Abstract
During the past 5 years there has been an increasing body of literature describing the roles cardiac myosin binding protein C (cMyBP-C) phosphorylation play in regulating cardiac function and heart failure. cMyBP-C is a sarcomeric thick filament protein that interacts with titin, myosin and actin to regulate sarcomeric assembly, structure and function. Elucidating the function of cMyBP-C is clinically important because mutations in this protein have been linked to cardiomyopathy in more than sixty million people worldwide. One function of cMyBP-C is to regulate cross-bridge formation through dynamic phosphorylation by protein kinase A, protein kinase C and Ca2+-calmodulin-activated kinase II, suggesting that cMyBP-C phosphorylation serves as a highly coordinated point of contractile regulation. Moreover, dephosphorylation of cMyBP-C, which accelerates its degradation, has been shown to associate with the development of heart failure in mouse models and in humans. Strikingly, cMyBP-C phosphorylation presents a potential target for therapeutic development as protection against ischemic–reperfusion injury, which has been demonstrated in mouse hearts. Also, emerging evidence suggests that cMyBP-C has the potential to be used as a biomarker for diagnosing myocardial infarction. Although many aspects of cMyBP-C phosphorylation and function remain poorly understood, cMyBP-C and its phosphorylation states have significant promise as a target for therapy and for providing a better understanding of the mechanics of heart function during health and disease. In this review we discuss the most recent findings with respect to cMyBP-C phosphorylation and function and determine potential future directions to better understand the functional role of cMyBP-C and phosphorylation in sarcomeric structure, myocardial contractility and cardioprotection.
Keywords: Cardiac myosin binding protein-C, Contractile protein, Protein phosphorylation, Heart failure, Cardioprotection
1. Introduction
Ischemia, myocardial infarction (MI) and heart failure (HF) constitute a growing health and economic problem. An estimated 80 million American adults have one or more types of cardiovascular disease [1]. Of these, HF afflicts approximately six million people in the U.S. each year at an estimated cost of $37 billion. HF is associated with diminished responsiveness of the β-adrenergic receptor (β-AR), loss of cardiac contractility, abnormalities in Ca2+ handling and altered cardiac contractile protein phosphorylation [2–5]. Recent studies involving a variety of cardiovascular diseases have shown that cardiac dysfunction might result from changes in the phosphorylation states of key cardiac contractile regulatory proteins [6–9]. For example, alteration in phosphorylation of contractile protein is involved in regulating myocardial dysfunction [10]. Sarcomeres are the functional unit of contraction in cardiac muscles, which consist of thick and thin filament proteins (Fig. 1). Several proteins that are post-translationally modified in the cardiac sarcomere have now been identified, including cardiac myosin binding protein-C (cMyBP-C), cardiac troponin I (cTnI), cardiac troponin T (cTnT), α-tropomyosin (α-TM) and the myosin light chain (MLC) [4,11–14]; however, in many cases the functional consequences of the post-translational modification are not known. cMyBP-C is a potential target for α- and β-adrenergic signaling in the thick filaments that are targeted by various kinases [15]. In heart muscle, phosphorylation of cMyBP-C has been shown to play an important role in sarcomeric structure and function, and protection of the heart from ischemic-reperfusion (I-R) injury [16,17]. The objectives of this review are to discuss the role(s) of cMyBP-C phosphorylation during normal heart function and development of HF, and to identify potential areas of research that will help us understand the functional role(s) that cMyBP-C plays in vivo.
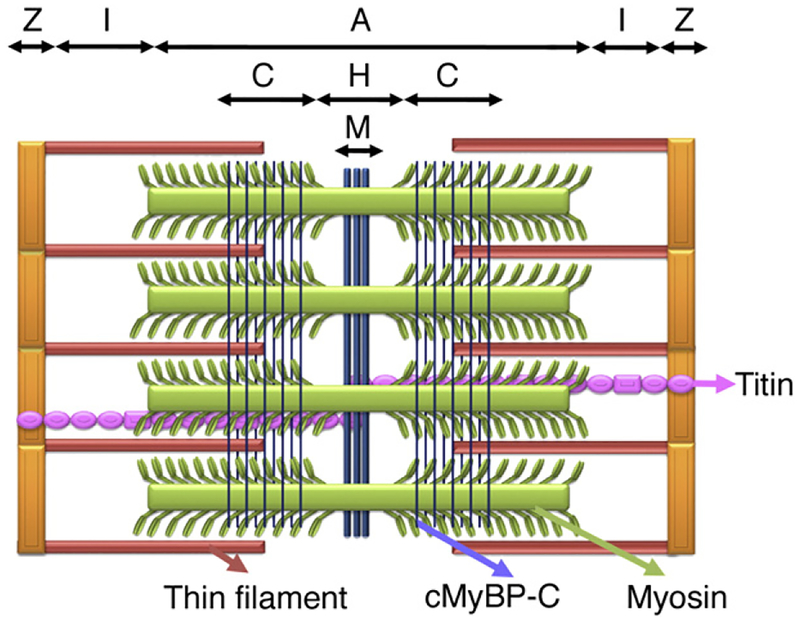
Fig. 1.
Schematic diagram of the cardiac sarcomere and arrangement of cMyBP-C. The sarcomere is the basic functional unit of a cardiac muscle’s cross-striated myofibril, which is defined as the boundary between two neighboring Z-lines. The I-band, A-band, H-zone and M-lines are shown. The thick filament protein is composed of myosin, cMyBP-C and titin. cMyBP-C is localized in the inner two-thirds of the A-band, i.e., the C-zone. cMyBP-C is oriented perpendicularly to the long axis of the myosin filaments. The two groups of 7–9 strips of cMyBP-C bands on either side of the H-zone give a characteristic doublet appearance within the A-band. The thin filament is composed of actin monomers, troponins (cTnT and cTnI) and α-TM, which is connected to nebulin in the I-bands.
2. Cardiac myosin binding protein-C
cMyBP-C is a 140-kDa protein that was originally identified 30 years ago as a contaminant during myosin preparation [18]. Later it was recognized as a protein associated with the thick filament found in vertebrate hearts [19]. In the sarcomere, cMyBP-C is localized in two groups that are separated by a bare H-zone within the inner two-thirds of the A-band, in the C-zone (Fig. 1). In skeletal muscles, skeletal MyBP-C is located in the A-band along a series of 7 to 9 transverse stripes that are spaced 43 nm apart in the C-zone [20]. Recently, similar striping patterns of cMyBP-C arrangement were shown in cardiac muscles [21]. The two groups of 7–9 cMyBP-C bands on either side of the H-zone give a characteristic doublet appearance within the A-band of a sarcomere [22]. Myosin tails form the thick filament backbone with their heads pointing outwards. The heads are arranged along three-stranded helices with three pairs of heads at each 14.3-nm layer and an axial repeat of 43 nm. The defined spacing of these stripes dictates that only every third level of myosin heads in the C-zone is associated with a cMyBP-C molecule [20]. Thus, cMyBP-C might be activated only when the central part of the sarcomere undergoes thick–thin filament interaction, i.e., during contraction when the myosin-actin interaction extends into the central core of the A-band. cMyBP-C comprises 2% of the contractile protein in the heart. Some evidences show that cMyBP-C plays a role in myofibril organization and assembly during myofibrillogenesis and in regenerating muscle cells [23,24]. Mice hearts in which cMyBP-C was knocked out (−/−) [25] and ablated by C-terminal truncation (t/t) [26] have abnormal sarcomere structure, with the contractile units lacking M-lines and alterations in thick filament size. These mice lacking cMyBP-C are viable, but develop cardiomyopathies and contractile dysfunction at 3 months of age, indicating that cMyBP-C is not necessary for sarcomere formation during embryogenesis and basal cardiac function but is needed for proper sarcomere organization and normal cardiac function. Moreover, heterozygous mice with either cMyBP-C knockout (+/−) or C-terminal truncation (+/t) showed regular sarcomere organization and normal cardiac function, suggesting haploinsufficiency due to a null allele alone is not adequate to trigger HCM disease. A detail genotype–phenotype correlation of all cMyBP-C animal models is shown in Table 1.
Table 1.
Animal models testing the roles of cMyBP-C.
| Genotype | Description | Phenotype | References |
|---|---|---|---|
| Expression of an N-terminal truncated cMyBP-C lacking the myosin and titin binding domains that mimic a class of HCM mutation in MYBPC3 | TG mice expressing the mutant cMyBP-C using the cardiac-specific a-MyHC promoter | TG mice showed HCM phenotype with contractile dysfunction, altered sarcomeric structure, increased myofilament Ca2+ sensitivity and decreased maximum relative power output. | Yang et al. [58] Yang et al. [108] |
| Expression of full-length wild-type cMyBP-C with an N-terminal Myc tag | TG mice expressing the cMyBP-C wild-type using the cardiac-specific α-MyHC promoter | This mouse model served as a control with Myc-tag with no phenotype apparent. | Yang et al. [58] Sadayappan et al. [74] |
| Replacement of myosin and titin binding domains (exon 30) in cMyBP-C by novel residues, causing HCM disease in human | A knock-in mouse model (t/t) | Homozygous mice (t/t) showed DCM phenotype with contractile dysfunction, low systolic stiffness, arrhythmia, impaired Ca2+ cycling and disordered myofibrils. | McConnell et al. [26] Berul et al. [109] Song et al. [89] Palmer et al. [110] Sadayappan et al. [74] Nyland et al. [111] |
| Lacking only the myosin binding site in cMyBP-C | TG mice expressing the mutant cMyBP-C using the cardiac-specific α-MyHC promoter | TG mice showed HCM phenotype with contractile dysfunction, abnormal sarcomere patterning and decreased heart rate in response to exercise. This model supports the dominant negative effects of a poison polypeptide. | Yang et al. [53] |
| Lacking C0 domain; determining the role of C0 domain | A knock-in mouse model that carries N-terminal-shortened cMyBP-C | The absence of phenotype, normal incorporation of mutant protein into sarcomere, and contractile enhancement with increased myofilament Ca2+ sensitivity were reported | Witt et al. [103] |
| Lacking exons 3 to 10 | A systematic cMyBP-C knock-out mouse model (−/−) | Homozygous mice showed HCM phenotype with contractile dysfunction. Myofilament showed increased force, velocity, power output and stretch activation, but decreased Ca2+ response. Studies concluded that cMyBP-C is not essential for cardiac development and basal cardiac function, but is required for normal sarcomeric organization, muscle activation and cardiac function. | Harris et al. [25] Korte et al. [112] Stelzer et al. [113] Brickson et al. [114] Colson et al. [115] Luther et al. [21] |
| Lacking the phosphorylation motif “LAGAGRRTS” | TG mice expressing the mutant cMyBP-C using the cardiac-specific α-MyHC promoter | The absence of phenotype, increased maximal Mg2+-ATPase activity, phosphorylation of cTnI and phospholamban were observed. | Yang et al. [75] |
Two HCM mutations expressing C-terminal truncated cMyBP-C;
|
TG Drosophila models expressing human C-terminal truncated cMyBP-Cs using the UAS-GAL4 system | TG flies showed sarcomeric structural abnormalities and deregulated Ca2+ signaling. | Vu Manh et al. [116] |
| cMyBP-C phospho-ablation | TG mice expressing the mutant cMyBP-C in which phosphorylation of Ser-273, Ser-282 and Ser-302 were ablated | TG mice showed cardiac hypertrophy with contractile dysfunction and altered sarcomeric structure. When TG mice crossed into cMyBP-C null (t/t) background, they failed to rescue the (t/t) phenotype. | Sadayappan et al. [74] Sadayappan et al. [76] Nagayama et al. [117] |
| cMyBP-C phospho-ablation | TG mice expressing the mutant cMyBP-C in which phosphorylation of Ser-273, Ser-282 and Ser-302 were ablated | TG mice exhibited both systolic and diastolic dysfunction as well as cardiac hypertrophy. When TG mice crossed into knockout (−/−) background, they failed to rescue the (−/−) phenotype. | Tong et al. [22] |
| cMyBP-C phospho-mimetic | TG mice expressing the mutant cMyBP-C in which Ser-273, Ser-282 and Ser-302 sites were mutated to mimic constitutive phosphorylation | No phenotype was apparent. When TG mice crossed into (t/t) background, they rescued the (t/t) phenotype. TG mouse hearts showed cardioprotection from I-R injury. TG mice showed improved contractile function under a β-MyHC background. | Sadayappan et al. [17] Sadayappan et al. [76] |
| A cMyBP-C point mutation, exon 6 (G>A), causing HCM disease | A knock-in mouse model carrying a point mutation, which results in truncation | Homozygous knock-in mice showed cardiac hypertrophy with contractile dysfunction and interstitial fibrosis. Heterozygous knock-in mice had no major phenotype. Studies concluded that both nonsense-mediated mRNA decay and UPS regulate cMyBP-C mutant protein turnover that play an important role in HCM pathogenesis. | Vignier et al. [55] |
cMyBP-C belongs to the intracellular immunoglobulin (Ig) super family and, like other sarcomeric proteins such as titin, myosin and M-protein, is composed of repeating domains of Ig and fibronectin type-3. cMyBP-C has three isoforms: fast-skeletal, slow-skeletal and cardiac, each of which is encoded by a separate gene. Skeletal and cardiac isoforms are expressed exclusively in skeletal and cardiac muscle, respectively. The cardiac isoform differs from the skeletal isoforms due to an extra Ig domain at the N-terminus (C0), a phosphorylation domain (M) in between C1 and C2 [27], and an inserted loop of 28 residues within the C5 domain (Fig. 2). Although the C0 domain is unique to cMyBP-C, its function and specific role in heart muscle is still not known. In addition, the C0–C1 domains contain a Pro-Ala-rich region that is similar to other actin-binding proteins [28]. Recently, in addition to being the target of post-translational modifications, a functional role of the M domain was identified: it is not simply a linker between C1 and C2, rather, it interacts with actin [29,31]. The amino acid sequences within the M domain are highly conserved between mouse and human. Importantly, cMyBP-C interacts with the S2 region of myosin at its C1-M-C2 domains [30] which is regulated by the phosphorylation of the M domain. cMyBP-C also binds to actin at the C1 and M domains [31], which may be important for the arrangement of actin-myosin filaments in sarcomeres. The precise arrangement of cMyBP-C in the sarcomere is yet to be systematically elucidated, though there are two models, a trimeric collar model and a strut model, that have been proposed regarding the arrangement of cMyBP-C and how it interacts with myosin and titin at its C-terminus. The collard model consists of three cMyBP-C molecules that form a trimeric collar arrangement around the thick filament where the C5–C10 domains wrap around the width of the MyHC with interactions occurring between C5–C8 and C7–C10 of the overlapping domains every 43 nm in the C zone of the sarcomere [32–34]. This model has been supported by experiments, which have demonstrated interactions between the C5 and C8 domains [35], as well as between C7 and C10 [32] in cMyBP-C and fast skeletal MyBP-C, but not in slow skeletal MyBP-C [35]. In support of the strut model, image analysis of electron micrographs have produced three-dimensional models that suggest that three or more of cMyBP-C’s C-terminal domains are oriented along the axis of the thick filament as support rods [36]. Both models do not predict the arrangement of N-terminal C0–C4 domains, but propose that the N-terminal domains are allowed to extend into the space between MLCs and MyHCs where they are able to interact with the myosin S2 region and reach the thin filament [28,37]. Though the collar model is currently forward, it certainly requires more structural data to fully confirm the model’s correctness [38].
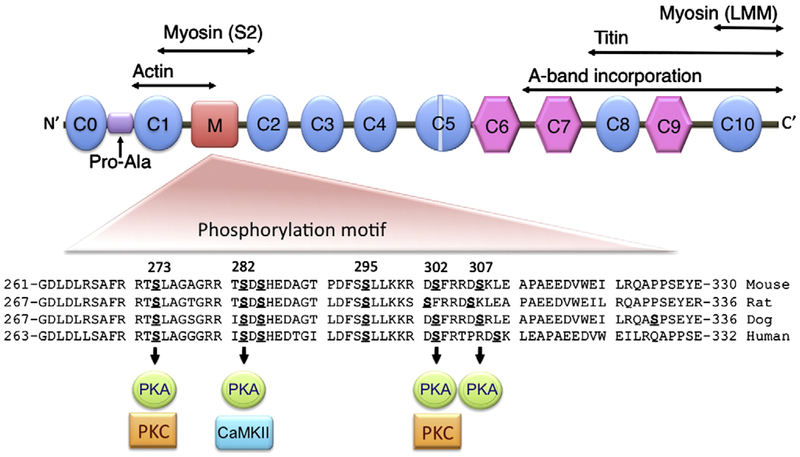
Fig. 2.
General domain structure of mouse cMyBP-C and identified phosphorylated serines. There are eight Ig (oval) and three fibronectin type-3 domains (hexagonal) numbered C0 to C10. The Pro-Ala–rich region (Pro-Ala), phosphorylated M domain, a 28-amino acid cardiac-specific insertion within the C5 domain and regions that interact with actin, myosin and titin sites are shown. The M domain contains multiple phosphorylation sites that can be phosphorylated by PKA, CaMKII and PKC. The amino acid sequences are conserved between species in the M domain. Domains 7–10 participate in binding of myosin and titin, which are necessary for cMyBP-C stability and sarcomere organization.
3. cMyBP-C mutations and human cardiomyopathy
cMyBP-C has defined roles in both the structural assembly and stability of the sarcomere, as well as in the modulation of contraction [17]. Much of our knowledge about cMyBP-C has emerged after it was confirmed that mutations in the cMyBP-C gene (MYBPC3) cause cardiomyopathies [39,40]. Cardiomyopathies that are associated with mutations in eight genes encoding sarcomeric proteins, including MYBPC3, are a frequent cause of HF (Table 2). Cardiomyopathies are currently categorized into the following four classes [41]: arrhythmogenic right ventricular cardiomyopathy, DCM, HCM and restrictive cardiomyopathy. The first HCM mutations in MYBPC3 were identified in 1995 [39,40] and to date 150 mutations have been published (Table 2), second only to the number characterized in the MYH7 gene.
Table 2.
List of sarcomeric genes and their mutations that associate with cardiomyopathies.
| Symbol | Gene | HCM | DCM | Total | Account |
|---|---|---|---|---|---|
| MYH7 | β-cardiac myosin heavy chain | 194 | 13 | 207 | 45% |
| MYBPC3 | Cardiac myosin binding protein-C | 149 | 1 | 150 | 33% |
| TNNT2 | Cardiac troponin T | 31 | 6 | 37 | 8% |
| TNNI3 | Cardiac troponin I | 27 | 1 | 28 | 6% |
| TPM1 | α-Tropomyosin | 11 | 2 | 13 | 3% |
| ACTC | α-Actin | 7 | 2 | 9 | 2% |
| MYL2 | Regulatory myosin light chain | 10 | 0 | 10 | 2% |
| MYL3 | Essential myosin light chain | 5 | 0 | 5 | 1% |
| Total mutations | 434 | 25 | 459 | 100% |
MYBPC3 is the second most frequent gene associated with the inherited cardiomyopathies worldwide. Source: Sarcomere Protein Gene Mutation Database: http://cardiogenomics.med.harvard.edu/genes/gene-list.
MYBPC3 consists of 24,000 base pairs of genomic DNA with 35 exons that encode a protein of 1,274 amino acids. Mutations in MYBPC3 are found in 33% of all cardiomyopathy cases (Table 2), of which 70% are predicted to have proteins truncated at the C-terminus that lack myosin- and/or titin-binding sites [39,40,42]. MYBPC3 mutations are generally associated with incomplete disease penetrance and late disease onset (after 40 years of age) [43,44]. Various clinical studies have shown that mutations in MYBPC3 are the most common cause of HCM when an identifiable sarcomeric protein mutation was found (18.2% in Mayo Clinic, 14.5% in Harvard Medical School, 26% in France, 18% in Germany, 21.7% in Sweden and 24% in Finland in a composite meta-analysis of five countries: the United States, France, Germany, Sweden and Finland [45]). The meta-analysis concludes that MYBPC3 mutations are the most common genetic cause of HCM with increased polymorphic variants. Surprisingly, in South Asian countries approximately 4% of the total population has a polymorphic deletion of 25-base pairs in MYBPC3 at the C-terminus that can cause HCM and DCM [46]. Prevalence of this mutation is high: it has been estimated that more than 60 million people are at risk worldwide [46,47], underscoring the necessity of understanding the role of cMyBP-C in the pathogenesis of HF. The 25-base pair deletion causes a translational reading frame shift in exon 33 [47], which results in the replacement of 62 regular residues with 55 novel amino acids in the C10 domain; however, this alteration was not able to be detected in tissue samples obtained from patients [46]. These data show the introduction of novel amino acids in the C10 domain decrease the stability of mutant cMyBP-C, suggesting that alterations in the C10 domain will affect cMyBP-C stability in the sarcomere. Therefore, it will be important to determine in vivo the critical function of the C-terminal domains that mediate cMyBP-C’s interaction with myosin and titin for its stability, sarcomeric structure and contractile function.
The functional and pathological consequences of most of the MYBPC3 mutations are not characterized fully, because neither mutant peptides nor normal levels of cMyBP-C have been found in cardiac tissue from affected HCM patients [48]. Literature show that the majority of patients with C-terminal truncation mutations in cMyBP-C show reduced level of total cMyBP-C [44,49,50], suggesting that the pathologic mechanism could be haploinsufficiency. Even HCM patients with missense MYBPC3 mutations show reduced total cMyBP-C levels, supporting the hypothesis that haploinsufficiency is the disease mechanism [50]. The absence of a misformed protein could be the result of inefficient synthesis or rapid degradation. If the truncated cMyBP-C protein cannot contribute to filament assembly in the sarcomere it could be rapidly degraded. Studies have shown that truncated cMyBP-C is preferentially degraded by the ubiquitin–proteasome system, which, in turn, might competitively inhibit the breakdown of other ubiquitin–proteasome system substrates [46,51,52]. The data also provide direct evidence that the mutant MYBPC3 allele can act as a null allele leading to haploinsufficiency of cMyBP-C in the sarcomere [49]. Furthermore, when various C-terminally truncated cMyBP-Cs were overexpressed in fetal cardiomyocytes, or in transgenic (TG) mice, they had markedly lower protein levels and sarcomeric incorporation than those with normal cMyBP-C [53]. These data are compatible with a “null-allele” mechanism and suggest that the truncated cMyBP-C is rapidly degraded, which might be promoted by the lack of biomolecular interaction between the truncated forms of cMyBP-C and myosin [54]. Haploinsufficiency might also be explained through the nonsense-mediated mRNA decay pathway, which reduces the transcript level of mRNAs that carry nonsense and frameshift mutations. In support of this, Vignier et al. [55] recently showed that the nonsense-mediated mRNA decay pathway is involved in the decay of mutant cMyBP-C mRNA at the cellular level, resulting in haploinsufficiency of cMyBP-C. This finding suggests that both nonsense-mediated mRNA decay and the ubiquitin–proteasome system play a role in contractile protein regulation. In contrast, other studies suggest a dominant-negative or poison polypeptide effect on sarcomere organization resulting from MYBPC3 mutations [56,57]. Many different strategies are being used to understand the pathogenesis of the cMyBP-C mutants that cause HCM and DCM. These include the use of mouse models of HCM and DCM in which cMyBP-C is knocked out [25] or modified [26,53,55,58], respectively (Table 1), but the mechanism of the disease process has not yet been defined. Research on the involvement of MYBPC3 in cardiomyopathies is now a highly competitive research field, evidenced by the current plethora of publications, reviews and editorials [33,59–63].
The C1 and C2 domains and the connecting M domain possess numerous HCM mutations of varying penetrance, stressing the importance of this region. Two of these mutations, G278E and G279A, occur within the conserved, cardiac-specific LAGGGRRIS sequence, in close proximity to two of the phosphorylation sites [64], suggesting that cMyBP-C phosphorylation has clinically important functions. It was first reported in patients with atrial fibrillation that the level of cMyBP-C phosphorylation at Ser-282, but not the level of cTnI phosphorylation, was decreased in association with impaired atrial contractility and arrhythmogenesis [65]. Furthermore, decreased cMyBP-C phosphorylation at Ser-282 was present in human patients with end-stage HF [10]. These studies were further extended to patients with HCM and showed diminished levels of cMyBP-C phosphorylation at Ser-282 in both end-stage HF and HCM [4]. In contrast, a recent study reported that total cMyBP-C content was reduced by 33% in heterozygous HCM patients with frameshift mutations in MYBPC3, and cMyBP-C phosphorylation at Ser-282 was unaffected; however, cTnI phosphorylation was reduced by 84% compared with controls [49]. Although conflicting, these studies clearly demonstrate that a divergent phosphorylation pattern exists among contractile proteins during the development of HF.
4. Phosphorylation and function of cMyBP-C
Myofilament protein phosphorylation represents a point of convergence for complex signaling events that ultimately result in cardioprotection and changes in contractile function [66]. A unique feature of cMyBP-C in cardiac muscle is that it has multiple phosphorylation sites in the M domain. Gautel et al. [27] reported the presence of three putative phosphorylation sites in the M domain at Ser-273, Ser-282 and Ser-302 that were phosphorylated by PKA (Fig. 2). They also showed the importance of the LAGGGRRIS sequence for the monophosphorylation of Ser-282 by PKA. Mohamed et al. [67] then defined three phosphorylation sites (Ser-273, Ser-282, and Ser-302), all of which are phosphorylated by PKA but only two of which (Ser-273 and Ser-302) are phosphorylated by PKC. Recent studies have identified a new PKA phosphorylation site at Ser-307 In vitro in murine cMyBP-C [68]. Independently, another study using rat neonatal cardiomyocytes described the Ser-295, Ser-315 and Ser-320 phosphorylation sites in rat cMyBP-C [69]. A study using a myocardial stunning model described the regulation of phosphorylation at Ser-279, Ser-288, Ser-290, Ser-308, Ser-313 and Ser-331 sites in canine cMyBP-C [70] (Fig. 2). Protein kinase D (PKD), another kinase that regulates contractile function, also phosphorylates the M domain [71] at unknown residue(s) that are unlikely to include Ser-282 [72]. A phosphorylated Ser-1169 site was also reported as the link between the C9 and C10 domains [67]. However, the physiological significance of these sites need to be defined using both In vitro and in vivo approaches.
In addition to cMyBP-C’s role in maintaining the structural arrangement of the sarcomere [28,33], phosphorylation of cMyBP-C appears to play a critical role in the protein’s ability to regulate force generation by modulating thick–thin filament interaction [22,73]. Several laboratories have focused on determining the phosphorylation state and ensuing function of cMyBP-C, with two independent groups showing that phosphorylation of cMyBP-C is essential for maintaining sarcomeric integrity and normal cardiac function in vivo [22,74]. The level of cMyBP-C phosphorylation decreases in mouse models during I-R injury; pathologic hypertrophy and development of HF in calcineurin TG and muscle Lim protein-knockout mice [74]; myocardial stunning [16,70]; and in patients with atrial fibrillation [65], HCM and HF [4,49]. The role of cMyBP-C phosphorylation in cardiac function was explored in four mouse models (Table 1) in which three phosphorylation sites (Ser-273, Ser-282 and Ser-302) were mutated either to non-phosphorylatable alanines (Ala) [22,74] or to phospho-mimetic aspartic acids (Asp) [17], and Ser-282 was ablated [75]. Ala replacement resulted in depressed cardiac function, and the protein was unable to rescue the cMyBP-C-null phenotype, unlike the wild-type cMyBP-C [22,74], suggesting that cMyBP-C phosphorylation is necessary for normal cardiac function. Conversely, TG mice expressing phospho-mimetic cMyBP-C developed cardioprotection from I-R injury with relatively conserved cardiac function and reduced cellular damage [17,76], underscoring the importance of cMyBP-C phosphorylation in maintaining normal myocardial function. In addition, dephosphorylation of cMyBP-C was associated with its degradation [16,17,70]. After I-R injury, total phosphorylation of cMyBP-C is decreased; most notably in the tri-phosphorylated species [16,74]. The reduced phosphorylation is accompanied by increased cleavage of cMyBP-C, which is associated with thick filament disruption, reduction in actomyosin cross-bridges and contractile dysfunction [16,17]. Investigators are actively seeking ways to detect degraded cMyBP-C fragments in the circulatory system for potential use as a biomarker for acute MI [77]. Phosphorylation of cMyBP-C is inversely related to its proteolysis [17], suggesting that phosphorylation of cMyBP-C might protect against degradation, thereby preserving contractile function. The exact mechanism of cMyBP-C degradation and cardioprotection and the potential impact of cMyBP-C phosphorylation status on the diagnostic value of circulating fragments are yet to be characterized systematically.
4.1. Ca2+-dependent calmodulin kinase II
Ca2+-depedent calmodulin kinase II (CaMKII) can directly phosphorylate cMyBP-C at multiple sites [78]. Hierarchical phosphorylation patterns for cMyBP-C have been defined In vitro [27], and CaMKII-site phosphorylation at Ser-282 might be needed for phosphorylation of PKC sites (Ser-273 and Ser-302) [67,78,79]. In vitro, further studies showed that CaMKII’s ability to phosphorylate Ser-273 and Ser-302 is markedly reduced when Ser-282 is mutated to Ala, suggesting that hierarchical phosphorylation might be involved in cMyBP-C-mediated regulation of cardiac muscle contraction [27,67,78,79]. Furthermore, phosphorylation of cMyBP-C at Ser-282 might be associated with CaMKII activation during myocardial stunning [70]. Conversely, inhibition of CaMKII phosphorylation of cMyBP-C decreases contractility [80]. Ca2+ ions can influence the contraction of cardiac muscle by activating CaMKII, which may specifically phosphorylate Ser-282 [27,78,81] and modulate contractility by changing the thick filament structure [79]. In conclusion, effects of the CaMKII phosphorylation on cMyBP-C function in vivo remain undefined.
4.2. Protein kinase A
PKA can phosphorylate four sites at the N-terminus (Ser-273, Ser-282, Ser-302 and Ser-307) [67,68]. This region binds to the S2 segment of myosin [30,37,82,83] close to the lever arm domain. That interaction can be dynamically regulated by phosphorylation/dephosphorylation of cMyBP-C [30]. After phosphorylation of cMyBP-C by PKA, the thick filaments exhibited a loose structure [84] that prevents binding to myosin, thereby changing the maximum Ca2+-activated force. In other studies of intact myocardium in mouse models, PKA-phosphorylated cMyBP-C accelerated cross-bridge turnover rate under sub-maximum Ca2+ activation, which supports the premise that cMyBP-C functionally interacts with the S2 region to affect contractile function [85]. Electron microscopy of isolated thick filaments confirmed that phosphorylation of cMyBP-C causes the cross-bridges to move away from the thick-filament backbone [82]. Thus, cMyBP-C phosphorylation can change both filament orientation and contractile mechanics [27,83,86]. There is still controversy regarding whether phosphorylation of cMyBP-C plays a role in regulating the properties of contraction. Data show that cMyBP-C influences actomyosin Mg2+-ATPase activity and the kinetics of cross-bridge cycling, and its phosphorylation modifies actomyosin Mg2+-ATPase activity and the rate of relaxation [27,76,78,87]. However, the failure of the specific phosphorylation of cMyBP-C to alter the Ca2+ sensitivity of actomyosin ATPase activity in reconstituted contractile protein systems has been cited by some as strong evidence against a role for phosphorylation of cMyBP-C in regulating contraction [88]. Studies show that phosphorylated cMyBP-C is able to increase the proximity of myosin to actin and loosens the packing of the myosin rod [17,82]; this relieves the inhibitory influence of non-phosphorylated cMyBP-C on acto-myosin function. TG mice expressing cMyBP-C with non-phosphorylatable Ala (S273A, S282A and S302A) exhibited slower cross-bridge turnover kinetics, which negatively affected cardiac function without changes in Ca2+ sensitivity [22]. Interestingly, cMyBP-C null mouse hearts showed accelerated myofilament kinetics equivalent to fully phosphorylated cMyBP-C, providing additional support to the importance of cMyBP-Cmyosin interaction [85]. In addition, when phospho-mimetic cMyBP-C was placed into the β-myosin heavy chain (MyHC) background, the mice showed improved cardiac function without affecting myofilament Ca2+ sensitivity [76]. Furthermore, normalization of Ca2+ transients by ablation or inhibition of phospholamban failed to rescue the cMyBP-C mutant mice [89], suggesting that cMyBP-C phosphorylation might not affect myofilament Ca2+ sensitivity directly. Although cMyBP-C phosphorylation may not directly influence myofilament Ca2+ sensitivity [76], other evidence clearly underscores that cMyBP-C phosphorylation is directly involved in the thick and thin filament cooperative activation, maximal force development, regulating cross-bridge cycling rates, as well as functioning as a tether [22,76,85,90].
When cardiomyocytes were subjected to PKA phosphorylation, different effects on actomyosin ATPase activity occurred and were dependent upon whether α-MyHC or β-MyHC was present [91]. In adult rat hearts with N95% β-MyHC, there was no significant change in actomyosin ATPase activity associated with PKA-mediated phosphorylation of cMyBP-C and cTnI. However, phosphorylation in cardiomyocytes with α-MyHC increased actomyosin ATPase activity [91]. Interestingly, the positions of the cross-bridges are different in filaments containing α and β-MyHC. The impact of PKA-mediated phosphorylation of cMyBP-C in cardiac function might depend on the myosin isoform that is present. In the mammalian heart, two functionally distinct MyHC isoforms, termed V1 (α-MyHC) and V3 (β-MyHC), are present. Functional differences between α-MyHC and β-MyHC, such as shortening velocity, force generation and ATPase activity, have been well documented [92]. If phosphorylation of cMyBP-C is important for regulating the kinetics of heart muscle contraction, one might expect to see a difference in its effect on thick filaments containing predominantly α- or β-MyHC. With α-MyHC, phosphorylation leads to a decrease in flexibility of the cross-bridges, looser packing of the thick filament backbone and extension of the cross-bridge [82]. In contrast, the cross-bridges containing N95% β-MyHC have greater flexibility with nonphosphorylated cMyBP-C than cross-bridges containing α-MyHC, and treatment with PKA does not extend the cross-bridges or change their flexibility. Expression of the phospho-mimetic cMyBP-C in a β-MyHC background showed improved contractile function in vivo [76]. The data further show that cMyBP-C phosphorylation at PKA sites can activate β-MyHC kinetics and Mg2+-ATPase activity in comparison to the α-MyHC background. Although cMyBP-C phosphorylation has different effects in different myosin backgrounds, it preserves cardiac function from I-R injury regardless of the myosin isoform, suggesting that the cardioprotection mechanism operates through a different mechanism than that for different myosin isoforms. In conclusion, total phosphorylation of cMyBP-C by PKA has been shown to affect thick filament orientation and myosin kinetics, but how cMyBP-C phosphorylation coordinates with the phosphorylation of thin filament proteins such as cTnI needs to be determined in terms of Ca2+ sensitivity and thick and thin filament activation.
4.3. Protein kinase C
PKC-ɛ is known to phosphorylate cMyBP-C only at Ser-273 and Ser-302 [93,94], and is activated after MI [95]. Xiao et al. [94] showed that PKC-ɛ activity has been linked to Ser-302 phosphorylation status during HF. Numerous studies have suggested that PKC-ɛ activation is important in ischemic preconditioning of the myocardium [93,95], associated with altered Mg2+-ATPase activity and contractile velocity [96]. Evidence from other studies suggests that cMyBP-C phosphorylation by PKC influences actomyosin Mg2+-ATPase activity, the kinetics of cross-bridge cycling and the rate of relaxation [27,78,87,97]. PKC phosphorylates cTnI and cMyBP-C with a concomitant improvement in post-ischemic contractile function [93]. PKC-α- and PKC-ɛ-mediated phosphorylation of cTnI, cTnT and cMyBP-C showed decreased myofilament Ca2+ sensitivity, which might alter contractility of human myocardium [98]. However, the only report on the effects of PKC-mediated phosphorylation of cMyBP-C showed that it could cause a decrease in maximum ATPase activity without a shift in the Ca2+-ATPase activity relation In vitro [99]. Furthermore, PKD, which is activated downstream of PKC in many settings [100] also appears to target cMyBP-C at residues that remain to be identified [71].
5. cMyBP-C phosphorylation regulates thick and thin filament interaction
cMyBP-C’s ability to tether the thick and thin sarcomeric filaments orients the actin filament and myosin heads, which facilitates activation of cross-bridge cycling [101]. It is hypothesized that the capacity of cMyBP-C to interact with S2 myosin is modified by cMyBP-C phosphorylation such that when cMyBP-C is dephosphorylated it interacts strongly with myosin (Fig. 3), thereby preventing its forcegenerating interaction with actin [102]. Conversely, when cMyBP-C phosphorylation levels at four serine residues (Ser-273, Ser-282, Ser-302 and Ser-307) are increased, the interaction with myosin S2 region is lost and cMyBP-C can then associate with thin filament proteins, such as actin and α-tropomyosin (α-TM) [15]. However, there is no direct evidence showing a cMyBP-C phosphorylation-dependent on/off interaction with myosin in vivo. An interaction between the C0 domain and some part of the myosin cross-bridge has been hypothesized on the basis of data from a mutant cMyBP-C knock-in mouse model [103] in which the linker between the C0 and C1 domains was deleted. In that model, there was an increase in Ca2+ sensitivity to force production similar to that previously seen in cardiomyocytes depleted of cMyBP-C [104]. Truncation of cMyBP-C in the genetically manipulated mouse was thought to prevent the C0 domain from reaching out to interact with the myosin head. In contrast, sequence comparison between the C0 domain and myomesin suggests that the C0 domain contains a novel putative LMM binding site [56]. It is not known whether this interaction occurs and what its function is. To further complicate interpretation, recent evidence suggests that cMyBP-C might interact with actin at the N-terminus [31]. Co-sedimentation assays showed a low affinity of F-actin for cMyBP-C [105,106], indicating that any interaction with actin is unlikely to occur through a cardiac-specific C0 domain. Homology modeling with the essential light chain of myosin identified the Pro-Ala-rich linker preceding the C1 domain as the likely candidate that binds actin [28]. Furthermore, cMyBP-C fragments containing the C0 domain were shown to bind to actin, which indicates that it might contribute to the weak binding state by shifting the binding of the N-terminus of cMyBP-C between actin and myosin [102]. Other data suggest that the C0 and the Pro-Ala-rich C0–C1 linker region might bind to actin [28,56,103]. Conversely, Shaffer et al. [31] recently identified two new binding domains in cMyBP-C, the C1 and M domains that can bind with actin. C1 can bind independently of the phosphorylation state of the M domain, whereas the M domain only binds when it becomes dephosphorylated. However, the exact binding residues that interact with actin have not been identified in cMyBP-C.
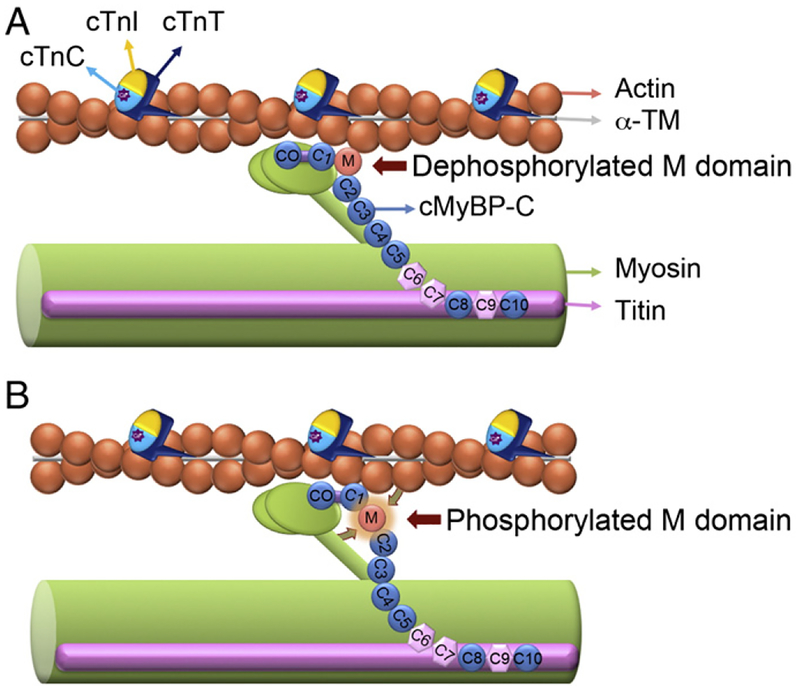
Fig. 3.
Dynamic phosphorylation of cMyBP-C’s M domain influences myosin–actin interactions. cMyBP-C facilitates myosin–actin interactions by tethering the fibers together and regulating the ability of the myosin head to bind to the actin filament during cross-bridge cycling. cMyBP-C interacts with the LMM of myosin at the C10 domain and titin at domains C8–C10. The C1-M-C2 domains have been shown to bind to myosin and actin. The capacity of these domains to interact is critical to overall sarcomeric integrity and function. The phosphorylatable M domain facilitates the role of cMyBP-C in cross-bridge formation. In the absence of M domain phosphorylation, the C1-M-C2 domains are tightly bound to myosin S2 region (A), whereas when M domain phosphorylated, C1-M-C2 domains and M domain release their interaction with myosin S2 region and actin, respectively, and changes in the thick filament orientation occur (B). C1 domain binds to actin irrespective of the M domain phosphorylation state. The dynamic phosphorylation of the M domain provides a point of control where the ordering of myosin heads and changes in force production can be regulated. Thin filament proteins include actin, α-TM, cTnI, cTnT and calcium (Ca2+)-binding protein cTnC are shown.
A crucial aspect of cMyBP-C’s behavior that is only beginning to be elucidated is the effects of the interaction between cMyBP-C and the MLC on the structure and function of the sarcomere. We hypothesize that there is a synergistic relationship between these two proteins, because when these two proteins are phosphorylated there is an increase in the ability of the myosin heads to interact with actin, which enhances cross-bridge formation [30,107]. TG mouse model expressing a cardiac specific nonphosphorylatable MLC demonstrated the necessity of MLC phosphorylation for normal cardiac function and response to β-AR stress [14], like cMyBP-C phosphorylation [74]. This model could be used in conjunction with TG cMyBP-C mouse models that have either phospho-mimetic [17] or -ablated [74] phosphorylation sites in order to determine the biophysical role of phosphorylation of cMyBP-C and the MLC independent of each other. Structural studies using C0–C2 peptides predict that cMyBP-C can bind with α-TM [29]. To determine the co-localization of the N-terminal region of the cMyBP-C with myosin and α-TM proteins, we performed immunofluorescence studies in neonatal rat cardiomyocytes using specific antibodies against cMyBP-C C0–C1, myosin and α-TM. Results showed that cMyBP-C’s N-terminal region co-localized with myosin and/or α-TM at the C-zone as well as in the rest of the A-band (Fig. 4). These data are consistent with the hypothesis that the cMyBP-C N-terminus can interact directly with myosin and α-TM, which may be critical to overall sarcomeric integrity and function [30]. The strength of the evidence for each of the cMyBP-C interactions with the thin filaments is variable, and the physiological consequences of thin filament interaction remain the subject of investigation. In addition, the exact cMyBP-C binding sites in actin and α-TM need to be clarified to define the nature of the cMyBP-C interaction with the thin filaments.
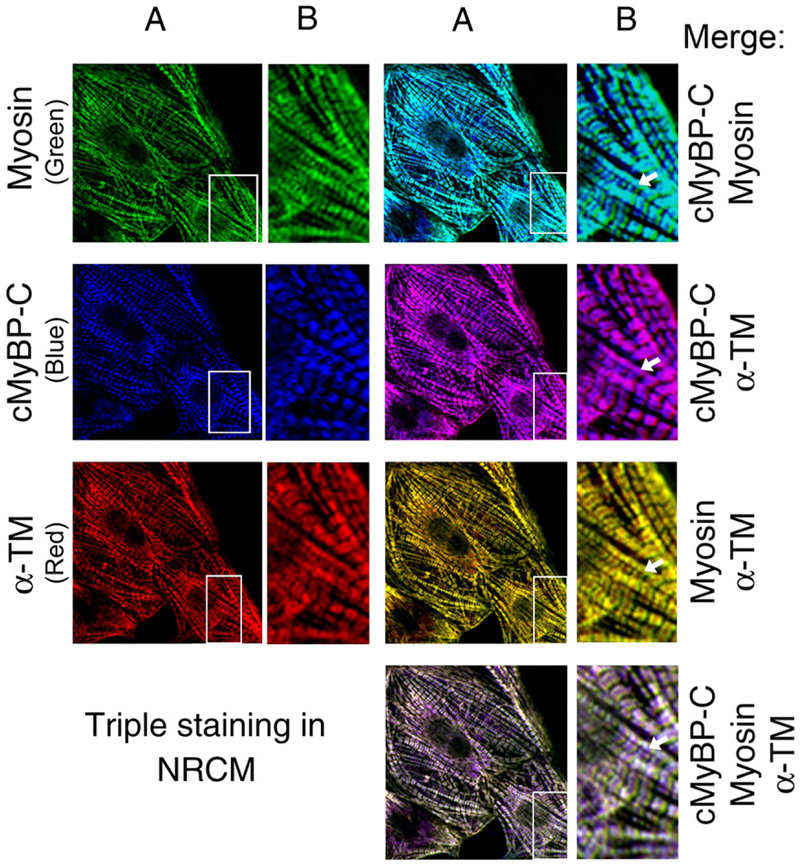
Fig. 4.
cMyBP-C connects thick and thin filaments. To determine the localization pattern of cMyBP-C along with thick and thin filament proteins, 3-day-old neonatal rat cardiomyocytes (NRCM) were used in chamber slides In vitro. The healthy NRCM were fixed and stained with the mouse monoclonal anti-α-myosin heavy chain (MF20, University of Iowa, Iowa), rabbit polyclonal anti-C0–C1 region of cMyBP-C [74] and sheep polyclonal anti-α-TM antibodies (Millipore, Billerica, MA). Secondary Alexa Fluor 488 goat anti-mouse IgG antibody for myosin (green color), Alexa Fluor Cy5 goat anti-rabbit IgG antibody for cMyBP-C (blue color) and Alexa Fluor 568 donkey anti-sheep IgG antibody for α-TM (red color) staining were used for immunofluorescence (Molecular Probes, Carlsbad, CA). Cardiomyocytes were analyzed with fluorescent microscopy (PCM-2000 scanner, Nikon, Melville, NY) and SimplePCI software (Compix Inc., Cranberry Township, PA). The rows of figures labeled B are enlarged from the inset in the rows of figures labeled A (60x magnification). Immunofluorescent staining of cMyBP-C (blue color) shows a typical doublet pattern of normal incorporation into the sarcomere. Co-staining with cMyBP-C with either myosin (green) or α-TM (yellow) show the co-localization of cMyBP-C with myosin (blue-green) and α-TM (pink). Co-localization of myosin and α-TM (yellow) confirm the presence of interaction between myosin and thin filament region. Triple staining shows that there is a defined merging of cMyBP-C, myosin and α-TM, but the presence of excess of pink (arrow) confirms the co-localization of cMyBP-C and α-TM. Nonoverlap regions are marked with arrows in respective panels. These data demonstrate that C0–C1 domains may directly interact with thin filament proteins via α-TM and thus connect thick and thin filaments.
6. Conclusion
cMyBP-C phosphorylation clearly has a direct effect on the heart’s contractile properties, sarcomere organization, and ability to tolerate I-R injury [17,76]. Clinically it might also be important to preserve cMyBP-C phosphorylation to attenuate the development of HF during I-R injury. Elucidating the link between cMyBP-C dysfunction and I-R injury and M domain-based cardioprotection represents a potential therapeutic avenue to improve myocardial contractility in the ischemic and failing heart. In addition, determining how cMyBP-C truncated proteins function (or do not function) during the development of sarcomeric dysfunction is critical in developing its potential as a therapeutic target. Although cMyBP-C is a substrate for multiple kinases in both the healthy and diseased heart, the different functional role(s) that each phosphorylation site has on myocardial contractility and their contribution to the thick and thin filament interaction need to be defined in order to establish the roles of cMyBP-C phosphorylation and function in the heart. In conclusion, cMyBP-C plays an important role in regulating the cardiac sarcomere, and future studies will require defining the phosphorylation and function of cMyBP-C in health and disease.
Acknowledgments
The authors apologize that they have not been able to mention every important study in this field due to space constraints. Preparation of this article was supported by an American Heart Association Scientist Development Grant (0830311N, Dr. Sadayappan).
Abbreviations:
- Ala
Alanine
- Asp
aspartic acid
- AR
adrenergic receptor
- cTn
cardiac troponin
- cMyBP-C
cardiac myosin binding protein-C
- CaMKII
Ca2+-depedent calmodulin kinase II
- DCM
dilated cardiomyopathy
- HCM
hypertrophic cardiomyopathy
- HF
heart failure
- I-R
ischemia-reperfusion
- Ig
immunoglobulin
- LMM
light meromyosin
- MyHC
myosin heavy chain
- MLC
myosin light chain
- MI
myocardial infarction
- PKA
protein kinase A
- PKC
protein kinase C
- TM
tropomyosin
- TG
transgenic
Footnotes
Disclosure statement
There are no unlabeled/unapproved uses of drugs or products, and no real or apparent conflicts of interest to report.
References
- [1].Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, et al. Heart disease and stroke statistics–2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2009;119:480–6. [DOI] [PubMed] [Google Scholar]
- [2].Yano M, Ikeda Y, Matsuzaki M. Altered intracellular Ca2+ handling in heart failure. J Clin Invest 2005;115:556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Messer AE, Jacques AM, Marston SB. Troponin phosphorylation and regulatory function in human heart muscle: dephosphorylation of Ser23/24 on troponin I could account for the contractile defect in end-stage heart failure. J Mol Cell Cardiol 2007;42:247–59. [DOI] [PubMed] [Google Scholar]
- [4].Jacques AM, Copeland O, Messer AE, Gallon CE, King K, McKenna WJ, et al. Myosin binding protein-C phosphorylation in normal, hypertrophic and failing human heart muscle. J Mol Cell Cardiol 2008;45:209–16. [DOI] [PubMed] [Google Scholar]
- [5].Hamdani N, Paulus WJ, van Heerebeek L, Borbely A, Boontje NM, Zuidwijk MJ, et al. Distinct myocardial effects of beta-blocker therapy in heart failure with normal and reduced left ventricular ejection fraction. Eur Heart J 2009;30: 1863–72. [DOI] [PubMed] [Google Scholar]
- [6].Sadayappan S, Robbins J. The death of transcriptional chauvinism in the control and regulation of cardiac contractility. Ann NY Acad Sci 2008;1123:1–9. [DOI] [PubMed] [Google Scholar]
- [7].Solaro RJ. Multiplex kinase signaling modifies cardiac function at the level of sarcomeric proteins. J Biol Chem 2008;283:26829–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hamdani N, Kooij V, van Dijk S, Merkus D, Paulus WJ, Remedios CD, et al. Sarcomeric dysfunction in heart failure. Cardiovasc Res 2008;77:649–58. [DOI] [PubMed] [Google Scholar]
- [9].Machackova J, Barta J, Dhalla NS. Myofibrillar remodeling in cardiac hypertrophy, heart failure and cardiomyopathies. Can J Cardiol 2006;22:953–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].El-Armouche A, Pohlmann L, Schlossarek S, Starbatty J, Yeh YH, Nattel S, et al. Decreased phosphorylation levels of cardiac myosin binding protein-C in human and experimental heart failure. J Mol Cell Cardiol 2007;43:223–9. [DOI] [PubMed] [Google Scholar]
- [11].Kobayashi T, Jin L, de Tombe PP. Cardiac thin filament regulation. Pflugers Arch 2008;457:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Warren CM, Arteaga GM, Rajan S, Ahmed RP, Wieczorek DF, Solaro RJ. Use of 2-D DIGE analysis reveals altered phosphorylation in a tropomyosin mutant (Glu54Lys) linked to dilated cardiomyopathy. Proteomics 2008;8:100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Marston SB, de Tombe PP. Troponin phosphorylation and myofilament Ca2+-sensitivity in heart failure: increased or decreased? J Mol Cell Cardiol 2008;45: 603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Scruggs SB, Hinken AC, Thawornkaiwong A, Robbins J, Walker LA, de Tombe PP, et al. Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation. J Biol Chem 2009;284:5097–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ge Y, Rybakova IN, Xu Q, Moss RL. Top-down high-resolution mass spectrometry of cardiac myosin binding protein-C revealed that truncation alters protein phosphorylation state. Proc Natl Acad Sci U S A 2009;106:12658–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Decker RS, Decker ML, Kulikovskaya I, Nakamura S, Lee DC, Harris K, et al. Myosin binding protein-C phosphorylation, myofibril structure, and contractile function during low-flow ischemia. Circulation 2005;111:906–12. [DOI] [PubMed] [Google Scholar]
- [17].Sadayappan S, Osinska H, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, et al. Cardiac myosin binding protein-C phosphorylation is cardioprotective. Proc Natl Acad Sci U S A 2006;103:16918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Offer G, Moos C, Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J Mol Biol 1973;74:653–76. [DOI] [PubMed] [Google Scholar]
- [19].Pepe FA, Drucker B. The myosin filament. III. C-protein. J Mol Biol 1975;99: 609–17. [DOI] [PubMed] [Google Scholar]
- [20].Craig R, Offer G. The location of C-protein in rabbit skeletal muscle. Proc R Soc Lond, B Biol Sci 1976;192:451–61. [DOI] [PubMed] [Google Scholar]
- [21].Luther PK, Bennett PM, Knupp C, Craig R, Padron R, Harris SP, et al. Understanding the organisation and role of myosin binding protein-C in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J Mol Biol 2008;384: 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tong CW, Stelzer JE, Greaser ML, Powers PA, Moss RL. Acceleration of crossbridge kinetics by protein kinase A phosphorylation of cardiac myosin binding protein-C modulates cardiac function. Circ Res 2008;103:974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schultheiss T, Lin ZX, Lu MH, Murray J, Fischman DA, Weber K, et al. Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J Cell Biol 1990;110:1159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ehler E, Rothen BM, Hammerle SP, Komiyama M, Perriard JC. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J Cell Sci 1999;112(Pt 10):1529–39. [DOI] [PubMed] [Google Scholar]
- [25].Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res 2002;90:594–601. [DOI] [PubMed] [Google Scholar]
- [26].McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, et al. Dilated cardiomyopathy in homozygous myosin binding protein-C mutant mice. J Clin Invest 1999;104:1235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gautel M, Zuffardi O, Freiburg A, Labeit S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J 1995;14:1952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Squire JM, Luther PK, Knupp C. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol 2003;331:713–24. [DOI] [PubMed] [Google Scholar]
- [29].Whitten AE, Jeffries CM, Harris SP, Trewhella J. Cardiac myosin-binding protein C decorates F-actin: implications for cardiac function. Proc Natl Acad Sci U S A 2008;105:18360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gruen M, Prinz H, Gautel M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein-C with myosin-S2 in an on-off fashion. FEBS Lett 1999;453:254–9. [DOI] [PubMed] [Google Scholar]
- [31].Shaffer JF, Kensler RW, Harris SP. The myosin binding protein-C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem 2009;284:12318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Moolman-Smook J, Flashman E, de Lange W, Li Z, Corfield V, Redwood C, et al. Identification of novel interactions between domains of myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circ Res 2002;91:704–11. [DOI] [PubMed] [Google Scholar]
- [33].Flashman E, Redwood C, Moolman-Smook J, Watkins H. Cardiac myosin binding protein-C: its role in physiology and disease. Circ Res 2004;94:1279–89. [DOI] [PubMed] [Google Scholar]
- [34].Flashman E, Watkins H, Redwood C. Localization of the binding site of the C-terminal domain of cardiac myosin binding protein-C on the myosin rod. Biochem J 2007;401:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Flashman E, Korkie L, Watkins H, Redwood C, Moolman-Smook JC. Support for a trimeric collar of myosin binding protein-C in cardiac and fast skeletal muscle, but not in slow skeletal muscle. FEBS Lett 2008;582:434–8. [DOI] [PubMed] [Google Scholar]
- [36].Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A 2008;105: 2386–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Winegrad S. Cardiac myosin binding protein-C. Circ Res 1999;84:1117–26. [DOI] [PubMed] [Google Scholar]
- [38].Robbins J, Benson DW. Structure–function relationships in myosin binding protein-C: taking off the blinders and collaring hypertrophic cardiomyopathy. Circ Res 2002;91:656–8. [DOI] [PubMed] [Google Scholar]
- [39].Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet 1995;11:434–7. [DOI] [PubMed] [Google Scholar]
- [40].Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet 1995;11:438–40. [DOI] [PubMed] [Google Scholar]
- [41].Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O’Connell J, et al. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation 1996;93:841–2. [DOI] [PubMed] [Google Scholar]
- [42].Carrier L, Bonne G, Bahrend E, Yu B, Richard P, Niel F, et al. Organization and sequence of human cardiac myosin binding protein-C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ Res 1997;80:427–34. [PubMed] [Google Scholar]
- [43].Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, et al. Mutations in the gene for cardiac myosin binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med 1998;338:1248–57. [DOI] [PubMed] [Google Scholar]
- [44].Moolman JA, Reith S, Uhl K, Bailey S, Gautel M, Jeschke B, et al. A newly created splice donor site in exon 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation 2000;101:1396–402. [DOI] [PubMed] [Google Scholar]
- [45].Van Driest SL, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Sarcomeric genotyping in hypertrophic cardiomyopathy. Mayo Clin Proc 2005;80:463–9. [DOI] [PubMed] [Google Scholar]
- [46].Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, et al. A common MYBPC3 (cardiac myosin binding protein-C) variant associated with cardiomyopathies in South Asia. Nat Genet 2009;41:187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Waldmuller S, Sakthivel S, Saadi AV, Selignow C, Rakesh PG, Golubenko M, et al. Novel deletions in MYH7 and MYBPC3 identified in Indian families with familial hypertrophic cardiomyopathy. J Mol Cell Cardiol 2003;35:623–36. [DOI] [PubMed] [Google Scholar]
- [48].Rottbauer W, Gautel M, Zehelein J, Labeit S, Franz WM, Fischer C, et al. Novel splice donor site mutation in the cardiac myosin binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization Of cardiac transcript and protein. J Clin Invest 1997;100:475–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, et al. Cardiac myosin binding protein-C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation 2009;119:1473–83. [DOI] [PubMed] [Google Scholar]
- [50].Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, et al. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res 2009;105:219–22. [DOI] [PubMed] [Google Scholar]
- [51].Sarikas A, Carrier L, Schenke C, Doll D, Flavigny J, Lindenberg KS, et al. Impairment of the ubiquitin–proteasome system by truncated cardiac myosin binding protein-C mutants. Cardiovasc Res 2005;66:33–44. [DOI] [PubMed] [Google Scholar]
- [52].Bahrudin U, Morisaki H, Morisaki T, Ninomiya H, Higaki K, Nanba E, et al. Ubiquitin–proteasome system impairment caused by a missense cardiac myosin binding protein-C mutation and associated with cardiac dysfunction in hypertrophic cardiomyopathy. J Mol Biol 2008;384:896–907. [DOI] [PubMed] [Google Scholar]
- [53].Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. In vivo modeling of myosin binding protein-C familial hypertrophic cardiomyopathy. Circ Res 1999;85:841–7. [DOI] [PubMed] [Google Scholar]
- [54].Flavigny J, Robert P, Camelin JC, Schwartz K, Carrier L, Berrebi-Bertrand I. Biomolecular interactions between human recombinant beta-MyHC and cMyBP-Cs implicated in familial hypertrophic cardiomyopathy. Cardiovasc Res 2003;60: 388–96. [DOI] [PubMed] [Google Scholar]
- [55].Vignier N, Schlossarek S, Fraysse B, Mearini G, Kramer E, Pointu H, et al. Nonsense-mediated mRNA decay and ubiquitin–proteasome system regulate cardiac myosin-binding protein-C mutant levels in cardiomyopathic mice. Circ Res 2009;105:239–48. [DOI] [PubMed] [Google Scholar]
- [56].Flavigny J, Souchet M, Sebillon P, Berrebi-Bertrand I, Hainque B, Mallet A, et al. COOH-terminal truncated cardiac myosin binding protein-C mutants resulting from familial hypertrophic cardiomyopathy mutations exhibit altered expression and/or incorporation in fetal rat cardiomyocytes. J Mol Biol 1999;294:443–56. [DOI] [PubMed] [Google Scholar]
- [57].Sato N, Kawakami T, Nakayama A, Suzuki H, Kasahara H, Obinata T. A novel variant of cardiac myosin binding protein-C that is unable to assemble into sarcomeres is expressed in the aged mouse atrium. Mol Biol Cell 2003;14: 3180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. A mouse model of myosin binding protein-C human familial hypertrophic cardiomyopathy. J Clin Invest 1998;102:1292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Oakley CE, Chamoun J, Brown LJ, Hambly BD. Myosin binding protein-C: enigmatic regulator of cardiac contraction. Int J Biochem Cell Biol 2007;39: 2161–6. [DOI] [PubMed] [Google Scholar]
- [60].Oakley CE, Hambly BD, Curmi PM, Brown LJ. Myosin binding protein-C: structural abnormalities in familial hypertrophic cardiomyopathy. Cell Res 2004;14: 95–110. [DOI] [PubMed] [Google Scholar]
- [61].Granzier HL, Campbell KB. New insights in the role of cardiac myosin binding protein-C as a regulator of cardiac contractility. Circ Res 2006;99:795–7. [DOI] [PubMed] [Google Scholar]
- [62].de Tombe PP. Myosin binding protein-C in the heart. Circ Res 2006;98:1234–6. [DOI] [PubMed] [Google Scholar]
- [63].Carrier L. Cardiac myosin binding protein-C in the heart. Arch Mal Coeur Vaiss 2007;100:238–43. [PubMed] [Google Scholar]
- [64].Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 2003;107:2227–32. [DOI] [PubMed] [Google Scholar]
- [65].El-Armouche A, Boknik P, Eschenhagen T, Carrier L, Knaut M, Ravens U, et al. Molecular determinants of altered Ca2+ handling in human chronic atrial fibrillation. Circulation 2006;114:670–80. [DOI] [PubMed] [Google Scholar]
- [66].Montgomery DE, Wolska BM, Pyle WG, Roman BB, Dowell JC, Buttrick PM, et al. alpha-Adrenergic response and myofilament activity in mouse hearts lacking PKC phosphorylation sites on cardiac TnI. Am J Physiol, Heart Circ Physiol 2002;282:H2397–2405. [DOI] [PubMed] [Google Scholar]
- [67].Mohamed AS, Dignam JD, Schlender KK. Cardiac myosin binding protein-C (MyBP-C): identification of protein kinase A and protein kinase C phosphorylation sites. Arch Biochem Biophys 1998;358:313–9. [DOI] [PubMed] [Google Scholar]
- [68].Shaffer JF, Jia W, Leary JA, S.P. H. PKA phosphorylates serine 307 of murine cardiac myosin binding protein-C In vitro. Biophys J 2008;96:500a. [Google Scholar]
- [69].Yuan C, Sheng Q, Tang H, Li Y, Zeng R, Solaro RJ. Quantitative comparison of sarcomeric phosphoproteomes of neonatal and adult rat hearts. Am J Physiol, Heart Circ Physiol 2008;295:H647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yuan C, Guo Y, Ravi R, Przyklenk K, Shilkofski N, Diez R, et al. Myosin binding protein-C is differentially phosphorylated upon myocardial stunning in canine and rat hearts: evidence for novel phosphorylation sites. Proteomics 2006;6: 4176–86. [DOI] [PubMed] [Google Scholar]
- [71].Haworth RS, Cuello F, Herron TJ, Franzen G, Kentish JC, Gautel M, et al. Protein kinase D is a novel mediator of cardiac troponin I phosphorylation and regulates myofilament function. Circ Res 2004;95:1091–9. [DOI] [PubMed] [Google Scholar]
- [72].Cuello F, Bardswell SC, Haworth RS, Yin X, Lutz S, Wieland T, et al. Protein kinase D selectively targets cardiac troponin I and regulates myofilament Ca2+ sensitivity in ventricular myocytes. Circ Res 2007;100:864–73. [DOI] [PubMed] [Google Scholar]
- [73].Palmer BM, Wang Y, Sadayappan S, Robbins J, Maughan DW. Phosphorylation sites of cardiac myosin binding protein-C are required for reduced calcium sensitivity of myofilaments due to protein kinase-A. Circulation 2006;114:II–66. [Google Scholar]
- [74].Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn II GW, et al. Cardiac myosin binding protein-C phosphorylation and cardiac function. Circ Res 2005;97:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Yang Q, Hewett TE, Klevitsky R, Sanbe A, Wang X, Robbins J. PKA-dependent phosphorylation of cardiac myosin binding protein-C in transgenic mice. Cardiovasc Res 2001;51:80–8. [DOI] [PubMed] [Google Scholar]
- [76].Sadayappan S, Gulick J, Klevitsky R, Lorenz JN, Sargent M, Molkentin JD, et al. Cardiac myosin binding protein-C phosphorylation in a {beta}-myosin heavy chain background. Circulation 2009;119:1253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jacquet S, Yin X, Sicard P, Clark J, Skanaganayagam GS, Mayr M, et al. Identification of cardiac myosin binding protein-C as a candidate biomarker of myocardial infarction by proteomic analysis. Mol Cell Proteomics in press; DOI: 10.1074/mcp.M900176-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Schlender KK, Bean LJ. Phosphorylation of chicken cardiac C-protein by calcium/calmodulin-dependent protein kinase II. J Biol Chem 1991;266:2811–7. [PubMed] [Google Scholar]
- [79].McClellan G, Kulikovskaya I, Winegrad S. Changes in cardiac contractility related to calcium-mediated changes in phosphorylation of myosin binding protein-C. Biophys J 2001;81:1083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tong CW, Gaffin RD, Zawieja DC, Muthuchamy M. Roles of phosphorylation of myosin binding protein-C and troponin I in mouse cardiac muscle twitch dynamics. J Physiol 2004;558:927–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hartzell HC, Glass DB. Phosphorylation of purified cardiac muscle C-protein by purified cAMP-dependent and endogenous Ca2+-calmodulin-dependent protein kinases. J Biol Chem 1984;259:15587–96. [PubMed] [Google Scholar]
- [82].Weisberg A, Winegrad S. Alteration of myosin cross bridges by phosphorylation of myosin binding protein-C in cardiac muscle. Proc Natl Acad Sci U S A 1996;93: 8999–9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Winegrad S. Myosin binding protein-C, a potential regulator of cardiac contractility. Circ Res 2000;86:6–7. [DOI] [PubMed] [Google Scholar]
- [84].Levine R, Weisberg A, Kulikovskaya I, McClellan G, Winegrad S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophys J 2001;81:1070–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Stelzer JE, Patel JR, Walker JW, Moss RL. Differential roles of cardiac myosin binding protein-C and cardiac troponin I in the myofibrillar force responses to protein kinase A phosphorylation. Circ Res 2007;101:503–11. [DOI] [PubMed] [Google Scholar]
- [86].Kunst G, Kress KR, Gruen M, Uttenweiler D, Gautel M, Fink RH. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res 2000;86:51–8. [DOI] [PubMed] [Google Scholar]
- [87].McClellan G, Weisberg A, Winegrad S. cAMP can raise or lower cardiac actomyosin ATPase activity depending on alpha-adrenergic activity. Am J Physiol 1994;267:H431–442. [DOI] [PubMed] [Google Scholar]
- [88].Hofmann PA, Lange III JH. Effects of phosphorylation of troponin I and C protein on isometric tension and velocity of unloaded shortening in skinned single cardiac myocytes from rats. Circ Res 1994;74:718–26. [DOI] [PubMed] [Google Scholar]
- [89].Song Q, Schmidt AG, Hahn HS, Carr AN, Frank B, Pater L, et al. Rescue of cardiomyocyte dysfunction by phospholamban ablation does not prevent ventricular failure in genetic hypertrophy. J Clin Invest 2003;111:859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Stelzer JE, Patel JR, Moss RL. Protein kinase A-mediated acceleration of the stretch activation response in murine skinned myocardium is eliminated by ablation of cMyBP-C. Circ Res 2006;99:884–90. [DOI] [PubMed] [Google Scholar]
- [91].Weisberg A, Winegrad S. Relation between crossbridge structure and actomyosin ATPase activity in rat heart. Circ Res 1998;83:60–72. [DOI] [PubMed] [Google Scholar]
- [92].Krenz M, Sanbe A, Bouyer-Dalloz F, Gulick J, Klevitsky R, Hewett TE, et al. Analysis of myosin heavy chain functionality in the heart. J Biol Chem 2003;278: 17466–74. [DOI] [PubMed] [Google Scholar]
- [93].Pyle WG, Chen Y, Hofmann PA. Cardioprotection through a PKC-dependent decrease in myofilament ATPase. Am J Physiol, Heart Circ Physiol 2003;285: H1220–1228. [DOI] [PubMed] [Google Scholar]
- [94].Xiao L, Zhao Q, Du Y, Yuan C, Solaro RJ, Buttrick PM. PKCepsilon increases phosphorylation of the cardiac myosin binding protein C at serine 302 both In vitro and in vivo. Biochemistry 2007;46:7054–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Inagaki K, Churchill E, Mochly-Rosen D. Epsilon protein kinase C as a potential therapeutic target for the ischemic heart. Cardiovasc Res 2006;70:222–30. [DOI] [PubMed] [Google Scholar]
- [96].Pyle WG, Lester JW, Hofmann PA. Effects of kappa-opioid receptor activation on myocardium. Am J Physiol, Heart Circ Physiol 2001;281:H669–678. [DOI] [PubMed] [Google Scholar]
- [97].Lim MS, Sutherland C, Walsh MP. Phosphorylation of bovine cardiac C-protein by protein kinase C. Biochem Biophys Res Commun 1985;132:1187–95. [DOI] [PubMed] [Google Scholar]
- [98].Kooij V, Boontje N, Zaremba R, Jaquet K, Dos Remedios C, Stienen GJ, et al. Protein kinase C alpha and epsilon phosphorylation of troponin and myosin binding protein-C reduce Ca(2+) sensitivity in human myocardium. Basic Res Cardiol 2009;105:289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Venema RC, Kuo JF. Protein kinase C-mediated phosphorylation of troponin I and C-protein in isolated myocardial cells is associated with inhibition of myofibrillar actomyosin MgATPase. J Biol Chem 1993;268:2705–11. [PubMed] [Google Scholar]
- [100].Avkiran M, Rowland AJ, Cuello F, Haworth RS. Protein kinase d in the cardiovascular system: emerging roles in health and disease. Circ Res 2008;102: 157–63. [DOI] [PubMed] [Google Scholar]
- [101].Colson BA, Bekyarova T, Locher MR, Fitzsimons DP, Irving TC, Moss RL. Protein kinase A-mediated phosphorylation of cMyBP-C increases proximity of myosin heads to actin in resting myocardium. Circ Res 2008;103:244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kulikovskaya I, McClellan G, Flavigny J, Carrier L, Winegrad S. Effect of myosin binding protein-C binding to actin on contractility in heart muscle. J Gen Physiol 2003;122:761–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Witt CC, Gerull B, Davies MJ, Centner T, Linke WA, Thierfelder L. Hypercontractile properties of cardiac muscle fibers in a knock-in mouse model of cardiac myosin binding protein-C. J Biol Chem 2001;276:5353–9. [DOI] [PubMed] [Google Scholar]
- [104].Hofmann PA, Greaser ML, Moss RL. C-protein limits shortening velocity of rabbit skeletal muscle fibres at low levels of Ca2+ activation. J Physiol 1991;439:701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Moos C. Fluorescence microscope study of the binding of added C protein to skeletal muscle myofibrils. J Cell Biol 1981;90:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yamamoto K, Moos C. The C-proteins of rabbit red, white, and cardiac muscles. J Biol Chem 1983;258:8395–401. [PubMed] [Google Scholar]
- [107].Ait Mou Y, le Guennec JY, Mosca E, de Tombe PP, Cazorla O. Differential contribution of cardiac sarcomeric proteins in the myofibrillar force response to stretch. Pflugers Arch 2008;457:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Yang Q, Osinska H, Klevitsky R, Robbins J. Phenotypic deficits in mice expressing a myosin binding protein-C lacking the titin and myosin binding domains. J Mol Cell Cardiol 2001;33:1649–58. [DOI] [PubMed] [Google Scholar]
- [109].Berul CI, McConnell BK, Wakimoto H, Moskowitz IP, Maguire CT, Semsarian C, et al. Ventricular arrhythmia vulnerability in cardiomyopathic mice with homozygous mutant myosin binding protein-C gene. Circulation 2001;104:2734–9. [DOI] [PubMed] [Google Scholar]
- [110].Palmer BM, Georgakopoulos D, Janssen PM, Wang Y, Alpert NR, Belardi DF, et al. Role of cardiac myosin binding protein-C in sustaining left ventricular systolic stiffening. Circ Res 2004;94:1249–55. [DOI] [PubMed] [Google Scholar]
- [111].Nyland LR, Palmer BM, Chen Z, Maughan DW, Seidman CE, Seidman JG, et al. Cardiac myosin binding protein-C is essential for thick-filament stability and flexural rigidity. Biophys J 2009;96:3273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Korte FS, McDonald KS, Harris SP, Moss RL. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res 2003;93:752–8. [DOI] [PubMed] [Google Scholar]
- [113].Stelzer JE, Dunning SB, Moss RL. Ablation of cardiac myosin binding protein-C accelerates stretch activation in murine skinned myocardium. Circ Res 2006;98: 1212–8. [DOI] [PubMed] [Google Scholar]
- [114].Brickson S, Fitzsimons DP, Pereira L, Hacker T, Valdivia H, Moss RL. In vivo left ventricular functional capacity is compromised in cMyBP-C null mice. Am J Physiol, Heart Circ Physiol 2007;292:H1747–1754. [DOI] [PubMed] [Google Scholar]
- [115].Colson BA, Bekyarova T, Fitzsimons DP, Irving TC, Moss RL. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J Mol Biol 2007;367:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Vu Manh TP, Mokrane M, Georgenthum E, Flavigny J, Carrier L, Semeriva M, et al. Expression of cardiac myosin binding protein-C (cMyBP-C) in Drosophila as a model for the study of human cardiomyopathies. Hum Mol Genet 2005;14:7–17. [DOI] [PubMed] [Google Scholar]
- [117].Nagayama T, Takimoto E, Sadayappan S, Mudd JO, Seidman JG, Robbins J, et al. Control of in vivo left ventricular contraction/relaxation kinetics by myosin binding protein-C: protein kinase A phosphorylation dependent and independent regulation. Circulation 2007;116:2399–408. [DOI] [PubMed] [Google Scholar]