Abstract
Primordial germ cells (PGCs) form during early embryogenesis with a supply of maternal mRNAs that contain shorter poly(A)-tails. How translation of maternal mRNAs is regulated during PGC development remains elusive. Here we describe a small molecule screen with zebrafish embryos that identified primordazine, a compound that selectively ablates PGCs. Primordazine’s effect on PGCs arises from translation repression through primordazine-response elements in the 3′UTRs. Systematic dissection of primordazine’s mechanism of action revealed that translation of mRNAs during early embryogenesis occurs by two distinct pathways, depending on the length of their poly(A)-tails. In addition to poly(A)-tail dependent translation (PAT), early embryos perform poly(A)-tail independent non-canonical translation (PAINT) via deadenylated 3′UTRs. Primordazine inhibits PAINT without inhibiting PAT, an effect that was also observed in quiescent but not in proliferating mammalian cells. These studies reveal that PAINT is an alternative form of translation in the early embryo and is indispensable for PGC maintenance.
Introduction
Major questions surround the mechanisms of translation in the early embryo before zygotic genome activation, where virtually all mRNAs are maternally supplied and exhibit short poly(A)-tails1. How such mRNAs are translated without the benefit of poly(A)-tails, and how the appropriate mRNAs are selected for translation, remain unknown. One possibility is that alternative translational mechanisms may exist in early embryos to circumvent the dependence on polyadenylation seen with canonical translation. We sought insight into translation in the early embryo by studying primordial germ cells (PGCs), which are among the first recognizable cell types to emerge during embryogenesis2.
In many animals, PGC development commences prior to the onset of maternal-to-zygotic transition (MZT), so the common transcriptional and epigenetic processes that govern much of embryonic development cannot be used to regulate early germ cell development. Instead, regulation of the stability, localization, and translation of maternally supplied mRNAs is thought to regulate the development of PGCs in zebrafish. Unfortunately, many of the factors contributing to these processes remain unknown. This may be due, at least in part, to the difficulty in acquiring PGCs in mammalian models. Although PGCs or PGC-like cells can be obtained from embryonic stem cells or induced pluripotent cells (iPS)3–7, current methods have very low efficiency. In addition, in vitro systems cannot replicate all the factors that orchestrate developmental processes of PGCs in a living organism, which are dominated by maternally supplied RNAs and proteins, as well as signals originating from somatic cells. The zebrafish is an excellent in vivo model for the study of PGCs since zebrafish embryos undergo rapid early development, and enable facile visualization of PGCs8,9.
In zebrafish embryos prior to MZT, mRNAs generally possess much shorter poly(A)-tails1. As a result, translation is thought to be repressed generally until the onset of cytoplasmic polyadenylation (CPA) which stimulates translation by increasing the length of poly(A)-tails on maternal mRNAs and allowing closed loop formation. Although CPA is an important process for translation during early embryogenesis, more than half of maternal mRNAs (~60%) do not undergo CPA10, raising the possibility that an alternative mechanism of translation, independent of poly(A)-tails, may exist in early embryos to allow for translation of deadenylated transcripts.
Here, we performed a chemical screen in zebrafish embryos with the goal of identifying small molecules that perturb establishment of the PGC lineage (Fig. 1a). We identified a small molecule we named primordazine that causes selective ablation of PGCs, with other cell types being seemingly unaffected. We discovered that primordazine’s underlying mechanism of action is translational repression, mediated by the 3′UTRs of specific genes. Moreover, we provide evidence that translation of mRNA during early embryo development is mediated by at least two distinct pathways: canonical, primordazine-insensitive translation in the case of polyadenylated mRNAs, and non-canonical, primordazine-sensitive translation in the case of deadenylated, primordazine-response element (PRE)-containing mRNAs. These PRE-containing target mRNAs, for example nanos3 and deadend1 (dnd1), are sequestered in abnormal granules upon primordazine treatment. Thus, phenotypic screening has identified a small molecule capable of disrupting PGC development, thereby uncovering a fundamental translational mechanism.
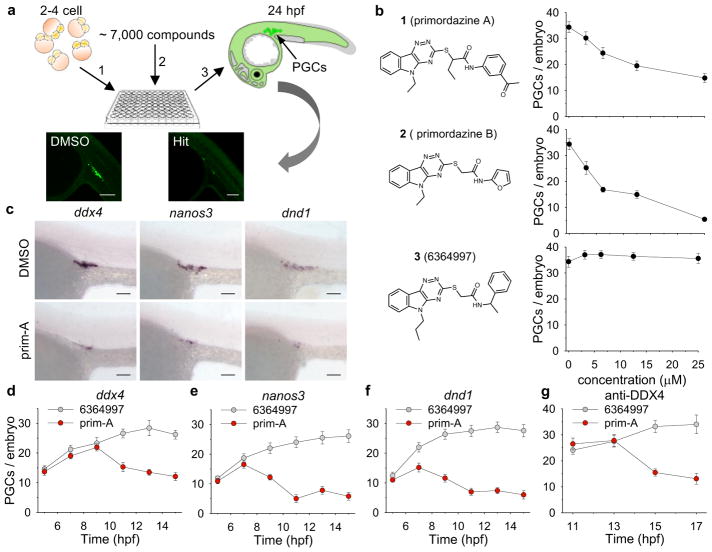
Figure 1. Discovery of small molecules, primordazine A and B, that ablate PGCs.
(a) Schematic diagram of chemical library screen. Approximately 10 zebrafish embryos at 0.5 hpf expressing EGFP in their PGCs were placed in each well of a 96 well plate. Compounds were applied to each well at ~ 10 μM and incubated for 24 h. The number of PGCs with EGFP expression was visually assessed. Hits were defined as compounds that reduced PGC numbers without visibly affecting other cell types. Sale bar, 100 μm. (b) Primordazine A and B were identified as inducing a dose-dependent decrease in the number of PGCs. However, 6364997 does not affect PGC number despite its structural similarity to primordazine. (n = 20 animals per condition for 1 and 2, n = 10 animals for 3) (c) Treatment with prim-A results in loss of PGCs labeled by WISH with three PGC markers, ddx4, nanos3, and dnd1 in embryos at 24 hpf. This experiment was repeated two times with similar results. Scale bar, 100 μm. Time course analysis of WISH for the three PGC markers, ddx4 (d), nanos3 (e), and dnd1 (f), shows that the number of PGCs begins to decrease around 9–10 hpf, while immunostaining for DDX4 indicates that PGC number drops from around 15 hpf (g). (d–g, n = 10 animals per condition/timepoint). Data in b and d–g is shown as mean ± s.e.m. (standard error of mean).
RESULTS
Discovery of primordazine, a compound that ablates PGCs
More than 7,000 structurally-diverse compounds were screened using embryos from a transgenic zebrafish line11 expressing EGFP in its PGCs. Two compounds caused disappearance of PGCs in the developing embryo without visibly causing changes in other cell types. Interestingly, the two compounds were structurally similar to each other, so we named them primordazine A (1) and B (2) (prim-A, prim-B). A third compound 3 (6364997) did not show any effect on PGCs despite structural similarity (Fig. 1b). PGC loss was confirmed by in situ hybridization (ISH) with PGC markers ddx412, nanos313, and deadend1 (dnd1)14 and by immunohistochemistry using anti-DDX4 antibody (Fig. 1c–g). We found that PGCs begin to disappear 10–12 hour post fertilization (hpf) (Fig. 1d–g). Although we occasionally observed a complete loss of PGCs by EGFP fluorescence or ISH, typically a few PGCs remained after primordazine treatment. Prim-A robustly decreased PGC numbers at 5–10 μM without noticeable toxicity (Supplementary Fig. 1). At 20 μM or higher concentrations, prim-A caused noticeable toxicity or developmental delay. Prim-B showed somewhat greater potency (Fig. 1b) with reduced toxicity compared to prim-A (Supplementary Fig. 2). In addition, we found that early treatment between 2 to 5 hpf was essential for primordazine’s activity (Supplementary Fig. 3), suggesting that primordazine’s effect on PGCs is mediated by a process occurring before 5 hpf. In addition, we tested whether the short exposure to primordazine during embryogenesis may impact sex determination or fertility in adult fish. Interestingly, we found that primordazine increases, in a dose-dependent way, the percentage of embryos that become male, consistent with previous studies showing that the number of PGCs affects sex determination in zebrafish15–18 (Supplementary Fig. 4). Both male and female fish treated with prim-B remain fertile, suggesting that the few residual PGCs are sufficient to maintain fertility.
To test if PGC loss is due to impairment of specification or migration, we examined PGCs at 3 hpf for specification and at 18 hpf for migration. No reduction in PGC number at 3 hpf and no ectopic PGCs at 18 hpf were observed, indicating that PGC loss does not result from deficits in specification or migration (Supplementary Fig. 5a). Since PGCs develop within mesoderm, we tested whether primordazine causes abnormal mesoderm by ISH for gsc19, ta20, and pax2a21, and found no abnormalities, suggesting that PGC loss after primordazine treatment is not due to defects in mesodermal patterning (Supplementary Fig. 5b).
Another possible mechanism of primordazine’s effect on PGCs could be a change of mRNA levels. To test this possibility, we performed RNA-seq with total RNA from embryos collected at 2 or 6 hpf with or without prim-B treatment. No difference in RNA levels between DMSO and prim-B conditions was observed. qRT-PCR analysis confirmed that RNA levels of dnd1, nanos3, and ddx4 were not significantly changed by prim-B. These results demonstrate that primordazine does not substantially alter RNA levels in embryos (Supplementary Fig. 6a–e). We also tested whether primordazine induces PGC apoptosis and found no positive TUNEL staining within the genital ridge in prim-A or -B treated embryos. Another active primordazine analog 5919059 (4) did show some TUNEL positivity, however, with random distribution, suggesting general toxicity rather than PGC apoptosis. Immunostaining for active caspase 3, a marker for apoptosis, was not observed in prim-B treated embryos at 12 hpf (Supplementary Fig. 7a–c). Furthermore, we found that injection of anti-apoptotic gene bcl2l mRNA or antisense morpholino oligonucleotide (MO) against pro-apoptotic gene tp53 failed to prevent PGC loss (Supplementary Fig. 7d,e). We also found that PGC loss is not caused by exposure to hydrogen peroxide, even at doses causing high embryonic mortality (Supplementary Fig. 8). Together, these results suggest that the PGC loss caused by primordazine is not due to apoptosis or typical mechanisms of toxicity.
Primordazine inhibits translation via PRE in the 3′UTR
To understand how primordazine causes a specific loss of PGCs, we tested whether primordazine inhibits translation of genes critical for PGC development, such as nanos313. We injected EGFP mRNA fused to the nanos3-3′UTR, causing EGFP to be expressed in PGCs. EGFP protein level was dramatically reduced by primordazine treatment (Fig. 2a) with the EGFP mRNA level remaining unchanged (Supplementary Fig. 6f). We therefore hypothesized that a sequence element in the nanos3-3′UTR could render translation of nanos3 sensitive to primordazine. To identify this sequence element, we tested various EGFP reporters. The nanos3-3′UTR contains a target sequence for miR-430 that facilitates nanos3 mRNA decay in somatic cells but not in PGCs22,23. However, we found that this site is not involved in primordazine function. We then tested a series of successive deletions across the 3′UTR and discovered a 40-nucleotide sequence (i.e. 40-2) as a sufficient element for primordazine’s response, which we named the primordazine-response element (PRE) (Fig. 2b; Supplementary Fig. 9). Although PRE-40-2 is sufficient to confer primordazine sensitivity, we utilized a larger 120-nucleotide (PRE-120) for most studies, because of its higher level of expression (Fig. 2c,d).
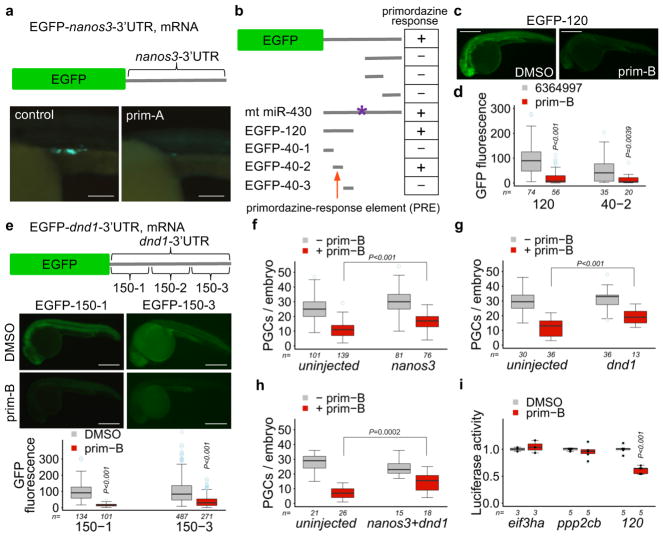
Figure 2. Primordazine inhibits translation via the 3′UTR of nanos3 or dnd1.
(a) The expression of EGFP mRNA fused to the nanos3-3′UTR was substantially reduced by Prim-A treatment. This experiment was repeated three times with similar results. Scale bar = 100 μm. (b) A series of mutations in the nanos3-3′UTR were tested for EGFP expression in response to prim-A. A 40 nucleotide sequence (40-2) was identified as a primordazine-response element (PRE) that renders mRNA susceptible to primordazine. The asterisk indicates a mutation at the miR-430 site. The bar is the region of nanos3-3′UTR fused to EGFP as a 3′UTR. This experiment was repeated two times with similar results. (c) Representative images show fluorescence of EGFP mRNA containing a 120-nucleotide sequence with DMSO or prim-B. This experiment was repeated five times with similar results. Sale bar, 500 μm. (d) Quantitative data show that the expression of EGFP mRNA containing PRE-120 or PRE-40-2 was dramatically decreased by prim-A treatment (n = number of animals). P values were calculated by two-sided Mann-Whitney U-test. (e) Primordazine significantly reduces the expression of EGFP containing 150-1 or 150-3 from dnd1-3′UTR (n = number of animals). Scale bar, 500 μm. P values were calculated by two-sided Mann-Whitney U-test. (f–i) Introduction of (f) nanos3, (g) dnd1, or (h) both slightly but significantly attenuates PGC loss by prim-B treatment (n = number of animals). (i) The 3′UTR of eif3ha or ppp2cb does not confer sensitivity to primordazine (n = independent experiments). Unpaired two-sided t-test (f–i). Box-and-whisker plots in d–i show a median (centerline), upper/lower quartiles (box limits), maximum/minimum (upper/lower whiskers), and light blue dots (outliers). 95% confidence intervals for d–i.
Next, we tested whether primordazine can inhibit EGFP translation via the 3′UTR of another PGC gene, dnd114. Among three EGFP reporters containing three different regions of the dnd1-3′UTR, EGFP-150-1 and -150-3 reporters showed significantly reduced EGFP protein levels upon prim-B treatment (Fig. 2e). While examining different 3′UTR fragments, we noticed that expression of EGFP reporters such as EGFP-120 and -150-1 could be observed throughout the entire body. Primordazine reduced translation of these constructs in somatic cells as effectively as in PGCs (Fig. 2c,e), indicating that the fundamental machinery allowing for primordazine’s action exists throughout the early embryo.
Given the ability of primordazine to repress translation via the 3′UTRs from nanos3 and dnd1, we examined whether exogenous expression of NANOS3 or DND1 could restore primordazine-induced PGC loss. Injection of polyadenylated mRNA of nanos3, dnd1, or both modestly but significantly attenuated primordazine’s effect on PGC loss (Fig. 2f–h), suggesting that PGC loss by primordazine is due in part to reduced translation of nanos3 and dnd1, and that there likely exist additional primordazine targets. Unfortunately, we could not monitor their protein levels by western blot because of the unavailability of antibodies for these proteins in zebrafish.
Next, we sought to determine whether or not primordazine is a general translational inhibitor. To that end, we tested the 3′UTRs of eif3ha or ppp2cb by cloning them into a luciferase reporter. eIF3h regulates the development of the nervous and cardiovascular systems24. Ppp2cb is a catalytic subunit of PP2A important for embryonic development and meiosis/mitosis25. Unlike 3′UTRs of nanos3 and dnd1, primordazine did not affect translation of mRNA bearing 3′UTRs from eif3ha or ppp2cb (Fig. 2i), indicating that primordazine is not a general translation inhibitor but rather acts via a specific subset of 3′UTRs.
Embryos exhibit two distinct forms of translation
Translation can be regulated by a variety of mRNA structural features including the 5′UTR, 3′UTR, 5′-cap, and the poly(A)-tail26–28. To test whether primordazine’s action involves translation initiation factors, we appended three different internal ribosome entry site (IRES) sequences from three viruses, EMCV, HCV, or CrPV, to Renilla luciferase mRNA containing PRE-120 (hereafter RLuc-120). The IRES sequences allow for translation independent of the 5′-cap. EMCV IRES requires all initiation factors (eIFs) except for eIF4E. HCV IRES requires fewer eIFs, whereas CrPV IRES does not depend on any eIFs26. Primordazine significantly reduced translation of all three IRES reporters (Fig. 3a). The independence of primordazine activity from the 5′-cap was further verified by the finding that primordazine blocked translation of RLuc-120 without a 5′-cap (Supplementary Fig. 10a). Additionally, we tested whether RNA helicase activity is important for primordazine’s action by injecting RLuc-120 mRNAs carrying 5′UTRs consisting of vector sequence alone, a structured β-globin stem-loop, or an unstructured (CAA)18 sequence which does not require unwinding for translation29,30. The translation from all three reporters showed a significant reduction upon prim-B treatment (Fig. 3b,c). These results indicate that primordazine inhibits translation independently of the 5′-cap, eIFs, and helicase activity.
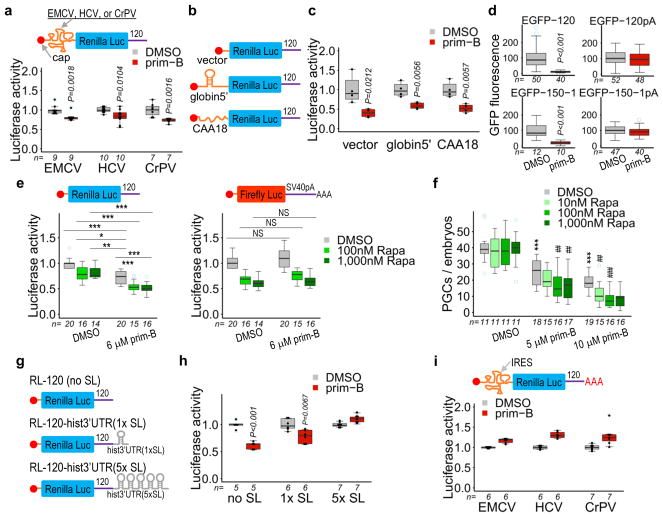
Figure 3. Primordazine inhibits Poly(A)-tail Independent Non-canonical Translation (PAINT) without inhibiting canonical, Poly(A)-tail-mediated Translation (PAT).
(a) The translation of RLuc-120 was significantly reduced by prim-B treatment despite the presence of an IRES in the 5′UTR (n = independent experiments). P value for EMCV was calculated by two-sided Mann-Whitney U-test. (b) Luciferase reporters that contain different 5′UTR structural elements. (c) Primordazine inhibits translation independently of RNA structure in the 5′UTR (n = 4, independent experiments). (d) EGFP-120 or EGFP-150-1 mRNAs without a poly(A)-tail exhibit a significant reduction in translation after prim-B treatment. However, the translation of the same mRNAs with a poly(A)-tail (EGFP-120pA or EGFP-150-1pA) is unaffected by prim-B treatment. n = number of animals. (e) Rapamycin treatment results in a reduction of the expression of RLuc-120, but to a lesser extent than prim-B. Cotreatment with rapamycin and prim-B causes a further reduction in luciferase translation. In comparison, prim-B does not affect translation of FLuc-SV40pA containing a polyadenylated SV40 poly(A) signal in the 3′UTR, while rapamycin substantially reduces it. In addition, cotreatment with rapamycin and prim-B does not show any additional effect on the translation of this reporter mRNA relative to rapamycin alone. (n = independent experiments) (f) Rapamycin synergistically reduces PGC number in the presence of prim-B, while rapamycin alone does not impact PGC maintenance (n = independent experiments). (g) Luciferase reporters with one (1× SL) or five repeats (5× SL) of stem loop from histone-3′UTR. (h) The presence of 5× SL completely abolishes primordazine’s inhibition on translation, while 1× SL exhibits a partial reduction in primordazine’s activity (n = independent experiments). (i) Polyadenylation of IRES RLuc-120 reporters confers resistance to primordazine (n = independent experiments). *,**,*** vs. DMSO; ##,### vs. prim-B; NS, not significant; * P<0.05; **,## P<0.01; ***,### P<0.001. Unpaired two-sided t-test with 95% confidence intervals (c–f, h, and i). Box-and-whisker plots in a, c–f, h, and i show a median (centerline), upper/lower quartiles (box limits), maximum/minimum (upper/lower whiskers), and light blue dots (outliers).
We next tested the influence of polyadenylation on primordazine activity. Remarkably, whereas reporters without polyadenylation were highly sensitive to primordazine-induced translational repression, the polyadenylated reporters were completely resistant to primordazine (Fig. 3d). We then asked if polyadenylation affects primordazine’s activity in a length-dependent manner or requires a certain minimal poly(A) length to obviate primordazine’s effectiveness. Using a series of EGFP-120 reporters containing various lengths of a poly(A)-tail, we found that the poly(A)-tail works in a length-dependent manner, with polyadenylation diminishing the effect of primordazine (Supplementary Fig. 11). Fitting the resulting curve identified 72 as the number of adenosines at which primordazine’s activity in translation inhibition is reduced by 50% (IA50). Together, these results indicate that the mechanism of primordazine’s action is independent of 5′-cap, eIFs, and helicase activity, but is highly dependent on the presence of a 3′ PRE and the absence of polyadenylation.
The dramatic effect of poly(A)-tail length on sensitivity to primordazine caused us to hypothesize that at least two forms of translation occur in the early embryo: canonical, primordazine-insensitive translation in the case of polyadenylated mRNAs, and non-canonical, primordazine-sensitive translation in the case of deadenylated, PRE-containing mRNAs. We therefore tested the interaction of primordazine with rapamycin, a known inhibitor of canonical translation that functions by inhibiting mTOR activity31,32.
At saturating concentrations of rapamycin, when canonical translation initiation was maximally inhibited by rapamycin, addition of prim-B resulted in substantial further reduction in translation of a deadenylated, PRE-containing reporter (Fig. 3e). However, when the luciferase reporter, FLuc-SV40pA, containing a polyadenylated SV40-poly(A) signal (SV40) in the 3′UTR was used, prim-B produced no translational inhibition beyond that achieved by rapamycin. These results provide additional evidence of two forms of translation in the early embryo. Primordazine does not affect the translation that can be inhibited by rapamycin, which is a canonical, poly(A)-tail dependent translation (PAT). However, translation of RLuc-120 involves a mechanism that is distinct from PAT and specifically inhibited by primordazine. We refer to this form of translation as PAINT (Poly(A)-tail Independent Non-canonical Translation).
Following this observation of distinct, additive effects of rapamycin and primordazine on translation inhibition, we postulated that cotreatment with rapamycin and primordazine should enhance primordazine’s effect on PGC loss. As predicted, embryos treated with prim-B and rapamycin possessed fewer PGCs compared to those treated with prim-B alone (Fig. 3f). This effect was not due to general toxicity, since we didn’t observe noticeable toxicity beyond a modest developmental delay (Supplementary Fig. 12). We also observed a similar synergistic effect of torin2, a direct mTOR inhibitor33 on PGC loss (Supplementary Fig. 13). Interestingly, treatment with rapamycin or torin2 alone did not affect PGC development, despite their inhibition of PAT, highlighting the importance of PAINT for PGC maintenance.
Closed-loop formation protects mRNA from primordazine
It is possible that polyadenylation protects mRNAs from primordazine through the actions of poly(A)-binding protein (PABP). Alternatively, polyadenylation may promote canonical translation by allowing formation of a canonical closed-loop structure. To differentiate between these possibilities, we exploited a histone-3′UTR that contains a stem loop that allows for translation by forming a canonical closed-loop without a poly(A)-tail, thus independent of PABP34. We tested RLuc-120 reporters containing one or five stem loops of histone-3′UTR and found that increasing numbers of stem loops confer proportional resistance to primordazine’s activity (Fig. 3g,h), suggesting that polyadenylation may protect transcripts from primordazine-mediated translational repression by enabling canonical closed-loop formation, not by recruiting PABP or via the poly(A)-tail per se. Additionally, we confirmed that the inability of primordazine to inhibit PAT is independent of eIFs and the 5′-cap (Fig. 3i; Supplementary Fig. 10b).
Collectively, these results provide evidence for the existence of an alternative, primordazine-sensitive form of translation during early embryogenesis (Supplementary Fig. 14). This poly(A)-tail independent, non-canonical translation (PAINT) is utilized by a subset of mRNAs with short poly(A)-tails. PAINT may involve recruitment of RNA binding proteins to specific sequence elements (e.g. PRE) in their deadenylated 3′UTRs. In contrast to PAT, PAINT can be inhibited by primordazine. Importantly, poly(A)-tail length is a major determinant of whether a transcript is translated by PAT or by PAINT.
Primordazine doesn’t act by cytoplasmic polyadenylation
How PAINT enables translation in the absence of poly(A)-tails remains unclear. Cytoplasmic polyadenylation (CPA) is known to increases the length of poly(A)-tails of a subgroup of mRNAs during early embryogenesis. One possible explanation for primordazine’s action could be that the PRE prioritizes its transcript for CPA, the resulting polyadenylation enables translation via PAT, and primordazine inhibits this process. To test this possibility, we injected embryos with cordycepin, an analog of adenosine that inhibits CPA. Despite its dramatic effect on embryonic development, PGC numbers were not affected (Supplementary Fig. 15a,b). In addition, we injected embryos with RLuc-120 reporter chemically modified with cordycepin (A0) and found that prim-B was still able to inhibit translation of the A0 reporter (Supplementary Fig. 15c).
Cytoplasmic polyadenylation elements (CPEs) are sequences that can target a transcript for CPA. We found that PRE-120 does contain two potential CPE sites. However, mutation of these CPEs didn’t impact primordazine’s action (Supplementary Fig. 15d,e). Furthermore, using an RNA-ligation mediated poly(A) test and by qRT-PCR using oligo(dT), we found that PRE-120 does not undergo CPA despite containing these two putative CPEs (Supplementary Fig. 15f,g). Endogenous dnd1 and nanos3 mRNAs retain short poly(A)-tail without CPA. By comparison, sox19b mRNA undergoes CPA, resulting in a significant increase in the length of its poly(A)-tail. Interestingly, ddx4 mRNA exists with relatively long poly(A)-tails without further lengthening by CPA. The long poly(A)-tails on ddx4 mRNA likely confers its resistance to primordazine treatment. Together, these results demonstrate that CPA is not a factor for PAINT or for primordazine’s mechanism of action.
Primordazine inhibits translation of a subset of mRNAs
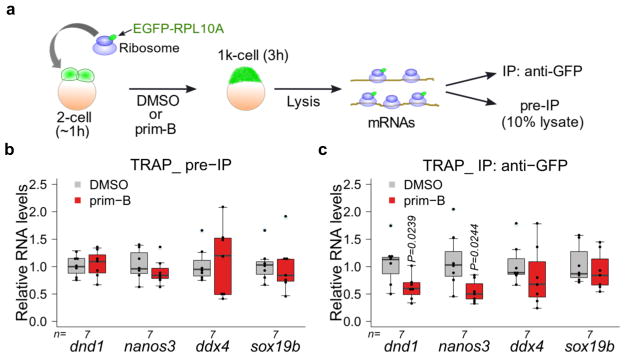
Next, we investigated whether primordazine inhibits translation of endogenous dnd1, nanos3, and ddx4 mRNAs. We utilized TRAP (Translating Ribosome Affinity Purification) to evaluate translation efficiency using a zebrafish TRAP transgenic line expressing egfp-rpl10a35 (Fig. 4a). We found that translation of dnd1 and nanos3 mRNAs was significantly reduced in the presence of prim-B with their mRNA levels unaffected. Meanwhile, translation of ddx4 mRNA was not affected by prim-B treatment (Fig. 4b,c). In addition, translation of sox19b mRNA, which undergoes CPA, was not affected by primordazine. Therefore, translation of the endogenous mRNAs for dnd1, nanos3, ddx4, and sox19b corresponded to translation of the reporters bearing their 3′ UTRs.
Figure 4. Primordazine inhibits translation of a subgroup of endogenous genes.
(a) Schematic of TRAP (Translating Ribosome Affinity Purification). Embryonic expression of EGFP-RPL10A is from maternally-provided mRNA from mothers carrying the transgene egfp-rpl10a under the control of the ef1α promoter. Embryos at the 1–2 cell stage were treated with DMSO or prim-B and collected at the 1k cell stage or 3 hpf. Resulting lysates were divided for pre-IP and IP with anti-GFP. Extracted RNAs were quantified by qRT-PCR. (b) The qRT-PCR shows that mRNA levels for dnd1, nanos3, ddx4, and sox19b in pre-IP samples were not significantly changed in response to prim-B. (c) Prim-B treatment significantly decreased levels of actively translated dnd1 and nanos3 mRNAs. n = independent experiments. In contrast, translation of ddx4 and sox19b mRNAs was not affected by prim-B treatment. Unpaired two-sided t-test with 95% confidence intervals (b and c). Box-and-whisker plots in b and c show a median (centerline), upper/lower quartiles (box limits), maximum/minimum (upper/lower whiskers), and light blue dots (outliers).
To uncover additional endogenous target genes for primordazine, we performed 2D-gel electrophoresis using Difference Gel Electrophoresis (Supplementary Fig. 16a,b). Several spots showed noticeable differences in quantity between DMSO and prim-B conditions. The identities of these spots were difficult to determine unambiguously by mass spectrometry due to contamination of yolk proteins and keratins and/or the presence of multiple proteins. Nevertheless, one spot was unambiguously identified as PGK1. Additionally, TRAP result showed that actively translated pgk1 mRNA is significantly reduced by prim-B treatment (Supplementary Fig. 16c). Consistent with these results, we also found in Subtelny et al.1 that the poly(A)-tail length of pgk1 mRNA remains very short up to 6 hpf (Supplementary Fig. 16d). Together, these results provide evidence that primordazine inhibits translation of a subgroup of endogenous mRNAs in early embryo. Systematic discovery of additional primordazine-sensitive mRNAs will be a focus of future studies.
Primordazine causes the formation of RNA granules
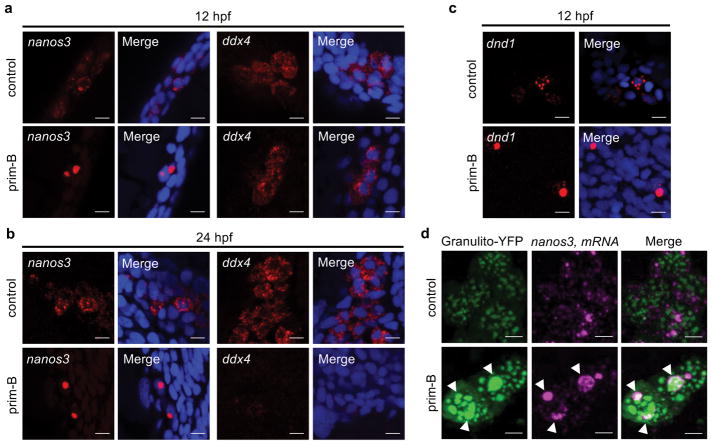
Since translational repression and RNA granule formation are often linked36,37 and abnormal RNA-protein granule formation is associated with many diseases38, we tested whether primordazine affects mRNA distribution by fluorescence ISH (FISH). We found that nanos3 mRNAs are diffusely distributed in untreated embryos but become localized to abnormally large granules upon prim-B treatment (Fig. 5a,b). We also observed similar granule formation for dnd1 mRNAs (Fig. 5c). In contrast, we did not observe ddx4 mRNA granules upon primordazine treatment, suggesting that mRNAs sensitive to primordazine-induced translational repression are also prone to granule formation.
Figure 5. Primordazine induces the formation of abnormal RNA granules.
(a,b) FISH reveals that nanos3 mRNAs form large granules in response to prim-B at 12 hpf (a) and 24 hpf (b), while ddx4 mRNAs did not show abnormal granules at 12 hpf (a) or largely disappeared at 24 hpf (b). Scale bar, 10 μm. (c) Similarly, dnd1 mRNAs form large granules in response to prim-B at 12 hpf. Scale bar, 10 μm. (d) mRNA of YFP fused to granulito, a germ cell granule marker, was injected to label germ cell granules. Prim-B treatment results in abnormally large granules among which some germ cell granules contain large nanos3 mRNA granules at 24 hpf. In contrast, germ cell granules in the absence of prim-B are uniformly distributed with most nanos3 mRNA residing outside. Scale bar, 5 μm. These experiments were repeated two times (a, c, and d) or three times (b) with similar results.
Given that polyadenylation prevents primordazine from inhibiting translation, we tested whether polyadenylation disturbs primordazine-induced RNA granule formation. To this end, we stained EGFP-120 or EGFP-120pA mRNA by FISH. We found that primordazine indeed induced RNA granule formation of EGFP-120 but not EGFP-120pA mRNAs without causing a substantial change in their mRNA levels (Supplementary Fig. 17). These results provide evidence for a link between translational repression and RNA granule formation.
We next attempted to characterize the location of primordazine-induced granules. We labeled germ granules by YFP fused to granulito, a germ granule marker39. Without primordazine, germ granules are relatively consistent in size and most nanos3 mRNAs are localized outside of germ granules. In comparison, prim-B treatment induced abnormally large germ granules, some of which colocalized with nanos3 mRNA granules, suggesting that primordazine-induced translational repression causes aberrant redistribution of germ granules and target mRNAs in PGCs (Fig. 5d).
Primordazine inhibits translation under select condition
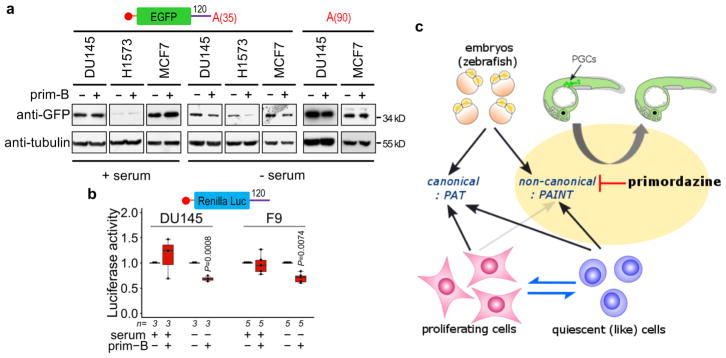
Beyond the early embryo, there are several settings in which transcription or translation is repressed, where alternative forms of translation, such as PAINT, might play a role in gene control. Cells in quiescence40,41 are thought to have transcription, translation, or both repressed, but translation of specific transcripts may be necessary for cell survival or regrowth. Consistent with this idea, we observed that translation of PRE-containing reporters without a poly(A)-tail was inhibited by primordazine in quiescent (like) but not in proliferating mammalian cells, while their mRNA levels remain unaffected (Fig. 6a,b; Supplementary Fig. 18), suggesting that PAINT may occur outside the early embryo in the context of specific cell states (Fig. 6c).
Figure 6. Primordazine inhibits PAINT in quiescent (like) but not in proliferating cells.
(a) EGFP-120 mRNA containing 35 adenosines were transfected in mammalian cancer cell lines. While prim-B treatment does not affect GFP levels in the proliferating cells, serum deprived quiescent (like) cells show a dramatic reduction in the translation of EGFP mRNA in response to prim-B. Note that this EGFP contains a myc tag in the N-terminal. This experiment was repeated two times with similar results. Full gels are shown in Supplementary Fig. 19. (b) The translation of RLuc-120 was measured in two mammalian cell lines in the presence or absence of serum. Primordazine treatment inhibits the translation of RLuc-120 mRNA only in serum free conditions. FLuc-SV40pA mRNA was used for normalization (n = independent experiments). Unpaired two-sided t-test. The box-and-whisker plot shows a median (centerline), upper/lower quartiles (box limits), and maximum/minimum (upper/lower whiskers). (c) Overview of the switch between canonical (PAT) and non-canonical (PAINT) translation in different cell states. While early zebrafish embryos have the ability to utilize both translation mechanisms, proliferating cells express proteins exclusively by canonical PAT. However, when cells switch into a quiescent (like) state, non-canonical (PAINT) becomes activated. Importantly, primordazine only inhibits non-canonical form of translation, PAINT, without inhibiting canonical PAT, leading to PGC loss in zebrafish embryos.
DISCUSSION
The formation of a closed-loop in mRNA is a critical rate-limiting step in canonical translation. Traditionally, a poly(A)-tail has been considered an obligatory structure for closed-loop formation and efficient translation. However, whether mRNAs with short or no poly(A)-tail can be effectively translated in vivo and whether poly(A)-tail independent translation plays an important role in physiology have not been clear. Here we describe a novel compound primordazine that specifically ablates PGCs by inhibiting poly(A)-tail independent non-canonical translation, PAINT, thereby providing in vivo evidence of the existence of PAINT and its physiological role in PGC maintenance. We also discovered that primordazine’s ability to inhibit translation does not depend on the 5′-cap, eIFs, or helicase activity, but rather, specific sequence regions in the deadenlyated 3′UTRs of target mRNAs are important for primordazine’s action. Therefore, we collectively named these sequence elements primordazine-response elements (PREs). Importantly, poly(A)-tail length appears to be a major determinant of whether a transcript is translated by canonical means (i.e. PAT) or by PAINT. Polyadenylation of deadenylated mRNAs that would normally be targets for primordazine-mediated translational inhibition redirects them for PAT, rendering them resistant to primordazine.
It is interesting to speculate why the embryo might employ a non-canonical form of translation such as PAINT. One possibility is that it allows select mRNAs to be translated via an alternative pathway during a period of general translational repression. Most mRNAs become translationally repressed42 during oocyte maturation, mainly due to deadenylation43,44. Canonical translation isn’t restored until mRNAs with long poly(A)-tails are generated by CPA45 or by zygotic transcription after MZT. Thus, genes such as nanos3 and dnd1, which must be translated early in embryogenesis, may not be efficiently translated by PAT due to their short poly(A)-tails, but may be targeted for PAINT through the PREs in their 3′UTRs. We expect that as embryos produce polyadenylated mRNAs after MZT, PAINT will become less active. This coordinated translation control may allow embryo to accomplish precise spatiotemporal development via translation of select mRNAs.
We have identified two genes, nanos3 and dnd1, master regulators of PGC identity, that are translated via PAINT, and isolated elements in their 3′UTRs that enable their non-canonical translation. Although we do not know how widespread the PAINT mechanisms are, we do know that primordazine-mediated translational inhibition can be observed in PGCs and somatic cells of the early embryo, suggesting that the mechanisms exist well beyond the PGC. Nevertheless, the most obvious phenotype resulting from inhibition of PAINT is a loss of PGCs. We speculate that PGCs may contain target genes for primordazine that are essential for PGC maintenance, whereas other cell types may have fewer or less essential genes that are targets for primordazine. In addition, it has been suggested that MZT may occur much later in PGCs than in somatic cells46, which may provide prolonged primordazine susceptibility of target mRNAs in PGCs. In the future, it will be interesting to examine in detail the effects of primordazine on somatic cells.
We found that cells in a quiescent-like state, but not those in proliferation, can utilize PAINT as an alternative form of translation (Fig. 6), perhaps because general repression of canonical translation enables PAINT to be detectable. Similarly, we speculate that cells in other translationally repressive states such as viral infection47 or stress conditions48–50 may utilize PAINT to translate select mRNAs. Future work will focus on discovering other conditions in which PAINT occurs, understanding the molecular machinery of PAINT, identifying additional genes undergoing PAINT, and defining the characteristics of PREs that enable a transcript to undergo PAINT. We believe a better understanding of the mechanisms for PAINT will shed important light on developmental processes and disease settings where cells need to utilize this non-canonical translational mechanism.
METHODS
Fish husbandry
Adult zebrafish (Danio rerio) of the wild type TuAB strain and the transgenic Tg(ddx4:EGFP)11 and Tg(xef1α:EGFP-rpl10a)35 lines were maintained in the fish facility at 28–29°C with a 14/10 hour light/dark cycle and were used for embryo production. 1:1–2 ratio of male and female fish were placed in a mating container and separated by a divider overnight. Embryos were collected after removing a divider in the morning and raised at 28.5°C in E3 solution (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) in the dark and staged following standard methods. All zebrafish experiments were approved by Massachusetts General Hospital and University of Utah Institutional Animal Care and Use Committees.
Animal experiments
Approximately 5–10 Tg(vasa/ddx4:EGFP) embryos, generated by Dr. Olsen’s lab11, at the one-cell stage were placed in 200 μl of E3 solution per well in 96 well plates which were not coated. 7,000 small molecules from the ChemBridge DiverSet library were added to each well at a concentration of ~ 10 μM prior to 1 hpf. The plate was incubated at 28°C in the dark overnight. The number of PGCs in embryos at ~ 24 hpf was visually assessed by EGFP fluorescence using a fluorescence stereomicroscope (Zeiss Discovery V8).
Small molecules
1 (primordazine A), 2 (primordazine B), 3 (6364997), and 4 (5919059) were obtained commercially (Chembridge Corp), and the identities and purities >95% were confirmed by LC-MS.
Cell lines
F9 cells were acquired from ATCC (CRL-1720). DU145, H1573, and MCF7 cells were obtained from MGH cancer center. All of mammalian cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 5% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. F9 cells were cultured on dishes or plates coated with 0.1% gelatin (EMD Millipore). Transient transfections for RNA transfection was performed using Stemfect RNA transfection kit (Stemgent) or Lipofectamine MessengerMax (Invitrogen) according to manufacturer’s protocol. To induce quiescent (like) cell state, cells were cultured in medium without serum about 24 h prior to transfection.
Plasmid construction and in vitro transcription
GFP constructs containing derivatives of the nanos3-3′UTR with deletion or point mutations was kindly provided by Dr. Kunio Inoue (Kobe University)23 and additional derivatives were made at EcoRI and XhoI sites. To make EGFP-120 mRNAs containing the different lengths of poly(A)-tails, the 120 nucleotide upstream of nanos3-3′UTR was inserted into EcoRI and XhoI sites of pTol2EGFPpA vector which was made in our lab by engineering pCMV-Tag3B vector. A poly(A)-tail with 5, 10, 20, 35, 50, 70, or 90 As was added into XhoI and MluI sites. The template plasmids were linearized by MluI and trimmed with Mung bean nuclease prior to in vitro transcription. The longer poly(A)-tail, ~200 As, was made by the poly(A) tailing kit (Ambion). The full length, 150-1, 150-2, and 150-3 3′UTRs of dnd1 were amplified by PCR with cDNA from 8 hpf zebrafish embryos and cloned into HindIII and XhoI sites of pTol2EGFPpA. To make luciferase reporters, 120 of nanos3-3′UTR or 150-1 of dnd1-3′UTR was subcloned into XbaI and NotI sites of pRL-SV40 (Promega) to make Renilla luciferase reporter constructs. For the normalization of input amount of mRNAs, Luc2, a derivative of Firefly luciferase, from pGL4.14 (Promega) was amplified and inserted into BamHI and EcoRI sites of pTol2EGFPpA, named pTol2Luc2pA. MluI digestion was performed to linearize Luc2 plasmid for in vitro transcription. Luc2 mRNAs were polyadenylated after in vitro transcription and coinjected with Rluc-120 or Rluc-150-1 mRNAs at 30 ng/μl. For the expression of dnd1 and nanos3, dnd1 (IMAGE clone ID: 7412116) was subcloned into EcoRI and SalI sites of pTol2mKate2pA which was engineered in our lab. Nanos3 (ATTC 10809379) was subcloned into EcoRI and XhoI sites of pCS2+. The plasmid for bcl2l expression was previously described51. The plasmid of granulito-YFP was a kind gift of Dr. Erez Raz39. To make Renilla reporters for the three IRESs, β-globin 5′UTR, and CAA(18), 120 nucleotides were amplified by PCR and inserted into XbaI and NotI sites of parental plasmids kindly provided by Dr. Martin Bushell (Medical Research Council)29. RL-120-hist3′UTR(1× SL) was made into HindIII and XhoI sites of RL-120 by inserting the PCR product of hist2h3c from a parental plasmid kindly provided by Dr. Jeremy Wilusz52. RL-120-hist3′UTR(5× SL) was generated by inserting 4× SL gBlock (Integrated DNA Technologies) into EcoRI and HindIII sites of RL-120-hist3′UTR(1× SL). To make RNAs for injection, the plasmids were linearized with a specific restriction enzyme that cuts right after a SV40 poly(A) signal, or a derivative of nanos3-3′UTR or dnd1-3′UTR, and in vitro transcribed using the T3, T7, or SP6 mMESSAGE mMACHINE kit (Ambion). mRNAs without a 5′-cap were made using MAXIscript kit (Ambion).
Microinjection of RNAs or morpholinos (MOs) into zebrafish embryos
MO for tp53 was obtained from GeneTools LLC (Corvallis, OR); tp53, gcgccattgctttgcaagaattg. 1 nl of 0.2 mM MO solution was injected into the yolk at the one cell stage embryos. For the expression of genes or GFP reporters, 1 nl of 100–500 ng/μl mRNA was injected into one cell stage embryos. For figure 5d, 1 nl of 80 ng/μl mRNA of EGFP-120 or EGFP-120pA was injected.
Luciferase assay
1 nl of the mixture of 200 ng/μl of a Renilla reporter mRNA and 20 ng/μl of Luc2 mRNA with a poly(A)-tail was injected into one cell stage embryos. 20–30 embryos at 6 hpf were collected into a 1.5 ml microtube. Embryos were washed with 1 ml of phosphate-buffered saline (PBS) and spun down for 30 sec. The supernatant was discarded by aspiration. The lysates were made by adding 3 μl of 1× passive lysis buffer per one embryo followed by homogenization with a plastic pestle. Upon centrifugation for 1 min, 20 μl of solution was transferred to a well of an opaque 96 well plate. Firefly and Renilla luciferase activities were measured using the Dual-Glo Luciferase Assay (Promega) in a VICTOR X3 plate reader (PerkinElmer). The Renilla luciferase activity was normalized by the Firefly luciferase activity.
RNA extraction and qRT-PCR
Approximately 30 embryos were collected at the indicated time point with or without drug treatment. Total RNA was extracted using Trizol (Invitrogen) or RNAzolRT (Molecular Research Center) according to manufacturer’s protocol. 1 μg of total RNA was used to synthesize cDNA using random hexamers and the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen). The real-time PCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems) and run on a 7500 Fast Real-Time PCR System machine (Applied Biosystems) in the fast mode. The relative amount of mRNAs was calculated by using the ΔΔCt (Ct, threshold cycle) method. The Ct value of ornithine decarboxylase 1 (odc1) was used for normalization. Primers are as follows: egfp forward 5′-tcaagatccgccacaacatc-3′, reverse 5′-gtgctcaggtagtggttgtc-3′; nanos3 forward 5′-tctgcagcttctgcaaacacaacg-3′, reverse 5′-gcagaatctcttggtgtgcgcttt-3′; dnd1 forward 5′-tctgcaggaatggatgcagaggaa-3′, reverse 5′-actcgtaaatggtgccgatgctct-3′; ddx4 forward 5′-acactgggagaagaggctttggaa-3′, reverse 5′-tttccactctcatcaccaccacca-3′; sox19b forward 5′-caacttcaccggtacgatctg-3′, reverse 5′-cactgctgctgtaggacatt-3′; pgk1 forward 5′-tggagtccctatgccagacaaatac-3′, reverse 5′-tgcacaggctttctccacatctg-3′; odc1 forward 5′-tgtccaatgaccgaaccctgatgt-3′, reverse 5′-ttgggtttcttgtgcagagttggc-3′ Rluc forward 5′-tggggtgcttgtttggcatttc-3′, reverse 5′-atcaggccattcatcccatgattc-3′; Luc2 forward 5′-cttcgaggaggagctattcttg-3′, reverse 5′-gtcgtacttgtcgatgagagtg-3′.
RNA-ligation mediated poly(A) test (RL-PAT)
2 μg of mRNA was treated with RNaseH (New England Biolabs) in the presence or absence of oligo(dT) and ligated with YJ-P1 primer (5′-ggtcactctatgatcgtacagc-3′) containing a 5′ phosphorylation and a 3′amino modification using T4 RNA Ligase (New England Biolabs). The ligated RNA was reverse transcribed with YJ-P′1(5′-gctgtacgatcatagagtgacc-3′) using Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen) according to manufacturer’s instructions. The PCR was performed using gene specific forward and YJ-P’1 as reverse primer and the Hot Star GoTaq Polymerase (Promega). Gene specific forward primers are as follows: egfp-F 5′-tcaagatccgccacaacatc-3′; dnd1-F 5′-tgagttgttttagtcagcctcatc-3′; nanos3-F 5′-ctacaggcgcaaccgcactc-3′; ddx4-F 5′-agctttgacctaaaggtgtttcc-3′; sox19b-F 5′-ttgctgccacaaagttcgttc-3′.
Translating Ribosome Affinity Purification (TRAP)
Tg(xef1α:EGFP-rpl10a) transgenic line was obtained from Dr. Dougherty’s lab35. Embryos from this transgenic fish line express EGFP-rpl10a that is maternally deposited in the egg. Embryos at the 1–2 cell stage were treated with DMSO or 10 μM prim-B. 100 μg/ml cycloheximide (Acros Organics) was added to E3 solution 5 min prior to sample collection. 250 embryos at 3 hpf that showed bright EGFP fluorescence were pooled for each sample and E3 solution was removed. Embryos were snap frozen using liquid nitrogen. All polysome purification and mRNA extractions were perform as previously described with a slight modification35,53. In brief, embryos were lysed in lysis buffer containing 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.4), 5 mM MgCl2, 150 mM KCl, 1% NP-40, 0.5 mM DTT, 100 μg/ml cycloheximide, 80 U/ml Ribolock (Thermo Scientific), and protease inhibitor cocktail (Pierce). The lysates were cleared by centrifugation at > 13,000 rpm for 15 min. 10% of lysates were saved as pre-IP samples. The polysomes were immunoprecipitated using protein G magnetic beads (Pierce) which were coated with 100 μg of mixed anti-GFP antibodies (clones 19C8 and 19F7). Beads were washed four times with a high potassium buffer containing 20 mM HEPES (pH 7.4), 5 mM MgCl2, 350 mM KCl, 1% NP-40, 0.5 mM DTT, 100 μg/ml cycloheximide, 40 U/ml Ribolock. Bound mRNAs were treated with DNaseI and extracted using Trizol-LS (Invitrogen). RNA concentration was determined using Qubit 3.0 (Invitrogen). 200 ng of pre-IP and 15 ng of IP RNA samples were used to synthesize cDNA. The qRT-PCR was performed as described in RNA extraction and qRT-PCR.
Illumina RNA sequencing
30 embryos were collected at 2 or 6 hpf in the presence of DMSO or 10 μM prim-B. RNA was extracted using RNAzolRT and purified with Direct-zol RNA kit (Zymo Research). The extracted RNA samples were treated with Ribo-Zero Gold rRNA Removal kit (Illumina) to deplete ribosomal RNAs (rRNAs) according to manufacturer’s protocol. Illumina RNA sequencing libraries were constructed using SMARTer Stranded RNA-Seq kit (Clontech) according to manufacturer’s protocol. Samples were multiplexed and sequenced on Illumina HiSeq 2500 machines to produce single-end 50 bp reads. To remove rRNAs reads, raw reads were filtered by SortmeRNA54 using Silva database for Eukarya. The remaining reads were aligned to the zebrafish Zv9/danRer7 genome sequence using TopHat 2.1.0 with the default setting. Differential gene expression analysis was conducted with two biological replicates using SeqMonk Analyser (Babraham Bioinformatics) and the R Bioconductor packages of DESeq255 and edgeR56. All three different analyses failed to observe differentially expressed genes with statistical significance.
Whole mount in situ hybridization (WISH) and fluorescence ISH (FISH)
The following probes were used for WISH: ddx4/vasa12, nanos313, dnd114, gsc19, ta/ntl7, and pax2a21. Full length EGFP was used to make the probe. Anti-sense digoxigenin-labeled probes was obtained using DIG RNA labeling kit (Roche). Embryos at indicated post-fertilization time were dechorinated by 0.5–1 mg/ml of pronase and fixed in 4% paraformaldehyde (PFA) in PBS overnight at 4°C. Embryos were washed with PBS containing 0.1% Tween-20 (PBST). After prehybridization in Hyb+ solution (50% formamide, 5× SSC (1× SSC: 150 mM NaCl, 15 mM sodium citrate, pH 7.0), 0.1% Tween-20, 9 mM citric acid, 0.5 mg/ml yeast torula RNA, 0.1 mg/ml heparin, pH 6) for at least 2 hours at 65°C, embryos were then hybridized with 1–2 ng/μl of RNA probe in Hyb+ solution at 65°C overnight. Embryos were washed twice with Hyb− solution (Hyb+ solution without yeast torula RNA and heparin), twice with 2× SSC containing 0.1% Tween-20 (2× SSCT), twice with 0.2× SSC containing 0.1% Tween-20 (0.2× SSCT) at 65°C and incubated with MABT (0.1 M maleic acid, 150 mM NaCl, 0.1% Tween-20, pH 7.5) with 2 mg/ml BSA and 5% sheep or goat serum) for 2–3 hours at room temperature for blocking. For FISH, PBST containing 2 mg/ml BSA and 5% sheep or goat serum was used for blocking. The embryos were then incubated overnight at 4°C with anti-digoxigenin antibody conjugated with alkaline phosphatase or peroxidase for WISH (Roche, 11 093 274 910) or FISH (Roche, 11 207 733 910), respectively. After several times washing with MABT, the substrates NBT/BCIP (Roche) were incubated with the embryos until the staining was developed to an optimal level. Staining was washed twice with PBST, once with methanol, and three times with PBST. For FISH, embryos were washed once with PBS, incubated for 8 min in TSA plus with cyanine 3 solution (PerkinElmer), and washed five times with PBST.
2D gel electrophoresis (2D-GE)
At least 5,000 embryos for each condition were collected at about 5 hpf or between 40–50% epiboly after treatment of DMSO or 10 μm prim-B from 0.5 hpf. Dechorionating and deyolking embryos was carried out as previously described57. Cell pellets were frozen in liquid nitrogen until use. Cells were lysed in 100 μl of lysis buffer containing 10 mM HEPES (pH 7.4), 150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 5% glycerol, 0.5% Triton X-100, 0.2% SDS, and protease inhibitor cocktail (Pierce). After sonication, insoluble particles were removed by centrifugation at 13,000 rpm for 10 min. Protein concentration was determined by BCA assay (Pierce). 2D-GE was performed as shown in previous references57,58. Briefly, 50 μg of protein from each condition was processed with 2D Clean-up Kit (Bio-Rad), labeled by Cy3 or Cy5 dye (Lumiprobe) for DIGE (Difference Gel Electrophoresis), and merged after quenching with 10 mM lysine. The merged sample was subjected to the first dimension isoelectric focusing using 11 cm pH 3–10 non-linear IPG strip (Bio-Rad) in Protean IEF Cell power supply (Bio-Rad), followed by the second dimension SDS-PAGE with 10% polyacrylamide gels. DIGE gel images were obtained with a Typhoon Trio Laser scanner (GE Healthcare). To minimize experimental variation during DIGE, color swapping was carried out by switching Cy dyes between samples. Protein spots that showed noticeable difference in fluorescence intensity between the two Cy dyes were chosen for mass spectrometric analysis. For the preparative gels, 300 μg of protein sample was loaded onto 17 cm pH 3–10 non-linear IPG strip (Bio-Rad) and stained using an enhanced colloidal Coomassie blue. Spots of interest were excised and subjected to in-gel tryptic digestion for mass spectrometry.
Western blot, immunocytochemistry, and TUNEL
Cells transfected with EGFP reporter mRNAs were harvested one day after transfection. Cell lysates were washed with ice-cold PBS and lysed with RNA binding buffer which contains 10 mM HEPES pH7.4, 150 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 5% glycerol, and cocktails of protease inhibitor and phosphatase inhibitor (Roche). Proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 5% skim milk in Tris-buffered saline containing 0.05% Tween-20 (TBST), and incubated with the specific antibodies (1:5,000 dilution of anti-GFP (Torrey Pines Biolabs, TP401); 1:10,000 dilution of anti-Tubulin (EMD Millipore, 05-829)) in TBST containing 2% BSA at 4°C overnight. After washing three to four times, HRP-conjugated secondary antibody (1:5,000) in TBST containing 5% skim milk was applied and washed after 1 h. The blots were visualized by chemiluminescence using ECL Prime (GE Healthcare). Immunocytochemistry for activated caspase-3 was performed as previously described59. Briefly, embryos at 12 hpf were dechorionated with pronase, fixed with 4% PFA, permeabilized with ice-cold methanol followed by 1× PDT (1× PBST, 0.3% Triton-X, 1% DMSO), blocked in 1× PBST containing 2 mg/ml BSA and 10% FBS, and incubated with anti-activated caspase-3 (BD Biosciences, No. 559565) at 1:500 at 4°C overnight. After washing with 1× PDT embryos were labeled with Alexa 555 anti-rabbit. TUNEL was performed as previously described60.
Imaging and data analysis
The z-stack images of embryos for FISH or immunostaining were acquired at 40× objective on a Zeiss Axio Observer Z1 microscope using the LSM700 scanning system. Nuclei were stained with DAPI (4′, 6-diamidino-2′-phenylindol, dichloride, Thermo Scientific). All confocal images were processed into maximum intensity projections of z-stacks, unless otherwise stated. Confocal data were processed using Zen software (Zeiss) and FIJI/ImageJ (http://rsbweb.nih.gov/ij/). To obtain the images of WISH or of the wide field fluorescence for the whole embryos, Discovery V8 stereomicroscope (Zeiss) was used with a 6.3× objective. PGC numbers were counted by GFP fluorescence from dechorionated Tg(ddx4:EGFP) embryos which were gently flattened by a coverslip to spread out PGCs.
Statistics and reproducibility
All results are expressed as mean ± s.e.m. (standard error of mean), unless otherwise stated. Box-and-whisker plots show a median (centerline), upper/lower quartiles (box limits) and maximum/minimum (upper/lower whiskers). Statistical significance was determined using unpaired two-tailed t-test, unless otherwise stated. P < 0.05 was considered as statistically significant.
Data availability
The data that support the findings of this study including RNA-seq and proteomics analysis are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We thank S. Vasudevan, T. Shioda, S. Lee, N. Dyson, W. Miles, and Y. Yu for discussions related to this manuscript. Thanks to U. Kim (next generation sequencing core at Massachusetts General Hospital) for help with the RNA-seq process, to A. Gonzales and D. Harrison for technical assistance, and to C. Reilly for chemical validation. Funding was provided by NIH grants 5T32HL007208-37 (Y.N.J.), R01GM088040 (R.T.P.), USDA grant 2014-07998 (R.T.P.), the Charles and Ann Sanders MGH Scholar Award (R.T.P.), and the L. S. Skaggs Presidential Endowed Chair (R.T.P.).
Footnotes
AUTHOR CONTRIBUTIONS
Y.N.J., P.J.S., N.J-Y., and R.T.P. designed the study and experiments. Y.N.J., P.J.S., N.J-Y., and P.Y.L. performed the experiments. Y.N.J. and P.J.S. performed data analysis. P.Y.L. and Y.N.J. performed RNA-seq. Y.N.J. performed bioinformatics. S.J. synthesized chemicals. W.Y.H. and J.R.Y. generated knockout fish. M.Y., S.E.O., M.S., C.R.H., and S.A.C. contributed to 2D gel electrophoresis and proteomic analysis. Y.N.J. and R.T.P. wrote the paper. All authors commented on the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
P.J.S. and R.T.P. have applied for a patent for primordazine.
References
- 1.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strome S, Updike D. Specifying and protecting germ cell fate. Nat Rev Mol Cell Biol. 2015;16:406–416. doi: 10.1038/nrm4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai H, et al. In vitro and in vivo differentiation of induced pluripotent stem cells into male germ cells. Biochem Biophys Res Commun. 2013;433:286–291. doi: 10.1016/j.bbrc.2013.02.107. [DOI] [PubMed] [Google Scholar]
- 4.Geijsen N, et al. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature. 2004;427:148–154. doi: 10.1038/nature02247. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the Mouse Germ Cell Specification Pathway in Culture by Pluripotent Stem Cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi K, et al. Offspring from Oocytes Derived from in Vitro Primordial Germ Cell–like Cells in Mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- 7.Park TS, et al. Derivation of Primordial Germ Cells from Human Embryonic and Induced Pluripotent Stem Cells Is Significantly Improved by Coculture with Human Fetal Gonadal Cells. STEM CELLS. 2009;27:783–795. doi: 10.1002/stem.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raz E. Primordial germ-cell development: the zebrafish perspective. Nat Rev Genet. 2003;4:690. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- 9.Paksa A, Raz E. Zebrafish germ cells: motility and guided migration. Curr Opin Cell Biol. 2015;36:80–85. doi: 10.1016/j.ceb.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Winata CL, et al. Cytoplasmic polyadenylation-mediated translational control of maternal mRNAs directs maternal-to-zygotic transition. Development. 2018;145:dev159566. doi: 10.1242/dev.159566. [DOI] [PubMed] [Google Scholar]
- 11.Krøvel AV, Olsen LC. Expression of a vas::EGFP transgene in primordial germ cells of the zebrafish. Mech Dev. 2002;116:141–150. doi: 10.1016/s0925-4773(02)00154-5. [DOI] [PubMed] [Google Scholar]
- 12.Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2-and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 13.Köprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidinger G, et al. dead end, a Novel Vertebrate Germ Plasm Component, Is Required for Zebrafish Primordial Germ Cell Migration and Survival. Curr Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- 15.Dai X, Jin X, Chen X, He J, Yin Z. Sufficient Numbers of Early Germ Cells Are Essential for Female Sex Development in Zebrafish. PLoS ONE. 2015;10:e0117824. doi: 10.1371/journal.pone.0117824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzung KW, et al. Early Depletion of Primordial Germ Cells in Zebrafish Promotes Testis Formation. Stem Cell Rep. 2015;4:61–73. doi: 10.1016/j.stemcr.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegfried KR, Nüsslein-Volhard C. Germ line control of female sex determination in zebrafish. Dev Biol. 2008;324:277–287. doi: 10.1016/j.ydbio.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 18.Slanchev K, Stebler J, Cueva-Méndez G, de la Raz E. Development without germ cells: The role of the germ line in zebrafish sex differentiation. Proc Natl Acad Sci U S A. 2005;102:4074–4079. doi: 10.1073/pnas.0407475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulte-Merker S, et al. Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development. 1994;120:843–852. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- 20.Schulte-Merker S, van Eeden FJ, Halpern ME, Kimmel CB, Nusslein-Volhard C. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 1994;120:1009–1015. doi: 10.1242/dev.120.4.1009. [DOI] [PubMed] [Google Scholar]
- 21.Majumdar A, Lun K, Brand M, Drummond IA. Zebrafish no isthmus reveals a role for pax2.1 in tubule differentiation and patterning events in the pronephric primordia. Development. 2000;127:2089–2098. doi: 10.1242/dev.127.10.2089. [DOI] [PubMed] [Google Scholar]
- 22.Giraldez AJ, et al. Zebrafish MiR-430 Promotes Deadenylation and Clearance of Maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 23.Mishima Y, et al. Differential Regulation of Germline mRNAs in Soma and Germ Cells by Zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhuri A, Maitra U, Evans T. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc Natl Acad Sci. 2013;110:9818–9823. doi: 10.1073/pnas.1302934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tischer T, Hörmanseder E, Mayer TU. The APC/C Inhibitor XErp1/Emi2 Is Essential for Xenopus Early Embryonic Divisions. Science. 2012;338:520–524. doi: 10.1126/science.1228394. [DOI] [PubMed] [Google Scholar]
- 26.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 28.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meijer HA, et al. Translational Repression and eIF4A2 Activity Are Critical for MicroRNA-Mediated Gene Regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 30.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 32.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 33.Liu Q, et al. Discovery of 9-(6-Aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a Potent, Selective, and Orally Available Mammalian Target of Rapamycin (mTOR) Inhibitor for Treatment of Cancer. J Med Chem. 2011;54:1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tryon RC, Pisat N, Johnson SL, Dougherty JD. Development of translating ribosome affinity purification for zebrafish. genesis. 2013;51:187–192. doi: 10.1002/dvg.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramaswami M, Taylor JP, Parker R. Altered Ribostasis: RNA-Protein Granules in Degenerative Disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strasser MJ, et al. Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Dev Biol. 2008;8:58. doi: 10.1186/1471-213X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valcourt JR, et al. Staying alive. Cell Cycle. 2012;11:1680–1696. doi: 10.4161/cc.19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKnight JN, Boerma JW, Breeden LL, Tsukiyama T. Global Promoter Targeting of a Conserved Lysine Deacetylase for Transcriptional Shutoff during Quiescence Entry. Mol Cell. 2015;59:732–743. doi: 10.1016/j.molcel.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, et al. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Belloc E, Méndez R. A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature. 2008;452:1017–1021. doi: 10.1038/nature06809. [DOI] [PubMed] [Google Scholar]
- 44.Pasternak M, Pfender S, Santhanam B, Schuh M. The BTG4 and CAF1 complex prevents the spontaneous activation of eggs by deadenylating maternal mRNAs. Open Biol. 2016;6:160184. doi: 10.1098/rsob.160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivshina M, Lasko P, Richter JD. Cytoplasmic Polyadenylation Element Binding Proteins in Development, Health, and Disease. Annu Rev Cell Dev Biol. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. [DOI] [PubMed] [Google Scholar]
- 46.Siddiqui NU, et al. Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012;13:R11. doi: 10.1186/gb-2012-13-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu H, et al. N-Terminal Domain of Feline Calicivirus (FCV) Proteinase-Polymerase Contributes to the Inhibition of Host Cell Transcription. Viruses. 2016;8:199. doi: 10.3390/v8070199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreev DE, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baird TD, et al. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol Biol Cell. 2014;25:1686–1697. doi: 10.1091/mbc.E14-02-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richter JD, Coller J. Pausing on Polyribosomes: Make Way for Elongation in Translational Control. Cell. 2015;163:292–300. doi: 10.1016/j.cell.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlueter PJ, Sang X, Duan C, Wood AW. Insulin-like growth factor receptor 1b is required for zebrafish primordial germ cell migration and survival. Dev Biol. 2007;305:377–387. doi: 10.1016/j.ydbio.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kramer MC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015;29:2168–2182. doi: 10.1101/gad.270421.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heiman M, et al. A Translational Profiling Approach for the Molecular Characterization of CNS Cell Types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Link V, Shevchenko A, Heisenberg CP. Proteomics of early zebrafish embryos. BMC Dev Biol. 2006;6:1. doi: 10.1186/1471-213X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshigi M, Pronovost SM, Kadrmas JL. Interactions by 2D Gel Electrophoresis Overlap (iGEO): a novel high fidelityapproach to identify constituents of protein complexes. Proteome Sci. 2013;11:21. doi: 10.1186/1477-5956-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorrells S, Toruno C, Stewart RA, Jette C. Analysis of Apoptosis in Zebrafish Embryos by Whole-mount Immunofluorescence to Detect Activated Caspase 3. J Vis Exp JoVE. 2013 doi: 10.3791/51060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Ham TJ, Mapes J, Kokel D, Peterson RT. Live imaging of apoptotic cells in zebrafish. FASEB J. 2010;24:4336–4342. doi: 10.1096/fj.10-161018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study including RNA-seq and proteomics analysis are available from the corresponding authors upon reasonable request.