Abstract
Objective:
To characterize the scientific reproducibility of biomedical research studies by query and analysis of semantic provenance graphs generated from provenance metadata terms extracted from PubMed articles.
Methods.
We develop a new semantic provenance graph generation algorithm that uses a provenance ontology developed as part of the Provenance for Clinical and Health Research (ProvCaRe) project. The ProvCaRe project has processed and extracted provenance metadata from more than 1.6 million full text articles from the PubMed database.
Results.
The semantic provenance graph generation algorithm is evaluated using provenance terms extracted from 75 selected articles describing sleep medicine research studies. In addition, we use eight provenance queries to evaluate the quality of semantic provenance graph generated by the new algorithm.
Conclusion.
The ProvCaRe project has created a unique resource to characterize the reproducibility of biomedical research studies and the semantic provenance graph generation algorithm enables users to effectively query and analyze the provenance metadata in the ProvCaRe knowledge repository.
Keywords: Knowledge Bases, Biomedical Ontologies, Health Information Systems
Introduction
Provenance metadata describes the history or origin of data, therefore it plays a critical role in supporting reproducibility of results from scientific experiments. Scientific reproducibility has received increasing attention in the biomedical research community, including clinical research, drug development, and basic science research, with publication of the U.S. National Institutes of Health (NIH) “Rigor and Reproducibility guidelines” [6; 15]. Reproducibility of results from biomedical research studies is critical to ensure that valid results are used to design new research studies, develop new drugs with less likelihood of failing during clinical trials, and ensure allocation of limited funding resources to rigorously designed research studies [13; 21]. Provenance metadata represents essential information about research studies, including protocols, data acquisition methods, and statistical analysis models.
However, there has been limited work in the use of provenance metadata to support reproducibility in biomedical research. Therefore, as part of the Provenance for Clinical and Health Research (ProvCaRe) project, we are developing a framework for using semantic provenance metadata for characterizing the reproducibility of biomedical research studies [23]. We have developed a provenance-focused natural language processing (NLP) pipeline to extract provenance metadata terms from more than 1.6 million full-length articles in the National Center for Biotechnology Information (NCBI) PubMed database consisting of 28 million citations [1]. The extracted provenance information are stored in a structured form in the ProvCaRe knowledge repository.
The ProvCaRe knowledge repository is a unique resource for sudying and characterizing the reproducibility of biomedical research studies using a systematic and exhaustive approach. Provenance metadata is intuitively modeled as a graph consisting of nodes and edges, for example ResearchStudy → hadCohortSizeOf → 5861(participants), where ResearchStudy and 5861(participants) are nodes and hadCohortSizeOf is an edge [8]. The World Wide Web Consortium (W3C), which is the organization for Web technology standards, developed the PROV specifications to facilitate interoperability and interchage of provenance metadata in data management and Web-based information systems [16; 19]. The PROV specifications can be extended to biomedical domain to model provenance as a graph with formal semantics with terms mapped to ontologies. We used this approach to generate semantic provenance information in the ProvCaRe knowledge repository with terms mapped to a provenance ontology to support queries related to scientific reproducibility.
In this paper, we investigate the generation of provenance graphs from the structured provenance information stored in the ProvCaRe knowledge repository. In particular, we describe a new method to transform structured provenance information into a W3C Resource Description Framework (RDF)-based semantic provenance graph. We also describe an approach to query and analyze the provenance graph using a test case of provenance information from 75 selected articles.
Background
Resources
W3C PROV Specifications
The W3C PROV specifications consists of: (1) the core model called PROV-DM, which defines a common terminology system for representing interoperable provenance metadata [19]; (b) the PROV Ontology (PROV-O) that represents PROV-DM in the W3C Web Ontology Language (OWL2) [12; 16]; and a set of constraints (PROV-Constraints) to define valid forms of provenance conforming to the PROV specifications [4]. We extended PROV-O in the ProvCaRe project to model provenance terms associated with biomedical research studies. In particular, we extended the three core concepts of PROV-O, namely “prov:Agent”, “prov:Entity”, “prov:Activity”; and its properties, namely “prov:wasDerivedFrom”, “prov:wasGeneratedBy”, and “prov:used”. The namespace prov refers to “http://www.w3.org/ns/prov#”.
ProvCaRe Knowledge Repository
The provenance terms extracted by the ProvCaRe NLP pipeline is currently stored in the ProvCaRe knowledge repository as set of three related provenance terms (for a detailed description of the NLP pipeline, we refer to [23]). For example, we stored provenance terms extracted from an article describing study on sleep-disordered breathing and hypertension by O’Connor et al. [20] as “hypertension status of participants”, “was ascertained at”, “second follow-up exam” in the ProvCaRe knowledge repository. However, this representation of provenance metadata has some limitations, for example (1) it is verbose (making it difficult to query efficiently); and (2) it cannot be modeled as a RDF graph structure. Given a user query, “How was the hypertension status of participants determined in this research study?”, the ProvCaRe query interface uses keyword-based matching together with ontology-based “expansion” of query expression for retrieving query results. Therefore, we address these challenges by developing a new algorithm to map the provenance information in the ProvCaRe repository to a semantic provenance graph.
Related Work
Provenance metadata has long been used in fine arts, library management, and computer science to track art items, literary resources, and data elements. In particular, provenance metadata plays a critical role in ensuring data quality [3] and reproducibility of experiment results [17]. Provenance terms describing the context of biomedical research experiments, for example sample size of the experiment, randomization techniques used, protocol used for selection of participants in a research study cohort, are critical for enabling the reproducibility of research study results [15].
Although, previous work in biomedical research domain had limited or no focus on provenance metadata for reproducibility, a number of guidelines and best practices have been adopted to improve reporting of research studies to support reproducibility. For example, the NIH “Rigor and Reproducibility guidelines” [10], and the Ontology for Clinical Research (OCRe) project, which has developed a formal model of clinical research studies [24]. Projects that have explored the role of provenance in scientific reproducibility in other domains include work by Chirigati [5] and [18]. Many projects have also explored querying of provenance graphs using query language for workflow provenance [2] and Semantic Web technologies such as SPARQL [17; 22].
To the best of our knowledge, ProvCaRe is first project to generate semantic provenance graph from unstructured text to support queries for scientific reproducibility.
Methods
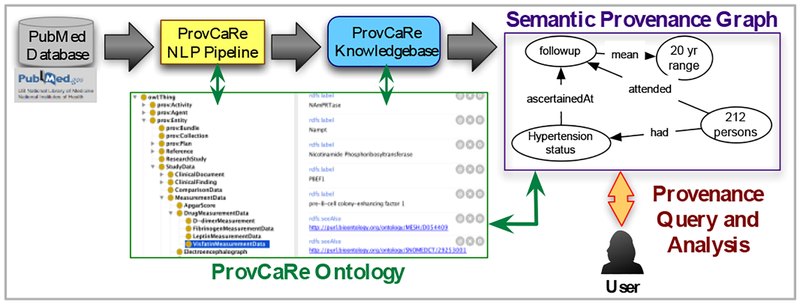
An overview of the ProvCaRe platform with different components is shown in Figure 1 with a provenance-focused NLP pipeline, storage of provenance triples in the knowledge repository, and the creation of semantic provenance graphs. The ProvCaRe ontology plays a central role as the reference knowledge model during both the extraction of provenance metadata and generation of provenance graphs.
Figure 1:
Architecture of the ProvCaRe framework with components, including the ProvCaRe ontology.
ProvCaRe ontology for biomedical provenance metadata
The ProvCaRe ontology has been developed by extending the classes and properties of the W3C PROV ontology using description logic OWL2 [16]. The two key design objectives of the ProvCaRe ontology are: (1) modeling the essential components of a research study, and (2) incorporating terms from existing domain ontologies for terms specific to different medical disciplines to provide appropriate level of granularity for provenance terms. Example terms modeled in the ProvCaRe ontology to meet the first objective are: the method used to conduct the study, the instruments used to record or analyze data in the study, and the different types of data used in the research study. Similarly, to meet the second objective, the ProvCaRe ontology currently models terms specific to sleep medicine, endocrinology, and neurological disorders. Example terms in the ProvCaRe ontology include, polysomnogram (sleep medicine), electroencephalogram (neurology), and Hyperaldosteronism (endocrinology). These terms are mapped to existing biomedical ontologies, such as SNOMED CT, National Cancer Institute (NCI) Thesaurus, and Gene Ontology (GO).
These mappings allow the provenance terms mapped to ProvCaRe ontology classes to reference terms in existing biomedical ontologies, which supports interoperability of the ProvCaRe knowledgebase with existing biomedical databases, such as NCBI Gene and Protein database. The ProvCaRe NLP pipeline uses the ProvCaRe ontology to identify and extract provenance terms from full-text articles in the PubMed database. At present, the ProvCaRe ontology consists of about 300 classes and 40 object properties. The ProvCaRe ontology plays a key role in the generation of the semantic provenance graphs from the provenance terms as it provides the schematic model to represent the nodes and edges of the graph structure.
Generating semantic provenance graph
The W3C Resource Description Framework (RDF) is a widely used Semantic Web technology for representing graph structures [11]. A RDF graph consists of “subject”, “predicate”, and “object”. For example, ResearchStudy → wasApprovedBy → EthicsCommittee, where ResearchStudy is the “subject”, wasApprovedBy is the “predicate”, and EthicsCommittee is the “object” of the RDF triple. Two or more RDF triples can be aggregated to form a RDF graph. The RDF graphs can be queried using the W3C SPARQL query language that also supports reasoning to infer new knowledge from RDF graphs.
We use RDF syntax to generate the semantic provenance graph in the ProvCaRe project. Figure 2 illustrates a new algorithm developed in the ProvCaRe project to transform structured provenance metadata extracted from unstructured text into graph structures with “subject”, “predicate”, and “object” mapped to ontology terms. The algorithm uses the output of the ProvCaRe NLP pipeline, which consists of the constituents of the sentence such as nouns, verb (“chunked sentence”), and output of the named entity recognition (NER) as listed in Line 1. The provenance-focused NER module has an important feature that uses the ProvCaRe ontology together with existing biomedical NLP resources such as the Unified Medical Language System (UMLS)-based MetaMap and the National Center for Biomedical Ontologies (NCBO) annotator for entity recognition [14]. This comprehensive approach to provenance NER ensures that the ProvCaRe NLP pipeline is able to effectively identify and extract provenance terms from unstructured text.
Figure 2:
The algorithm used to generate semantic provenance graph from provenance terms stored in the ProvCaRe knowledgebase
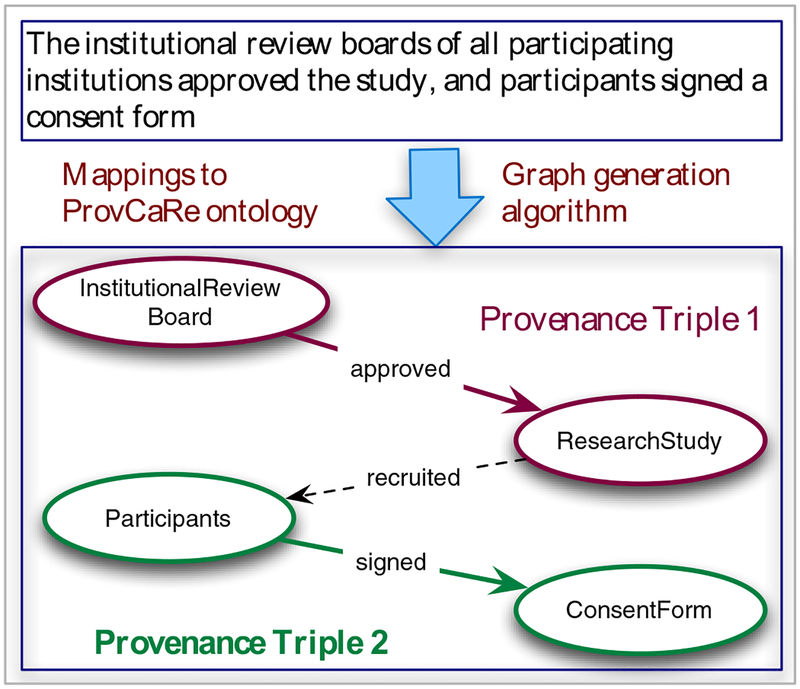
In Line 3, 4, and 5 of the graph generation algorithm (Figure 2), we use mappings between structured provenance and ProvCaRe ontology classes to generate concise representation of provenance terms, which can be used to generate components of the provenance graph. For example, the sentence from an article by O’Connor et al. describing a prospective cohort study on sleep disordered breathing and hypertension [20], “The institutional review boards of all participating institutions approved the study, and participants signed a consent form” describes provenance of ethical approval and participant consent for the research study. The graph generation algorithm following the steps described in Figure 2 generated a semantic provenance graph (Figure 3).
Figure 3:
An example semantic provenance graph generated from provenance terms extracted from a sentence in an article describing a prospective cohort study by O’Connor et al.
It is important to note the differences between the verbose structured provenance information generated by the ProvCaRe NLP pipeline and the concise graph representation generated by the new algorithm. For example, the provenance RDF graph supports querying by graph query languages, including SPARQL and use of reasoning techniques based on RDF semantics [9]. An important feature of the semantic provenance graph generation algorithm is its ability to correctly identify different constituents of a sentence containing provenance terms and generate one or more provenance RDF triples that accurately represents the information content of a sentence. As shown in Figure 3, the algorithm correctly generates two provenance triples representing: (1) the formal approval for the study; and (2) the consent forms signed by the study participants.
These individual provenance triples can be linked or aggregated together to form the corresponding semantic provenance graph for analysis. This approach allows the ProvCaRe query feature to support graph traversal-based queries. For example, using the predicate “recruited” to link the two provenance RDF triples in Figure 3, a graph traversal query can identify that a research study used consent forms as part of the recruiting protocol.
Query and analysis of provenance graphs
As we discussed earlier, a key objective of the ProvCaRe project is to query and analyze these semantic provenance graphs to characterize the reproducibility of research studies. Therefore, the ProvCaRe project developed a Web-based user interface to support user queries that search for research studies associated with their research hypothesis. The query results include the provenance metadata extracted from the research studies associated with the user query. This provenance query and analysis interface is accessible at https://provcare.case.edu, which allows users to explore the ProvCaRe knowledge repository.
The current version of the ProvCaRe query module uses a “keyword-based search” together with ontology-driven “query expansion” approach to query the structured provenance information in the ProvCaRe knowledgebase. As part of our ongoing work, we are implementing a SPARQL query engine in ProvCaRe to query the semantic provenance graph generated by the new graph generation algorithm described in this paper. In the next section, we evaluate the results of the graph generation algorithm.
Results
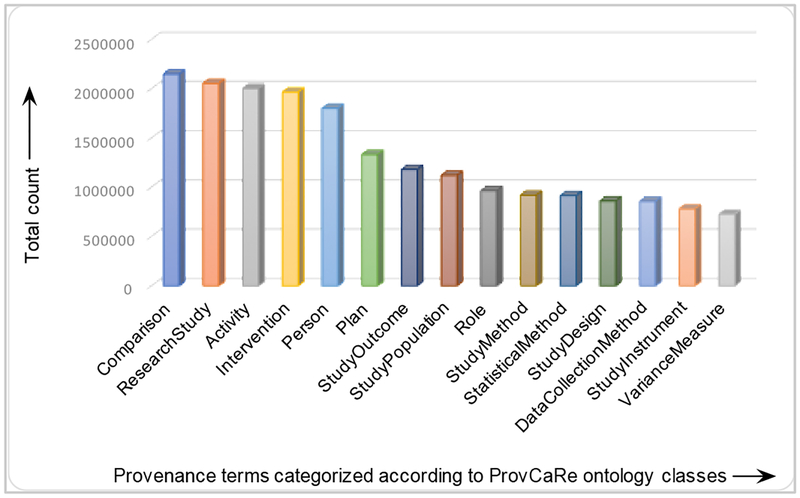
We used a corpus of 75 articles describing sleep medicine research studies for evaluation of the graph generation algorithm. This corpus of articles was selected from sleep medicine reearch studies that have made their data available as part of the National Sleep Research Resource (NSRR) [7]. However, we note that as part of the ProvCaRe project, we have processed more than 1.6 million full text articles available in the PubMed database. For example, Figure 4 illustrates the distribution of 35 million terms extracted from the 1.6 million articles categorized according to a select number of ProvCaRe ontology classes (terms are organized by their frequency in the figure). Provenance terms describing the design of research studies, for example “comparative” research studies have the highest frequency, whereas provenance terms describing the statistical data analysis term “variance” has the lowest frequency.
Figure 4:
Distribution of provenance terms from 1.6 million full text articles corresponding to the ProvCaRe ontology classes
Evaluation of the graph generation algorithm and properties of the semantic provenance graph
In Table 1, we describe the characteristics of semantic provenance graph generated from the corpus of 75 papers with 10,875 provenance RDF triples generated from more than 30,000 sentences. The 33,783 nodes of the provenance RDF triples were mapped to various biomedical ontologies, for example the term Population was mapped to SNOMED CT and the term Study was mapped to the ProvCaRe ontology. It is interesting to note that more than 6900 sentences generated more than 1 provenance RDF triple, which is a key feature of the semantic provenance graph generation algorithm as it accurately reflects the information content of the sentence.
Table 1–
Properties of semantic provenance graph generated from 75 full-text research articles
| Type of characterization | Count |
|---|---|
| Number of provenance RDF triples | 10,875 |
| Provenance terms mapped to ontology classes | 33,783 |
| Total number of sentences processed from 75 papers | 30,450 sentences |
| Total number of sentences with more than 1 provenance RDF triple generated | 6917 sentences |
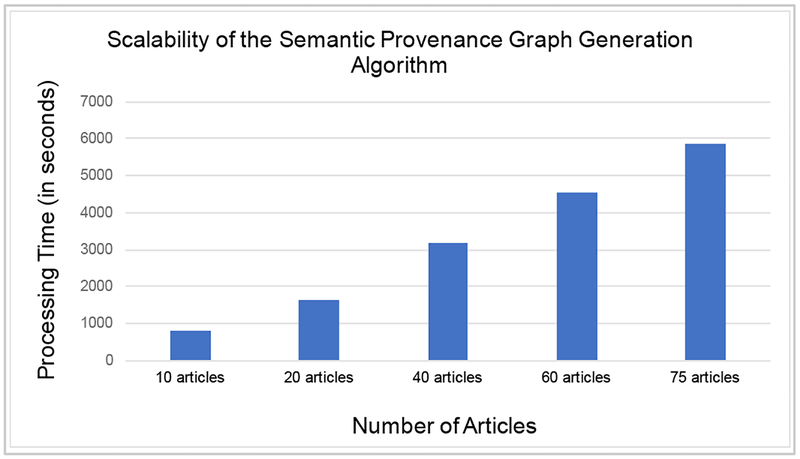
An important feature of the semantic provenance graph generation algorithm is scalability in terms of time required to process large number of provenance terms. Figure 5 shows the performance of the algorithm for five sets of articles with increasing size. The results show that the algorithm scales almost linearly as the number of articles increases. As part of our ongoing work, we are further optimizing the performance of the algorithm as we aim to process all the 35 million provenance terms extracted from 1.6 million articles currently stored in the ProvCaRe knowledge repository.
Figure 5:
The performance of the semantic provenance graph generation algorithm profiled over five different sets of articles
Qualitiative evaluation of the semantic provenance graph
In addition to the quantitative analysis of the provenance triples generated by the semantic provenance graph generation algorithm, we qualitatively evaluated the semantic provenance graph generated for a research study described in the article by O’Connor et al. (this article was selected at random from the corpus of 75 articles). We used a set of eight queries represent important contextual information about not only this specific research study reported by O’Connor et al., but also other biomedical research studies. The queries are listed below:
Q1. What were the inclusion and exclusion criteria?
Q2. How were subjects assigned to groups?
Q3. How was blood pressure measured?
Q4. What was the definition of hypertension?
Q5. What method was used to perform sleep studies?
Q6. How were apnea-hypopnea indices categorized?
Q7. When were repeat blood pressure measurements checked?
Q8. How was the data analyzed?
The objectives of evaluating these queries over the provenance graph generated from the provenance metadata was to evaluate the quality of the semantic provenance graph. The query results were manually reviewed and were found to closely correspond to the provenance information available in the original article by O’Connor et al. We propose to perform a systematic evaluation of the provenance graph query results using the sematic provenance graph generated from the 1.6 million articles in future.
Discussion
The use of the semantic provenance graph structure to underpin the ProvCaRe knowledge repository is expected to significantly improve query and analysis features available to users. This will in turn enable the biomedical research community to query and analyze provenance metadata reported by various research studies in the context of scientific reproducibility. However, there are several practical challenges that need to be addressed before the ProvCaRe knowledge repository can play a greater role in characterizing the reproducibility of research studies. These challenges include the lack of detailed description of a research study in published articles, for example protocol related to data collection, data analysis, and validation of study hypothesis.
This challenge can be addressed by making available the provenance information associated with research studies in public metadata repositories. Together with existing data repositories, these provenance metadata repositories can support reproducibility of published studies. An important challenge in the ProvCaRe project is the scalability of graph query operations as we progressively convert all the provenance information in the ProvCaRe knowledgebase into RDF provenance graph. We note that the ProvCaRe query interface includes a novel provenance-based ranking feature that allows users to assign weight values to ProvCare ontology terms, which is used to rank query results. Together with the provenance-based ranking feature and time complexity of graph operations, the current performance of the ProvCaRe query interface requires significant optimization to support real-time user interactions. We are exploring the use of customized indexing techniques to address this challenge.
Conclusions
We demonstrated the practical use of provenance metadata extracted from full-text articles in the PubMed database to characterize the reproducibility of research studies. In particular, we described the development and application of a new graph generation algorithm to transform verbose provenance metadata extracted by the ProvCaRe NLP pipeline into semantic provenance graphs, which can support query and analysis of provenance metadata. The ProvCaRe knowledgebase with provenance extracted from more than 1.6 million full-text articles in the PubMed database is a unique resource to characterize the scientific reproducibility of biomedical research studies and its representation as a semantic provenance graph will allow users to perform knowledge discovery tasks using ontology-driven reasoning techniques.
Acknowledgements
This work is supported by the NIH-NIBIB Big Data to Knowledge 1U01EB020955 grant, and NSF grant# 1636850.
References
- [1].PubMed, https://www.ncbi.nlm.nih.gov/pubmed/.
- [2].Anand MK, Bowers S, Ludäscher B, Techniques for efficiently querying scientific workflow provenance graphs, in: Proceedings of the 13th international Conference on Extending Database Technology, Manolescu SSI, Teubner J, Kitsuregawa M, Leger A, Naumann F, Ailamaki A, Ozcan F, ed., ACM, New York, NY, Lausanne, Switzerland, 2010, pp. 287–298. [Google Scholar]
- [3].Buneman P, Davidson S, Data provenance - the foundation of data quality, in, 2010.
- [4].Cheney J, Missier P, Moreau L, Constraints of the PROV Data Model, in: W3C Recommendation, World Wide Web Consortium W3C, 2013. [Google Scholar]
- [5].Chirigati FS, Shasha DE, Freire J, ReproZip: Using Provenance to Support Computational Reproducibility, in: Theory and Practice of Provenance (TaPP), 2013. [Google Scholar]
- [6].Collins FS, Tabak LA, Policy: NIH plans to enhance reproducibility, Nature 505 (2014), 612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dean DA, Goldberger AL, Mueller R, Kim M, Rueschman M, Mobley D, Sahoo SS, Jayapandian CP, Cui L, Morrical MG, Surovec S, Zhang GQ, Redline S, Scaling up scientific discovery in sleep medicine: the National Sleep Research Resource., SLEEP 39 (2016), 1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gil Y, Cheney J, Groth P, Hartig O, Miles S, Moreau L, Pinheiro da Silva P, Coppens S, Garijo D, Manuel Gomez J, Missier P, Myers J, Sahoo SS, Zhao J, Provenance xg final report, in: W3C Technical Report, W3C, 2010. [Google Scholar]
- [9].Hayes P, RDF Semantics, in: W3C Recommendation, McBride B, ed., World Wide Web Consortium, 2004. [Google Scholar]
- [10].N.I.o. Health, Principles and Guidelines for Reporting Preclinical Research, in, 2016.
- [11].Herman I, Adida B, Sporny M, Birbeck M, RDFa 1.1 Primer - Second Edition, in: W3C Working Group Note, World Wide Web Consortium (W3C), 2013. [Google Scholar]
- [12].Hitzler P, Krötzsch M, Parsia B, Patel-Schneider PF, Rudolph S, OWL 2 Web Ontology Language Primer, in: W3C Recommendation, World Wide Web Consortium W3C, 2009. [Google Scholar]
- [13].Ioannidis JPA, Why Most Published Research Findings Are False, PLoS Med 2 (2005), e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jonquet C, Shah NM, Musen MA, The Open Biomedical Annotator, in: AMIA Summit on Translat Bioinformatics, AMIA, San Francisco, 2009, pp. 56–60. [PMC free article] [PubMed] [Google Scholar]
- [15].Landis SC, Amara SG, Asadullah K, Austin CP, Blumenstein R, Bradley EW, Crystal RG, Darnell RB, Ferrante RJ, Fillit H, Finkelstein R, Fisher M, Gendelman HE, Golub RM, Goudreau JL, Gross RA, Gubitz AK, Hesterlee SE, Howells DW, Huguenard J, Kelner K, Koroshetz W, Krainc D, Lazic SE, Levine MS, Macleod MR, McCall JM, Moxley RT 3rd, Narasimhan K, Noble LJ, Perrin S, Porter JD, Steward O, Unger E, Utz U, Silberberg SD, A call for transparent reporting to optimize the predictive value of preclinical research, Nature 490 (2012), 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lebo T, Sahoo SS, McGuinness D, PROV-O: The PROV Ontology, in: W3C Recommendation, World Wide Web Consortium W3C, 2013. [Google Scholar]
- [17].Missier P, Sahoo SS, Zhao J, Goble C, Sheth A, Janus: from Workflows to Semantic Provenance and Linked Open Data, in: IPAW 2010, Troy, NY, 2010. [Google Scholar]
- [18].Moreau L, Provenance-based reproducibility in the semantic web., Web Semantics: Science, Services and Agents on the World Wide Web 9 (2011), 202–221. [Google Scholar]
- [19].Moreau L, Missier P, PROV Data Model (PROV-DM), in: W3C Recommendation, World Wide Web Consortium W3C, 2013. [Google Scholar]
- [20].O’Connor GT, Caffo B, Newman AB, Quan SF, Rapoport DM, Redline S, Resnick HE, Samet J, Shahar E, Prospective study of sleep-disordered breathing and hypertension: the Sleep Heart Health Study, Am J Respir Crit Care Med 179 (2009), 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Prinz F, Schlange T, Asadullah K, Believe it or not: how much can we rely on published data on potential drug targets?, Nature Reviews Drug Discovery 10 (2011), 712. [DOI] [PubMed] [Google Scholar]
- [22].Sahoo SS, Semantic Provenance: Modeling, Querying, and Application in Scientific Discovery, Ph.D., Wright State University, 2010. [Google Scholar]
- [23].Sahoo SS, Valdez J, Kim M, Rueschman M, Redline S, ProvCaRe: Characterizing Scientific Reproducibility of Biomedical Research Studies using Semantic Provenance Metadata, International Journal of Medical Informatics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sim I, Tu SW, Carini S, Lehmann HP, Pollock BH, Peleg M, Wittkowski KM, The Ontology of Clinical Research (OCRe): an informatics foundation for the science of clinical research., Journal of Biomedical Informatics 52 (2014), 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]