Abstract
Array-based sensor ‘chemical nose/tongue’ platforms are inspired by the mammalian olfactory system. Multiple sensor elements in these devices selectively interact with target analytes, producing a distinct pattern of response and enabling analyte identification. This approach offers unique opportunities relative to ‘traditional’ highly specific sensor elements such as antibodies. Array-based sensors excel at distinguishing small changes in complex mixtures, and this capability is being leveraged for chemical biology studies and clinical pathology, enabled by a diverse toolkit of new molecular, bioconjugate and nanomaterial technologies. Innovation in the design and analysis of arrays provides a robust set of tools for advancing biomedical goals, including precision medicine.
Keywords: array-based sensing, biomedicine, cancer cell detection, protein detection, diagnosis
Graphical Abstract:
Array-based sensing provides new tools for biosensing.

1. Introduction
Sensors are an integral part of everyday life, monitoring our health and wellbeing and maintaining our safety. Chemical sensors are an important subclass that use recognition elements and transducers to detect and quantify important molecules. But beyond the latest smart technologies, perhaps the most powerful chemical-focused sensory tool that we rely upon to keep us safe and healthy is our olfactory system – smell.
A ‘chemical nose’ sensor is broadly defined as an array-based system that uses synthetic molecules and/or materials to mimic the mammalian olfactory systems.1 The synthetic model of the natural systems can be created and tuned to specific sensing challenges. In this review, we use the term ‘chemical nose’ to describe the array-based sensing approach. However, it is conceptually equivalent between ‘chemical nose’ and ‘chemical tongues or E-tongues’. Unlike specific sensing, the chemical nose works on the principle of selective binding between an analyte and an array of cross-reactive receptors to generate distinct responses for each analyte. The responses can then be read out and linked back to the analyte through pattern recognition. Therefore, the same chemical nose can detect multiple analytes with relatively few sensor elements, by returning multiple distinct patterns from the array.
A major difference between chemical nose sensing and more traditional specific sensing is that chemical nose uses the simultaneous interaction of multiple analytes with multiple sensor elements to recognize the overall changes to the make-up of complex mixtures, rather than identifying specific elements within them.2 Thus, this array-based approach is particularly powerful in sensing complex bioanalytes. The data-rich outputs of array-based sensing methods are becoming widely adopted by the analytical community, due to the increase in capabilities with statistical and cheminformatic techniques in analysis, as well as the recognition that many complex sensing challenges cannot be solved with conventional analytical tools.3
Array-based sensors have now been applied in a broad range of applications including explosives4 and volatile-organic compound (VOC) detection,5 environmental monitoring6 and anti-counterfeiting technologies.7 Several thorough reviews have been written, particularly by the groups of Walt and Suslick, and detail the history and chemical space of the technology.8,2a
In this review we focus on the emerging use of chemical noses in biology and medicine, where their performance in complex mixtures demonstrates their unique and useful capabilities.9 We will first outline briefly the design and operation of cross-reactive arrays for sensing. We will examine the particular materials utilized for the platforms, and the statistical analyses used to realize their output. In the following sections, we will review recent studies where profiling of biological samples has been achieved to great effect. In particular, we will focus on two key areas - protein sensing and cell biology. In each case we will examine the potential application of sensor arrays in biomedical R&D and translation to the clinic, focusing on oncology as well as other pathologies. Finally, we will offer some insights on future directions for chemical noses in biological sensing.
2. Design, Construction and Analysis of Arrays
2.1. Designing arrays for Biosensing – the Basic Components
There are two essential processes involved in biosensing: analyte recognition and signal transduction. For array-based sensing a final component is required, namely pattern recognition. Each recognition element in the sensor array is designed to ensure that interactions will occur with the analytes being studied (selectivity). However, because the recognition elements are not specific to one analyte, cross-reactivity also occurs between receptors. The recognition event (or lack thereof) between analyte and each receptor in the array then needs to be transduced to a measurable outcome such as an electric, fluorescent, or colorimetric signal. Finally, the response from each recognition element must be collected and combined for each analyte and then analyzed using statistical methodologies for classification or identification.
2.2. Building Arrays for Biosensing – Material Choices
Chemical-nose arrays for biosensing require the ability to interface synthetic organic and inorganic materials with biomolecules. Purely biological array elements often suffer from poor signal transduction or instability under ambient conditions. By careful choice of materials and integration of synthetic elements, more reliable and stable arrays can be created.
Nanomaterials are particularly useful for biosensing due to their unique physiochemical properties that can be used for novel recognition and transduction processes.10 Nanoparticles have highly tunable size and surface functionalities where one or multiple interacting ligands can serve as the recognition elements.11 This synthetic control enables the attachment of custom recognition elements in a modular fashion. Coupled with the high surface-to-volume ratio of smaller size, nanoparticles provide more interaction sites leading to less sensor material required and enhanced sensitivity.12 Furthermore, the optoelectronic properties of many nanoparticles allow them to double as (part of) the transduction system, by simultaneously acting as a platform for a recognition element and providing an electromagnetic output that can be induced or modified on a sensor binding event. Examples include the plasmon-induced fluorescence quenching commonly observed with gold nanoparticles (AuNPs), the dependence of the plasmon band position of AuNPs on size and aggregation,13 and the tunable fluorescence emission of metal nanoclusters.14
Macromolecules provide many of the same attributes as nanomaterials, by functioning both as a recognition element and platform for the reporter. Synthetic macromolecules, including conjugated and non-conjugated polymers and macrocycles have been integrated into sensor arrays15 as well as systems utilizing biological polymers (peptides and nucleotides).16 Conjugated polymers have been developed extensively for array-based sensing due to their bright, tunable emission, with a key requirement being the development of water-soluble variants for interfacing with biological systems.17 Biological polymers have been applied to exploit their inherent biological compatibility and specificity and further engineered to introduce the desired cross-reactivity. Examples include the use of synthetic peptides as generalized substrates for enzyme sensing.18 Through design of a peptides with differing sequences, the peptides can interact with many different members of a particular family of enzymes (e.g clotting factors). The small differences in activity that occur between each enzyme and each of the peptides generates the cross-reactivity needed to distinguish between members of that enzyme family.19 In addition, oligopeptides have also demonstrated utility in cell membrane sensing, with designed patterns of charge and hydrophobicity/hydrophilicity.20
Finally, small molecule fluorophores, fluorescent proteins and molecular chromophores are also good candidates for creating sensor arrays, either in conjunction with polymers or nanoparticles as for most of the examples discussed in this review, or as arrays in their own right.6
2.3. Transduction of the Recognition Event
Transduction in chemical noses has most commonly been achieved by colorimetric and fluorometric detection, although electrochemical read-out,21 as well as spectroscopic fingerprinting techniques (Raman,22 IR,23 and chiral spectroscopy24) have also been applied. Recently, other less common examples of solution/solid phase biosensing arrays such as chemiluminescent25, SPR technique26 and microcantilevers27 have also been reported. Colorimetric detection has been achieved most simply with dye color changes on binding ions, small molecules and biomacromolecules.8,28 A second approach has been to use the strong plasmonic absorbance band of AuNPs. The red color of spherical AuNPs is sensitive to their surface modification and will red shift when particles aggregate or increase in size.29 By modifying the surface of the AuNPs with cross-reactive binding moieties, this selective color change effect can be induced with groups of analytes. Successful implementation of this transduction strategy has been demonstrated with detection of bacterial species,30 toxic metal ions,31 and proteins.32 Recently, a third style of colorimetric array has emerged, using opal-like photonic crystal structures that display strong color changes based on their chemistry and interaction with various analytes.33
Fluorometric transduction is widely applied in biosensor arrays thanks to its high sensitivity, and widespread adoption in biology.34 Fluorometric sensing can either be turn-on or turn-off, caused by separating a quencher and fluorophore or bringing them together with the analyte respectively.35 AuNPs in particular have found broad application as quenchers for such systems.36 Another option is to employ a fluorescence color change mediated by energy transfer (often Forster resonance energy transfer – FRET). Using FRET strategies, several research groups have designed selective sensor arrays.37 A final fluorometric transduction mechanism is via the use of environmentally responsive dyes. Many fluorophores such as Nile Red, merocyanines, and BODIPY display fluorescence intensity and emission wavelength shifts on changes to the polarity, protic strength or viscosity of their local environment, enabling their use in cross-reactive arrays.38
One key advantage of fluorometric sensing over colorimetric is the ability to multiplex several different color outputs in a single measurement – three or more fluorophores can be interrogated in a single sensor array, without the need for physical separation.39 This leads to high-throughput sensor arrays that require much less sample to analyze.40
2.4. Statistical Data Analysis Methods
The conversion of patterns generated by cross-reactive arrays into interpretable data is at the heart of the array-based sensor, i.e. ‘classification’. Classification can be achieved in the case of specific single-element sensor by visual recognition, such as in the case of a pregnancy-test style assay or antibody-based ELISA (enzyme-linked immunosorbent assay).41 However, in cross-reactive arrays the patterns can be harder to interpret visually and so generally some degree of computer processing is required. To achieve classification with a sensor array, the transduced signals are tabulated alongside any information on the sample composition (what ‘class’ it fits e.g. a cell line, a protein family or a disease state) so that patterns from unknown samples can be cross-referenced with samples of a known class. The aim is to reduce many tens or hundreds of values in the transduced signals, derived from each element for the cross-reactive array, into a simpler, smaller vector that summarizes the most distinguishing features of the pattern, and can be easily compared with other samples for the purpose of analysis.42 The statistical methodologies that are applied for this purpose have been reviewed previously,8,43 so we will only briefly outline some possibilities here.
Two families of classification algorithms exist - unsupervised learning and supervised learning. Unsupervised techniques attempt to process the data presented without any class information from the user, in an attempt to find elements of the array that best separate the data (Principle Component Analysis - PCA) or find data that naturally cluster together (Hierarchical Clustering Analysis - HCA).44 These techniques work well for identifying trends in large array datasets and for identifying which sensing elements in a multi-sensor array contribute the most to the discrimination of analytes.
Supervised techniques are a crucial part of array-based sensing, enabling the sensing readout from the transduced signals. A primary ‘training’ dataset from samples of known class is used to create an algorithm that can organize the samples into one of the defined classes. Once this model has been trained it can be tested, using a secondary, independent set of data, withholding the class information from the model. The success of the algorithm at classifying this second dataset gives an indicator for the accuracy of the model at identifying true unknown samples. In the simplest cases (e.g. Linear Discriminant Analysis - LDA) an unknown is classified into one of the known groupings based on simple linear fits. By applying more advanced supervised algorithms, more complex discriminatory patterns can be generated that not only identify unknown samples that are a match to one of the original training data classes, but also those that are similar but different to any of the original classes, or samples that fall well outside any known class.20b Examples of such algorithms include support vector machines (SVM),44 or artificial neural networks (ANN),22 but a detailed discussion of these machine learning techniques is beyond the scope of this review.
3. Array-based Sensors for Biomedical Applications
Since the advent of reliable enzymatic and antibody panels, arrays have been applied to biomedical sciences for the simultaneous detection of multiple analytes using specific receptors, resulting in lectin,45 aptamer,46 and affimer47 arrays. However, the specificity of such systems, while theoretically high, may often be much lower than assumed, particularly where an unpredictable cross-reactivity might occur.48 Recent work on a direct comparison between two genetic marker panels using specific DNA sensing has observed significant variability between independent methods purporting to measure the same DNA sequences. Furthermore, the diagnoses inferred from the DNA biomarker results were contradicted by the outcome of a well-known protein biomarker assay performed on the same samples.49 Other issues including the stability of biological sensor constructs (such as antibodies) and complicated attachment of transduction elements hinder translation of laboratory techniques to the clinic.50
In comparison, selective array-based sensors operating in a biological environment face a different, but very particular set of challenges. For example, the need for water-solubility, and stability conferred by careful materials selection, as discussed in Section 2. One approach to this challenge is to utilize transducers linked to engineered biomaterials that confer the desirable degree of bio-specificity while maintaining the essential cross-reactivity.51 Through this strategy, successful biosensor arrays can be created with high sensitivity. These arrays are able to detect small changes in the composition of complex mixture without the need for prior biomarker discovery and isolation steps.52
3.1. From Buffer to Biological Matrices for Protein Detection
The first demonstrations of arrays for biomedical science focused on the detection and discrimination of individual proteins. Much of the work combines a fluorogenic sensor with a variety of recognition elements that create differential fluorescence patterns when bound/not bound to the target protein in solution. As an excellent proof-of-concept, a wide range of different proteins have been successfully detected using fluorescent metal complexes,53 nanoclusters,54 quantum dots,55 or colorimetric gold nanoparticle aggregation.32 However, it needs to be noted that while many examples exist of array-based sensing in buffer or water,56 fewer groups have tested their systems in biologically relevant media.
In addressing this challenge, biological matrices such as serum have been incorporated into sensing protocols. An early pioneer work detected analyte proteins in ‘spiked’ serum through an AuNP and fluorescent protein array.57 The positively charged headgroups on the AuNPs electrostatically attracted a negative fluorescent protein (FP) and quenched its fluorescence. If the FP was displaced by another negatively charged protein (e.g. serum albumin, transferrin or fibrinogen) then the FP lit up. By tuning the AuNP surface functionality, the strength of the AuNP-FP and analyte interactions was varied to create a sensor array. Using the AuNP array, five major serum proteins could be discriminated at various concentrations and in various ratios (Figure 2).
Figure 2.
Protein detection in serum using array-based sensors. (a) Structure of cationic Au-NPs. (b) Sensing scheme of AuNP-GFP array with serum proteins, where the addition of proteins causes differential release of GFP. (c) Differentiation of five major serum proteins in spiked human serum samples. Adapted with permission from Reference [57]. Copyright 2009 Nature Publishing Group
In other examples, two or more fluorescent metal nanocluster cores with different surface functionality have been used as the fluorometric recognition/transduction element, rather than rely on a two-part NP/FP combination. The binding of the protein to the nanocluster modulates both the fluorescence intensity and the emission wavelength (the color) giving two channels to be probed per sensor element, improving the discrimination (Figure 3).58 Work by Ouyang and co-workers used two of these color-changing particles to discriminate between proteins in buffer, but also serum collected from patients, some diagnosed with hepatoma. Ouyang and coworkers have also applied their fluorescent particles to a 1D PAGE assay for protein biomarker detection.59 Recently, Xu et al. reported a near infrared sensor array of which dual ligand co-functionalized gold nanoclusters were decorated with amino acids. This platform not only discriminated proteins at nanomolar concentrations but also serum samples collected from different stages of breast cancer patients as well as healthy people.60 Similar sensing strategies have also been tested with human urine samples as well as disease sera from patients.61
Figure 3.
Demonstration of the use of fluorescent gold nanoclusters derived from collagen (Col) and Macerozyme-10 (Mac) in protein sensing. (a) Differential changes in optical signals upon interaction with target proteins in aqueous solution - lysozyme (Lys), human serum albumin (HSA), egg white albumin (EA), pepsin (Pep), hemoglobin (Hb), trypsin (Try), catalase (CAT), and transferrin (Tf). (b) Fluorescence emission shifts in both intensity and wavelength upon binding with select proteins. Adapted with permission from Reference [58]. Copyright 2014 American Chemical Society.
3.2. Targeting Specific Biological Challenges in Chemical Biology and Pathology
Moving beyond nanomaterials, synthetic macromolecules have been used to discriminate proteins in a variety of settings with biological relevance.62 Many of these works also include non-specific biological domains as part of their sensor elements to tailor the cross-reactivity of their arrays to the targets of interest.
Combinatorial fluorescent molecular probes are emerging to be powerful in generating pattern recognition arrays.63 Recently, Margulies and coworkers reported an elegant multipart fluorescent probe featuring three fluorophores including a DNA aptamer hairpin, and two additional protein recognition sites based on small molecules (shown in Figure 4).64 The protein recognition sites were selected for known non-specific binding interactions with disease associated protein families, such as glutathione-S-transferases (GSTs), matrix metalloproteases (MMPs) and platelet-derived growth factors (PDGFs). Upon binding different variants of the three classes of proteins, the fluorophores responded through changes in color and intensity, due to solvatochromic and FRET processes. The probe was further applied to sense target proteins in human urine as biomarkers for various kidney and liver diseases. It performed well, even in the presence of competing serum proteins. Interestingly, the multiplexed probe was also demonstrated as a sensor operating inside cells for exploring inhibition pathways. The strategy of utilizing molecular fluorescent probes has also been implemented to detect and differentiate amyloid beta aggregates that play a crucial role in Alzheimer’s disease.65
Figure 4.
Discrimination of proteins using a multifunctional small molecule bearing responsive fluorophores. (a) Schematic illustration of sensor construction. 3 protein binding moieties (EA- ethacrynic amide, MT – marimastat and Apt – a DNA aptamer) and 4 fluorophores (indicated by *s – nitrobenzoxadiazole, nile red, cyanine 5.5 and cyanine 7) were added to a cis-amino proline scaffold to form the sensor. (b) Fluorescence patterns generated by sensors after adding tested proteins. (c) LDA classification of enriched proteins in urine samples for different glutathione-S-transferases (GSTs), matrix metalloproteases (MMPs) and platelet-derived growth factors (PDGFs). Adapted from Reference [64]. Copyright 2017 Nature Publishing Group.
Another approach to small molecule probes was demonstrated by Waters and coworkers, who applied a combinatorial supramolecular approach to sense post-translational modifications of histones.66 They created a dynamic sensor array by combining a thiolated-aromatic, bridging two aromatic molecules to form a supramolecular pocket. This pocket was then mixed with a dye (lucigenin) to form a host-guest complex. The working principle of such sensor is that the histone can bind the supramolecular sensor, causing displacement of the dye, and a corresponding fluorescence change. Different modifications of the histone affected the binding strength between the sensor and histone, resulting in changes to the amount of dye displaced and fluorescent pattern generation. The combinatorial sensor also distinguished between different post-translational methylation, acetylation and phosphorylation patterns.
Anslyn et al. monitored the post-translational phosphorylation and subsequent activation of kinases in cell lysates as a tool for studying the kinase signaling and apoptosis pathways67. As with the work of Waters, Anslyn created a dynamic supramolecular array with three components: a hydrogen-bonding molecule, a Zn2+ complex mounted in various configurations on a three-armed supramolecular receptor complex and two cross-reactive peptide recognition sequences for their target. This three-armed structure hosted a coumarin dye for transduction. With a seven-sensor array created from this combinatorial library, the activity of four MAP kinases could be monitored. More importantly, the authors also demonstrated that in identical cell lines triggered to undergo different kinase signaling pathways, differing phosphorylation could be ‘fingerprinted’ with the sensor array.
In a recent work we have extended the use of array-based sensing using polymers into diagnostics. As shown in Figure 5, a responsive sensor array was generated that responded to proteins through solvatochromism and energy transfer using three dyes on a polymer backbone and measured the fluorometric response of the dye mixed with human serum. The sensor array was tested against blood samples from a cohort of healthy patients and those with liver fibrosis (n=60), and discrimination was possible, with 80% accuracy versus the standard test.68 This study signposts the potential applications of array-based serum diagnostics for early warning systems or classification of diseases that present as serum proteome modification.
Figure 5.
Liver fibrosis diagnosis using array-based sensing strategy. (a) Schematic illustration of polymer-based array sensing for serum proteome to distinguish between fibrotic and nonfibrotic patients. (b) Polymer structure featuring three responsive fluorophores. (c) Potential working principle of environmental polymers. Adapted with permission from Reference [68]. Copyright 2017 Wiley-VCH.
4. Applications of Arrays for Phenotyping Cells
Beyond sensing proteins in cell lysates (demonstrated in Section 3), the complex surface composition of intact living cells makes them excellent targets for array-based sensing. In particular, cell phenotyping is emerging as an important tool for understanding and treating cancers, with rapid assessment of cell-behavior in response to environmental stimuli. Cell sensing is a broad area, where both prokaryotes (specifically bacteria) and eukaryotes have been classified. Since bacterial sensing has been recently reviewed69, in this section of review, we will focus on eukaryotic cells with highlights of recent advances and applications in oncology.
4.1. Cancer Detection and Progression
Early studies on sensing cancer cells looked to provide diagnostic information through cell surface profiles.70 Sensors were constructed to detect the differences between non-cancerous, cancerous and metastatic cell lines. We applied a similar approach to the protein sensing work discussed in Section 3 (Figure 2), leveraging the fluorescence turn-on interaction between cationic AuNPs with different surface functionalities and anionic fluorescent polymers or proteins to create arrays capable of distinguishing the three categories.71
Other materials such as gold nanoclusters have also been explored for cancer cell surface sensing. Tao and coworkers have recently reported separation of ten triple-negative breast cancer cell lines from multiple patients, with varying degrees of metastasis, using dual-ligand functionalized gold nanoclusters (Figure 6).72 Anslyn and coworkers created an array from nine thiazole-orange labelled peptides, tuned to interact with various cell-surface features, and a piece of double-stranded DNA that intercalated the thaizole orange modifying the fluorescence of the array in-situ on the cell surface. Multivariate analysis on the array data showed that it could distinguish eight cancer cell lines form different parts of the body. Most interestingly, by applying a support vector machine (SVM) to their data, a ninth cell line not included in the test set could be detected and differentiated.20b
Figure 6.
Breast cancer cell line sensing with gold nanoclusters. (a) Schematic illustration of the dual-ligand functionalized gold nanoclusters sensor array. (a) (b) LDA classification of 10 breast cancer cell lines. Adapted with permission from Reference [72]. Copyright 2018 Elsevier.
Wu et al. recently built an electrical impedance array based on non-specific functionalization of graphene electrodes that not only differentiated differing cancer cell lines but could also detect a model for circulating tumor cells (CTCs) at low concentrations, i.e MCF-7 spiked into matrices containing peripheral blood mononuclear cells.73 CTCs are strongly linked with metastasis and poor prognoses. Once CTCs exit the primary tumor site and enter the bloodstream they become hard to detect by conventional antibody-based techniques.74 This study represents a step towards using signature-based sensing in challenging oncological scenarios.
As noted in all the works above, cancer is not a static disease. Each stage is associated with significant geno- and phenotypic changes in the cells. Much of the work described has focused on cells with significant genotypic differences; differentiating different cancerous cell lines or between metastatic and non-metastatic cell lines. However, in many areas, arrays are tested against multiple cell lines with differing genetic backgrounds as well as varying degrees of metastasis. The changes arising from the different genetic background will also contribute to sensor response, potentially overwhelming any detection of metastasis. Thus, it is important to also test against isogenic cell lines with minimum genetic diversity to isolate these cell-line features.
4.2. Beyond Cell Classification – High Content Screening of Chemotherapeutics
A very different application of array-based cell sensing from diagnostics is to monitor the response and death of cells when exposed to an environmental stimulus such as a drug. Rapid determination of drug mechanism is a key step in therapeutic discovery,75 and cell-based screening methods are increasingly used to facilitate the process.76 However, many of the cell-based assays are limited by the multi-step processing of cells prior to analyses, and an incomplete understanding of biomolecular pathways for correlating drug and response.77 To demonstrate the utility of array-based sensors in this area, a cationic benzyl-functionalized AuNP was complexed with three different anionic FPs. When exposed to cells, the AuNP interacts with cell surfaces and releases FPs into solution, turning-on fluorescent signals for profiling cell surfaces. The cell lines studied were exposed to one of 15 different anti-cancer drugs featuring seven different drug mechanisms. The sensing results showed distinct patterns for each treated cell line, which could be further clustered into the hypothesized drug mechanism, and novel drugging routes were also elucidated (Figure 7).78
Figure 7.
The use of array-based sensor in profiling drug mechanisms of chemotherapeutics. (a) Complexation of sensor array. (b) Workflow for drug screening using nanoparticle-based arrays. (c) Classification of 7 different drug mechanisms. Adapted with permission from Reference [78]. Copyright 2015 Nature Publishing Group.
5. Summary and Outlook
Cross-reactive arrays provide unique capabilities for identifying changes in complex biological mixtures. The hypothesis-free nature of these systems facilitates exploration of sensing space and has already been used to discriminate analytes difficult or impossible to differentiate using standard approaches. The ‘nose’ approach has been used to ‘fingerprint’ proteins, cell types and even the presence or absence of disease. Although highly specific biological arrays will provide a cornerstone of bioanalysis and pathology, it is clear that selective cross-reactive arrays are playing an increasingly important role in biomedicine.
The performance of cross-reactive sensor arrays continues to improve, as more advanced designs and improved statistical analyses are brought to bear on challenging biological problems. Fundamental discoveries in pathology, cell biology and physiology have been discussed here and research is now looking to address three challenging areas.
The first major challenge is to combine the outputs of the sensor array with advances in highly specific sensors, utilizing antibodies, aptamers and other engineered affinity proteins (e.g. affimers) to understand exactly what the arrays are responding to. In this way biomarker discovery can lead to better arrays and arrays can lead to better biomarker discovery.
A second challenge is to improve the quality of data and analysis. Once an array has demonstrated its potential for e.g. fingerprinting a disease, larger scale screening and rigorous repetition will be required to translate the technology to clinic. With larger datasets, more robust statistical analysis can be employed, and more reliable conclusions can be drawn. The statistics applied to arrays are becoming better understood by the community, and it is promising to see advanced techniques such as SVM becoming popular – however care should be taken to apply the correct classification tool and avoid violating key assumptions of the classification algorithms used (see reference79 for a discussion of this with respect to LDA and SVM).80
Finally, it is time to begin moving array-based sensing out of the lab.81 Suslick et al. have had success at creating hand held devices for their colorimetric arrays,82 but the solvated arrays used in many of the biosensing examples above will require careful engineering of microfluidic polydimethylsiloxane and paper-based systems.83 This development, coupled with advances in mobile reading technologies,84 will lead to simple, robust point-of-care diagnostics.
Specific array-based sensors have been part of analytical science for a long time, but now selective cross-reactive ‘chemical nose’ sensors are making real headway. We have illustrated here that biology and medicine will be areas where these arrays can become a highly disruptive technology. By moving beyond tried and tested sensing challenges and working with bioscientists and clinicians, the ‘chemical nose’ has a promising future in biomedical technologies.
Figure 1.
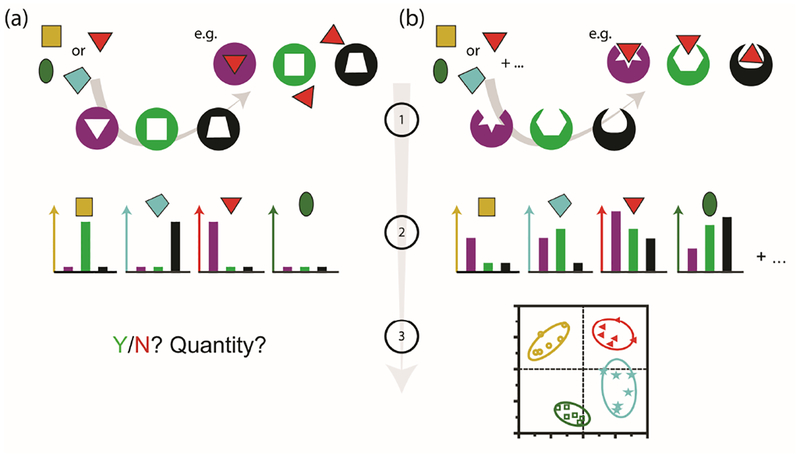
An overview of chemical nose sensing – moving beyond N receptors for N analytes. (a) A traditional specific sensor and (b) a cross-reactive (but still selective) array-based sensor. In (a) one element can interact with one analyte transducing single responses with N receptors needed to measure N analytes. For (b), each element in a mixture interacts in different ways with a cross-reactive array. The transduction of the interactions leads to pattern generation for the combination of elements. The patterns are then processed, and it is possible to detect more analytes than there are elements.
Acknowledgements
V.M.R. acknowledges support from the NIH (GM077173, CA207932 and AI AI134770) Y.G. receives funding from Chemistry Biology Interface program (GM008515) at University of Massachusstat Amhertst. W.J.P acknowledges the Royal Society for an International Exchange Grant, the University of Glasgow for an LKAS Fellowship and the Killam Foundation for a Postdoctoral Research Fellowship.
Biographies
Yingying Geng received her B.Sc. in Biochemistry and Molecular Biology from University of Richmond, VA in 2014. She is currently pursuing her Ph.D. at the Molecular and Cellular Biology/Department of Chemistry at the University of Massachusetts Amherst, under the supervision of Professor Vincent M. Rotello. Her current research interest focuses on the use of nanomaterials and polymers for cell-surface profiling.

William Peveler studied chemistry at the University of Oxford and latterly University College London, where he completed his PhD in 2015 on the topic of “arrays for explosives detection”. Subsequently he undertook postdoctoral work with Professor William Rosenberg and Professor Claire Carmalt at UCL, which included time in the lab of Professor Vincent Rotello. Most recently he has been working at the University of British Columbia with Dr Russ Algar, before returning to the UK to take up an independent LKAS Fellowship at the University of Glasgow.

Vincent Rotello is the Charles A. Goessmann Professor of Chemistry and a University Distinguished Professor at the University of Massachusetts Amherst. He is a Fellow of both the American Association for the Advancement of Science (AAAS) and of the Royal Society of Chemistry (U.K.). He is currently the Editor in Chief of Bioconjugate Chemistry and is on the Editorial Board of 14 other journals. His research program focuses on using synthetic organic chemistry to engineer the interface between the synthetic and biological worlds, and spans the areas of devices, polymers, and nanotechnology, with over 530 peer-reviewed publications to date. He is actively involved in bionanotechnology, and his research includes programs in delivery, imaging, diagnostics and nanotoxicology.

References
- [1].Guerrini L, Garcia-Rico E, Pazos-Perez N, Alvarez-Puebla RA, ACS Nano 2017, 11, 5217–5222. [DOI] [PubMed] [Google Scholar]
- [2].a) Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, Walt DR, Chem. Rev 2000, 100, 2595–2626; [DOI] [PubMed] [Google Scholar]; b) Lavigne JJ, V Anslyn E, Angew. Chem. Int. Ed 2001, 40, 3118–3130. [DOI] [PubMed] [Google Scholar]
- [3].Peveler WJ, Yazdani M, Rotello VM, ACS Sensors 2016, 1, 1282–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Peveler WJ, Roldan A, Hollingsworth N, Porter MJ, Parkin IP, ACS Nano 2016, 10, 1139–1146; [DOI] [PubMed] [Google Scholar]; b) Diehl KL, Anslyn EV, Chem. Soc. Rev 2013, 42, 8596. [DOI] [PubMed] [Google Scholar]
- [5].a) Zhang Z, Kim DS, Lin C-Y, Zhang H, Lammer AD, Lynch VM, Popov I, Miljanić OŠ, Anslyn EV, Sessler JL, J. Am. Chem. Soc 2015, 137, 7769–7774; [DOI] [PubMed] [Google Scholar]; b) Lim SH, Feng L, Kemling JW, Musto CJ, Suslick KS, Nat. Chem 2009, 1, 562–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu Y, Minami T, Nishiyabu R, Wang Z, Anzenbacher P, J. Am. Chem. Soc 2013, 135, 7705–7712. [DOI] [PubMed] [Google Scholar]
- [7].a) Han J, Ma C, Wang B, Bender M, Bojanowski M, Hergert M, Seehafer K, Herrmann A, Bunz UHF, Chem 2017, 2, 817–824; [Google Scholar]; b) Li Z, Suslick KS, ACS Sensors 2018, 3, 121–127. [DOI] [PubMed] [Google Scholar]
- [8].Askim JR, Mahmoudi M, Suslick KS, Chem. Soc. Rev 2013, 42, 8649–8682. [DOI] [PubMed] [Google Scholar]
- [9].Peveler WJ, Algar WR, ACS Chem. Biol 2018, acschembio.7b01022. [DOI] [PubMed] [Google Scholar]
- [10].Bigdeli A, Ghasemi F, Golmohammadi H, Abbasi-Moayed S, Nejad MAF, Fahimi-Kashani N, Jafarinejad S, Shahrajabian M, Hormozi-Nezhad MR, Nanoscale 2017, 9, 16546–16563. [DOI] [PubMed] [Google Scholar]
- [11].Hötzer B, Medintz IL, Hildebrandt N, Small 2012, 8, 2297–2326. [DOI] [PubMed] [Google Scholar]
- [12].a) Bunz UHF, Rotello VM, Angew. Chemie Int. Ed 2010, 49, 3268–3279; [DOI] [PubMed] [Google Scholar]; b) Daniel MCM, Astruc D, Chem. Rev 2004, 104, 293–346; [DOI] [PubMed] [Google Scholar]; c) Radwan SH, Azzazy HM, Expert Rev. Mol. Diagn 2009, 9, 511–524. [DOI] [PubMed] [Google Scholar]
- [13].Saha K, Agasti S, Kim C, Li X, Rotello V, Chem. Rev 2012, 112, 2739–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen LY, Wang CW, Yuan Z, Chang HT, Anal. Chem 2015, 87, 216–229. [DOI] [PubMed] [Google Scholar]
- [15].a) Freudenberg J, Hinkel F, Jänsch D, Bunz UHF, Top. Curr. Chem 2017, 375, 67; [DOI] [PubMed] [Google Scholar]; b) You L, Zha D, Anslyn EV, Chem. Rev 2015, 115, 7840–7892. [DOI] [PubMed] [Google Scholar]
- [16].Sapsford KE, Berti L, Medintz IL, Angew. Chemie Int. Ed 2006, 45, 4562–4589. [DOI] [PubMed] [Google Scholar]
- [17].Liu B, Bazan GC, Chem. Mater 2004, 16, 4467–4476. [Google Scholar]
- [18].Algar WR, Ancona MG, Malanoski AP, Susumu K, Medintz IL, ACS Nano 2012, 6, 11044–11058. [DOI] [PubMed] [Google Scholar]
- [19].a) Shults MD, Janes KA, Lauffenburger DA, Imperiali B, Nat. Methods 2005, 2, 277–284; [DOI] [PubMed] [Google Scholar]; b) Zamora-Olivares D, Kaoud TS, Jose J, Ellington A, Dalby KN, V Anslyn E, Angew. Chem. Int. Ed 2014, 53, 14064–14068. [DOI] [PubMed] [Google Scholar]
- [20].a) Han J, Cheng H, Wang B, Braun MS, Fan X, Bender M, Huang W, Domhan C, Mier W, Lindner T, et al. , Angew. Chemie Int. Ed 2017, 56, 15246–15251; [DOI] [PubMed] [Google Scholar]; b) Gade AM, Meadows MK, Ellington AD, Anslyn EV, Org. Biomol. Chem 2017, 15, 9866–9874. [DOI] [PubMed] [Google Scholar]
- [21].Yáñez-Sedeño P, Campuzano S, Pingarrón JM, Anal. Chim. Acta 2017, 960, 1–17. [DOI] [PubMed] [Google Scholar]
- [22].Kasera S, Herrmann LO, del Barrio J, Baumberg JJ, Scherman OA, Sci. Rep 2014, 4, 6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Baker MJ, Trevisan J, Bassan P, Bhargava R, Butler HJ, Dorling KM, Fielden PR, Fogarty SW, Fullwood NJ, Heys KA, et al. , Nat. Protoc 2014, 9, 1771–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tullius R, Platt GW, Khosravi Khorashad L, Gadegaard N, Lapthorn AJ, Rotello VM, Cooke G, Barron LD, Govorov AO, Karimullah AS, et al. , ACS Nano 2017, 11, 12049–12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shahrajabian M, Hormozi-Nezhad MR, Sci. Rep 2016, 6, 32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choi S, Huang S, Li J, Chae J, Lab Chip 2011, 11, 3681–3688. [DOI] [PubMed] [Google Scholar]
- [27].Wang P, Pei H, Wan Y, Li J, Zhu X, Su Y, Fan C, Huang Q, Nanoscale 2012, 4, 6739–6742. [DOI] [PubMed] [Google Scholar]
- [28].Carey JR, Suslick KS, Hulkower KI, Imlay JA, Imlay KRC, Ingison CK, Ponder JB, Sen A, Wittrig AE, J. Am. Chem. Soc 2011, 133, 7571–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Srivastava S, Frankamp BL, Rotello VM, Chem. Mater 2005, 17, 487–490. [Google Scholar]
- [30].Li B, Li X, Dong Y, Wang B, Li D, Shi Y, Wu Y, Anal. Chem 2017, 89, 10639–10643. [DOI] [PubMed] [Google Scholar]
- [31].Fahimi-Kashani N, Hormozi-Nezhad MR, Anal. Chem 2016, 88, 8099–8106. [DOI] [PubMed] [Google Scholar]
- [32].Wei X, Wang Y, Zhao Y, Chen Z, Biosens. Bioelectron 2017, 97, 332–337. [DOI] [PubMed] [Google Scholar]
- [33].Fenzl C, Hirsch T, Wolfbeis OS, Angew. Chem. Int. Ed 2014, 53, 3318–3335. [DOI] [PubMed] [Google Scholar]
- [34].Das S, Powe AM, Baker GA, Valle B, El-Zahab B, Sintim HO, Lowry M, Fakayode SO, McCarroll ME, Patonay G, et al. , Anal. Chem 2012, 84, 597–625. [DOI] [PubMed] [Google Scholar]
- [35].Wu D, Sedgwick AC, Gunnlaugsson T, Akkaya EU, Yoon J, James TD, Chem. Soc. Rev 2017, 46, 7105–7123. [DOI] [PubMed] [Google Scholar]
- [36].a) Le Reste L, Hohlbein J, Gryte K, Kapanidis AN, Biophys. J 2012, 102, 2658–2668; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kapur A, Aldeek F, Ji X, Safi M, Wang W, Del Cid A, Steinbock O, Mattoussi H, Bioconjug. Chem 2017, 28, 678–687. [DOI] [PubMed] [Google Scholar]
- [37].a) Rana S, Elci SG, Mout R, Singla AK, Yazdani M, Bender M, Bajaj A, Saha K, Bunz UHF, Jirik FR, et al. , J. Am. Chem. Soc 2016, 138, 4522–4529; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chatzimarkou A, Chatzimitakos TG, Kasouni A, Sygellou L, Avgeropoulos A, Stalikas CD, Sensors Actuators B Chem 2018, 258, 1152–1160. [Google Scholar]
- [38].a) Klymchenko AS, Acc. Chem. Res 2017, 50, 366–375; [DOI] [PubMed] [Google Scholar]; b) MacNevin CJ, Gremyachinskiy D, Hsu C-W, Li L, Rougie M, Davis TT, Hahn KM, Bioconjug. Chem 2013, 24, 215–223; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nomura W, Ohashi N, Okuda Y, Narumi T, Ikura T, Ito N, Tamamura H, Bioconjug. Chem 2011, 22, 923–930. [DOI] [PubMed] [Google Scholar]
- [39].Klostranec JM, Xiang Q, Farcas GA, Lee JA, Rhee A, Lafferty EI, Perrault SD, Kain KC, Chan WCW, Nano Lett 2007, 7, 2812–2818. [DOI] [PubMed] [Google Scholar]
- [40].Massey M, Kim H, Conroy EM, Algar WR, J. Phys. Chem. C 2017, 121, 13345–13356. [Google Scholar]
- [41].a) Fang C-C, Chou C-C, Yang Y-Q, Wei-Kai T, Wang Y-T, Chan Y-H, Anal. Chem 2018, 90, 2134–2140. [DOI] [PubMed] [Google Scholar]; b) Banerjee R, Jaiswal A, Analyst 2018, 143, 1970–1996. [DOI] [PubMed] [Google Scholar]
- [42].Bishop CM, in Pattern Recognition and Machine Learning, Springer, New York, 2006. [Google Scholar]
- [43].Stewart S, Ivy MA, Anslyn EV, Chem. Soc. Rev 2014, 43, 70–84. [DOI] [PubMed] [Google Scholar]
- [44].Gokcen I, Peng J, in Lect. Notes Comput. Sci (: Yakhno T), Springer Berlin Heidelberg, Berlin, Heidelberg, 2002, pp. 104–113. [Google Scholar]
- [45].Hirabayashi J, Yamada M, Kuno A, Tateno H, Chem. Soc. Rev 2013, 42, 4443–4458. [DOI] [PubMed] [Google Scholar]
- [46].Eissa S, Zourob M, Sci. Rep 2017, 7, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Johnson A, Song Q, Ko Ferrigno P, Bueno PR, Davis JJ, Anal. Chem 2012, 84, 6553–6560. [DOI] [PubMed] [Google Scholar]
- [48].a) Saper CB, J. Histochem. Cytochem 2009, 57, 1–5; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Stewart S, Syrett A, Pothukuchy A, Bhadra S, Ellington A, Anslyn E, Chembiochem 2011, 12, 2021–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].a) Torga G, Pienta KJ, JAMA Oncol 2017, DOI 10.1001/jamaoncol.2017.4027; [DOI] [Google Scholar]; b) Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, Collisson EA, Divers SG, Hoon DSB, Kopetz ES, et al. , PLoS One 2015, 10, e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lowe D, Dudgeon K, Rouet R, Schofield P, Jermutus L, Christ D, in Adv. Protein Chem. Struct. Biol, 2011, pp. 41–61. [DOI] [PubMed] [Google Scholar]
- [51].Hatai J, Motiei L, Margulies D, J. Am. Chem. Soc 2017, 139, 2136–2139. [DOI] [PubMed] [Google Scholar]
- [52].Tomita S, Niwa O, Kurita R, Anal. Chem 2016, 88, 9079–9086. [DOI] [PubMed] [Google Scholar]
- [53].Hewitt SH, Wilson AJ, Chem. Commun 2017, 53, 12278–12281. [DOI] [PubMed] [Google Scholar]
- [54].Yuan Z, Du Y, Tseng Y-T, Peng M, Cai N, He Y, Chang H-T, Yeung ES, Anal. Chem 2015, 87, 4253–4259. [DOI] [PubMed] [Google Scholar]
- [55].Chen S, Wei L, Chen X-W, Wang J-H, Anal. Chem 2015, 87, 10902–10909. [DOI] [PubMed] [Google Scholar]
- [56].You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM, Nat. Nanotechnol 2007, 2, 318–323. [DOI] [PubMed] [Google Scholar]
- [57].De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM, Nat. Chem 2009, 1, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Xu S, Lu X, Yao C, Huang F, Jiang H, Hua W, Na N, Liu H, Ouyang J, Anal. Chem 2014, 86, 11634–11639. [DOI] [PubMed] [Google Scholar]
- [59].Liu H, Ma L, Xu S, Hua W, Ouyang J, J. Mater. Chem. B 2014, 2, 3531–3537. [DOI] [PubMed] [Google Scholar]
- [60].Xu S, Gao T, Feng X, Fan X, Liu G, Mao Y, Yu X, Lin J, Luo X, Biosens. Bioelectron 2017, 97, 203–207. [DOI] [PubMed] [Google Scholar]
- [61].a) Xu S, Wu Y, Sun X, Wang Z, Luo X, J. Mater. Chem. B 2017, 5, 4207–4213; [DOI] [PubMed] [Google Scholar]; b) Xu S, Su Z, Zhang Z, Nie Y, Wang J, Ge G, Luo X, J. Mater. Chem. B 2017, 5, 8748–8753. [DOI] [PubMed] [Google Scholar]
- [62].Han J, Wang B, Bender M, Pfisterer J, Huang W, Seehafer K, Yazdani M, Rotello VM, Rotello CM, Bunz UHF, Polym. Chem 2017, 8, 2723–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rout B, Motiei L, Margulies D, Synlett 2014, 25, 1050–1054. [Google Scholar]
- [64].Pode Z, Peri-Naor R, Georgeson JM, Ilani T, Kiss V, Unger T, Markus B, Barr HM, Motiei L, Margulies D, Nat. Nanotechnol 2017, 12, 1161–1168. [DOI] [PubMed] [Google Scholar]
- [65].Hatai J, Motiei L, Margulies D, J. Am. Chem. Soc 2017, 139, 2136–2139. [DOI] [PubMed] [Google Scholar]
- [66].Peacor BC, Ramsay CM, Waters ML, Chem. Sci 2017, 8, 1422–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zamora-Olivares D, Kaoud TS, Dalby KN, V Anslyn E, J. Am. Chem. Soc 2013, 135, 14814–14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Peveler WJ, Landis RF, Yazdani M, Day JW, Modi R, Carmalt CJ, Rosenberg WM, Rotello VM, Adv Mater 2018. doi: 10.1002/adma.201800634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chen J, Andler SM, Goddard JM, Nugen SR, Rotello VM, Chem. Soc. Rev 2017, 46, 1272–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Le NDB, Yazdani M, Rotello VM, Nanomedicine (Lond). 2014, 9, 1487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].a) Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM, Proc. Natl. Acad. Sci. U.S.A 2009, 106, 10912–10916; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM, Chem. Sci 2010, 1, 134–138. [Google Scholar]; c) Rana S, Le NDB, Mout R, Duncan B, Elci SG, Saha K, Rotello VM, ACS Cent. Sci 2015, 1, 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tao Y, Li M, Auguste DT, Biomaterials 2017, 116, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wu L, Ji H, Guan Y, Ran X, Ren J, Qu X, NPG Asia Mater. 2017, 9, e356. [Google Scholar]
- [74].a) Martin OA, Anderson RL, Narayan K, MacManus MP, Nat. Rev. Clin. Oncol 2017, 14, 32–44; [DOI] [PubMed] [Google Scholar]; b) Mohme M, Riethdorf S, Pantel K, Nat. Rev. Clin. Oncol 2017, 14, 155–167. [DOI] [PubMed] [Google Scholar]
- [75].a) Editorial Nat. Med 2010, 16, 347–347;20376007 [Google Scholar]; b) Gonzalez de Castro D, Clarke PA, Al-Lazikani B, Workman P, Clin. Pharmacol. Ther 2013, 93, 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fishman MC, Porter JA, Nature 2005, 437, 491–493. [DOI] [PubMed] [Google Scholar]
- [77].a) O’Brien PJ, Basic Clin. Pharmacol. Toxicol 2014, 115, 4–17; [DOI] [PubMed] [Google Scholar]; b) Jiang H, Pritchard JR, Williams RT, Lauffenburger DA, Hemann MT, Nat. Chem. Biol 2011, 7, 92–100; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schirle M, Bantscheff M, Kuster B, Chem. Biol 2012, 19, 72–84. [DOI] [PubMed] [Google Scholar]
- [78].Rana S, Le NDB, Mout R, Saha K, Tonga GY, Bain RES, Miranda OR, Rotello CM, Rotello VM, Nat. Nanotechnol 2015, 10, 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Li Z, Fang M, LaGasse MK, Askim JR, Suslick KS, Angew. Chem. Int. Ed 2017, 56, 9860–9863. [DOI] [PubMed] [Google Scholar]
- [80].a) Boeker P, Sens. Actuator B Chem 2014, 204, 2–17; [Google Scholar]; b) Kuhn M, Johnson K, Significance 2014, 11, 35–37. [Google Scholar]
- [81].Wolfbeis OS, Angew. Chem. Int. Ed 2013, 52, 9864–9865. [DOI] [PubMed] [Google Scholar]
- [82].Askim JR, Suslick KS, Anal. Chem 2015, 87, 7810–7816. [DOI] [PubMed] [Google Scholar]
- [83].a) López-Marzo AM, Merkoçi A, Lab Chip 2016, 16, 3150–3176; [DOI] [PubMed] [Google Scholar]; b) Il Hong J, Chang B-Y, Lab Chip 2014, 14, 1725–1732. [DOI] [PubMed] [Google Scholar]
- [84].a) Petryayeva E, Algar WR, RSC Adv 2015, 5, 22256–22282; [Google Scholar]; b) Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A, Lab Chip 2012, 12, 2678; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ballard ZS, Shir D, Bhardwaj A, Bazargan S, Sathianathan S, Ozcan A, ACS Nano 2017, 11, 2266–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]