Abstract
The Cu(I)- or Ag(I)-catalyzed cycloaddition between 8-ethynyladenine or guanine nucleosides and TMSN3 gave 8-(1-H-1,2,3-triazol-4-yl) nucleosides in good yields. On the other hand, reactions of 5-ethynyluracil or cytosine nucleosides with TMSN3 led to the chemoselective formation of triazoles via Cu(I)-catalyzed cycloaddition or vinyl azides via Ag(I)-catalyzed hydroazidation. These nucleosides with a minimalistic triazolyl modification showed excellent fluorescent properties with 8-(1-H-1,2,3-triazol-4-yl)-2′-deoxyadenosine (8-TrzdA), exhibiting a quantum yield of 44%. The 8-TrzdA 5′-triphosphate was incorporated into duplex DNA containing a one-nucleotide gap by DNA polymerase β.
Graphical Abstract
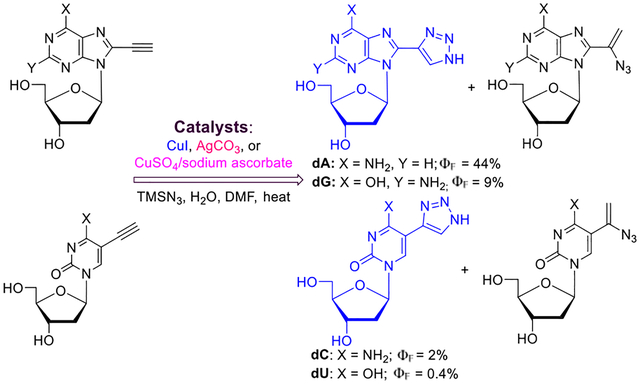
The natural nucleic acid has weak fluorescent properties.1 However, modified nucleoside analogues with fluorescent properties serve as powerful tools to study nucleic acid interactions, activities, and structures.2–9 The nucleoside derivatives with fluorescent nucleobases have been recently reviewed.10 The criteria4 for designing fluorescent nucleosides are (i) sensitiveness to the microenvironment, (ii) emission at long wavelengths, (iii) high quantum efficiency, and (iv) minimalistic modifications, which cause the least amount of distortion of natural hydrogen bonding.
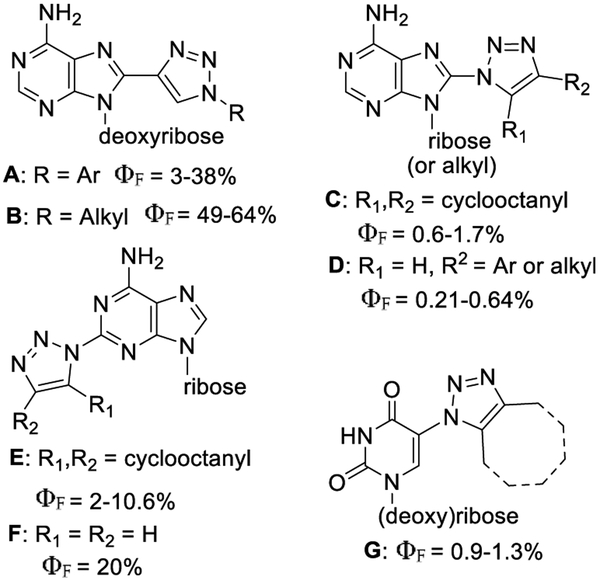
Fluorescent properties of nucleosides bearing triazolyl modifications varied mostly by the (i) position of nucleobases, at which a triazolyl unit is attached, (ii) site of triazolyl attachment to the nucleobase (N1 vs C4), and (iii) additional substitutions either at nucleobase or triazolyl units. The syntheses of the N-substituted triazolyl analogues are mainly based on CuAAC and strain promoted click chemistry.6,11–18 For example, the 8-(1H-1,2,3-triazol-4-yl)adenosine derivatives A and B (Figure 1), with triazolyl carbon attached to the C8 position of the purine ring, have quantum yields up to 64%,14,19 while (1,2,3-triazol-1-yl)adenosines C and D, with triazolyl nitrogen attached to the C8 position of purine, display significantly lower quantum yields (0.21–1.7%).6,15 The 2-(1,2,3-triazol-1-yl)adenosine analogues E and F with a triazole moiety at the C2 position have a moderate to relatively high quantum yield (2–20%).6,11 The 5-azidouracil nucleosides click with cyclooctyne substrates, generating 5-(1,2,3-triazol-1-yl) nucleosides G for the living cell fluorescent imaging.6
Figure 1.
Structures and quantum yields of the selected triazoles
So far, only a few examples of nucleosides modified with the minimalistic (N-unsubstituted) (1H-1,2,3-triazol-4-yl) unit are reported. Thus, 5-(1H-1,2,3-triazol-4-yl)-2′-deoxyuridine (5-TrzdU) was synthesized by microwave-assisted CuAAC between the corresponding 5-ethynyluracil and in situ generated pivaloyloxymethyl (POM) azide followed by N-deprotection20 or by the nucleobase-exchange reaction.21 N-POM-protected 5-TrzdU was incorporated into DNA via solid-phase synthesis.20 The 5-TrzdU20 and its analogues17,22 stack in the major groove and increase the stability of the duplex DNA.
Strategies developed for the synthesis of N-unsubstituted triazoles include: (i) CuI-catalyzed [3+2] cycloaddition of terminal alkynes and trimethylsilyl azide (TMSN3),23 (ii) Pd-24 or p-toluenesulfonic acid-catalyzed25 cycloaddition between activated alkene and NaN3, and (iii) deprotection of N-substituted triazoles.12,26,27 Herein, we report the catalyst-dependent cycloaddition of acetylenic nucleosides with TMSN3 for the synthesis of 5-pyrimidine and 8-purine nucleosides modified with a minimalistic N-unsubstituted 1,2,3-triazol-4-yl moiety and their fluorescent properties.
Synthesis.
Treatment of 8-ethynyl-2′-deoxyadenosine 1a28 with TMSN3 in the presence of CuI as a catalyst23 gave 8-(1H-1,2,3-triazol-4-yl)-2′-deoxyadenosine (8-TrzdA, 2a; method A; Table 1, entry 1). Analogous treatment of TBDMS-protected 2′-deoxyadenosine 1b28 with TMSN3 yielded protected 8-TrzdA (2b, 47%; entry 2). 8-Ethynyl-2′-deoxyguanosine 1c29 under analogous conditions provided 8-(1H-1,2,3-triazol-4-yl)-2′-deoxyguanosine (8-TrzdG, 2c; entry 3). Thus, cycloaddition of the 8-ethynylpurine nucleosides with TMSN3 in the presence of CuI produced triazoles as sole products.
Table 1.
Synthesis of 8-(1H-1,2,3-Triazol-4-yl)purine Nucleosides
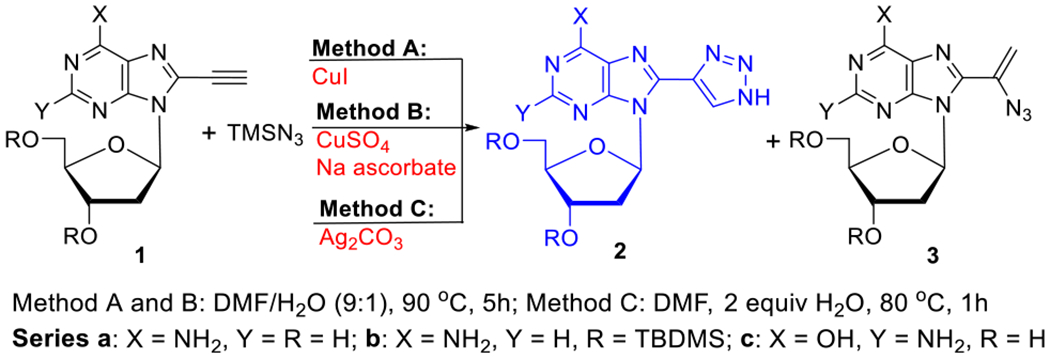 | ||||||
|---|---|---|---|---|---|---|
| entry | compound 1 | method | compound 2 | yielda [%] | compound 3 | yielda [%] |
| 1 | 1a | A | 2a | 48 (52b) | 3a | 0 |
| 2 | 1b | A | 2b | 47 | 3b | 0 |
| 3 | 1c | A | 2c | 31 (36b) | 3c | 0 |
| 4 | 1a | B | 2a | 62 | 3a | 0 |
| 5 | 1b | B | 2b | 71 | 3b | 0 |
| 6 | 1c | B | 2c | 52 | 3c | 0 |
| 7 | 1a | C | 2a | 58 | 3a | 15 |
| 8 | 1b | C | 2b | 55 | 3b | 0 |
| 9 | 1c | Cc | 2c | 60 | 3c | 0 |
Isolated yields. Products were purified on a silica gel column.
Products were purified on RP-HPLC.
4 h.
To avoid both loss of catalytic ability of Cu(I) and colorization of the reaction mixture, we found that treatment of 1a with TMSN3 in the presence of in situ generated Cu(I) from CuSO4/sodium ascorbate gave 2a with a 62% isolated yield (method B, entry 4). Similar treatment of 1b and 1c with TMSN3 afforded triazoles 2b (entry 5) and 2c (entry 6) in higher yields than that by method A.
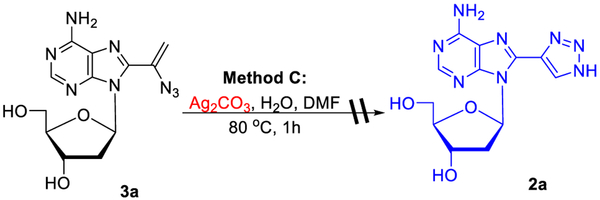
We also found that Ag2CO3-catalyzed hydroazidation of 1a with TMSN3 (in DMF with 2 equiv of H2O) produced 2a (58%) in addition to the vinyl azide 3a (15%; method C, entry 7). Analogous treatment of 1b provided 2b without a detected trace of vinyl azide 3b (entry 8). Subjection 1c to method C for 4 h also yielded 2c as the sole product (entry 9). It is noteworthy that Ag2CO3-catalyzed reactions between arylacetylenes and TMSN3 provide vinyl azides as main products.30,31
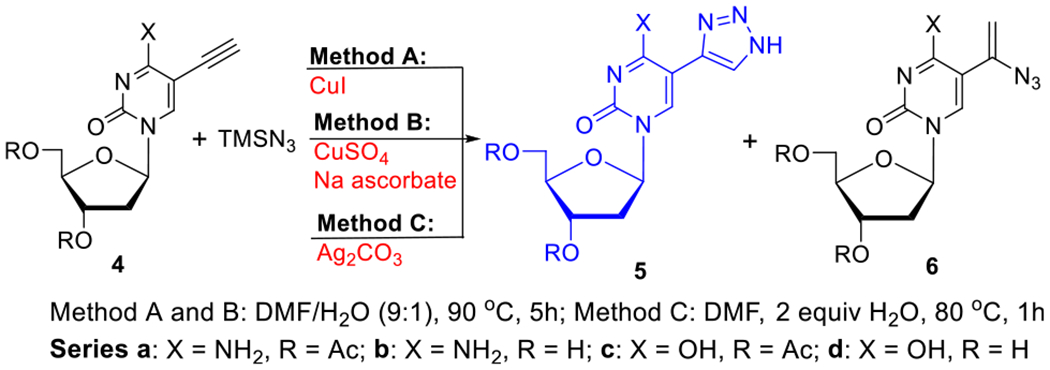
Transition-metal-catalyzed cycloaddition of TMSN3 with alkyne has been extended into the 5-ethynyl pyrimidine nucleosides 4a–d (methods A–C, Table 2). Thus, CuI-catalyzed cycloaddition of 3′,5′-di-O-acetyl-5-ethynyl-2′-deoxycytidine 4a32 yielded 5-(1H-1,2,3-triazol-4-yl) product 5a (method A; 45%, entry 1), while unprotected 2′-deoxycytidine substrate 4b32 provided 5-(1H-1,2,3-triazol-4-yl)-2′-deoxycyti-dine 5b (5-TrzdC, entry 2). Protected 4c33 and unprotected 5-ethynyl-2′-deoxyuridine 4d33 gave 5-(1H-1,2,3-triazol-4-yl) products 5c (65%, entry 3) and 5d (5-TrzdU; 62%, entry 4). It appears that H2O is required for the reaction to proceed23 since the mixture of DMF/H2O (9:1) as a solvent gave the best yields of products 5a–d. Hydroazidation of 4d in the presence of 2 equiv of H2O in DMF gave 5d (50%, entry 5) as a sole product, showing that a stoichiometric amount of H2O was sufficient for cycloaddition to occur.
Table 2.
Synthesis of 5-(1H-1,2,3-Triazol-4-yl) and 5-(1-Azidovinyl)pyrimidine Nucleosides
 | ||||||
|---|---|---|---|---|---|---|
| entry | compound 4 | method | compound 5 | yielda [%] | compound 6 | yielda [%] |
| 1 | 4a | A | 5a | 45 | 6a | 0 |
| 2 | 4b | A | 5b | 48b | 6b | 0 |
| 3 | 4c | A | 5c | 65 | 6c | 0 |
| 4 | 4d | A | 5d | 62 | 6d | 0 |
| 5 | 4d | Ac | 5d | 50 | 6d | 0 |
| 6 | 4a | B | 5a | 65 | 6a | 0 |
| 7 | 4b | B | 5b | 60 | 6b | 0 |
| 8 | 4c | B | 5c | 82 | 6c | 0 |
| 9 | 4d | B | 5d | 72 | 6d | 0 |
| 10 | 4a | C | 5a | 7 | 6a | 51 |
| 11 | 4c | C | 5c | 0 | 6c | 52 |
Isolated yields. Products were purified on a silica gel column.
Yield was 53% when 5b was purified on RP-HPLC.
H2O (2 equiv).
The CuSO4/sodium ascorbate-catalyzed cycloaddition of TMSN3 to alkynes 4a–d produced 5a–d in higher yields (60–82%; method B, entries 6–9) than by method A. However, treatment of alkyne 4a with TMSN3 in the presence of Ag2CO3 provided triazole 5a in a low yield (7%), affording instead the 5-(1-azidovinyl)cytosine nucleoside 6a34 as a major product (51%, entry 10). The analogous hydroazidation of 4c produced 5-(1-azidovinyl) 6c (52%) as the sole product (entry 11). It appears that paths for these reactions between nucleoside alkynes and TMSN3 depends not only on the catalyst used but also on the nature of the nucleobases, to which the alkyne group is attached, leading selectively to triazoles or vinyl azides.
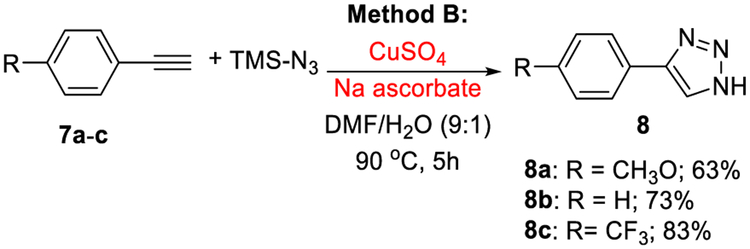
The CuSO4/sodium ascorbate-catalyzed synthesis of triazoles from alkynes and TMSN3 (method B) has a general character as illustrated with p-substituted phenylacetylenes 7a–c (Scheme 1). Thus, aryl alkyne 7a with EDG (CH3O) gave triazole product 8a (63%), while aryl alkyne 7c with EWG (CF3) yielded triazole 8c in a higher yield of 83%. Electron-withdrawing groups (EWGs) are known to promote the formation of triazoles in higher yields.23,35
Scheme 1.
Synthesis of p-Substituted Phenyl Triazoles by Method B
In summary, N-unsubstituted triazolyl analogues of the four natural bases of DNA were prepared by treatment of 5-ethynylpyrimidine or 8-ethynylpurine 2′-deoxynucleosides with TMSN3 in the presence of CuI or CuSO4/sodium ascorbate as a catalyst. Interestingly, Ag-catalyzed reactions of TMSN3 with 8-alkyne purines gave mainly triazole products 2a–c (Table 1), while with 5-alkyne pyrimidines provided mainly Markovnikov addition products 6a or 6c (Table 2), probably due to electronic differences between pyrimidine and purine imidazole rings. NMR studies showed that triazolyl nucleosides 2 and 5 are stable in D2O/DMSO-d6 solution (500 μL/50 μL; 2 mg/mL) at 37 °C for 72 h.
A reaction mechanism for the formation of triazoles might involve [3+2] cycloaddition or an initial formation of vinyl azide followed by 1,5-electrocyclization and tautomerization. However, attempts to validate the latter mechanism by treatment with 8-(1-azidovinyl)-2′-deoxyadenosine 3a using method C failed to produce the desired triazole 2a (Scheme 2). Also treatment of 5-(1-azidovinyl)-2′-deoxyuridine 6d using method B did not produce triazole 5d. Thus, the formation of the triazolyl unit involving [3+2] cycloaddition23 of alkyne with in situ generated HN3 seems to be plausible (Figure S1).
Scheme 2.
Attempted Conversion of Vinyl Azide to Triazole
Enzymatic Incorporation of 8-TrzdATP into DNA.
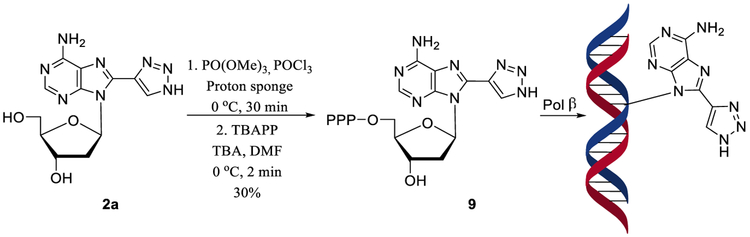
Treatment of 8-TrzdA 2a with POCl3 in the presence of a proton sponge36 followed by the addition of tributylammonium pyrophosphate (TBAPP) and tributylamine (TBA) yielded 8-TrzdATP 9 (Scheme 3).
Scheme 3.
Synthesis of 8-TrzdATP and Its Incorporation into DNA by Human DNA Polymerase β (Pol β)
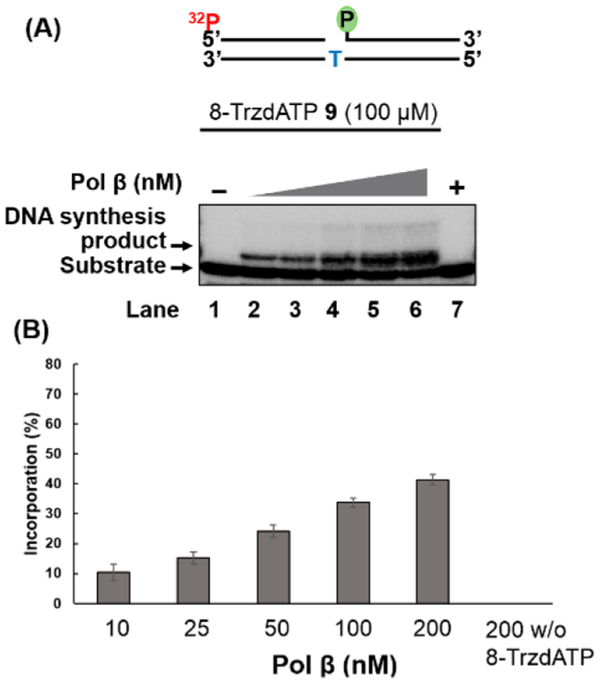
We also examined the incorporation of the triazolyl nucleotides into an oligonucleotide DNA substrate containing a nucleotide gap by pol β. The pol β efficiently incorporated 8-TrzdATP 9 into the one-nucleotide gap substrate containing a phosphate group on the downstream strand (Figure 2, Table S3). Incorporation of 8-TrzdATP increased with increasing concentrations of pol β (10–200 nM; lanes 2–6). Ten nM pol β was sufficient to insert 8-TrzdATP into the DNA to generate ~10% of the DNA synthesis product (lane 2). The 200 nM concentration of pol β resulted in 40% of incorporation of 8-TrzdATP (lane 6). Incorporation of 9 was extended by pol β in duplex DNA in the presence of dGTP (Figure S4). Polymerase-catalyzed incorporation of the 8-substituted purine nucleotides is known to depend on the size of the substituent and preference for syn/anti conformation of the base.37,38
Figure 2.
(A) Incorporation of 8-TrzdATP 9 into duplex DNA containing a one-nucleotide gap by pol β. Substrates were 32P-labeled at the 5′-end of the upstream strand of the substrate. Lane 1 indicates substrate only. Lanes 2–6 indicate the DNA synthesis product at increasing concentrations of pol β (10–200 nM). Lane 7 indicates the reaction with pol β without 8-TrzdATP. (B) Bar chart illustrating the quantification of the pol β DNA synthesis product.
Fluorescent Properties.
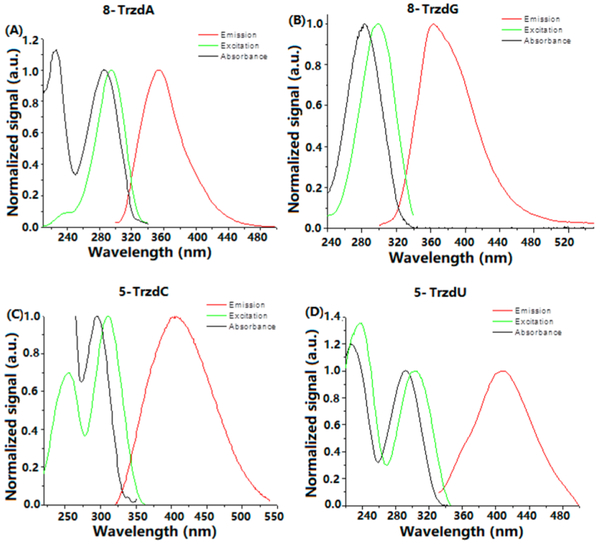
The normalized fluorescence emission, absorption, and excitation spectra for 1H-1,2,3-triazol-4-yl nucleosides in methanol are shown in Figure 3. Their photophysical data are summarized in Table 3. The 8-TrzdA 2a with the C4 of triazolyl attaching to the C8 of adenine exhibits the highest quantum yield (ΦF) of 44% and emits at 300–480 nm with the maximum emission at 355 nm. The silyl-protected 8-TrzdA analogue 2b has a ΦF of 48% (Table S1). The emission of 8-TrzdG 2c starts at 300 nm but extends to 540 nm, and the maximum emission is at 364 nm with a ΦF of 9%. Emission λmax of 2a is pH- and solvent-independent (Figure S2, Table S2).
Figure 3.
Normalized fluorescence emission, absorption, and excitation spectra for (A) 8-TrzdA, (B) 8-TrzdG, (C) 5-TrzdC, and (D) 5-TrzdU in MeOH. Absorption and excitation spectra were normalized to one at the absorption band of the lowest energy.
Table 3.
Photophysical Data for Compounds 2a, 2c, 5b, and 5da
| 2a | 2c | 5b | 5d | |
|---|---|---|---|---|
| εmax (M−1 cm−1) | 17950 | 14800 | 18750 | 12400 |
| λmax (abs) (nm) | 287 | 283 | 295 | 291 |
| λmax (exc) (nm) | 295 | 299 | 311 | 238 |
| λmax (exc) (nm) | 235 | 254 | 302 | |
| λmax (emi) (nm) | 355 | 364 | 407 | 408 |
| Stokes shift (nm) | 68 | 65 | 112 | 117 |
| ΦF | 0.44 | 0.09 | 0.02 | 0.004 |
| τ1 (ns) | 0.69 | 0.61 | 0.10 | 0.14 |
| τ2 (ns) | 2.07 | 3.22 | 4.35 | 1.06 |
| τaverage (ns) | 1.55 | 2.47 | 3.72 | 0.805 |
| f1 (%) | 37 | 28 | 15 | 27 |
| f2 (%) | 63 | 71 | 85 | 73 |
In MeOH
The 5-pyrimidine analogues 5-TrzdU 5d and 5-TrzdC 5b showed a larger Stokes shift of ~110 nm with a maximum emission approximately at 408 nm and lower quantum yields. The 5-TrzdC emits at 320–550 nm (ΦF = 2%), while 5-TrzdU emits at 320–500 nm (ΦF = 0.4%). Acetyl-protected 5a and 5c have similar fluorescent properties (Table S1). The emission spectra of 5b show a strong pH dependence in aqueous phosphate buffer with a bathochromic shift of the emission band maximum from 378 nm (pH 4.0) to 435 nm (pH 7.0) and to 433 nm (pH 12.0). Emission maxima in DMSO and ACN are similar to those observed in phosphate buffer at a neutral and basic pH (Figure S3, Table S2).
All triazoles showed biphasic fluorescence decay. The triazoles present a fast lifetime of 0.1–4.4 ns (Table 3). Compound 5b shows the longest lifetime of 4.35 ns and longest average lifetime of 3.7 ns. Compounds 2a (63%), 2c (71%), 5b (85%), and 5d (73%) showed a larger contribution of the long lifetime (τ2).
Fluorescent N-unsubstituted 1,2,3-triazol-4-yl deoxynucleo-sides have been synthesized by catalyst-dependent reactions between 5-ethynylpyrimidine or 8-ethynylpurine nucleosides with TMSN3. CuI or CuSO4/sodium ascorbate-catalyzed cycloaddition gave triazoles products for both purine and pyrimidine nucleosides. In contrast, Ag2CO3-catalyzed reactions with 8-ethynylpurine nucleosides produced 8-triazolyls, whereas 5-ethynylpyrimidine nucleosides followed the hydroazidation pathway to give 5-(1-azidovinyl) as a major product. The triazoles showed good fluorescent properties with 8-TrzdA, exhibiting the highest quantum yield of 44%. 8-TrzdATP was incorporated into duplex DNA containing a one-nucleotide gap by human DNA polymerase β. Metabolic incorporation of 8-purine and 5-pyrimidine 1,2,3-triazol-4-yl nucleosides into DNA for fluorescent imaging and their antiviral and anticancer evaluation will be published elsewhere.39
EXPERIMENTAL SECTION
General Information.
1H NMR spectra at 400 MHz and 13C NMR at 100.6 MHz were recorded in DMSO-d6 unless otherwise noted. All chemical shift values are reported in parts per million (ppm) and referenced to the residual solvent peaks of DMSO-d6 (2.50 ppm) for 1H NMR and DMSO-d6 (39.52 ppm) peaks for 13C NMR spectra, with coupling constant (J) values reported in hertz (Hz). HRMS were obtained in TOF (ESI) mode. TLC was performed on Merck silica gel 60-F254, and products were detected with 254 nm light. Merck silica gel 60 (230–400 mesh) was used for column chromatography. All reagents and solvents were purchased from commercial suppliers and used without further purification. Experimental procedures for the fluorescent characterization of the triazolyl nucleosides and protocols for the incorporation of 8-TrzdATP 9 by human DNA polymerase are detailed in the Supporting Information.
8-(1H-1,2,3-Triazol-4-yl)-2′-deoxyadenosine (2a).
Procedure A: CuI as a Catalyst.
The stirred solution of 1a28 (27.5 mg, 0.1 mmol) in DMF/H2O (1 mL, 9:1, v/v) was degassed with Ar for 15 min. TMSN3 (26.3 μL, 23 mg, 0.2 mmol) and CuI (1 mg, 0.005 mmol) were then added, and the resulting mixture was further degassed for another 5 min and was stirred at 90 °C for 5 h. After the mixture cooled to ambient temperature, the volatiles were evaporated and the residue was column chromatographed (CHCl3/MeOH, 100:0 → 85:15) to give 2a (15.0 mg, 48%): UV (MeOH) λmax 203, 228, 287 nm (ε 15 900, 16 050, 17 950), λmin 213, 250 nm (ε 13 800, 5100); 1H NMR δ 2.20 (ddd, J = 12.9, 5.9, 1.7 Hz, 1H), 3.12–3.19 (m, 1H), 3.49–3.55 (m, 1H), 3.67–3.72 (m, 1H), 3.90 (q, J = 3.7 Hz, 1H), 4.46–4.51 (m, 1H), 5.27 (d, J = 3.7 Hz, 1H), 5.77–5.80 (m, 1H), 7.07 (t, J = 7.2 Hz, 1H), 7.51 (s, 2H), 8.13 (s, 1H), 8.44 (s, 1H), 15.72 (s, 1H); 13C{1H} NMR δ 38.1, 62.4, 71.6, 85.9, 88.4, 119.4, 130.6, 137.8, 141.6, 149.8, 152.1, 156.1; HRMS (ESI) m/z calcd for C12H14N8O3Na [M + Na]+ 341.1081, found 341.1062.
Note that purification of the crude reaction mixture on RP-HPLC (solvent A, 100% ACN; solvent B, 5% ACN/H2O; gradient 0% A → 15% A in 30 min, flow rate = 2 mL/min) gave 2a (52%).
Procedure B: CuSO4/Sodium Ascorbate as a Catalyst.
The stirred solution of 1a (27.5 mg, 0.1 mmol) and CuSO4·5H2O (2.5 mg, 0.01 mmol) in DMF/H2O (1 mL, 9:1, v/v) was degassed with Ar for 15 min. TMSN3 (26.3 μL, 23 mg, 0.2 mmol) and sodium ascorbate (4 mg, 0.02 mmol) were then added, and the resulting mixture was further degassed for another 5 min and was stirred at 90 °C for 5 h. After the mixture was cooled to ambient temperature, the volatiles were evaporated and the residue was column chromatographed (CHCl3/MeOH, 100:0 → 85:15) to give 2a (19.7 mg, 62%) with the spectroscopic data as described above.
Procedure C: Ag2CO3 as a Catalyst.
Ag2CO3 (2.8 mg, 0.01 mmol) was added to a solution of 1a (27.5 mg, 0.1 mmol), TMSN3 (26.3 μL, 23 mg, 0.2 mmol), and H2O (3.6 μL, 3.6 mg, 0.2 mmol) in DMF (1 mL). The resulting mixture was stirred at 80 °C for 1 h. After the mixture cooled to ambient temperature, the volatiles were evaporated under the reduced pressure and the residue was column chromatographed (CHCl3/MeOH, 100:0 → 85:15) to give 3a (4.8 mg, 15%, see below for spectroscopic characterization) followed by 2a (18.4 mg, 58%).
3′,5′-Di-O-tert-butyldimethylsilyl-8-(1H-1,2,3-triazol-4-yl)-2′-deoxyadenosine (2b).
Treatment of 1b28 (201.6 mg, 0.4 mmol) with CuI by Procedure A (column chromatography; hexane/EtOAc 50:50 → 0:100) gave 2b (103.2 mg, 47%): UV (MeOH) λmax 225, 285 nm (ε 17 500, 14 100), λmin 247 nm (ε 3900); 1H NMR δ −0.13 (s, 3H), −0.07 (s, 3H), 0.12 (s, 6H), 0.77 (s, 9H), 0.90 (s, 9H), 2.20–2.27 (m, 1H), 3.57–3.66 (m, 2H), 3.77 (dd, J = 9.0, 4.7 Hz, 1H), 3.88 (dd, J = 10.8, 6.0 Hz, 1H), 4.88 (“q”, J = 4.2 Hz, 1H), 7.01 (t, J = 6.5 Hz, 1H), 7.36 (s, 2H), 8.13 (s, 1H), 8.41 (s, 1H); 13C{1H} NMR δ −5.6, −5.5, −4.9, −4.7, 17.8, 17.9, 25.6, 25.7, 36.4, 62.3, 72.2, 84.5, 86.6, 119.3, 130.6, 138.0, 142.0, 150.2, 152.3, 156.0; HRMS (ESI) m/z calcd for C24H43N8O3Si2 [M + H]+ 547.2991, found 547.3004.
Treatment of 1b (201.6 mg, 0.4 mmol) with CuSO4/sodium ascorbate by Procedure B (column chromatography; hexane/EtOAc 50:50 → 0:100) gave 2b (155.6 mg, 71%).
Treatment of 1b (201.6 mg, 0.4 mmol) with Ag2CO3 by Procedure C (column chromatography; hexane/EtOAc 50:50 → 0:100) gave 2b (120.4 mg, 55%).
8-(1H-1,2,3-Triazol-4-yl)-2′-deoxyguanosine (2c).
Treatment of 1c29 (29.1 mg, 0.1 mmol) with CuI by Procedure A (column chromatography; CHCl3/MeOH, 95:5 → 80:20) gave 2c (10.2 mg, 31%): UV (MeOH) λmax 205, 283 nm (ε 14 450, 14 800); λmin 238 (ε 2600); 1H NMR δ 2.07–2.13 (m, 1H), 3.12–3.19 (m, 1H), 3.50 (dd, J = 11.7, 5.3 Hz, 1H), 3.65 (dd, J = 11.8, 4.8 Hz, 1H), 3.78–3.81 (m, 1H), 4.38–4.44 (m, 1H), 5.05 (“s”, 1H), 5.18 (d, J = 3.7 Hz, 1H), 6.43 (s, 2H), 8.34 (t, J = 7.8 Hz, 1H), 10.83 (s, 1H), 15.42 (s, 1H); 13C{1H} NMR δ 37.4, 62.3, 71.4, 84.8, 88.0, 117.6, 128.8, 138.8, 151.9, 153.1, 154.0, 156.5; HRMS (ESI) m/z calcd for C12H15N8O4 [M + H]+ 335.1211, found 335.1214.
Note that purification of the crude reaction mixture on RP-HPLC (C18, A, 100% ACN; B, 5% ACN/H2O; 0% A → 15% A in 30 min, flow rate = 2 mL/min) gave 2c (36%).
Treatment of 1c (29.1 mg, 0.1 mmol) with CuSO4/sodium ascorbate by Procedure B (column chromatography; CHCl3/MeOH, 95:5 → 80:20) gave 2c (26.0 mg, 52%).
Treatment of 1c (29.1 mg, 0.1 mmol) with Ag2CO3 by Procedure C (column chromatography; CHCl3/MeOH, 95:5 → 80:20) gave 2c (30.1 mg, 60%).
8-(1-Azidovinyl)-2′-deoxyadenosine (3a):
4.8 mg, 15%; Procedure C, see above; 1H NMR δ 2.19 (ddd, J = 13.1, 6.3, 2.3 Hz, 1H), 3.20–3.27 (m, 1H), 3.47–3.54 (m, 1H), 3.67 (dt, J = 11.8, 4.0 Hz, 1H), 3.89 (q, J = 4.1 Hz, 1H), 4.46–4.51 (m, 1H), 5.31 (d, J = 4.1 Hz, 1H), 5.40 (d, J = 2.0 Hz, 1H), 5.46 (dd, J = 8.1, 4.0 Hz, 1H), 5.56 (d, J = 2.0 Hz, 1H), 6.36 (dd, J = 8.1, 6.4 Hz, 1H), 7.56 (s, 2H), 8.15 (s, 1H); 13C{1H} NMR δ 37.4, 62.1, 71.3, 85.6, 88.4, 108.7, 118.6, 134.2, 143.9, 149.5, 152.8, 156.4; HRMS (ESI) m/z calcd for C12H14N8O3Na+ [M + Na]+ 341.1081, found 341.1088.
3′,5′-Di-O-Acetyl-5-(1H-1,2,3-triazol-4-yl)-2′-deoxycyti-dine (5a).
Treatment of 4a40 (134.0 mg, 0.4 mmol) with CuI by Procedure A (column chromatography; CHCl3/MeOH, 100:0 → 90:10) to give 5a (67.6 mg, 45%): UV (MeOH) λmax 208, 238, 293 nm (ε 13 050, 9400, 4150), λmin 225, 271 nm (ε 8350, 3150); 1H NMR δ 1.99 (s, 3H), 2.08 (s, 3H), 2.36 (ddd, J = 14.1, 5.8, 2.0 Hz, 1H), 2.44–2.47 (m, 1H), 4.20–4.23 (m, 1H), 4.26–4.35 (m, 2H), 5.20–5.22 (m, 1H), 6.21 (dd, J =7.6, 6.2 Hz, 1H), 7.67 (s, 1H), 7.90 (s, 1H), 8.07 (s, 1H), 8.24 (s, 1H), 15.28 (s, 1H); 13C{1H} NMR δ 20.5, 20.7, 36.6, 63.7, 74.3, 81.6, 86.0, 97.2, 126.9 (HMQC showed a cross peak to proton at 8.24 ppm), 139.8, 153.6, 162.4, 170.0, 170.2; HRMS (ESI) m/z calcd for C15H19N6O6+ [M + H]+ 379.1361, found 379.1372.
Treatment of 4a (134.0 mg, 0.4 mmol) with CuSO4/sodium ascorbate by using Procedure B (column chromatography; CHCl3/MeOH, 100:0 → 90:10) gave 5a (97.6 mg, 65%).
5-(1H-1,2,3-Triazol-4-yl)-2′-deoxycytidine (5b).
Treatment of 4b40 (25.1 mg, 0.1 mmol) with CuI by using Procedure A (column chromatography; CHCl3/MeOH, 100:0 → 80:20) gave 5b (13.9 mg, 48%): UV (MeOH) λmax 207, 238, 296 nm (ε 18 750, 13 900, 5500), λmin 224, 273 nm (ε 11 900, 3500); 1H NMR δ 2.08–2.14 (m, 1H), 2.21 (ddd, J = 13.2, 6.1, 4.7 Hz, 1H), 3.59–3.66 (m, 1H), 3.68–3.75 (m, 1H), 3.82 (q, J = 3.3 Hz, 1H), 4.24–4.30 (m, 1H), 5.24 (s, 1H), 5.29 (s, 1H), 6.18 (t, J = 6.1 Hz, 1H), 7.68 (s, 1H), 7.80 (s, 1H), 8.07 (s, 1H), 8.60 (s, 1H), 15.18 (s, 1H); 13C{1H} NMR δ 41.4, 61.2, 70.0, 85.9, 88.0, 96.9, 126.8, 140.6, 142.0, 154.2, 162.7; HRMS (ESI) m/z calcd for C11H15N6O4+ [M + H]+ 295.1149, found 295.1160.
Note that the purification of the crude reaction mixture on RP-HPLC (C18, A, 100% ACN; B, 5% ACN/H2O; 0% A → 15% A in 30 min, flow rate = 2 mL/min) gave 5b (53%).
Treatment of 4b (25.1 mg, 0.1 mmol) with CuSO4/sodium ascorbate by using Procedure B (column chromatography; CHCl3/MeOH, 100:0 → 80:20) gave 5b (17.4 mg, 60%).
3′,5′-Di-O-Acetyl-5-(1H-1,2,3-triazol-4-yl)-2′-deoxyuridine (5c).
Treatment of 4c33 (33.6 mg, 0.1 mmol) with CuI by using Procedure A (column chromatography; CHCl3/MeOH, 100:0 → 92:8) gave 5c (24.6 mg, 65%): UV (MeOH) λmax 231, 293 nm (ε 10 700, 9600), λ min 258 nm (ε 2900); 1H NMR δ 2.08 (s, 3H), 2.13 (s, 3H), 2.36–2.46 (m, 2H), 4.22–4.26 (m, 2H), 4.27–4.31 (m, 1H), 5.21–5.26 (m, 1H), 6.25 (t, J = 6.3 Hz, 1H), 8.18 (s, 1H), 8.31 (s, 1H), 11.82 (s, 1H), 15.19 (s, 1H); 13C{1H} NMR δ 20.7, 20.8, 36.7, 63.8, 74.2, 81.7, 84.9, 105.5, 128.2 (HMQC showed a cross peak to proton at 8.18 ppm), 135.8, 149.6, 161.2, 170.1, 170.4; HRMS (ESI) m/z calcd for C15H18N5O7+ [M + H]+ 380.1201, found 380.1208.
Treatment of 4c (33.6 mg, 0.1 mmol) with CuSO4/sodium ascorbate by using Procedure B (column chromatography; CHCl3/MeOH, 100:0 → 92:8) gave 5c (31.0 mg, 82%).
5-(1H-1,2,3-Triazol-4-yl)-2′-deoxyuridine (5d).
Treatment of 4d33 (25.1 mg, 0.1 mmol) with CuI by using Procedure A (column chromatography; CHCl3/MeOH, 100:0 → 85:15) gave 5d20,21 (18.4 mg, 62%): UV (MeOH) λmax 231, 292 nm (ε 12 400, 11 450), λmin 259 nm (ε 3700); 1H NMR δ 2.18 (dd, J = 6.3, 4.7 Hz, 2H), 3.55–3.63 (m, 2H), 3.84 (q, J = 3.4 Hz, 1H), 4.25–4.31 (m, 1H), 5.04 (“s”, 1H), 5.29 (d, J = 4.1 Hz, 1H), 6.22 (t, J = 6.6 Hz, 1H), 8.14 (s, 1H), 8.49 (s, 1H), 11.68 (s, 1H), 15.10 (s, 1H); 13C{1H} NMR δ 39.9, 61.3, 70.6, 84.7, 87.6, 105.1, 131.8, 135.8, 137.0, 149.7, 161.2; HRMS (ESI) m/z calcd for C11H14N5O5+, [M + H]+ 296.0989, found 296.0983.
Treatment of 4d (25.1 mg, 0.1 mmol) with CuI by using modified Procedure A with 2 equiv of H2O and DMF as a solvent (column chromatography; CHCl3/MeOH, 100:0 → 85:15) gave 5d (14.9 mg, 50%).
Treatment of 4d (25.1 mg, 0.1 mmol) with CuSO4/sodium ascorbate by using Procedure B (column chromatography; CHCl3/MeOH, 100:0 → 85:15) gave 5d (21.2 mg, 72%).
3′,5′-Di-O-Acetyl-5-(1-azidovinyl)-2′-deoxycytidine (6a).
Treatment of 4a (134.0 mg, 0.4 mmol) with Ag2CO3 by using Procedure C (column chromatography; CHCl3/MeOH, 100:0 → 90:10) gave 6a (75.6 mg, 51%) followed by 5a (10.4 mg, 7%). Compound 6a: 1H NMR (CDCl3) δ 2.02–2.14 (m, 1H), 2.08 (s, 3H), 2.10 (s, 3H), 2.75 (ddd, J = 14.3, 5.5, 2.0 Hz, 1H), 4.31–4.41 (m, 3H), 5.03 (d, J = 1.5 Hz, 1H), 5.11 (d, J = 1.4 Hz, 1H), 5.21 (dt, J = 6.3, 1.8 Hz, 1H), 5.83 (s, 2H) 6.28 (dd, J = 8.0, 5.6 Hz, 1H), 7.80 (s, 1H); 13C[1H] NMR (CDCl3) δ 20.8, 21.0, 39.0, 64.0, 74.4, 82.9, 86.8, 101.8, 103.0, 139.9, 140.2, 154.3, 162.7, 170.4, 170.6; HRMS (ESI) m/z calcd for C15H19N6O6+ [M + H]+ 379.1361, found 379.1355.
3′,5′-Di-O-Acetyl-5-(1-azidovinyl)-2′-deoxyuridine (6c).
Treatment of 4c (67.2 mg, 0.2 mmol) with Ag2CO3 by using Procedure C (hexane/EtOAc 50:50) gave 6c41 (41.2 mg, 52%): 1H NMR δ 2.07 (s, 6H), 2.33–2.46 (m, 2H), 4.24–4.29 (m, 3H), 5.06 (“s”, 1H), 5.19–5.21 (m, 1H), 6.00 (“s”, 1H), 6.14 (t, J = 6.4 Hz, 1H), 7.83 (s, 1H), 11.73 (s, 1H); 13C[1H] NMR δ 20.4, 20.8, 36.7, 63.8, 74.2, 81.8, 85.4, 101.6, 107.3, 136.9, 138.3, 149.3, 160.9, 170.1, 170.2; HRMS (ESI) m/z calcd for C15 H18N5O7+ [M + H]+ 380.1201, found 380.1195.
4-(4-Methoxyphenyl)-1H-1,2,3-triazole (8a).
Treatment of 7a (13.2 mg, 0.1 mmol) with CuSO4/sodium ascorbate by using Procedure B (column chromatography; hexane/EtOAc, 100:0 → 70:30) gave 8a23,42 (11 mg, 63%): 1H NMR δ 3.80 (s, 3H), 7.02 (d, J = 8.8 Hz, 2H), 7.78 (d, J = 8.8 Hz, 2H), 8.22 (s, 1H); 13C{1H} NMR δ 55.6, 114.5, 114.8, 123.3, 127.4, 145.8, 159.7; HRMS (ESI) m/z calcd for C9H10N3O+ [M +H]+ 176.0818, found 176.0822.
4-Phenyl-1H-1,2,3-triazole (8b).
Treatment of 7b (20 mg, 0.2 mmol) with CuSO4/sodium ascorbate by using Procedure B (column chromatography; hexane/EtOAc, 100:0 → 70:30) gave 8b23,42 (21 mg, 73%): 1H NMR δ 7.39 (t, J = 7.2 Hz, 1H), 7.50 (t, J = 7.5 Hz, 2H), 7.86 (d, J = 7.3 Hz, 2H), 8.34 (s, 1H); 13C{1H} NMR δ 126.0, 127.7, 128.5, 129.4, 130.8, 145.6; HRMS (ESI) m/z calcd for C8H8N3+ [M + H]+ 146.0713, found 146.0708.
4-(4-Trifluoromethylphenyl)-1H-1,2,3-triazole (8c).
Treatment of 7c (17 mg, 0.1 mmol) with CuSO4/sodium ascorbate by using Procedure B (column chromatography; hexane/EtOAc, 100:0 → 70:30) gave 8c42 (17.6 mg, 83%): 1H NMR δ 7.82 (d, J = 8.4 Hz, 2H), 8.10 (d, J = 8.1 Hz, 2H), 8.54 (s, 1H); 13C{1H} NMR δ 124.7 (q, J = 271.8 Hz), 126.4 (q, J = 3.8 Hz), 126.5, 127.9, 128.8 (q, J = 31.9 Hz), 135.0, 144.7; 19F NMR δ −1.0; HRMS (ESI) m/z calcd for C9H7F3N3+ [M +H]+ 214.0587, found 214.0593.
8-(1H-1,2,3-Triazol-4-yl)-2′-deoxyadenosine 5′-triphosphate (8-TrzdATP, 9).
POCl3 (28 μL, 46 mg, 0.3 mmol) was added to a stirred solution of 8-TrzdA 2a (48 mg, 0.15 mmol) and proton sponge (80 mg, 0.375 mmol) in (MeO)3PO (2 mL) at 0 °C. The resulting mixture was stirred at 0 °C for 30 min; then tributylammomium pyrophosphate solution in DMF (0.5 M; 1.5 mL, 0.75 mmol) followed by Bu3N (106.8 μL, 83.4 mg, 0.45 mmol) were added, and stirring was continued at 0 °C for 2 min. The reaction was quenched by adjusting the pH to 7.5 with 2 M TEAB buffer. The residue was dissolved in H2O (5 mL) and was extracted with EtOAc (3 × 5 mL). The water layer was evaporated and coevaporated with a mixture of EtOH/H2O (1:1, 5 mL). The residue was column chromatographed (DEAE–Sephadex, TEAB 0.1 M → 0.6 M) and the appropriate fractions were evaporated in a vacuum and coevaporated 5 times with a mixture of EtOH/H2O (1:1, 10 mL) to give 8-TrzdATP triethylammonium salt 9 (33.4 mg, 30%): 1H NMR (D2O) δ 2.33 (ddd, J = 13.4, 6.5, 3.9, 1H), 3.22–3.26 (m, 1H), 4.04–4.10 (m, 1H), 4.14 (“q”, J = 5.2 Hz, 1H), 4.18–4.24 (m, 1H), 4.66 (“quint”, J = 3.9 Hz, 1H), 6.74 (t, J = 7.8 Hz, 1H), 8.20 (s, 1H), 8.41 (s, 1H); 31P NMR (D2O) δ –23.22 (t, J = 21.0 Hz, 1Pβ), −11.34 (d, J = 21.0 Hz, 1Pα), −10.22 (d, J = 21.0 Hz, 1Pγ); 13C{1H} NMR (D2O) δ 36.2, 65.2, 70.6, 84.3, 84.8, 118.6, 128.8, 136.1, 143.0, 149.9, 152.6, 155.0; HRMS m/z calcd for C12H16N8O12P3 [M – H]– 557.0106, found 557.0091.
Supplementary Material
ACKNOWLEDGMENTS
Z.W. and A.H.H. are recipients of the FIU Dissertation Year and Presidential Fellowships, respectively. This work was partially supported by NIH (S.F.W., SC1CA138176; Y.L., ES023569).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.joc.8b03135.
NMR spectra, mechanistic study, and fluorescent characterization (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Daniels M; Hauswirth W Fluorescence of the Purine and Pyrimidine Bases of the Nucleic Acids in Neutral Aqueous Solution at 300 K. Science 1971, 171, 675–677. [DOI] [PubMed] [Google Scholar]
- (2).Holz B; Klimasauskas S; Serva S; Weinhold E 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res 1998, 26, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Seo YJ; Ryu JH; Kim BH Quencher-Free, End-Stacking Oligonucleotides for Probing Single-Base Mismatches in DNA. Org. Lett 2005, 7, 4931–4933. [DOI] [PubMed] [Google Scholar]
- (4).Greco NJ; Tor Y Simple Fluorescent Pyrimidine Analogues Detect the Presence of DNA Abasic Sites. J. Am. Chem. Soc 2005, 127, 10784–10785. [DOI] [PubMed] [Google Scholar]
- (5).Srivatsan SG; Tor Y Fluorescent Pyrimidine Ribonucleotide: Synthesis, Enzymatic Incorporation, and Utilization. J. Am. Chem. Soc 2007, 129, 2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zayas J; Annoual M; Das JK; Felty Q; Gonzalez WG; Miksovska J; Sharifai N; Chiba A; Wnuk SF Strain Promoted Click Chemistry of 2- or 8-Azidopurine and 5-Azidopyrimidine Nucleosides and 8-Azidoadenosine Triphosphate with Cyclooctynes. Application to Living Cell Fluorescent Imaging. Bioconjugate Chem 2015, 26, 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Srivatsan SG; Tor Y Using an emissive uridine analogue for assembling fluorescent HIV-1 TAR constructs. Tetrahedron 2007, 63, 3601–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Tanpure AA; Pawar MG; Srivatsan SG Fluorescent Nucleoside Analogs: Probes for Investigating Nucleic Acid Structure and Function. Isr. J. Chem 2013, 53, 366–378. [Google Scholar]
- (9).Wilhelmsson LM Fluorescent nucleic acid base analogues. Q. Rev. Biophys 2010, 43, 159–183. [DOI] [PubMed] [Google Scholar]
- (10).Xu W; Chan KM; Kool ET Fluorescent nucleobases as tools for studying DNA and RNA. Nat. Chem 2017, 9, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ozols K; Cirule D; Novosjolova I; Stepanovs D; Liepinsh E; Bizdena E; Turks M Development of N6-methyl-2-(1,2,3-triazol-1-yl)-2′-deoxyadenosine as a novel fluorophore and its application in nucleotide synthesis. Tetrahedron Lett 2016, 57, 1174–1178. [Google Scholar]
- (12).Kavoosi S; Rayala R; Walsh B; Barrios M; Gonzalez WG; Miksovska J; Mathivathanan L; Raptis RG; Wnuk SF Synthesis of 8-(1,2,3-triazol-1-yl)-7-deazapurine nucleosides by azide–alkyne click reactions and direct CH bond functionalization. Tetrahedron Lett 2016, 57, 4364–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lawson CP; Dierckx A; Miannay F-A; Wellner E; Wilhelmsson LM; Grotli M Synthesis and photophysical characterisation of new fluorescent triazole adenine analogues. Org. Biomol. Chem 2014, 12, 5158–5167. [DOI] [PubMed] [Google Scholar]
- (14).Dyrager C; Börjesson K; Dinér P; Elf A; Albinsson B; Wilhelmsson LM; Grøtli M Synthesis and Photophysical Characterisation of Fluorescent 8-(1H-1,2,3-Triazol-4-yl)adenosine Derivatives. Eur. J. Org. Chem 2009, 1515–1521. [Google Scholar]
- (15).Redwan IN; Bliman D; Tokugawa M; Lawson C; Grøtli M Synthesis and photophysical characterization of 1- and 4-(purinyl)triazoles. Tetrahedron 2013, 69, 8857–8864. [Google Scholar]
- (16).Bag SS; Gogoi H Design of “Click” Fluorescent Labeled 2′-deoxyuridines via C5-[4-(2-Propynyl(methyl)amino)]phenyl Acetylene as a Universal Linker: Synthesis, Photophysical Properties, and Interaction with BSA. J. Org. Chem 2018, 83, 7606–7621. [DOI] [PubMed] [Google Scholar]
- (17).Kumar P; Hornum M; Nielsen LJ; Enderlin G; Andersen NK; Len C; Hervé G; Sartori G; Nielsen P High-Affinity RNA Targeting by Oligonucleotides Displaying Aromatic Stacking and Amino Groups in the Major Groove. Comparison of Triazoles and Phenyl Substituents. J. Org. Chem 2014, 79, 2854–2863. [DOI] [PubMed] [Google Scholar]
- (18).Hong SW; Hwang GT Photophysical Study of 2-Fluorenyl-1,2,3-Triazole-labeled 2′-Deoxyuridine and Its Oligonucleotide. Bull. Korean Chem. Soc 2018, 39, 78–83. [Google Scholar]
- (19).Dierckx A; Dinér P; El-Sagheer AH; Kumar JD; Brown T; Grøtli M; Wilhelmsson LM Characterization of photophysical and base-mimicking properties of a novel fluorescent adenine analogue in DNA. Nucleic Acids Res 2011, 39, 4513–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kočalka P; Andersen NK; Jensen F; Nielsen P Synthesis of 5-(1,2,3-Triazol-4-yl)-2′-deoxyuridines by a Click Chemistry Approach: Stacking of Triazoles in the Major Groove Gives Increased Nucleic Acid Duplex Stability. ChemBioChem 2007, 8, 2106–2116. [DOI] [PubMed] [Google Scholar]
- (21).Hatano A; Kurosu M; Yonaha S; Okada M; Uehara S One-pot approach to functional nucleosides possessing a fluorescent group using nucleobase-exchange reaction by thymidine phosphorylase. Org. Biomol. Chem 2013, 11, 6900–6905. [DOI] [PubMed] [Google Scholar]
- (22).Hornum M; Djukina A; Sassnau A-K; Nielsen P Synthesis of new C-5-triazolyl-functionalized thymidine analogs and their ability to engage in aromatic stacking in DNA : DNA and DNA : RNA duplexes. Org. Biomol. Chem 2016, 14, 4436–4447. [DOI] [PubMed] [Google Scholar]
- (23).Jin T; Kamijo S; Yamamoto Y Copper-Catalyzed Synthesis of N-Unsubstituted 1,2,3-Triazoles from Nonactivated Terminal Alkynes. Eur. J. Org. Chem 2004, 3789–3791. [Google Scholar]
- (24).Barluenga J; Valdés C; Beltrán G; Escribano M; Aznar F Developments in Pd Catalysis: Synthesis of 1H-1,2,3-Triazoles from Sodium Azide and Alkenyl Bromides. Angew. Chem., Int. Ed 2006, 45, 6893–6896. [DOI] [PubMed] [Google Scholar]
- (25).Quan X-J; Ren Z-H; Wang Y-Y; Guan Z-H p-Toluenesulfonic Acid Mediated 1,3-Dipolar Cycloaddition of Nitroolefins with NaN3 for Synthesis of 4-Aryl-NH-1,2,3-triazoles. Org. Lett 2014, 16, 5728–5731. [DOI] [PubMed] [Google Scholar]
- (26).Kalisiak J; Sharpless KB; Fokin VV Efficient Synthesis of 2-Substituted-1,2,3-triazoles. Org. Lett 2008, 10, 3171–3174. [DOI] [PubMed] [Google Scholar]
- (27).Cha H; Lee K; Chi DY Synthesis of N-unsubstituted 1,2,3-triazoles via aerobic oxidative N-dealkylation using copper (II) acetate. Tetrahedron 2017, 73, 2878–2885. [Google Scholar]
- (28).Reddy MR; Shibata N; Kondo Y; Nakamura S; Toru T Design, Synthesis, and Spectroscopic Investigation of Zinc Dodecakis-(trifluoroethoxy)phthalocyanines Conjugated with Deoxyribonucleosides. Angew. Chem 2006, 118, 8343–8346. [DOI] [PubMed] [Google Scholar]
- (29).Shinohara Y; Matsumoto K; Kugenuma K; Morii T; Saito Y; Saito I Design of environmentally sensitive fluorescent 2′-deoxyguanosine containing arylethynyl moieties: Distinction of thymine base by base-discriminating fluorescent (BDF) probe. Bioorg. Med. Chem. Lett 2010, 20, 2817–2820. [DOI] [PubMed] [Google Scholar]
- (30).Liu Z; Liao P; Bi X General Silver-Catalyzed Hydroazidation of Terminal Alkynes by Combining TMS-N3 and H2O: Synthesis of Vinyl Azides. Org. Lett 2014, 16, 3668–3671. [DOI] [PubMed] [Google Scholar]
- (31).For a review on the chemistry of vinyl azides see:; Hu B; DiMagno SG Reactivities of vinyl azides and their recent applications in nitrogen heterocycle synthesis. Org. Biomol. Chem 2015, 13, 3844–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Suzol SH; Howlader AH; Wen Z; Ren Y; Laverde EE; Garcia C; Liu Y; Wnuk SF Pyrimidine Nucleosides with a Reactive (β-Chlorovinyl)sulfone or (β-Keto)sulfone Group at the C5 Position, Their Reactions with Nucleophiles and Electrophiles, and Their Polymerase-Catalyzed Incorporation into DNA. ACS Omega 2018, 3, 4276–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Liang Y; Suzol SH; Wen Z; Artiles AG; Mathivathanan L; Raptis RG; Wnuk SF Uracil Nucleosides with Reactive Group at C5 Position: 5-(1-Halo-2-sulfonylvinyl)uridine Analogues. Org. Lett 2016, 18, 1418–1421. [DOI] [PubMed] [Google Scholar]
- (34).For studies on the generation of aminyl and iminyl radicals from 5-(1-azidovinyl)pyrimidinyl nucleosides and their potential application as radiosensitizers, see:; Wen Z; Peng J; Tuttle PR; Ren Y; Garcia C; Debnath D; Rishi S; Hanson C; Ward S; Kumar A; Liu Y; Zhao W; Glazer PM; Liu Y; Sevilla MD; Adhikary A; Wnuk SF Electron-Mediated Aminyl and Iminyl Radicals from C5 Azido-Modified Pyrimidine Nucleosides Augment Radiation Damage to Cancer Cells. Org. Lett 2018, 20, 7400–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Journet M; Cai D; Kowal JJ; Larsen RD Highly efficient and mild synthesis of variously 5-substituted-4-carbaldehyde-1,2,3-triazole derivatives. Tetrahedron Lett 2001, 42, 9117–9118. [Google Scholar]
- (36).Kovács T; Ötvös L Simple synthesis of 5-vinyl- and 5-ethynyl-2′-deoxyuridine-5′-triphosphates. Tetrahedron Lett 1988, 29, 4525–4528. [Google Scholar]
- (37).For selected papers on the polymerase-catalyzed incorporation of 8-substituted purine nucleotides into DNA, see:; (a) Cahova H; Pohl R; Bednarova L; Novakova K; Cvacka J; Hocek M Synthesis of 8-bromo-, 8-methyl- and 8-phenyl-dATP and their polymerase incorporation into DNA. Org. Biomol. Chem 2008, 6, 3657–3660 [DOI] [PubMed] [Google Scholar]; (e.g., 8-bromo dATP = good substrate, 8-methyl dATP = moderate substrate, and 8-phenyl dATP = poor substrate).; (b) Kamath-Loeb AS; Hizi A; Kasai H; Loeb LA Incorporation of the Guanosine Triphosphate Analogs 8-Oxo-dGTP and 8-NH2- dGTP by Reverse Transcriptases and Mammalian DNA Polymerases.J. Biol. Chem 1997, 272, 5892–5898. [DOI] [PubMed] [Google Scholar]; (8-NH2-dGTP is a better substrate than 8-Oxo-dGTP.)
- (38).For selected papers on the effects of the 8-substituted purine nucleotides on polymerases-catalyzed DNA synthesis, see:; (a) Vivet-Boudou V; Isel C; Sleiman M; Smyth R; Ben Gaied N; Barhoum P; Laumond G; Bec G; Götte M; Mak J; Aubertin A-M; Burger A; Marquet R 8-Modified-2’-Deoxyadenosine Analogues Induce Delayed Polymerization Arrest during HIV-1 Reverse Transcription. PLoS One 2011, 6, e27456. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chen LS; Sheppard TL Chain Termination and Inhibition of Saccharomyces cerevisiae Poly(A) Polymerase by C-8-modified ATP Analogs. J. Biol. Chem 2004, 279, 40405–40411. [DOI] [PubMed] [Google Scholar]; (c) Jäger S; Rasched G; Kornreich-Leshem H; Engeser M; Thum O; Famulok M A Versatile Toolbox for Variable DNA Functionalization at High Density. J. Am. Chem. Soc 2005, 127, 15071–15082. [DOI] [PubMed] [Google Scholar]
- (39).N-Unsubstituted triazolyl nucleosides might enhance the anticancer activity of cisplatin by augmenting binding between cisplatin and the triazole moiety. For studies on the complexation of triazoles with cisplatin, see:; (a) Komeda S; Lutz M; Spek AL; Chikuma M; Reedijk J Inorg. Chem 2000, 39, 4230–4236. [DOI] [PubMed] [Google Scholar]; (b) Komeda S; Lutz M; Spek AL; Yamanaka Y; Sato T; Chikuma M; Reedijk JJ Am. Chem. Soc 2002, 124, 4738–4746. [DOI] [PubMed] [Google Scholar]
- (40).Dodd DW; Swanick KN; Price JT; Brazeau AL; Ferguson MJ; Jones ND; Hudson RHE Blue fluorescent deoxycytidine analogues: convergent synthesis, solid-state and electronic structure, and solvatochromism. Org. Biomol. Chem 2010, 8, 663–666. [DOI] [PubMed] [Google Scholar]
- (41).Kumar R; Wiebe LI; Knaus EE Synthesis of 5-(1-azidovinyl) and 5-[2-(1-azirinyl)] analogs of 2′-deoxyuridine. Can. J. Chem 1996, 74, 1609–1615. [Google Scholar]
- (42).Lee H; Lee JK; Min S-J; Seo H; Lee Y; Rhee H Copper(I)-Catalyzed Synthesis of 1,4-Disubstituted 1,2,3-Triazoles from Azidoformates and Aryl Terminal Alkynes. J. Org. Chem 2018, 83, 4805–4811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.