Abstract
Light-sheet microscopy is an imaging approach that offers unique advantages for a diverse range of neuroscience applications. Unlike point-scanning techniques such as confocal and two-photon microscopy, light-sheet microscopes illuminate an entire plane of tissue, while imaging this plane onto a camera. Although early implementations of light sheet were optimized for longitudinal imaging of embryonic development in small specimens, emerging implementations are capable of capturing light-sheet images in freely moving, unconstrained specimens and even the intact in vivo mammalian brain. Meanwhile, the unique photobleaching and signal-to-noise benefits afforded by light-sheet microscopy’s parallelized detection deliver the ability to perform volumetric imaging at much higher speeds than can be achieved using point scanning. This review describes the basic principles and evolution of light-sheet microscopy, followed by perspectives on emerging applications and opportunities for both imaging large, cleared, and expanded neural tissues and high-speed, functional imaging in vivo.
Keywords: light sheet, microscopy, GCaMP, tissue clearing, functional imaging
1. INTRODUCTION
The past decade has seen the rapid growth of diverse approaches for the fluorescent labeling of in vivo cells. In particular, genetically encoded fluorescent proteins can now be expressed in specific cell types, enabling detailed structural mapping and connectivity tracing (Chalfie et al. 1994). A growing palette of fluorescent proteins has also been engineered to change fluorescence in response to calcium and a host of other cellular signaling molecules, enabling unprecedented optical readouts of cellular function (Akerboom et al. 2012, Chen et al. 2013, Dana et al. 2016, Gong et al. 2015, Heim et al. 2007, Kralj et al. 2012, Patriarchi et al. 2018). Combined with optogenetics, which permits the optical manipulation of cellular activity, and the evolution of a wide range of model organisms, from worms and flies to mice and marmosets, we now have the potential to optically interrogate complete brain circuits and nervous systems in real time (Boyden et al. 2005, Packer et al. 2015).
The parallel development and maturation of multiphoton microscopy fueled this optical revolution, providing the ability to visualize the activity of single cells up to around 1 mm deep into the scattering mammalian brain (Denk et al. 1990, Horton et al. 2013, Kobat et al. 2009). Diverse innovations have increased imaging speeds and fields of view and have enabled combined imaging and photomanipulation in awake, behaving animals performing complex tasks (Grewe et al. 2010, Nadella et al. 2016, Packer et al. 2015, Sofroniew et al. 2016, Weisenburger & Vaziri 2018). However, the desire to see even more—to extend views from two-dimensional (2D) planes of neurons to large 3D volumes spanning the entire brain—has prompted the exploration of alternative microscopy approaches capable of even higher throughput cellular-level imaging in the living brain.
Light-sheet microscopy is an approach that was first proposed over 100 years ago (Siedentopf & Zsigmondy 1902). However, modern cameras, lasers, small model organisms such as worms, fish, and flies, and modern tissue-clearing techniques have unlocked the potential of this approach for diverse neuroscience applications (Keller & Ahrens 2015). Here we explain the basic principles of light-sheet microscopy and its unique benefits over point-scanning methods. The evolution of modern light-sheet configurations is then reviewed, with a focus on approaches being refined for neuroscience applications. We then discuss the promise of light-sheet approaches for imaging large, cleared, and expanded samples and emerging single-objective light-sheet methods that are enabling high-speed, volumetric functional imaging in challenging samples such as the in vivo mammalian brain (Bouchard et al. 2015, Hillman et al. 2018).
2. THE ADVANTAGES OF LIGHT-SHEET MICROSCOPY
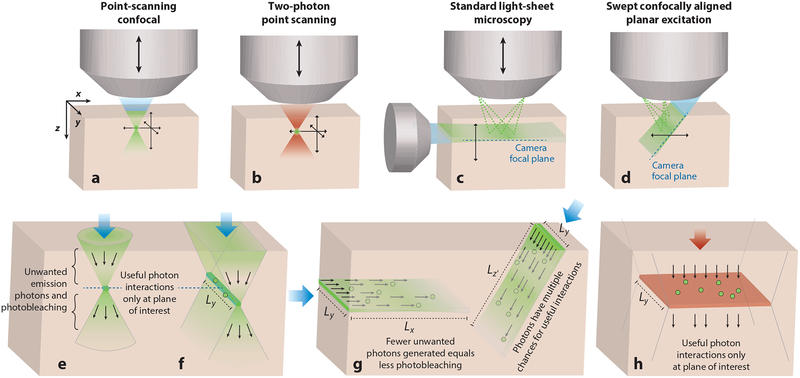
Early microscopes achieved cellular imaging by thinly slicing tissues. To obtain cellular resolution imaging in intact or in vivo tissues, microscopy methods must achieve optical sectioning of the tissue, whereby an image is formed of a single layer or subregion despite the presence of surrounding tissue. The most common method for in vivo optical sectioning is laser point scanning, as used in conventional confocal and two-photon microscopy (Denk et al. 1990, White et al. 1987). In laser point scanning, a small point of focused light is moved sequentially to different 3D positions, while an image is formed by assigning the signal detected at each point to its position in 3D space (Figure 1a,b).
Figure 1.
Microscopy approaches for optical sectioning: (a) point-scanning confocal, (b) two-photon point scanning, (c) conventional light-sheet microscopy, and (d) oblique light-sheet (e.g. swept confocally aligned planar excitation) microscopy. In all panels, blue (or red) shading indicates excitation light, and green indicates fluorescence emission. Arrows indicate the direction of scanning to form a 3D image. (e) Depiction of the useful versus unused light generated in point scanning. (f) A confocal line-scanning approach to lateral parallelization of illumination. (g) Conventional and oblique light-sheet geometries that more efficiently form a sheet with photons have multiple chances to generate useful interactions with fluorophores along their path, while generating less extraneous fluorescence and thus photobleaching. (h) Depiction of wide-field, temporal-focusing, two-photon microscopy.
Light-sheet microscopy takes a different approach to optical sectioning. In a light-sheet microscope, the plane of interest is illuminated with a thin sheet of light, while a camera is carefully focused onto this plane (Figure 1c,d). A 3D volumetric image can be formed by moving this coaligned excitation and detection plane relative to the sample. Many consider light-sheet microscopy as a method that is primarily suitable for imaging nonscattering samples. However, light-sheet microscopy delivers significant benefits over point scanning in terms of simplicity, light efficiency, low phototoxicity, and the potential for high volumetric imaging speeds, while the resolution benefits of its theta geometry extend its utility to imaging scattering tissues (Engelbrecht & Stelzer 2006, Stelzer & Lindek 1994). The physical basis of these advantages is detailed below.
2.1. The Speed Limitations of Point Scanning
Despite its major contributions to biomedical research, point-scanning microscopy is reaching fundamental speed limits for applications requiring fast volumetric imaging. To attain higher volumetric imaging speeds, the point of light needs to be scanned faster and faster. As point scanning rates increase, the integration time available to collect signal per pixel becomes shorter, requiring highly sensitive detectors and brighter laser illumination to collect sufficient signal to form an image. To image a volume of 512 × 512 × 100 (Lx × Ly × Lz) voxels at 10 volumes per second (VPS), the integration time per voxel would reach tp = 4 ns, the fluorescence lifetime of green fluorescent protein (Pepperkok et al. 1999). The corresponding pixel sample rate of 262 MHz is also over three times faster than the pulse rate of 80-MHz Ti:sapphire lasers commonly used for two-photon microscopy. This configuration would also require a scanner moving at a more than 500-kHz line rate, yet current resonant scanners are limited to around 24-kHz line rates (Table 1, case 1).
Table 1.
Comparison of scanning, integration time, and total power between different point, multipoint, and light-sheet configurations
| Case | Configuration | x pixels | y pixels | z pixels | Pixel rate (MHz) | Frame rate (Hz) | Volume rate (Hz) | Integration time/pix (ns) | x scan speed (Hz) | z scan speed (Hz) | Power per pixel (W) | Estimated total power (W) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Point-scanning confocal | 512 | 512 | 100 | 262 | 1,000 | 10 | 4 | 512,000 | 10 | Pp | Pp |
| 2 | Line-scan confocal | 512 | 512 | 100 | 262 | 1,000 | 10 | 1,953 | 1,000 | 10 | Pp/512 | Pp |
| 3 | Fast light sheet (xy) | 512 | 512 | 100 | 262 | 1,000 | 10 | 1,000,000 | 0 | 10 | Pp/5122 | ~Pp/512 |
| 4 | Standard resonant confocal | 512 | 512 | 100 | 8 | 30 | 0.3 | 127 | 15,360 | 0.3 | Pp/33 | Pp/33 |
Properties are compared to the parameters of a modern 30 frames per second resonant scanning confocal microscope. Abbreviation: Pp, instantaneous power per pixel.
2.2. Light Sheet: Increased Integration Time and Better Light Efficiency
A logical extension is to increase imaging speed by parallelizing the detection of signal from multiple regions of tissue at the same time. These implementations require more complex hardware configurations to maintain optical sectioning but include multispot scanning confocal and two-photon microscopy (Bewersdorf et al. 1998, Gräf et al. 2005, Watson et al. 2009), line-scanning confocal microscopy (Castellano-Munoz et al. 2012, Rajadhyaksha et al. 1999), and spinning-disk confocal microscopy (Gräf et al. 2005).
Considering the example above, if we now illuminate all spots along a lateral line corresponding to Ly = 512 pixels in parallel (Figure 1f), for the same volume rate, the system’s Lx scanner now only needs to move at 1,000 lines per second, while the integration time per voxel will be tL = Lytp = 512 × tp = 2 μs. Furthermore, if the point-scanning case uses power Pp, the equivalent energy per pixel (power × time) can be delivered in the line-scanning case by simply distributing Pp across the length of the line (yielding PL = Pp/512 illumination per pixel), thanks to the 512-times longer integration time (Table 1, case 2). Nevertheless, a 2-μs integration time is equivalent to sampling at 500 kHz per channel on a 512-element array detector, which is beyond the reach of current technologies.
Now consider a light sheet that can illuminate an entire x-y plane of the sample (while still achieving optical sectioning). For the same example as above, an Lx × Ly = 512 × 512 plane would only require a scanner in the Lz direction, moving at only 10 lines per second, while the integration time per pixel would now be tLS = LxLytp = 512 × 512 × tp = 1 ms. This readout rate (1 kHz frame rate for 512 × 512 pixels) is feasible with the latest intensified, high-speed cameras. Although cameras are not considered to be as sensitive and low noise as the modern detectors used in point-scanning microscopy, integration time is a very important factor. Point-scanning detectors must have low noise for very short per-pixel integration times (on the order of megahertz or more rates). However, the longer (kilohertz rate) integration times needed for light-sheet microscopes are an excellent match for the noise-equivalent powers of modern scientific CMOS cameras. Readout rates of newer intensified cameras are now exceeding 1.3-GHz pixel rates, far above what is currently possible with point scanning.
In terms of illumination power, one might expect that Lx × Ly more power is needed to illuminate a plane of area A = Lx × Ly (Choi et al. 2012). However, in the case of a low numerical aperture (NA) light sheet, photons propagate along the direction of the sheet (Figure 1g). As fluorescence cross sections are typically low, each photon is afforded repeated opportunities for fluorophore excitation along its path (Hillman et al. 2018). A light sheet can thus provide near-equivalent irradiance at any voxel in the illuminated plane by only extending illumination over the Ly direction. This effect means that, accounting for the much longer integration time available, a factor closer to Pp/Lx total power is required to form a light sheet that will deposit equivalent energy per pixel to the two cases above (Hillman et al. 2018) (Table 1, case 3). This lower total power requirement reduces reliance on higher power lasers, while also reducing the overall light dose to tissue, detailed further below.
2.3. Extending Analysis to Two-Photon Excitation
The benefits of multispot scanning do not directly extrapolate to two-photon microscopy, because spreading laser power P across Ly points will reduce the two-photon signal produced by 1/(Ly)2 (Denk et al. 1990). Total laser power cannot simply be increased, as the laser power available for two-photon microscopy is limited by available pulsed laser sources as well as thermal effects in living tissues: Average powers exceeding 200 mW at the sample have been shown to cause brain heating, while pulse energies of 2 nJ are considered to be the upper limit for fluorophore saturation (Picot et al. 2018, Podgorski & Ranganathan 2016). In addition, lateral parallelization of illumination in two-photon microscopy typically degrades axial sectioning and penetration depth, requiring complex additions such as temporal focusing to restrict excitation to a narrower axial range (Dana & Shoham 2012, Vaziri & Shank 2010) (Figure 1h).
Although these limitations make high-speed two-photon light-sheet microscopy seem improbable, light-sheet microscopy’s more efficient use of illumination power, combined with longer integration times that permit the use of lower repetition rate and thus higher peak-power lasers, supports its feasibility within thermal damage and saturation limits (for further modeling and analysis of these effects, see Hillman et al. 2018, Schrodel et al. 2013, Truong et al. 2011).
2.4. Selective Plane Illumination Significantly Decreases Phototoxicity
All of the comparisons above assume the same energy deposited per voxel, but the instantaneous power levels per voxel vary significantly between each case. Recent results have suggested that for fluorescent proteins, photobleaching half-lives shorten with higher incident power (Cranfill et al. 2016). This factor implies that collecting fluorescence over long integration times with lower-power illumination (as in light-sheet microscopy) could result in significantly less photobleaching than depositing an equivalent amount of energy (power × time) with a shorter integration time and higher incident power (as with fast point scanning). This effect may in part explain the lower degree of photobleaching that is observed with spinning-disk confocal compared to single-point scanning (Hillman et al. 2018).
In addition to the advantages above, another contributor to the low phototoxicity of light-sheet microscopy is its selective illumination of only the plane being imaged at any given point in time. This key feature of light sheets is shown in Figure 1g, as compared to high-NA point and line scanning in Figure 1e,f. High-NA point and line excitation concentrates photons at a particular depth, and yet all of the light reaching this depth has already traveled through tissues above the plane of interest, and most of it continues into the tissue beyond that plane. This extraneous light significantly contributes to photodamage of the parts of the sample that are not being imaged during volumetric imaging: Each additional depth plane N acquired in point or line scanning exposes all the other planes of the volume to the near-equivalent dose of light, as if they had been imaged N times each. In light-sheet microscopy, photons are mostly constrained to traveling in the plane that is being imaged such that imaging each additional plane of a volume does not reexpose planes in the rest of the volume. This property dramatically reduces the overall exposure of the tissue volume compared to imaging different depth planes using confocal techniques. Readers are referred to Hillman et al. (2018) for detailed modeling of this effect.
Combining all these factors together, light-sheet illumination provides very significant benefits for in vivo imaging, providing the possibility of high-speed imaging of volumes with good signal-to-noise ratios, as well as significantly reduced photobleaching compared to point-scanning methods.
3. THE EVOLUTION OF MODERN LIGHT-SHEET MICROSCOPY
3.1. Early Light-Sheet Configurations and Applications
Although several early studies recognized the benefits of using light sheets for microscopy (Fuchs et al. 2002, Voie & Spelman 1995), Huisken et al.’s (2004) paper revealed the exciting potential of light-sheet microscopy for imaging small, living organisms such as Drosophila and zebrafish embryos. The selective plane illumination microscopy (SPIM) system presented by Huisken et al. was composed of a detection objective lens and a second lens positioned at 90° to the first to create a sheet of light aligned with the detection objective’s focal plane. The sample was embedded in a small agarose cylinder and held between the two orthogonal optical elements. Volumetric imaging was achieved by physically translating the agarose cylinder through the light-sheet plane, and the sample was rotated to obtain views from different illumination directions. Although it was not optimized for fast imaging speeds, the system’s low phototoxicity permitted longitudinal imaging of the sample over many hours of embryonic development.
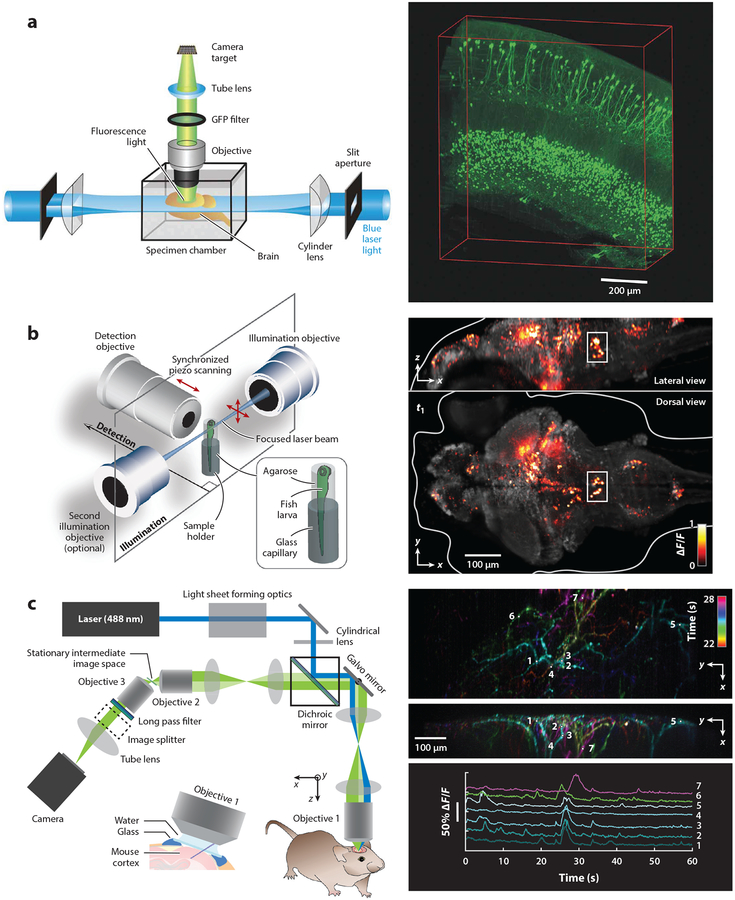
Dodt et al.’s (2007) paper demonstrated another key application for light-sheet microscopy by imaging large, cleared (ex vivo) samples. Termed ultramicroscopy, this approach employed a similar orthogonal light sheet and detection approach but was optimized to image entire cleared mouse brains by using much larger light sheets and imaging chambers, as shown in Figure 2a.
Figure 2.
Examples of light-sheet approaches for neuroscience applications. (a) Demonstration of ultramicroscopy for light-sheet imaging of cleared samples (left), including green fluorescent protein (GFP)-expressing neurons in the mouse brain (right). Panel a adapted with permission from Dodt et al. (2007). (b) Whole-brain imaging of calcium activity (GCaMP) in a zebrafish larva at around 1 volume per second (right). (Left) The system used scanned light sheets incident from both sides of the sample, with the detection lens moved by a piezo actuator, to maintain focus on the sheet. The fish was positioned in an agarose tube between the three objective lenses. Panel b adapted with permission from Ahrens et al. (2013). (c) Swept confocally aligned planar excitation (SCAPE) imaging of GCaMP activity in apical dendrites of layer 5 neurons in awake, behaving mouse cortex at 10 volumes per second. (Right, top and middle) Time-color-encoded firing events over a 6-s period shown as maximum-intensity projections over z and x. (Right, bottom) Raw time courses of fluorescence change (ΔF/F) extracted from the locations indicated in the right, top and middle panels, demonstrating minimal photobleaching and a high signal-to-noise ratio. Panel c adapted with permission from Hillman et al. (2018).
3.2. Improvements to Sample Geometries and Spatial Resolution
Extensions of these first approaches included adding additional optical paths for illumination and the detection of fluorescence from different directions to reduce the need for sample rotation, although multiobjective approaches often made sample positioning even more challenging (Keller et al. 2008, 2010; Tomer et al. 2012). Other configurations were proposed to circumvent this constraint such as inverted SPIM, which mounted the orthogonal excitation and detection arms at 45° to horizontal, permitting imaging of intact samples from above or below (Wu et al. 2011).
A wide range of additional innovations extended the imaging resolution and 3D field of view for diverse applications of light-sheet microscopy. These improvements included varying the in-plane propagation direction of the light sheet to reduce striping (Huisken & Stainier 2007); beam shaping and adaptive optics (Dalgarno et al. 2012, Lindek et al. 1996, Liu et al. 2018, Royer et al. 2016, Wilding et al. 2016); and the use of Bessel beams (Fahrbach & Rohrbach 2010, Planchon et al. 2011), Airy beams (Vettenburg et al. 2014), lattice sheets (Chen et al. 2014), beam-waist scanning (Dean et al. 2015), two-photon excitation (Lavagnino et al. 2013, Truong et al. 2011), structured illumination (Keller et al. 2010, Mertz & Kim 2010), and nonlinear photoactivation (Cella Zanacchi et al. 2013). Additional methods have incorporated deconvolution and computational reconstruction of multiple projections to yield higher isotropic resolution (Chhetri et al. 2015, Wu et al. 2016), although the routine deconvolution of large, high-speed data sets remains very computationally demanding. Multiple studies have also demonstrated light-sheet approaches for subcellular imaging (Gustavsson et al. 2018, Liu et al. 2018).
3.3. Removing the Need for Sample Translation Unlocks Faster Imaging Speeds
Despite these innovations, the volumetric imaging speed of many SPIM implementations has remained limited by the need for the sample to be physically translated through the coaligned light sheet and camera detection plane. To overcome this limitation, implementations were developed that used galvanometric mirrors to translate the light sheet through the sample. However, as the illumination plane was now moving, the detection objective’s focal plane also needed to be varied to keep it aligned with the illuminated plane. This detection refocusing can be achieved by physically moving the detection objective (e.g., using a piezo) (Ahrens et al. 2013), by using optical components such as an electrically tunable lens in the detection path to adjust the focal plane without moving the detection objective (Fahrbach et al. 2013), or by extending the detection objective’s depth of field (Tomer et al. 2015). These advances led to speed improvements sufficient to capture neuronal calcium activity throughout the brain of a living zebrafish larva (Ahrens et al. 2013), as shown in Figure 2b.
3.4. Single-Objective Light-Sheet Approaches for High-Speed In Vivo Microscopy
In parallel, multiple different light-sheet approaches were proposed that did not rely on multiple objective lenses around the sample. For example, objective coupled planar illumination (OCPI) microscopy generates a sheet of light using an optic mounted rigidly to the side of a high-NA objective lens (Holekamp et al. 2008). OCPI microscopy was developed by Holekamp et al. (2008) to image neuronal activity via calcium-sensitive dyes across the layers of the living mouse vomeronasal organ. The rigid coupling of the detection objective lens and light-sheet-generating optics enables volumetric acquisition via the physical movement of both the lens and optics along the objective’s axis, rather than requiring independent movement or translation of the sample. This approach provided an early demonstration that light sheets could enable cellular-level functional imaging in intact, living, scattering neural tissue. In a related approach, Engelbrecht et al. (2010) created a miniaturized SPIM geometry using gradient-index lenses and a prism mirror, which could be inserted into the living brain for optically sectioned imaging.
Another technique, oblique-plane microscopy, developed by Dunsby (2008), generated an oblique light sheet using a single objective lens and detected fluorescent light back through the same lens while maintaining a sufficient angle between illumination and detection to generate a light-sheet imaging condition. Bouchard et al. (2015) subsequently demonstrated swept confocally aligned planar excitation (SCAPE) microscopy, which combined a scanning galvanometer mirror with oblique plane illumination to enable the excitation sheet to be swept laterally across the sample, while maintaining the focus of the camera on the moving plane. SCAPE microscopy provides a way to acquire very high-speed light-sheet volumetric imaging through a single, stationary objective lens, while also requiring no movement of the sample. This approach is detailed in the next section.
4. PERSPECTIVES ON EMERGING APPLICATIONS OF LIGHT-SHEET MICROSCOPY FOR MODERN NEUROSCIENCE
Here, we highlight two key light-sheet microscopy applications that are emerging in neuroscience: (a) the use of light sheets for imaging intact, cleared, and expanded neural tissues and (b) light-sheet imaging for high-speed, volumetric functional imaging of indicators of cellular activity in vivo. These two applications are described in more detail below.
4.1. Light-Sheet Imaging of Cleared and Expanded Tissues
Early tissue clearing techniques dehydrated samples, replacing water with oils that matched the refractive index of lipid membranes to reduce scattering (Dodt et al. 2007). Recent advances in tissue clearing have enabled the in situ preservation of rich biochemical information as well as fluorescent proteins, improved labeling during postprocessing, improved clearing performance, and decreased tissue processing time and complexity (Chung & Deisseroth 2013, Kim et al. 2015, Murakami et al. 2018, Renier et al. 2014, Yang et al. 2014). Improvements in in vivo labeling strategies that can trace specific cells, circuits, and cell types have further increased the utility of this approach, providing a bridge between in vivo recordings of cellular function and behavior and the subcellular-level structures and connectivity of the brain (Chatterjee et al. 2018, K.H. Chen et al. 2015, Zolnik et al. 2017).
A further recent advance is tissue expansion (F. Chen et al. 2015), which can uniformly expand tissue while preserving its labeling and structure. This expansion enables features that are usually below a size resolvable with light microscopy to be visualized in intact specimens. Recent work has combined expansion microscopy with superresolution lattice light-sheet microscopy to enable near-nanoscale imaging of fluorescent contrast in mice and Drosophila brains (Gao et al. 2018).
However, both cleared and expanded samples place significant demands on 3D imaging throughput. Resonant-scanning confocal microscopy approaches can require 20 hours or more to image a single cleared mouse brain at approximately 1-μm resolution. Expanded tissues can be even larger, while also being delicate. These samples can span a wide range of refractive indices and can contain multicolor labeling or even require sequential staining and labeling.
4.1.1. Instrumentation for cleared-tissue light-sheet imaging.
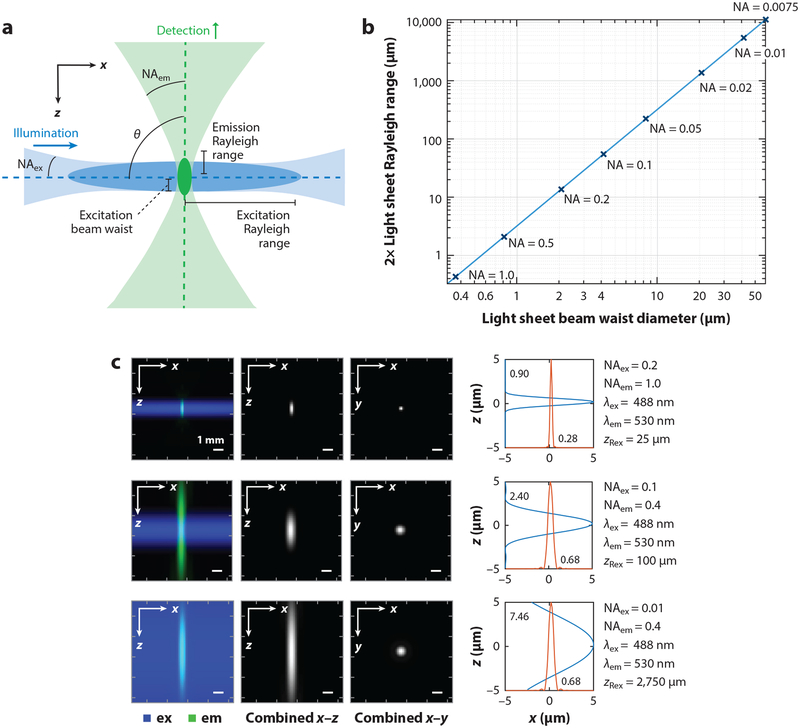
Light-sheet microscopy promises substantial throughput advantages for imaging large, cleared, and expanded tissues. Despite the availability of a range of commercial systems, challenges remain that require further innovation. Firstly, the effects of Gaussian beam illumination impose challenging constraints on field of view versus axial resolution (see the sidebar titled Resolution and Field of View: The Battle with Gaussian Beams; Figure 3a,b). For example, a 0.0075-NA light sheet whose Rayleigh range spans a 10-mm mouse brain would have a 60-μm beam waist at its center. However, as shown in Figure 3c, this effect can be offset by maximizing detection NA (see the sidebar titled Detection Numerical Aperture and Light Scattering).
Figure 3.
Determinants of spatial resolution in light-sheet microscopy. (a) A typical light-sheet geometry combines the light sheet’s Gaussian beam properties with the PSF of the detection optics (see also Equation 1), with resolution depending on the NA of both. (b) A lookup table of Gaussian beam properties for different NA foci shows how the beam’s minimum waist diameter scales with its R (the distance along the sheet at which the waist is × the minimum beam waist), modeled for 488 nm in water, refractive index n = 1.33. (c) Plots show simulated PSFs based on Equation 1 using a single-beam PSF model (PSF_Generator) by Kirshner et al.=(2013) (left). Plots show intensity cross sections through the combined PSF over z and x directions (right). Abbreviations: em, emission; ex, excitation; NA, numerical aperture; PSF, point spread function; R, Rayleigh range.
However, even if a thin, uniform light sheet could be generated, a second constraint is the ability to sample an image of this sheet with sufficient pixel density. A typical ~2,000 × 2,000-pixel scientific CMOS camera imaging a 10-mm field of view would only have a 20-μm-per-pixel sampling density—insufficient to resolve cell bodies or neural processes. Cameras with much higher numbers of pixels, and accompanying low magnification, and high-NA objective lenses will be needed for this single-shot approach to be successful for most applications.
Consequently, most modern light-sheet systems for imaging large, cleared samples rely on some form of tiling image stitching, zooming in on smaller regions of interest to get both a higher density sampling and a narrower Rayleigh range of the light sheet. Others are implementing higher NA beam waist scanning, in some cases reading out only the rows of the camera corresponding to where the waist is narrow (Dean et al. 2015, Migliori et al. 2018), although this approach decreases the light efficiency of the system.
Unfortunately, modes where data must be tiled introduce many complexities, from how to hold the sample securely and move it precisely during prolonged acquisitions, to how to handle the merging and stitching of very large data sets without artifacts (a single-color 12 × 10 × 8 mm mouse brain data set at 1-μm sampling density with 16-bit precision would be around 2 TB in size).
Further challenges are posed by the liquids surrounding cleared samples, which must be contained in a chamber that permits good optical coupling of both excitation and emission light from multiple directions without distortions from refractive index mismatches between air, glass, and the liquid’s (and sample’s) refractive index (see Figure 2a).
Rotated inverted-SPIM and single-objective light-sheet approaches combined with stage-scanning are showing promise for imaging cleared tissues with large lateral extents and can employ dipping lenses to maximize NA and reduce interface distortions. However, despite the availability of new longer working distance, high-NA objective lenses, working distance can be a limiting factor. Some clearing media are also proving damaging to costly dipping objective lenses, while the difference between the high-refractive index media used for standard clearing and the lower refractive index of water around expanded tissues combine to make it challenging to engineer a light-sheet system that can serve all needs.
Thus, although light-sheet microscopy holds significant promise for cleared-tissue imaging, further innovation is required before high-throughput tissue processing and imaging will be efficient and fully automated.
4.1.2. Data visualization and analysis.
Light-sheet data collected on cleared tissues are often presented as 3D rendered movies, generated in commercial visualization packages. However, as tissue processing and imaging throughput accelerate, the scope of questions that can be addressed with these approaches will increase, and data analysis will need to become more automated and quantitative. New algorithms to vectorize connectivity and extract features such as cell density, projection density, spine density, and interactions between cell types from these enormous data sets are needed, on par with analysis efforts developed for modern electron microscopy connectomics (Kim et al. 2014). Fortunately, modern machine learning and deep learning–based analysis have significant potential for both automated segmentation and image registration. These approaches can also be leveraged to connect in vivo properties or variables to difficult-to-discern structural features or interrelationships in these huge data sets. Such methods will soon be ready for light-sheet data if imaging methods can reach sufficient imaging speeds and performance standardization.
4.2. Fast Light-Sheet Imaging of Neuronal Calcium Dynamics
The use of in vivo two-photon microscopy for imaging cellular function in the intact brain is well established. Yet improvements in genetically encoded fluorescent calcium indicators, as well as increasingly diverse animal models and transfection methods, are providing strong demand for higher-speed 3D imaging than point scanning can deliver. Although light-sheet microscopes can surpass the integration time and photodamage limitations of high-speed point scanning, traditional multiobjective light-sheet implementations have limited utility for imaging the intact, behaving brain.
Single-objective light-sheet methods have overcome this limitation and make it possible to image in a geometry identical to a confocal or two-photon microscope, opening up applications in awake, behaving animals and freely moving small organisms and the ability to perform, for example, simultaneous electrophysiology, pharmacology, or behavioral recordings. SCAPE microscopy delivers two additional important advantages over point-scanning methods. First, SCAPE’s objective lens remains completely stationary during 3D image formation, removing the chance of disturbing sensitive or reactive samples and enabling integration of additional light paths for sample observation or patterned optogenetic manipulation, for example. SCAPE’s second advantage is its ability to capture volumetric images at very high speeds via both its unique sheet-scanning and -descanning optical path and its efficient use of camera pixel readouts.
4.2.1. SCAPE instrumentation.
The optical layout of a basic SCAPE system is shown in Figure 2c. The sample is illuminated by an oblique light sheet [extending over the y and z′ (i.e., oblique-z) axes] emerging from the primary objective lens. Fluorescence light generated by this sheet illumination is collected back through the same objective lens and reimaged to an intermediate oblique imaging plane, leveraging the symmetry of the optical system (Botcherby et al. 2007). This intermediate image can be relayed to focus onto an obliquely aligned camera (Dunsby 2008) such that the camera is aligned with the light-sheet plane, as in other light-sheet systems. A key aspect of SCAPE is that to form a 3D volumetric image, a galvanometer mirror is used to scan the oblique light sheet laterally across the sample (across x), while light returning from the sample is also reflected off the same moving galvanometer. This mechanism de-scans the returning light such that the intermediate image of the light sheet remains stationary and focused on the camera. As a result, there is no requirement to synchronize separate movements of the light sheet and detection plane—each camera image represents a different (xn, y, z′) image, sampling a rhomboid-shaped volume in the sample.
The galvanometer mirror only needs to scan at a line rate equal to the volume-imaging rate (e.g., 10 lines per second for 10 VPS). Because images are oriented in y-z′, the number of rows on the camera corresponds to the number of depths in the sample, a parameter that can be adjusted to the required penetration depth and/or Rayleigh range of the oblique light sheet. A standard scientific CMOS camera can read out 100 rows at over 2,000 frames per second, providing the ability to capture 200 x-planes across the sample at 10 VPS. Faster cameras can permit even faster volumetric imaging, as there is no mechanical limitation to imaging at speeds of over 300 VPS, while the light efficiencies of light-sheet microscopy permit good signal to noise, even at acquisition speeds over 10,000 frames per second. The system’s x-y field of view is typically 1 × 1 mm using a 20×, 1.0-NA water immersion objective with a 2-mm working distance. The system’s optical resolution depends on the angle between the light sheet and detection cone (see Figure 3a) and the detection NA, which is constrained to around 50% or less of the primary objective’s NA, owing to the need for image rotation (Dunsby 2008). Magnification of the intermediate image plane onto the camera is varied to change the micrometer-per-pixel sampling density, while a color image splitter can be used to enable side-by-side acquisition of dual fluorophores in parallel.
4.2.2. Emerging applications of high-speed volumetric microscopy in neuroscience.
High-speed light-sheet systems have significant potential to push past the bottlenecks faced by point-scanning microscopy for neuroscience applications. The growing palette of dynamic functional fluorescent indicators is rapidly extending beyond calcium to include dopamine and soon membrane potentials, signals that could require imaging at 200–1,000 VPS.
Figure 2c shows SCAPE data capturing the calcium dynamics of apical dendrites in the awake, behaving mouse brain at 10 VPS, equivalent to a two-photon microscope imaging at a 156-MHz pixel rate. These data were acquired using single-photon excitation at 488 nm and are thus limited beyond depths of around 300 μm. However, red-shifted and two-photon implementations are proving feasible and could lead to an order-of-magnitude improvement in sample density for in vivo volumetric imaging in the mammalian cortex.
Beyond the mouse brain, significantly higher volumetric imaging speeds also permit imaging of previously intractable samples such as freely crawling Drosophila larvae or Caenorhabditis elegans worms (Vaadia et al. 2019). The entire nervous system of these samples can fit within the field of view of a microscope, and the movement of their bodies is their primary behavior. The current best choice for imaging these samples is spinning-disk confocal microscopy, which can require samples to be tracked using a moving stage, require physical z-translation, and be constrained by significant photobleaching of the sample during behavioral experiments. Single-objective light-sheet techniques such as SCAPE bring the potential for much faster imaging of a new generation of neuroscience samples where 3D movement not only can be tolerated (and registered) but can be a measurable in the experiment, while the functional signaling of cells throughout the sample can be tracked and extracted in real time. Such applications can extend to looking at signaling during the mechanical deformation of tissues and signaling that affects physical movement (such as in the heart), as well as the dynamics of flow, dilations, and the physical migration of cells through tissues.
4.2.3. Data analysis.
The data analysis challenges presented by these new kinds of high-speed 3D data are necessarily diverse. Dimensionality reduction methods that turn a movie of flashing cells into an interpretable map of cell locations and their activity patterns are applicable to fast 3D light-sheet data (Pnevmatikakis et al. 2016). However, the move from two to three dimensions, as well as the large size of the data sets (at least 1 TB/h of recording), requires innovations for both 3D image registration and handling such large data. Improved cross-validation of these processing methods is especially important given the difficulty of checking outputs in such large data sets that require unsupervised and automated analysis for the fastest possible throughput.
For broader applications, a wide array of analysis tools is required, particularly 4D denoising and robust 3D tracking and signal extraction. Fortunately, emerging deep learning methods are showing significant promise for these applications, although again careful implementations that permit recognition of patterns across data, while remaining feasible in terms of memory allocation, require continued innovation. Finally, the visualization and interpretation of results beyond information extraction require interfacing with neurotheory to leverage these new observables into the decoding of the complex relationships between neural network activity and behavior.
5. CONCLUSIONS
The past 5 years have seen massive growth in optical indicators and animal models and gradual progress towards faster volumetric imaging. Light-sheet microscopy provides a potential major leap forward in our capabilities for fast volumetric microscopy in diverse areas of neuroscience.
RESOLUTION AND FIELD OF VIEW: THE BATTLE WITH GAUSSIAN BEAMS.
A light sheet is a flattened beam of light that can illuminate a thin plane of a sample. Such a sheet can be generated using a cylindrical lens that leaves light collimated in one direction (y), while forming a low-NA focus in the other (x). But a low-NA Gaussian focus is not strictly a sheet; it gradually narrows along z to a waist and then expands again. A higher-NA sheet will have a narrower waist but a shorter range over which the waist remains narrow (its Rayleigh range). A lower-NA sheet will have a thicker waist but will have a longer Rayleigh range (see Figure 3a,b). This trade-off makes it challenging to obtain both high-resolution optical sectioning and a large field of view, while xyz resolution will never be uniform over the volume being imaged. Many attempts have been made to overcome this Gaussian effect (see Section 3.2), including using Bessel beams, which have a narrower waist over a much longer z-range but are confounded by side lobes (Planchon et al. 2011). Another approach is to scan a higher-NA beam waist along the sheet’s axial direction (Dean et al. 2015), yet this approach sacrifices some light efficiency and integration time benefits. Overcoming these properties of Gaussian beams to create thin, uniform light sheets will be key to future improvements to the performance of light-sheet systems.
DETECTION NUMERICAL APERTURE AND LIGHT SCATTERING.
Although light-sheet thickness is often considered to be equal to a light-sheet microscope’s optical sectioning resolution, the combined point spread function (PSF) of a light-sheet microscope is given by
| 1. |
where h(x,y,z) is the PSF of the illumination (ill), detection (det), or combined (LS) system and θ is the angle between the sheet and detection cone (Stelzer & Lindek 1994) (Figure 3a). As demonstrated in Figure 3c, for a thick light sheet, the system’s axial resolution can be constrained by the tighter axial range of a higher numerical aperture (NA) detection PSF. In all cases, x-y resolution also improves with higher NA detection, as with conventional microscopy. This effect may also explain why light-sheet microscopy can be effective for imaging into scattering tissues. Scattering degrades the shape of the light sheet and also blurs the image that reaches the camera. However, the detection PSF will still contribute to the optical sectioning of the system, even if the light sheet is broadened, suggesting strong benefits from maximizing detection NA. Unfortunately, the detection NA of most light-sheet systems is limited by either geometric or immersion medium constraints or is reduced by image rotation in single-objective approaches (Dunsby 2008). Other approaches to overcoming scattering include structured illumination and adaptive optics as well as exploiting the lower scattering and absorption properties of tissues at longer wavelengths, achievable with red-shifted fluorophores or two-photon excitation in the infrared.
ACKNOWLEDGMENTS
We acknowledge the contributions of our many collaborators who have guided us in understanding how to make optimized light-sheet microscopy for a range of applications. We also acknowledge the contributions of Hillman lab members, including Kripa Patel, Citlali Perez-Campos, Matthew Bouchard, Srinidhi Bharadwaj, Grace Lee, and Wenxuan Liang, to the development of SCAPE microscopy, its variants, and its application to diverse samples. We acknowledge grant support from the US National Institutes of Health BRAIN initiative (5U01NS094296, UF1NS108213, U19NS104649, and R01HL131438), the National Science Foundation (graduate fellowships to Bouchard and Patel and CAREER CBET-0954796 to E.M.C.H), the Simons Foundation Collaboration on the Global Brain, the US Department of Defense Multidisciplinary University Research Initiative (W911NF-12-1-0594), the Kavli Institute for Brain Science, the Columbia-Coulter Translational Research Partnership, and the Coulter Foundation.
Footnotes
DISCLOSURE STATEMENT
SCAPE intellectual property is licensed to Leica Microsystems for commercial development. All authors have a potential financial conflict of interest relating to SCAPE microscopy.
LITERATURE CITED
- Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. 2013. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods 10:413–20 [DOI] [PubMed] [Google Scholar]
- Akerboom J, Chen T-W, Wardill TJ, Tian L, Marvin JS, et al. 2012. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci 32:13819–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewersdorf J, Pick R, Hell SW. 1998. Multifocal multiphoton microscopy. Opt. Lett 23:655–57 [DOI] [PubMed] [Google Scholar]
- Botcherby EJ, Juskaitis R, Booth MJ, Wilson T. 2007. Aberration-free optical refocusing in high numerical aperture microscopy. Opt. Lett 32:2007–9 [DOI] [PubMed] [Google Scholar]
- Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, et al. 2015. Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat. Photonics 9:113–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. 2005. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci 8:1263–68 [DOI] [PubMed] [Google Scholar]
- Castellano-Munoz M, Peng AW, Salles FT, Ricci AJ. 2012. Swept field laser confocal microscopy for enhanced spatial and temporal resolution in live-cell imaging. Microsc. Microanal 18:753–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella Zanacchi F, Lavagnino Z, Faretta M, Furia L, Diaspro A. 2013. Light-sheet confined super-resolution using two-photon photoactivation. PLOS ONE 8:e67667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. 1994. Green fluorescent protein as a marker for gene-expression. Science 263:802–5 [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Sullivan HA, MacLennan BJ, Xu R, Hou Y, et al. 2018. Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat. Neurosci 21:638–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, et al. 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346:1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. 2015. Expansion microscopy. Science 347:543–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. 2015. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, et al. 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499:295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetri RK, Amat F, Wan Y, Hockendorf B, Lemon WC, Keller PJ. 2015. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat. Methods 12:1171–78 [DOI] [PubMed] [Google Scholar]
- Choi H, Tzeranis DS, Cha JW, Clemenceau P, de Jong SJG, et al. 2012. 3D-resolved fluorescence and phosphorescence lifetime imaging using temporal focusing wide-field two-photon excitation. Opt. Express 20:26219–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Deisseroth K. 2013. CLARITY for mapping the nervous system. Nat. Methods 10:508–13 [DOI] [PubMed] [Google Scholar]
- Cranfill PJ, Sell BR, Baird MA, Allen JR, Lavagnino Z, et al. 2016. Quantitative assessment of fluorescent proteins. Nat. Methods 13:557–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno HIC, Cizmar T, Vettenburg T, Nylk J, Gunn-Moore FJ, Dholakia K. 2012. Wavefront corrected light sheet microscopy in turbid media. Appl. Phys. Lett 100:191108 [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, et al. 2016. Sensitive red protein calcium indicators for imaging neural activity. eLife 5:e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Shoham S. 2012. Remotely scanned multiphoton temporal focusing by axial grism scanning. Opt. Lett 37:2913–15 [DOI] [PubMed] [Google Scholar]
- Dean KM, Roudot P, Welf ES, Danuser G, Fiolka R. 2015. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys. J 108:2807–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. 1990. Two-photon laser scanning fluorescence microscopy. Science 248:73–76 [DOI] [PubMed] [Google Scholar]
- Dodt H-U, Leischner U, Schierloh A, Jahrling N, Mauch CP, et al. 2007. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4:331–36 [DOI] [PubMed] [Google Scholar]
- Dunsby C 2008. Optically sectioned imaging by oblique plane microscopy. Opt. Express 16:20306–16 [DOI] [PubMed] [Google Scholar]
- Engelbrecht CJ, Stelzer EHK. 2006. Resolution enhancement in a light-sheet-based microscope (SPIM). Opt. Lett 31:1477–79 [DOI] [PubMed] [Google Scholar]
- Engelbrecht CJ, Voigt F, Helmchen F. 2010. Miniaturized selective plane illumination microscopy for high-contrast in vivo fluorescence imaging. Opt. Lett 35:1413–15 [DOI] [PubMed] [Google Scholar]
- Fahrbach FO, Rohrbach A. 2010. A line scanned light-sheet microscope with phase shaped self-reconstructing beams. Opt. Express 18:24229–44 [DOI] [PubMed] [Google Scholar]
- Fahrbach FO, Voigt FF, Schmid B, Helmchen F, Huisken J. 2013. Rapid 3D light-sheet microscopy with a tunable lens. Opt. Express 21:21010–26 [DOI] [PubMed] [Google Scholar]
- Fuchs E, Jaffe J, Long R, Azam F. 2002. Thin laser light sheet microscope for microbial oceanography. Opt. Express 10:145–54 [DOI] [PubMed] [Google Scholar]
- Gao R, Asano SM, Upadhyayula S, Pisarev I, Milkie DE, et al. 2018. Cortical column and whole brain imaging of neural circuits with molecular contrast and nanoscale resolution. bioRxiv 374140. 10.1101/374140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, et al. 2015. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350:1361–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräf R, Rietdorf J, Zimmermann T. 2005. Live cell spinning disk microscopy In Microscopy Techniques, ed. Rietdorf J, pp. 57–75. Berlin: Springer; [DOI] [PubMed] [Google Scholar]
- Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. 2010. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat. Methods 7:399–405 [DOI] [PubMed] [Google Scholar]
- Gustavsson AK, Petrov PN, Lee MY, Shechtman Y, Moerner WE. 2018. 3D single-molecule super-resolution microscopy with a tilted light sheet. Nat. Commun 9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim N, Garaschuk O, Friedrich MW, Mank M, Milos RI, et al. 2007. Improved calcium imaging in transgenic mice expressing a troponin C-based biosensor. Nat. Methods 4:127–29 [DOI] [PubMed] [Google Scholar]
- Hillman EMC, Voleti V, Patel K, Li W, Yu H, et al. 2018. High-speed 3D imaging of cellular activity in the brain using axially-extended beams and light sheets. Curr. Opin. Neurobiol 50:190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holekamp TF, Turaga D, Holy TE. 2008. Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron 57:661–72 [DOI] [PubMed] [Google Scholar]
- Horton NG, Wang K, Kobat D, Clark CG, Wise FW, et al. 2013. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics 7:205–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisken J, Stainier DY. 2007. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett 32:2608–10 [DOI] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK. 2004. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305:1007–9 [DOI] [PubMed] [Google Scholar]
- Keller PJ, Ahrens MB. 2015. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 85:462–83 [DOI] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Santella A, Khairy K, Bao Z, et al. 2010. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat. Methods 7:637–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. 2008. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322:1065–69 [DOI] [PubMed] [Google Scholar]
- Kim JS, Greene MJ, Zlateski A, Lee K, Richardson M, et al. 2014. Space-time wiring specificity supports direction selectivity in the retina. Nature 509:331–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Cho JH, Murray E, Bakh N, Choi H, et al. 2015. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. PNAS 112:E6274–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner H, Aguet F, Sage D, Unser M. 2013. 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J. Microsc 249:13–25 [DOI] [PubMed] [Google Scholar]
- Kobat D, Durst ME, Nishimura N, Wong AW, Schaffer CB, Xu C. 2009. Deep tissue multiphoton microscopy using longer wavelength excitation. Opt. Express 17:13354–64 [DOI] [PubMed] [Google Scholar]
- Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. 2012. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat. Methods 9:90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavagnino Z, Zanacchi FC, Ronzitti E, Diaspro A. 2013. Two-photon excitation selective plane illumination microscopy (2PE-SPIM) of highly scattering samples: characterization and application. Opt. Express 21:5998–6008 [DOI] [PubMed] [Google Scholar]
- Lindek S, Cremer C, Stelzer EH. 1996. Confocal theta fluorescence microscopy with annular apertures. Appl. Opt 35:126–30 [DOI] [PubMed] [Google Scholar]
- Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, et al. 2018. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science 360:eaaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J, Kim J. 2010. Scanning light-sheet microscopy in the whole mouse brain with HiLo background rejection. J. Biomed. Opt 15:016027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliori B, Datta MS, Dupre C, Apak MC, Asano S, et al. 2018. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol. 16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami TC, Mano T, Saikawa S, Horiguchi SA, Shigeta D, et al. 2018. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci 21:625–37 [DOI] [PubMed] [Google Scholar]
- Nadella KM, Ros H, Baragli C, Griffiths VA, Konstantinou G, et al. 2016. Random-access scanning microscopy for 3D imaging in awake behaving animals. Nat. Methods 13:1001–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HW, Hausser M. 2015. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods 12:140–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, et al. 2018. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360:eaat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperkok R, Squire A, Geley S, Bastiaens PIH. 1999. Simultaneous detection of multiple green fluorescent proteins in live cells by fluorescence lifetime imaging microscopy. Curr. Biol 9:269–74 [DOI] [PubMed] [Google Scholar]
- Picot A, Dominguez S, Liu C, Chen IW, Tanese D, et al. 2018. Temperature rise under two-photon optogenetic brain stimulation. Cell Rep. 24:1243–53.e5 [DOI] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, et al. 2011. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8:417–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, et al. 2016. Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89:285–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgorski K, Ranganathan G. 2016. Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol 116:1012–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajadhyaksha M, Anderson RR, Webb RH. 1999. Video-rate confocal scanning laser microscope for imaging human tissues in vivo. Appl. Opt 38:2105–15 [DOI] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. 2014. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159:896–910 [DOI] [PubMed] [Google Scholar]
- Royer LA, Lemon WC, Chhetri RK, Wan Y, Coleman M, et al. 2016. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol 34:1267–78 [DOI] [PubMed] [Google Scholar]
- Schrodel T, Prevedel R, Aumayr K, Zimmer M, Vaziri A. 2013. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nat. Methods 10:1013–20 [DOI] [PubMed] [Google Scholar]
- Siedentopf H, Zsigmondy R. 1902. Uber Sichtbarmachung und Größenbestimmung ultramikoskopischer Teilchen, mit besonderer Anwendung auf Goldrubingläser. Ann. Phys 315:1–39 [Google Scholar]
- Sofroniew NJ, Flickinger D, King J, Svoboda K. 2016. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife 5:e14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer EHK, Lindek S. 1994. Fundamental reduction of the observation volume in far-field light-microscopy by detection orthogonal to the illumination axis: confocal theta microscopy. Opt. Commun 111:536–47 [Google Scholar]
- Tomer R, Khairy K, Amat F, Keller PJ. 2012. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9:755–63 [DOI] [PubMed] [Google Scholar]
- Tomer R, Lovett-Barron M, Kauvar I, Andalman A, Burns VM, et al. 2015. SPED light sheet microscopy: fast mapping of biological system structure and function. Cell 163:1796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. 2011. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods 8:757–60 [DOI] [PubMed] [Google Scholar]
- Vaadia R, Li W, Voleti V, Singhania E, Hillman EMC, Grueber WB. 2019. Characterization of proprioceptive system dynamics in behaving Drosophila larvae using high-speed volumetric microscopy. Curr. Biol 29(6):935–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri A, Shank CV. 2010. Ultrafast widefield optical sectioning microscopy by multifocal temporal focusing. Opt. Express 18:19645–55 [DOI] [PubMed] [Google Scholar]
- Vettenburg T, Dalgarno HI, Nylk J, Coll-Llado C, Ferrier DE, et al. 2014. Light-sheet microscopy using an Airy beam. Nat. Methods 11:541–44 [DOI] [PubMed] [Google Scholar]
- Voie AH, Spelman FA. 1995. Three-dimensional reconstruction of the cochlea from two-dimensional images of optical sections. Comput. Med. Imaging Graph. 19:377–84 [DOI] [PubMed] [Google Scholar]
- Watson BO, Nikolenko V, Yuste R. 2009. Two-photon imaging with diffractive optical elements. Front. Neural Circuits 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenburger S, Vaziri A. 2018. A guide to emerging technologies for large-scale and whole-brain optical imaging of neuronal activity. Annu. Rev. Neurosci 41:431–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Amos WB, Fordham M. 1987. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J. Cell Biol. 105:41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding D, Pozzi P, Soloviev O, Vdovin G, Sheppard CJ, Verhaegen M. 2016. Pupil filters for extending the field-of-view in light-sheet microscopy. Opt. Lett 41:1205–8 [DOI] [PubMed] [Google Scholar]
- Wu Y, Chandris P, Winter PW, Kim EY, Jaumouille V, et al. 2016. Simultaneous multiview capture and fusion improves spatial resolution in wide-field and light-sheet microscopy. Optica 3:897–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Ghitani A, Christensen R, Santella A, Du Z, et al. 2011. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. PNAS 108:17708–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, et al. 2014. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158:945–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnik TA, Sha F, Johenning FW, Schreiter ER, Looger LL, et al. 2017. All-optical functional synaptic connectivity mapping in acute brain slices using the calcium integrator CaMPARI. J. Physiol 595:1465–77 [DOI] [PMC free article] [PubMed] [Google Scholar]