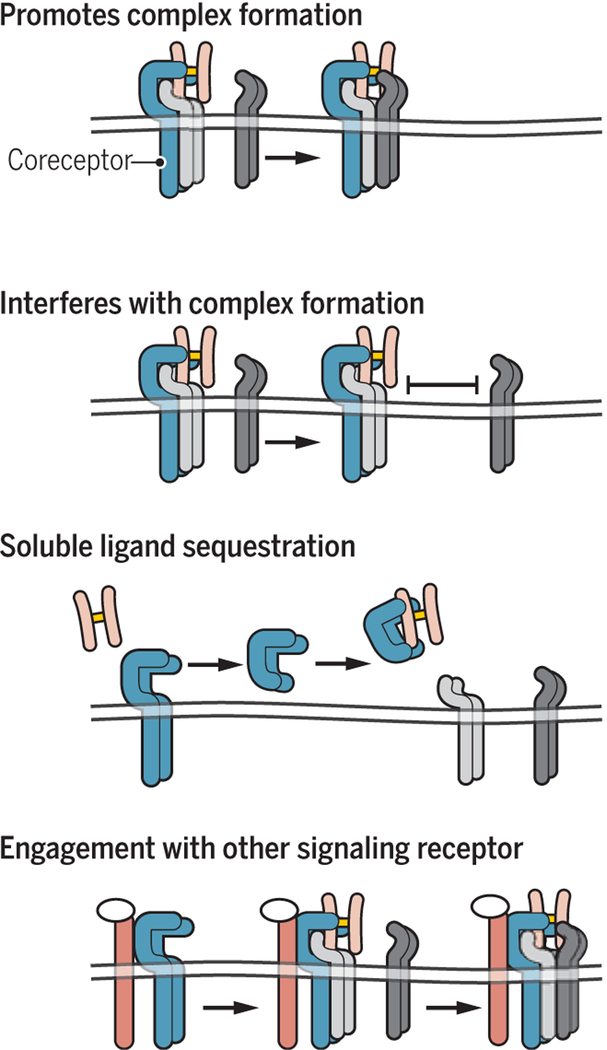
Figure 3. Roles of coreceptors in TGF-β family ligand binding to heteromeric complexes of type II (green) and type I receptors.
The membrane-anchored coreceptors (pink) are generally present as dimers at the cell surface and can promote ligand binding to the receptors (top left), as shown for betaglycan, which enhances binding of TGF-β2 to the TGF-β receptors, or for crypto which enables nodal binding to activin receptors, and nodal signaling. Alternatively, association of coreceptors with type II receptors (green) can interfere with the formation of complexes between type II and type I (blue) receptors (top right), as reported for the roles of RGMs as BMP coreceptors. Co-receptors, such as betaglycan and endoglin, can also be cleaved at the cell surface, resulting in the release of their ectodomains (bottom left). The released ectomains retain their affinity for ligand, resulting in ligand sequestration and repression of signaling activation. Co-receptors also provide opportunities to coordinate activation of distinct signaling pathways (bottom right). Thus, betaglycan can bind bFGF in addition to TGF-β and coordinately regulate FGF and TGF-β signaling.
Credit: Veronica Falconieri Hays/Science Signaling
Endoglin is also a single-pass transmembrane protein, but lacks glycosaminoglycan side chains, and is expressed at the cell surface as a disulfide-bonded dimer (61, 75, 77). Endoglin is expressed as two isoforms with different cytoplasmic domains and functions (81). Its preferential expression on endothelial cells suggests roles in vasculogenesis and angiogenesis (77, 82). Endoglin binds TGF-β1 and -β3, and promotes TGF-β signaling through ALK-1, a predominant BMP type I receptor, while inhibiting signaling through the type I receptor TβRI, also known as ALK-5 (61, 75). Endoglin also binds activin A, BMP-7 and BMP-2, but does so only by associating with the corresponding type II receptors (75). In contrast to these ligands, endoglin binds BMP-9 and BMP-10 with high affinity, suggesting a natural physiological role in the presentation of these ligands to their receptors and in the vascular roles of these ligands (77, 82, 83). Like betaglycan, the ectodomain of endoglin can be proteolytically released, and may thus serve as a soluble scavenger (84).
The membrane-associated, glycosylphosphatidylinositol (GPI)-linked “repulsive guidance molecules” (RGMs) act as BMP coreceptors. Three related RGMs, RGMa, RGMb (also known as DRAGON) and RGMc (also known as hemojuvelin), have been identified in mammalian cells. They interact with neogenin, a netrin receptor with neuronal function, yet also bind BMPs and promote BMP binding to the type II BMP receptors (13, 85–88). How RGMs promote BMP binding to receptor complexes and enhance BMP signaling remains to be clarified, although they are thought to control endocytosis of the ligand-receptor complexes in a clathrin-dependent manner, thus promoting BMP-induced Smad activation (13, 85, 86). How RGM-mediated clustering of neogenin with BMP receptors controls neogenin signaling and function also remains to be elucidated.
Some coreceptors help define the ligand specificity, as illustrated with the epidermal growth factor-Cripto-1/FRL-1/Cryptic (EGF-CFC) family protein Cripto, a GPI-anchored membrane protein. Cripto has affinity for some type I receptors, most notably ActRIB (ALK-4), which in combination with the type II receptor ActRII or ActRIIB, mediates activin signaling. Cripto enables Nodal binding to the activin receptor complex, thus activating Nodal signaling, whereas its absence promotes activin signaling and prevents Nodal signaling (75, 89, 90). Cripto also binds TGF-β and prevents its binding to the TβRI receptor (91). Cripto may have additional roles in integrating signaling pathways, including EGF receptor and Wnt signaling (92).
BAMBI (acronym for “BMP and activin membrane-bound inhibitor”), a transmembrane type I receptor with a truncated cytoplasmic domain without kinase activity, interacts with type I receptors, binds TGF-β, activins and BMPs, and thus dampens the responses to TGF-β ligands through interference with functional receptor complexes (75, 93). The induction of BAMBI expression in response to TGF-β suggests that it acts in a negative feedback loop of TGF-β signaling (75, 93), yet BAMBI also represses BMP signaling in development (93, 94).
Besides enhancing or attenuating, or helping to specify, ligand binding to their receptors, coreceptors also determine the functional availability of the receptors and receptor compartmentalization at the cell surface, and the intracellular routing and fate of the ligand-receptor complexes. Such functions are expected to be major determinants of the signaling response, but have only been minimally studied, and are therefore less appreciated (61, 75). Additional cell surface proteins that do not act as coreceptors may control the cell surface distribution of the TGF-β family receptors or their assembly in multiprotein complexes that act as signaling centers, possibly allowing crosstalk between TGF-β family receptors and other signaling receptors and mediators. For example, vascular endothelial (VE-) cadherin can interact with the TβRII and TβRI receptors as well as endoglin, and promotes TGF-β signaling in endothelial cells (95). The tight junction protein occludin associates with the TβRI receptor and promotes the localization of TGF-β receptor complexes at tight junctions (96). Also, the hyaluronan receptor CD44 associates with the TβRI receptor, and hyaluronan promotes TGF-β-induced Smad activation by the TβRI receptor, while TβRI-mediated phosphorylation of the short cytoplasmic tail of CD44 enables anchoring of CD44 complexes to the actin cytoskeleton, and promotes cell migration (97, 98). Finally, TGF-β and/or BMP receptors can associate with RTKs (98–100), again illustrating possible functional interactions in signaling centers that control TGF-β family receptor signaling and enable signaling crosstalk between different receptor types.
POSTTRANSLATIONAL CONTROL OF FUNCTIONAL RECEPTOR AVAILABILITY
In addition to secreted, ligand-interacting proteins and membrane-associated coreceptors that help define the ligand binding, post-translational mechanisms further control the functional availability of TGF-β family receptors at the cell surface (Fig. 2). TGF-β family receptors are modified by N-glycosylation at Asn, and possibly O-glycosylated at serine and/or threonine, in their ectodomains (61, 101). Whereas ectodomain glycosylation can regulate the transport to the cell surface, the stability, and/or ligand binding of other transmembrane receptors, little is known about its roles in the control of TGF-β family receptors (102, 103). N-glycosylation facilitates transport of the TβRII receptor to the cell surface (102), and impaired glycosylation decreases ligand binding to TβRII (104). Consequently, inhibition of N-glycosylation reduces TGF-β responsiveness (102). Additionally, ectodomain N-glycosylation enhances the binding of BMP-2 to the BMP-RII but not the ActRII or ActRIIB type II receptors (103). High glucose (meaning 25 mM the amount associated with hyperglycemia) promotes N- and O-glycosylation of proteins (105, 106), raising the possibility that it might also promote TGF-β or BMP receptor glycosylation and thus increase ligand binding.
Ectodomain shedding also controls the functional availability of the cell surface TGF-β receptors, and, consequently, TGF-β responsiveness. The transmembrane metalloprotease TACE, also known as ADAM17, cleaves TβRI, but not the TβRII receptor, in response to ERK or p38 MAPK signaling (107, 108). Consequently, mitogenic and inflammatory stimuli that activate either pathway dampen TGF-β signaling by attenuating the TβRI responses, without decreasing ligand binding to TβRII (107, 108). TACE-mediated ectodomain shedding of TβRI is followed, through the activity of γ-secretase, by intracellular release of the TβRI cytoplasmic domain, which translocates into the nucleus where it controls transcription of target genes (109, 110).
The TACE-mediated decrease in TGF-β responsiveness illustrates that the sensitivity to TGF-β family proteins depends on the abundance of functional receptors at the cell surface. Thus, processes that enhance the abundance of cell surface TGF-β receptors increase TGF-β responsiveness (107). A large pool of the TβRII and TβRI receptors resides inside cells, enabling rapid receptor transport to the cell surface to enhance the sensitivity to TGF-β and autocrine TGF-β signaling (111, 112). Glucose at 25 mM and insulin both induce a rapid transport of intracellular TβRII and TβRI receptors to the cell surface (111, 112). This induction results from rapid activation of AKT, which phosphorylates the intracellular, membrane-associated Rab guanosine triphosphatase (GTPase)-activating protein (RabGAP) AS160, thus relieving the intracellular retention and promoting cell surface transport of both TβRII and TβRI (112), similarly to AKT-mediated cell surface transport of the glucose transporter Glut4 (113–115). This mechanism positions AKT activation as a central regulator of TGF-β responsiveness, and enables other AKT-activating ligands, such as growth factors that act through RTKs, to enhance TGF-β responsiveness. This mechanism may be highly relevant in diabetes-associated hyperglycemia, which is treated with insulin, a major inducer of AKT activation, and in cancers, which often show increased Akt activation. Whether similar mechanisms control the responsiveness of other TGF-β family receptors at the cell surface remains to be seen. Because also TGF-β induces AKT activation (see further below), TGF-β itself induces, through a positive feedback loop, an increase in cell surface abundance of TGF-β receptors, thus enabling signal amplification.
The functional availability of TGF-β family receptors is also extensively regulated by poly-ubiquitylation and, consequently, degradation, involving several ubiquitylating enzymes and multiple de-ubiquitylases (DUBs) (61, 116, 117). Proteins are ubiquitylated on lysine through the sequential activities of ubiquitin-activating E1, ubiquitin-conjugating E2, and a plethora of E3 ubiquitin ligases that provide target selectivity, including HECT-type E3 ligases (116, 118). Among these, Smurf1 and Smurf2, identified for their ability to ubiquitylate Smads, target the TβRI receptors for poly-ubiquitylation. They are recruited by Smad7 to the TβRI receptors, enabling them to ubiquitylate TβRI and promote receptor degradation (119, 120). Other HECT-like E3 ligases, such as NEDD4L, also known as NEDD4–2, and WWP1, also target TβRI for ubiquitylation and degradation (121, 122). The differential contributions of the different E3 ligases, and their roles in the degradation of other TGF-β family receptors remain to be clarified.
Substrate ubiquitylation is balanced by ubiquitin removal by DUBs. Among the DUBs that have been implicated in the control of receptor availability, USP4, USP11, USP15 and UCH37 deubiquitylate TβRI, directly or through the association either of Smad7 with TβRI or of tumor necrosis factor (TNF) receptor-associated factor 4 (TRAF4) with the TβRII-TβRI receptor complex (116, 117). DUB associations prevent receptor degradation, thus sustaining TGF-β receptor abundance and responsiveness (116, 117). Phosphorylation of USP4 by AKT enhances the association of USP4 with cell surface TβRI, and USP4-mediated TβRI de-ubiquitylation (123). As with the E3 ubiquitin ligases, further studies need to define the context-dependent roles, regulation and contributions of the different DUBs to the functional availability of TGF-β family receptors. Finally, neddylation, which results in the covalent attachment of a single ubiquitin-like NEDD8 polypeptide, controls the stability of the TβRII but not the TβRI receptor. TβRII neddylation is mediated by the NEDD8 E2 conjugating enzyme Ubc12 and the E3 ligase c-Cbl, and stabilizes the receptor, presumably by opposing ubiquitylation and degradation while promoting TβRII internalization, and thus enhancing TGF-β signaling (124). Neddylation is reversed by isopeptidases that remove NEDD8 from its substrate; thus, the de-neddylase NEDP1 has been shown to attenuate TβRII neddylation (124).
LIGAND-INDUCED RECEPTOR ACTIVATION
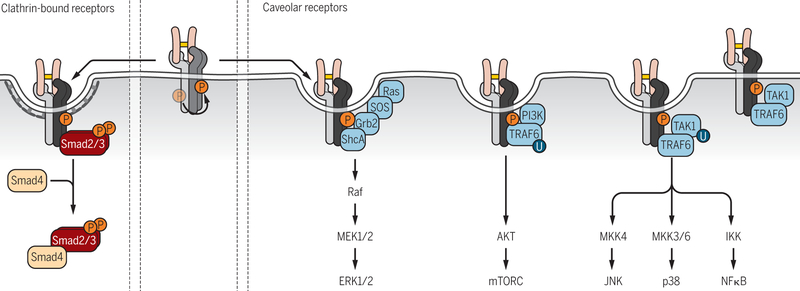
Signaling initiates when ligand binds the complex of two type II and two type I receptors at the cell surface. In polarized epithelial cells, the cell surface receptors localize basolaterally, leaving the apical surface insensitive to TGF-β stimulation (125, 126). In non-polarized cells or cells with front-rear polarity, the cell surface receptors are more equally distributed, with high levels at sites of cell contacts and at the leading edge of migrating cells, or at the tips of filopodia and cytonemes (60, 61). The type I and type II TGF-β receptors are continuously internalized, both in the absence and in response to TGF-β (127), and it is often assumed that TGF-β proteins do not induce receptor internalization, in contrast to growth factor-induced RTK internalization. Observations, however, argue for TGF-β-induced receptor internalization that results in decreased cell surface abundance and might lead to increased receptor degradation (128–131). Studies of receptor routing may be affected by the basal, autocrine TGF-β signaling, and the distribution of cell surface receptors in clathrin-associated nascent endosomes versus, predominantly, caveolar lipid raft compartments (128–130, 132), and perhaps more importantly from insufficient antibody quality to track endogenous receptor routing.
Ligand contacts with all four ectodomains induce conformational changes at the ligand-receptor interface, and stabilize the receptor association, thus bringing their cytoplasmic domains in close proximity (13, 63). This stabilization enables the type II receptors to phosphorylate the type I receptors in their juxtamembrane GS domain at serines and a threonine, which then induces conformational changes that release the 12 kd immunophilin FKBP12 from the GS domain (133, 134). This dissociation relieves the inhibitory interaction of the kinase domain with the GS domain, and activates the kinase of the type I receptors (63, 134). FKBP12 association with the type I receptors prevents inadvertent signaling activation in the absence of ligand (133, 135).
Also the inhibitory Smad6 and Smad7 help silence the type I receptors in the absence of ligand (136); this occurs through association with the GS domains and likely other sequences of the cell surface type I receptors (63, 136). The ligand-induced conformational changes in the receptor complex (63) enable the arginine methyltransferase PRMT1, which is associated with the type II receptors, to methylate Smad6 or Smad7, resulting in their dissociation from the type I receptors (137, 138). Thus, PRMT1-mediated methylation of inhibitory Smads and their dissociation from the type I receptors precede and enable the recruitment of the effector Smads for subsequent activation through C-terminal phosphorylation (63, 136, 137). It is not known how Smad6 and/or Smad7 methylation temporally and functionally relates to type I receptor phosphorylation and activation, nor whether this process controls non-Smad signaling.
Ligand binding also induces phosphorylation at other receptor sites, besides the GS-domain of the type I receptors. Their roles have remained poorly characterized, but allow additional regulation and functions (61, 63). TGF-β family proteins induce type II receptor autophosphorylation (61, 67), similarly to growth factor-induced RTKs, and enable the type I receptors kinase to phosphorylate the type II receptors (61); yet these processes remain to be dissected. Autophosphorylation at Ser213 and Ser249 enhances the kinase activity of TβRII, whereas autophosphorylation at Ser416 attenuates it (139). Phosphorylation on Tyr, either through autophosphorylation (67) or by Src (140), enhances the TβRII activity (67). Ligand binding also induces TβRI phosphorylation on tyrosine, likely through autophosphorylation (68), and on serine and threonine outside the GS domain. Tyrosine phosphorylation of the receptors may generate docking sites for additional signaling mediators. Thus, TGF-β induces recruitment of ShcA, through its phosphotyrosine-binding PTΒ and SH2 domains, to the tyrosine-phosphorylated TβRI or TβRII, thereby initiating ERK/MAPK signaling (68, 140). TGF-β-induced TβRI phosphorylation also promotes Lys389 sumoylation of TβRI, which facilitates the recruitment and activation of effector Smads, and Smad-mediated signaling (141).
Consistent with the reversible nature of phosphorylation, several phosphatases target TGF-β family receptors. GADD34, a regulatory subunit of the protein phosphatase 1 (PP1) complex can associate with Smad7, and thus recruit the PP1 catalytic subunit to TβRI for dephosphorylation (142). Also the PP2A phosphatase complex can associate with type I receptors and modulate their signaling, although its effect may depend on the identity of the receptor-interacting B subunit of PP2A (143, 144). Additionally, the tyrosine phosphatase TCPTP was found to tyrosine-dephosphorylate TβRII after its recruitment by α1β1 integrin to TβRII (145). Finally, in Xenopus oocytes, the association of the protein phosphatase Dullard with BMP receptor complexes results in BMPRIA dephosphorylation, poly-ubiquitylation and degradation (146). The prevalence of phosphorylation and dephosphorylation at an array of cytoplasmic domain sites in the tetrameric receptor complexes stands in contrast with how little we know about the regulation of receptor activities and functions by (de)phosphorylation.
Various transmembrane proteins and receptors are functionally regulated by acetylation on Lys (147, 148), or methylation on Lys or Arg (149); however, these modes of post-translational regulation are less studied than the control of protein function by phosphorylation. It is not known whether TGF-β family receptors are regulated by these modifications.
SMADS AS SIGNALING EFFECTORS
Upon ligand-induced receptor activation, Smads (Fig. 4) transmit signals from the receptors into the nucleus to repress or activate target gene expression. While TGF-β family proteins also induce non-Smad signaling, the Smads are seen as the major TGF-β family transducers; they direct changes in gene expression and uniquely act as signaling effectors to the TGF-β family proteins. Thus, upon ligand binding, the type I receptors that are activated by type II receptors in turn activate effector Smads through phosphorylation of their C-terminal two serines. These “receptor-activated Smads” (R-Smads) then dissociate from the receptors, and combine with Smad4 to form complexes that translocate into the nucleus, where they cooperate with high affinity DNA-binding transcription factors and coregulators to activate or repress target genes (150–152) (Fig. 5, A and B). This model resonates because it seems simple and linear; however, it allows for substantial versatility, largely because of its dependence on functional association with other transcription factors, many of which act as effectors of other signaling pathways (discussed below), as well as through its roles in target repression/activation outside of a strictly transcriptional manner (Fig. 5, B-F; also discussed in more detail below).
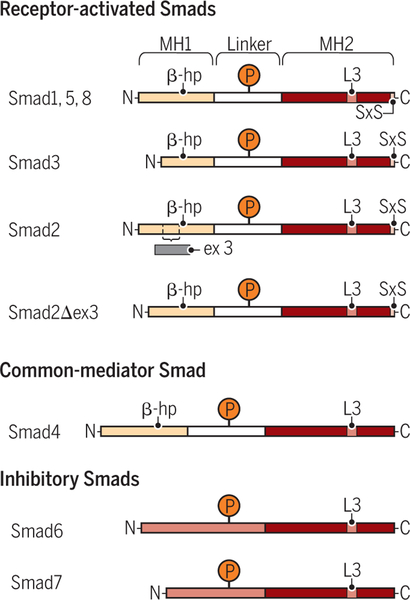
Figure 4. Schematic comparison of the simplified structures of R-Smads (Smad1, Smad2, Smad3, Smad5 and Smad8), Smad4, and inhibitory Smads (Smad6 and Smad7).
The R-Smads and Smad4 have two conserved domains, the MH1 (brown-grey) and MH2 (dark orange) domains, separated by a variable serine- and proline-rich linker region (light grey). The linker region is targeted for phosphorylation (P) by various signaling kinases that thus control the stabilities and functions of the Smads. A β-hairpin (β-hp; arrow head), which in Smad2 is interrupted by a sequence encoded by exon 3 but is maintained in the Smad2Δ3 variant, enables MH1 domain binding to DNA. The inhibitory Smads lack an MH1 domain, and have a long and variable sequence (light orange) preceding the MH2 domain. This sequence is thought to be structurally versatile depending on post-translational modifications and protein interactions. The positively charged L3 loop in the MH2 domain mediates association with the activated type I receptors and with other Smads. The R-Smads, but not Smad4 and the inhibitory Smads, have a conserved C-terminal SXS motif that is phosphorylated by the activated type I receptor, resulting in R-Smad activation.
Credit: Veronica Falconieri Hays/Science Signaling
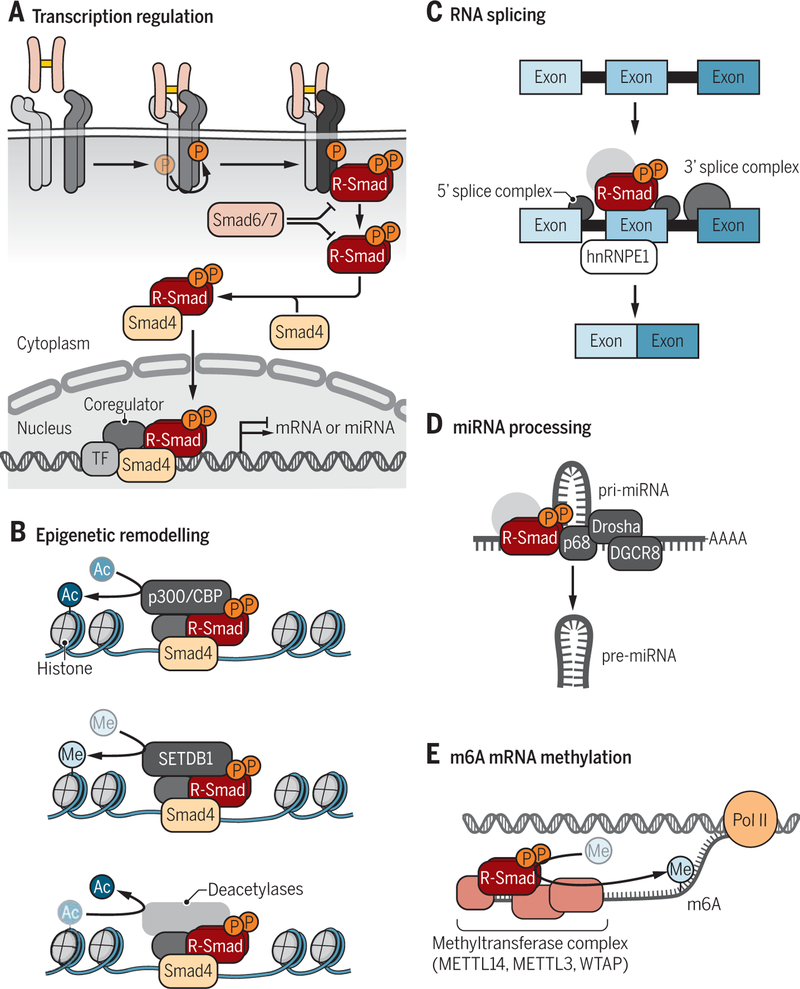
Figure 5. Smad-dependent regulation of gene expression.
(A) Simplified model of TGF-β-induced R-Smad activation leading to Smad-mediated activation of gene expression. Signaling is initiated by TGF-β binding to a heteromeric complex of type II (green) and type I (blue) receptors, resulting in activation of the type I receptors (dark blue) and C-terminal R-Smad phosphorylation. The activated R-Smads dissociate from the type I receptors, form a complex with Smad4 and the R-Smad/Smad4 complexes translocate into the nucleus, where they regulate gene expression with transcription factors (TF) and coregulators. Inhibitory Smads (Smad6 and Smad7) interfere with functional Smad activation, by associating with type I receptors, thus preventing R-Smad activation, or by interfering with the complex formation of R-Smads with Smad4. (B) Activated R-Smad/Smad4 complexes associate and cooperate with high affinity DNA-binding transcription factors to activate or repress the transcription of genes into mRNA or microRNA precursors. (C) Activated R-Smad/Smad4 complexes recruit histone modifying enzymes, resulting in chromatin remodeling. Recruitment of the p300 acetyltransferase, which commonly acts as transcription coactivator for Smad complexes, confers H3K9 acetylation, whereas recruitment of the SETDB1methyltransferase induces H3K9 methylation and thus represses transcription. Smad-mediated recruitment of histone deacetylases leads to histone deacetylation (not depicted). (D) TGF-β-activated Smad complexes regulate mRNA splicing in association with hnRNPE1. (E) Activated R-Smads direct miRNA processing through association with the p68 RNA helicase in complex with Drosha RNAse. (F) Activated Smad2 or Smad3 can associate with m6A methyltransferase complexes to promote methylation of mRNA.
Credit: Veronica Falconieri Hays/Science Signaling
Structure and functional activation of Smads.
The mammalian genome encodes eight Smads, including the inhibitory Smads, Smad6 and Smad7, which, as mentioned above, control receptor stability. Smad2 and Smad3 act as R-Smads for activin and TGF-β signaling, whereas Smad1, Smad5 and Smad8 mediate responses to BMPs and GDFs (151, 152). Which Smads are activated by the less studied TGF-β family ligands has not been fully resolved, nor have the functions of Smad8 been well defined. Smad4, the common partner for the activated R-Smads in trimeric Smad complexes, does not require activation by type I receptors (151, 152). The R-Smads and Smad4 have a conserved N-terminal MH1 and C-terminal MH2 domain (151) (Fig. 4). Their MH1 domains have nuclear localization signals and, with the exception of the most commonly expressed form of Smad2, a β hairpin structure that enables Smad binding to DNA (63, 151–153). The L3 loop in the MH2 domains mediates the R-Smad association with the type I receptors, and thus helps specify the ligand-induced R-Smad activation, yet also mediates Smad-Smad interactions in trimeric Smad complexes (63). The MH2 domains in all R-Smads, but not Smad4 or the inhibitory Smad6 and Smad7, are followed by a short sequence with two C-terminal serines that are phosphorylated by type I receptors (151, 154) (Fig. 4). A variable linker region between the MH1 and MH2 domains has phosphorylation sites for several kinases, including some that function in other signaling pathways (Fig. 4). When phosphorylated, these sites serve as docking sites for other proteins, and define the stability, nuclear localization and other properties of these Smads (154). Thus, linker regions of the R-Smads serve as hubs for functional crosstalk with other kinase-driven signaling pathways, complementing the crosstalk at regulatory sequences of target genes (Fig. 6). Consequently, each R-Smad has distinct properties and is individually regulated. Adding to this complexity, several Smads are expressed as differently spliced isoforms (155). For example, R-Smads with deletions in their linker domains (155–157) and a variant of Smad8 lacking the C-terminal two serines required for its activation (157) have been reported. The commonly studied Smad2 lacks DNA binding capacity, due to a sequence insert that disrupts the β sheet in the MH1 domain, whereas the Smad2β isoform lacks this insert and thus binds DNA similarly to Smad3 (63, 158, 159) (Fig. 4).
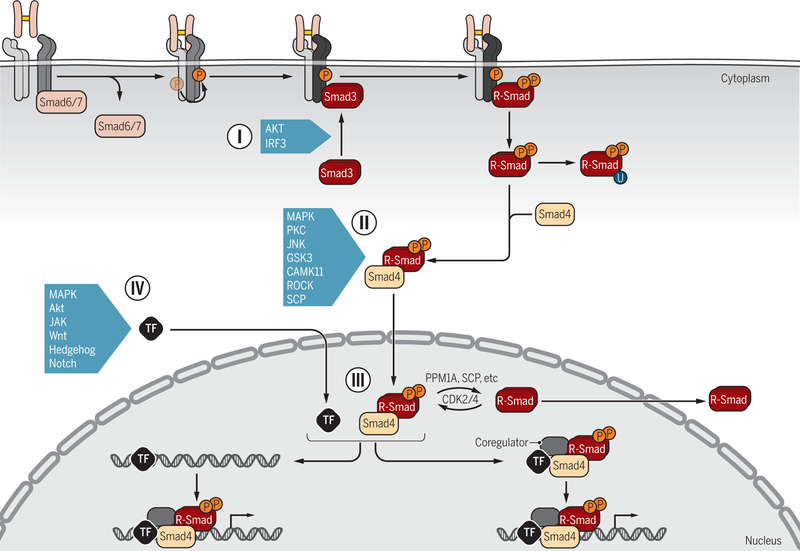
Figure 6. Signaling crosstalk through posttranslational control of Smad activation and functions.
R-Smad association with the receptors (I) is controlled by inhibitory Smad6 and/or Smad7, which prevent R-Smad access to the activated type I receptors (dark blue). Additionally, upon activation in response to various signaling pathways, AKT and IRF3 bind to Smad3 and thus attenuate Smad3 binding to activated type I receptors. Various signaling pathways that act through kinases target the linker regions of R-Smads for phosphorylation (II), with the possibility for further regulation by subsequent dephosphorylation, and thus control the subcellular localization, stability and function of Smads. Smads can also be poly-ubiquitylated, leading to degradation, and in some cases targeted linker phosphorylation is a prerequisite for subsequent poly-ubiquitylation and degradation. Some kinases and a phosphatase are listed as examples. In the nucleus (III), phosphorylation and dephosphorylation by kinases and phosphatases further regulate the Smad activities. Direct transcriptional activation or repression of target genes requires association of Smad complexes with DNA-binding transcription factors (TFs) and coregulators (IV). Smads have been shown to associate with a wide variety of TFs, depending on the signaling status of the cells and the targeted gene. Extensive signaling crosstalk occurs at the level of Smad-complex association with DNA-binding TFs, because they are also regulated by phosphorylation or other modifications in response to signaling pathways. Some examples are listed. Such crosstalk may occur before binding of Smad-TF complexes to regulatory gene sequences or after formation of the DNA-binding nucleoprotein complexes.
Credit: Veronica Falconieri Hays/Science Signaling
Ligand-induced activation of R-Smads results from their recruitment to activated type I receptors, dependent on prior dissociation of FKBP12 from the type I receptor GS domains (133–135), and Arg methylation and dissociation of the inhibitory Smad6 and/or Smad7 from the receptors (137, 138). Thus, ligand-induced phosphorylation of the type I receptor’s GS domain by the type II receptor kinase switches its affinity from FKBP12 binding to R-Smad recruitment (134). The association of R-Smads with the receptor complexes then enables the type I receptor kinases to phosphorylate R-Smads at two C-terminal serines, thus promoting the dissociation of the two activated R-Smads from the receptors and their association with Smad4 in trimeric complexes (63, 151, 160, 161). R-Smad recruitment and activation occur in association with clathrin-mediated endocytosis (131). Dab2, which associates with the TGF-β type I receptor, promotes clathrin-mediated endocytosis of the receptor complexes; thus, increased Dab2 expression enhances TGF-β-induced Smad activation (162, 163). Conversely, antiviral RLR signaling results in activation of IRF-3, which then associates with Smad3, thus preventing its recruitment to TβRI and attenuating TGF-β-induced Smad3 activation and gene expression. This signaling crosstalk depends on structural similarities between interferon-regulatory factor 3 (IRF3) and Smads that enable their association (164).
The selectivity in Smad activation is dictated by the type I receptors to which the R-Smads are recruited. The major type I receptors for activin and TGF-β, ActRIB and TβRI, activate Smad2 and Smad3, while the major BMP type I receptors, ALK-1, ALK-2 (also known as ActRIA), BMPRIA and BMPRIB, activate Smad1 and Smad5 (61, 63, 151). The R-Smad association involves the GS domain and L45 loop between kinase subdomains 4 and 5 of the type I receptors, and the MH2 domains of the R-Smads (63, 165–168). Phosphorylation of the GS domain by the type II receptors enables this negatively charged surface of the type I receptor to interface with a basic patch in the MH2 domain, whereas a cluster of four amino acids in the receptor’s L45 loop provides receptor-Smad specificity by interfacing with complementary amino acids in the L3 loop of the R-Smad MH2 domain (63). The membrane-anchored cytoplasmic SARA or structurally and functionally related Hgs/Hrs proteins facilitate and stabilize the association of activated Smad2 and Smad3 with type I receptors, and thus promote TGF-β-induced Smad2 and Smad3 activation (63, 131, 132, 151, 169–171). Endofin may have a similar function in the activation of BMP-specific R-Smads (172, 173). Conformational changes upon C-terminal phosphorylation of the receptor-associated R-Smads enable the dissociation of the activated R-Smads from the receptors and SARA, and promote R-Smad association with Smad4 in trimeric Smad complexes (63, 161, 174).
The trimeric assembly of Smad complexes is mediated by the MH2 domains and phosphorylated C-termini of R-Smads (161, 174). Heterotrimeric complexes of two R-Smads and one Smad4 are thought to be the most common and predominant functional complexes. Association of Smad4 with two activated R-Smads creates three distinct binding interfaces that are likely to define protein interactions in transcription complexes (63, 161, 174). Smad complexes are often assumed to have two identical R-Smads; however, the inclusion of two different R-Smads provides further functional versatility. Thus, TGF-β-induced heterotrimers of Smad2, Smad3 and Smad4 are abundantly formed, but differ functionally from complexes that contain pairs of Smad2 or Smad3, and target distinct genes (175). Also trimers that consist of only activated R-Smads have been observed (160, 175), and complexes of two Smad3s and one Smad2 are prevalent in TGF-β-treated hepatoma cells, as are complexes of one R-Smad and two Smad4 molecules (175). BMPs and GDFs may similarly induce the formation of trimeric complexes of Smad1 and/or Smad5 with Smad4. Additional diversity is achieved when the receptor complexes comprise two distinct type I receptors, e.g. TβRI (also known as ALK-5) and ALK-1, resulting in coordinated activation of Smad2 and Smad3 as well as Smad1 and Smad5 in response to TGF-β. Mixed Smad complexes that combine R-Smads activated by distinct type I receptors target distinct genes that are not controlled by complexes of two identical R-Smads with Smad4 (176, 177).
R-Smads and Smad4 shuttle between the cytoplasm and nucleus, but the activated Smad complexes are mostly in the nucleus (151, 178). Nuclear localization sequences are found in the MH1 domains of several Smads, and Smad interactions with distinct importins or nucleoporins suggest differential regulation of nuclear import depending on the Smad, and involving C-terminal R-Smad phosphorylation (151, 178–181). Also nuclear export sequences have been identified in some Smads, and nuclear export of the individual Smads may be differentially regulated by different exportins and associated Ran GTPases (182–185). Additionally, the cytoplasmic retention of the Smads may be regulated, possibly involving microtubules and associated motor proteins (186, 187). Signaling crosstalk, including signals that confer changes in Smad linker phosphorylation, further defines the cytoplasmic and nuclear retention and subcellular Smad movement (151, 154, 178). A better appreciation of the extensive regulation of the subcellular Smad distribution and ligand-induced nuclear Smad accumulation is required to understand the dynamics and termination of Smad-mediated transcription responses.
Smads control transcription and histone modifications and enable signaling crosstalk at target genes.
Direct transcriptional activation or repression of target genes requires association of Smad complexes with transcription factors that bind specific DNA sequences in proximal promoter regions or enhancers with high affinity (Fig. 5, A and B). Whether Smads activate or repress transcription depends on the associated transcription factors and coregulators, the target gene, the physiological state of the cell and trimeric composition of the Smad complexes. The extensive diversity of transcription factors with which Smads associate explains the large number of target genes, and inability to reliably predict a consensus sequence for activin-, TGF-β- or BMP-responsive genes (150, 152, 188, 189).
With the exception of the most prevalent Smad2 isoform, all R-Smads and Smad4 can bind DNA directly through an 11 amino acid β-hairpin in their MH1 domains that positions itself in the major groove of the DNA helix (63, 152, 153). Optimal DNA sequences have been proposed for Smad binding, and are named Smad binding elements (SBEs). For Smad3 and Smad4, this sequence is AGAC or its reverse complement GTCT (152, 190–192), while Smad1 and Smad5 prefer GGCGCC (152, 193). These sequences, which have been experimentally validated, are therefore often thought of as predictive of Smad binding. Additional ChIP analyses, however, revealed preferential binding of all R-Smads and Smad4 to GC-rich sequences that approximate a consensus sequence GGCGC or GGCCG, not only making Smad binding sites less stringent, but also removing the divergence between Smad¾ and Smad1/5/8 binding sequences (194). As Smad complexes act through cooperation with transcription factors that bind specific DNA sequences, their binding to proximal promoter sequences is primarily dictated by the associated transcription factor. Without this association, Smads bind DNA only weakly, meaning with a Kd of around 1 × 10−7 M (153), whereas sequence-specific transcription factors often bind DNA with Kds of 10−9 to 10−11 M. Hence, tandem or palindromic repeats of SBEs are required to enable Smad binding with high affinity or to direct Smad-mediated transcription in reporter assays. The juxtaposition of Smad complexes with associated DNA sequence-specific transcription factors raises two scenarios for ligand-induced recruitment of Smad complexes to regulatory gene sequences, meaning either the high-affinity DNA-binding transcription factor associates with proximal promoter sequences prior to Smad recruitment, or a Smad complex is coordinately recruited with the high-affinity DNA binding transcription factor (152). Either way, the juxtaposition of a Smad binding site and a high affinity DNA sequence for the associated transcription factor is expected to cooperatively increase the DNA binding affinities of both components. This cooperation not only facilitates Smad recruitment, but also enhances the DNA binding of the Smad-associated transcription factor in response to TGF-β (195, 196).
The number and diversity of DNA-binding transcription factors with which Smads both physically and functionally associate are staggering, and a structural basis for association with Smad complexes is not apparent. Smads often associate with their partner through MH2 domains, yet MH1 domains provide the interface with many other partners, or both MH1 and MH2 domains are required (152, 188). This extensive versatility of Smads in partnering with DNA binding transcription factors can only be explained through structural versatility at protein interfaces, which remains to be explored.
Among the Smad-associated transcription factors are many that are regulated in their DNA binding and/or transactivation function by signaling pathways, and thus serve as signaling effectors (197), e.g. in response to RTK activation or inflammation pathways. For example, Smad3-Smad4 complexes associate and cooperate with c-Jun in the AP1 transcription complex (198) or with ATF3 (199), which are activated in response to growth factors, cytokines and stresses through other pathways, including MEK-MAPK signaling. Additionally, Smads cooperate with p53, when phosphorylated in response to RTK activation (200). Other Smad-associated transcription factors are effectors of signaling pathways that control cell differentiation (197). Thus, Smads can interact with the transcription factors TCF or LEF, which are activated by Wnt signaling, enabling extensive crosstalk between Smad and Wnt signaling at promoter sequences (201). Similar scenarios promote the crosstalk of Smad signaling with Notch and Hedgehog signaling in the control of target gene expression (197, 202–204). Consequently, the cooperation of Smad complexes with DNA-binding specific transcription factors imposes a dependence of the Smad activity on signaling crosstalk with other pathways that control the functions of these transcription factors and coregulators (Fig. 6, IV). Conversely, activated Smads also control cell responses downstream of signaling pathways that activate or control the functions of transcription factors that associate with Smad complexes.
The association and cooperation of Smads with DNA binding transcription factors also explain the regulation of cell differentiation and tissue patterning by TGF-β family proteins during development. Thus, TGF-β family signaling enables cooperation of Smads with pluripotency-associated or lineage-specific, DNA-binding transcription factors, promoting, repressing or facilitating differentiation initiation or progression along many if not all lineages. Besides the interactions of Smads with effectors of Wnt, Hedgehog and Notch signaling, activated Smads also cooperate with Sox, GATA, Runx and myogenic bHLH transcription factors in diverse lineages (49, 205, 206).
The combination of Smad4 with two R-Smads in Smad complexes enables Smad4 to function as transcription coactivator that stabilizes the association of the activated R-Smads with the histone acetyltransferases CBP or p300, and thus enhances R-Smad-mediated transactivation (207). Consistent with its role in gene responses to all TGF-β family proteins, Smad4 inactivation is often thought to abolish Smad-mediated gene expression changes. This is not the case. Smad4 is required for or strongly enhances R-Smad-mediated transcription activation of many genes, consistent with its role as coactivator, yet many target genes are still activated by R-Smads in the absence of Smad4, albeit often to a much lower extent than in the presence of Smad4 (208). Hence, the requirement for Smad4 depends on the target gene, and presumably the cell type and signaling physiology, with its role ranging from obligatory to unnecessary. R-Smad-mediated transcription without Smad4 contribution is consistent with the TGF-β-induced formation of R-Smad trimers, such as Smad2:Smad3:Smad3 complexes (175).
Interactions of Smad complexes with corepressors and coactivators define the level of transcription activation and functions of Smads as transcription activators or repressors (Fig. 5A-C). Many of these cofactors modify the histone acetylation and/or methylation, directly or indirectly, enabling Smads to direct epigenetic changes (Fig. 5C). The association of the C-terminally phosphorylated R-Smad sequence with the histone acetyl transferase p300 (152), and stabilization of this association by Smad4 (152, 207), position p300 and Smad4 as coactivators of R-Smads. These interactions allow Smads to direct p300 to acetylate histone 3 (209, 210). The enzymatic activity of p300 is required for Smad2-mediated transcription activation, suggesting that chromatin remodeling is required for transcription activation by Smads (210). The histone acetyltransferases GCN5 and P/CAF have also been shown to associate with TGF-β-activated Smads and potentiate Smad signaling (211, 212). TGF-β-activated Smad complexes can also recruit histone methyltransferases, rather than histone acetyltransferases. Recruitment of Suv39h1 (213) or SETDB1 (214) confer TGF-β-induced H3K9 methylation and transcription repression at promoter sequences of the Il2 and Snai1 genes, respectively (187,188), while Smad3-mediated recruitment of the H3K4 methyltransferase SET9 targets the ACTA2 gene (215). Differential recruitment of histone acetyltransferases versus methyltransferases can provide a balance in Smad-mediated transcription activation versus repression (214) (Fig. 5C). Smad complexes can also recruit histone deacetylases, either directly, as in the case of the HDAC4 or HDAC5 recruitment by Smad3 to the osteocalcin promoter (209), or through recruitment of a corepressor such as TGIF, which in turn recruits histone deacetylases (216), thus possibly inhibiting the recruitment of the p300 acetyltransfereases. HDAC4 or HDAC5 recruitment mediates Smad-mediated transcription repression (209), whereas TGIF recruitment dampens transcription activation (216). Ski and the Ski-related SnoN, renamed SKIL, strongly repress the activated Smad complexes at target promoters (152, 217). They attenuate Smad-activated transcription through association with the Smad complexes and corepressors such as NCOR or SIN3A, thus preventing p300 recruitment (217). Also Evi-1, which cooperates with the corepressor CtBP and ZNF451, and thus blocks recruitment of p300 to Smad complexes, acts as corepressor to repress Smad-mediated transcription activation (218–220). The E3 ubiquitin ligase TIF1γ (also known as TRIM33 or ectodermin) also interacts with Smad complexes, but its proposed roles in Smad signaling seem conflicting (152, 221, 222). While TIF1γ and Smad4 association with the R-Smads were seen as mutually exclusive (221), TIF1γ recruitment to Smad complexes following its association with acetylated (and possibly methylated) histone 3 tails results in Smad4 mono-ubiquitylation (222). The TIF1γ association may allow for further chromatin remodeling that facilitates binding of Smad complexes to enhancers of target genes (221, 222). Finally, Smad complexes can repress transcription by interfering with nucleoprotein complex formation at gene promoters (223, 224).
Smads direct RNA processing
Many genes are transcribed into several mRNAs due to differential RNA splicing of the primary transcript, and thus encode protein isoforms with alternate functions. This regulation may depend on the cell type, and may change as cells differentiate from one cell type to another, e.g. during epithelial-mesenchymal transition (EMT) (225, 226). TGF-β induces changes in RNA splicing through Smad-mediated repression of genes encoding splicing regulators, such as the epithelial splicing regulatory factors ESRP1 and ESRP2 (227). TGF-β signaling also directly controls RNA splicing through association of Smad3 with the primary transcript (Fig. 5D). Indeed, in response to TGF-β, Smad3 was shown to associate with the RNA-binding protein PCBP1, also known as hnRNPE1, and thus directs changes in splicing of the CD44 pre-mRNA, yielding functionally distinct CD44 isoforms (228).
Small noncoding miRNAs control gene expression through association with partially complementary sequences that are often located in 3’UTRs of mRNAs, leading to mRNA degradation or direct inhibition of translation. miRNA genes are transcribed by RNA polymerase II into pri-miRNAs that are then processed through cleavage into pre-miRNAs and give rise to mature miRNAs (229). Smads directly activate or repress miRNA gene expression through association with DNA sequence-specific transcription factors, in a similar way as in the control of protein encoding genes (151, 230) (Fig. 5B). They also indirectly control the generation of miRNAs, by inducing the expression of transcription factors that activate or repress miRNA genes (151, 230), as seen with e.g. TGF-β-induced repression of miR-200 expression through Smad3-mediated activation of ZEB1 expression (231). Smad complexes also direct the maturation of miRNAs (Fig. 5E). Indeed, TGF-β- and BMP-activated Smads were seen to associate with a Smad-binding dsRNA motif in select pri-miRNAs and with the p68 RNA helicase in complex with Drosha RNAse, and thus promote the pri-miRNA processing into miRNAs (151, 232, 233). Since individual miRNAs target a large number of mRNAs, the TGF-β-induced, Smad-mediated control of pri-miRNA expression and processing is bound to have substantial effects. Additionally, longer LncRNAs that also control the expression of a variety of genes, either transcriptionally or post-transcriptionally, contribute to the TGF-β response. The expression of some of these is under the direct control of TGF-β/Smad signaling (230, 234).
Finally, reminiscent of Smad-mediated epigenetic changes, Smad2 and/or Smad3 were shown to associate with the METTL3-METLL14-WAPP complex, which N6-methylates adenosine in RNA (Fig. 5F). Smad-mediated recruitment of this m6A-methyltransferase complex at targeted nascent pre-mRNAs results in their co-transcriptional adenosine methylation, consequently promoting their degradation, and decreased expression of encoded proteins (235).
Inhibitory Smads attenuate Smad activation and control transcription
Smad6 and Smad7, and their homologs, Dad in Drosophila and TAG68 in C. elegans, inhibit ligand-induced activation of R-Smads (136). These inhibitory Smads have an MH2 domain similar to all other Smads, but distinguish themselves structurally by their lack of an MH1 domain. Similarly to Smad4, they are not known to require functional activation, and lack the two C-terminal serines that in R-Smads are phosphorylated by type I receptors. The long N-terminal sequence of inhibitory Smads does not represent a domain, yet has motifs for post-translational modifications or protein interactions (136). Its seemingly unstructured nature suggests variable conformations depending on protein associations and post-translational modifications, and functional versatility. Inhibitory Smads localize in the nucleus and cytoplasm, and TGF-β and BMP induce their rapid export from the nucleus (236, 237), raising the as yet unanswered question which signal from the receptor induces this translocation. A sequence preceding the MH2 domain helps define the subcellular localization (238).
Although less characterized than the effector Smads, the inhibitory Smads have several roles in the control of TGF-β family signaling. Smad6 is thought to predominantly inhibit BMP signaling, while Smad7 acts in both BMP and TGF-β signaling (136). Inhibitory Smads associate with the type I receptors, primarily through their MH2 domains, similarly to the effector Smads, but also involving the N-terminal sequence, and this association needs to be undone to enable R-Smad recruitment and activation (136, 238). As mentioned, ligand-induced Arg methylation of Smad6 and/or Smad7 by PRMT1 promotes their dissociation, enabling recruitment and activation of R-Smads (137, 138). Since they also function in non-Smad signaling activation (136) ( see further), inhibitory Smad association with the type I receptor may balance activation of Smads versus non-Smad pathways. The association of inhibitory Smads with type I receptors also allows for recruitment of E3 ubiquitin ligases, and, consequently, receptor ubiquitylation and proteasome-mediated receptor degradation (136), resulting in decreased TGF-β responsiveness, as already discussed.
The inhibitory Smads repress ligand-induced R-Smad activation at two levels (136). Their association with type I receptors through interfaces that primarily involve their MH2 domains prevents R-Smad recruitment through their MH2 domains to type I receptors (136, 239, 240). This competition for receptor association is easily seen as a basis for inhibition of ligand-induced R-Smad activation by inhibitory Smads (136) . Inhibitory Smads also associate with R-Smads through their MH2 domains, similarly to the interaction of activated R-Smads with each other or Smad4. In this scenario, the competition of an inhibitory Smad with Smad4 for binding to R-Smads represses the formation of R-Smad complexes with Smad4 as transcription coactivator at target genes (241). Such inhibitory Smad complexes are consistent with the roles of the MH2 domains in complex formation, and lack of C-terminal phosphorylation in both Smad4 and the inhibitory Smads.
Consistent with their predominant nuclear localization, inhibitory Smads are also likely to act as transcription factors in the control of target gene expression (136). Several observations provide the basis for this assertion. First, the participation of Smad6 or Smad7 in heteromeric complexes with R-Smads allows for recruitment of inhibitory Smads by R-Smads to regulatory DNA sequences of target genes (136, 242). Association of Smad6 with the corepressor CtBP and recruitment of Smad6 to the Id1 gene repress BMP-induced Id1 expression (243), while Smad6 was also seen to recruit histone deacetylases (244). Furthermore, linking Smad7 to a DNA binding sequence confers transcription repression (245). These observations support the notion that inhibitory Smads may direct transcription repression of target genes. Which genes are directly controlled by inhibitory Smads and the underlying mechanisms need to be defined.
Signaling crosstalk and posttranslational control of Smad functions
In addition to functional association with DNA binding transcription factors that are themselves controlled by signaling pathways (Fig. 6, IV), Smads receive extensive crosstalk through post-translational modifications by other signaling effectors (Fig. 6, II and III) that thus define the behavior and activities of the Smads (154, 197).
Various kinases, including some that act in MAP kinase pathways, and cyclin-dependent kinases, phosphorylate R-Smads on serines and/or threonines in their linker regions (154) (Fig. 6, II and III). With a presumed unstructured nature of Smad linker regions, their phosphorylation can create conformational interfaces for recruitment and association with other proteins. Phosphorylation at linker sites enhances or attenuates the nuclear translocation and activities of R-Smads, depending on the kinase and phosphorylation site(s), and additional post-translational modifications (154). Consequently, R-Smad activation by C-terminal phosphorylation is by no means the only determinant of Smad activity, and scenarios are easily envisioned of increased R-Smad activities in response to non-TGF-β family ligands. Smad4 and the inhibitory Smads are also targeted by other kinases (154), albeit most likely less extensively than the R-Smads.
C-terminal and linker phosphorylation of R-Smads can be reversed by dephosphorylation. Among the phosphatases that have been invoked, PPMA1/PP2Cα dephosphorylates the C-terminal serines of Smad2 and Smad3, as well as Smad1 and Smad5, thus dampening Smad-mediated transcription and promoting nuclear export of R-Smads (246, 247). MTMR4 also C-terminally dephosphorylates, and thus deactivates Smad2 and Smad3 (248), and SCP4 dephosphorylates BMP-activated Smad1 and Smad5 (249). In contrast, the SCP phosphatases SCP1, SCP2 and SCP3 remove the linker phosphorylation of Smad2 and Smad3, as well as Smad1 (250, 251). The control of Smad activities through phosphorylation by kinases that function in other pathways is extensive and diverse (154, 197) (Fig. 6, II and III), but what signaling pathways promote Smad dephosphorylation is unclear.
The stability and availability of Smads are extensively regulated by poly-ubiquitylation on lysine, which targets Smads for proteasomal degradation, and opposing deubiquitylases (154, 252). This level of control affects the R-Smads at steady state and after C-terminal activation, as well as Smad4 and the inhibitory Smads, although most studies relate to R-Smads (154, 252). R-Smad ubiquitylation and degradation lead to signal attenuation and termination (154, 252), although to which extent R-Smads are targeted for degradation or deubiquitylation remains unclear. Various E3 ubiquitin ligases, i.e. the enzymes that provide ubiquitylation specificity, were shown to ubiquitylate R-Smads (154, 252). Among these, the HECT family E3 ligases Smurf1 and Smurf2 are best studied, and target TGF-β-activated Smad2 and Smad3, and BMP-activated Smad1 and Smad5 (253, 254). TGF-β-activated Smad3 is also ubiquitylated by SCF family E3 ubiquitin ligases, as well as the CHIP E3 ligase and nuclear Arkadia/RNF111 E3 ligase, leading to Smad3 degradation (255–257). Also Smad4 ubiquitylation is controlled by several E3 ubiquitin ligases, including Smurf and SCF E3 ligases (258), whereas the stabilities of inhibitory Smads are controlled through ubiquitylation by Smurf1 and Smurf2, Arkadia and several other E3 ligases (154). Conversely, Smad de-ubiquitylation opposes ubiquitin-mediated degradation, and thus stabilizes the Smad levels. The diversity of E3 ligases and deubiquitylases illustrate an extensive network of control mechanisms that define the functional Smad availability and contribute to Smad signal termination in a context-dependent manner (117).
Although little is known about how signaling controls Smad ubiquitylation and de-ubiquitylation, Smad phosphorylation has been linked to ubiquitylation and degradation (154). For example, the Wnt signaling effector GSK3β, as well as CDK8 and CDK9 phosphorylate activated Smad2 and Smad3 in the linker region, and the phosphorylated motif is then recognized by the E3 ubiquitin ligase Nedd4L, resulting in ubiquitylation and degradation. CDK8 and CDK9 also phosphorylate the Smad1 linker, leading to recognition of Smad1 by Smurf1 or Nedd4L, and Smad1 degradation (154, 259, 260). Additionally, MH1 domain phosphorylation of non-activated Smad3 by GSK3β leads to ubiquitylation and degradation (261). Further insight will come from learning how Smad ubiquitylation and stability relate to the subcellular localization of Smads, their activation state and control by other pathways.
Attachments of a single ubiquitin or SUMO polypeptide also control the activities of R-Smads, Smad4 and inhibitory Smads. Unlike poly-ubiquitylation, mono-ubiquitylation and sumoylation do not lead to degradation, but mediate protein-protein interactions, and/or control subcellular localization and stability (262, 263). Smad2 and Smad3 are mono-ubiquitylated in response to TGF-β, albeit with different consequences. Smad2 mono-ubiquitylation enhances its association with the TβRI receptor (264), whereas Smad3 mono-ubiquitylation in its MH1 domain interferes with Smad3 binding to DNA (265), and Smad3 mono-ubiquitylation in its MH2 domain, after phosphorylation by CDK8 or CDK9, interferes with Smad complex formation (266). Smad4 mono-ubiquitylation at Lys507 in its MH2 domain promotes trimer formation with Smad3, and TGF-β-induced transcription, yet also enhances its nuclear export (267), whereas Smad4 mono-ubiquitylation by TRIM33/TIF1γ at another lysine in its MH2 domain interferes with complex formation with Smad2 or Smad3 (268). Smad6 mono-ubiquitylation impairs its binding to the BMP type I receptor (269). Smad3 and Smad4 were found to be sumoylated (270, 271), but the effects of Smad sumoylation need to be better defined; stabilization, protection against ubiquitylation, and effects on subcellular localization are likely possibilities (238).
Smad2, Smad3 and Smad4 can also be poly-ADP-ribosylated in their MH1 domains by PARP-1, thus interfering with DNA binding of Smad4 and Smad-mediated transcription (272). TGF-β also induces MH2 domain acetylation of Smad2 and Smad3. Their acetylation occurs in the Smad complexes following association with the acetyltransferases CBP or p300, and enhances Smad-mediated transcription (273). Also Smad7 can be acetylated in the nucleus by p300, albeit not in the MH2 domain. In this case, the acetylation at two lysines prevents their ubiquitylation, allowing acetylation to stabilize Smad7 (274). Smad7 acetylation is reversed by the HDAC1 and SRT1 deacetylases, thus enhancing Smurf1-mediated ubiquitylation and degradation (273, 275). Finally, BMP- or TGF-β-induced Arg methylation of Smad6 and Smad7 by PRMT1 induces their dissociation from type I receptors, enabling R-Smad activation (137, 138), as discussed, and recruitment of the H3K4 methyltransferase SET9 results in Smad7 methylation, promotes Smad7 association with the E3 ubiquitin ligase Arkadia, and, consequently, destabilizes Smad7 (276).
Context-dependent regulation of gene expression by Smads
As is apparent from the mechanisms underlying Smad signaling, Smad complexes not only direct target gene transcription that then leads to secondary gene expression changes, but also control mRNA splicing, miRNA expression and processing, and epigenetic changes. Functional interactions of Smads with transcription factors define the dependence of gene responses on signaling cooperation, while post-translational mechanisms control the availability, properties and behavior of Smads. With further diversity in Smad complex formation, the Smad pathway is a highly nuanced, context-dependent and versatile pathway that controls gene expression.
Several scenarios illustrate far-reaching effects of Smad signaling in gene reprogramming. Direct Smad-mediated changes in transcription of genes encoding splicing mediators (227), combined with direct effects of Smads on hnRNA processing (228), reprogram the diversity of mRNAs and encoded protein isoforms with diverse functional properties. “Self-enabling” scenarios direct the Smad-mediated expression of transcription factors with which Smad complexes subsequently cooperate to activate or repress target gene transcription. For example, TGF-β-induced, Smad¾-directed transcription of genes encoding EMT transcription factors, such as Snail1, enable subsequent cooperation of Smad¾ complexes with these transcription factors in driving EMT-associated gene reprogramming (277–279). Conceptually similarly, activin-induced activation of Mixer expression by Smad2/4 complexes enables cooperation of Smad2/4 with Mixer in targeting genes (152, 280, 281), and the TGF-β-induced activation of ATF3 expression enables cooperation of Smad¾ complexes with ATF3 in the TGF-β-induced repression of Id1 (199).
In Smad-mediated gene expression, the cooperation of Smad complexes with individual DNA-binding transcription factors defines gene synexpression groups, meaning sets of coordinately regulated target genes that similarly respond to the same signaling crosstalk. For example, cooperation of TGF-β-activated Smad¾ complexes with Snail1 confers coordinate regulation of genes during EMT (279), while Smad associations with myogenic bHLH transcription factors coordinately repress myogenic differentiation genes (191, 205, 282). Self-enabling scenarios and Smad-mediated control of gene synexpression groups enable TGF-β family proteins to direct gene reprogramming and differentiation of most, if not all, lineages through cooperation of Smads with lineage-defining master transcription factors (191, 205).
Finally, Smad-mediated control is often transient, not only because of the transient nature of Smad activation, but also because TGF-β-, activin- and BMP-induced Smad complexes activate negative feedback loops through the expression of proteins that subsequently repress Smad activation or Smad-mediated transcription. For example, TGF-βs, activins and BMPs induce, through Smad complexes, the expression of Smad6 and/or Smad7 that subsequently repress ligand-induced R-Smad activation (136). Also, TGF-β induces the expression of SnoN, which then inhibits Smad-mediated transcription activation, yet also controls unrelated processes (217).
NON-SMAD SIGNALING PATHWAYS
Whereas Smad activation defines TGF-β family signaling, TGF-β family receptors also activate other signaling pathways, including the MAPK and phosphoinositide 3-kinase (PI3K)-AKT-mammalian target of rapamycin (mTOR) pathways, which are well-studied mediators of RTKs (283) (Fig. 7). Intuitively, one might attribute these pathways to tyrosine phosphorylation by the receptors, a notion supported by the mechanism of TGF-β-induced ERK/MAPK signaling (68), yet activation of some non-Smad signaling effectors appears to not require the type I receptor kinase activity (283). The activation levels of these non-Smad pathways in response to TGF-β or BMPs are generally lower than their activation by growth factors that act through RTKs. This may relate to the low efficiency tyrosine phosphorylation by dual specificity kinases, but is also consistent with the much lower levels of cell surface receptors, when compared to RTKs. The variability in non-Smad signaling pathway activation also suggests the existence of functionally different receptor complexes (Fig. 7). TGF-β-induced ERK/MAPK and AKT signaling emanate from caveolar compartments (284, 285), similarly to activation by RTKs, and Smad signaling associates with clathrin-mediated endocytosis (131). TGF-β family proteins additionally often induce a delayed activation of non-Smad pathways that may result from Smad-mediated gene expression of transcription factors, growth factors that act through RTKs, or miRNAs that control signaling.
Figure 7. TGF-β receptors activate Smad signaling and non-Smad signal transduction pathways.
Smad-mediated signaling occurs in association with nascent clathrin-dependent endosomal compartments, while receptor-induced non-Smad signaling pathways emanate from caveolar compartments or are not yet known to associate with either type of compartment. TGF-β-induced ERK/MAPK pathway activation occurs in caveolar lipid raft compartments and requires ShcA. TGF-β-induced PI3K-AKT signaling has also been shown to emanate from caveolar lipid raft compartments and to require ShcA, raising the possibility that both pathways initiate from the same receptor complexes. However, TGF-β-induced AKT activation was also shown in different cells to require TRAF6 and to not require the TβRI kinase activity, suggesting that it initiates from different receptor complexes, as shown. TGF-β-induced p38 MAPK and JNK activation has been shown to require TRAF6 and to be initiated by TAK1 activation, while TGF-β-induced p38 MAPK activation has been localized to cholesterol-rich lipid raft compartments, suggesting the existence of distinct complexes for TGF-β-induced p38MAPK, JNK and NFκB signaling, as shown. Whether Smad7 association with the type I receptor facilitates, is required for or antagonizes TGF-β-induced p38 MAPK activation is unclear because of seemingly conflicting reports.
Credit: Veronica Falconieri Hays/Science Signaling
Activation of the ERK MAP kinase pathway
TGF-β and BMPs often induce ERK/MAPK signaling within 5–10 min, with similar kinetics as RTKs, albeit at lower levels (286, 287). The activation of this pathway by TGF-β depends on ShcA (68). TGF-β induces p52ShcA recruitment, through its phosphotyrosine-binding SH2 and PTΒ domains, to the tyrosine-phosphorylated TβRI receptor, and ShcA phosphorylation on serine and tyrosine (68). As with RTKs, tyrosine phosphorylation of ShcA enables activation of ERK/MAPK signaling, while the roles of the extensive serine phosphorylation (68) remains uncharacterized. ShcA was also seen to associate with the TβRII receptor as a result of TβRII phosphorylation on tyrosine by Src (140). TGF-β-induced ShcA phosphorylation on tyrosine promotes the formation of ShcA-Grb2-SOS complexes and Ras activation, leading to MEK1 and/or MEK2, and ERK/MAPK activation, similarly to activation of this pathway by tyrosine kinases (68, 288). It should be noted that Ras activation leading to ERK/MAPK activation was the first identified TGF-β signaling readout (286, 289), prior to the discovery of Smads. ShcA is also required for BMP-induced ERK/MAPK activation (290), suggesting related scenarios for BMP-induced MEK½-ERK/MAPK activation. Whether ShcB, ShcC and ShcD exert similar roles has not yet been shown.
TGF-β receptor complexes that activate ERK/MAPK signaling differ from those that activate Smads. TGF-β-induced ERK/MAPK signaling occurs in caveolar lipid-raft microdomains (284), whereas Smad signaling associates with clathrin-mediated endocytosis (131, 132). Furthermore, p52ShcA association with TβRI precludes Smad recruitment, since p52ShcA and Smad3 compete for TβRI association (291). Consequently, p52ShcA not only initiates ERK/MAPK pathway activation, but also represses TGF-β-induced Smad3 activation, and ShcA expression defines the balance between TGF-β-induced Smad and ERK/MAPK signaling (291).
Activation of the ERK/MAPK pathway, or any other non-Smad pathway, by TGF-β family receptors complements Smad signaling and contributes to physiological TGF-β family responses. Such contributions, for example to the control of cell differentiation by TGF-β family proteins, depend on coordinated regulation of signaling pathways, and are illustrated with the roles of ERK/MAPK signaling in TGF-β-induced EMT that is driven by Smad-mediated gene responses (292). TGF-β-induced ERK/MAPK signaling is expected to induce the same responses as RTK-induced ERK/MAPK pathway activation, although differences in subcellular receptor localization may lead to different responses. ERK/MAPK phosphorylates and controls the functions of transcription factors that cooperate with Smads in the activation or repression of TGF-β/Smad target genes, and additionally controls, through phosphorylation of the R-Smad linker regions, the nuclear import and functions of the R-Smads, as discussed.
Activation of p38 MAPK and JNK pathway signaling through TAK1
TGF-β family proteins also induce rapid activation of the c-Jun N-terminal kinase (JNK) and p38 MAPK pathways (283). Similarly to inflammatory cytokines, TGF-β induces JNK activation through phosphorylation by the MAPK kinase (MAPKK) named MKK4 (293, 294), and p38 MAPK activation by the related MAPK kinases MKK3 or MKK6 (295, 296). These MKKs are activated through phosphorylation by the MAPKK kinase TAK1 (TGF-β-activated kinase, also known as MAP3K7) (283, 297, 298). In spite of its name, TAK1 signaling is not restricted to TGF-β; it also activates JNK and p38 MAPK signaling in response to inflammation or stress signals, including interleukin-1, TNF and Toll-like receptor (TLR) ligands (298). Other MAPKK kinases have been implicated in TGF-β-induced p38 MAPK and/or JNK activation (283) , but their roles are less characterized.
TGF-β induces TAK1 recruitment to TGF-β receptor complexes, through association with the RING domain E3 ubiquitin ligases TRAF6 or TRAF4 (299–301). This association induces intramolecular TRAF6 or TRAF4 polyubiquitylation, allowing either to serve as a scaffold for TAK1 activation. TAK1 is then ubiquitylated by TRAF6 or TRAF4 at a site that does not lead to degradation but is required for its activation, resulting in JNK and p38 MAPK activation (283, 300, 301). This mechanism of pathway activation does not require direct phosphorylation by the type I receptor kinases (299, 300), and resembles JNK and p38 MAPK activation by the interleukin-1 receptor and Toll-like receptors, which also recruit TRAF6 to activate TAK1 (302). It is not known whether TRAF6 and TRAF4 are differentially recruited or activated, or induce different responses, whether BMP-induced JNK and p38 MAPK activation result from a similar mechanism, and whether other MAPKK kinases that activate these pathways similarly act through TRAFs. TRAF6-mediated TAK1 ubiquitylation can also lead to activation of IKK (inhibitor of κB kinase), and thus to nuclear factor κB (NFκB) signaling, in response to TGF-β (303), similarly to NFκB activation by interleukin-1, TNF or TLR ligands (304).
Smad6 and Smad7 also control JNK and p38 MAPK pathway activation (283) . The reported findings are confusing. An attractive scenario involves Smad7 as scaffold for TAK1, MKK3 and p38 MAPK that enables TGF-β-induced p38 MAPK phosphorylation and activation (305), but other observations argue against a role of Smad6 or Smad7 as facilitators of JNK and p38 MAPK pathway activation. For example, Smad6 and Smad7 can inhibit BMP-induced p38 MAPK activation (306), and Smad6 was reported to inhibit TGF-β-induced TAK1, JNK and p38 MAPK activation (307).
Similarly to ERK/MAPK activation, TGF-β-induced JNK and p38 MAPK activation occur in lipid raft and caveolar microdomains (284). This is consistent with the predominant presence of TRAF6, and the TRAF6-mediated NFκB signaling in response to IL-1 or TLR signaling, in such microdomains (308, 309). Athough localized in the same types of compartments, TGF-β receptor complexes that activate JNK and p38 MAPK are most likely distinct from those that activate ERK/MAPK signaling (Fig. 7), since TGF-β-induced ERK/MAPK signaling has not been linked to inhibitory Smads and does not involve TRAF-mediated TAK1 recruitment. How they relate to lipid-raft caveolar receptor complexes that are ubiquitylated and degrade through association of Smurf2 to Smad7 remains unclear. Better characterization of these complexes and the roles of inhibitory Smads in TGF-β- or BMP-induced non-Smad signaling is needed.
The roles of JNK, p38 MAPK and/or NFκB signaling in many processes, such as gene expression, inflammation responses, cell differentiation and cell death, make it likely that their activation contributes to TGF-β responses. Supporting this notion, blocking p38 MAPK activation, silencing TRAF6 expression or interfering with TAK1 function prevent TGF-β- or BMP-induced apoptosis (296, 299, 300, 305–307). p38 MAPK activation and TRAF6 or TRAF4 expression or ubiquitylation are also required for TGF-β-induced EMT (284, 300, 301, 310).
PI3K-AKT-mTOR signaling
Growth factors and cytokines that act through RTKs or receptor-associated tyrosine kinases induce AKT activation through PI3K, a dimer of a regulatory and a catalytic subunit (311). AKT controls many processes and promotes cell survival and proliferation. AKT phosphorylates and thus activates mTOR, which, among other responses, promotes protein synthesis and cell metabolism, is required for cell migration, and phosphorylates AKT in a positive feedback loop (311, 312). TGF-βs and BMPs rapidly induce AKt and mTOR activation, often followed by delayed, indirect activation, as seen with other non-Smad signaling pathways (283, 313, 314).
In several cell systems, TGF-β-induced AKT activation requires the TβRI kinase, and the regulatory PI3-kinase subunit, p85, was seen to associate constitutively with the TβRII receptor, and with TβRI in response to TGF-β (315). These findings await further characterization, but suggest similarities with RTKs. However, AKT can be activated through ubiquitylation by TRAF6 (316), and TGF-β can induce p85 ubiquitylation by TGF-β receptor-associated TRAF6, resulting in its association with the TβRI receptor, and activation of PI3 kinase and AKT (317). In this scenario, TGF-β-induced PI3 kinase and AKT activation does not require the TβRI kinase (317). These observations suggest alternate mechanisms of TGF-β-induced AKT activation, perhaps dependent on cell type. BMP-induced AKT activation has not been biochemically defined.
TGF-β-induced AKT activation can initiate from caveolar receptor complexes (285). Silencing caveolin 1 or ShcA expression abolishes TGF-β-induced AKT activation (285, 291), suggesting that the same receptor complexes activate ERK/MAPK and AKT in response to TGF-β. In contrast, TRAF6-mediated PI3K and AKT activation in response to TGF-β is reminiscent of TGF-β-induced, TRAF6-mediated JNK and p38 MAPK pathway activation, perhaps linking AKT activation to TGF-β receptors that activate the JNK and p38 MAPK pathways.
The substantial activation of the PI3K-AKT-mTOR pathway by TGF-β or BMPs suggests major contributions of this pathway to TGF-β family responses (283). Among these, Akt regulates transcription factors, in some cases through phosphorylation, thus attenuating degradation. Many of these control cell differentiation and cooperate with Smad complexes in activating or repressing target genes in response to TGF-β family signals. For example, TGF-β-activated Smad3/4 complexes induce Snail1 expression, which initiates EMT and then cooperate with Snail1 in the control of target genes that elaborate the EMT program (277–279). Furthermore, AKT activation stabilizes Snail1 and thus enhances Snail1 activity and EMT (318). Additional crosstalk between TGF-β-induced AKT activation and Smad complexes explains the requirement for AKT activation in TGF-β-induced EMT. AKT also phosphorylates FOXO transcription factors and thus attenuates their nuclear import and cooperation with Smad complexes in activating genes encoding the cyclin-dependent kinase (CDK) inhibitors p15Ink4B and p21Cip1. This crosstalk may help protect the cells against TGF-β-induced growth arrest and apoptosis (319, 320). Also mTOR contributes to TGF-β family responses. For example, mTOR activity is required for TGF-β-induced cell motility, and directs the TGF-β-induced increase in protein synthesis and cell size (313, 321). AKT also associates with Smad3, and thus attenuates Smad3 activation and Smad3-mediated TGF-β responses (322, 323), yet AKT activation was also reported to stabilize Smad1 and Smad3, thus enhancing TGF-β and BMP responses (261, 324). Finally, phosphorylation of the RabGAP AS160 by AKT promotes transport of TGF-β receptors to the cell surface, thus increasing TGF-β sensitivity in response to AKT activation (112).
JAK-STAT activation
TGF-β and BMP signaling exert crosstalk with cytokine signaling through functional associations of activated Smad complexes with STAT transcription factors that are activated through phosphorylation by receptor-associated Janus kinases (JAKs). This mechanism is fully consistent with the general mode of Smad-mediated transcription control through association of Smads with DNA sequence-specific transcription factors at regulatory gene sequences, and thus controls a subset of STAT-activated target genes. Such crosstalk has been illustrated with the cooperation of BMP and LIF signaling through Smad1-STAT3 complexes (325), and of TGF-β and interleukin-6 signaling through Smad3-STAT3 complexes (326); however, Smad3 has also been shown to antagonize STAT-mediated transcription (327, 328). Adding to this level of crosstalk, TGF-β also induces JAK-STAT signaling, leading to activation of JAK1-STAT3 signaling in hepatic stellate cells (329), and JAK2 in fibroblasts (330). JAK1 that associates constitutively with the TβRI receptor directly activates STAT3 signaling in response to TGF-β in hepatic stellate cells and fibroblasts, independently of Smad signaling (331). Like several other non-Smad signaling pathways, such direct TGF-β-induced JAK1-STAT3 activation is context-dependent, and followed by a second wave of JAK1-STAT3 signaling that depends on new protein synthesis (331). TGF-β-induced JAK1-STAT3 signaling is required for or contributes to a subset of TGF-β target genes, consistent with the crosstalk between Smads and STATs in the control of gene expression. Cooperation of Smad with JAK-STAT signaling plays essential roles in cell differentiation, immune responses and fibrosis.
Less characterized, but not less functional non-Smad pathways
Again similarly to RTKs or receptor-associated tyrosine kinases, TGF-β family ligands induce a rapid activation of Rho-like GTPases that control cytoskeletal dynamics and cell motility, and contribute to changes in gene expression (283). The mechanisms of their activation by TGF-β family receptors need to be defined, but may depend on the cell type or even cell line. For example, TGF-β induces transient RhoA activation in epithelial cells within 5 min (332, 333), and BMP rapidly induces activation of RhoA and Rho-associated kinase in mesenchymal cells during cell spreading (334). Remarkably, TGF-β induces repression of RhoA activity at tight junctions of epithelial cells during TGF-β-induced epithelial junction dissolution (335), even though RhoA activation is required for EMT. It is therefore likely that RhoA activation and function are controlled at the subcellular level, depending on the physiological state of the cells. TGF-β also induces activation of Rho-like proteins, such as Rac1 and Cdc42 (336), and Cdc42 was shown to associate with TGF-β receptor complexes (96). Furthermore, association of the BMP-RII receptor with LIMK kinase 1 with cooperation from Cdc42 inactivates cofilin, and regulates BMP-induced changes in actin dynamics (337, 338). As with other non-Smad pathways, delayed activation of RhoA and Cdc42 is seen in response to TGF-β ligands, and can result from Smad-mediated expression of some regulators, as apparent with the TGF-β-induced expression of the RhoA guanine exchange factor (GEF) NET1 (339).
TGF-β ligands can also induce other pathways that are not considered as major effector pathways of RTKs (283). For example, TGF-β was seen to induce protein kinase A (PKA) activation in association with Smad complexes (340). TGF-β-induced NFκB signaling is also observed, and might be due to IKK activation (303), as discussed, although other scenarios may also be plausible. Additionally, TGF-β can induce activation the c-Abl tyrosine kinase (341), whereas c-Abl was seen to associate with and phosphorylate the BMP-RIA receptor (342).
Adding to this complexity, the cytoplasmic domain of TβRI can directly regulate transcription (109, 110). ERK and p38 MAP kinase pathways induce TACE to proteolytically release the TβRI ectodomain, thus decreasing TGF-β responsiveness (107). Similarly to Notch processing, this cleavage can be followed in cancer cells by intracellular release of the TβRI cytoplasmic domain, resulting from cleavage by γ-secretase (109). In response to TGF-β, TGF-β receptor-associated TRAF6 recruits presenilin, a component of the γ-secretase complex, to TβRI, activates presenilin through polyubiquitylation, and ubiquitylates the TβRI cytoplasmic domain prior to its intramembranous cleavage. The ubiquitylated TβRI cytoplasmic domain then translocates into the nucleus, associates with the coactivator p300, and promotes the activation of genes that facilitate cell invasion (109, 110). Whether these genes are direct targets for TGF-β/Smad signaling, and the intracellular domain cooperates with Smad complexes is unknown.
Finally, time-dependent dissection of early TGF-β-induced changes in the phospho-proteome revealed a plethora of phosphorylation changes that do not require Smad-mediated gene expression (343). Many, if not most, might result from activation of Smad-independent kinases and kinase pathways, yet others might result from direct phosphorylation by the ligand-activated type II and/or type I receptors.
Concluding remarks
As apparent from this introduction to TGF-β family signaling, it has become apparent that the mere expression of the ligands and signaling mediators does not allow predictions about the cellular responses to TGF-β family members, and that a simple linear pathway of TGF-β family signaling through Smads does not and cannot account for the TGF-β family responses. Ligands, often thought to act as homodimers, may physiologically and developmentally function as heterodimers. With the identities of the receptors resolved for several ligands, it has become apparent that cells exquisitely regulate TGF-β responsiveness at the receptor level, and that much remains to be learned about the control of functional receptor availability and activation through posttranslational modifications in response to other pathways. With much emphasis on Smad signaling, we have come to appreciate their extensive functional versatility and dependence on other signaling pathways. How they control epigenetic modifications and RNA processing deserves additional exploration. Finally, the ability of TGF-β family ligands to directly activate non-Smad signaling pathways complements Smad signaling and is integral to the cell responses to these ligands. Hopefully, this introduction provides a basis and a reference to those who enter this inescapable yet exciting area of research on TGF-β family signaling.
Gloss:
The transforming growth factor-β (TGF-β) family of secreted proteins is implicated in the control of widely diverse physiologies, and its dysregulation with disease. In this review, which contains 7 figures and 343 references, we describe the exquisite nature of TGF-β family signaling in its roles in diverse and context-specific cellular behaviors. These pathways are far from linear and still far from being fully illuminated.
Acknowledgments:
As our knowledge on TGF-β family signaling mechanisms has expanded to the extent that it is daunting to review this field, it is no longer possible to critically review and give proper credit to the many reports that have been published. We apologize to all researchers who may feel that their research was not considered in this review, and thank the extensive research community who has contributed to our understanding of how TGF-β family proteins exert their many activities.
Funding:
Research in the Derynck laboratory has been funded by grants from the National Institutes of Health, predominantly from the National Cancer Institute.
Footnotes
Competing Interests:
The authors have no competing interests. R.D. is a Founder of Pliant Therapeutics, and a consultant to Genentech Inc. and NGM Biopharmaceuticals Inc.
References and Notes
- 1.Derynck R, Miyazono K, The Biology of the TGF-β Family. (Cold Spring Harbor Laboratory Press, New York, ed. Derynck Rik, Miyazono Kohei 2016), pp. 1164. [Google Scholar]
- 2.Morikawa M, Derynck R, Miyazono K, TGF-β and the TGF-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol 8, a021873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moses HL, Roberts AB, Derynck R, The discovery and early days of TGF-β: a historical perspective. Cold Spring Harb Perspect Biol 8, a021865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, Roberts AB, Sporn MB, Goeddel DV, Human transforming growth factor-β complementary DNA sequence and expression in normal and transformed cells. Nature 316, 701–705 (1985). [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Alexander PB, Wang XF, TGF-β family signaling in the control of cell proliferation and survival. Cold Spring Harb Perspect Biol 9, a022145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seoane J, Gomis RR, TGF-β family signaling in tumor suppression and cancer progression. Cold Spring Harb Perspect Biol 9, a022277 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikushima H, Miyazono K, TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer 10, 415–424 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Massagué J, TGF-β in cancer. Cell 134, 215–230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim KK, Sheppard D, Chapman HA, TGF-β1 signaling and tissue fibrosis. Cold Spring Harb Perspect Biol 10, a022293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacFarlane EG, Haupt J, Dietz HC, Shore EM, TGF-β family signaling in connective tissue and skeletal diseases. Cold Spring Harb Perspect Biol 9, a022269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin CH, Emergence, development and diversification of the TGF-β signalling pathway within the animal kingdom. BMC Evol Biol 9, 28 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Zhang S, Wang X, Novel signaling interface constituted with membrane receptor-like kinases emerged from the study of interaction and transphosphorylation of BRI1 and BAK1. Curr Top Med Chem 17, 2393–2400 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Hinck AP, Mueller TD, Springer TA, Structural biology and evolution of the TGF-β family. Cold Spring Harb Perspect Biol 8, a022103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Airaksinen MS, Saarma M, The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci 3, 383–394 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Airaksinen MS, Holm L, Hätinen T, Evolution of the GDNF family ligands and receptors. Brain Behav Evol 68, 181–190 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Mullican SE, Lin-Schmidt X, Chin CN, Chavez JA, Furman JL, Armstrong AA, Beck SC, South VJ, Dinh TQ, Cash-Mason TD, Cavanaugh CR, Nelson S, Huang C, Hunter MJ, Rangwala SM, GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat Med 23, 1150–1157 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Chang CC, Sun Z, Madsen D, Zhu H, Padkjaer SB, Wu X, Huang T, Hultman K, Paulsen SJ, Wang J, Bugge A, Frantzen JB, Norgaard P, Jeppesen JF, Yang Z, Secher A, Chen H, Li X, John LM, Shan B, He Z, Gao X, Su J, Hansen KT, Yang W, Jorgensen SB, GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat Med 23, 1158–1166 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Murray-Rust J, McDonald NQ, Blundell TL, Hosang M, Oefner C, Winkler F, Bradshaw RA, Topological similarities in TGF-β2, PDGF-BB and NGF define a superfamily of polypeptide growth factors. Structure 1, 153–159 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Robertson IB, Rifkin DB, Regulation of the bioavailability of TGF-β and TGF-β-related proteins. Cold Spring Harb Perspect Biol 8, a021907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritsch C, Sawala A, Harris R, Maartens A, Sutcliffe C, Ashe HL, Ray RP, Different requirements for proteolytic processing of bone morphogenetic protein 5/6/7/8 ligands in Drosophila melanogaster. J Biol Chem 287, 5942–5953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama T, Marques G, Wharton KA, A large bioactive BMP ligand with distinct signaling properties is produced by alternative proconvertase processing. Sci Signal 5, ra28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson EN, Wharton KA, Alternative cleavage of the bone morphogenetic protein (BMP), Gbb, produces ligands with distinct developmental functions and receptor preferences. J Biol Chem 292, 19160–19178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray AM, Mason AJ, Requirement for activin A and transforming growth factor-β1 pro-regions in homodimer assembly. Science 247, 1328–1330 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Assoian RK, Komoriya A, Meyers CA, Miller DM, Sporn MB, Transforming growth factor-β in human platelets. Identification of a major storage site, purification, and characterization. J Biol Chem 258, 7155–7160 (1983). [PubMed] [Google Scholar]
- 25.Namwanje M, Brown CW, Activins and inhibins: roles in development, physiology, and disease. Cold Spring Harb Perspect Biol 8, a021881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mason AJ, Hayflick JS, Ling N, Esch F, Ueno N, Ying SY, Guillemin R, Niall H, Seeburg PH, Complementary DNA sequences of ovarian follicular fluid inhibin show precursor structure and homology with transforming growth factor-β. Nature 318, 659–663 (1985). [DOI] [PubMed] [Google Scholar]
- 27.Ling N, Ying SY, Ueno N, Shimasaki S, Esch F, Hotta M, Guillemin R, Pituitary FSH is released by a heterodimer of the beta-subunits from the two forms of inhibin. Nature 321, 779–782 (1986). [DOI] [PubMed] [Google Scholar]
- 28.Little SC, Mullins MC, Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11, 637–643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuerer C, Nostro MC, Constam DB, Nodal.Gdf1 heterodimers with bound prodomains enable serum-independent nodal signaling and endoderm differentiation. J Biol Chem 289, 17854–17871 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montague TG, Schier AF, Vg1-Nodal heterodimers are the endogenous inducers of mesendoderm. eLife 6, e28183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daopin S, Piez KA, Ogawa Y, Davies DR, Crystal structure of transforming growth factor-β2: an unusual fold for the superfamily. Science 257, 369–373 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Schlunegger MP, Grütter MG, An unusual feature revealed by the crystal structure at 2.2 A resolution of human transforming growth factor-β2. Nature 358, 430–434 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Mittl PR, Priestle JP, Cox DA, McMaster G, Cerletti N, Grütter MG, The crystal structure of TGF-β3 and comparison to TGF-β2: implications for receptor binding. Protein Sci 5, 1261–1271 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP, Crystal structure of the human TβR2 ectodomain-TGF-β3 complex. Nat Struct Biol 9, 203–208 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Wei Z, Salmon RM, Upton PD, Morrell NW, Li W, Regulation of bone morphogenetic protein 9 (BMP9) by redox-dependent proteolysis. J Biol Chem 289, 31150–31159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stockis J, Colau D, Coulie PG, Lucas S, Membrane protein GARP is a receptor for latent TGF-β on the surface of activated human Treg. Eur J Immunol 39, 3315–3322 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM, GARP (LRRC32) is essential for the surface expression of latent TGF-β on platelets and activated FOXP3+ regulatory T cells. Proc Natl Acad Sci U S A 106, 13445–13450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockis J, Dedobbeleer O, Lucas S, Role of GARP in the activation of latent TGF-β1. Mol Biosyst 13, 1925–1935 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Liénart S, Merceron R, Vanderaa C, Lambert F, Colau D, Stockis J, van der Woning B, De Haard H, Saunders M, Coulie PG, Savvides SN, Lucas S, Structural basis of latent TGF-β1 presentation and activation by GARP on human regulatory T cells. Science 362, 952–956 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Qin Y, Garrison BS, Ma W, Wang R, Jiang A, Li J, Mistry M, Bronson RT, Santoro D, Franco C, Robinton DA, Stevens B, Rossi DJ, Lu C, Springer TA, A milieu molecule for TGF-β required for microglia function in the nervous system. Cell 174, 156–171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SJ, McPherron AC, Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci U S A 98, 9306–9311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA, Latent TGF-β structure and activation. Nature 474, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mi LZ, Brown CT, Gao Y, Tian Y, Le VQ, Walz T, Springer TA, Structure of bone morphogenetic protein 9 procomplex. Proc Natl Acad Sci U S A 112, 3710–3715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang C, Agonists and antagonists of TGF-β family ligands. Cold Spring Harb Perspect Biol 8, a021923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wohl AP, Troilo H, Collins RF, Baldock C, Sengle G, Extracellular regulation of bone morphogenetic protein activity by the microfibril component fibrillin-1. J Biol Chem 291, 12732–12746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakai LY, Keene DR, Renard M, De Backer J, FBN1: the disease-causing gene for Marfan syndrome and other genetic disorders. Gene 591, 279–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munger JS, Sheppard D, Cross talk among TGF-β signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol 3, a005017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong X, Zhao B, Iacob RE, Zhu J, Koksal AC, Lu C, Engen JR, Springer TA, Force interacts with macromolecular structure in activation of TGF-β. Nature 542, 55–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budi EH, Duan D, Derynck R, Transforming growth factor-β receptors and Smads: regulatory complexity and functional versatility. Trends Cell Biol 27, 658–672 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Le VQ, Iacob RE, Tian Y, McConaughy W, Jackson J, Su Y, Zhao B, Engen JR, Pirruccello-Straub M, Springer TA, Tolloid cleavage activates latent GDF8 by priming the pro-complex for dissociation. EMBO J 37, 384–397 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ge G, Hopkins DR, Ho WB, Greenspan DS, GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol 25, 5846–5858 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konrad L, Luers GH, Volck-Badouin E, Keilani MM, Laible L, Aumuller G, Hofmann R, Analysis of the mRNA expression of the TGF-β family in testicular cells and localization of the splice variant TGF-β2B in testis. Mol Reprod Dev 73, 1211–1220 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Brazil DP, Church RH, Surae S, Godson C, Martin F, BMP signalling: agony and antagony in the family. Trends Cell Biol 25, 249–264 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Blitz IL, Cho KW, Chang C, Twisted gastrulation loss-of-function analyses support its role as a BMP inhibitor during early Xenopus embryogenesis. Development 130, 4975–4988 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Scott IC, Blitz IL, Pappano WN, Maas SA, Cho KW, Greenspan DS, Homologues of Twisted gastrulation are extracellular cofactors in antagonism of BMP signalling. Nature 410, 475–478 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Chang C, Holtzman DA, Chau S, Chickering T, Woolf EA, Holmgren LM, Bodorova J, Gearing DP, Holmes WE, Brivanlou AH, Twisted gastrulation can function as a BMP antagonist. Nature 410, 483–487 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Harrington AE, Morris-Triggs SA, Ruotolo BT, Robinson CV, Ohnuma S, Hyvonen M, Structural basis for the inhibition of activin signalling by follistatin. EMBO J 25, 1035–1045 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers KW, Schier AF, Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol 27, 377–407 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Roy S, Hsiung F, Kornberg TB, Specificity of Drosophila cytonemes for distinct signaling pathways. Science 332, 354–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kornberg TB, Distributing signaling proteins in space and time: the province of cytonemes. Curr Opin Genet Dev 45, 22–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heldin C-H, Moustakas A, Signaling receptors for TGF-β family members. Cold Spring Harb Perspect Biol 8, a022053(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheifetz S, Weatherbee JA, Tsang ML, Anderson JK, Mole JE, Lucas R, Massagué J, The transforming growth factor-β system, a complex pattern of cross-reactive ligands and receptors. Cell 48, 409–415 (1987). [DOI] [PubMed] [Google Scholar]
- 63.Chaikuad A, Bullock AN, Structural basis of intracellular TGF-β signaling: receptors and Smads. Cold Spring Harb Perspect Biol 8, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wieser R, Wrana JL, Massagué J, GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J 14, 2199–2208 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S, The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Rask-Andersen M, Zhang J, Fabbro D, Schioth HB, Advances in kinase targeting: current clinical use and clinical trials. Trends Pharmacol Sci 35, 604–620 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Lawler S, Feng X-H, Chen RH, Maruoka EM, Turck CW, Griswold-Prenner I, Derynck R, The type II transforming growth factor-β receptor autophosphorylates not only on serine and threonine but also on tyrosine residues. J Biol Chem 272, 14850–14859 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, Derynck R, TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J 26, 3957–3967 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dhanasekaran N, Premkumar Reddy E, Signaling by dual specificity kinases. Oncogene 17, 1447–1455 (1998). [DOI] [PubMed] [Google Scholar]
- 70.Ehrlich M, Gutman O, Knaus P, Henis YI, Oligomeric interactions of TGF-β and BMP receptors. FEBS Lett 586, 1885–1896 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Gilboa L, Wells RG, Lodish HF, Henis YI, Oligomeric structure of type I and type II transforming growth factor β receptors: homodimers form in the ER and persist at the plasma membrane. J Cell Biol 140, 767–777 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang W, Jiang Y, Wang Q, Ma X, Xiao Z, Zuo W, Fang X, Chen YG, Single-molecule imaging reveals transforming growth factor-β-induced type II receptor dimerization. Proc Natl Acad Sci U S A 106, 15679–15683 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheifetz S, Bassols A, Stanley K, Ohta M, Greenberger J, Massagué J, Heterodimeric transforming growth factor β. Biological properties and interaction with three types of cell surface receptors. J Biol Chem 263, 10783–10789 (1988). [PubMed] [Google Scholar]
- 74.De Crescenzo G, Pham PL, Durocher Y, O’Connor-McCourt MD, Transforming growth factor-β (TGF-β) binding to the extracellular domain of the type II TGF-β receptor: receptor capture on a biosensor surface using a new coiled-coil capture system demonstrates that avidity contributes significantly to high affinity binding. J Mol Biol 328, 1173–1183 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Nickel J, ten Dijke P, Mueller TD, TGF-β family co-receptor function and signaling. Acta Biochim Biophys Sin 50, 12–36 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Bilandzic M, Stenvers KL, Betaglycan: a multifunctional accessory. Mol Cell Endocrinol 339, 180–189 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Sugden WW, Siekmann AF, Endothelial cell biology of endoglin in hereditary hemorrhagic telangiectasia. Curr Opin Hematol 25, 237–244 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Dong M, How T, Kirkbride KC, Gordon KJ, Lee JD, Hempel N, Kelly P, Moeller BJ, Marks JR, Blobe GC, The type III TGF-β receptor suppresses breast cancer progression. J Clin Invest 117, 206–217 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez-Casillas F, Payne HM, Andres JL, Massagué J, Betaglycan can act as a dual modulator of TGF-β access to signaling receptors: mapping of ligand binding and GAG attachment sites. J Cell Biol 124, 557–568 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andres JL, DeFalcis D, Noda M, Massagué J, Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J Biol Chem 267, 5927–5930 (1992). [PubMed] [Google Scholar]
- 81.Velasco S, Alvarez-Munoz P, Pericacho M, Dijke PT, Bernabeu C, Lopez-Novoa JM, Rodriguez-Barbero A, L- and S-endoglin differentially modulate TGFβ1 signaling mediated by ALK1 and ALK5 in L6E9 myoblasts. J Cell Sci 121, 913–919 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Goumans MJ, Zwijsen A, ten Dijke P, Bailly S, Cold Spring Harb Perspect Biol 10, a031989 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castonguay R, Werner ED, Matthews RG, Presman E, Mulivor AW, Solban N, Sako D, Pearsall RS, Underwood KW, Seehra J, Kumar R, Grinberg AV, Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. J Biol Chem 286, 30034–30046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hawinkels LJ, Kuiper P, Wiercinska E, Verspaget HW, Liu Z, Pardali E, Sier CF, ten Dijke P, Matrix metalloproteinase-14 (MT1-MMP)-mediated endoglin shedding inhibits tumor angiogenesis. Cancer Res 70, 4141–4150 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Siebold C, Yamashita T, Monnier PP, Mueller BK, Pasterkamp RJ, RGMs: Structural insights, molecular regulation, and downstream signaling. Trends Cell Biol 27, 365–378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian C, Liu J, Repulsive guidance molecules (RGMs) and neogenin in bone morphogenetic protein (BMP) signaling. Mol Reprod Dev 80, 700–717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rajagopalan S, Deitinghoff L, Davis D, Conrad S, Skutella T, Chédotal A, Mueller BK, Strittmatter SM, Neogenin mediates the action of repulsive guidance molecule. Nat Cell Biol 6, 756–762 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY, Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet 38, 531–539 (2006). [DOI] [PubMed] [Google Scholar]
- 89.Yeo C, Whitman M, Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell 7, 949–957 (2001). [DOI] [PubMed] [Google Scholar]
- 90.Yan YT, Liu JJ, Luo Y, Haltiwanger CE,RS, Abate-Shen C, Shen MM, Dual roles of Cripto as a ligand and coreceptor in the nodal signaling pathway. Mol Cell Biol 22, 4439–4449 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gray PC, Shani G, Aung K, Kelber J, Vale W, Cripto binds transforming growth factor β (TGF-β) and inhibits TGF-β signaling. Mol Cell Biol 26, 9268–9278 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nagaoka T, Karasawa H, Castro NP, Rangel MC, Salomon DS, Bianco C, An evolving web of signaling networks regulated by Cripto-1. Growth Factors 30, 13–21 (2012). [DOI] [PubMed] [Google Scholar]
- 93.Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massagué J, Niehrs C, Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature 401, 480–485 (1999). [DOI] [PubMed] [Google Scholar]
- 94.Tsang M, Kim R, de Caestecker MP, Kudoh T, Roberts AB, Dawid IB, Zebrafish nma is involved in TGFβ family signaling. Genesis 28, 47–57 (2000). [DOI] [PubMed] [Google Scholar]
- 95.Rudini N, Felici A, Giampietro C, Lampugnani M, Corada M, Swirsding K, Garre M, Liebner S, Letarte M, ten Dijke P, Dejana E, VE-cadherin is a critical endothelial regulator of TGF-β signalling. EMBO J 27, 993–1004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL, High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 (2005). [DOI] [PubMed] [Google Scholar]
- 97.Bourguignon LY, Singleton PA, Zhu H, Zhou B, Hyaluronan promotes signaling interaction between CD44 and the transforming growth factor beta receptor I in metastatic breast tumor cells. J Biol Chem 277, 39703–39712 (2002). [DOI] [PubMed] [Google Scholar]
- 98.Porsch H, Mehic M, Olofsson B, Heldin P, Heldin C-H, Platelet-derived growth factor beta-receptor, transforming growth factor β type I receptor, and CD44 protein modulate each other’s signaling and stability. J Biol Chem 289, 19747–19757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jin W, Yun C, Kim HS, Kim SJ, TrkC binds to the bone morphogenetic protein type II receptor to suppress bone morphogenetic protein signaling. Cancer Res 67, 9869–9877 (2007). [DOI] [PubMed] [Google Scholar]
- 100.Sammar M, Stricker S, Schwabe GC, Sieber C, Hartung A, Hanke M, Oishi I, Pohl J, Minami Y, Sebald W, Mundlos S, Knaus P, Modulation of GDF5/BRI-b signalling through interaction with the tyrosine kinase receptor Ror2. Genes Cells 9, 1227–1238 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Wells RG, Yankelev H, Lin HY, Lodish HF, Biosynthesis of the type I and type II TGF-β receptors. Implications for complex formation. J Biol Chem 272, 11444–11451 (1997). [DOI] [PubMed] [Google Scholar]
- 102.Kim YW, Park J, Lee HJ, Lee SY, Kim SJ, TGF-β sensitivity is determined by N-linked glycosylation of the type II TGF-β receptor. Biochem J 445, 403–411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lowery JW, Amich JM, Andonian A, Rosen V, N-linked glycosylation of the bone morphogenetic protein receptor type 2 (BMPR2) enhances ligand binding. Cell Mol Life Sci 71, 3165–3172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goetschy JF, Letourneur O, Cerletti N, Horisberger MA, The unglycosylated extracellular domain of type-II receptor for transforming growth factor-β. A novel assay for characterizing ligand affinity and specificity. Eur J Biochem 241, 355–362 (1996). [DOI] [PubMed] [Google Scholar]
- 105.Fukami K, Asano E, Ueda M, Sekiguchi F, Yoshida S, Kawabata A, High glucose induces N-linked glycosylation-mediated functional upregulation and overexpression of Cav3.2 T-type calcium channels in neuroendocrine-like differentiated human prostate cancer cells. J Pharmacol Sci 133, 57–60 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Alisson-Silva F, Freire-de-Lima L, Donadio JL, Lucena MC, Penha L, Sa-Diniz JN, Dias WB, Todeschini AR, Increase of O-glycosylated oncofetal fibronectin in high glucose-induced epithelial-mesenchymal transition of cultured human epithelial cells. PloS One 8, e60471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu C, Xu P, Lamouille S, Xu J, Derynck R, TACE-mediated ectodomain shedding of the type I TGF-β receptor downregulates TGF-β signaling. Mol Cell 35, 26–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu P, Liu J, Sakaki-Yumoto M, Derynck R, TACE activation by MAPK-mediated regulation of cell surface dimerization and TIMP3 association. Sci Signal 5, ra34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mu Y, Sundar R, Thakur N, Ekman M, Gudey SK, Yakymovych M, Hermansson A, Dimitriou H, Bengoechea-Alonso MT, Ericsson J, Heldin C-H, Landström M, TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer. Nat Commun 2, 330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gudey SK, Sundar R, Mu Y, Wallenius A, Zang G, Bergh A, Heldin C-H, Landström M, TRAF6 stimulates the tumor-promoting effects of TGFβ type I receptor through polyubiquitination and activation of presenilin 1. Sci Signal 7, ra2 (2014). [DOI] [PubMed] [Google Scholar]
- 111.Wu L, Derynck R, Essential role of TGF-β signaling in glucose-induced cell hypertrophy. Dev Cell 17, 35–48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Budi EH, Muthusamy BP, Derynck R, The insulin response integrates increased TGF-β signaling through Akt-induced enhancement of cell surface delivery of TGF-β receptors. Sci Signal 8, ra96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zaid H, Antonescu CN, Randhawa VK, Klip A, Insulin action on glucose transporters through molecular switches, tracks and tethers. Biochem J 413, 201–215 (2008). [DOI] [PubMed] [Google Scholar]
- 114.Mafakheri S, Chadt A, Al-Hasani H, Regulation of RabGAPs involved in insulin action. Biochem Soc Trans 46, 683–690 (2018). [DOI] [PubMed] [Google Scholar]
- 115.Jiang ZY, Zhou QL, Coleman KA, Chouinard M, Boese Q, Czech MP, Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc Natl Acad Sci U S A 100, 7569–7574 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang J, Zhang X, Xie F, Zhang Z, van Dam H, Zhang L, Zhou F, The regulation of TGF-β/SMAD signaling by protein deubiquitination. Protein & cell 5, 503–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu S, de Boeck M, van Dam H, ten Dijke P, Regulation of the TGF-β pathway by deubiquitinases in cancer. Int J Biochem Cell Biol 76, 135–145 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Bernassola F, Karin M, Ciechanover A, Melino G, The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 (2008). [DOI] [PubMed] [Google Scholar]
- 119.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL, Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol Cell 6, 1365–1375 (2000). [DOI] [PubMed] [Google Scholar]
- 120.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K, Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J Biol Chem 276, 12477–12480 (2001). [DOI] [PubMed] [Google Scholar]
- 121.Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, Imamura T, NEDD4–2 (neural precursor cell expressed, developmentally down-regulated 4–2) negatively regulates TGF-β (transforming growth factor-β) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-β type I receptor. Biochem J 386, 461–470 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K, Negative regulation of transforming growth factor-β (TGF-β) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23, 6914–6923 (2004). [DOI] [PubMed] [Google Scholar]
- 123.Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu CX, ten Dijke P, USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat Cell Biol 14, 717–726 (2012). [DOI] [PubMed] [Google Scholar]
- 124.Zuo W, Huang F, Chiang YJ, Li M, Du J, Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, Deng H, Feng X-H, Luo S, Gao C, Chen YG, c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-β type II receptor. Mol Cell 49, 499–510 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Murphy SJ, Dore JJ, Edens M, Coffey RJ, Barnard JA, Mitchell H, Wilkes M, Leof EB, Differential trafficking of transforming growth factor-β receptors and ligand in polarized epithelial cells. Mol Biol Cell 15, 2853–2862 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yin X, Murphy SJ, Wilkes MC, Ji Y, Leof EB, Retromer maintains basolateral distribution of the type II TGF-β receptor via the recycling endosome. Mol Biol Cell 24, 2285–2298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mitchell H, Choudhury A, Pagano RE, Leof EB, Ligand-dependent and -independent transforming growth factor-β receptor recycling regulated by clathrin-mediated endocytosis and Rab11. Mol Biol Cell 15, 4166–4178 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen YG, Endocytic regulation of TGF-β signaling. Cell Res 19, 58–70 (2009). [DOI] [PubMed] [Google Scholar]
- 129.Ehrlich M, Endocytosis and trafficking of BMP receptors: Regulatory mechanisms for fine-tuning the signaling response in different cellular contexts. Cytokine Growth Factor Rev 27, 35–42 (2016). [DOI] [PubMed] [Google Scholar]
- 130.Dore JJ Jr., Yao D, Edens M, Garamszegi N, Sholl EL, Leof EB, Mechanisms of transforming growth factor-β receptor endocytosis and intracellular sorting differ between fibroblasts and epithelial cells. Mol Biol Cell 12, 675–684 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL, Distinct endocytic pathways regulate TGF-β receptor signalling and turnover. Nat Cell Biol 5, 410–421 (2003). [DOI] [PubMed] [Google Scholar]
- 132.Le Roy C, Wrana JL, Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol 6, 112–126 (2005). [DOI] [PubMed] [Google Scholar]
- 133.Huse M, Chen YG, Massague J, Kuriyan J, Crystal structure of the cytoplasmic domain of the type I TGF β receptor in complex with FKBP12. Cell 96, 425–436 (1999). [DOI] [PubMed] [Google Scholar]
- 134.Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massagué J, The TGF β receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell 8, 671–682 (2001). [DOI] [PubMed] [Google Scholar]
- 135.Chen YG, Liu F, Massagué J, Mechanism of TGFβ receptor inhibition by FKBP12. EMBO J 16, 3866–3876 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miyazawa K, Miyazono K, Regulation of TGF-β Family Signaling by Inhibitory Smads. Cold Spring Harb Perspect Biol 9, a022095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu J, Wang AH, Oses-Prieto J, Makhijani K, Katsuno Y, Pei M, Yan L, Zheng YG, Burlingame A, Brückner K, Derynck R, Arginine Methylation Initiates BMP-Induced Smad Signaling. Mol Cell 51, 5–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Katsuno Y, Qin J, Oses-Prieto JA, Wang H, Jackson-Weaver O, Zhang T, Lamouille S, Wu J, Burlingame ALL, Xu J, Derynck R, Arginine methylation of Smad7 by PRMT1 in TGF-β-induced epithelial-mesenchymal transition and epithelial stem cell generation. J Biol Chem 293, 130590–13072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Luo K, Lodish HF, Positive and negative regulation of type II TGF-β receptor signal transduction by autophosphorylation on multiple serine residues. EMBO J 16, 1970–1981 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Galliher AJ, Schiemann WP, Src phosphorylates Tyr284 in TGF-β type II receptor and regulates TGF-β stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res 67, 3752–3758 (2007). [DOI] [PubMed] [Google Scholar]
- 141.Kang JS, Saunier EF, Akhurst RJ, Derynck R, The type I TGF-β receptor is covalently modified and regulated by sumoylation. Nat Cell Biol 10, 654–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X, GADD34-PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. J Cell Biol 164, 291–300 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Griswold-Prenner I, Kamibayashi C, Maruoka EM, Mumby MC, Derynck R, Physical and functional interactions between type I transforming growth factor β receptors and Bα, a WD-40 repeat subunit of phosphatase 2A. Mol Cell Biol 18, 6595–6604 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Batut J, Schmierer B, Cao J, Raftery LA, Hill CS, Howell M, Two highly related regulatory subunits of PP2A exert opposite effects on TGF-β/Activin/Nodal signalling. Dev 135, 2927–2937 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen X, Wang H, Liao HJ, Hu W, Gewin L, Mernaugh G, Zhang S, Zhang ZY, Vega-Montoto L, Vanacore RM, Fassler R, Zent R, Pozzi A, Integrin-mediated type II TGF-β receptor tyrosine dephosphorylation controls SMAD-dependent profibrotic signaling. J Clin Invest 124, 3295–3310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Satow R, Kurisaki A, Chan TC, Hamazaki TS, Asashima M, Dullard promotes degradation and dephosphorylation of BMP receptors and is required for neural induction. Dev Cell 11, 763–774 (2006). [DOI] [PubMed] [Google Scholar]
- 147.Verdin E, Ott M, 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol 16, 258–264 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Drazic A, Myklebust LM, Ree R, Arnesen T, The world of protein acetylation. Biochim Biophys Acta 1864, 1372–1401 (2016). [DOI] [PubMed] [Google Scholar]
- 149.Biggar KK, Li SS, Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol 16, 5–17 (2015). [DOI] [PubMed] [Google Scholar]
- 150.Massagué J, TGFbeta signalling in context. Nat Rev Mol Cell Biol 13, 616–630 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hata A, Chen YG, TGF-β signaling from receptors to Smads. Cold Spring Harb Perspect Biol 8, a022061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hill CS, Transcriptional Control by the SMADs. Cold Spring Harbor perspectives in biology 8, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP, Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94, 585–594 (1998). [DOI] [PubMed] [Google Scholar]
- 154.Xu P, Lin X, Feng X-H, Posttranslational regulation of Smads. Cold Spring Harb Perspect Biol 8, a022087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tao S, Sampath K, Alternative splicing of SMADs in differentiation and tissue homeostasis. Dev Growth Differ 52, 335–342 (2010). [DOI] [PubMed] [Google Scholar]
- 156.Kjellman C, Honeth G, Jarnum S, Lindvall M, Darabi A, Nilsson I, Edvardsen K, Salford LG, Widegren B, Identification and characterization of a human Smad3 splicing variant lacking part of the linker region. Gene 327, 141–152 (2004). [DOI] [PubMed] [Google Scholar]
- 157.Cheng KH, Ponte JF, Thiagalingam S, Elucidation of epigenetic inactivation of SMAD8 in cancer using targeted expressed gene display. Cancer Res 64, 1639–1646 (2004). [DOI] [PubMed] [Google Scholar]
- 158.Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K, Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem 274, 703–709 (1999). [DOI] [PubMed] [Google Scholar]
- 159.Liu L, Liu X, Ren X, Tian Y, Chen Z, Xu X, Du Y, Jiang C, Fang Y, Liu Z, Fan B, Zhang Q, Jin G, Yang X, Zhang X, Smad2 and Smad3 have differential sensitivity in relaying TGFβ signaling and inversely regulate early lineage specification. Sci Rep 6, 21602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K, Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J 17, 4056–4065 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, de Caestecker M, Lin K, Structural basis of heteromeric Smad protein assembly in TGF-β signaling. Mol Cell 15, 813–823 (2004). [DOI] [PubMed] [Google Scholar]
- 162.Shapira KE, Hirschhorn T, Barzilay L, Smorodinsky NI, Henis YI, Ehrlich M, Dab2 inhibits the cholesterol-dependent activation of JNK by TGF-β. Mol Biol Cell 25, 1620–1628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hocevar BA, Smine A, Xu XX, Howe PH, The adaptor molecule Disabled-2 links the transforming growth factor β receptors to the Smad pathway. EMBO J 20, 2789–2801 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xu P, Bailey-Bucktrout S, Xi Y, Xu D, Du D, Zhang Q, Xiang W, Liu J, Melton A, Sheppard D, Chapman HA, Bluestone JA, Derynck R, Innate antiviral host defense attenuates TGF-β function through IRF3-mediated suppression of Smad signaling. Mol Cell 56, 723–737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Feng X-H, Derynck R, A kinase subdomain of transforming growth factor-β (TGF-β) type I receptor determines the TGF-β intracellular signaling specificity. EMBO J 16, 3912–3923 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massagué J, Determinants of specificity in TGF-β signal transduction. Genes Dev 12, 2144–2152 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lo RS, Chen YG, Shi Y, Pavletich NP, Massagué J, The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF-β receptors. EMBO J 17, 996–1005 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engström U, Heldin C-H, Funa K, ten Dijke P, The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett 434, 83–87 (1998). [DOI] [PubMed] [Google Scholar]
- 169.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL, SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95, 779–791 (1998). [DOI] [PubMed] [Google Scholar]
- 170.Miura S, Takeshita T, Asao H, Kimura Y, Murata K, Sasaki Y, Hanai JI, Beppu H, Tsukazaki T, Wrana JL, Miyazono K, Sugamura K, Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol Cell Biol 20, 9346–9355 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu G, Chen YG, Ozdamar B, Gyuricza CA, Chong PA, Wrana JL, Massagué J, Shi Y, Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science 287, 92–97 (2000). [DOI] [PubMed] [Google Scholar]
- 172.Shi W, Chang C, Nie S, Xie S, Wan M, Cao X, Endofin acts as a Smad anchor for receptor activation in BMP signaling. J Cell Sci 120, 1216–1224 (2007). [DOI] [PubMed] [Google Scholar]
- 173.Goh JB, Wallace DF, Hong W, Subramaniam VN, Endofin, a novel BMP-SMAD regulator of the iron-regulatory hormone, hepcidin. Sci Rep 5, 13986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massagué J, Shi Y, Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-β signaling. Mol Cell 8, 1277–1289 (2001). [DOI] [PubMed] [Google Scholar]
- 175.Lucarelli P, Schilling M, Kreutz C, Vlasov A, Boehm ME, Iwamoto N, Steiert B, Lattermann S, Wasch M, Stepath M, Matter MS, Heikenwalder M, Hoffmann K, Deharde D, Damm G, Seehofer D, Muciek M, Gretz N, Lehmann WD, Timmer J, Klingmüller U, Resolving the combinatorial complexity of Smad protein complex formation and its link to gene expression. Cell Syst 6, 75–89 e11 (2018). [DOI] [PubMed] [Google Scholar]
- 176.Daly AC, Randall RA, Hill CS, Transforming growth factor β-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol Cell Biol 28, 6889–6902 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ramachandran A, Vizan P, Das D, Chakravarty P, Vogt J, Rogers KW, Müller P, Hinck AP, Sapkota GP, Hill CS, TGF-β uses a novel mode of receptor activation to phosphorylate SMAD1/5 and induce epithelial-to-mesenchymal transition. eLife 7, e31756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hill CS, Nucleocytoplasmic shuttling of Smad proteins. Cell Res 19, 36–46 (2009). [DOI] [PubMed] [Google Scholar]
- 179.Kurisaki A, Kose S, Yoneda Y, Heldin C-H, Moustakas A, Transforming growth factor-β induces nuclear import of Smad3 in an importin-β1 and Ran-dependent manner. Mol Biol Cell 12, 1079–1091 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xiao Z, Latek R, Lodish HF, An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene 22, 1057–1069 (2003). [DOI] [PubMed] [Google Scholar]
- 181.Xu L, Alarcon C, Col S, Massagué J, Distinct domain utilization by Smad3 and Smad4 for nucleoporin interaction and nuclear import. J Biol Chem 278, 42569–42577 (2003). [DOI] [PubMed] [Google Scholar]
- 182.Xiao Z, Brownawell AM, Macara IG, Lodish HF, A novel nuclear export signal in Smad1 is essential for its signaling activity. J Biol Chem 278, 34245–34252 (2003). [DOI] [PubMed] [Google Scholar]
- 183.Kurisaki A, Kurisaki K, Kowanetz M, Sugino H, Yoneda Y, Heldin C-H, Moustakas A, The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol Cell Biol 26, 1318–1332 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dai F, Lin X, Chang C, Feng X-H, Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-β signaling. Dev Cell 16, 345–357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chen F, Lin X, Xu P, Zhang Z, Chen Y, Wang C, Han J, Zhao B, Xiao M, Feng X-H, Nuclear export of Smads by RanBP3L regulates bone morphogenetic protein signaling and mesenchymal stem cell differentiation. Mol Cell Biol 35, 1700–1711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Batut J, Howell M, Hill CS, Kinesin-mediated transport of Smad2 is required for signaling in response to TGF-β ligands. Dev Cell 12, 261–274 (2007). [DOI] [PubMed] [Google Scholar]
- 187.Jin Q, Gao G, Mulder KM, Requirement of a dynein light chain in TGFβ/Smad3 signaling. J Cell Physiol 221, 707–715 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Feng X-H, Derynck R, Specificity and versatility in TGF-β signaling through Smads. Annu Rev Cell Dev Biol 21, 659–693 (2005). [DOI] [PubMed] [Google Scholar]
- 189.Massagué J, Seoane J, Wotton D, Smad transcription factors. Genes Dev 19, 2783–2810 (2005). [DOI] [PubMed] [Google Scholar]
- 190.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA, Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565–576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Morikawa M, Koinuma D, Miyazono K, Heldin CH, Genome-wide mechanisms of Smad binding. Oncogene 32, 1609–1615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE, Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell 1, 611–617 (1998). [DOI] [PubMed] [Google Scholar]
- 193.Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, Miyazono K, ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucl Acids Res 39, 8712–8727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Martin-Malpartida P, Batet M, Kaczmarska Z, Freier R, Gomes T, Aragon E, Zou Y, Wang Q, Xi Q, Ruiz L, Vea A, Marquez JA, Massagué J, Macias MJ, Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat Commun 8, 2070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Feng X-H, Lin X, Derynck R, Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-β. EMBO J 19, 5178–5193 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R, TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J 20, 2254–2272 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Luo K, Signaling Cross talk between TGF-β/Smad and other signaling pathways. Cold Spring Harb Perspect Biol 9, a022137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhang Y, Feng X-H, Derynck R, Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394, 909–913 (1998). [DOI] [PubMed] [Google Scholar]
- 199.Kang Y, Chen CR, Massagué J, A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell 11, 915–926 (2003). [DOI] [PubMed] [Google Scholar]
- 200.Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S, Integration of TGF-β and Ras/MAPK signaling through p53 phosphorylation. Science 315, 840–843 (2007). [DOI] [PubMed] [Google Scholar]
- 201.Attisano L, Labbé E, TGFβ and Wnt pathway cross-talk. Cancer Metastasis Rev 23, 53–61 (2004). [DOI] [PubMed] [Google Scholar]
- 202.Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U, Ibanez CF, Cross-talk between the Notch and TGF-β signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol 163, 723–728 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP, Integration of TGF-β/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J 23, 1155–1165 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nye MD, Almada LL, Fernandez-Barrena MG, Marks DL, Elsawa SF, Vrabel A, Tolosa EJ, Ellenrieder V, Fernandez-Zapico ME, The transcription factor GLI1 interacts with SMAD proteins to modulate transforming growth factor β-induced gene expression in a p300/CREB-binding protein-associated factor (PCAF)-dependent manner. J Biol Chem 289, 15495–15506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mullen AC, Wrana JL, TGF-β family signaling in embryonic and somatic stem-cell renewal and differentiation. Cold Spring Harb Perspect Biol 9, a022186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Grafe I, Alexander S, Peterson JR, Snider TN, Levi B, Lee B, Mishina Y, TGF-β family signaling in mesenchymal differentiation. Cold Spring Harb Perspect Biol 10, a022202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Feng X-H, Zhang Y, Wu RY, Derynck R, The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev 12, 2153–2163 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Levy L, Hill CS, Smad4 dependency defines two classes of transforming growth factor-β (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol 25, 8108–8125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kang JS, Alliston T, Delston R, Derynck R, Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J 24, 2543–2555 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS, Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J 25, 4490–4502 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Itoh S, Ericsson J, Nishikawa J, Heldin C-H, ten Dijke P, The transcriptional co-activator P/CAF potentiates TGF-β/Smad signaling. Nucl Acids Res 28, 4291–4298 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kahata K, Hayashi M, Asaka M, Hellman U, Kitagawa H, Yanagisawa J, Kato S, Imamura T, Miyazono K, Regulation of transforming growth factor-β and bone morphogenetic protein signalling by transcriptional coactivator GCN5. Genes Cells 9, 143–151 (2004). [DOI] [PubMed] [Google Scholar]
- 213.Wakabayashi Y, Tamiya T, Takada I, Fukaya T, Sugiyama Y, Inoue N, Kimura A, Morita R, Kashiwagi I, Takimoto T, Nomura M, Yoshimura A, Histone 3 lysine 9 (H3K9) methyltransferase recruitment to the interleukin-2 (IL-2) promoter is a mechanism of suppression of IL-2 transcription by the transforming growth factor-β-Smad pathway. J Biol Chem 286, 35456–35465 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Du D, Katsuno Y, Meyer D, Budi EH, Chen SH, Koeppen H, Wang H, Akhurst RJ, Derynck R, Smad3-mediated recruitment of the methyltransferase SETDB1/ESET controls Snail1 expression and epithelial-mesenchymal transition. EMBO Rep 19, 135–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Shuttleworth VG, Gaughan L, Nawafa L, Mooney CA, Cobb SL, Sheerin NS, Logan IR, The methyltransferase SET9 regulates TGFB1 activation of renal fibroblasts via interaction with SMAD3. J Cell Sci 131, (2018). [DOI] [PubMed] [Google Scholar]
- 216.Wotton D, Lo RS, Lee S, Massagué J, A Smad transcriptional corepressor. Cell 97, 29–39 (1999). [DOI] [PubMed] [Google Scholar]
- 217.Deheuninck J, Luo K, Ski and SnoN, potent negative regulators of TGF-β signaling. Cell Res 19, 47–57 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Izutsu K, Kurokawa M, Imai Y, Maki K, Mitani K, Hirai H, The corepressor CtBP interacts with Evi-1 to repress transforming growth factor β signaling. Blood 97, 2815–2822 (2001). [DOI] [PubMed] [Google Scholar]
- 219.Alliston T, Ko TC, Cao Y, Liang YY, Feng X-H, Chang C, Derynck R, Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J Biol Chem 280, 24227–24237 (2005). [DOI] [PubMed] [Google Scholar]
- 220.Feng Y, Wu H, Xu Y, Zhang Z, Liu T, Lin X, Feng XH, Zinc finger protein 451 is a novel Smad corepressor in transforming growth factor-β signaling. J Biol Chem 289, 2072–2083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, Studer L, Mark W, Patel DJ, Massagué J, A poised chromatin platform for TGF-β access to master regulators. Cell 147, 1511–1524 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS, Recruitment of TIF1γ to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell 43, 85–96 (2011). [DOI] [PubMed] [Google Scholar]
- 223.Liu D, Black BL, Derynck R, TGF-β inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev 15, 2950–2966 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Liu D, Kang JS, Derynck R, TGF-β-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J 23, 1557–1566 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, Xing Y, Carstens RP, Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol Cell Biol 36, 1704–1719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pradella D, Naro C, Sette C, Ghigna C, EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 16, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, Saitoh M, TGF-β drives epithelial-mesenchymal transition through δEF1-mediated downregulation of ESRP. Oncogene 31, 3190–3201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tripathi V, Sixt KM, Gao S, Xu X, Huang J, Weigert R, Zhou M, Zhang YE, Direct regulation of alternative splicing by SMAD3 through PCBP1 is essential to the tumor-promoting role of TGF-β. Mol Cell 64, 549–564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shen J, Hung MC, Signaling-mediated regulation of microRNA processing. Cancer Res 75, 783–791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Janakiraman H, House RP, Gangaraju VK, Diehl JA, Howe PH, Palanisamy V, The long (lncRNA) and short (miRNA) of It: TGFβ-mediated control of RNA-binding proteins and noncoding RNAs. Mol Cancer Res 16, 567–579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ahn SM, Cha JY, Kim J, Kim D, Trang HT, Kim YM, Cho YH, Park D, Hong S, Smad3 regulates E-cadherin via miRNA-200 pathway. Oncogene 31, 3051–3059 (2012). [DOI] [PubMed] [Google Scholar]
- 232.Davis BN, Hilyard AC, Lagna G, Hata A, SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56–61 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A, Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell 39, 373–384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH, A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 (2014). [DOI] [PubMed] [Google Scholar]
- 235.Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, Kadiwala J, Hubner NC, de Los Mozos IR, Sadee C, Lenaerts AS, Nakanoh S, Grandy R, Farnell E, Ule J, Stunnenberg HG, Mendjan S, Vallier L, The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature 555, 256–259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Itoh S, Landström M, Hermansson A, Itoh F, Heldin C-H, Heldin NE, ten Dijke P, Transforming growth factor β1 induces nuclear export of inhibitory Smad7. J Biol Chem 273, 29195–29201 (1998). [DOI] [PubMed] [Google Scholar]
- 237.Itoh F, Asao H, Sugamura K, Heldin CH, ten Dijke P, Itoh S, Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads. EMBO J 20, 4132–4142 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hanyu A, Ishidou Y, Ebisawa T, Shimanuki T, Imamura T, Miyazono K, The N domain of Smad7 is essential for specific inhibition of transforming growth factor-β signaling. J Cell Biol 155, 1017–1027 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mochizuki T, Miyazaki H, Hara T, Furuya T, Imamura T, Watabe T, Miyazono K, Roles for the MH2 domain of Smad7 in the specific inhibition of transforming growth factor-β superfamily signaling. J Biol Chem 279, 31568–31574 (2004). [DOI] [PubMed] [Google Scholar]
- 240.Kamiya Y, Miyazono K, Miyazawa K, Smad7 inhibits transforming growth factor-β family type I receptors through two distinct modes of interaction. J Biol Chem 285, 30804–30813 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hata A, Lagna G, Massagué J, Hemmati-Brivanlou A, Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev 12, 186–197 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhang S, Fei T, Zhang L, Zhang R, Chen F, Ning Y, Han Y, Feng XH, Meng A, Chen YG, Smad7 antagonizes transforming growth factor β signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol 27, 4488–4499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lin X, Liang YY, Sun B, Liang M, Shi Y, Brunicardi FC, Feng X-H, Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol Cell Biol 23, 9081–9093 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bai S, Cao X, A nuclear antagonistic mechanism of inhibitory Smads in transforming growth factor-β signaling. J Biol Chem 277, 4176–4182 (2002). [DOI] [PubMed] [Google Scholar]
- 245.Yan X, Pan J, Xiong W, Cheng M, Sun Y, Zhang S, Chen Y, Yin Yang 1 (YY1) synergizes with Smad7 to inhibit TGF-β signaling in the nucleus. Sci China Life Sci 57, 128–136 (2014). [DOI] [PubMed] [Google Scholar]
- 246.Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng X-H, PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell 125, 915–928 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Duan X, Liang YY, Feng XH, Lin X, Protein serine/threonine phosphatase PPM1A dephosphorylates Smad1 in the bone morphogenetic protein signaling pathway. J Biol Chem 281, 36526–36532 (2006). [DOI] [PubMed] [Google Scholar]
- 248.Yu J, Pan L, Qin X, Chen H, Xu Y, Chen Y, Tang H, MTMR4 attenuates transforming growth factor β (TGFβ) signaling by dephosphorylating R-Smads in endosomes. J Biol Chem 285, 8454–8462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zhao Y, Xiao M, Sun B, Zhang Z, Shen T, Duan X, Yu PB, Feng XH, Lin X, C-terminal domain (CTD) small phosphatase-like 2 modulates the canonical bone morphogenetic protein (BMP) signaling and mesenchymal differentiation via Smad dephosphorylation. J Biol Chem 289, 26441–26450 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sapkota G, Knockaert M, Alarcon C, Montalvo E, Brivanlou AH, Massagué J, Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-β pathways. J Biol Chem 281, 40412–40419 (2006). [DOI] [PubMed] [Google Scholar]
- 251.Wrighton KH, Willis D, Long J, Liu F, Lin X, Feng X-H, Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-β signaling. J Biol Chem 281, 38365–38375 (2006). [DOI] [PubMed] [Google Scholar]
- 252.Lönn P, Moren A, Raja E, Dahl M, Moustakas A, Regulating the stability of TGFβ receptors and Smads. Cell Res 19, 21–35 (2009). [DOI] [PubMed] [Google Scholar]
- 253.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH, A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400, 687–693 (1999). [DOI] [PubMed] [Google Scholar]
- 254.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R, Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A 98, 974–979 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K, Miyazono K, Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol Biol Cell 12, 1431–1443 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Xin H, Xu X, Li L, Ning H, Rong Y, Shang Y, Wang Y, Fu XY, Chang Z, CHIP controls the sensitivity of transforming growth factor-β signaling by modulating the basal level of Smad3 through ubiquitin-mediated degradation. J Biol Chem 280, 20842–20850 (2005). [DOI] [PubMed] [Google Scholar]
- 257.Mavrakis KJ, Andrew RL, Lee KL, Petropoulou C, Dixon JE, Navaratnam N, Norris DP, Episkopou V, Arkadia enhances Nodal/TGF-β signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol 5, e67 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Liang M, Liang YY, Wrighton K, Ungermannova D, Wang XP, Brunicardi FC, Liu X, Feng X-H, Lin X, Ubiquitination and proteolysis of cancer-derived Smad4 mutants by SCFSkp2. Mol Cell Biol 24, 7524–7537 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massagué J, Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139, 757–769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Aragon E, Goerner N, Zaromytidou AI, Xi Q, Escobedo A, Massagué J, Macias MJ, A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev 25, 1275–1288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang XF, Axin and GSK3-β control Smad3 protein stability and modulate TGF-β signaling. Genes Dev 22, 106–120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Swatek KN, Komander D, Ubiquitin modifications. Cell Res 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Rosonina E, Akhter A, Dou Y, Babu J, Sri Theivakadadcham VS, Regulation of transcription factors by sumoylation. Transcription 8, 220–231 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Bai Y, Yang C, Hu K, Elly C, Liu YC, Itch E3 ligase-mediated regulation of TGF-β signaling by modulating Smad2 phosphorylation. Mol Cell 15, 825–831 (2004). [DOI] [PubMed] [Google Scholar]
- 265.Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, Enzo E, Moro S, Polo S, Dupont S, Cordenonsi M, Piccolo S, USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol 13, 1368–1375 (2011). [DOI] [PubMed] [Google Scholar]
- 266.Tang LY, Yamashita M, Coussens NP, Tang Y, Wang X, Li C, Deng CX, Cheng SY, Zhang YE, Ablation of Smurf2 reveals an inhibition in TGF-β signalling through multiple mono-ubiquitination of Smad3. EMBO J 30, 4777–4789 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Moren A, Hellman U, Inada Y, Imamura T, Heldin CH, Moustakas A, Differential ubiquitination defines the functional status of the tumor suppressor Smad4. J Biol Chem 278, 33571–33582 (2003). [DOI] [PubMed] [Google Scholar]
- 268.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S, FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 (2009). [DOI] [PubMed] [Google Scholar]
- 269.Zhang X, Zhang J, Bauer A, Zhang L, Selinger DW, Lu CX, ten Dijke P, Fine-tuning BMP7 signalling in adipogenesis by UBE2O/E2–230K-mediated monoubiquitination of SMAD6. EMBO J 32, 996–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Imoto S, Sugiyama K, Muromoto R, Sato N, Yamamoto T, Matsuda T, Regulation of transforming growth factor-β signaling by protein inhibitor of activated STAT, PIASy through Smad3. J Biol Chem 278, 34253–34258 (2003). [DOI] [PubMed] [Google Scholar]
- 271.Liang M, Melchior F, Feng XH, Lin X, Regulation of Smad4 sumoylation and transforming growth factor-β signaling by protein inhibitor of activated STAT1. J Biol Chem 279, 22857–22865 (2004). [DOI] [PubMed] [Google Scholar]
- 272.Lönn P, van der Heide LP, Dahl M, Hellman U, Heldin C-H, Moustakas A, PARP-1 attenuates Smad-mediated transcription. Mol Cell 40, 521–532 (2010). [DOI] [PubMed] [Google Scholar]
- 273.Simonsson M, Heldin CH, Ericsson J, Grönroos E, The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem 280, 21797–21803 (2005). [DOI] [PubMed] [Google Scholar]
- 274.Gronroos E, Hellman U, Heldin CH, Ericsson J, Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell 10, 483–493 (2002). [DOI] [PubMed] [Google Scholar]
- 275.Kume S, Haneda M, Kanasaki K, Sugimoto T, Araki S, Isshiki K, Isono M, Uzu T, Guarente L, Kashiwagi A, Koya D, SIRT1 inhibits transforming growth factor β-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. J Biol Chem 282, 151–158 (2007). [DOI] [PubMed] [Google Scholar]
- 276.Elkouris M, Kontaki H, Stavropoulos A, Antonoglou A, Nikolaou KC, Samiotaki M, Szantai E, Saviolaki D, Brown PJ, Sideras P, Panayotou G, Talianidis I, SET9-mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep 15, 2733–2744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A, HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem 283, 33437–33446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL, Crystal RG, de Herreros AG, Moustakas A, Pettersson RF, Fuxe J, A SNAIL1-SMAD¾ transcriptional repressor complex promotes TGF-β mediated epithelial-mesenchymal transition. Nat Cell Biol 11, 943–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lamouille S, Xu J, Derynck R, Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Germain S, Howell M, Esslemont GM, Hill CS, Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev 14, 435–451 (2000). [PMC free article] [PubMed] [Google Scholar]
- 281.Kunwar PS, Zimmerman S, Bennett JT, Chen Y, Whitman M, Schier AF, Mixer/Bon and FoxH1/Sur have overlapping and divergent roles in Nodal signaling and mesendoderm induction. Dev 130, 5589–5599 (2003). [DOI] [PubMed] [Google Scholar]
- 282.Hernandez-Hernandez JM, Garcia-Gonzalez EG, Brun CE, Rudnicki MA, The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin Cell Dev Biol 72, 10–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zhang YE, Non-Smad signaling pathways of the TGF-β family. Cold Spring Harb Perspect Biol 9, a022129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zuo W, Chen YG, Specific activation of mitogen-activated protein kinase by transforming growth factor-β receptors in lipid rafts is required for epithelial cell plasticity. Mol Biol Cell 20, 1020–1029 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Meyer C, Godoy P, Bachmann A, Liu Y, Barzan D, Ilkavets I, Maier P, Herskind C, Hengstler JG, Dooley S, Distinct role of endocytosis for Smad and non-Smad TGF-β signaling regulation in hepatocytes. J Hepatol 55, 369–378 (2011). [DOI] [PubMed] [Google Scholar]
- 286.Yan Z, Winawer S, Friedman E, Two different signal transduction pathways can be activated by transforming growth factor β1 in epithelial cells. J Biol Chem 269, 13231–13237 (1994). [PubMed] [Google Scholar]
- 287.Lai CF, Cheng SL, Signal transductions induced by bone morphogenetic protein-2 and transforming growth factor-β in normal human osteoblastic cells. J Biol Chem 277, 15514–15522 (2002). [DOI] [PubMed] [Google Scholar]
- 288.McKay MM, Morrison DK, Integrating signals from RTKs to ERK/MAPK. Oncogene 26, 3113–3121 (2007). [DOI] [PubMed] [Google Scholar]
- 289.Mulder KM, Morris SL, Activation of p21ras by transforming growth factor β in epithelial cells. J Biol Chem 267, 5029–5031 (1992). [PubMed] [Google Scholar]
- 290.Chang SF, Chang TK, Peng HH, Yeh YT, Lee DY, Yeh CR, Zhou J, Cheng CK, Chang CA, Chiu JJ, BMP-4 induction of arrest and differentiation of osteoblast-like cells via p21 CIP1 and p27 KIP1 regulation. Mol Endocrinol 23, 1827–1838 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Muthusamy BP, Budi EH, Katsuno Y, Lee MK, Smith SM, Mirza AM, Akhurst RJ, Derynck R, ShcA protects against epithelial-mesenchymal transition through compartmentalized inhibition of TGF-β-induced Smad activation. PLoS Biol 13, e1002325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Derynck R, Muthusamy BP, Saeteurn KY, Signaling pathway cooperation in TGF-β-induced epithelial-mesenchymal transition. Curr Opin Cell Biol 31, 56–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Engel ME, McDonnell MA, Law BK, Moses HL, Interdependent SMAD and JNK signaling in transforming growth factor-β-mediated transcription. J Biol Chem 274, 37413–37420 (1999). [DOI] [PubMed] [Google Scholar]
- 294.Hocevar BA, Brown TL, Howe PH, TGF-β induces fibronectin synthesis through a c-Jun N-terminal kinase-dependent, Smad4-independent pathway. EMBO J 18, 1345–1356 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Hanafusa H, Ninomiya-Tsuji J, Masuyama N, Nishita M, Fujisawa J, Shibuya H, Matsumoto K, Nishida E, Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-β-induced gene expression. J Biol Chem 274, 27161–27167 (1999). [DOI] [PubMed] [Google Scholar]
- 296.Yu L, Hébert MC, Zhang YE, TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J 21, 3749–3759 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K, Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270, 2008–2011 (1995). [DOI] [PubMed] [Google Scholar]
- 298.Sakurai H, Targeting of TAK1 in inflammatory disorders and cancer. Trends Pharmacol Sci 33, 522–530 (2012). [DOI] [PubMed] [Google Scholar]
- 299.Sorrentino A, Thakur N, Grimsby S, Marcusson A, von Bulow V, Schuster N, Zhang S, Heldin C-H, Landström M, The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol 10, 1199–1207 (2008). [DOI] [PubMed] [Google Scholar]
- 300.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE, TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-β. Mol Cell 31, 918–924 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Zhang L, Zhou F, Garcia de Vinuesa A, de Kruijf EM, Mesker WE, Hui L, Drabsch Y, Li Y, Bauer A, Rousseau A, Sheppard KA, Mickanin C, Kuppen PJ, Lu CX, ten Dijke P, TRAF4 promotes TGF-β receptor signaling and drives breast cancer metastasis. Mol Cell 51, 559–572 (2013). [DOI] [PubMed] [Google Scholar]
- 302.Wu H, Arron JR, TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays 25, 1096–1105 (2003). [DOI] [PubMed] [Google Scholar]
- 303.Hamidi A, von Bulow V, Hamidi R, Winssinger N, Barluenga S, Heldin CH, Landström M, Polyubiquitination of transforming growth factor β (TGFβ)-associated kinase 1 mediates nuclear factor-κB activation in response to different inflammatory stimuli. J Biol Chem 287, 123–133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB, TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J Mol Biol 326, 105–115 (2003). [DOI] [PubMed] [Google Scholar]
- 305.Edlund S, Bu S, Schuster N, Aspenström P, Heuchel R, Heldin NE, ten Dijke P, Heldin C-H, Landström M, Transforming growth factor-β1 (TGF-β)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-β-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell 14, 529–544 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kimura N, Matsuo R, Shibuya H, Nakashima K, Taga T, BMP2-induced apoptosis is mediated by activation of the TAK1-p38 kinase pathway that is negatively regulated by Smad6. J Biol Chem 275, 17647–17652 (2000). [DOI] [PubMed] [Google Scholar]
- 307.Jung SM, Lee JH, Park J, Oh YS, Lee SK, Park JS, Lee YS, Kim JH, Lee JY, Bae YS, Koo SH, Kim SJ, Park SH, Smad6 inhibits non-canonical TGF-β1 signalling by recruiting the deubiquitinase A20 to TRAF6. Nat Commun 4, 2562 (2013). [DOI] [PubMed] [Google Scholar]
- 308.Ha H, Kwak HB, Lee SK, Na DS, Rudd CE, Lee ZH, Kim HH, Membrane rafts play a crucial role in receptor activator of nuclear factor κB signaling and osteoclast function. J Biol Chem 278, 18573–18580 (2003). [DOI] [PubMed] [Google Scholar]
- 309.Oakley FD, Smith RL, Engelhardt JF, Lipid rafts and caveolin-1 coordinate interleukin-1β (IL-1β)-dependent activation of κ by controlling endocytosis of Nox2 and IL-1β receptor 1 from the plasma membrane. J Biol Chem 284, 33255–33264 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Sundar R, Gudey SK, Heldin CH, Landström M, TRAF6 promotes TGFβ-induced invasion and cell-cycle regulation via Lys63-linked polyubiquitination of Lys178 in TGFβ type I receptor. Cell Cycle 14, 554–565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT, The PI3K pathway in human disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Saxton RA, Sabatini DM , mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 313.Lamouille S, Derynck R, Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol 178, 437–451 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Gamell C, Osses N, Bartrons R, Ruckle T, Camps M, Rosa JL, Ventura F, BMP2 induction of actin cytoskeleton reorganization and cell migration requires PI3-kinase and Cdc42 activity. J Cell Sci 121, 3960–3970 (2008). [DOI] [PubMed] [Google Scholar]
- 315.Yi JY, Shin I, Arteaga CL, Type I transforming growth factor β receptor binds to and activates phosphatidylinositol 3-kinase. J Biol Chem 280, 10870–10876 (2005). [DOI] [PubMed] [Google Scholar]
- 316.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, Hur L, Grabiner BC, Lin X, Darnay BG, Lin HK, The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hamidi A, Song J, Thakur N, Itoh S, Marcusson A, Bergh A, Heldin CH, Landström M, TGF-β promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85α. Sci Signal 10, eaal4186 (2017). [DOI] [PubMed] [Google Scholar]
- 318.Wang H, Fang R, Wang XF, Zhang F, Chen DY, Zhou B, Wang HS, Cai SH, Du J, Stabilization of Snail through AKT/GSK-3β signaling pathway is required for TNF-α-induced epithelial-mesenchymal transition in prostate cancer PC3 cells. Eur J Pharmacol 714, 48–55 (2013). [DOI] [PubMed] [Google Scholar]
- 319.Zhang X, Tang N, Hadden TJ, Rishi AK, Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta 1813, 1978–1986 (2011). [DOI] [PubMed] [Google Scholar]
- 320.Seoane J, Le HV, Shen L, Anderson SA, Massagué J, Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223 (2004). [DOI] [PubMed] [Google Scholar]
- 321.Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R, TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci 125, 1259–1273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Remy I, Montmarquette A, Michnick SW, PKB/Akt modulates TGF-β signalling through a direct interaction with Smad3. Nat Cell Biol 6, 358–365 (2004). [DOI] [PubMed] [Google Scholar]
- 323.Conery AR, Cao Y, Thompson EA, Townsend CM Jr., Ko TC, Luo K, Akt interacts directly with Smad3 to regulate the sensitivity to TGF-β induced apoptosis. Nat Cell Biol 6, 366–372 (2004). [DOI] [PubMed] [Google Scholar]
- 324.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massagué J, Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25, 441–454 (2007). [DOI] [PubMed] [Google Scholar]
- 325.Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T, Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284, 479–482 (1999). [DOI] [PubMed] [Google Scholar]
- 326.Yamamoto T, Matsuda T, Muraguchi A, Miyazono K, Kawabata M, Cross-talk between IL-6 and TGF-β signaling in hepatoma cells. FEBS Lett 492, 247–253 (2001). [DOI] [PubMed] [Google Scholar]
- 327.Cocolakis E, Dai M, Drevet L, Ho J, Haines E, Ali S, Lebrun JJ, Smad signaling antagonizes STAT5-mediated gene transcription and mammary epithelial cell differentiation. J Biol Chem 283, 1293–1307 (2008). [DOI] [PubMed] [Google Scholar]
- 328.Wang G, Yu Y, Sun C, Liu T, Liang T, Zhan L, Lin X, Feng XH, STAT3 selectively interacts with Smad3 to antagonize TGF-β signalling. Oncogene 35, 4388–4398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Liu Y, Liu H, Meyer C, Li J, Nadalin S, Konigsrainer A, Weng H, Dooley S, ten Dijke P, Transforming growth factor-β (TGF-β)-mediated connective tissue growth factor (CTGF) expression in hepatic stellate cells requires Stat3 signaling activation. J Biol Chem 288, 30708–30719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Dees C, Tomcik M, Palumbo-Zerr K, Distler A, Beyer C, Lang V, Horn A, Zerr P, Zwerina J, Gelse K, Distler O, Schett G, Distler JH, JAK-2 as a novel mediator of the profibrotic effects of transforming growth factor β in systemic sclerosis. Arthritis Rheum 64, 3006–3015 (2012). [DOI] [PubMed] [Google Scholar]
- 331.Tang LY, Heller M, Meng Z, Yu LR, Tang Y, Zhou M, Zhang YE, Transforming growth factor-β (TGF-β) directly activates the JAK1-STAT3 axis to induce hepatic fibrosis in coordination with the SMAD pathway. J Biol Chem 292, 4302–4312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL, Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell 12, 27–36 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Edlund S, Landström M, Heldin C-H, Aspenström P, Transforming growth factor-β-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell 13, 902–914 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Wang YK, Yu X, Cohen DM, Wozniak MA, Yang MT, Gao L, Eyckmans J, Chen CS, Bone morphogenetic protein-2-induced signaling and osteogenesis is regulated by cell shape, RhoA/ROCK, and cytoskeletal tension. Stem Cells Dev 21, 1176–1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL, Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 307, 1603–1609 (2005). [DOI] [PubMed] [Google Scholar]
- 336.Wilkes MC, Murphy SJ, Garamszegi N, Leof EB, Cell-type-specific activation of PAK2 by transforming growth factor β independent of Smad2 and Smad3. Mol Cell Biol 23, 8878–8889 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Foletta VC, Lim MA, Soosairajah J, Kelly AP, Stanley EG, Shannon M, He W, Das S, Massague J, Bernard O, Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J Cell Biol 162, 1089–1098 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Lee-Hoeflich ST, Causing CG, Podkowa M, Zhao X, Wrana JL, Attisano L, Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO J 23, 4792–4801 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Shen X, Li J, Hu PP, Waddell D, Zhang J, Wang XF, The activity of guanine exchange factor NET1 is essential for transforming growth factor-β-mediated stress fiber formation. J Biol Chem 276, 15362–15368 (2001). [DOI] [PubMed] [Google Scholar]
- 340.Wang L, Zhu Y, Sharma K, Transforming growth factor-β1 stimulates protein kinase A in mesangial cells. J Biol Chem 273, 8522–8527 (1998). [DOI] [PubMed] [Google Scholar]
- 341.Daniels CE, Wilkes MC, Edens M, Kottom TJ, Murphy SJ, Limper AH, Leof EB, Imatinib mesylate inhibits the profibrogenic activity of TGF-β and prevents bleomycin-mediated lung fibrosis. J Clin Invest 114, 1308–1316 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Kua HY, Liu H, Leong WF, Li L, Jia D, Ma G, Hu Y, Wang X, Chau JF, Chen YG, Mishina Y, Boast S, Yeh J, Xia L, Chen GQ, He L, Goff SP, Li B, c-Abl promotes osteoblast expansion by differentially regulating canonical and non-canonical BMP pathways and p16INK4a expression. Nat Cell Biol 14, 727–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.D’Souza RC, Knittle AM, Nagaraj N, van Dinther M, Choudhary C, ten Dijke P, Mann M, Sharma K, Time-resolved dissection of early phosphoproteome and ensuing proteome changes in response to TGF-β. Sci Signal 7, rs5 (2014). [DOI] [PubMed] [Google Scholar]