Abstract
Cyanobacterial carboxysomes encapsulate carbonic anhydrase and ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). Genetic deletion of the major structural proteins encoded within the ccm operon in Synechococcus sp. PCC 7002 (ΔccmKLMN) disrupts carboxysome formation and significantly affects cellular physiology. Here we employed both metabolite pool size analysis and isotopically nonstationary metabolic flux analysis (INST-MFA) to examine metabolic regulation in cells lacking carboxysomes. Under high CO2 environments (1%), the ΔccmKLMN mutant could recover growth and had a similar central flux distribution as the control strain, with the exceptions of moderately decreased photosynthesis and elevated biomass protein content and photorespiration activity. Metabolite analyses indicated that the ΔccmKLMN strain had significantly larger pool sizes of pyruvate (> 18 folds), UDPG (uridine diphosphate glucose), and aspartate as well as higher levels of secreted organic acids (e.g., malate and succinate). These results suggest that the ΔccmKLMN mutant is able to largely maintain a fluxome similar to the control strain by changing in intracellular metabolite concentrations and metabolite overflows under optimal growth conditions. When CO2 was insufficient (0.2%), provision of acetate moderately promoted mutant growth. Interestingly, the removal of microcompartments may loosen the flux network and promote RuBisCO side-reactions, facilitating redirection of central metabolites to competing pathways (i.e., pyruvate to heterologous lactate production). This study provides important insights into metabolic regulation via enzyme compartmentation and cyanobacterial compensatory responses.
Keywords: Compensatory response, INST-MFA, Microcompartment, Mixotrophic, Photorespiration, RuBisCO
1. Introduction
Cyanobacteria are promising production systems to meet renewable resource demands because they are photosynthetic, genetically tractable, and capable of rapid growth. Photosynthetic organisms assimilate carbon using ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). However, in addition to its slow carboxylation kinetics, RuBisCO also oxygenates ribulose-1,5-bisphosphate (RuBP) producing 2-phosphoglycolate (2PG), a compound that is toxic to cells at high concentrations (Kern et al., 2011). Therefore, cyanobacteria have evolved CO2 concentrating mechanisms (CCM) around RuBisCO, encapsulating the enzyme within clathrate structures comprised of multiple proteins that form microcompartments referred to as carboxysomes. Carboxysomes result from protein-protein interactions between structural proteins (ccmKLMNO) and RuBisCO (rbcL, rbcS), which promotes the formation of an icosahedral shell that also encases carbonic anhydrase (CA, cccA) (Price et al., 1992; Cameron et al., 2013). The hierarchy of protein-protein interactions required for carboxysome assembly has previously been elucidated (Cameron et al., 2013). These polyhedral structures concentrate and increase CO2 availability to RuBisCO (Cai et al., 2009; Dou et al., 2008), without saturating CA (Mangan and Brenner, 2014).
Bacteria, as simple systems, are presumed to have little or no spatial subcellular organization, and thus the cytosol is assumed to be homogenous. However, evidence of metabolons (a multi-enzyme complex formed by non-covalent bonds between enzymes in a metabolic pathway) in bacteria exists (Ishikawa et al., 2004; Broddrick et al., 2016), indicating that enzymes may be more self-organized than previously suspected (Sweetlove and Fernie, 2018). Carboxysome compartmentalization and photosystem co-localization on cyanobacterial membranes are an obvious exception, and the advanced spatial organization confers enhanced growth and productivity. A small fraction of un-encapsulated RuBisCO is also localized outside of carboxysomes, often in association with the cell periphery near the thylakoid membranes in procarboxysome assembly centers (Agarwal et al., 2009; Cameron et al., 2013). It is possible that a cascade of enzymes proximal to carboxysomes might form metabolons that channel CO2 fixation reactions. The direct passing of metabolites among cascade enzymes may uniquely contribute to metabolic regulation (Abernathy et al., 2017b). In eukaryotes, the formation of metabolons are associated with cellular organelles and membrane compartments, such as the mitochondria or peroxisomes (Graham et al., 2007). In prokaryotes, evidence suggests that enzyme proximity channeling may be widely present in central pathways (Abernathy et al., 2019). Cyanobacterial RuBisCO is the starting point for reduced carbon metabolism in the cell, and the encapsulating carboxysome may offer an anchoring point for cascade enzymes. Removal of carboxysomes can change the cellular locations of both RuBisCO and downstream enzymes (Fig. 1A). Currently, it is unclear how the redistribution of enzymes in space affects the metabolic network.
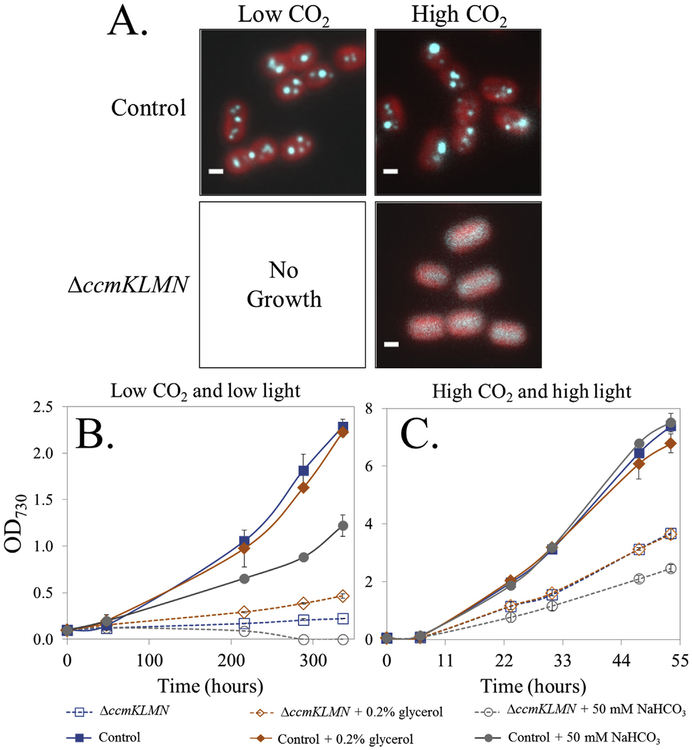
Fig. 1. Strain physiology under high and low CO2 and light conditions.
(A) Representative fluorescence images of the Synechococcus PCC 7002 control and ΔccmKLMN. Chlorophyll fluorescence is red, and GFP-labeled RuBisCO is cyan. Scale bars are 1 μm. (B) Growth under 0.2% CO2 and 25 μmol m−2•s−1. (C) Growth under 1% CO2 and 125 μmol m−2•s−1. Glycerol at a concentration of 0.2% (w/w) or sodium bicarbonate (NaHCO3) at a concentration of 50 mM were added to cultures to investigate photo-mixotrophic growth rates.
Isotopic experiments are regularly used to investigate pathway activity (Antoniewicz et al., 2007; Boyle et al., 2017) and metabolite channeling (Shearer et al., 2005; Williams et al., 2011; Zhang et al., 2017). Isotopically nonstationary metabolic flux analysis (INST-MFA) uses time-dependent mass isotopomer distributions (MID) of free metabolites to estimate fluxes through photosynthetic metabolism (Young et al., 2011; Abernathy et al., 2017a; Hendry et al., 2017; Xiong et al., 2015a; Ma et al., 2014). The same labeling experiments can also identify heterogeneous intracellular environments (i.e. metabolically inactive pools of central metabolites) (Abernathy et al., 2017a). In this study, INST-MFA on a carboxysome (ΔccmKLMN) mutant offers a strategy to understand Synechococcus 7002 metabolism in the absence of an innate microcompartment. Specifically, a ΔccmKLMN mutant was generated (Gordan et al., 2016), eliminating the protein structure around RuBisCO (Fig. 1). Due to the lack of a carbon concentrating mechanism, the mutant was grown under 1% v/v CO2 environments (hereby referred to as the high CO2 condition). The mutant phenotypes were measured and compared to the control strain, including spatial locations of RuBisCO, flux organizations, metabolite concentrations, biomass compositions, labeling dynamics of cascade metabolites, and photosynthetic capabilities. The results obtained here provide holistic insights into the effect of microcompartments on cyanobacterial metabolism.
2. Materials and methods
2.1. Strains and cultivation conditions
The Synechococcus PCC 7002 control strain and ΔccmKLMN mutant were cultured and maintained in A+ medium without citrate (18 g/L NaCl, 0.6 g/L KCl, 1 g/L NaNO3, 5 g/L MgSO4•7H2O, 0.05 g/L KH2PO4, 0.37 g/L CaCl2, 0.03 Na2EDTA•2H2O, 1 g/L Tris HCl, 0.555 mM H3BO3, 2.3 μM ZnCl2, 0.21 μM MoO3, 3 μM NH4Fe(SO4)2•12 H2O, 22 μM MnCl2, 0.012 μM CuSO4, 0.05 CoCl2, 0.0004 g/L vitamin B12) (Stevens et al., 1973). 100 μg/mL kanamycin or 25 μg/mL gentamicin were added to the ΔccmKLMN and control strain cultures, respectively. Concentrated glycerol, acetate, pyruvate, and sodium bicarbonate stocks were filtered and added to denoted cultures for studying carbon substrate effects. The same temperature (37 °C), shaking speed (200 rpm), and culture volume (20 mL of medium in 125 mL shake flasks) were used in all experiments. Cultivation occurred at either 0.2% v/v CO2 or 1% v/v CO2 with continuous light. In order to balance the minimal CO2 fixation that occurs in the low CO2 condition (0.2%), the light intensity was adjusted to 25 μmol of photons m−2•s−1. To assess the metabolism in the high CO2 condition (1%), light intensity was scaled linearly to 125 μmol of photons m−2•s−1. During cell growth, optical density was measured at 730 nm (OD730).
2.2. Synechococcus PCC 7002 ΔccmKLMN mutant generation and imaging
The Synechococcus PCC 7002 mutant was generated previously (Gordan et al., 2016) and kindly provided by the Pfleger group (University of Wisconsin-Madison). Briefly, the control strain was created by inserting RbcL-GFP in the SYNPCC7002_A2842 neutral site along with a gentamicin resistance cassette. The ΔccmKLMN mutant was generated by replacing ccmKLMN with a kanamycin resistance cassette along with RbcL-GFP (Gordan et al., 2016). The GFP marker enabled us to confirm the presence or absence of carboxysomes in the control and ΔccmKLMN, respectively (Fig. 1). Fluorescence microscopy images were acquired on a custom Nikon TiE inverted wide-field ORCA Flash 4.0 V2+ Digital sCMOS camera (Hamamatsu) using a Nikon CFI60 Plan Apochromat Lamda 100× oil immersion objective (1.45 N.A.) using an excitation wavelength of 488 nm (GFP) or 640 nm (Cy5/chlorophyll) with an LED source (Spectra X Light Engine, Lumencor, Beverton, OR) and emission through standard GFP and Cy5 filter sets. Prior to imaging, cells were grown in ambient or high CO2 conditions in an AL-41L4 Environmental Chamber (Percival Scientific, Perry, IA) maintained at 37 °C with continuous illumination (~150 μmol photons m−2 s−1) provided by cool white fluorescent lamps.
2.3. Dynamic labeling experiments
INST-MFA was conducted at 1% v/v CO2 with 125 μmol of photons m−2•s−1 to investigate phenotypic differences between the control and the mutant strains. For each experiment, cultures were inoculated from a seed culture to an OD730 of 0.05 in A+ medium and grown to exponential phase. Prior to the labeling experiment, a 2 mL aliquot of saturated NaH13CO3 (> 98% purity, Sigma Aldrich, St Louis) was injected into the medium for a final concentration of 4 g/L of NaH13CO3. Following the 13C-pulse, the cultures were quenched at different time intervals (30, 60, 120, 180, 300, 150, 600, 900 s) with ice cold medium and a liquid nitrogen bath (Abernathy et al., 2017a). The time-courses samples were centrifuged for 5 min (8000×g, ~0 °C). Biomass pellets were then frozen at −80 °C until metabolite extraction and analysis. Biological duplicates were used to generate standard deviations for experimentally measured MID values. Relative metabolite pool sizes were analyzed by an MS isotopomer ratio approach using fully labeled cell extracts as internal standards, specifically, fully labeled E. coli K-12 (cultured with uniformly labeled 13C glucose and 13C sodium bicarbonate). The estimation of metabolite pool sizes was performed by mixing a known amount of 13C labeled E. coli biomass with unlabeled cyanobacteria (three biological replicates). Then, the mixtures were extracted for LC-MS analysis. The isotopic ratio of each metabolite (labeled vs. unlabeled) was normalized by the amount of E. coli and cyanobacterial biomass, respectively (Abernathy et al., 2017a).
2.4. Metabolite extraction and analysis
The pelleted samples were extracted with a chloroform–methanol method (Ma et al., 2014). Supernatant aliquots were taken for analysis by lyophilizing 5 mL of medium. All samples were re-suspended in 1 mL of ddH2O for LC-MS/MS. Ion-pairing LC–MS/MS was performed at the Proteomics and Mass Spectrometry Facility, Donald Danforth Plant Science Center, St. Louis. Succinic acid in the supernatant was precisely quantified using an UV-method enzyme kit from R-Biopharm (Darmstadt, Germany) with A+ medium as controls (Table S1).
2.5. Isotopically nonstationary 13C-MFA
A cyanobacterial metabolic network was constructed with carbon atom transitions, which included the Calvin cycle, the oxidative pentose phosphate pathway, the TCA pathway, anaplerotic pathways, and a grouped photorespiration pathway (Table S2). A lumped biomass equation based on biomass composition analysis and biochemical equations was used to fit the model to biomass production. The INCA platform used a custom MATLAB ODE solver for a least squares regression to fit the experimentally determined free metabolite labeling data for Synechococcus PCC 7002 control and the ΔccmKLMN mutant (Young, 2014). INCA provided a goodness of fit via a Chi square statistical test as well as 95% confidence intervals of all estimated parameters by evaluating the sensitivity of the sum of squared residuals to parameter variations. Dilution parameters from INCA were used to account for labeling dilutions due to metabolically inactive pools (Young et al., 2011).
2.6. Biomass composition analysis
The cells were harvested during exponential growth by centrifugation, washed with a 0.9% NaCl solution and then freeze-dried. Protein and amino acid compositional analyses were performed by the Molecular Structure Facility, University of California (Davis, CA). Total carbohydrate was measured using the Anthrone Method, as described in our previous paper (Abernathy et al., 2017a). Fatty acid content and profile analyses were performed by Microbial ID (Newark, USA). Chlorophyll, pigments, RNA, DNA, and inorganics were estimated from previously published literature (Hendry et al., 2017). The biomass measurements were used to fine-tune the biomass equation in the INCA model. p values were calculated using a two-tailed student’s t-test.
2.7. GC–MS analysis of proteinogenic amino acids from mixotrophic cultures
To assess the capability of Synechococcus 7002 to uptake glycerol, acetate, or pyruvate, unlabeled seed cultures were grown at 1% CO2 and used to inoculate A+ medium (5% v/v ratio). The A+ medium was supplemented with either 25 mM glycerol-2-13C (second position labeled), 25 mM fully labeled acetate-13C2, or 25 mM pyruvate-3-13C (third position labeled). The 13C-cultures were transferred to either 1% CO2 or 0.2% CO2 conditions and grown until mid-log phase. The labeling in proteinogenic amino acids was then analyzed using a TBDMS method (You et al., 2012).
2.8. Measuring photosynthetic parameters and oxygen evolution
Cells were harvested during log growth and adjusted based on cell numbers. The chlorophyll fluorescence parameter, maximum quantum yield (FV/FM) of PSII, was measured using a fluorescence fluorometer, FL-200 (Photon Systems Instruments, Brno, Czech Republic). Cells were dark adapted for 90 s before F0 and FM were measured using red LED measuring light. Using a custom-built Clark-type electrode, light induced oxygen evolution was measured for 2 min at 37 °C following the addition of 10 mM NaHCO3 as described by a previous protocol (Ungerer et al., 2018). For respiratory O2 uptake, O2 consumption was measured for 2 min under dark conditions. Chlorophyll a content was measured by removing one milliliter of cell culture, extracting with methanol, and measuring the resulting absorbance at 665 and 652 nm as determined by a previous method (Porra et al., 1989).
3. Results
3.1. Growth comparison between the control strain and ΔccmKLMN mutant
The Synechococcus control strain and the ΔccmKLMN mutant were both grown at high CO2 (1% v/v) and under continuous light (125 μmol photons m−2•s−1) (Fig. 1 B&C). The control and ΔccmKLMN mutant growth rates in the exponential phase were 0.11 ± 0.005 h−1 and 0.085 ± 0.005 h−1, respectively. In the high CO2 concentration, the addition of NaHCO3 reduced mutant growth while there was no growth effect on the control strain. Under low CO2 conditions (0.2% v/v CO2), inclusion of 50 mM NaHCO3 reduced growth rate for the control strain. When the CO2 concentration was 0.2% or below, the ΔccmKLMN mutant exhibited little growth and the addition of 50 mM NaHCO3 was harmful to mutant survival. Under high CO2 conditions, neither the control strain nor the ΔccmKLMN strain utilized glycerol to promote cell growth. The mutant showed slight growth with glycerol under low CO2 circumstances. These observations confirmed the deleterious impact on cyanobacterial growth if the cyanobacterial ability to concentrate CO2 in microcompartments is compromised.
3.2. Biomass composition analysis
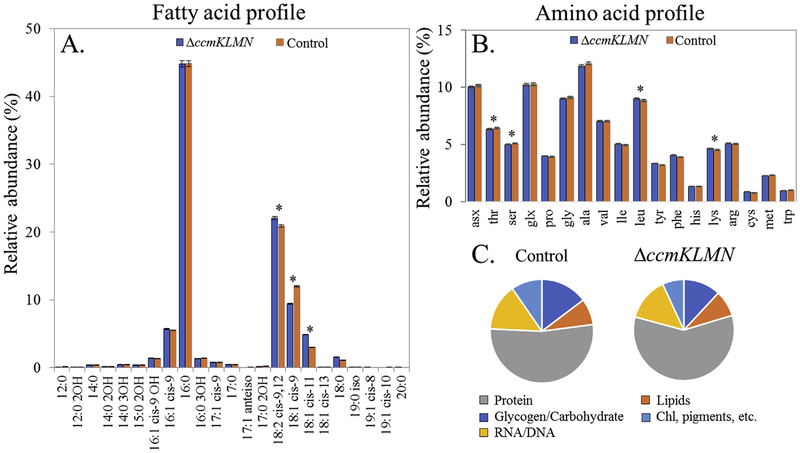
The biomass compositions (Fig. 2) were measured to reveal the differences in carbon partitioning between the control strain and the ΔccmKLMN mutant. The fatty acid profile was compositionally similar between the two strains, with the exception of statistically significant (p value < 0.05) increases of 18:2 cis-9,12 and 18:1 cis-11 fatty acids and a decrease of 18:1 cis-9 fatty acid in the ΔccmKLMN mutant. Protein-based amino acid profiles were also similar, with the exceptions of slightly elevated levels of leucine and lysine and slightly reduced levels of serine and threonine in the mutant. The biomass of the ΔccmKLMN strain exhibited a higher protein fraction (p value < 0.01), 51.2 ± 1.7 wt% protein of dry cell weight (DCW), compared to the control strain (43.7 ± 0.4%). The total glycogen/carbohydrate and lipid content were comparable with the ΔccmKLMN mutant comprised of 10.4 ± 3.1 wt% carbohydrate and 7.3 ± 3.0 wt% lipids and the control strain containing 12.3 ± 3.4 wt % carbohydrate and 6.8 ± 4.9 wt% lipids. The remaining components were expected to be chlorophyll and other pigments, soluble metabolites, and ashes.
Fig. 2. Biomass compositional analysis of Synechococcus PCC 7002 control and ΔccmKLMN mutant.
(A) Fatty acid profile found in each strain as a relative abundance of the total fatty acid content. (B) Amino acid profile found in each strain as the relative mole percentage. asx: aspartate and asparagine; glx: glutamate and glutamine. (C) Macromolecule biomass composition as a percentage of dry cell weight. * indicates a significant difference (p value < 0.05). Both control and mutant strains were cultivated in high CO2/light conditions.
3.3. Flux topology in PCC 7002 control and ΔccmKLMN mutant strains
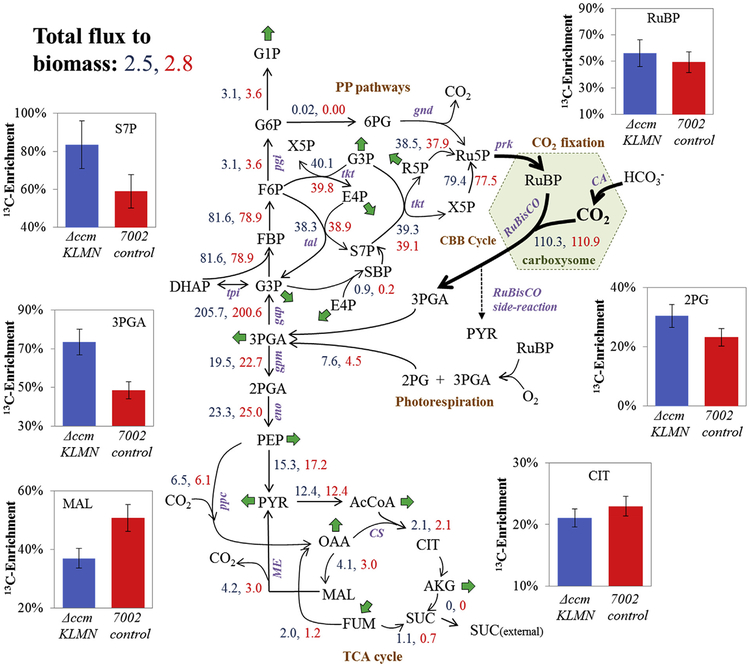
Using the biomass constraints as a sink reaction in the metabolic model, MFA was performed with the control and ΔccmKLMN mutant strains under the high CO2 and high light conditions. Fig. 3 represents a simplified flux network and is scaled with net fluxes normalized to a CO2 uptake of 100 units using a network derived from annotated genes found in the KEGG database. The full model can be found in Table S2. The INST-MFA model produced statistically acceptable sum-of-squared-residual fits for both the ΔccmKLMN mutant and the control strain (Tables S3 and S4, respectively).
Fig. 3. Synechococcus PCC 7002 control and ΔccmKLMN flux maps as determined by INCA (under high CO2/light conditions.
Net fluxes are normalized to a CO2 uptake of 100 mmol and reported as mmol/gDCW/h. ΔccmKLMN flux values are reported in blue, and PCC 7002 control flux values in red. Green arrows represent biomass synthesis reactions. Percentages of total 13C enrichment at steady state of key metabolites are represented by bar graphs. Total 13C enrichment was calculated at isotopic steady state. Standard deviations are based off biological replicates (n = 2). The complete metabolic network and full flux results can be found in Tables S2–S4.
Based on the INST-MFA analysis, both the control and mutant strains showed similar flux topology under high light and high CO2 conditions. The Calvin cycle in PCC 7002 made use of ~110 mmol CO2. Due to the malic shunt (Malate→Pyruvate + CO2) and pyruvate dehydrogenase (Pyruvate→AcCoA + CO2), ~15% of the fixed carbon was lost. The resulting biomass synthesis was 2.5 gDCW for ΔccmKLMN mutant and 2.8 gDCW for the control strain. Nearly 11% of the carbon flux from CO2 was directed to citrate synthesis for initiating the TCA pathway, while major carbon flux was cycled through the CBB cycle to regenerate RuBP. The relative fluxes through most central metabolic pathways (including the glycolysis, the TCA and the Calvin cycle pathways) were statistically indistinguishable between the ΔccmKLMN strain and the mutant (despite small differences in fluxes directed towards biomass synthesis and extracellular organic acids). This result indicated that, under sufficient light/CO2, ΔccmKLMN largely maintains a similar flux network to the control strain.
Compared to the other model cyanobacterium strains, Synechocystis 6803 (Young et al., 2011) and Synechococcus 7942 (Jazmin et al., 2017), PCC 7002 showed subtle metabolic network differences. In the TCA cycle, PCC 7002 lacks fumarate hydratase which interconverts malate and fumarate such that fumarate is solely generated as a byproduct of purine biosynthesis (Fig. 3). PCC 7002 also lacks the typical γ-aminobutyric acid shunt to close the TCA cycle. However, the TCA cycle can be functionally connected via the succinic semialdehyde bypass route (Zhang et al., 2016). Under our growth condition, the flux from AKG to succinate was close to zero and thus the TCA pathway was mainly used for biosynthesis. Furthermore, phosphoketolase, phosphate acetyltransferase, and acetate kinase are not present in the PCC 7002 genome annotation. Thus PCC 7002 may not be able to generate acetyl-CoA from the pentose phosphate pathway. In contrast, Synechocystis 6803 has a phosphoketolase shunt that splits xylulose-5-phosphate to acetyl phosphate and G3P (note: acetyl phosphate can be converted into acetyl-CoA or acetate to promote bio-production) (Knoop et al., 2013; Xiong et al., 2015b; Chwa et al., 2016; Moriyama et al., 2014). As INST-MFA is limited in its ability to distinguish between these pathways, we were unable to determine any non-annotated homologues or new enzymes in PCC 7002 central pathways. These enzymes may further fine-tune the flux network.
3.4. Photorespiration and photosynthesis
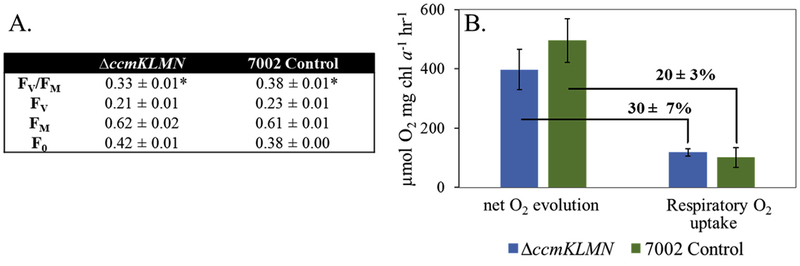
Carboxysome knock-out may impact photorespiration and photo-system activities. Photorespiration occurs when RuBisCO oxygenates RuBP to 2PG and 3-phosphoglycerate (3PGA). 2PG can be metabolized to glyoxylate and then to glycine, serine, and glycerate. In the control strain, photorespiration can be repressed by the carboxysomes which provide an efficient CO2 concentrating mechanism. The flux map showed (Fig. 3) that the control strain produced 4.5 mol of 2PG per 100 C-moles of biomass under the high light/high CO2 condition. Best estimates of photorespiratory flux for the ΔccmKLMN mutant indicated a net flux of 7.6 mol of 2PG per 100 C-moles of carbon fixation. This increase in photorespiration is consistent with increased free RuBisCO without a carboxysome’s physical protection against O2 (Cameron et al., 2013). To investigate the functions of the photosystems, chlorophyll fluorescence (FV/FM) under high CO2 conditions was measured for both strains (Fig. 4a). The FV/FM ratio determines the quantum efficiency of photosystem II (PSII). Here, the variable fluorescence (FV) was increased in the control strain compared to the ΔccmKLMN mutant while the maximum fluorescence (FM) remained similar. Thus, there was a 15% higher FV/FM ratio in the control strain (i.e., higher photosynthetic efficiency in the control strain). Furthermore, photosynthesis activity is associated with O2 evolution. The ΔccmKLMN mutant exhibited a lower net O2 evolution than the control strain (397 ± 68 μmol O2/mg chl a/hr vs 496 ± 74 μmol O2/mg chl a/hr). But respiratory O2 uptake by the control and the mutant strains was identical under dark condition (Fig. 4b), which accounted for 30 ± 7% of the O2 evolution in the ΔccmKLMN strain and 20 ± 3% in the control strain. In cyanobacteria, respiratory O2 uptake is due to photorespiration, oxidative phosphorylation (Wan et al., 2017), and the Mehler reactions (formation of H2O2 by photosynthesis) (Allahverdiyeva et al., 2011). Therefore, photo-respiration/photosynthesis analyses suggested that removal of carboxysomes moderately increased cyanobacterial photorespiration and decreased the photosystem efficiency under high CO2 growth conditions.
Fig. 4. Comparison of photosynthetic efficiency in PCC 7002 control and ΔccmKLMN mutants.
(A): PSII activities (FV/FM). (B): O2 evolution and uptake. F0: initial fluorescence; Fv: variable fluorescence; FM: maximum fluorescence. Technical (n = 2) and biological replicates (n = 3) were used for standard deviations. *p < 0.001. Percentages represent the respiratory O2 uptake (as measured in the dark) as a fraction of the total net O2 evolution.
3.5. Change in pool sizes in response to carboxysome knockout
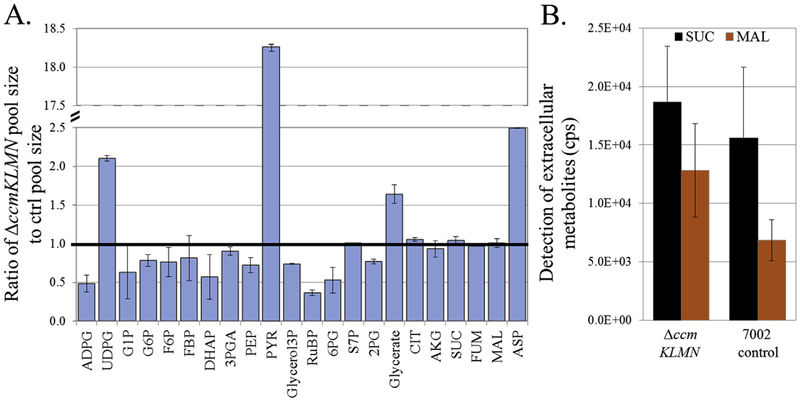
Metabolite concentrations impact reaction free energy and flux organization (Park et al., 2016). This study further investigated ΔccmKLMN compensatory responses at metabolite levels (Heise et al., 2015). Pool sizes were measured using fully 13C labeled E. coli cell extracts as an internal standard (Fig. 5a). The ΔccmKLMN mutant had significantly increased levels of intracellular pyruvate compared to the control (18-folds higher). The mutant also had significantly higher levels of intracellular UDPG (2.1-fold), aspartate (2.5-fold), and glycerate (1.6-fold) and reduced levels of ADPG (0.5-fold). UDPG was also more rapidly labeled in the ΔccmKLMN mutant (Fig. S1), which may be beneficial for sucrose synthesis (Qiao et al., 2018). The metabolite pool size of RuBP was lower and the levels of 2PG and 3PGA were slightly decreased compared to the control. The ΔccmKLMN mutant also had an increased pool size and labeling rate of glycerate (a downstream photorespiration intermediate), which provides further evidence for the elevation of photorespiration flux. In the ΔccmKLMN mutant, the flux from PEP to OAA (aspartate precursor) was expected to decrease in comparison to the control strain (Hackenberg et al., 2012). However, free aspartate accumulated in the mutant, possibly due to a slower anabolism for utilizing free amino acids. Increased expression of nblA1, involved in degradation of the phycobilisome light-harvesting apparatus, has been previously observed in the cyanobacteria lacking carboxysomes (Hackenberg et al., 2012). This suggests that protein turnover could also influence metabolite pools in the ΔccmKLMN. Overflow metabolites in the supernatant were also detected by LC-MS. The ΔccmKLMN mutant excreted a small amount of organic acids (e.g., succinate and malate) (Fig. 5b). We utilized a quantitative enzyme-based spectroscopic assay and found that the ΔccmKLMN mutant produced extracellular succinate up to 2.8 ± 0.3 mg/L in the late growth phase (Table S1).
Fig. 5. Comparison of metabolite pool size between Synechococcus PCC 7002 control and ΔccmKLMN.
(A) Relative pool size comparisons between the two strains. A y-axis of 1 represents equal metabolite concentrations while a y-axis > 1 indicates the factor of pool size in ΔccmKLMN greater than PCC 7002 control. The relative pool size was normalized using a13C-labeled internal standard and cell density. (B) Succinate and malate in the culture supernatant were detected by LC-MS, and the peak intensity (counts per second, cps) was proportional to their concentrations. Exact succinate concentration was further quantified using an enzyme kit (Table S1).
Intriguingly, 13C-pulse experiments indicated that central metabolites in the mutant strain had high 13C-enrichments at the end of tracing experiments (Fig. 3 and Fig. S1). For example, 13C-enrichments in ASP, UDPG, and glycerate from the mutant reached > 80% at isotopic steady state while enrichment in same metabolites from the control strain was between 40 and 60% (see discussion section for further explanations).
4. Discussion
4.1. Compensatory response in cyanobacteria conserves flux networks
Carboxysomes are key microcompartments in cyanobacteria for carbon concentration and assimilation. The loss of carboxysomes significantly impairs cell growth under atmospheric and low CO2 conditions. Under high CO2 concentration (1% v/v), the ΔccmKLMN mutant maintains flux ratios in central pathways similar to the control strain. In previous reports, quantitative RT-PCR and microarrays were used to monitor the abundance of gene transcripts in both ΔccmM and wild type cyanobacterial cells grown in high CO2 conditions (Emlyn-Jones et al., 2006; Hackenberg et al., 2012). Despite large transcriptional changes seen in genes involved in photosynthesis, high-light stress, and N assimilation in the ΔccmKLMN mutant, little-to-no transcriptional change was detected in its carbon uptake (i.e., genes encoding carbon transporter systems and transcriptional regulators), the Calvin Cycle, and other central metabolic pathways. These reports indicate that these central metabolic pathways are likely post-transcriptionally regulated (Hackenberg et al., 2012). The loss of the CCM impairs the carbon supply and energy-balance, leading to significant accumulations of intracellular metabolites (i.e., UDPG, glycerate, and pyruvate) as well as secretion of succinate and malate. The CCM is also closely tied to allosteric metabolites (Burnap et al., 2015), and a loss of carboxysomes may cue metabolite flux shifts towards free amino acids (i.e. aspartate). Macromolecule partitioning in biomass was also changed and the ΔccmKLMN mutant contains a higher protein content. Potentially, the ΔccmKLMN mutant may accumulate more stress proteins than the wild type strain (Hackenberg et al., 2012).
Free RuBisCO was reported to produce a small level of pyruvate instead of 3PGA via beta elimination of a phosphate ion and oxidative stress conditions (e.g., H2O2) increased this RuBisCO based pyruvate formation (Andrews and Kane, 1991). It has also been proposed that the loss of the carboxysome limits cyanobacterium ability to dissipate excess light energy. The resulting stresses (e.g., production of reactive oxygen species) on ΔccmKLMN mutant may lead to a significant increase of intracellular pyruvate accumulation through this RuBisCO side-reaction. Additionally, high-expression of nblA1, involved in degradation of the phycobilisome light-harvesting apparatus, was also observed in the cyanobacterial strain lacking carboxysomes (Hackenberg et al., 2012). This suggests that protein turnover in the ΔccmKLMN can be another source of pyruvate as well as free aspartate. Furthermore, the ΔccmKLMN mutant had lower [ATP]/[AMP] than the control strain (Fig. S2). An elevated ratio of [NADPH]/[NADP] was also observed in the ΔccmKLMN mutant (Fig. S2), which may indicate saturated terminal electron acceptors for photosynthesis and thus lead to lower net O2 evolution (Wang et al., 2016). In summary, ΔccmKLMN mutant demonstrated a suboptimal balance between the rates of photosynthesis and carbon fixation after spatial re-organizations of RubisCO. However, the compensatory responses at different cellular processes as well as metabolite overflow were able to restore a conserved flux network under high CO2 conditions (Cano et al., 2018).
4.2. Using carboxysome-deficient mutants to redirect carbon fluxes
Cyanobacterial species are promising photobiorefineries (Yu et al., 2015). Often, cyanobacterial engineering efforts target pathways that act as carbon sinks (Xiong et al., 2015a). For example, blocking the synthesis of glycogen is considered a useful strategy for rerouting carbon flux towards bioproduction (Ungerer et al., 2018; Hendry et al., 2017). However, such a strategy does not necessarily increase product titers (Li et al., 2014). In this study, the ΔccmKLMN mutant was able to increase UDPG pool sizes (by two-fold) as well as UDPG synthesis rates (Fig. 5 and Fig. S1). As UDPG is the precursor for sucrose and other valuable polysaccharides like heparosan (Sarnaik et al., 2019), the ΔccmKLMN mutant can potentially be utilized for production of value-added products. Additionally, the ΔccmKLMN mutant under high CO2 growth conditions accumulates pyruvate and organic acids, while largely maintaining a similar growth rate as the control strain. Common metabolic engineering strategies for increasing pyruvate concentration typically include N-deprivation and knocking out glycogen synthesis, but both methods cause significant growth inhibition (Benson et al., 2016; Jackson et al., 2015). Thus, knocking out carboxysomes may represent an alternative strategy to increase intracellular pyruvate (Figs. 3 and 5). For example, the removal of carboxysomes from a lactate producing PCC 7002 strain was recently shown to increase productivity by 24% and to increase titer by 125% (Clark et al., 2018). As carboxysome-free cyanobacterial mutants offer a unique method of for bio-containment of genetically engineered cell factories, these findings offer additional benefits for using a ΔccmKLMN mutant as a phototrophic platform.
4.3. Impact of carboxysome loss on photomixotrophic metabolism
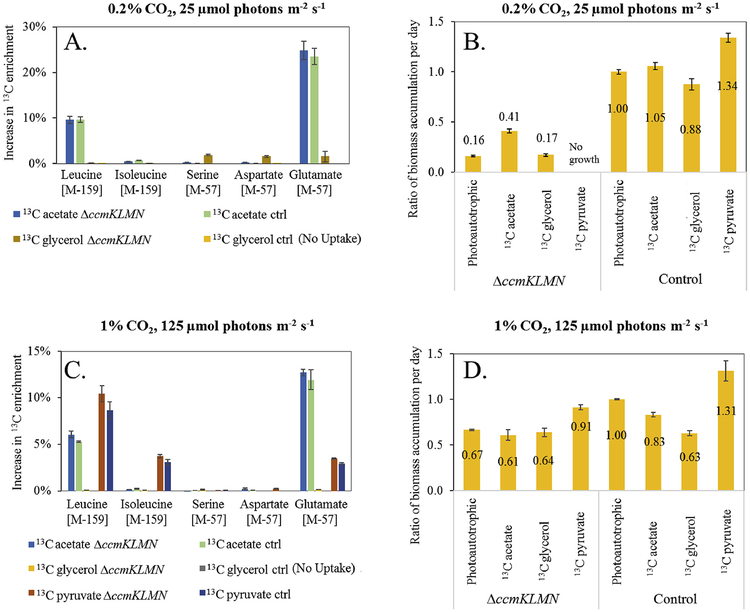
To probe photomixotrophic metabolism in the WT and carboxysome mutant, labeled glycerol, acetate, and pyruvate were used for isotopic experiments (Fig. 6). Both strains incorporated acetate and resulted in labeled Leu (derived from acetyl-CoA) and Glu (derived from ketoglutarate), which indicates that organic carbons are incorporated to metabolites of the TCA cycle. The incorporation of the 13C-carbon from acetate labeling into leucine but not isoleucine suggests PCC 7002 uses a threonine dependent pathway rather than the citramalate pathway for isoleucine synthesis (Wu et al., 2010). Under the high CO2 conditions, a mixotrophic environment had minimal impact on ΔccmKLMN mutant growth. Under the low CO2 condition, acetate supplementation rescued growth of the ΔccmKLMN mutant and resulted in the incorporation of acetate carbons into the biomass (revealed by isotopic labeling of amino acids). The increase of acetate usage benefitted photomixotrophic metabolism when cyanobacterial carboxysomes were disrupted. Glycerol was also found to be incorporated into biomass in the ΔccmKLMN mutant under the low CO2 condition. Literature has indicated that glycerol increased NAD(P)H in PCC 7002 cells growing at atmospheric conditions (Ludwig and Bryant, 2012). Here, neither strain utilized significant amounts of labeled glycerol under high CO2 conditions. Finally, exogenous pyruvate can be co-utilized by both strains for synthesis of some amino acids (such as leucine, isoleucine and glutamate). However, pyruvate impaired ΔccmKLMN mutant survival under low light and low CO2 conditions.
Fig. 6. Steady-state proteinogenic amino acid labeling from labeled glycerol, acetate and pyruvate to monitor photomixotrophic growth of PCC 7002 control (ctrl) and ΔccmKLMN.
GC-MS data for proteinogenic amino acids under photomixotrophic conditions with 25 mM labeled carbon. Standard deviations are estimated based on two biological replicates (n = 2). Percent 13C increase is graphed (i.e., evidence of organic carbon utilization). (A) Labeling of amino acids and (B) Ratio of biomass accumulation under mixotrophic and low light/CO2 conditions. (C) Labeling of amino acids and (D) Ratio of biomass accumulation under mixotrophic and high light/CO2 conditions. Note: “ratio of biomass accumulation per day” is defined as: .
4.4. Insights into microcomponent based flux regulation
Presence of bacterial metabolons or metabolite channeling may cause a heterogeneous mixture of intracellular metabolites. In INST-MFA, dilution parameters are typically introduced to aid in model fitting due to observations that metabolites never reach fully isotopic enrichment. These fitted dilution parameters for multiple metabolites represent unlabeled biomass reflux or heterogeneous metabolite pools that arise from enzyme localization and/or metabolite channeling. Removal of carboxysomes resulted in higher steady state 13C enrichment in central metabolites (particularly sugar phosphate metabolites) and smaller dilution parameters from INST-MFA. This indicates that metabolite pools become more homogeneous after disruption of metabolons. Recent literature has focused on metabolite channeling via enzyme clusters as a way for cells to regulate fluxes and cell homeostasis (Zhang et al., 2017; Sweetlove and Fernie, 2018; Abernathy et al., 2017b). In cyanobacteria, the Calvin cycle enzymes may channel intermediates through enzyme co-localization, as evidenced through ultra-centrifugation and immunoelectron microscopy studies (Agarwal et al., 2009). Disruption of native channels and redistribution of enzymes may decrease inactive metabolite pools, leading to more 13C enrichment in some metabolites at the end of pulse-tracing experiments. The channeling concept offers a new method for loosening metabolic rigidity and for flux redirection towards products. Deciphering channeling associated flux regulation can improve microbial feedstock utilization and biomanufacturing prospects (Abernathy et al., 2017b).
5. Conclusions
Carboxysome removal in PCC 7002 alters the spatial organization of CO2 assimilation. Enzyme organization and protein-protein interactions affect flux networks, but the quantitative details remain poorly understood. The information provided here on cell metabolons/channeling can improve INST-MFA modeling results and minimize the isotopic dilution effect. The outcomes from this study also offers insights into micro-compartment based metabolic regulations. Future studies are needed to elucidate the rules dictating the formation of metabolons or metabolite channeling in central pathways, and as such will enable bio-designers to add an extra level of metabolic control to current methods that rely mostly on gene copy number, transcriptional and translational regulations.
Supplementary Material
Acknowledgements
This work was supported by the Department of Energy, DESC0012722, the USDA-ARS, and the National Science Foundation, NSF-MCB #1616820 and NSF-DBI #1427621; the latter provided support for the acquisition of the LC-MS/MS used in this project. The authors would like to thank Dr. Himadri Pakrasi, Dr. Justin Ungerer, Dr. Marcus Foston and Virginia Johnson for their assistance in lab equipment and measuring photosynthesis/respiratory parameters.
Abbreviations:
- 2PG
2-phosphoglycolate
- 2PGA
2-phosphoglycerate
- 3PGA
3-phosphoglycerate
- 6PG
6-phosphogluconate
- ACA(or AcCoA)
acetyl-CoA
- AKG
α-ketoglutarate
- ADPG
adenosine diphosphate glucose
- CARB
carbohydrate
- CIT
citrate
- DHAP
dihydroxyacetone phosphate
- E4P
erythrose-4-phosphate
- F6P
fructose-6-phosphate
- FBP
fructose bisphosphate
- FUM
fumarate
- G1P
glucose-1-phosphate
- G6P
glucose-6-phosphate
- G3P
glyceraldehyde-3-phosphate
- GA
glycerate
- gDCW
gram dry cell weight
- GOX
glyoxylate
- MAL
malate
- OAA
oxaloacetate
- PEP
phosphoenolpyruvate
- PYR
pyruvate
- R5P
ribose-5-phosphate
- RuBisCO
ribulose-1,5-bisphosphate carboxylase/oxygenase
- Ru5P
ribulose-5-phosphate
- RuBP
ribulose-1,5-bisphosphate
- S7P
sedoheptulose-7-phosphate
- SBP
sedoheptulose-1,7-bisphosphate
- SUC
succinate
- UDPG
uridine diphosphate glucose
- X5P
xyulose-5-phosphate
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymben.2019.04.010.
References
- Abernathy MH, Yu J, Fa M, Liberton M, Ungerer J, Hollinshead WD, Gopalakrishnan S, He L, Maranas CD, Pakrasi HB, Allen DK, Tang YJ, 2017a. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol. Biofuels 10, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy MH, He L, Tang Y, 2017b. Channeling in native microbial pathways: implications and challenges for metabolic engineering. Biotechnol. Adv 35 (6). [DOI] [PubMed] [Google Scholar]
- Abernathy MH, Zhang Y, Hollinshead WD, Wang G, Baidoo EEK, Liu T, Tang YJ, 2019. Comparative studies of glycolytic pathways and channeling under in vitro and in vivo modes. AIChE J 65, 483–490. [Google Scholar]
- Agarwal R, Ortleb S, Sainis JK, Melzer M, 2009. Immunoelectron microscopy for locating Calvin cycle enzymes in the thylakoids of Synechocystis 6803. Mol. Plant 2(1), 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahverdiyeva Y, Ermakova M, Eisenhut M, Zhange P, Pichaud P, Hagemann M, Cournac L, Aro EM, 2011. Interplay between flavodiiron proteins and photorespiration in Synechocystis sp. PCC 6803. J. Biol. Chem 286, 24007–24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TJ, Kane HJ, 1991. Pyruvate is a by-product of catalysis by ribulosebisphosphate carboxylase/oxygenase. J. Biol. Chem 266 (15), 9447–9452. [PubMed] [Google Scholar]
- Antoniewicz MR, Kraynie DF, Laffend LA, Gonzalez-Lergier J, Kelleher JK, Stephanopoulos G, 2007. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab. Eng 9 (3), 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson PJ, Purcell-Meyerink D, Hocart CH, Truong TT, James GO, Rourke L, Djordjevic MA, Blackburn SI, Price GD, 2016. Factors altering pyruvate excretion in a glycogen storage mutant of the cyanobacterium, Synechococcus PCC 7942. Front. Microbiol 7, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle NR, Sengupta N, Morgan JA, 2017. Metabolic flux analysis of heterotrophic growth of Chlamydomonas reinhardtii. PLoS One 12 (5), e0177292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broddrick JT, Rubin BE, Welkie DG, Du N, Mih N, Diamond S, Lee JJ, Golden SS, Palsson BO, 2016. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc. Natl. Acad. Sci. U.S.A 113 (51), e8344–e8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap RL, Hagemann M, Kaplan A, 2015. Regulation of CO2 concentrating mechanism. Life 5 (1), 348–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA, 2013. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155 (5), 1131–1140. [DOI] [PubMed] [Google Scholar]
- Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S, 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to functions as a CO2 leakage barrier. PLoS One 4 (10), e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RL, Gordon GC, Bennett NR, Lyu H, Root TW, Pfleger BF, 2018. High-CO2 requirement as a mechanism for the containment of genetically modified cyanobacteria. ACS Synth. Biol 7 (2), 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano M, Holland SC, Artier J, Burnap RL, Ghirardi M, Morgan JA, Yu J, 2018. Glycogen synthesis and metabolite overflow contribute to energy balancing in cyanobacteria. Cell Rep 23, 667–672. [DOI] [PubMed] [Google Scholar]
- Chwa JW, Kim WJ, Sim SJ, Um Y, Woo HM, 2016. Engineering of a modular and synthetic phosphoketolase pathway for photosynthetic production of acetone from CO2 in Synechococcus elongatus PCC 7942 under light and aerobic condition. Plant Biotechnol. J 14 (8), 1768–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Heinhorst S, Williams EB, Murin CD, Shively JM, Cannon GC, 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts a diffusional barrier for CO2. J. Biol. Chem 283 (16), 10377–10384. [DOI] [PubMed] [Google Scholar]
- Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM, 2006. A Synechococcus PCC7942 ΔccmM (cyanophyceae) mutant pseudoreverts to air growth without regaining carboxysomes. J. Phycol 42 (4), 769–777. [Google Scholar]
- Gordan GC, Korosh TC, Cameron JC, Markley AL, Begemann MB, Pfleger BF, 2016. CRISPR interference as titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. Strain PCC 7002. Metab. Eng 38, 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JWA, Williams TCR, Morgan M, Fernie AR, Ratcliffe G, Sweetlove LJ, 2007. Glycolytic enzymes associate dynamically with mitrochondria in response to respiratory demand and support substrate channeling. Plant Cell 19, 3723–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg C, Huege J, Engelhardt A, Wittink F, Laue M, Matthijs HC, Kopka J, Bauwe H, Hagemann M, 2012. Low-carbon acclimation in carboxysome-less and photorespiratory mutants of the cyanobacterium Synechocystis sp. Strain PCC 6803. Microbiology 158, 398–413. [DOI] [PubMed] [Google Scholar]
- Heise R, Fernie AR, Stitt M, Nikoloski Z, 2015. Pool size measurements facilitate the determination of fluxes at branching points in non-stationary metabolic flux analysis: the case of Arabidopsis thaliana. Front. Plant Sci 6, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry JI, Prasannan C, Ma F, Mollers B, Jaiswal D, Digmurti M, Allen DK, Frigaard NU, Dasgupta S, Wangikar PP, 2017. Rerouting of carbon flux in a glycogen mutant of cyanobacteria assessed via isotopically non-stationary 13C metabolic flux analysis. Biotechnol. Bioeng 114 (10), 2298–2308. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Tsuchiya D, Oyama T, Tsunaka Y, Morikawa K, 2004. Structural basis for channeling mechanism of a fatty acid β-oxidation multienzyme complex. EMBO J 23, 2745–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, Eaton-Rye JJ, Bryant DA, Posewitz MC, Davies FK, 2015. Dynamics of photosynthesis in a glycogen-deficient glgC mutant of Synechococcus sp. Strain PCC 7002. Appl. Environ. Microbiol 81, 6210–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazmin LJ, Xu Y, Cheah YE, Adebiyi AO, Johnson CH, Young JD, 2017Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab. Eng 42, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern R, Bauwe H, Hagemann M, 2011. Evolution of enzymes involved in the photo-respiratory 2-phosphoglycolate cycle from cyanobacteria via algae toward plants. Photosynth. Res 109, 103–114. [DOI] [PubMed] [Google Scholar]
- Knoop H, Grundel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R, 2013. Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol 9 (6), e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shen CR, Liao JC, 2014. Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942. Photosynth. Res 120 (3), 301–310. [DOI] [PubMed] [Google Scholar]
- Ludwig M, Bryant DA, 2012. Synechococcus sp. Strain PCC 7002 transcriptome: acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front. Microbiol 3, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Jazmin LJ, Young JD, Allen DK, 2014. Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc. Natl. Acad. Sci. U.S.A 111 (47), 16967–16972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan NM, Brenner MP, 2014. Systems analysis of the CO2 concentrating mechanism in cyanobacteria. eLife 02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, Tajima N, Sekine K, Sato N, 2014. Characterization of three putative xylulose 5-phosphate/fructose 6-phosphate phosphoketolases in the cyanobacterium Anabaena sp. PCC 7120. Biosci. Biotechnol. Biochem 79 (5), 767–774. [DOI] [PubMed] [Google Scholar]
- Park JO, Rubin SA, Xu YF, Amador-Nogues D, Fan J, Shlomi T, Rabinowitz JD, 2016. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol 12, 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE, 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. [Google Scholar]
- Price GD, Coleman JR, Badger MR, 1992. Association of carbonic anhydrase activity with carboxysomes isolated from cyanobacterium Synechococcus PCC7942. Plant Physiol 100 (2), 784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao C, Duan Y, Zhang M, Hagemann M, Luo Q, Lu X, 2018. Effects of reduced and enhanced glycogen pools on salt-induced sucrose production in sucrose-secreting strain of Synechococcus elongatus PCC 7942. Appl. Environ. Microbiol 84 (2) e02023–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnaik A, Abernathy MH, Han X, Ouyang Y, Xia K, Chen Y, Cress B, Zhang F, Lali A, Pandit R, Linhardt RJ, Tang YJ, Koffas MAG, 2019. Metabolic engineering of cyanobacteria for photoautotrophic production of heparosan, a pharmaceutical compound. Algal Res 37, 57–63. [Google Scholar]
- Shearer G, Lee JC, Koo JA, Kohl Dh, 2005. Quantitative estimation of channeling from early glycolytic intermediates to CO in intact Escherichia coli. FEBS J 272 (13), 3260–3269. [DOI] [PubMed] [Google Scholar]
- Stevens SE, Patterson COP, Myers J, 1973. The production of hydrogen peroxide by blue-green algae: a survey. J. Phycol 9, 427–430. [Google Scholar]
- Sweetlove LJ, Fernie AR, 2018. The role of dynamic enzyme assemblies and substrate channeling in metabolic regulation. Nat. Commun 9, 2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J, Lin PC, Chen HY, Pakrasi HB, 2018. Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. mBio 9 (1) e02327–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan N, DeLorenzo DM, He L, You L, Immethun CM, Wang G, Baidoo EEK, Hollinshead WD, Keasling JD, Moon TS, Tang YJ, 2017. Cyanobacterial carbon metabolism: fluxome plasticity and oxygen dependence. Biotechnol. Bioeng 114 (7). [DOI] [PubMed] [Google Scholar]
- Wang X, Liu W, Xin C, Zheng Y, Cheng Y, Sun S, Li R, Zhu XG, Dai SY, Rentzepis PM, Yuan JS, 2016. Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natal. Acad. Sci. USA 113 (50), 14225–14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TCR, Sweetlove LJ, Ratcliffe RC, 2011. Capturing metabolite channeling in metabolic flux phenotypes. Plant Physiol 157 (3), 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Zhang B, Feng X, Rubens JR, Huang R, Hicks LM, Pakrasi HB, Tang YJ, 2010. Alternative isoleucine synthesis pathway in cyanobacterial species. Microbiology 156 (2), 596–602. [DOI] [PubMed] [Google Scholar]
- Xiong W, Morgan JA, Ungerer J, Wang B, Maness PC, Yu J, 2015a. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Native Plants 1, 15053. [Google Scholar]
- Xiong W, Lee TC, Rommelfanger S, Gjersing E, Cano M, Maness PC, Ghirardi M, Yu J, 2015b. Phosphoketolase pathway contributes to carbon metabolism in cyanobacteria. Native Plants 2, 15187. [DOI] [PubMed] [Google Scholar]
- You L, Page L, Feng X, Berla B, Pakrasi HB, Tang YJ, 2012. Metabolic pathway confirmation and discovery through 13C-labeling of proteinogenic amino acids. J. Vis. Exp 59, e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Shastri AA, Stephanopoulos G, Morgan JA, 2011. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab. Eng 13 (6), 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, 2014. INCA: a computational platform for isotopically nonstationary metabolic flux analysis. Bioinformatics 30 (9), 1333–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Liberton M, Cliften PF, Head RD, Jacobs JM, Smith RD, Koppenaal DW, Brand JJ, Pakrasi HB, 2015. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep 5, 8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Qian X, Chang S, Dismukes GC, Bryant DA, 2016. Natural and synthetic variants of the tricarboxylic acid cycle in cyanobacteria: introduction of the GABA shunt into Synechococcus sp. PCC 7002. Front. Microbiol 7, 1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Beard KFM, Swart C, Bergmann S, Krahnert I, Nikoloski Z, Graf A, Ratcliffe RG, Sweetlove LJ, Fernie AR, Obata T, 2017. Protein-protein interactions and metabolite channeling plant tricarboxylic acid cycle. Nat. Commun 16(8), 15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.