Abstract
Congenital malformations, or structural birth defects, are now the leading cause of infant mortality in the United States and Europe [1, 2]. Of the congenital malformations, congenital heart disease (CHD) is the most common [1, 2]. Thus, a molecular understanding of heart development is an essential goal for improving clinical approaches to CHD. However, CHDs are commonly a result of genetic defects that manifest themselves in a spatial and temporal manner during the early stages of embryogenesis, leaving them mostly intractable to mass spectrometry-based analysis. Here we describe the technologies and advancements in the field of mass spectrometry over the past few years that have begun to provide insights into the molecular and cellular basis of CHD and prospects for these types of approaches in the future.
Introduction
Congenital heart disease (CHD) is the leading cause of infant mortality in the United States and Europe [1,2]. Since the initiating event in the majority of CHDs takes place during early development of the human heart, studies to understand CHD etiology have relied on vertebrate model systems, most notably the mouse [3–10]. Much effort has therefore gone into identifying the genes and networks required for these stages of normal mouse development at the RNA level [11–19]. However, only 10% of RNAs that show 2-fold or more changes in levels of expression are associated with alterations in protein abundance. Conversely, changes in protein levels are often not associated with changes in RNA levels [20–23]. It is therefore evident that research into the molecular roots of CHD should involve direct assessments of protein expression levels and interaction networks.
Tandem mass spectrometry (MS/MS)-based analysis has proven invaluable in studying the temporal and spatial distribution of proteins during development in a range of animal model systems [19,24–29]. This type of proteomic-based approach allows not only for the generation of a compendium of proteins of a given cell type at a given stage of development, but also provides information for the characterization of post-translational modifications (PTMs). Hence, these type of approaches also may identify the growth factor signaling pathways that control a protein’s function. By using multiple or parallel reaction monitoring, it is further possible to determine the precise amount of a protein in a given cell or tissue type [30,31]. Despite these powerful advantages, proteomic-based approaches still face limitations, most notably the amount of material that is required for an in-depth analysis from small tissues that express modest levels of a given protein relative for example, to tissue culture cells. Thus, the field of cardiac biology has been largely limited to analysis of serum and plasma biomarkers (e.g. [32–34]).
To circumvent this issue, technologies, procedures, and workflows have been developed to increase the efficiency of protein recovery from murine tissue and cell types, as well as the application of proteomics to surrogate systems which may in principle mimic human cardiac differentiation. These systems include cardiomyocyte differentiation of embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), as well as direct reprogramming of differentiated cells (e.g. cardiac fibroblasts) into cardiomyocytes.
Proteomic-Based Approaches in Embryonic Heart Tissue
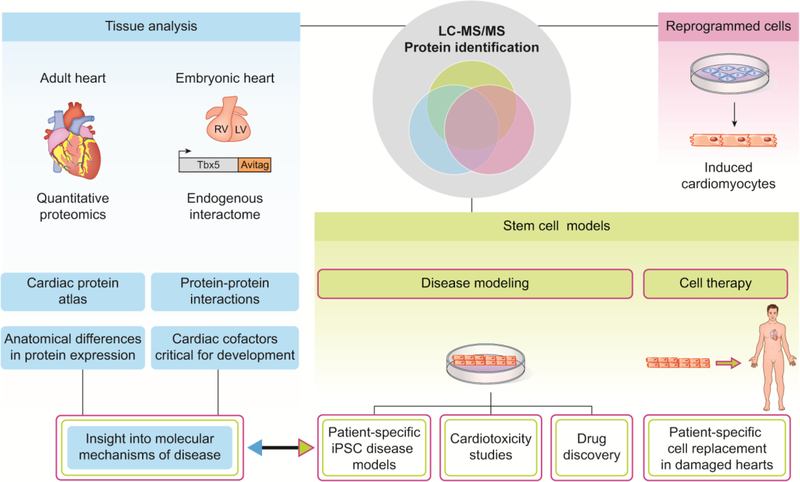
The identification and characterization of endogenous proteins and protein complexes in vivo under physiological conditions are essential to gain a basic understanding of normal cardiac development and the pathology of CHD [19]. However, use of these approaches has been limited due to lack of optimized mass spectrometry-based protocols and workflows for analysis of such small samples in early-stage tissues and embryos. To identify endogenous interactomes utilizing targeted MS, approaches have focused on optimization of protein extraction buffers and cell/tissue lysis conditions, as well as increasing the efficiency of immuno-isolation. One approach has been to tag the endogenous protein through homologous recombination with the Avi-tag [35–37]. The Avi-tag is an artificial epitope tag that combines the minimal invasiveness of a small peptide tag with the specificity and strength of the biotin-streptavidin attachment, the strongest non-covalent peptide-ligand interaction in nature, exceeding any antibody-antigen interaction [38–41]. Therefore, the approach offers high-affinity and high-specificity isolation of the targeted protein (Figure 1). This type of approach has proven highly effective in the isolation of the transcription factor TBX5, mutations in which cause Holt-Oram syndrome [42–45]. This study demonstrated that TBX5 interacts biochemically and genetically with the nucleosome remodeling and deacetylase (NuRD) transcriptional repressor complex, thus defining a TBX5-NuRD interaction essential to cardiac development [37].
Figure 1. Schematic of proteomic based approaches in cardiac tissue and cells.
Mass spectrometry based approaches in cardiac tissue have led to the generation of a global protein atlas of the healthy human heart. The isolation of endogenous interactomes utilizing gene recombination in the mouse has identified cardiac cofactors that are critical for embryonic development. Cell based systems that produce de novo cardiomyocytes have multiple applications to treating human disease including engraftment into the heart, drug discovery, and cardiotoxicity studies. All of these approaches can provide insight into the molecular mechanism of disease.
Differentiation of Embryonic Stem Cells Into Cardiomyocytes.
In vivo developmental biology systems have identified some of the molecular mechanisms that underlie CHD. However, the utility of these systems for proteomic-based analysis is limited by the amount of tissue one can obtain from a given species at a given developmental stage. Furthermore, due to species-specific differences in various aspects of cardiac biology and development, findings from many systems cannot be applied directly to human biology. This is notably relevant to those incidences of cardiac-related deaths attributed to the limited ability of the damaged heart tissue to regenerate [46–48]. The production of de novo cardiomyocytes, either by human ESCs or human iPSCs, or direct reprogramming, holds the potential for cardiac repair, novel cardiac drug discovery, identification of drug response predictors, and elucidation of the molecular mechanisms during development that underlie cardiac diseases [49–51].
The first of these technologies pursued was the identification and propagation of ESCs, first in the mouse and later in human. ESCs, derived from the inner cell mass of mammalian blastocysts, have the ability to grow indefinitely while maintaining pluripotency [52–54]. Reports on the in vitro differentiation of mouse ESCs into cardiac progenitors demonstrated that these cells can give rise to multiple cardiac cell lineages [55–58]. Subsequent studies showed effective differentiation of human ESCs into the cardiac lineages (Figure 2) [59–62]. By combining these types of approaches with the Avi-tag/BirA system, it was possible to use targeted MS to isolate the temporal changes in the TBX20 interactome [63,64]. Loss of function mutations in TBX20 can cause dilated cardiomyopathy, atrial septal defects, or mitral valve disease, while gain of function mutations of TBX20 have been reported in patients with Tetralogy of Fallot (i.e. pulmonary outflow tract obstruction, ventricular septal defects, overriding aortic root, and right ventricular hypertrophy) [65–69]. Analysis of TBX20 during ESC cardiomyocyte differentiation showed a temporal regulation of the TBX20 interactome and led to the identification of CASZ1 [70,71] as a TBX20 interacting protein, a cardiac interaction that when disrupted leads to dilated cardiomyopathy [63,64].
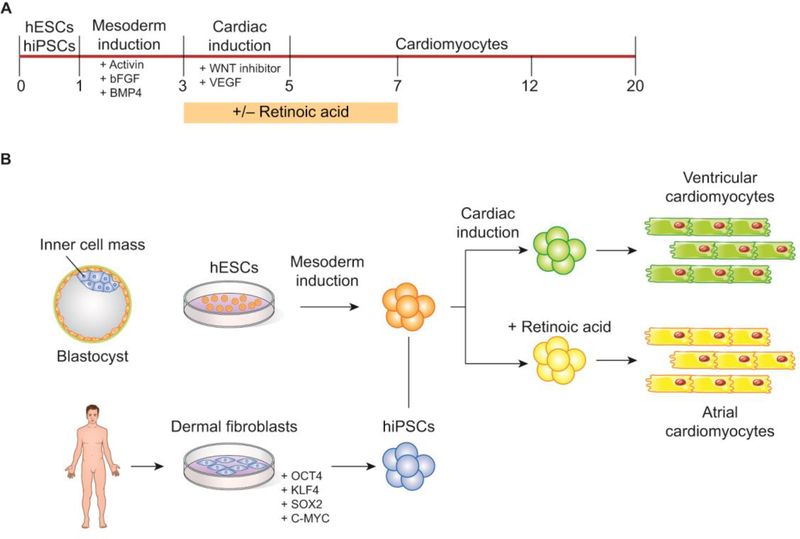
Figure 2. Pluripotent stem cells differentiated into cardiomyocytes.
(A) Cardiomyocyte production timeline. (B) Embryonic stem cells harvested from the inner cell mass of a blastocyst are induced into the mesodermal and cardiac lineages with the treatment of various growth factors. Fibroblasts harvested from the dermis of a human patient are reprogrammed into pluripotent stem cells with the addition of 4 defined factors: OCT4, KLF4, SOX2, and C-MYC. These cells are then induced into the mesodermal and cardiac lineages. Production of atrial-specific cardiomyocytes requires treatment with Retionic Acid at defined stages of differentiation while cultures without Retionic Acid differentiate into ventricular cardiomyocytes.
In addition to providing an in vitro system for analyzing temporal protein regulation, ESC differentiation led to the expectation that human ESCs may provide an understanding of cardiac disease mechanisms and therefore lead to effective therapeutic treatments for patients. A recent study utilizing mouse and human ESCs combined with a shot gun liquid chromatography LC-MS/MS approach identified 246 cell surface markers during key stages of mesoderm specification and early cardiac development. This led to the identification of FZD4 as a marker of lateral plate mesoderm, further enhancing cardiomyocyte enrichment [72]. However, the use of human embryos faces many ethical controversies that hinder the application of human ESCs. In addition, it is difficult to generate patient or disease-specific ESCs, which would greatly aid in their effective application. Therefore, new technologies were pursued that could meet all of these goals and greatly minimize the ethical implications.
Differentiation of Induced Pluripotent Stem Cells Into Cardiomyocytes
iPSC technology reprograms a fully differentiated somatic cell (usually taken from dermal fibroblasts) into a pluripotent stem cell that retains all of the genetic characteristics of its host (e.g. a human patient). iPSC generation requires transduction with four defined transcription factors: Oct3/4, Sox2, Klf4, and c-Myc [73,74]. iPSCs can then be differentiated into functional cardiomyocytes (cardiac troponin T-positive cells) utilizing embryonic growth factor signals that induce mesoderm and subsequent cardiac specification [75]. Until recently, most of the human iPSC-to-cardiomyocyte studies have produced mixed cardiovascular populations that contain ventricle-like cells together with pacemaker and atrial-like cells [76–78]. Atrial and ventricular cardiomyocytes derive from different mesoderm populations and consequently exhibit distinct molecular and functional profiles essential for their diverse physiological roles in the heart (Figure 2) [79,80].
To effectively model and treat diseases that affect specific regions of the heart (such as atrial fibrillation), it is essential to develop differentiation strategies that promote the generation of each of the cardiomyocyte subtypes [80]. Importantly, Lee et al. [80] showed in cardiac-differentiated human iPSCs that retinoic acid signaling at the mesoderm stage of development is required for atrial specification. This work was expanded upon by Cyganek et al. [79] to quantitatively analyze the proteomes of atrial versus ventricular human iPSC-derived cardiomyocytes (iPSC-aCMs and iPSC-vCMs) using stable-isotope labeling by amino acids in cell culture (SILAC) and LC-MS/MS. Analysis of equal portions of SILAC-labeled iPSC-aCMs and unlabeled iPSC-vCMs, and vice versa, allowed calculation of abundance ratios based on their mass differences and subsequently displayed the protein expression differences between the 2 cardiomyocyte subtypes. The authors identified 3,568 proteins present in their samples, 94 of which showed significantly higher expression and 178 of which showed significantly lower expression in iPSC-aCMs compared with iPSC-vCMs. To validate their findings, the authors compared their data to a previous study of the proteomes of human fetal [81] and adult [82] atrial and ventricular tissues and found an enrichment of atrial proteins in the iPSC-aCMs whereas iPSC-vCMs showed enrichment in ventricle-related proteins [79,81,82]. Significantly, they discovered a subset of differentially expressed proteins that were observed in the iPSC-aCMs and vCMS, as well as in the fetal tissues, that were not differentially expressed in the adult tissues, which suggests that these genes are important during cardiac development [79].
Direct Reprogramming of Fibroblasts Into Induced Cardiomyocytes (iCM)
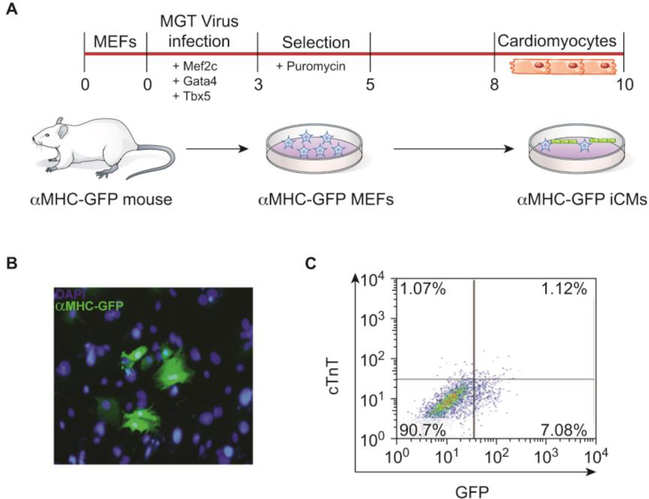
Direct reprogramming is the process of converting fibroblasts into cardiomyocyte-like cells (iCMs) without an embryonic/pluripotent intermediate, but rather through a direct transformation of cell types [83,84]. In mouse fibroblasts this can be achieved by retroviral overexpression of three cardiac lineage-specific transcription factors, Mef2C (M), Gata4 (G), and Tbx5 (T) (MGT) (Figure 3) [83–95]. Proteomic analysis and quantitation of proteins using isobaric labeling with tandem mass tags (TMT) demonstrated systematic and temporally distinct alterations in the levels of specific functional classes of proteins during the initial 72 hours of reprogramming. Surprisingly, few if any of these proteins are cardiac-related but rather are extracellular matrix proteins, translation factors, and chromatin-binding proteins [96]. New questions stemming from these findings include how expression of these classes of proteins bypasses the embryonic steps and when in the process a fibroblast cell first expresses markers of or becomes a true cardiomyocyte [97].
Figure 3. Direct cardiomyocyte reprogramming system.
(A) In vitro reprogramming timeline and experimental schematic. Mouse embryonic fibroblasts are harvested from αMHC-GFP transgenic mice which only express green fluorescent protein in mature cardiomyocytes. Fibroblasts are exposed to retroviral overexpression of the cardiac-specific transcription factors, Mef2C, Gata4, and Tbx5 (MGT). Induced cardiomyocytes can be observed after 8 days in culture following selection for cells that successfully incorporated the MGT virus. (B) Immunostaining analysis of induced cardiomyocytes at day 10 of culture shows GFP positive cells which marks mature cardiomyocytes and DAPI to mark nuclei. (C) FACS analysis of αMHC-GFP positive cells and cardiac troponin T (cTnT) positive cells, an additional mature cardiomyocyte marker, illustrates the reprogramming efficiency.
Perspectives
Though great progress has been made in applying proteomic mass spectrometry-based approaches to cardiac development and disease, the field is still in its infancy and is plagued by the absence of large in-depth data sets from specific stages of heart development or from tissue derived from models of human disease states. Recently, two studies utilizing quantitative mass spectrometry in human fetal and adult hearts generated a global protein atlas of the healthy human heart [81,82]. Lu et al identified cardiac proteins expressed in human atria and ventricles during fetal development [81]. This work was further enhanced by Doll et al [82], which generated a proteomic map from 16 different anatomical cardiac regions as well as 3 different cardiac cell types. These two studies have created valuable datasets that can be used for comparison to disease states (Figure 1).
Data from these types of approaches during embryonic development are emerging, but the utility of the results can be limited by not knowing which proteins and protein pathways are conserved and which have diverged between human and vertebrate model systems such as the mouse and, in industry, the pig (Sus scrofa). Furthermore, though surrogate systems have proven invaluable in assessing cardiomyocyte maturation, it is still not known what type of cardiomyocyte is produced through ESC or iPSC differentiation or through direct reprogramming. These resources and issues will need to be addressed in the coming years.
Finally, the majority of proteomic approaches to date have focused on whole tissue or cardiomyocytes, while it is now apparent that cardiac fibroblasts also play an essential role in heart development and disease [84,98,99]. Moreover, there appears to be many more cell types in the heart than initially believed [84,100]. Thus, it will be necessary to expand current technologies [101] as well as develop new approaches that allow isolation and characterization of pure populations of these cell types.
Acknowledgments
The authors acknowledge support from the NIH (RO1 HD089275, HL126509 and, HL127640) during the writing of this review. We are extremely grateful to Christopher Slagle for critical reading of the manuscript.
Footnotes
Conflict of interest statement
Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Uncategorized References
- 1.Dolk H, Loane M, Garne E: The prevalence of congenital anomalies in europe. Adv Exp Med Biol (2010) 686(349–364. [DOI] [PubMed] [Google Scholar]
- 2.Heron M, Hoyert DL, Murphy SL, Xu J, Kochanek KD, Tejada-Vera B: Deaths: Final data for 2006. Natl Vital Stat Rep (2009) 57(14):1–134. [PubMed] [Google Scholar]
- 3.Liu X, Kim AJ, Reynolds W, Wu Y, Lo CW: Phenotyping cardiac and structural birth defects in fetal and newborn mice. Birth defects research (2017) 109(10):778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirzaei H, Di Biase S, Longo VD: Dietary interventions, cardiovascular aging, and disease: Animal models and human studies. Circ Res (2016) 118(10):1612–1625. [DOI] [PubMed] [Google Scholar]
- 5.Shirakabe A, Ikeda Y, Sciarretta S, Zablocki DK, Sadoshima J: Aging and autophagy in the heart. Circ Res (2016) 118(10):1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdurrachim D, Luiken JJ, Nicolay K, Glatz JF, Prompers JJ, Nabben M: Good and bad consequences of altered fatty acid metabolism in heart failure: Evidence from mouse models. Cardiovascular research (2015) 106(2):194–205. [DOI] [PubMed] [Google Scholar]
- 7.Duncker DJ, Bakkers J, Brundel BJ, Robbins J, Tardiff JC, Carrier L: Animal and in silico models for the study of sarcomeric cardiomyopathies. Cardiovascular research (2015) 105(4):439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiedler LR, Maifoshie E, Schneider MD: Mouse models of heart failure: Cell signaling and cell survival. Curr Top Dev Biol (2014) 109(171–247. [DOI] [PubMed] [Google Scholar]
- 9.Weiss RM, Miller JD, Heistad DD: Fibrocalcific aortic valve disease: Opportunity to understand disease mechanisms using mouse models. Circ Res (2013) 113(2):209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ralphe JC, de Lange WJ: 3d engineered cardiac tissue models of human heart disease: Learning more from our mice. Trends Cardiovasc Med (2013) 23(2):27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sanchez-Danes A, Moignard V, Dubois C, Paulissen C, Kinston S, Gottgens B et al. : Defining the earliest step of cardiovascular lineage segregation by single-cell rna-seq. Science (2018) 359(6380):1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurent F, Girdziusaite A, Gamart J, Barozzi I, Osterwalder M, Akiyama JA, Lincoln J, Lopez-Rios J, Visel A, Zuniga A, Zeller R: Hand2 target gene regulatory networks control atrioventricular canal and cardiac valve development. Cell Rep (2017) 19(8):1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G, Xu A, Sim S, Priest JR, Tian X, Khan T, Quertermous T, Zhou B, Tsao PS, Quake SR, Wu SM: Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev Cell (2016) 39(4):491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vedantham V, Galang G, Evangelista M, Deo RC, Srivastava D: Rna sequencing of mouse sinoatrial node reveals an upstream regulatory role for islet-1 in cardiac pacemaker cells. Circ Res (2015) 116(5):797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, Bruneau BG et al. : Single-cell resolution of temporal gene expression during heart development. Dev Cell (2016) 39(4):480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLaughter DM, Christodoulou DC, Robinson JY, Seidman CE, Baldwin HS, Seidman JG, Barnett JV: Spatial transcriptional profile of the chick and mouse endocardial cushions identify novel regulators of endocardial emt in vitro. J Mol Cell Cardiol (2013) 59(196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnette DN, VandeKopple M, Wu Y, Willoughby DA, Lincoln J: Rna-seq analysis to identify novel roles of scleraxis during embryonic mouse heart valve remodeling. PLoS One (2014) 9(7):e101425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.den Hartogh SC, Wolstencroft K, Mummery CL, Passier R: A comprehensive gene expression analysis at sequential stages of in vitro cardiac differentiation from isolated mesp1-expressing-mesoderm progenitors. Sci Rep (2016) 6(19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slagle CE, Conlon FL: Emerging field of cardiomics: High-throughput investigations into transcriptional regulation of cardiovascular development and disease. Trends Genet (2016) 32(11):707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie L, Wu G, Culley DE, Scholten JC, Zhang W: Integrative analysis of transcriptomic and proteomic data: Challenges, solutions and applications. Crit Rev Biotechnol (2007) 27(2):63–75. [DOI] [PubMed] [Google Scholar]
- 21.Bonaldi T, Straub T, Cox J, Kumar C, Becker PB, Mann M: Combined use of rnai and quantitative proteomics to study gene function in drosophila. Mol Cell (2008) 31(5):762–772. [DOI] [PubMed] [Google Scholar]
- 22.Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP: Defining the consequences of genetic variation on a proteome-wide scale. Nature (2016) 534(7608):500–505.This was a system wide study quantifying proteins and transcripts on an unbiased genome wide scale in mouse liver. The authors compare RNA and protein levels as a function of genetic background.
- 23.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C: Global signatures of protein and mrna expression levels. Mol Biosyst (2009) 5(12):1512–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baxi AB, Lombard-Banek C, Moody SA, Nemes P: Proteomic characterization of the neural ectoderm fated cell clones in the xenopus laevis embryo by high-resolution mass spectrometry. ACS Chem Neurosci (2018).This paper utilized single cell proteomics to show that changes in protein composition are associated with cell fate in Xenopus.
- 25.Lombard-Banek C, Portero EP, Onjiko RM, Nemes P: New-generation mass spectrometry expands the toolbox of cell and developmental biology. Genesis (2017) 55(1-2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Chen J, Hou F, Feng Y, Zhang R: Itraq-based quantitative proteomic analysis reveals the lateral meristem developmental mechanism for branched spike development in tetraploid wheat (triticum turgidum l.). BMC Genomics (2018) 19(1):228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blundon MA, Schlesinger DR, Parthasarathy A, Smith SL, Kolev HM, Vinson DA, Kunttas-Tatli E, McCartney BM, Minden JS: Proteomic analysis reveals apc-dependent post-translational modifications and identifies a novel regulator of beta-catenin. Development (2016) 143(14):2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombard-Banek C, Reddy S, Moody SA, Nemes P: Label-free quantification of proteins in single embryonic cells with neural fate in the cleavage-stage frog (xenopus laevis) embryo using capillary electrophoresis electrospray ionization high-resolution mass spectrometry (ce-esi-hrms). Mol Cell Proteomics (2016) 15(8):2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry RA, Singh T, Kuo YM, Biester A, O’Keefe A, Lee S, Andrews AJ, O’Reilly AM: Quantitative measurement of histone tail acetylation reveals stage-specific regulation and response to environmental changes during drosophila development. Biochemistry (2016) 55(11):1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebhardt HA, Root A, Sander C, Aebersold R: Applications of targeted proteomics in systems biology and translational medicine. Proteomics (2015) 15(18):3193–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Picotti P, Aebersold R: Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat Methods (2012) 9(6):555–566. [DOI] [PubMed] [Google Scholar]
- 32.Herrington DM, Mao C, Parker SJ, Fu Z, Yu G, Chen L, Venkatraman V, Fu Y, Wang Y, Howard TD, Jun G et al. : Proteomic architecture of human coronary and aortic atherosclerosis. Circulation (2018) 137(25):2741–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho JE, Lyass A, Courchesne P, Chen G, Liu C, Yin X, Hwang SJ, Massaro JM, Larson MG, Levy D: Protein biomarkers of cardiovascular disease and mortality in the community. J Am Heart Assoc (2018) 7(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Israr MZ, Heaney LM, Suzuki T: Proteomic biomarkers of heart failure. Heart Fail Clin (2018) 14(1):93–107. [DOI] [PubMed] [Google Scholar]
- 35.Conlon FL, Miteva Y, Kaltenbrun E, Waldron L, Greco TM, Cristea IM: Immunoisolation of protein complexes from xenopus. Methods Mol Biol (2012) 917(369–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greco TM, Miteva Y, Conlon FL, Cristea IM: Complementary proteomic analysis of protein complexes. Methods Mol Biol (2012) 917(391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, Temple B, Yang XH, Wilczewski CM, Davis IJ, Cristea IM et al. : The cardiac tbx5 interactome reveals a chromatin remodeling network essential for cardiac septation. Dev Cell (2016) 36(3):262–275.This paper identified the TBX5 endogenous cardiac transctional complex uder physiological conditions using Avi-tag technologies. This studies showed that TBX5 biochemicallly and genetically interacts with the NuRD complex and demonstrated that mutations disrupting this interaction are associated with the cardaic disease Holt Oram syndrome.
- 38.van Werven FJ, Timmers HT: The use of biotin tagging in saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res (2006) 34(4):e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bayer EA, Wilchek M: The use of the avidin-biotin complex as a tool in molecular biology. Methods Biochem Anal (1980) 26(1–45. [DOI] [PubMed] [Google Scholar]
- 40.Maine GN, Li H, Zaidi IW, Basrur V, Elenitoba-Johnson KS, Burstein E: A bimolecular affinity purification method under denaturing conditions for rapid isolation of a ubiquitinated protein for mass spectrometry analysis. Nat Protoc (2010) 5(8):1447–1459. [DOI] [PubMed] [Google Scholar]
- 41.Roesli C, Neri D, Rybak JN: In vivo protein biotinylation and sample preparation for the proteomic identification of organ- and disease-specific antigens accessible from the vasculature. Nat Protoc (2006) 1(1):192–199. [DOI] [PubMed] [Google Scholar]
- 42.Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D et al. : Holt-oram syndrome is caused by mutations in tbx5, a member of the brachyury (t) gene family. Nat Genet (1997) 15(1):21–29. [DOI] [PubMed] [Google Scholar]
- 43.Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc Straceski J, Renault B et al. : Mutations in human cause limb and cardiac malformation in holt-oram syndrome. Nat Genet (1997) 15(1):30–35. [DOI] [PubMed] [Google Scholar]
- 44.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG: A murine model of holt-oram syndrome defines roles of the t-box transcription factor tbx5 in cardiogenesis and disease. Cell (2001) 106(6):709–721. [DOI] [PubMed] [Google Scholar]
- 45.Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG et al. : A molecular pathway including id2, tbx5, and nkx2–5 required for cardiac conduction system development. Cell (2007) 129(7):1365–1376. [DOI] [PubMed] [Google Scholar]
- 46.Pasumarthi KB, Field LJ: Cardiomyocyte cell cycle regulation. Circ Res (2002) 90(10):1044–1054. [DOI] [PubMed] [Google Scholar]
- 47.Poss KD, Wilson LG, Keating MT: Heart regeneration in zebrafish. Science (2002) 298(5601):2188–2190. [DOI] [PubMed] [Google Scholar]
- 48.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S et al. : Evidence for cardiomyocyte renewal in humans. Science (2009) 324(5923):98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burridge PW, Keller G, Gold JD, Wu JC: Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell (2012) 10(1):16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mordwinkin NM, Burridge PW, Wu JC: A review of human pluripotent stem cell-derived cardiomyocytes for high-throughput drug discovery, cardiotoxicity screening, and publication standards. J Cardiovasc Transl Res (2013) 6(1):22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J, Li H, Trounson A, Wu JC, Nioi P: Combining hipscs and human genetics: Major applications in drug development. Cell Stem Cell (2017) 21(2):161–165. [DOI] [PubMed] [Google Scholar]
- 52.Evans MJ, Kaufman MH: Establishment in culture of pluripotential cells from mouse embryos. Nature (1981) 292(5819):154–156. [DOI] [PubMed] [Google Scholar]
- 53.Martin GR: Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A (1981) 78(12):7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM: Embryonic stem cell lines derived from human blastocysts. Science (1998) 282(5391):1145–1147. [DOI] [PubMed] [Google Scholar]
- 55.Maltsev VA, Rohwedel J, Hescheler J, Wobus AM: Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech Dev (1993) 44(1):41–50. [DOI] [PubMed] [Google Scholar]
- 56.Kattman SJ, Huber TL, Keller GM: Multipotent flk−1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell (2006) 11(5):723–732. [DOI] [PubMed] [Google Scholar]
- 57.Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G: Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc Natl Acad Sci U S A (2005) 102(37):13170–13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wobus AM, Wallukat G, Hescheler J: Pluripotent mouse embryonic stem cells are able to differentiate into cardiomyocytes expressing chronotropic responses to adrenergic and cholinergic agents and ca2+ channel blockers. Differentiation (1991) 48(3):173–182. [DOI] [PubMed] [Google Scholar]
- 59.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ et al. : Human cardiovascular progenitor cells develop from a kdr+ embryonic-stem-cell-derived population. Nature (2008) 453(7194):524–528. [DOI] [PubMed] [Google Scholar]
- 60.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L: Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest (2001) 108(3):407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ: Human embryonic stem cells develop into multiple types of cardiac myocytes: Action potential characterization. Circ Res (2003) 93(1):32–39. [DOI] [PubMed] [Google Scholar]
- 62.Xu C, Police S, Rao N, Carpenter MK: Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res (2002) 91(6):501–508. [DOI] [PubMed] [Google Scholar]
- 63.Kaltenbrun E, Greco TM, Slagle CE, Kennedy LM, Li T, Cristea IM, Conlon FL: A gro/tle-nurd corepressor complex facilitates tbx20-dependent transcriptional repression. J Proteome Res (2013) 12(12):5395–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kennedy L, Kaltenbrun E, Greco TM, Temple B, Herring LE, Cristea IM, Conlon FL: Formation of a tbx20-casz1 protein complex is protective against dilated cardiomyopathy and critical for cardiac homeostasis. PLoS Genet (2017) 13(9):e1007011.This paper demonstrated that the TBX20 interactome is temporally regulated and that mutations disrupting an interaction with CASZ1 are associated with human cardiomyopathy.
- 65.Posch MG, Gramlich M, Sunde M, Schmitt KR, Lee SH, Richter S, Kersten A, Perrot A, Panek AN, Al Khatib IH, Nemer G et al. : A gain-of-function tbx20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. Journal of medical genetics (2010) 47(4):230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, Castro ML, Butler TL, Hyun C, Guo G, Otway R, Mackay JP et al. : Mutations in cardiac t-box factor gene tbx20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet (2007) 81(2):280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu C, Shen A, Li X, Jiao W, Zhang X, Li Z: T-box transcription factor tbx20 mutations in chinese patients with congenital heart disease. European journal of medical genetics (2008) 51(6):580–587. [DOI] [PubMed] [Google Scholar]
- 68.Qian L, Mohapatra B, Akasaka T, Liu J, Ocorr K, Towbin JA, Bodmer R: Transcription factor neuromancer/tbx20 is required for cardiac function in drosophila with implications for human heart disease. Proceedings of the National Academy of Sciences of the United States of America (2008) 105(50):19833–19838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hammer S, Toenjes M, Lange M, Fischer JJ, Dunkel I, Mebus S, Grimm CH, Hetzer R, Berger F, Sperling S: Characterization of tbx20 in human hearts and its regulation by tfap2. Journal of cellular biochemistry (2008) 104(3):1022–1033. [DOI] [PubMed] [Google Scholar]
- 70.Dorr KM, Amin NM, Kuchenbrod LM, Labiner H, Charpentier MS, Pevny LH, Wessels A, Conlon FL: Casz1 is required for cardiomyocyte g1-to-s phase progression during mammalian cardiac development. Development (Cambridge, England) (2015) 142(11):2037–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christine KS, Conlon FL: Vertebrate castor is required for differentiation of cardiac precursor cells at the ventral midline. Dev Cell (2008) 14(4):616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoon C, Song H, Yin T, Bausch-Fluck D, Frei AP, Kattman S, Dubois N, Witty AD, Hewel JA, Guo H, Emili A et al. : Fzd4 marks lateral plate mesoderm and signals with norrin to increase cardiomyocyte induction from pluripotent stem cell-derived cardiac progenitors. Stem Cell Reports (2018) 10(1):87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S: Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell (2007) 131(5):861–872. [DOI] [PubMed] [Google Scholar]
- 74.Yamanaka S: Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell (2007) 1(1):39–49. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ: Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res (2009) 104(4):e30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA et al. : Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature (2014) 510(7504):273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C et al. : Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol (2007) 25(9):1015–1024. [DOI] [PubMed] [Google Scholar]
- 78.Caspi O, Huber I, Kehat I, Habib M, Arbel G, Gepstein A, Yankelson L, Aronson D, Beyar R, Gepstein L: Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J Am Coll Cardiol (2007) 50(19):1884–1893. [DOI] [PubMed] [Google Scholar]
- 79.Cyganek L, Tiburcy M, Sekeres K, Gerstenberg K, Bohnenberger H, Lenz C, Henze S, Stauske M, Salinas G, Zimmermann WH, Hasenfuss G et al. : Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight (2018) 3(12).This paper identified the proteins present in ventricular cardiomyocytes and atrial cardiomyocytes from iPSCs providing a comprehensive understanding of the molecular and functional identities of these two cardiac subtypes. This will provide a useful dataset to chamber-specific disease modeling and drug testing.
- 80.Lee JH, Protze SI, Laksman Z, Backx PH, Keller GM: Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell (2017) 21(2):179–194 e174. [DOI] [PubMed] [Google Scholar]
- 81.Lu ZQ, Sinha A, Sharma P, Kislinger T, Gramolini AO: Proteomic analysis of human fetal atria and ventricle. J Proteome Res (2014) 13(12):5869–5878.This paper generated a quantitative proteomic atlas for human cardiac tissue harvested during fetal development. It is a useful dataset for cardiac development and congenital heart disease studies.
- 82.Doll S, Dressen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, Meier F, Deutsch MA, Lahm H, Lange R, Krane M et al. : Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun (2017) 8(1):1469.This paper generated a comprehensive protein map of the health human adult heart including the identification of proteins expressed in 16 different anatomical locations and multiple cardiac cell types. This is an extremely useful dataset for cardiac disease comparisons.
- 83.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D: Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell (2010) 142(3):375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA et al. : Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature (2012) 485(7400):599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qian L, Berry EC, Fu JD, Ieda M, Srivastava D: Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc (2013) 8(6):1204–1215. [DOI] [PubMed] [Google Scholar]
- 86.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D: In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature (2012) 485(7400):593–598.The above 4 references demonstrated that fibroblast cells can be directly reprogrammed into induced cardiomyocytes with treatment of 3 key cardiac transcription factors. Furthermore, the authors show that they can aid in cardiac injury repair.
- 87.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ: Microrna-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circulation research (2012) 110(11):1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U: A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. Journal of molecular and cellular cardiology (2012) 53(3):323–332. [DOI] [PubMed] [Google Scholar]
- 89.Chen JX, Krane M, Deutsch MA, Wang L, Rav-Acha M, Gregoire S, Engels MC, Rajarajan K, Karra R, Abel ED, Wu JC et al. : Inefficient reprogramming of fibroblasts into cardiomyocytes using gata4, mef2c, and tbx5. Circulation research (2012) 111(1):50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K, Nishiyama T et al. : Mir-133 promotes cardiac reprogramming by directly repressing snai1 and silencing fibroblast signatures. The EMBO journal (2014) 33(14):1565–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Inagawa K, Miyamoto K, Yamakawa H, Muraoka N, Sadahiro T, Umei T, Wada R, Katsumata Y, Kaneda R, Nakade K, Kurihara C et al. : Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of gata4, mef2c, and tbx5. Circulation research (2012) 111(9):1147–1156. [DOI] [PubMed] [Google Scholar]
- 92.Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD: Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. Journal of molecular and cellular cardiology (2013) 60(97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, Bursac N, Leong KW: Transcription factors myocd, srf, mesp1 and smarcd3 enhance the cardio-inducing effect of gata4, tbx5, and mef2c during direct cellular reprogramming. PloS one (2013) 8(5):e63577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, Tohyama S et al. : Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proceedings of the National Academy of Sciences of the United States of America (2013) 110(31):12667–12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN: Reprogramming of human fibroblasts toward a cardiac fate. Proceedings of the National Academy of Sciences of the United States of America (2013) 110(14):5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sauls K, Greco TM, Wang L, Zou M, Villasmil M, Qian L, Cristea IM, Conlon FL: Initiating events in direct cardiomyocyte reprogramming. Cell Rep (2018) 22(7):1913–1922.Using a quantitative mass spectrometry-based proteomic approach, the authors identified the temporal global changes in protein abundance that occur during initial phases of induced cardiomyocyte reprogramming.
- 97.Reid A, Tursun B: Transdifferentiation: Do transition states lie on the path of development? Curr Opinion in Systems Biology (2018) 11(1):18–23.This review focuses on the intermediate states that a cell passes through during transdifferentiation. An understanding of these intermediate states are crucial to direct cardiac reprogramming.
- 98.Tallquist MD: Cardiac fibroblasts: From origin to injury. Curr Opin Physiol (2018) 1(75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ivey MJ, Kuwabara JT, Pai JT, Moore RE, Sun Z, Tallquist MD: Resident fibroblast expansion during cardiac growth and remodeling. J Mol Cell Cardiol (2018) 114(161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tallquist MD, Molkentin JD: Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol (2017) 14(8):484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amin NM, Greco TM, Kuchenbrod LM, Rigney MM, Chung MI, Wallingford JB, Cristea IM, Conlon FL: Proteomic profiling of cardiac tissue by isolation of nuclei tagged in specific cell types (intact). Development (Cambridge, England) (2014) 141(4):962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]