Abstract
Alzheimer’s disease (AD) is characterized by beta-amyloid (Aβ) accumulation, phosphorylated tau (p-tau) formation, hyper-activation of glial cells, and neuronal loss. The mechanisms of AD pathogenesis, however, remain poorly understood partially due to the lack of relevant human models that can comprehensively recapitulate multistage-intercellular interactions in human AD brains. Here, we present a new 3D human AD tri-culture model by using neurons, astrocytes, and microglia in a 3D microfluidic platform (3D hNeuroGliAD). Our model provided key representative AD features: Aβ aggregation, p-tau accumulation and neuroinflammatory activity. In particular, the model mirrored microglial recruitment, neurotoxic activities such as axonal cleavage and NO release damaging AD neurons and astrocytes. Our model will serve to facilitate the development of more precise human brain models for basic mechanistic studies in neural-glial interactions and drug discovery.
Keywords: 3D human brain culture model, 3D brain chip, 3D organoid, Alzheimer’s disease, amyloid beta, phosphorylated tau, microfluidics, microglia, astrocyte, neuroinflammation, axotomy, neuronal loss
Introduction
Alzheimer’s disease (AD) is the leading cause of age-related neurodegeneration affecting over 5.2 million people in the United States alone1. Our knowledge regarding mechanisms underlying the AD pathogenesis has greatly improved in the last decades, but still there is no cure1. Moreover, many unanswered questions have remained regarding the pathogenic cascade of AD including reactive gliosis and the related neuronal damages2–4. Recent advances in cell reprograming allow the study of human neurons derived from AD patient fibroblasts5–14. These fibroblast-derived neurons share the genetic material of AD patients and provide an excellent tool to recapitulate the pathogenic cascades of AD in human brain environment10–14. Indeed, familial/sporadic AD neurons derived from human induced pluripotent stem cells (iPSCs) exhibited relatively high levels of Aβ40 and phosphorylated-tau (p-tau) compared to controls14. Furthermore, we previously reported a unique 3D culture model of AD using human neural progenitor cells overexpressing APP and PSEN1 with human familial AD (FAD) mutations15,16. When seeded in a novel 3D culture system, these cells demonstrated robust extracellular aggregates of Aβ (Aβ-plaque-like) and Aβ-induced pathological tau aggregation (NFT-like)15–18. While this 3D model system successfully recapitulate key pathological hallmarks of AD, it did not contain a neuroinflammatory component, which is the essential pathological events observed in human AD patients and AD mouse models.
The next challenge is the generation of a human AD culture model that includes neuroinflammation, a key component of neurodegenerative disorders commonly induced through activation of microglia and astrocytes. The sustained activation of microglia results in a chronic neuroinflammatory response and increased production of proinflammatory cytokines, such as TNF-α and IL-1β19. Of relevance, recent genome-wide association studies have uncovered several risk-associated genes in the development of sporadic AD. Most of these risk genes are either expressed by microglia or associated with their reactivity, including CD3320, BIN121, CR122, TREM223, and CLU24. As such, there is a pressing need for the development of organotypic-models to detail cellular mechanisms and recapitulate the complex neural-glial interactions in AD.
Here, we present an engineered model of neuron (Neu)-astrocyte (AC)-microglia (MG) interaction in AD environment (Neu+AC+MG AD) that enables the study of human microglia recruitment, neuroinflammatory response and neuron/astrocyte damages. To achieve this, we built a human tri-culture model system in which neuron-astrocyte differentiated first, and then added adult microglia cells at different stages of AD. This new 3D human AD tri-culture model system demonstrated Aβ aggregation and p-tau formation along with increased chemokines/cytokines including CCL2, IL8, TNF-α and IFN-γ. Notably, 3D Neu+AC AD culture induced microglia recruitment and led to marked neuron/astrocyte loss. Together, these results suggest that our new 3D human AD tri-culture system provide a valid model to investigate complex neuroinflammatory molecular mechanisms underlying AD pathology and may contribute to finding new therapeutic targets.
Results
Generation of human AD tri-culture model (Neu+AC+MG AD) composed of neuron, astrocyte and microglia using a microfluidic platform.
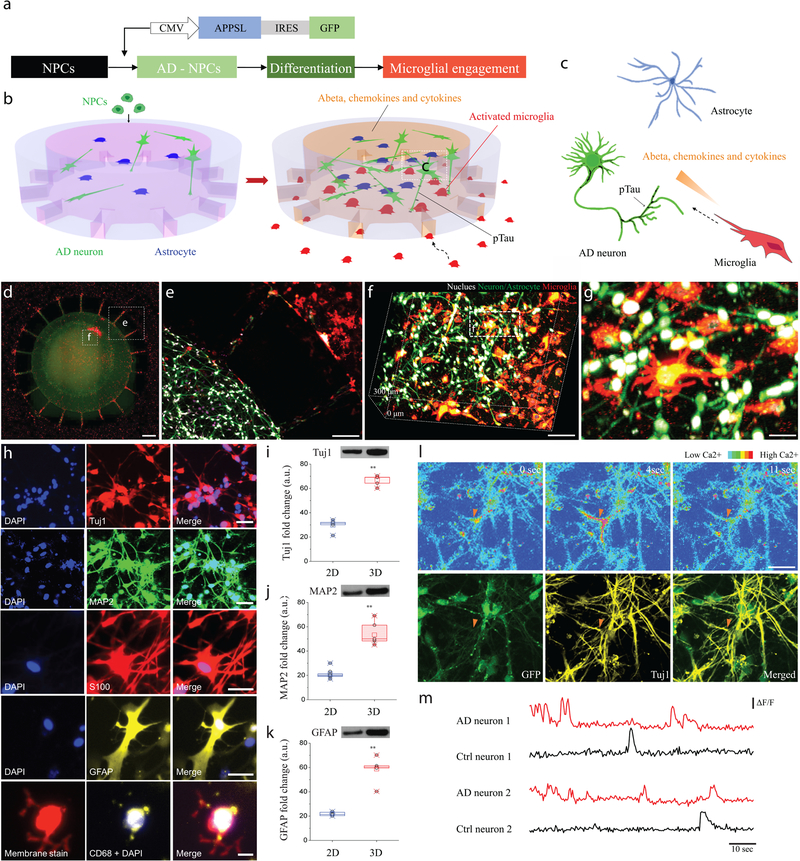
The AD brain platform used here consists of a microfluidic device containing two chambers mimicking the in vivo AD environment to observe interactions between neurons/astrocytes and microglia. The central chamber was loaded with the 3D AD neuron/astrocyte differentiated cells while the angular chamber was loaded with adult microglia (Fig. 1a–c). Human neural progenitor cells (hNPCs) that produce high levels of Aβ were developed through overexpression of a variant of the human beta-amyloid precursor protein (APP) containing FAD mutations with both K670N/M671L (Swedish) and V717I (London) FAD mutations (APPSL) as previously published (Supplementary Fig. 1)15. These transduced cells with control-GFP (neuron+astrocyte or Neu+AC) or APPSL-GFP (AD neuron+astrocyte or Neu+AC AD) lentiviral particles were enriched on the basis of GFP signals through fluorescence-activated cell sorting (FACS). Representative fluorescence microphotographs of differentiated ReN cells showed stable expression for more than 3 weeks (control-GFP: Neu+AC and APPSL-GFP: Neu+AC AD, Supplementary Fig. 1). The central and angular chambers were linked by migration channels (10 × 50 × 500 μm in height x width x length) forming gradients of soluble factors. The long and thin migration channels prevented inadvertent entrance of inactivated microglia through mechanical constraints. Using this platform, we observed the migration of microglial cells, at single cell resolution, in real-time, for extended periods (Fig. 1d). Human neural progenitor cells, differentiated into neuron and astrocyte (green) were successfully tri-cultured with adult microglia (red) (Figure 1d–g). These observations demonstrated that our 3D microfluidic device enabled the formation of physiologically relevant human brain tissue-mimetic 3D structures (Supplementary Fig.2).
Figure 1. Construction of a 3D organotypic human AD culture model (3D NeuroGliAD: 3D Neu+AC+MG AD): a tri-culturing system of AD neurons, astrocytes differentiated from human neural progenitor cells (hNPCs), and human adult microglia in a 3D microfluidic platform.
(a) Schematic of differentiation of NPCs to AD Neuron/astrocyte and microglial engagement. Schematics describe multicellular 3D layouts in (b) a microfluidic human AD culture model and (c) a human AD brain tissue. (d) Fluorescent Microphotographs show the layout of human AD neuron+astrocyte (green) in a central chamber (CC) and microglia (red) in an angular chamber (AC). (e) Microglia are recruited across micro-channels between the CC and the AC by soluble factors from the AD culture cells. (f-g) Representative confocal microphotographs in the CC highlight the physiological 3D cellular engagement of neurons (green), astrocyte (green), and microglia (red) with nuclear staining (white). (h) Immunofluorescent microphotographs validate the differentiated neurons (blue: nucleus) with class III beta-tubulin Tuj1 (red) and MAP2 (green), astrocytes (blue: nucleus) with S100 (red) and GFAP (yellow), and the recruited microglia (blue: nucleus, red: pre-stained membrane) with CD68 (yellow). (i-k) Western blots assay demonstrates the increased expression of neuronal (Tuj1; left; df: t=9.105, df=5.24, R squared = 0.9405, F = 41.68, p=0.0009, ndevice= 20 from 3 week 1:1 thick culture condition; MAP2; middle; df: t=25.64, df=7.526, R squared = 0.9887, F = 1.67, p=0.0027, ndevice= 20 from 3 week 1:1 thick culture condition) and astrocyte markers (GFAP; right; df: t=12.58, df=7.828, R squared = 0.9529, F = 1.349, p=0.0019, ndevice= 20 from 3 week 1:1 thick culture condition) of the 3D culture system compared to a 2D culture after 3-week of differentiation. Data are normalized to undifferentiated hNPCs. All experiments were repeated ≥ 3 times. (l) Time-lapse microphotographs show calcium signaling with Cal590 fluorescent dyes from firing neurons (top, red triangle) and are compared with immunostained neuronal marker of Tuj1 in yellow and membrane in green (bottom). (m) Normalized values (ΔF/F) from neurons representative for control and the AD model after 3-week of differentiation indicate functionally connected neuronal activities All experiments were repeated ≥3 times: unpaired, two-sided Student’s t-test; **P < 0.01; All parameters are presented as the mean ± SEM. Scale bars: 250 μm in (d, e), 100 μm in (f), 40 μm in (g), 40 μm for Tuj1 and MAP2, 20 μm for S100, GFAP and CD68 in (h), 50 μm in (l), respectively.
To demonstrate hNPCs differentiation in 3D cultures (Supplementary Video 1&2), fixed cells were analyzed by immunofluorescence for the presence of neuronal/astrocyte markers. As previously reported, human hNPCs robustly expressed class III beta-tubulin (Tuj1), microtubule-associated protein 2 (MAP2), glial fibrillary acidic protein (GFAP) and calcium-binding protein (S100, S100A6 and S100β) in week 3 (Fig. 1h, Supplementary Fig.3–4)15. Activated microglia, as demonstrated by CD68 expression, were also observed in tri-culture conditions in the 3D Neu+AC+MG AD culture model (Fig. 1h). Neuronal (Tuj1, MAP2) and glial (GFAP) markers in undifferentiated and 3-week differentiated AD ReN cells were quantified by Western blot analysis (Fig. 1i–k). Western blot analysis for neuronal markers Tuj1 and MAP2, and the astrocytic marker GFAP demonstrated that neuronal and astrocyte differentiation of NPCs was significantly promoted by 2.1-fold, 2.9-fold and 2.3-fold, respectively, in 3D culture in microfluidic devices compared to 2D cultures. These differences may reflect the ability of 3D environments to promote differentiation and/or maturation of neural progenitor cells providing a high surface area for growth25,26.
We next characterized action potential-evoked calcium dynamics in individual neurons (Fig. 1l) filled with Cal-59027. The recorded individual neurons were immunoassayed with the neuronal marker Tuj1 to confirm that the calcium dynamics was from neurons. The high-magnification microphotographs of the soma of stained neurons displayed characteristic ring-shape appearance arising from a lower fluorescence level in the nucleus compared with the surrounding cytoplasm (Supplementary Fig. 5). We analyzed the spontaneous activity of these neurons by combining calcium activation, neuronal marker, fast-time lapse imaging (50 frames/time) and found that single-action potential-evoked calcium transients were discernable with a good signal-to-noise ratio (Fig. 1m & Supplementary Video. 3)27. Interestingly, 3D Neu+AC AD showed a significant increase of neuronal activity levels could be observed when compared with week-matched control (3D Neu+AC) in the 3-week-old cells. Furthermore, the fraction of neurons that were hyperactive (>6 transients per 60 seconds) was markedly larger in 9-week-old 3D Neu+AC AD than in 3-week-old 3D Neu+AC AD, while the fraction of silent neurons (0 transients per 30 seconds) was not different between 3D Neu+AC AD at week 3 and week 9 (Supplementary Fig. 6 & Video. 4–5). Taken together, these data clearly demonstrate the generation of functional neuron/astrocyte cell models in our 3D microfluidic culture system.
Increased soluble Aβ, inflammatory cytokines/chemokines and phosphorylated tau (p-tau) in the 3D Neu+AC AD model
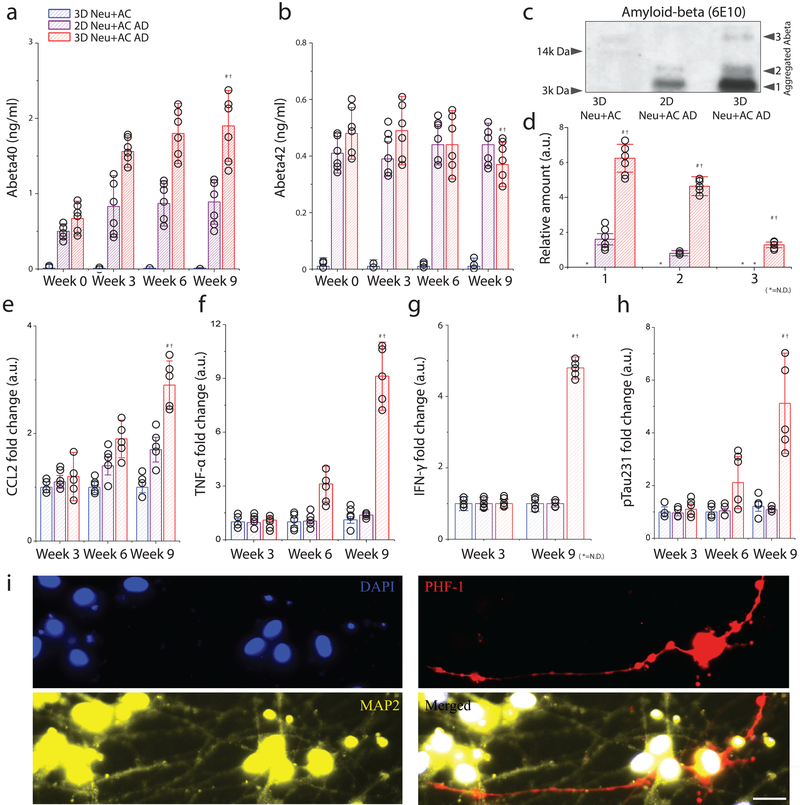
The levels of soluble 40-amino acid and 42-amino acid Aβ isoforms (Aβ40 and Aβ42) were determined in conditioned media collected after 3, 6, and 9-weeks of differentiation in either 2D or 3D cultures of control neuron/astrocyte (Neu+AC) or AD neuron/astrocyte (Neu+AC AD) cells. Aβ40 (10.2-fold and 1.7-fold) and Aβ42 (8.2-fold and 1.1-fold) levels were compared to the control 3D Neu+AC and 2D Neu+AC AD (Fig. 2a and b) at week 3. Furthermore, at week 9, 3D Neu+AC AD was associated with significantly higher accumulations of Aβ40; 78-fold and 2.8-fold compared to levels observed in 3D Neu+AC and 2D Neu+AC AD, respectively. Interestingly, the soluble Aβ42week9/ Aβ42week0 ratio in 3D Neu+AC AD was significantly decreased (≈ 20%), indicating that the 3D culture conditions may increase the aggregation of Aβ42 (Fig. 2a and b, Supplementary Fig.7&8). This observation was further validated through Western blot analysis of the various forms of aggregated Aβ in whole-lysates (Fig. 2c and 2d) with increased aggregated forms of Aβ in 3D Neu+AC AD28. These observations are consistent with an accelerated Aβ accumulation due to limited diffusion in 3D culture conditions and diffusion of AD neuron-derived Aβ into the media in 2D cultures as previously reported15,16.
Figure 2. Recapitulation of pathological AD signatures: amyloid beta, phosphorylated tau, and IFN-γ in the 3D human AD neuron model (3D Neu+AC AD).
(a) Soluble Aβ 40 and (b) Aβ 42 were measured after 0-, 3-, 6-, and 9-week of differentiation in a 3D control (3D Neu+AC; blue), a 2D AD model (2D Neu+AC AD; purple), and a 3D AD model (3D Neu+AC AD; red), respectively. ndevice= 6 from 3 week 1:1 thick culture condition (c) Western blot analysis confirms the presence of SDS-resistant Aβ multimeric forms in 3D Neu+AC AD. All experiments were repeated ≥3 times (d) Quantification of elevated levels of multiple forms shown in Fig. 3c (F(DFn,DFd) = F(2,9) = 530.6, SS= 6.438). Noticeable increases of (e) CCL2 (R square =0.9804, F (2, 12) = 300.6), (f) TNF-α were observed in the 3D Neu+AC AD model (R square=0.9953, F (2, 12) = 1271). (g) IFN-γ (R square=0.7584, F (2, 12) = 18.84), (h) soluble p-tau (pSer231 tau, R square=0.9235, F (2, 12) = 72.39), and (i) p-tau accumulation (PHF-1; red) in neuronal cell body and neurites (green) were observed only after 9 weeks in the 3D Neu+AC AD model. All experiments were repeated ≥3 times: One-way ANOVA with Tukey–Kramer test; Statistical significance is denoted by # 3D Neu+AC week9 vs 3D Neu+AC ADweek9, † 2D Neu+AC+MG ADweek9 vs 3D Neu+AC+MG ADweek9, P < 0.001 with numberdevice = 5 in (a-h). Scale bar: 25 μm in (i).
To determine whether our 3D Neu+AC AD secretes CCL2 (MCP-1), a chemokine up-regulated in AD brains, we compared the level of CCL2 in various stages of the AD culture models29. We found that 3D Neu+AC AD (1800 pg/ml) showed 2.9-fold and 1.3-fold more CCL2 secretion in conditioned media compared to 3D Neu+AC (620 pg/ml) and 2D Neu+AC AD (1384 pg/ml), respectively (Fig.2e). To further explore pathology development in 3D cultured AD neurons/astrocyte, chemokine and cytokine secretion by 3D Neu+AC, 2D Neu+AC AD, and 3D Neu+AC AD cells was determined. First, we evaluated TNF-α levels in the culture medium and found TNF-α levels increased by 2.8-fold and by 8.2-fold in week 6 (61 pg/ml) and week 9 (180 pg/ml) in 3D Neu+AC AD, respectively (Fig.2f), while moderate increases were observed in 2D Neu+AC AD. Surprisingly, IFN-γ was exclusively secreted by 3D Neu+AC AD cells (9.2 pg/ml) but not detected in the other conditions tested (Fig.2g).
Regarding tau alterations30, ELISA analysis of soluble phosphorylated tau (pSer231 tau; phosphorylated at Serine 231) in 3D Neu+AC AD cells showed that pSer231 tau levels increased by 2.2-fold and 5.1-fold at weeks 6 (271 pg/ml) and 9 (630 pg/ml), respectively, compared to 3D Neu+AC (Fig.2h). The presence of p-tau phosphorylation at Ser396/Ser404 and Ser202/Thr206 was also confirmed by immunofluorescent staining using p-tau antibody PHF-1 and AT8, respectively, in both neuritic and soma bodies of 3D Neu+AC AD cells (Fig.2i and Supplementary Fig.9).
3D Neu+AC AD model induced microglial migration, phenotype changes and secretion of pro-inflammatory factors
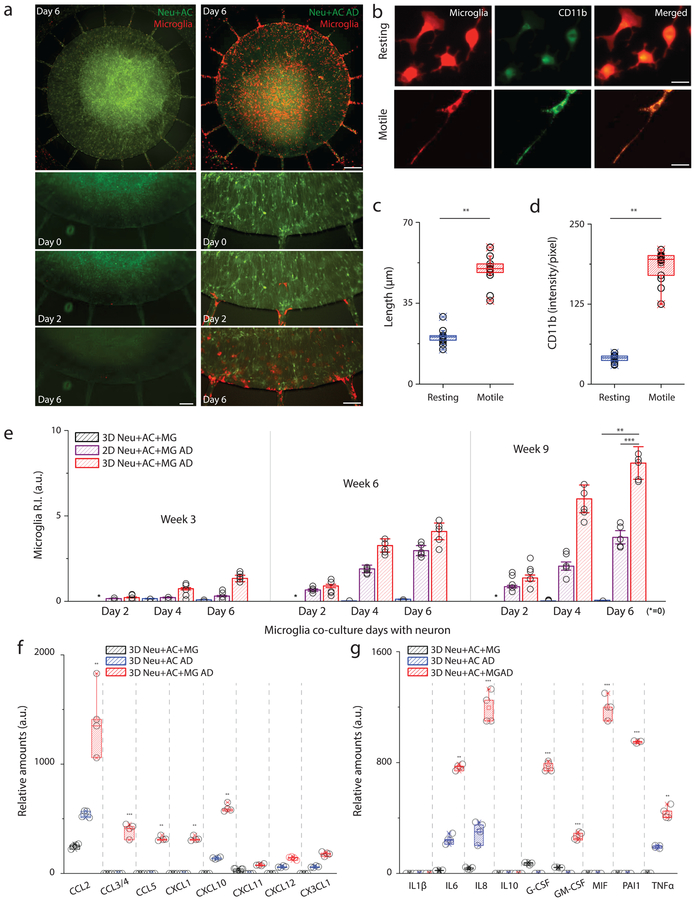
To test the hypothesis that neurons/astrocytes in AD environments induce activation/recruitment of microglial cells, we plated microglial cells in the angular chamber at Day 0 of week 3, week 6, and week 9. To characterize the migration and phenotype changes of microglia, individual microglial cells were monitored for seven days using time-lapse imaging microscopy (Fig. 3a). In the first 24 hours, microglia in the angular chamber showed widely branched filopodia in all directions, a morphology typically associated with ‘resting’ microglia. Furthermore, most of the cells remained stationary during that period. Morphological changes and microglial migration towards the central chamber seeded with 3D Neu+AC AD was observed following cell seeding after 48 hrs (Fig. 3b). As determined by image analysis, microglial cells became elongated with significantly increased cell length and microglial cell body area (Fig. 3b and c). Furthermore, exposure of microglia to 3D Neu+AC AD induced up-regulation of microglial activation markers such as CD11b (Fig. 3d). The migration rate of microglial cells in the 3D Neu+AC+MG AD environment was significantly higher compared with those in the other conditions tested: 3D Neu+AC+MG or 2D Neu+AC+MG AD. The microglial recruitment index (recruitment index, R.I., Supplementary Fig.10) was 8.1 ± 0.78% in 3D Neu+AC+MG AD cells compared with 4.2 ± 2.3% in 2D Neu+AC+MG AD cells and 0.9 ± 1.2% in 3D Neu+AC+MG cells (Fig. 3e). Interestingly, neutralization of CCL2 chemokine signaling using an anti-CCL2 antibody reduced the ability of microglial to migrate in response to Neu+AC AD co-culture conditions (Supplementary Fig. 11). Other factors, such as ATP31 (Supplementary Fig. 12) may also contribute to microglial recruitment in our system. Notably, in the 3D Neu+AC+MG AD model, the aged cultures (week 9) led to the recruitment of more microglia cells as compared to early stage cultures. 3D Neu+AC+MG AD and 2D Neu+AC+MG AD culture platforms had comparable effects on maximal microglial migration speed (5.0 ± 0.91 vs. 4. 3 ± 0.81, data not shown). The observed speed of migration was similar with that previously observed in AD environments with soluble and bound Aβ conditions15
Figure 3. Activation of microglial inflammation: morphogenesis, marker expression, recruitment and inflammatory mediator release.
(a) Time-lapse microphotographs show microglial (red) recruitment by AD neuron+astrocyte (Neu+AC AD, green) compare to a control (Neu+AC, green). (b) Microglia are ramified (resting) on day 0 in the annular chamber (AC), become elongated (motile) on day 2, and migrate along the micro-channels toward the central chamber (CC). All experiments were repeated ≥3 times. Both the (c) length and (d) level of CD11b expression increase in the motile microglia (F (1, 14) = 564.7), All experiments were repeated ≥3 times: unpaired, two-sided Student’s t-test, ncells= 10 at Day4 culture condition. (e) Tri-culture in the 3D AD model (red, 3D Neu+AC+MG AD) leads to a dramatic increase in microglia recruitment as measured by the total number of microglia accumulated in the CC compared to the 3D control (blue, 3D Neu+AC+MG) and the 2D AD model (purple, 2D Neu+AC+MG AD) (R-square=0.9745, F (2, 12) = 228.9), All experiments were repeated ≥3 times: unpaired, Two-way ANOVA tests, ndevice= 5 at each culture condition. The 3D Neu+AC+MG AD also produced discernable amounts of (f) chemokines and (g) pro-inflammatory soluble factors (F (7, 21) = 115.6). All experiments were repeated ≥3 times, Two-way ANOVA tests, ndevice= 4 at each culture condition; Statistical significance is denoted by **P < 0.001, ***P < 0.0001 with numberdevice = 5 in (a), numbercell = 100 in (b, c, d), numberdevice = 5 in (e, f, g), respectively. n.s. stands for non-significant. All parameters are presented as the mean ± SEM. Scale bars: 25 μm (left) and 200 μm (right) in (a), 10 μm in (b), respectively.
Next, we set out to identify the cytokines and chemokines that regulate the microglial migration/activation in our 3D AD models. For this purpose, we first measured the secreted cytokine/chemokine levels in 3D Neu+AC and 3D Neu+AC AD models without microglial cells. Conditioned cell culture medium were collected from the week 9 models and levels of 36 cytokines were analyzed. Interestingly, protein levels of CCL2, CXCL10 and CX3CL1 were significantly increased in 3D Neu+AC AD compared to the control 3D Neu+AC model (Fig. 3f). In contrast, no changes in CCL3/4, CCL5 and CXCL1 expression were found in either culture model. Next, we assessed how inflammatory signaling was impacted by adding microglial cells to the 3D Neu+AC AD model. AD-related neuroinflammatory responses, such as chemokine release (Fig. 3f) and pro-inflammatory factor secretion (Fig. 3g, Supplementary Fig.13), were generally increased by the addition of microglial cells. Indeed, IL-6, IL-8 and TNF-α concentrations were elevated by 2.2-, 2.7- and 1.3-fold, respectively, compared to 3D Neu+AC AD alone. In addition, activated microglia induced the release of multiple chemokines including CCL2 (2.1-fold), CCL5 (26-fold), CXCL10 (2.6-fold), CXCL12 (1.2-fold) in 3D Neu+AC+MG AD compared to the 3D Neu+AC AD. We also observed the unique production of the leukocyte growth factors GM-CSF and G-CSF, which typically are not produced by neurons. MIF and PAI-1, important biomarkers of tau hyper-phosphorylation in late-stage AD mouse model, were greatly increased in 3D Neu+AC+MG AD (Fig 3g)31. Secretion of inflammtory cytokines such as IL-1β and IL-17 and anti-inflammatory markers such as IL-1RA, IL-10, and TGF-ß was very limited or below detectable levels. These data suggest that microglia elicit broad inflammatory signaling in response to our 3D Neu+AC AD model.
Recruited microglia are toxic to AD neurons and astrocytes
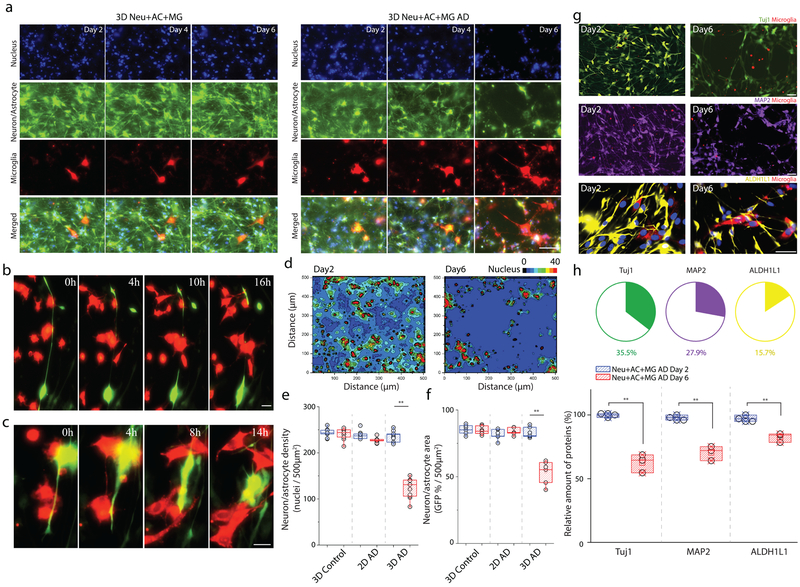
Next, we assessed the extent of cellular damage mediated by microglia in our 3D Neu+AC+MG AD culture model. On day 2, after adding microglial cells to 9-week-differentiated 3D Neu+AC AD system, we observed morphological changes of microglial cells in co-localized with 3D Neu+AC AD cells expressing GFP signals (Fig. 4a, Supplementary Video. 6). After day 4, we observed slight decreases in neuron/astrocyte surface area, including cell body and axon, when co-localized with microglia (Fig. 4a). Axonal damages were also observed with co-localized microglial cells (Fig. 4b, Supplementary Fig. 14 & Video.7). Time-lapse imaging also indicated that AD neurons undergo changes in cell morphology, including retraction of the neurites toward the main cell bodies (Fig.4c, Supplementary Video.8). To quantitatively analyzed the cellular damages, we next performed image analysis by converting nucleus stained images into a dimension matrix to determine nucleus density in tri-culture conditions with microglia via heat map contour (Fig.4d, Supplementary Fig. 15). At day 2, cells exhibited relatively moderate density across ROI regions. However, at day 6, a marked decrease of cellular density was observed around nearby microglia cells (Fig. 4e). Indeed, after day 6 of tri-culture with microglial cells, there was a 21% reduction in neuron+astrocyte survival counted by nucleus (Fig. 4e) and a marked reduction of about 37% in neuron+astrocyte surface area (Fig. 4f) when tri-cultured with microglia compared to 9-week-old 3D Neu+AC AD only (Supplementary Fig. 15). To elucidate and quantify how microglia influence the induction of neuronal and/or astrocyte loss, we analyzed 9-week-old 3D Neu+AC+MG AD at day 2 and day 6 post- microglial cell recruitment using immunostaining and immunoblotting to compare and quantify the loss of each cell type (Fig. 4g and h). TUJ1/MAP2 (neurons) and ALDH1L1 (astrocyte) markers were used for the quantification of the neuron and astrocyte loss, respectively. Our data indicated broad losses of neurons (Tuj1: 35.5%, MAP2: 27.9%) and astrocytes (15.7%) across all 3D Neu+AC+MG AD model systems tested, whereas the number of microglia (CD68-positive) remained the same (Fig. 5e). Our results, therefore, suggest that the addition of microglial cells promotes distinct changes in the number and morphology of neurons/astrocytes, which is consistent with the neuropathological findings previously demonstrated in the 5xfAD mouse model32.
Figure 4. Exacerbated neuronal damage through the interactions with reactive microglial cells.
Time-lapse microphotographs highlight the neurotoxic interactions among recruited microglia (red) and AD neurons/astrocytes (green) with nucleus staining (blue) in a real time: (a) gradual depletion of neurons/astrocytes from day 2 to day 6, (b) axonal cleavage, (c) and retraction of neuronal neurite (16 hours of observation) in microglial co-localizing regions of interest (ROI) of 3D Neu+AC+MG AD model compared to a control of 3D Neu+AC+MG. (d) Localized neuronal damage proximate to the recruited microglia is visualized on a contour heat map. All experiments were repeated ≥3 times Neuronal loss is quantified by measuring (e) the number of nucleus, (f) GFP-overlaying surface area of neurons/astrocytes in the 3D Neu+AC (3D control), the 2D Neu+AC AD (2D AD), and 3D Neu+AC AD (3D AD) in the absence (blue) and the presence of microglia (red). nimages= 20 & ndevice= 5 at each culture condition (g) Cell types are identified by immunostaining specific markers (Tuj1: green, MAP2: purple, ALDH1L1: yellow) and (h) the amounts of surviving neurons/astrocytes are compared between day 2 and day 6 after microglial recruitment (F (1, 6) = 95.12). Two-way ANOVA with Tukey–Kramer test; Statistical significance is denoted by **, P<0.0001 with numberimages = 20 in (d-f), numberdevice = 5 in (d-f, h), respectively. All parameters are presented as the mean ± SEM. Scale bars: 80 μm in (a), 10 μm in (b), 10 μm in (c), 80 μm (Tuj1 and MAP2) 10 μm (GFAP) in (g), respectively.
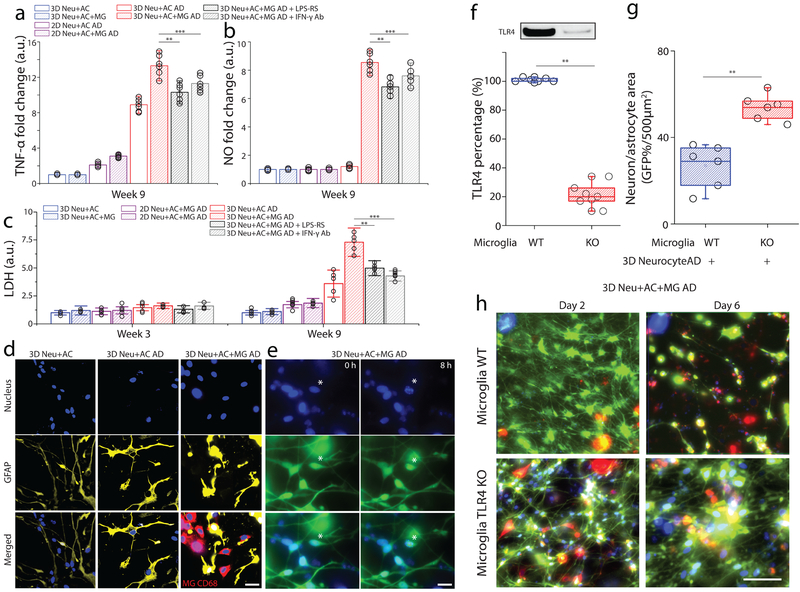
Figure 5. Assessment of neurotoxic neuron-glia interactions mediated by TLR4 and IFN-γ receptor.
In a tri-culture, 3D Neu+AC+MG AD express elevated levels of (a) TNF-α and (b) NO compared to the control (3D Neu+AC+MG and 2D/3D Neu+AC AD) and treatment using either TLR4 antagonist or blocking IFN-γ antibody reduced the concentrations of TNF-α and NO (R-square=0.9024, F(DFn, DFd)=1.061(4, 10), F= 23.11), All experiments were repeated ≥3 times, Two-way ANOVA. (c) In the 3D Neu+AC+MG AD after 9-week of differentiation, neurotoxic interactions were mimicked by LDH concentrations (R-square=0.7104, F (1, 14) = 15.65); the damage was attenuated through inhibition using a TLR4 antagonist and a blocking IFN-γ antibody (f, R-square=0.8954, F (1, 14) = 22.61). (d) Immunostaining of the astrocyte marker GFAP (yellow) and microglial CD68 (red) demonstrated the activation of astrocytes and microglia in 3D Neu+AC AD and 3D Neu+AC+MG AD compared to 3D Neu+AC. (e) Apoptotic cells with fragmented nuclei observed after 8h in 3D Neu+AC+MG AD. (f) Immunoblot and quantification of proteins isolated from WT and TLR4 knock-down microglia. Fold change was determined by normalizing TLR4 band intensity to Actin band (R-square=0.8934, F (1, 3) = 21.65). Reduced neuron+astrocyte loss was (g) measured with GFP-overlaying surface area (R-square=0.8240, F (1, 3) = 19.25) and (h) visualized with fluorescent microphotographs in 3D Neu+AC AD with TLR4-KO microglia. All experiments were repeated ≥3 times. Two-way ANOVA test; Statistical significance is denoted by # IFN-γ vs IFN-γ + Aβ, † Aβ vs IFN-γ + Aβ, #* IFN-γ + Aβ vs IFN-γ + Aβ + TLR4 Ab, †* IFN-γ + Aβ vs IFN-γ + Aβ + LPS-RS P<0.0002 and **p<0.001, ***p<0.0001with numberwell = 5 in (a, b), numberdevice = 5 in (c, d, e), respectively. All parameters are presented as the mean ± SEM. Scale bars: 10 μm in (d), 5 μm in (i) 40 μm in (i), respectively.
Recruited microglia induced neuronal loss through a IFN-γ and TLR4-dependent mechanism
To test whether cytokines and chemokines contribute to neuron and astrocyte loss, we evaluated the contribution of IFN-γ and TLR4 on the production of neurotoxic mediators in our tri-culture system (Fig. 5a). TNF-α and NO concentrations were measured in the presence of either neutralizing or blocking antibodies to IFN-γ or TLR4, respectively33. Recruited microglia in the 3D Neu+AC+MG AD model exhibited a 2.3-fold increase in TNF-α concentrations compared to the 3D Neu+AC AD model without microglia (Fig. 5b). NO levels were also increased by 9.1-fold in the 3D Neu+AC+MG AD model, while not detectable in the 2D Neu+AC+MG AD model possibly due to lack of IFN-γ as in this system (Fig. 2g and 5c). Addition of either TLR4 or IFN-γ neutralizing antibodies reduced TNF-α and NO concentrations demonstrating that the neuron/astrocyte loss in 3D Neu+AC+MG AD is dependent on microglial activation. We also measured lactate dehydrogenase (LDH) release in conditioned media as a biochemical marker of cell death (Fig. 5d). Notably, incubation with the TLR4 antagonist (LPS-RS) significantly decreased LDH levels in the 3D Neu+AC+MG AD culture model (Fig. 5d). These data suggest that the microglia-induced neuronal loss in the 3D Neu+AC+MG AD model, occurs at least partially via IFN-γ− and TLR4 -dependent mechanisms, in agreement with the other models33.
We also observed increased GFAP signals and altered astrocyte morphology in the 3D Neu+AC AD and the Neu+AC+MG AD models, suggestive of astrogliosis (Fig. 5e). Both 3D Neu+AC AD and Neu+AC+MG AD induced astrogliosis in the week 9 model, as indicated by increased GFAP expression. Furthermore, astrocyte activation was concomitant with activation of microglia as indicated by increases in CD68 immunoreactivity and thickening of their processes indicative of a shift in state toward a more activated amoeboid morphology (Fig. 5e). Additionally, we observed that some of the neuronal loss was associated with the activation of apoptotic pathways as an observation of fragmented nuclei after 8h in 3D Neu+AC+MG AD (Fig. 5f).
Knocking down TLR4 expression provided protection against neurotoxic microglial activation
Since we observed a reduction in TNF-α and NO concentrations following the addition of TLR4 blocking antibodies, we hypothesized that TLR4 activity is required for microglial activation. Thus, we investigated whether selective knock-down of TLR4 receptor expression can similarly reduce the inflammatory status of microglia in our AD models. To test this, we used lentiviral particles to knock-down TLR4 in WT microglial cells (TLR4 KD) and plated these cells in the angular chamber. TLR4 KD microglial cells exhibited an 79% decrease in TLR4 compared to WT microglial cells (Fig. 5g). Knocking down TLR4 in microglial cells protected neuron and astrocyte loss (2.3-fold) compared to WT microglial cells in the 3D Neu+AC+MG AD systems.
Human iPSC-derived AD neurons/astrocytes replicated microglial recruitment, activation and neuron/astrocyte death in tri-culture model system
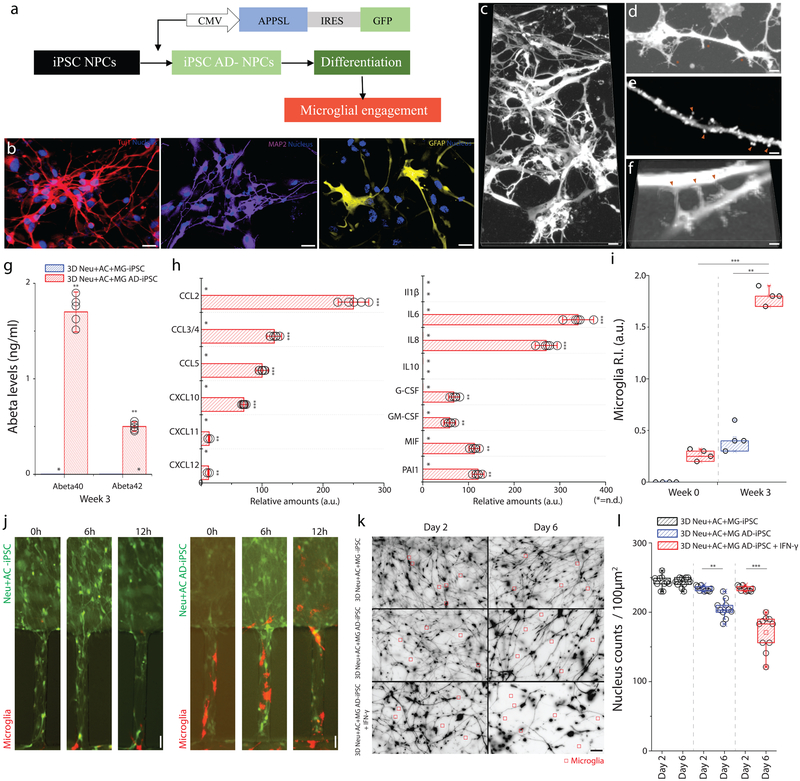
To confirm that the proposed mechanism can be replicated in other cultured human neuron/astrocyte, we newly generated human induced pluripotent stem cell (iPSC)-derived neural stem cells (NSC) expressing high levels of pathogenic Aβ species using the similar techniques used for the ReNcell VM-based 3D AD models (Fig. 6a). Human NSCs, stably expressing APPSL/GFP (AD-iPSC) or GFP alone (control iPSC), were plated and differentiated for 3 weeks under 3D culture conditions. In these culture conditions, we also confirmed that cells were differentiated into neurons and astrocytes, respectively, as determined by the Tuj1, MAP2 and GFAP immunofluorescence staining (Fig. 6b and c). To further examine the development of synapses-like structures, dendritic protrusions at week 0 and week 3 were compared (Fig. 6d and e). Especially, we imaged these protrusions making connections in between synapses-like structures in the volume (Fig. 6f). As expected, the 3D Neu+AC+MG AD-iPSC culture produced high levels of Aβ40 (1.75ng/ml) and Aβ42 (0.45ng/ml) (Fig. 6g). We also analyzed the cytokine secretion profile at week 3. Incubation of 3D Neu+AC AD-iPSC for 3 weeks led to increased cytokine/chemokine secretion. Indeed, the release of inflammation-related CCL2, CCL5, IL6, IL8 and MIF were upregulated following the addition of microglial cells (Fig. 6h). Interestingly, IL-1β, IL-6, IL-10, and TNF-α were not detected in the 3D Neu+AC AD-iPSC cultures without microglia (Fig. 6h). Importantly, microglial migration/recruitment was largely elevated in the 3D Neu+AC+MG AD-iPSC as compared to 3D Neu+AC+MG control-iPSC (Fig. 6i–j). We next treated 3D Neu+AC+MG AD-iPSC with IFN-γ and compared the neuron/astrocyte loss between AD and control cells3D Neu+AC+MG AD-iPSC showed significantly higher levels of neuron/astrocyte loss while the IFN-γ neutralizing antibody treatments protected neuron/astrocyte loss (Fig. 6k–l). These data further validated our findings on glial activation and the role of cytokines in 3D tri-culture models with iPSC-derived human neuron/astrocyte cells.
Figure 6. Neuron-glia interactions recapitulated with human iPSC-derived AD neurons/astrocytes.
(a) Schematics describe the procedure used to generate iPSC-AD NPCs and illustrate the plating. (b) Immunofluorescent microphotographs validate the differentiated neuron (blue: nucleus) with class III beta-tubulin Tuj1 (red) and MAP2 (purple), astrocytes (blue: nucleus) with GFAP (yellow) following 3-week of differentiation. (c) Representative microphotographs of differentiated neurons/astrocytes show (d) protrusion of dendritic spines, (e) synaptic boutons, and (f) connections. (g) 3D Neu+AC+MG AD-iPSC express elevated levels of Aβ40 and Aβ42 compared to the control (3D Neu+AC+MG -iPSC) (F (2, 4) = 125.6), All experiments were repeated ≥3 times. (h) The 3D Neu+AC+MG AD-iPSC also produces discernable amounts of chemokines (left) and pro-inflammatory soluble factors (right) (F (1, 22) = 321.3). (i-j) Tri-culture in 3D Neu+AC+MG AD-iPSC (red bars in i, right microphotographs in j) leads to a dramatic increase in microglia recruitment as measured by the total number of microglia accumulated in the CC compared to the 3D control (blue bars in i, left microscopic images in j) (R-square=0.7954, F (2, 12) = 199.7), All experiments were repeated ≥3 times: unpaired, Two-way ANOVA tests, ndevice= 5 at each culture condition. (k) Representative microscopic images show the reduced density of neurons/astrocytes tri-cultured with microglia (red square) after microglial engagement for 2 days and 6 days. (l) The amounts of surviving neurons/astrocytes are measured in 3D Neu+AC AD-iPSC treated with IFN-γ compared with untreated controls (3D Neu+AC+MG -iPSC, 3D Neu+AC AD-iPSC) ((R-square=0.9731, F (1, 10) = 17.45)). Two-way ANOVA test; Statistical significance is denoted by **p<0.001, ***p<0.0001 with numberdevice = 5 in (g, h, i). All parameters are presented as the mean ± SEM. Scale bars: 20 μm in (b, c, k), 5 μm in (d, e, f), 10 μm in (j), respectively.
In conclusion, we were able to recapitulate dynamic neural-glial interactions in a novel 3D human tri-culture (Neu+AC+MG) model system. This model was generated by differentiating genetically modified human neural progenitor cells into neurons and astrocytes, expressing AD pathological hallmarks: Aβ aggregation, p-tau formation, and chemokine/cytokines such as CCL2, TNF-α and IFN-γ. We achieved this by co-culturing on a microfluidic platform, and by tri-culturing human adult microglia in separated angular chambers to observe microglia recruitments with AD neuron and astrocytes. The relevance of this tri-culture model lies in the demonstration of physiologically relevant neural-glial interactions: microglial morphogenesis, recruitment, and the release of proinflammatory cytokines and leukocyte-chemokines regulated by AD neurons and astrocytes, and microglial neurotoxic activation contributing to neuron/astrocyte damage. These findings carry important implications for studies aimed at a deeper understanding of the gliosis and pathogenesis of AD and screens aimed at finding new therapeutic molecules for preventing and treating AD.
Discussion
Relevant in vitro models of human AD brain must be capable of recapitulating all three-pillars of AD pathology: Aβ accumulation, phosphorylated tau aggregation, and neuroinflammation. Human neurons derived from Alzheimer’s disease patients have shown elevated levels of toxic Aβ species and phosphorylated tau. However, current AD neuronal models do not include neuroinflammatory changes that are mediated by microglial cells. To overcome this limitation, we developed a tri-culture model system of neuron-astrocyte-microglia that can achieve microglial recruitment, secretion of pro-inflammatory cytokines/chemokines, and neuron/astrocyte loss. The generation of this in vitro human AD culture model system will provide more physiologically relevant system to address key pathological features in AD.
Derived from 3D culture platforms, the microfluidic model presented a more physiologically relevant brain environment characterized by increased expression of neuronal (Tuj1, MAP2) and astrocyte (GFAP) markers as compared two 2D systems, in agreement with previous reports15. Maturation of these neurons/astrocytes in 3D could result from several mechanisms, (i) materials whose properties closely mimic those of the in vivo extracellular matrix (ECM) have been shown to promote differentiation of progenitor cells into the mature phenotypes25, and/or (ii) the 3D environment provides a high surface area for growth and differentiation26. Importantly, in contrast to previously developed AD-derived iPSC that only showed early AD stage signatures, our 3D tri-culture model at week 9 mimics late stage AD markers i.e., robust increases in phosphorylated tau formation. Moreover, 3D tri-culture model secretes cytokines/chemokines such as TNF-α, IFN-γ and MCP-1. The clinical relevance of these findings is supported by a marked upregulation of TNF-α in APP AD transgenic mice and by an increased expression of IFN-γ in AD brain tissue. IFN-γ is released predominantly by Aβ activated immune cells34,33 and TNF-α activated astrocytes35–37 and bind to IFN-γ receptors that are abundant and functionally present on microglia38. Taken together, these findings provide evidence that the combination of neuron-astrocyte-microglia 3D cultures accelerates pathogenic cascades of AD, thus representing a suitable model for the study of AD mechanisms.
The role of chemokines in the recruitment of microglia in AD brain is not fully known. To identify the mechanism(s) of microglial accumulation into AD brain, we added adult microglia cells in a separate angular chamber connected to the pre-cultured neuronal-astrocyte central chamber. Microglial recruitment was significantly higher in the 3D Neu+AC+MG AD as compared to 3D control or 2D AD microenvironments. Although the higher recruitment in the 3D Neu+AC+MG AD neuron is likely due to a combination of many factors, three major elements contributed to the increase in microglia recruitment. First, Aβ accumulation affected the increase of microglia recruitment. In our previous study, we demonstrated that soluble Aβ served as a ‘recruiting signal’ for microglia39. 3D Neu+AC+MG AD was cultured for 9 weeks, thus, greater levels of Aβ were present in either 3D gel- or conditioned media. Second, 3D Neu+AC AD secreted key recruiting chemokines, such as CCL2/MCP-1, which is upregulated in human Alzheimer’s disease brains. CCL2/MCP-1 is produced mainly by cultured microglia29 and astrocytes40. Indeed, neutralizing anti- CCL2/MCP-1 antibodies significantly inhibited the migration of microglia. CXCL10 induced migration and a proinflammatory phenotype in cultured microglia and T cells41. Finally, in conditioned media from 6- and 9-week 3D Neu+AC+MG AD cultures showed increased ATP levels that likely promote microglial activation or chemotaxis42. Microglia also express several purinoceptors, including a Gi-coupled subtype that has been implicated in ATP- and ADP-mediated migration in vitro43. An increase of extracellular ATP levels during AD progression has been reported in a murine model of the disease44. These data highlight the relevance of our 3D Neu+AC+MG AD neuron model for the study of key factors driving microglia recruitment or the expression study of surface markers contributing to migration. Moreover, 3D Neu+AC AD-secreted Aβ and MCP-1 can both generate molecular gradients affecting microglial activation and alter their phenotype39. Changes in their phenotypes showed the trend for CD11b up-regulation and correlated more closely with measures of the “M1”-like phenotype45.
In addition to neurotoxic soluble factors, activated microglia have been shown to contribute to neuronal synapse engulfment by physical contact in AD mouse models, including C1q upregulation via the classical complement system46. In our 3D culture model, we observed physical interruptions between neurons and microglia i.e., cleaved axons and retraction of neurite with co-localized microglia. While we could not determine whether microglia regulate synapses in our model, this observation demonstrated that our model will allow the study of neuronal loss associated with microglia physical interactions.
It should be noted that an immortalized microglia cell line, transformed with simian virus 40 (SV40) T antigen, was used in our studies. Extending these studies using human iPSC-derived microglia as well as primary microglia will be helpful to confirm our findings. Future studies incorporating AD risk-associated innate immune genotypes, e.g. in CD33 and TREM2, in 3D mixed neural-glial cultures should be another interesting application.
In conclusion, our 3D AD culture model represents a significant improvement over current in vitro human AD models by adding inflammatory activity. Although physiologically relevant in vivo studies will ultimately be needed to confirm the clinical utility of our model, success of this 3D tri-culture modeling should foster a great understanding of the molecular mechanisms underlying the gliosis and pathogenesis, and more effective screening of therapies aimed at slowing, stopping or reversing neurodegeneration in AD.
Online Methods
Media and reagents of neural progenitor cells (NPCs)
ReN cell VM human neural progenitor cells (NPCs) were purchased from EMD Millipore (Billerica, MA, USA). The cells were plated onto BD Matrigel® (BD Biosciences, San Jose, CA, USA) coated T25 cell culture flasks (BD Biosciences, San Jose, CA, USA) and maintained in DMEM/F12 (Life Technologies, Grand Island, NY, USA) media supplemented with 2 mg heparin (StemCell Technologies, Vancouver, Canada), 2% (v/v) B27 neural supplement (Life Technologies, Grand Island, NY, USA), 20 mg EGF (Sigma-Aldrich, St Louis, MO, USA), 20 mg bFGF (Stemgent, Cambridge, MA, USA) and 1% (v/v) penicillin/streptomycin/amphotericin-B solution (Lonza, Hopkinton, MA, USA) in a CO2 cell culture incubator. Cell culture media were changed every 3 days until cells were confluent. For 2D neuron/astrocyte differentiation, the cells were plated onto Matrigel®-coated microfluidic devices with DMEM/F12 differentiation media supplemented with 2 mg heparin, 2% (v/v) B27 neural supplement, and 1% (v/v) penicillin/streptomycin/amphotericin-B solution without growth factors. One half volume of the differentiation media was changed every 3–4 days for 3–9 weeks. LPS-RS was purchased from InvivoGen (San Diego, CA). LPS-RS was resuspended in dimethylsulphoxide (DMSO; 0.1 mg in 10 ml) followed by 1:10 dilution in differentiation culture medium. This stock solution of 22.2 mM was then diluted for cell treatment experiments.
Viral infection of NPCs
To transduce the NPCs with the lentiviral constructs, 50–100 μl viral solution (1 × 106 TU ml−1, MOI=4) were added to 85% confluent proliferating ReN cells in 6-well dishes, incubated for 24 h, and washed three times to stop the infection. The expression of the infected genes was confirmed by GFP expression by fluorescence microscopy and western blot analysis.
3D cell cultures and differentiation of NPCs
For 3D cultures, BD Matrigel® (BD Biosciences) was mixed with the cells (10 × 106 cells per ml). The final cell concentration for the mixture was approximately 5 × 106 cells per ml (1:1 3D thick culture for confocal and westernblot) and 2 × 106 cells per ml (1:5 3D thin culture for immunostaining and recruitment). Ten microliter of cell mixtures were transferred into the microfluidic device using pre-chilled pipettes. The microfluidic devices were incubated for 1h at 37°C during which 3D gels (100–600 μm) formed and media were changed. The 3D-plated cells were differentiated for 3–9 weeks depending on the experiments; media was changed every 3–4 days. For drug treatments, differentiation media containing either 1mM LPS-RS diluted in DMSO or the concentration of DMSO were added to 3–9-week differentiated 3D AD neuron-microglia cells and the 3D cultures maintained for an additional 1 week.
Expansion, viral infection and differentiation of hiPSC-NPCs
Human iPSC derived neural progenitor cells (hiPSC-NPCs) were purchased from EMD Millipore (SCR131). The cells were plated onto BD Matrigel® (BD Biosciences, San Jose, CA, USA) coated 6 well plate (BD Biosciences, San Jose, CA, USA) and expanded in expansion medium (EMD Millipore, SCM004) supplemented with FGF-2 (20ng/ml) and L-Glutamine (2 mM) in a CO2 cell culture incubator. To transduce the hiPSCs with the lentiviral constructs, 50–100 μl viral solution (1 × 106 TU ml−1, MOI=4) were added to 85% confluent proliferating hiPSCs in 6-well dishes, incubated for 48 h, and washed three times to stop the infection. The expression of the infected genes was confirmed by GFP expression by fluorescence microscopy and followed by FACS sorting. To differentiate, hiPSC-NPCs were prepared approximately 10 × 106 cells per ml of Matrigel®. Ten microliter of cell mixtures were transferred into the microfluidic device using pre-chilled pipettes. The microfluidic devices were incubated for 1h at 37°C during which 3D gels (100–600 μm) formed and media were changed in differentiation media (EMD Millipore, SCM017). The 3D-plated cells were differentiated for 3 weeks; media was changed every 3–4 days.
FACS enrichment of the transduced NPCs and iPSC-NPCs
The infected ReN cells were washed with DPBS and then incubated with Accutase (Millipore) for 5 min. The cell pellets were resuspended in PBS supplemented with 2% serum replacement solution (Life Technologies) and 2% B27, and then passed through a cell strainer filter (70 mm Nylon, BD Biosciences). The cell concentrations were adjusted to 2 × 106 cells per ml and then enriched by using FACSAria cell sorter (Flow Cytometry Core Facility, Charlestown Navy Yard, MGH)). GFP channels were used to detect the expression of the transduced genes in the individual cells. The sorted/enriched cells were maintained in normal proliferation media.
Microfluidic Device Fabrication
Negative photoresists, SU-8 50 and SU-8 100 (MicroChem, Newton, MA, USA), were sequentially patterned using standard lithography on a 4” silicon wafer to create a mold for cell migration channels of 50 μm in height and chemokine compartments of 100 μm in height. A mixture of a base and a curing agent with a 10:1 weight ratio (SYLGARD 184 A/B, Dow corning, Midland, MI, USA) was poured onto the SU-8 mold and cured for one hour at room temperature under vacuum and, subsequently, cured for more than 3 hours in an oven at 80 °C. The cured poly dimethyl-siloxane (PDMS) replica was peeled off from the mold and holes were punched for fluid reservoirs. Arrayed holes were also laser-cut (Zing 24, Epilog Laser, Golden, CO, USA) into a thin PDMS membrane of 250 μm in thickness (HT 6240, Bisco Silicones, Elk Grove, IL, USA) and an acrylic plate of 6 mm in thickness. The machined membrane and the plate were glued together using uncured PDMS and incubated at 80 °C overnight. This assembly was irreversibly bonded first to the PDMS replica using oxygen plasma at 50 mW, 5 cm, for 30 seconds (PX-250, March Plasma Systems, Petersburg, FL, USA), and later to a glass-bottomed Uni-Well plate (MGB001-1-2-LG, Matrical Bioscience, Spokane, WA, USA). Immediately after the bonding, 10 μL of poly (l-lysine) solution (PLL, M.W. 70,000–150,000, 1.0 mg/mL, Sigma-Aldrich Co. LLC, St. Louis, MO, USA) was injected into each platform and incubated for 2 hours at a room temperature to promote cellular adhesion. PLL-treated surface was rinsed with autoclaved and 0.2 μm filtered water (AM9920, Life Technologies, Grand Island, NY, USA).
Calcium Imaging
To assess neural activity, cellular calcium dynamic was monitored using Rhod-2 (Life Technologies), a Ca2+ indicator. Neuron-cultured samples were incubated for 30 min at 37°C in artificial buffered cerebrospinal fluid (i.e., 25 mM NaHCO3, 25 mM D-glucose, 125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 1 mM MgCl2•6H2O, and 2 mM CaCl2•2H2O) supplemented with 2 M of OGB-1 diluted in pluronic F-127 20% solution/DMSO (Sigma). After incubation, the samples were washed twice with fresh artificial buffered cerebrospinal fluid and incubated for 30 min at 37°C. Fluorescence intensity dynamics was measured using time-lapse imaging (Texas red filter cube; 20X objective; Nikon microscope). at a 30ms frame rates. Raw intensity values were extracted using the Nikon software suite, and relative changes in fluorescence intensity to baseline ((F-F0)/F0) were regarded as neuronal calcium signals.
Microglia preparation
The immortalized human microglia-SV40 cell line derived from primary human microglia was purchased from Applied Biological Materials, Inc. (ABM, Inc.) and cultured in Prigrow III medium supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin in type I collagen-coated T25-flasks (ABM, Inc.). Before the experiment, cells were washed using medium without serum and the cell membrane was labeled with red fluorescent dye (PKH26PCL, Sigma-Aldrich). Briefly, after centrifugation (400 g for 5 minutes), the cells were re-suspended in 1 mL of Diluent C (G8278, Sigma-Aldrich) and immediately mixed with 4 μL of dye solution (PKH26PCL, Sigma-Aldrich). The cell/dye mixture was incubated at room temperature for 4 minutes and periodically mixed by pipetting to achieve a bright, uniform, and reproducible labeling. After the incubation, the staining was stopped by adding an equal volume (1 mL) of 1% BSA in PBS and incubating for 1 minute to remove excess dye. Unbound dye was washed by centrifugation and suspending cells in culture medium (106 cells/ml). Ten μL of the cell suspension was injected into each platform and 100 μL of a culturing medium was added into side and central extra wells. The loaded 3D micro-devices were then incubated at 37°C supplied with 5 % CO2.
Time-lapse imaging
After microglia loading, cells were recorded using time-lapse imaging using a fully automated Nikon TiE microscope with a heated incubator to 37°C and 5 % CO2 (10x magnification; Micro Device Instruments, Avon, MA, USA). To achieve accurate cell tracking, the maximum time resolution of acquisition was 1 frame per second.
ELISA
Aβ40 and Aβ42 levels were mainly measured by Human/Rat Aβ ELISA Kit from Wako (Osaka, Japan). The conditioned media from un- differentiated or differentiated ReN cells were collected and diluted by 1:10 or 1:100 with a dilution buffer provided by the manufacturer. A Synergy 2 ELISA plate reader (BioTek, Winooski, VT, USA) was used to quantify Aβ40 and Aβ42 ELISA signals.
IFN-γ (DIF50) secretion were quantified in supernatants by Human Quantikine ELISA Kits (R&D Systems, Oxon, UK) according to manufactures instructions.
Human cytokine array
The Human Cytokine Array (ARY005) kits were purchased and used to simultaneously detect relative expression levels of 36 human cytokines (R&D Systems, Oxon, UK). Conditioned medias from each sample, 0.5 ml respectively, were mixed with a cocktail of biotinylated detection antibodies. The mixture was then incubated with the Human Cytokine Array Panel A membrane. A wash step was needed to remove unbound material, and then Streptavidin-HRP and chemiluminescent detection reagents were added. Light was produced at each spot in proportion to the amount of analyte bound. The positive signals on developed films were identified by placing the transparency overlay on the ChemiDoc imaging system (Bio-Rad) and aligning it with the reference spots in three corners of each array. Pixel density was analyzed with ImageJ (Wayne Rasband, NIH).
Aβ preparation.
Soluble Aβ monomer was prepared by dissolving a synthesized clear human Aβ, 1–42 (4349-v, Peptide Institute, Inc., Japan) and a FITC-labeled human Aβ,1–42 (A-1119, rPeptide, Bogart, GA, USA) in DMSO at 1 mg/mL for a monomer form. Although Aβ pretreatment has generally been conducted in HFIP, because of recent reports of HFIP toxicity Aβ was directly dissolved in DMSO according to manufacturer’s recommendations (Peptide Institute, Inc., Japan). Indeed, the slow evaporation of HFIP may trigger the formation of Aβ oligomers, which could hasten fibril formation in combination with the humidity in air. The dissolved Aβ monomer solution was further diluted in PBS at 0.1 mg/mL and stored up to a week at 4°C or incubated in a shaker at 37°C to prepare oligomeric forms.
Immunostaining
For immunofluorescent stains, we rinsed the cells and 3D cultures twice with PBS (phosphate buffered saline). Cells were then fixed through a RT 15–30 min incubation in fresh 4% paraformaldehyde aqueous solution (157–4, ElectronMicroscopy Sciences) followed by rinsing twice with PBS. Cells were permeabilized through incubation in 0.1% Triton X-100 in PBST (phosphate buffered saline with 0.1% tween®20) for 15 minutes at RT. Cell on-specific binding was blocked through overnight incubation in 3% human serum albumin in PBST at 4 °C. After a 24-hours incubation with the primary antibody solutions at 4°C, the cells were washed five times. The following antibodies (and dilutions) were used: anti-p-tau antibody (1:40, AT8, Thermo Scientific, MN1020); anti-PHF (1:1000, A gift from P. Davies, Albert Einstein College of Medicine); anti-GFAP antibody (1:500, Neuromap, N206A/8), anti-MAP2 antibody (1:200, Cell Signaling Technology, 4542); anti-CD68 (1:100, Cell Signaling, 76437); anti-cd11b (1:100, Life Technologies, NB110–89474); anti-S100 (1:400, Abcam, ab868); anti-S100A6 (1:200, Cell Signaling, D3H3W); anti-S100B (1:500, Abcam, Ab218515); anti-ALDH1L1 (1:200, EMD Millipore, MABN495).
Western blot analysis
Protein lysates (15–75 μg) were resolved on 12% Bis-Tris or 4–12% gradient Bis/Tris gels (Life Technologies) and the proteins were transferred to nitrocellulose membranes (Bio-Rad). For amyloid-ß western blot analysis, the membranes were cross-linked with 0.5% glutaraldehyde solution before blocking. Western blot results were visualized using enhanced chemiluminescence (ECL). Signals were captured using ChemiDoc imaging system (Bio-Rad) and quantified using ImageJ software (NIH). The following primary antibodies were used (dilutions): anti- Aβ (1:100, 6e10; Signet); anti-Tuj1 (1:200, Abcam, ab24629); anti-MAP2 (1:200, Cell Signaling, 4542); anti-GFAP (1:100, Neuromap, N206A/8); anti-p-tau AT8 (1:30, Thermo Scientific, MN1020), p-tau PHF-1 (1:200, Abcam, ab66275), CD68 (1:200, BD Bioscience, 556059), CD11b (1:200, EMD Millipore, MM_NF-MABF515), anti-ALDH1L1 (1:200, EMD Millipore), Cy5 anti-mouse secondary antibody (1:400, Jackson Immunoresearch, 715-175-150).
Statistical analysis
All statistical analysis were performed by using Graphpad prism software. Either two-tailed Student’s t-test when comparing two groups/conditions or one-way ANOVA followed by post hoc test when comparing 3 or more groups/conditions were used. Data are expressed as means ± SEM, and a P-value ≤ 0.05 was considered significant. Special symbols (**, ***, #, †, #* and †*) to denote significant differences between two-group or three group analysis. Statistical analysis are detailed in figure legends. No statistical methods were used to predetermine sample sizes, and experiments were repeated to adequately reduce confidence intervals and avoid errors in statistical testing. Data collection and analysis were not performed blind to the conditions of the experiments. Randomization was not performed, and exclusions were not made.
Supplementary Material
Acknowledgements.
We appreciate Dr. Luisa Quinti (MGH) for sharing the preliminary results regarding 3D neuron/microglia co-culture system, Dr. Marc Busche (MGH) for helpful guidance of the calcium imaging and Dr. Se Hoon Choi (MGH) for helpful discussion regarding the data interpretation interpretation and Dr. You Jung Kang (UNCC) for critical reviewing our manuscript. This work was supported by the NIH/NIA (P01 AG015379 and RF1 AG048080 to DYK and RET; R01 AG014713 to D.Y.K.), Pioneering Funding Award funded by Cure Alzheimer’s Fund (CAF, H.C., D.Y.K., R.E.T.), BrightFocus Foundation (D.Y.K.), Duke Energy Special Initiatives funded by Charlotte Research Institute (CRI, H.C.) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1A6A3A03019848, J.P.).
Footnotes
Competing Financial Interests Statement
The authors declare no competing interests.
References
- 1.2016 Alzheimer’s disease facts and figures. Alzheimer’s & Dementia 12, 459–509 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Tanzi RE & Bertram L Twenty Years of the Alzheimer’s Disease Amyloid Hypothesis: A Genetic Perspective. Cell 120, 545–555 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Karran E & De Strooper B The amyloid cascade hypothesis: are we poised for success or failure? Journal of Neurochemistry 139, 237–252 (2016). [DOI] [PubMed] [Google Scholar]
- 4.A Armstrong R A critical analysis of the ‘amyloid cascade hypothesis’. Folia Neuropathologica 3, 211–225 (2014). [PubMed] [Google Scholar]
- 5.Paquet D et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Tubsuwan A et al. Generation of induced pluripotent stem cells (iPSCs) from an Alzheimer’s disease patient carrying a L150P mutation in PSEN-1. Stem Cell Research 16, 110–112 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Moore S et al. APP Metabolism Regulates Tau Proteostasis in Human Cerebral Cortex Neurons. Cell Reports 11, 689–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muratore CR et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Human Molecular Genetics 23, 3523–3536 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sproul AA et al. Characterization and Molecular Profiling of PSEN1 Familial Alzheimer’s Disease iPSC-Derived Neural Progenitors. PLOS ONE 9, e84547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo T et al. Modeling Alzheimer’s Disease with iPSCs Reveals Stress Phenotypes Associated with Intracellular Aβ and Differential Drug Responsiveness. Cell Stem Cell 12, 487–496 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Yagi T et al. Establishment of Induced Pluripotent Stem Cells from Centenarians for Neurodegenerative Disease Research. PLOS ONE 7, e41572 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch P et al. Presenilin-1 L166P Mutant Human Pluripotent Stem Cell–Derived Neurons Exhibit Partial Loss of γ-Secretase Activity in Endogenous Amyloid-β Generation. The American Journal of Pathology 180, 2404–2416 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Shi Y et al. A human stem cell model of early Alzheimer’s disease pathology in Down syndrome. Science Translational Medicine 4, 124ra29–124ra29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Israel MA et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 482, 216–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi SH et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YH et al. A 3D human neural cell culture system for modeling Alzheimer’s disease. Nat Protoc 10, 985–1006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Avanzo C et al. Alzheimer’s in 3D culture: Challenges and perspectives. Bioessays 37, 1139–1148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi SH, Kim YH, Quinti L, Tanzi RE & Kim DY 3D culture models of Alzheimer’s disease: a road map to a “cure-in-a-dish”. Mol Neurodegener 11, 75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heneka MT et al. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griciuc A et al. Alzheimer’s Disease Risk Gene CD33 Inhibits Microglial Uptake of Amyloid Beta. Neuron 78, 631–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butovsky O et al. Identification of a unique TGF-β–dependent molecular and functional signature in microglia. Nature Neuroscience 17, 131–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crehan H, Hardy J & Pocock J Blockage of CR1 prevents activation of rodent microglia. Neurobiol Dis 54, 139–149 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Guerreiro R et al. TREM2Variants in Alzheimer’s Disease. New England Journal of Medicine 368, 117–127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q, Li Y, Cyras C, Sanan DA & Cordell B Isolation and characterization of apolipoproteins from murine microglia. Identification of a low density lipoprotein-like apolipoprotein J-rich but E-poor spherical particle. Journal of Biological Chemistry 275, 31770–31777 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z-N et al. Layered hydrogels accelerate iPSC-derived neuronal maturation and reveal migration defects caused by MeCP2 dysfunction. Proceedings of the National Academy of Sciences 113, 3185–3190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han S et al. Three-dimensional extracellular matrix-mediated neural stem cell differentiation in a microfluidic device. Lab Chip 12, 2305–2308 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Tischbirek C, Birkner A, Jia H, Sakmann B & Konnerth A Deep two-photon brain imaging with a red-shifted fluorometric Ca2+ indicator. Proc Natl Acad Sci USA 112, 11377–11382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khoury El JB et al. CD36 Mediates the Innate Host Response to β-Amyloid. The Journal of Experimental Medicine 197, 1657–1666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khoury El J et al. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nature Medicine 13, 432–438 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Brion JP Neurofibrillary Tangles and Alzheimer’s Disease. European Neurology 40, 130–140 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Li S-Q et al. Deficiency of macrophage migration inhibitory factor attenuates tau hyperphosphorylation in mouse models of Alzheimer’s disease. J Neuroinflammation 12, 177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spangenberg EE et al. Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-β pathology. Brain 139, 1265–1281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papageorgiou IE et al. TLR4-activated microglia require IFN-γ to induce severe neuronal dysfunction and death in situ. Proceedings of the National Academy of Sciences 113, 212–217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Browne TC et al. IFN-γ Production by amyloid β-specific Th1 cells promotes microglial activation and increases plaque burden in a mouse model of Alzheimer’s disease. J. Immunol 190, 2241–2251 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Garwood CJ, Pooler AM, Atherton J, Hanger DP & Noble W Astrocytes are important mediators of Aβ-induced neurotoxicity and tau phosphorylation in primary culture. Cell Death & Disease 2, e167–e167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White JA, Manelli AM, Holmberg KH, Van Eldik LJ & LaDu MJ Differential effects of oligomeric and fibrillar amyloid-β1–42 on astrocyte-mediated inflammation. Neurobiol Dis 18, 459–465 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Xiao BG & Link H IFN-gamma production of adult rat astrocytes triggered by TNF-alpha. Neuroreport 9, 1487–1490 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Hashioka S, Klegeris A, Schwab C, Yu S & McGeer PL Differential expression of interferon-γ receptor on human glial cells in vivo and in vitro. Journal of Neuroimmunology 225, 91–99 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Cho H et al. Microfluidic Chemotaxis Platform for Differentiating the Roles of Soluble and Bound Amyloid-β on Microglial Accumulation. Scientific Reports 3, 169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smits HA et al. Amyloid-β-induced chemokine production in primary human macrophages and astrocytes. Journal of Neuroimmunology 127, 160–168 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Baruch K et al. Breaking immune tolerance by targeting Foxp3(+) regulatory T cells mitigates Alzheimer’s disease pathology. Nature Communications 6, 7967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes SE et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nature Neuroscience 9, 1512–1519 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Dou Y et al. Microglial migration mediated by ATP-induced ATP release from lysosomes. Cell Research 22, 1022–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez de Lara MJ & Pintor J Presence and release of ATP from the retina in an Alzheimer’s disease model. J. Alzheimers Dis. 43, 177–181 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Johansson JU et al. Suppression of Inflammation with Conditional Deletion of the Prostaglandin E2 EP2 Receptor in Macrophages and Brain Microglia. J Neurosci 33, 16016–16032 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong S et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.