INTRODUCTION
The development of the mouse embryo is a dynamic process that requires the spatial and temporal coordination of multiple cell types as they migrate, proliferate, undergo apoptosis, and differentiate to form complex structures. However, the confined nature of embryos as they develop in utero limits our ability to observe these morphogenetic events in vivo. Previous work has used fixed samples and histological methods such as immunofluorescence or in situ hybridization to address expression or localization of a gene of interest within a developmental time line. However, such methods do not allow us to follow the complex, dynamic movements of individual cells as the embryo develops. Genetic manipulation methods now allow us to label virtually any cell type or protein of interest fluorescently, providing powerful insights into morphogenetic events at cellular and subcellular resolutions. The development of ex vivo embryo culture methods combined with high-resolution imaging now provides a strong platform for observing morphogenetic events as they occur within the developing embryo. In this article, we discuss the advantages of live embryo imaging for observing dynamic morphogenetic events in vivo.
RELATED INFORMATION
Protocols for Preparation of Rat Serum for Culturing Mouse Embryos (Garcia et al. 2011a), Preparation of Postimplantation Mouse Embryos for Imaging (Garcia et al. 2011b), and Time-Lapse Imaging of Postimplantation Mouse Embryos (Garcia et al. 2011c) are also available.
OVERVIEW
Mouse embryonic development is a highly dynamic process requiring the coordination of multiple cellular events over the course of gestation. Over a short time period, cells undergo a number of cellular processes including proliferation, programmed cell death, migration, and differentiation, and are assigned roles that are necessary for the subsequent development of the organism. Over the years, advances in genetic manipulation of the mouse genome and high-throughput screens of chemically induced genetic mutations have allowed for the identification of hundreds of genes that play key roles in mouse embryonic development. Understanding the fundamental roles of these genes in vivo relies on the accurate assessment of the phenotype at the cellular level.
In many organisms that develop external to the mother, such as zebrafish, Drosophila, and Xenopus, live imaging has been used to determine the relationships between cells that are involved in rapid morphogenetic processes (Lichtman and Fraser 2001; Dickinson 2006; Keller et al. 2008; Nowotschin et al. 2009). Because development is a dynamic process that is constantly adapting to environmental cues, spatiotemporal information about cellular behaviors can be gained through serial observation in vivo. However, because mouse embryos develop in utero, most of what is known about cellular behaviors stems from embryos collected and fixed at several time points. An assessment of expression patterns and/or subcellular localization using immunohistological methods in serial sections can be helpful in determining gene functions during development, but complex spatiotemporal cellular dynamics cannot be captured in these static images. Therefore, a more direct approach must be used to fully understand the role of specific genes and proteins in the regulation of cellular function. Although culturing and imaging preimplantation embryos have been possible for some time, postimplantation embryos have been traditionally roller cultured, which does not permit time-lapse analysis of embryonic development. To circumvent these limitations, we have adapted known static culture methods for culture onto a microscope stage, which allows dynamic imaging of embryos from gastrulation until early organogenesis (Jones et al. 2002, 2005; Rhee et al. 2006; Aulehla et al. 2008; Kwon et al. 2008). Whole-embryo culture, combined with stable fluorescent markers such as fluorescent proteins, provides a powerful tool for understanding the cellular consequences of genetic manipulation in mice.
IMAGING EARLY MOUSE DEVELOPMENT
The developmental stages of the mouse embryo have been described in great detail in many volumes (Theiler 1989; Rugh 1990; Kaufman 1992), and there are strong efforts to digitize atlases of fixed stages of development, making them more accessible and interactive (Davidson et al. 1997; Burger et al. 2004; MacKenzie-Graham et al. 2004; Kaufman and Richardson 2005). Atlases and descriptions of mouse development have been invaluable for understanding wild-type as well as mutant phenotypes, but information about dynamic processes such as cell lineage and cell-fate specification, cell migration, and morphogenetic processes cannot be fully revealed from static images. Thus, to have a much more detailed and accurate understanding of the cellular behaviors in developing organisms, methods for manipulating embryos ex vivo have been developed.
Protocols for culturing preimplantation embryos have provided great insight into early cleavage events and have been the cornerstone of transgenic and gene knockout technologies (Nagy et al. 2003). However, imaging early postimplantation development ex vivo is decidedly more difficult. Embryos at these stages require a rich supply of freshly prepared medium in the form of serum to substitute for maternal nutrients, and they have been traditionally grown in roller culture (Tam 1998). The constant movement of the interface between the media surface and the air in roller cultures promotes gas exchange, and because the flow rate of gas is minimal, evaporation is limited. This article and its related protocols delineate how similar conditions can be used to culture mouse embryos in static culture on the stage of a microscope for ex vivo time-lapse imaging. This procedure is recommended for mouse embryos from 6.5 to 10.0 days post coitum (dpc) for up to 24 h. After 10.0 dpc, embryos become far more difficult to maintain using this method because of the inefficiency of nutrient transfer to the embryo and because of embryonic size limitations. However, a serum-free protocol for growing midgestation-stage embryos in culture has been described (Moore-Scott et al. 2003; Zwerts et al. 2007). These stages have also been studied successfully by labeling cells in embryos that are allowed to develop exo utero (Muneoka et al. 1986; Serbedzija et al. 1992; Makiko et al. 2008).
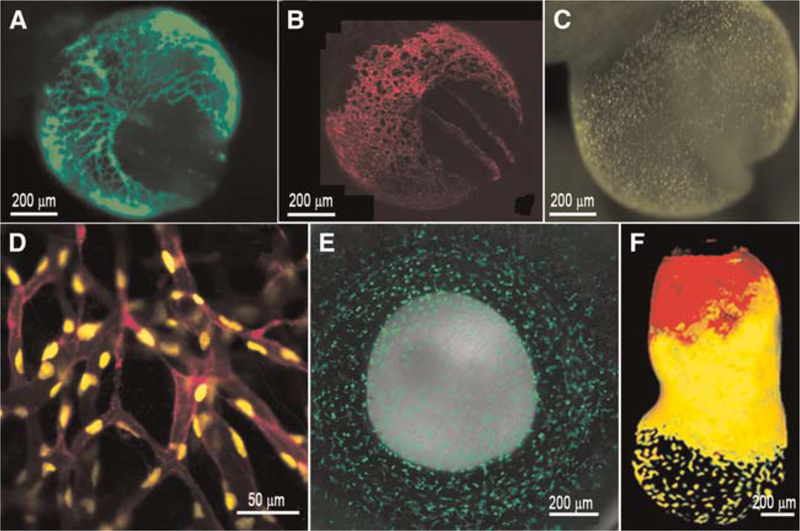
Many of the cell-labeling techniques developed for other developmental models are applicable to mouse embryos grown and imaged in static culture, including fluorescent dye injection, tissue grafting (Quinlan et al. 2008), and retroviral labeling. The strength of the mouse model, however, is the ability to manipulate the genome in such a way that genetically encoded fluorescent proteins can be expressed in specific cells or tissues (Fig. 1). In fact, several genetic strains have been engineered to express fluorescent proteins in defined embryonic regions. For example, the Oct4 promoter has been used to drive green fluorescence in primordial germ cells (Anderson et al. 2000), the β-globin promoter has been used to express green fluorescent protein (GFP) only in primitive erythroblasts (Fig. 1A; Dyer et al. 2001), the Afp and Ttr promoters can be used for specific labeling of the visceral endoderm (Fig. 1F; Kwon et al. 2006, 2008), the Flk1 promoter has been used to label endothelial cells in a nuclear (Stuart et al. 2005) and membrane-localized (Larina et al. 2009b) manner (Fig. 1B,C), and the c-fms promoter can be used to image macrophage cell lineages (Fig. 1E; Sasmono et al. 2003). In addition many knockins, where fluorescent protein sequences are inserted into a locus to disrupt gene function, allow researchers to combine fluorescence imaging with mutant analysis (e.g., the Nkx2.5GFP/GFP) (Biben et al. 2000), which expresses GFP in the embryonic and adult heart, or the Tbx6 mutant, which labels the presomitic mesoderm and newly formed somites [Hadjantonakis et al. 2008]).
FIGURE 1.
Fluorescent transgene expression in mouse embryos. (A) β-globin marks primitive erythroblasts in circulation within the yolk sac and embryo proper at 8.5 dpc. (B,C) The Flk1 promoter is used to drive membrane mCherry (B) and nuclear YFP (C) in endothelial cells of the developing embryo at 8.5 dpc. (D) A close-up image of vessels expressing YFP and mCherry in distinct subcellular localizations. (E) GFP expression driven by the cfms promoter in the neonatal eye (postnatal day 1), marking macrophages. (F) Expression of a double transgenic embryo at 7.0 dpc expressing GFP and red fluorescent protein (RFP) in cells derived from the visceral endoderm. (F, Reprinted from Hadjantonakis et al. 2008 with the permission from Elsevier © 2008.)
Fluorescent proteins that are specifically localized to distinct subcellular regions have also been produced (Fig. 1D). In some cases, localization of the protein is conferred by engineering fusion proteins or by tagging with sequences recognized by protein trafficking machinery. With the advent of convenient tools for multispectral analysis and linear unmixing, such as those available on models of Zeiss laser-scanning confocal microscopes, it is now possible to image labels of multiple colors without spectral bleed-through, even if the emission spectra are highly overlapping (Dickinson et al. 2001, 2003). Preparation of Postimplantation Mouse Embryos for Imaging (Garcia et al. 2011b) and Time-Lapse Imaging of Postimplantation Mouse Embryos (Garcia et al. 2011c) provide instructions for the application of static embryo culture techniques with time-lapse imaging for an in vivo assessment of dynamic cellular functions during embryonic development.
EXAMPLES OF LIVE IMAGING OF MOUSE EMBRYOS
Cell Behavior and Endoderm Morphogenesis in the Mouse Gastrula
Before gastrulation, the mouse embryo is separated into a bilaminar, cup-shaped structure comprised of the visceral endoderm and the epiblast. The visceral endoderm is fated to give rise to the extraembryonic visceral yolk sac, whereas the epiblast will give rise to the embryo proper and extraembryonic mesoderm of the yolk sac (Hogan and Zaret 2002). Older models of endoderm morphogenesis during gastrulation postulated that visceral endoderm cells are displaced proximally to extraembryonic regions via definitive endoderm ingression through the primitive streak (Lawson et al. 1986). However, the morphogenetic events that regulate this process had not been assessed with a high degree of spatial or temporal resolution in vivo until recently (Kwon et al. 2008). Using two transgenic mouse lines marking cells of the visceral endoderm (Afp::GFP and Ttr::RFP; see Fig. 1F; Kwon et al. 2006, 2008) and static embryo culture techniques (Kwon et al. 2008), the investigators were able to observe a dynamic series of events that resulted in the mixing of both the visceral and definitive endoderm lineages during gastrulation and gut endoderm formation. A novel model of endoderm morphogenesis via intercalation of the two lineages thus emerged. Three-dimensional ex vivo time lapses using Afp::GFP-positive (α-fetoprotein) embryos starting at late streak (7.0 dpc) revealed an initially uniform expression of GFP corresponding to the visceral endoderm overlying both the extraembryonic ectoderm and epiblast. Within the next 2 h, GFP-negative regions appeared within the visceral endoderm and increased in size over the next 6 h. The investigators concluded that the GFP-negative gaps present in visceral endoderm represent GFP-negative cells (from the definitive endoderm) intercalating within cells of the visceral endoderm. To complement these in vivo embryo experiments, the investigators verified intercalation within the visceral and definitive endoderm using genetic and immunohistological approaches. This use of static embryo culture and three-dimensional (3D) time-lapse imaging revealed the dynamic dispersal and intercalation of definitive and visceral endoderm, providing a new model for the morphogenesis of endoderm during early postimplantation development.
Imaging Vascular Development and Cardiac Function
The culture of 8.5-dpc embryos has been used to visualize the formation of the early cardiovascular system (Jones et al. 2005; Lucitti et al. 2007; Larina et al. 2009b). At ~7.5 dpc, isolated clusters of blood, surrounded by endothelial cells, form in the yolk sac of the embryo. These clusters expand and interconnect to form a random network known as the capillary plexus. This plexus connects to the heart and, as the heart begins to beat, the plexus is remodeled into a more mature, tree-like vascular system (Flamme et al. 1997). Using a mouse that expresses GFP driven by the β-globin promoter (Dyer et al. 2001), it is possible to image the formation of primitive erythroblasts and visualize changes in the vasculature. Endothelial cells can also be observed directly using a mouse that expresses GFP driven by the Tie2 promoter (Motoike et al. 2000) and two mouse lines expressing nuclear yellow fluorescent protein (YFP) (Fraser et al. 2005) and membrane-localized mCherry (Larina et al. 2009b) under the control of the Flk1 promoter. These markers can be used to image erythroblast differentiation and the formation of the capillary plexus, as well as the subsequent remodeling of the capillary plexus (see Fig. 1A–C). In addition to observing changes in morphology, the α-globin::GFP mice have also been used to quantify flow velocities in the yolk sac during development (for more information on methods for blood velocity quantification, see Jones et al. 2004; Lucitti et al. 2007; Larina et al. 2011). Finally, new advancements in optical coherence tomography and the application of embryo culture techniques have allowed for the assessment of mouse embryonic development and quantification of blood flow without the use of fluorescent markers or dyes (Larina et al. 2009a).
IMAGING SETUP
Control of the Culture Chamber Environment
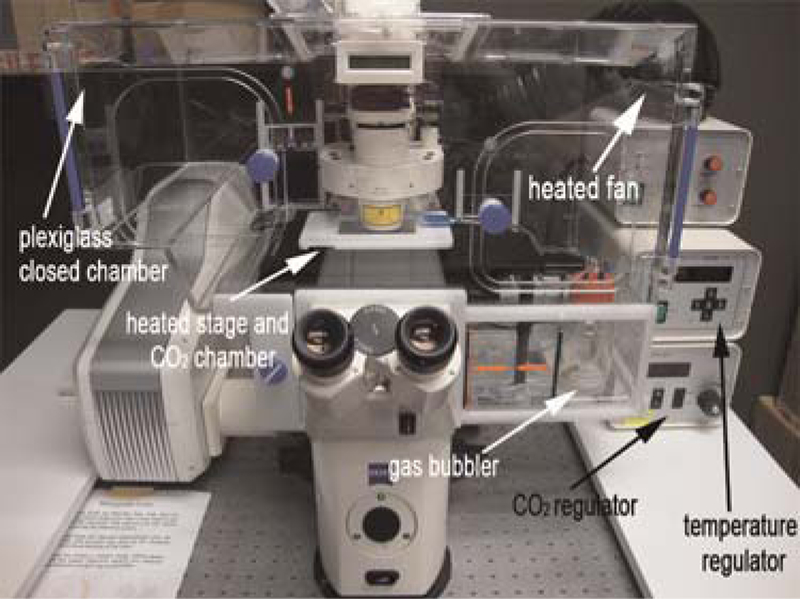
Proper environmental control of the microscope stage is probably the single most important factor for successful culturing of embryos. Mammalian embryos are especially sensitive to small changes in temperature (~1˚C) and slight amounts of evaporation. To keep the embryos at 37˚C, either construct a heater box that will fit around the microscope stage or purchase one from a commercial manufacturer (e.g., Zeiss) (Fig. 2). A commercial heater box is capable of precisely regulating both temperature and CO2 content throughout the culture in both the ambient air and the stage itself, tasks that a homemade box may not be able to do with the same level of precision.
FIGURE 2.
Example of a commercial heater box. Static culture system for time-lapse imaging of postimplantation mouse embryos. The microscope and static culture are housed in a temperature-(37°C) and CO2-regulated Plexiglas chamber. The gas mixture for the culture is humidified using a bubbler and an inlet to the heated stage allows for ample gas exchange during static culture.
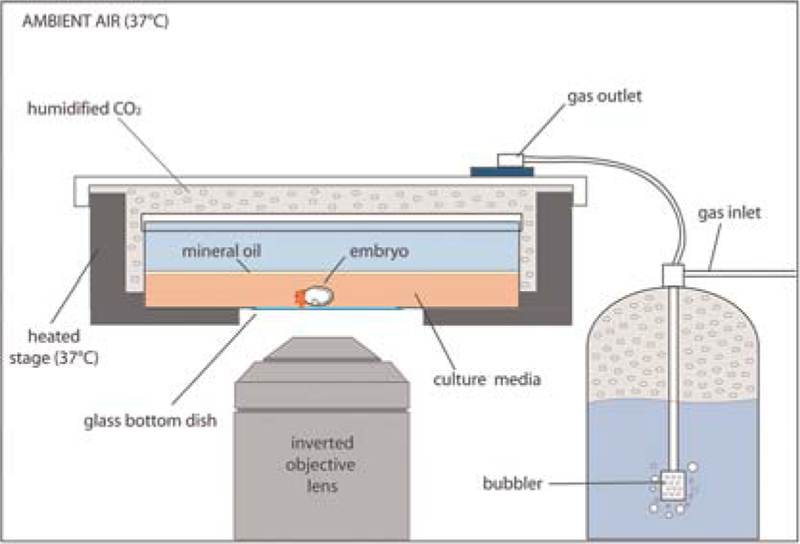
For imaging on an inverted microscope, embryos are routinely cultured in Lab-Tek two-well chambers with glass coverslip bottoms (Nunc) or 35-mm dishes with glass coverslip bottoms (MatTek). In commercial heater boxes, gas is fed into the culture environment through a heated stage and a humidified gas chamber that fits over the microscope stage (Fig. 3). With a self-constructed heater box, humidified CO2 created via a gas-wash bubbler is fed into the culture environment using 1/8-in tubing. The gas inlet into the culture chamber is created as follows. Use a soldering iron or a hot syringe needle to make a small hole in the chamber lid. This allows gas to be introduced into the culture chamber from the gas-washing bottle outlet. Place a small male-male barbed polypropylene fitting (1/16 × 1/8-in; Cole-Parmer Instrument Company) into this hole. The external end of the fitting will attach to the gas-washing bottle via appropriate tubing. Use silicon grease to ensure that the gas inlet and outlet are airtight.
FIGURE 3.
Schematic of heated culture chamber on an inverted microscope. The embryo is positioned inside a 37°C heated stage and covered with a gas outlet, allowing for humidified CO2 gas exchange within the chamber.
The gas requirements for mammalian embryos change depending on the stage of embryonic development. The gas consists of 5% CO2, variable oxygen concentration, and balanced nitrogen. For embryos between 4.5 dpc and 6.5 dpc, the oxygen should be kept at 95%; embryos between 7.5 dpc and 9.5 dpc develop well with 20% oxygen. Because mammalian embryos are sensitive to the slightest amount of evaporation, several steps can be taken to prevent evaporation. First, the gas is humidified through a gas-washing bottle. Additional optional steps include applying a thin layer of mineral oil over the surface of the medium and/or sealing the chamber using silicon grease or Teflon tape. Concerns about evaporation may vary depending on the geographic location of the experiment. In cities with high humidity year round, application of mineral oil and silicon grease is unnecessary. However, in more arid locales, prevention of evaporation is critical for a successful culture.
DISCUSSION
A major advantage of ex vivo optical imaging of mouse embryos is the ability to resolve cellular and subcellular events in a spatiotemporal fashion as they occur in development. These culture and imaging techniques allow scientists to assess the role of specific molecules in the context of a living, developing embryo, during both normal development and after genetic or pharmacological alterations. Although other imaging methods, such as ultrasound, optical coherence tomography, and magnetic resonance imaging, can provide insights into in utero development, these methods lack the cellular and subcellular resolution offered by confocal imaging. However, specific contrast agents are continuously being developed for these methods, and improvements in both resolution and contrast are being made at a rapid pace. In addition, the number of fluorescent probes and transgenic reporter mice used to observe dynamic cellular functions extends into the thousands and is increasing with each year, making mouse models advantageous for in vivo assessment of cellular function.
Although new transgenic animals are being developed to observe specific subsets of cells and tissues, these efforts are often laborious and expensive. In some instances, this limitation can be surmounted with the use of transgenic mice that ubiquitously express photoactivatable/photoconvertible fluorescent proteins (Nowotschin et al. 2009). Fluorescence from these proteins can either be activated or converted by laser light to track migrating cells, or follow morphogenetic movements in whole tissues, without the need to construct a new transgenic animal. In addition, there are a multitude of photoactivatable fluorescent proteins that can be combined with other transgenic markers.
One caveat to imaging mammalian embryos in culture is that they are more technically difficult to culture than other traditional models for imaging development, such as explant cultures. Mammalian embryos are especially sensitive to fluctuations in temperature, medium content, and physical pressure. A constant temperature must be maintained at 37°C–38°C to ensure proper development. High-quality rat serum is essential for growth, and at 8.5 dpc, even slight pressure on the yolk sac impairs circulation, preventing the embryos from turning and, hence, developing at a normal rate. The sensitivity of mammalian embryos to culture conditions can impact the results, but time-lapse analysis provides a wealth of dynamic data, so multiple trials are often worth the data generated by a few good movies.
Movement or embryonic drift during time-lapse imaging can be problematic in any organism, making it difficult to track cells and take quantitative measurements. Of the immobilization techniques described in Preparation of Postimplantation Mouse Embryos for Imaging (Garcia et al. 2011b) (Step 8), only the holding pipette offers significant control over the orientation of the embryo. In addition, the significant growth of the mammalian embryo between 7.5 and 9.5 dpc makes it difficult to keep all areas of interest in focus; choose the best objective lens for the resolution and magnification required. Furthermore, many algorithms are available for realigning imaging sequences, which can improve the information gained from such movies (Megason and Fraser 2003; Rupp et al. 2004; Megason 2009) and can be readily used in image processing software, such as Imaris (Bitplane). This software lets the user correct for drift over time, eliminating all objects that move out of frame over the course of a movie. Although the resulting movie may be significantly smaller as a result of drift, the amount of data recovered after drift correction is still valuable, especially when quantifying cell migration events.
The cup-shaped configuration of the yolk sac and turning and/or movement of the mouse embryo also make imaging and culturing techniques more complex. The curvature of the embryo can increase the number of optical sections needed to image a given region, but users risk the chance of exposing their samples to too much light, which can be harmful to the embryo. Thus, for each type of experiment, the number of optical sections scanned, fluorophores imaged, and intensity of the laser power used must be carefully balanced for optimal 3D, high-resolution imaging with minimal phototoxicity. Cellular movement, particularly migratory events, is also more complicated to assess, as cells tend to move across different optical sections over the course of a time-lapse imaging sequence. In addition, as the embryo grows, the radius of curvature is in constant flux. These problems are not isolated to mouse embryos, however, and can be overcome using 3D software for reconstructing and projecting the data and/or multiphoton imaging for improved depth penetration.
Dynamic imaging of mouse development is an important tool for a very robust genetic and developmental system. Whole-embryo culture and imaging of mammalian embryos combines molecular biology, genomics, and microscopy to understand the interplay of intercellular communication and the signaling components important for orchestrating complex developmental events.
REFERENCES
- Anderson R, Copeland TK, Schöler H, Heasman J, Wylie C. 2000. The onset of germ cell migration in the mouse embryo. Mech Dev 91: 61–68. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wiegraebe W, Baubet V, Wahl MB, Deng C, Taketo M, Lewandoski M, Pourquie O. 2008. A β-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol 10: 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, et al. 2000. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2–5. Circ Res 87: 888–895. [DOI] [PubMed] [Google Scholar]
- Burger A, Davidson D, Baldock R. 2004. Formalization of mouse embryo anatomy. Bioinformatics 20: 259–267. [DOI] [PubMed] [Google Scholar]
- Davidson D, Bard J, Brune R, Burger A, Dubreuil C, Hill W, Kaufman M, Quinn J, Stark M, Baldock R. 1997. The mouse atlas and graphical gene-expression database. Semin Cell Dev Biol 8: 50–517. [DOI] [PubMed] [Google Scholar]
- Dickinson ME. 2006. Multimodal imaging of mouse development: Tools for the postgenomic era. Developmental Dynamics 235: 2386–2400. [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Bearman G, Tille S, Lansford R, Fraser SE. 2001. Multispectral imaging and linear unmixing add a whole new dimension to laser scanning fluorescence microscopy. Biotechniques 31: 1272, 1274–1276, 1278. [DOI] [PubMed] [Google Scholar]
- Dickinson ME, Simbuerger E, Zimmermann B, Waters CW, Fraser SE. 2003. Multiphoton excitation spectra in biological samples. J Biomed Optics 8: 329–338. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Farrington SM, Mohn D, Munday JR, Baron MH. 2001. Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development 128: 1717–1730. [DOI] [PubMed] [Google Scholar]
- Flamme I, Frolich T, Risau W. 1997. Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J Cell Physiol 173: 206–210. [DOI] [PubMed] [Google Scholar]
- Fraser ST, Hadjantonakis A-K, Sahr KE, Willey S, Kelly OG, Jones EAV, Dickinson ME, Baron MH. 2005. Using a histone yellow fluorescent protein fusion for tagging and tracking endothelial cells in ES cells and mice. Genesis 42: 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MD, Udan RS, Hadjantonakis A-K, Dickinson ME. 2011a. Preparation of rat serum for culturing mouse embryos. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5593. [DOI] [PMC free article] [PubMed]
- Garcia MD, Udan RS, Hadjantonakis A-K, Dickinson ME. 2011b. Preparation of postimplantation mouse embryos for imaging. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5594. [DOI] [PMC free article] [PubMed]
- Garcia MD, Udan RS, Hadjantonakis A-K, Dickinson ME. 2011c. Timelapse imaging of postimplantation mouse embryos. Cold Spring Harb Protoc doi: 10.1101/pdb.prot5595. [DOI] [PMC free article] [PubMed]
- Hadjantonakis A-K, Pisano E, Papaioannou VE. 2008. Tbx6 regulates left/right patterning in mouse embryos through effects on nodal cilia and perinodal signaling. PLoS ONE 3: e2511. doi: 10.1371/journal.pone.0002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BLM, Zaret KS. 2002. Development of the endoderm and its tissue derivatives In Mouse development: Patterning, morphogenesis, and organogenesis (ed. Rosant J, Tam P), pp. 301–330. Academic Press, New York. [Google Scholar]
- Jones EAV, Crotty D, Kulesa PM, Waters CW, Baron MH, Fraser SE, Dickinson ME. 2002. Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis 34: 228–235. [DOI] [PubMed] [Google Scholar]
- Jones EAV, Baron MH, Fraser SE, Dickinson ME. 2004. Measuring hemodynamic changes during mammalian development. Am J Physiol Heart Circ Physiol 287: H1561–H1569. [DOI] [PubMed] [Google Scholar]
- Jones EA, Baron MH, Fraser SE, Dickinson ME. 2005. Dynamic in vivo imaging of mammalian hematovascular development using whole embryo culture. Methods Mol Med 105: 381–394. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. 1992. The atlas of mouse development. Academic Press, New York. [Google Scholar]
- Kaufman MH, Richardson L. 2005. 3D reconstruction of the vessels that enter the right atrium of the mouse heart at Theiler Stage 20. Clin Anat 18: 27–38. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. 2008. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322: 1065–1069. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Fraser SE, Eakin S, Mangano M, Isern J, Sahr KE, Hadjantonakis A-K, Baron MH. 2006. Tg(Afp-GFP) expression marks primitive and definitive endoderm lineages during mouse development. Dev Dynam 235: 2549–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis A-K. 2008. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell 15: 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larina IV, Ivers S, Syed S, Dickinson ME, Larin KV. 2009a. Hemodynamic measurements from individual blood cells in early mammalian embryos with Doppler swept source OCT. Opt Lett 34: 986–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larina IV, Shen W, Kelly OG, Hadjantonakis AK, Baron MH, Dickinson ME. 2009b. A membrane associated mCherry fluorescent reporter line for studying vascular remodeling and cardiac function during murine embryonic development. Anat Rec 292: 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larina IV, Garcia MD, Vadakkan T, Larin K, Dickinson ME. 2011. Imaging mouse embryonic cardiovascular development In Imaging in developmental biology: A laboratory manual (ed. Sharpe and Wong), pp. 659–668. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. 1986. Cell fate and cell lineage in the endoderm of the presomite mouse embryo, studied with an intracellular tracer. Dev Biol 115: 325–339. [DOI] [PubMed] [Google Scholar]
- Lichtman JW, Fraser SE. 2001. The neuronal naturalist: Watching neurons in their native habitat. Nat Neurosci 4: 1215–1220. [DOI] [PubMed] [Google Scholar]
- Lucitti JL, Jones EAV, Huang C, Chen J, Fraser SE, Dickinson ME. 2007. Vascular remodeling of the mouse yolk sac requires hemodynamic force. Development 134: 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Lee EF, Dinov ID, Bota M, Shattuck DW, Ruffins S, Yuan H, Konstantinidis F, Pitiot A, Ding Y, et al. 2004. A multimodal, multidimensional atlas of the C57BL/6J mouse brain. J Anat 204: 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiko Y, Toshihisa H, Hiroki O. 2008. Mouse exoutero development system: Protocol and troubleshooting. Congenital Anomalies 48: 183–187. [DOI] [PubMed] [Google Scholar]
- Megason SG. 2009. In toto imaging of embryogenesis with confocal time-lapse microscopy. Meth Mol Biol 546: 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, Fraser SE. 2003. Digitizing life at the level of the cell: High-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mech Dev 120: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Moore-Scott BA, Gordon J, Blackburn CC, Condie BG, Manley NR. 2003. New serum-free in vitro culture technique for midgestation mouse embryos. Genesis 35: 164–168. [DOI] [PubMed] [Google Scholar]
- Motoike T, Loughna S, Perens E, Roman BL, Liao W, Chau TC, Richardson CD, Kawate T, Kuno J, Weinstein BM, et al. 2000. Universal GFP reporter for the study of vascular development. Genesis 28: 75–81. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Wanek N, Bryant SV. 1986. Mouse embryos develop normally exoutero. J Exp Zool 239: 289–293. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. 2003. Manipulating the mouse embryo: A laboratory manual, 3 rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Nowotschin S, Eakin GS, Hadjantonakis A-K. 2009. Live-imaging fluorescent proteins in mouse embryos: Multidimensional, multi-spectral perspectives. Trends Biotechnol 27: 266–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan GA, Khoo PL, Wong N, Trainor PA, Tam PP. 2008. Cell grafting and labeling in postimplantation mouse embryos. Methods Mol Biol 461: 47–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JM, Pirity MK, Lackan CS, Long JZ, Kondoh G, Takeda J, Hadjan-tonakis A-K. 2006. In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis 44: 202–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugh R 1990. The mouse. Oxford University Press, Oxford, UK. [Google Scholar]
- Rupp PA, Czirok A, Little CD. 2004. avb3 integrin-dependent endothelial cell dynamics in vivo. Development 131: 2887–2897. [DOI] [PubMed] [Google Scholar]
- Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, Ostrowski MC, Himes SR, Hume DA. 2003. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101: 1155–1163. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner M, Fraser SE. 1992. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development 116: 297–307. [DOI] [PubMed] [Google Scholar]
- Stuart TF, Hadjantonakis A-K, Kenneth ES, Stephen W, Olivia GK, Elizabeth AVJ, Mary ED, Margaret HB. 2005. Using a histone yellow fluorescent protein fusion for tagging and tracking endothelial cells in ES cells and mice. Genesis 42: 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP. 1998. Postimplantation mouse development: Whole embryo culture and micro-manipulation. Int J Dev Biol 42: 895–902. [PubMed] [Google Scholar]
- Theiler K 1989. The house mouse: Atlas of embryonic development. Springer-Verlag, New York. [Google Scholar]
- Zwerts F, Lupu F, De Vriese A, Pollefeyt S, Moons L, Altura RA, Jiang Y, Maxwell PH, Hill P, Oh H, et al. 2007. Lack of endothelial cell surviving causes embryonic defects in angiogenesis, cardiogenesis, and neural tube closure. Blood 109: 4742–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]