Abstract
The discovery of bacteria in the female urinary bladder has fundamentally changed current dogma regarding the urinary tract and related urinary disorders. While prior research has characterized many of the bacterial components of the female urinary tract, the viral fraction of this community is largely unknown. Viruses within the human microbiota far outnumber bacterial cells, with the most abundant viruses being those that infect bacteria (bacteriophages). Similar to observations within the microbial communities (microbiota) of the gut and oral cavity, preliminary surveys of the urinary tract and bladder microbiota indicate a rich diversity of uncharacterized bacteriophage (phage) species. Phages are vital members of microbiota, playing critical roles in shaping bacterial metabolism and community structure. Here, we review the current knowledge of phages within the urinary tract and their possible contribution to urinary tract health. Furthermore, as the natural predators of bacteria, phages have garnered renewed interest in their use as antimicrobial agents. Both historical and recent applications of phage therapy for lower urinary tract infections and other disorders are discussed.
The Bladder Microbiome
The Human Microbiome Project (HMP) revolutionized our understanding of bacteria that inhabit the human body 1. The bladder, however, was not included in the HMP publications, which focused on the oral cavity, nasal cavity, skin, gastrointestinal tract, and vagina 1,2. In the absence of a urinary tract infection (UTI), it was long thought that the bladder was sterile 3. This dogma resulted, in part, from the wide use of standard clinical microbiology urine culture protocols, which are designed to detect common fast-growing pathogens with basic nutrient needs and no aversion to oxygen (especially Escherichia coli). Thus, the standard protocol does not detect anaerobes, slow growers, or bacteria with complex needs. Using an enhanced urine culture method called expanded quantitative urine culture (EQUC) and/or DNA sequencing methods, diverse bacterial and fungal species have been detected in standard culture-negative urine obtained directly from the bladder via transurethral catheterization (herein catheterization) or suprapubic aspiration 4–12 (for a review, see Whiteside et al. 13). These and other studies of the bladder microbiome and microbiota have revealed associations of bladder bacteria with post-operative UTIs 14,15, urgency urinary incontinence (UUI) 8,9,11, and response to overactive bladder treatment 16. Some bacteria are even associated with the lack of symptoms and protection against post-instrumentation UTI 8–10. These results suggest that the bladder may possess its own protective microbiota and that dysbiosis results in disorders, e.g. UTI and UUI (for recent reviews, see Brubaker & Wolfe 17; Mueller et al. 18). A recent effort to generate a genomic catalogue of bacteria isolated from the bladder has revealed that the genomes of bladder bacteria are quite distinct from bacteria isolated from the gut, but somewhat similar to those of the vagina 19.
Viruses and the Human Microbiome Project
Although the HMP focused on characterizing the bacterial fraction of the human microbiota, sequencing of some viral genomes was unavoidable, both because viral DNA was present in the samples and because prophage DNA was present within the bacteria 20. Subsequent to the original initiative, bacterial and viral communities within the five niches studied in the HMP have been extensively investigated (for a review, see Lloyd-Price et al. 21), most notably the communities inhabiting the gastrointestinal tract. These viral communities include both eukaryotic viruses as well as viruses that infect bacteria or bacteriophage (phage). The gut virome (viral component of the microbiome) has been the focus of numerous studies 22–27 (for a review, see Manrique et al. 28), each leading to the same conclusion: phages are key members of the gut’s microbiota. Within the gut of healthy individuals, there exists a core phage community 27 and disruption of this core phage community (dysbiosis) has been associated with certain GI symptoms and disease 24,26–29. Most recently, seven phage taxa within the gut were found to be associated with type 2 diabetes, establishing a type 2 diabetes-specific gut phage community 30. Other studies have characterized the viromes of the other body sites from the HMP 31–36; the data from these studies are publicly available (Table 1). Like the gut, studies of these other body sites have identified associations between phage communities and patient symptoms/disease. For example, phage within the oral cavity have been linked to periodontitis 33. In contrast with the sites of the HMP, investigation of the phage communities has only recently begun.
Table 1.
Current number of viral sequences from virome studies of HMP anatomical sites (as of 4/2018). Clusters corresponds to genetically distinct groups. [Data retrieved from the IMG/VR system 90.]
| Airway | GI Tract | Oral | Skin | Urogenital tract/ vagina |
|
|---|---|---|---|---|---|
| Total # of Metagenomic Viral Contigs | 268 | 29107 | 48904 | 2461 | 422 |
| Unique Viral Clusters | 41 | 3465 | 5246 | 200 | 67 |
| Total # of Genomes | 107 | 510 | 886 | 491 | 101 |
Bacteriophages
Phages are ubiquitous viruses that infect bacteria; they are the most abundant biological entities, far exceeding even bacteria. Surveys of the marine environment have led to an estimated 1030 phages in the oceans alone, representing roughly 10 phages for each bacterial cell 37. In addition to their abundance within the marine environment 38, phages are prevalent within the soil 39,40 and freshwater 41. They have even been isolated from some of the most inhospitable conditions, including desert sands 42, sea ice 43, and the depths of the ocean 44. In addition to our surroundings, phages are abundant in and on plants 45 and within the bodies of insects and mammals 46,47. Indeed, the human gut alone is home to an estimated two trillion phages 48, greatly exceeding the number of eukaryotic viruses in our bodies 49. Although phages are unable to infect eukaryotic cells, there is evidence of direct interactions between phages and human cells 50,51. However, the full extent of direct and indirect effects of phages can only be imagined, as researchers have only just begun to explore the roles of phages in the human body, including the human immune and central nervous systems (for a review, see Barr 48).
There are three distinct, generally well-characterized life cycles for phage propagation and reproduction: lytic, lysogenic and chronic. All phages infect their bacterial host by binding to surface receptors, a process called adsorption (Fig. 1); these receptor binding proteins are often quite specific, leading to a narrow range of hosts (strains or species) that a particular phage can infect (see discussion in Koskella & Meaden 52). Following adsorption to the host cell, the phage injects its DNA or RNA genome into the host’s cytoplasm. In the lytic cycle, the phage genome replicates and phage proteins are synthesized. For dsDNA phages, DNA is inserted into the protein procapsid, while for ssDNA and ssRNA phages, the capsid is formed around the nucleic acid (Fig. 1). The bacterium’s cell wall breaks (“bursts”) (for a review, see Young 53), and the phage progeny disperse into the surrounding environment. While some phages are obligately lytic, others, called temperate, can alternate between the lysogenic and lytic cycles. In the lysogenic cycle, the phage genome is either integrated into the host genome or remains in the cytoplasm as a self-replicating plasmid 54 (Fig. 1). The phage genome (now called a prophage) generally replicates in synchrony with the host chromosome. Temperate phages, such as the model phage λ, are capable of going through the lytic and lysogenic cycles. Their prophages can remain dormant for generations until, often, an environmental cue, such as host starvation, change in nutrients, temperature 54,55, triggers entry into the lytic cycle – a process known as induction. This decision between the lysogenic and lytic cycle can also be determined by the recently reported phage-produced peptide communication system (the ‘arbitrium’ system), in which progeny phage lysogenize when this small-molecule is abundant within the environment 56. In addition to the lytic and lysogenic cycles, phages can reproduce by chronic infection; phages are shed from the bacterial cell without killing the host cell, e.g., the filamentous phage M13. While the majority of known phages can be associated with one of these three life cycles, additional modes of infection and reproduction have been described 55,57,58.
Fig. 1.
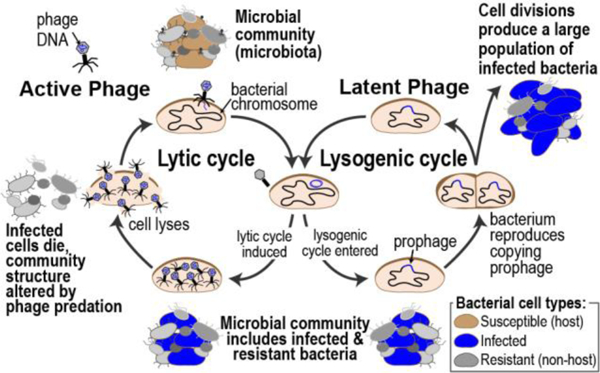
Lytic and lysogenic cycles of phages and their impact on microbiota
Given these multiple mechanisms of infection and persistence, it comes as no surprise that phages can have profound effects on microbial communities (Fig. 1). Phages can transform the microbial community through predation (lysis) 59–61. Furthermore, phages can drive bacterial diversity within a community 62–65, including adaptation in susceptible host species, e.g. loci associated with phage-resistance 66,67. Exposure to temperate phages can increase bacterial virulence (a process referred to as lysogenic conversion) 68,69 by, for example, encoding for toxins 70,71. Case reports detail shiga toxin (Stx)-producing E. coli strains, most commonly associated with enteric infections, found within the urine of individuals with UTIs 72–74. Thus, lysogeny can be beneficial for the bacterial host (for a review, see Harrison & Brockhurst 75). Because they integrate their genome into their host’s genome, some temperate phages can transfer genetic material from one cell to another (transduction); this process can benefit the recipient host cell. Indeed, temperate phages are well known to mediate horizontal gene transfer (HGT) and have helped spread virulence and/or resistance factors through bacterial communities (for a review, see Touchon et al. 76). Similarly, lytic phages can also transfer bacterial DNA 77. Data exists that support both frequent and infrequent phage-mediated spread of antibiotic resistance genes 78–81. Phage also can contribute to HGT indirectly; a recent study identified two superspreader phages 82. In this scenario, phage lysis spreads intact host plasmids, enabling HGT via transformation. Given the large genetic diversity present within phage communities 83, we have only begun to scratch the surface of the complexities of phage-host dynamics 84–86 (for a review, see Manrique et al. 28).
As phage genomes can be dsDNA, ssDNA, dsRNA, or ssRNA, linear or circular, and even segmented, sequencing phage populations often is limited by the genomic nucleic acid extraction protocol and may necessitate amplification prior to sequencing (for a review, see Hayes et al. 87). Nevertheless, whole genome sequencing technologies have enabled researchers to identify new phage species. In contrast to cellular organisms, there is no universal marker for phages because no gene is conserved within all phages. To identify phages, researchers often target genes that encode structural proteins as phylogenetic markers 88. These signature sequences, however, are far from comprehensive 89. Only a small fraction of phage sequence diversity is represented in extant sequence databases, and it is heavily biased for sequences of phages with DNA genomes that infect bacterial species routinely studied in the laboratory 84,90,91. Metagenomics has allowed us to explore the diversity of phages on Earth. Later in this review, we will take a closer look into the viral metagenomic studies of the lower urinary tract.
Viruses of the Urinary Tract
The urinary tract harbors a diverse eukaryotic viral community, including Adenoviruses, Anelloviruses, Papillomaviruses, and Polyomaviruses. Adenoviruseses can be detected in urine 92, and can range from limited, localized infections in otherwise healthy individuals to severe and potentially fatal infections in immunocompromised individuals (for a review, see Lion 93). Torque teno virus (TTV), also referred to as small anellovirus, has largely been studied in relation to immunodeficiency in transplant recipients (for a review, see Tan et al. 94). In the study of Rani et al. 95, mid-stream clean catch urine was collected from 22 kidney transplant recipients; whole genome sequencing was conducted for these urinary viromes and 108 different types of TTV were detected. The most prevalent eukaryotic virus in urine samples is human polyomavirus 1 (BK virus) and 2 (JC virus) 96. While both polyomaviruses appear to have little effect on healthy individuals, each can lead to nephropathy and hemorrhagic cystitis in immunocompromised populations (for a review, see Hirsch et al. 97 and Rinaldo et al. 98). Recently, we conducted metagenomic sequencing of the bladder microbiome (bacterial and viral fraction) of 30 individuals and were able to reconstruct the full JC virus genome in five of these samples 99. JC virus and other polyomaviruses have similarly been detected in other viromes from urine samples (obtained by an undescribed voided urine collection method) 100. Human papillomaviruses (HPV) also have been detected in voided urine 101 and bladder tissue 102,103. While certain HPV genotypes have been attributed to condylomata acuminatum of the bladder 104,105, these high-risk genotypes are rare. In one investigation of the urinary virome, 95% of the 20 subjects sampled included HPV sequences 106; for eight patients, these samples were collected via intermittent catheterization, and the others via an undescribed voided urine collection method. It is, however, estimated that eukaryotic viruses represent just a small fraction of the urinary virome 99,100,106,107.
Lytic phages have been isolated directly from urine on numerous occasions. In fact, the seminal work of the co-discoverer of bacteriophages, Félix d’ Hérelle, first isolated a phage from urine. While d’ Hérelle may not have known exactly what a phage was, this invisible microbe lysed the Shiga bacillus 108. A century later, two studies isolated phages capable of infecting Pseudomonas aeruginosa from urine samples 109,110. Transmission Electron Microscopy (TEM) of the isolated phages provided clues into the phages’ morphology, which includes a tailed siphophage 109 and two tailless phages 110. Our group has isolated a fourth Pseudomonas-infecting phage, induced from a bacterial isolate from a urine sample collected via catheterization (Johnson et al., in preparation). This phage is capable of lysing P. aeruginosa PAO1. Coliphages, or phages that infect E. coli, also have been isolated from voided and catheterized urine samples. From the routine plating of a patient’s urine sample (collection method unknown), Dallas & Kingsbery 111 found 100,000 CFU/ml of bacterial growth and, upon closer inspection, plaques. Four coliphages were isolated from clinical urine samples and their morphologies were determined (via TEM) to be siphophages 109. We isolated an additional seven coliphages from the bladder (from urine collected via catheterization) of four women with UUI 112. From the complete genomes of these seven coliphages, six resemble coliphages isolated from cattle slurry 113, while the sequence of the seventh (phage Wrath) most closely resembles a lysogenic Bacillus phage sequence. TEM images, including that in Fig. 2 for the coliphage Greed, also suggests the morphology of siphophages. The host-range of Greed was recently tested 114; in addition to its ability to lyse the laboratory strains E. coli C and K-12, it is also capable of infecting and lysing some E. coli strains isolated from urine samples, including the uropathogen E. coli CFT073.
Fig. 2.
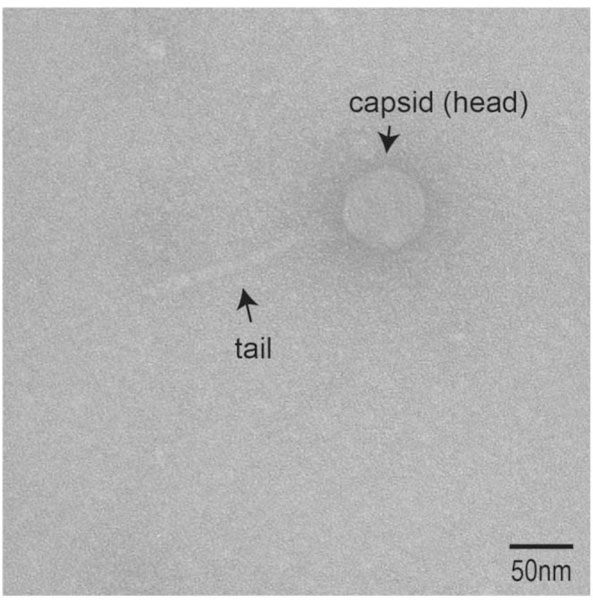
Bacteriophage Greed, isolated from catheterized urine microbiome sample.
The lytic phage population, however, includes but one part of the phage community within the bladder; recent evidence suggests that lysogenic phages are dominant within the gut microbial community 115. Within E. coli isolates from the bladder, several prophages have been identified 116. Although a lytic Gardnerella-infecting phage has yet to be isolated, numerous prophage sequences were identified within the genomes of four Gardnerella strains isolated from urine specimens obtained via catheterization from the bladders of adult women with UUI 117. Analysis of these four genomes and other publicly available Gardnerella genomes revealed that phage infections were pervasive within the urinary microbiota 117. We next expanded this examination of lysogenic phages to include 181 bacterial isolates, representative of the phylogenetic diversity within the bladder 118. These samples were collected from women both with and without lower urinary tract symptoms. Over 400 phage sequences were identified; the majority (86%) of these bacterial isolates harbored one or more lysogenic phages 118. Furthermore, many (57%) of the phages identified in this study 118 exhibited no sequence similarity to any known phages, indicative of a vast unexplored phage population residing in the bladder.
To date, three published studies have employed a metagenomic approach to sequence the viral fraction of the urinary microbiota. The first study, conducted by Santiago-Rodriguez et al. 106, sought to determine whether the urine viral community was affected by urinary tract health status. The viral fraction (eukaryotic viruses and extracellular phages) of urine collected from 10 individuals with and 10 individuals without a diagnosed UTI were sequenced. For each cohort, samples were collected either via catheterization or voided urine from five males and five females. Only 27% were homologous to known viruses, the majority of which represented phage genes 106, again hinting at a large unexplored phage community within the urinary tract. While the subsequent study of Thannesberger et al. 100 also found a large phage community, they concluded that the phage community primarily consisted of relatives of known species, the majority resembling Chlamydiamicroviruses, which infect Chlamydia spp. This study included two healthy individuals and four individuals with human cytomegalovirus (CMV) infections. However, information about how the urine was collected or demographics for the patients was not provided. The third published virome study sequenced urine samples collected (via the voided mid-stream clean catch method) from 14 male and eight female kidney transplant patients 95. While phages are likely present in these samples, this study did not mention phages and sequence data is not publicly available; instead, the authors focused solely on eukaryotic viruses.
Viral diversity within the urinary tract also has been studied by sequencing the entire urinary microbiome. Moustafa et al. 107 recently published a study in which metagenomic sequencing was performed for the urinary microbiome of 49 individuals (collected via clean-catch). As this study did not select for the viral fraction, most of the sequenced data corresponded to bacterial genetic material. Nevertheless, viral – primarily phage – sequences were detected 107. Like the study conducted by Santiago-Rodriguez et al. 106, this study also examined samples from individuals with UTIs and detected sequences homologous to those from phages that infect bacteria commonly found within the urinary tract and associated with UTIs, including Escherichia, Enterococcus, Lactobacillus and Pseudomonas 107. We recently took a similar approach, sequencing urine collected by catheterization from 10 asymptomatic women and 20 women with overactive bladder. Partial and complete viral genomes were reconstructed in 12 of the 30 samples sequenced, including the complete genomes of novel phage strains 99. Partial and complete phage genomes also exhibited sequence homology to previously characterized lytic or lysogenic phages that infect Gardnerella, Lactobacillus, and Streptococcus spp. In sequencing both the bacterial and viral members of the microbiota, associations between phages and their hosts can be inferred.
Challenges of Studying Phages in the Bladder
In contrast to the microbiota of the GI tract, oral cavity or vagina, the bladder has orders of magnitude less microbial biomass 7,119,120. DNA concentrations are often low, a challenge faced by both those studying the bacterial and viral constituents of the bladder 121. Thus, two of the metagenomic studies of the urinary virome employed amplification prior to sequencing 100,106. While efficient in increasing viral genomic material, these amplification methods have well documented biases 122. Perhaps of greater concern is the method in which urine is collected and the community the collected urine represents. In a previous work 5, the bacterial communities obtained via catheterization and suprapubic aspiration from women were compared, finding that both methods: (1) did not resemble the skin or vaginal microbiomes and (2) successfully avoided vulvo-vaginal contaminants. The method of urine collection is a frequently debated and investigated topic in the field 17,121,123, balancing invasive procedures during collection and the purity of the sample obtained. This debate is not unique to the bladder, urinary tract, or urine; biopsies and stool samples present quantitatively and qualitatively different stories for the gut 124 and methods for sampling the gut microbiota are still being refined 125,126. Studies of voided urine have routinely observed vaginal contamination of clean-catch samples 12,107. The virome studies of Santiago-Rodriguez et al. 106, Rani et al. 95, and Moustafa et al. 107 investigated voided urines either partially or entirely (in the case of the last two studies). Thus, it remains unknown if the viruses detected reside in the bladder and/or in/on the urethra, vagina, or skin. In contrast, catheterized urines have a lower probability of contaminants 127. Study of less invasive methods for collection is an ongoing pursuit 128. Until a less invasive method can be determined, investigations of the bladder virome should use urine obtained by catheterization or suprapubic aspirate.
Phages and Urinary Tract Health
Associations between phage communities, bacterial populations, and the human host are open questions within the field. Recent studies suggest that phages may contribute to human health 129, in particular the gut 24,26–29. These studies of the gut will likely illuminate studies of the bladder and other niches of the human body. Evidence has emerged recently suggesting a similar association between phage communities and bladder symptoms. We observed variation in the abundance of lysogenic phages in bacteria isolated from asymptomatic individuals and those with overactive bladder 118. Variation, determined via the beta diversity statistic and principle component analysis, was not detected in the extracellular phage populations of individuals with and without UTI symptoms 106. However, it is important to note that, in contrast to the gut phage communities, our understanding of the diversity of phages within the urinary tract has only just begun. Cataloguing the phage community in both asymptomatic and symptomatic individuals is a critical first step in understanding if and how phages contribute to urinary tract health. Further investigation of the phage-bacteria dynamics of the bladder and urinary tract could reveal indicators for early detection of symptoms.
Phages also may offer a defense to the human host. Studies of the gut communities have revealed unexpected ways in which phages interact with human cells, organs, and immune system. The prevalence of phages within the mucosal surfaces of the gut may confer a direct benefit to the human host, protecting the epithelium from bacteria 130. Recent evidence suggests that phages are in fact more virulent to bacteria when human cells are present 131. Furthermore, phages can interact directly with human cells. Studies have found that phages can bind to cancer cell membranes and inhibit or attenuate tumor growth 132. Although phages cannot infect eukaryotic cells, they can enter eukaryotic cells. A phage may be a stow-away, as a cell of an invasive bacterial species that harbors a phage could enter a eukaryotic cell 133,134. Alternatively, eukaryotic cells can take up free phages by endocytosis 133. A recent study showed that phages are capable of penetrating epithelial cell layers with an estimated 31 billion phage particles passing through these layers of the gut into the body daily 51. Within the human body, phages can modulate immune responses (for a review, see Górski et al. 135). Evidence includes phage-mediated inhibition of in vitro T-cell proliferation 50 and stimulation of humoral responses in mice 136. While the future of phage-mediated immunoregulation holds promise, the mechanisms by which phages interact with the immune system remains largely unknown.
There is growing appreciation of the therapeutic potential for modulating the human microbiome. Induction and release of temperate phages can lyse sensitive competitor strains or lysogenize other cells 137,138. Alternatively, an individual’s bacterial infection can be treated with obligately lytic phages, known as phage therapy. In the face of the increasing threat of antibiotic-resistant bacterial strains, phage therapy has regained interest (for a recent perspective, see Lin et al. 139). Phage therapy was a promising area of UTI treatment in the early 20th century. For instance, in a 1928 report, phages isolated from sewage were 90% efficient in lysing bacteria causing UTIs 140. While the US and Western Europe abandoned phage therapy when antibiotics became commercially available (amongst other reasons) 141, this area of research and treatment continued in Eastern European countries. Phage therapy is a publicly available treatment for individuals with UTIs in these countries. Within the scientific literature, in one study 142,41 E. coli and 9 Klebsiella pneumoniae strains isolated from individuals with UTIs were challenged with phages from collections from the Democratic Republic of Georgia. Only one E. coli isolate was resistant to the individual phages and phage cocktails tested, and one phage was capable of lysing all K. pneumoniae strains. Similar efficiencies have been observed for other bacterial species that cause UTI symptoms. A single patient, for whom antibiotics were unable to clear the root cause of the UTI -P. aeruginosa, was successfully treated with phages 143. A 2-year clinical trial recently concluded, and while results are not yet published, patients with UTIs were treated with either a bacteriophage, an antibiotic, or a placebo 144 (https://clinicaltrials.gov/ct2/show/NCT03140085). Phages have also been explored for their potential use in treating long-term catheters to minimize bacterial biofilm development and catheter blockage, which can cause catheter-associated UTIs (CAUTIs) (for a review, see Siddiq & Darouiche 145) in addition to chronic bacterial prostatitis (for review, see Letkiewicz et al. 146). Nevertheless, a better understanding of phage, microbiota, and human host interactions is imperative for the feasibility of phage therapy in Western medicine.
Conclusions
While whole genome sequencing and new enhanced culture methods have greatly benefited the study of the microorganisms within the bladder and the rest of the lower urinary tract, significant work remains. Although the literature reflects an ongoing debate surrounding the potential presence of vulvo-vaginal and/or skin bacterial contaminants in urine samples, here we begin the same conversation relative to the study of the virome. Most of the studies discussed here, with a few exceptions 106,112,117,118, have used voided urine for isolation of lytic phages or sequencing of the urinary virome. To the best of our knowledge, the phage populations of adjacent anatomical locations have yet to be investigated. As we have just begun to explore the phage communities within the urinary tract, such considerations must be kept in mind. More samples must be sequenced to determine if, like in the gut 27, a core phage community exists within the bladder, the urethra, peri-urethral and adjacent urogenital niches. Only through such efforts can we fully ascertain what a healthy and unhealthy community looks like. Is a shift from a lysogenic to a lytic lifestyle a cause or consequence of bacterial community dysbiosis or urinary symptoms? As sequencing costs continue to decline, studies such as that by Moustafa et al. 107 and Garretto et al. 99 are particularly powerful in capturing the dynamics between phages and their hosts.
Knowledge of the phage communities within the lower urinary tract and their role in urinary tract health is a vital first step in the development of new strategies to treat urinary symptoms and infections. Phage therapy has the potential to combat antibiotic resistant bacterial infections and other maladies, and anecdotal evidence of its success certainly warrants further investigation 147. Phage/drug cocktails are promising as well; for instance, such a cocktail was recently used to clear a vascular graft P. aeruginosa infection 148. Critical to effective and reliable phage therapy strategies, however, is the understanding of the extant beneficial microbiota. Phage therapies should ideally cause minimal to no disturbance of this community. In contrast to broad-spectrum antibiotics, phages can be directed very narrowly toward a specific bad player within the community. Given the observed novelty of many of the phages sequenced from urine and from the bladder 106,118, perhaps the genomes of the gate-keepers of urinary tract health have already been sequenced. Our understanding of the phage population of the urinary tract is in its infancy and future studies will likely open new areas of investigation.
Acknowledgements
This work was supported by the NIH (R01 DK104718 to A.J.W). A.G. is supported by the Carbon Research Fellowship at Loyola University Chicago. T.M.-E. was supported by a Loyola University Chicago Interdisciplinary Research Fellowship. We would like to thank Dr. Jason Shapiro for critical reading of the manuscript and assistance with generating the TEM image.
References
- 1.Huttenhower C et al. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aagaard K et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. The FASEB Journal 27, 1012–1022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas-White K, Brady M, Wolfe AJ & Mueller ER The bladder is not sterile: History and current discoveries on the urinary microbiome. Curr Bladder Dysfunct Rep 11, 18–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouts DE et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med 10, 174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfe AJ et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 50, 1376–1383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker L et al. Urinary bacteria in adult women with urgency urinary incontinence. Int Urogynecol J 25, 1179–1184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilt EE et al. Urine is not sterile: Use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 52, 871–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce MM et al. The female urinary microbiome: A comparison of women with and without urgency urinary incontinence. mBio 5, e01283–14-e01283–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce MM et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol 213, 347.e1–347.e11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas-White K, Fok C, Mueller ER, Wolfe AJ & Brubaker L Pre-operative urinary microbiome reveals post-operative urinary tract infection risk. Neurourol Urodynam 34, S21–S22 (2015). [Google Scholar]
- 11.Karstens L et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol 6, 78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ackerman AL & Underhill DM The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann Transl Med 5, 31–31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whiteside SA, Razvi H, Dave S, Reid G & Burton JP The microbiome of the urinary tract—a role beyond infection. Nat Rev Urol 12, 81–90 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Fok CS et al. Day of surgery urine cultures identify urogynecologic patients at increased risk for postoperative urinary tract infection. J Urol 189, 1721–1724 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Nienhouse V et al. Interplay between bladder microbiota and urinary antimicrobial peptides: Mechanisms for human urinary tract infection risk and symptom severity. PLoS ONE 9, e114185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas-White KJ et al. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J 27, 723–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brubaker L & Wolfe AJ Microbiota in 2016: Associating infection and incontinence with the female urinary microbiota. Nat Rev Urol 14, 72–74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller ER, Wolfe AJ & Brubaker L Female urinary microbiota. Curr Opin Urol 27, 282–286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas-White K Culturing of female bladder bacteria reveals an interconnected urogenital microbiome. Nat Commun 9, 8350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wylie KM et al. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol 12, 71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd-Price J, Abu-Ali G & Huttenhower C The healthy human microbiome. Genome Med 8, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minot S et al. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A 110, 12450–12455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minot S et al. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Res 21, 1616–1625 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norman JM et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim ES et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21, 1228–1234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogilvie LA & Jones BV The human gut virome: a multifaceted majority. Front Microbiol 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manrique P et al. Healthy human gut phageome. Proc Natl Acad Sci U S A 113, 10400–10405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manrique P, Dills M & Young M The human gut phage community and its implications for health and disease. Viruses 9, 141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner J et al. Bacteriophages in gut samples from pediatric Crohn’s Disease patients: Metagenomic analysis using 454 Pyrosequencing. Inflamm Bowel Dis 19, 1598–1608 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Ma Y, You X, Mai G, Tokuyasu T & Liu C A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome 6, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foulongne V et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS ONE 7, e38499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pride DT et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J 6, 915–926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ly M et al. Altered oral viral ecology in association with periodontal disease. MBio 5, e01133–01114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannigan GD et al. The human skin double-stranded DNA virome: topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome. MBio 6, e01578–01515 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannigan GD et al. Evolutionary and functional implications of hypervariable loci within the skin virome. PeerJ 5, e2959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pérez-Brocal V & Moya A The analysis of the oral DNA virome reveals which viruses are widespread and rare among healthy young adults in Valencia (Spain). PLoS ONE 13, e0191867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chibani-Chennoufi S, Bruttin A, Dillmann M-L & Brussow H Phage-Host Interaction: An Ecological Perspective. J Bacteriol 186, 3677–3686 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breitbart M, Bonnain C, Malki K & Sawaya NA Phage puppet masters of the marine microbial realm. Nat Microbiol 3, 754–766 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Williamson KE, Radosevich M & Wommack KE Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol 71, 3119–3125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson KE, Fuhrmann JJ, Wommack KE & Radosevich M Viruses in soil ecosystems: An unknown quantity within an unexplored territory. Annu Rev Virol 4, 201–219 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Bruder K et al. Freshwater Metaviromics and Bacteriophages: A Current Assessment of the State of the Art in Relation to Bioinformatic Challenges. Evol Bioinform Online 12, 25–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prestel E, Regeard C, Salamitou S, Neveu J & Dubow MS The bacteria and bacteriophages from a Mesquite Flats site of the Death Valley desert. Antonie Van Leeuwenhoek 103, 1329–1341 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Luhtanen A-M et al. Isolation and characterization of phage–host systems from the Baltic Sea ice. Extremophiles 18, 121–130 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Nigro OD et al. Viruses in the oceanic basement. mBio 8, e02129–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koskella B & Parr N The evolution of bacterial resistance against bacteriophages in the horse chestnut phyllosphere is general across both space and time. Philos Trans R Soc Lond B Biol Sci 370, 20140297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pramono AK et al. Discovery and complete genome sequence of a bacteriophage from an obligate intracellular symbiont of a cellulolytic protist in the termite gut. Microbes Environ 32, 112–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreno PS et al. Characterisation of the canine faecal virome in healthy dogs and dogs with acute diarrhoea using shotgun metagenomics. PLoS ONE 12, e0178433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barr JJ A bacteriophages journey through the human body. Immunol Rev 279, 106–122 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Navarro F & Muniesa M Phages in the human body. Front Microbiol 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Górski A et al. Bacteriophage translocation. FEMS Immunol Med Microbiol 46, 313–319 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Nguyen S et al. Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. mBio 8, e01874–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koskella B & Meaden S Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young R Phage lysis: Three steps, three choices, one outcome. J Microbiol 52, 243–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Little J Lysogeny, Prophage Induction, and Lysogenic Conversion. in Phages Their Role in Bacterial Pathogenesis and Biotechnology (eds. Waldor M, Friedman D & Adhya S) 37–54 (ASM Press, 2005). [Google Scholar]
- 55.Abedon ST Bacteriophage ecology: population growth, evolution, and impact of bacterial viruses. (Cambridge University Press, 2008). [Google Scholar]
- 56.Erez Z et al. Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calendar R The bacteriophages. (Oxford University Press, 2006). [Google Scholar]
- 58.Hobbs Z & Abedon ST Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic’. FEMS Microbiol Lett 363, (2016). [DOI] [PubMed] [Google Scholar]
- 59.Fuhrman JA Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Suttle CA Viruses in the sea. Nature 437, 356–361 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Clokie MRJ, Millard AD, Letarov AV & Heaphy S Phages in nature. Bacteriophage 1, 31–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buckling A & Rainey PB Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci 269, 931–936 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodriguez-Valera F et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7, 828–836 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Koskella B & Brockhurst MA Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev 38, 916–931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tellier A, Moreno-Gámez S & Stephan W Speed of adaptation and genomic footprints of host-parasite coevolution under arms race and trench warfare dynamics. Evolution 68, 2211–2224 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Braga LPP, Soucy SM, Amgarten DE, da Silva AM & Setubal JC Bacterial diversification in the light of the interactions with phages: The Genetic Symbionts and Their Role in Ecological Speciation. Front Ecol Evol 6, (2018). [Google Scholar]
- 67.Scanlan PD Bacteria–Bacteriophage Coevolution in the Human Gut: Implications for Microbial Diversity and Functionality. Trends Microbiol 25, 614–623 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Hosseinidoust Z, van de Ven TGM & Tufenkji N Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl Environ Microbiol 79, 6110–6116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wendling CC et al. Tripartite species interaction: eukaryotic hosts suffer more from phage susceptible than from phage resistant bacteria. BMC Evol Biol 17, 98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner PL & Waldor MK Bacteriophage control of bacterial virulence. Infect Immun 70, 3985–3993 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casas V & Maloy S Role of bacteriophage-encoded exotoxins in the evolution of bacterial pathogens. Future Microbiol 6, 1461–1473 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Starr M et al. Hemolytic-uremic syndrome following urinary tract infection with enterohemorrhagic Escherichia coli: case report and review. Clin Infect Dis 27, 310–315 (1998). [DOI] [PubMed] [Google Scholar]
- 73.Gadea M et al. Two cases of urinary tract infection caused by Shiga toxin-producing Escherichia coli O157:H7 strains. Rev Argent Microbiol 44, 94–96 (2012). [PubMed] [Google Scholar]
- 74.Toval F et al. Characterization of urinary tract infection-associated shiga toxin-producing Escherichia coli. Infect Immun 82, 4631–4642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harrison E & Brockhurst MA Ecological and evolutionary benefits of temperate phage: What does or doesn’t kill you makes you stronger. BioEssays 39, 1700112 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Touchon M, Moura de Sousa JA & Rocha EP Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr Opin Microbiol 38, 66–73 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Drexler H Transduction by bacteriophage T1. Proc Natl Acad Sci USA 66, 1083–1088 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Modi SR, Lee HH, Spina CS & Collins JJ Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abeles SR, Ly M, Santiago-Rodriguez TM & Pride DT Effects of Long Term Antibiotic Therapy on Human Oral and Fecal Viromes. PLoS ONE 10, e0134941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Enault F et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J 11, 237–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lood R, Ertürk G & Mattiasson B Revisiting antibiotic resistance spreading in wastewater treatment plants-Bacteriophages as a much neglected potential transmission vehicle. Front Microbiol 8, 2298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keen EC et al. Novel “Superspreader” bacteriophages promote horizontal gene transfer by transformation. mBio 8, e02115–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hatfull GF Dark matter of the biosphere: The amazing world of bacteriophage diversity. J Virol 89, 8107–8110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roux S, Hallam SJ, Woyke T & Sullivan MB Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J, Gao Y & Zhao F Phage-bacteria interaction network in human oral microbiome: Human oral virome. Environ Microbiol 18, 2143–2158 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Shapiro JW & Putonti C Gene Co-occurrence Networks Reflect Bacteriophage Ecology and Evolution. mBio 9, e01870–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hayes S, Mahony J, Nauta A & van Sinderen D Metagenomic approaches to assess bacteriophages in various environmental niches. Viruses 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kristensen DM et al. Orthologous gene clusters and taxon signature genes for viruses of prokaryotes. J Bacteriol 195, 941–950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Adriaenssens EM & Cowan DA Using signature genes as tools to assess environmental viral ecology and diversity. Appl Environ Microbiol 80, 4470–4480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paez-Espino D et al. IMG/VR: a database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res 45, D457–D465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paez-Espino D et al. Uncovering Earth’s virome. Nature 536, 425–430 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Echavarria M, Forman M, Ticehurst J, Dumler JS & Charache P PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J Clin Microbiol 36, 3323–3326 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lion T Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27, 441–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tan SK, Relman DA & Pinsky BA The human virome: Implications for clinical practice in transplantation medicine. J Clin Microbiol 55, 2884–2893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rani A et al. A diverse virome in kidney transplant patients contains multiple viral subtypes with distinct polymorphisms. Sci Rep 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang H et al. High incidence of JC viruria in JC-seropositive older individuals. J Neurovirol 8, 447–451 (2002). [DOI] [PubMed] [Google Scholar]
- 97.Hirsch HH, Kardas P, Kranz D & Leboeuf C The human JC polyomavirus (JCPyV): virological background and clinical implications. APMIS 121, 685–727 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Rinaldo CH, Tylden GD & Sharma BN The human polyomavirus BK (BKPyV): virological background and clinical implications. APMIS 121, 728–745 (2013). [DOI] [PubMed] [Google Scholar]
- 99.Garretto A, Thomas-White K, Wolfe AJ & Putonti C Detecting viral genomes in the female urinary microbiome. J Gen Virol 99, 1141–1146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thannesberger J et al. Viruses comprise an extensive pool of mobile genetic elements in eukaryote cell cultures and human clinical samples. The FASEB J 31, 1987–2000 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Tshomo U et al. Evaluation of the performance of Human Papillomavirus testing in paired urine and clinician-collected cervical samples among women aged over 30 years in Bhutan. Virol J 14, 74 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iwasawa A et al. Presence of human papillomavirus 6/11 DNA in condyloma acuminatum of the urinary bladder. Urol Int 48, 235–238 (1992). [DOI] [PubMed] [Google Scholar]
- 103.Karim RZ, Rose BR, Brammah S & Scolyer RA Condylomata acuminata of the urinary bladder with HPV 11. Pathology 37, 176–178 (2005). [DOI] [PubMed] [Google Scholar]
- 104.Chrisofos M, Skolarikos A, Lazaris A, Bogris S & Deliveliotis C HPV 16/18-associated condyloma acuminatum of the urinary bladder: first international report and review of literature. Int J STD AIDS 15, 836–838 (2004). [DOI] [PubMed] [Google Scholar]
- 105.Murray AJ, Bivalacqua TJ & Sopko NA Innumerable Condyloma Acuminatum tumors of the bladder. Urol Case Rep 12, 76–77 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Santiago-Rodriguez TM, Ly M, Bonilla N & Pride DT The human urine virome in association with urinary tract infections. Front Microbiol 6, 14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moustafa A et al. Microbial metagenome of urinary tract infection. Sci Rep 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.d’Herelle F Sur un microbe invisible antagoniste des bacilles dysenteriques. Comptes rendus Acad Sciences 165, 373–375 (1917). [Google Scholar]
- 109.Brown-Jaque M, Muniesa M & Navarro F Bacteriophages in clinical samples can interfere with microbiological diagnostic tools. Sci Rep 6, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jalil MB, Al-Hmudi HA, Al-Alsaad LA & Abdul-Hussein ZR Isolation and characterization of bacteriophages against multiple drug resistant Pseudomonas aeruginosa with using the bacteriophage as a therpy in the mice model. International Journal of Development Research 7, 11519 (2017). [Google Scholar]
- 111.Dallas SD & Kingsbery L Bacteriophage plaques on primary isolation media of a urine culture growing Escherichia coli. Clin Microbiol Newsl 19, 53–56 (1997). [Google Scholar]
- 112.Malki K et al. Seven Bacteriophages Isolated from the Female Urinary Microbiota. Genome Announc 4, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smith R, O’Hara M, Hobman JL & Millard AD Draft genome sequences of 14 Escherichia coli phages isolated from cattle slurry. Genome Announc 3, e01364–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Putonti C, Garretto A, Shapiro JW & Wolfe AJ The role of bacterial viruses in the female urinary microbiome. Female Pelvic Med Reconstr Surg 23, S39–S40 (2017). [Google Scholar]
- 115.Kim M-S & Bae J-W Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J 12, 1127–1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Price TK et al. Genome sequences and annotation of two urinary isolates of E. coli. Stand Genomic Sci 11, 79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Malki K et al. Genomes of Gardnerella Strains Reveal an Abundance of Prophages within the Bladder Microbiome. PLoS ONE 11, e0166757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miller-Ensminger T et al. Bacteriophages of the urinary microbiome. J Bacteriol 200, e00738–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Khasriya R et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clinl Microbiol 51, 2054–2062 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Price TK et al. The clinical urine culture: Enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol 54, 1216–1222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bao Y et al. Questions and challenges associated with studying the microbiome of the urinary tract. Ann Transl Med 5, 33–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yilmaz S, Allgaier M & Hugenholtz P Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat Methods 7, 943–944 (2010). [DOI] [PubMed] [Google Scholar]
- 123.Lewis DA et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol 3, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gangping L et al. Diversity of duodenal and rectal microbiota in biopsy tissues and luminal contents in healthy volunteers. J Microbiol Biotechnol 25, 1136–1145 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Kim D et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 5, 52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Panek M et al. Methodology challenges in studying human gut microbiota – effects of collection, storage, DNA extraction and next generation sequencing technologies. Sci Rep 8, 2045–2322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brubaker L & Wolfe A The urinary microbiota: a paradigm shift for bladder disorders? Curr Opin Obstet Gynecol 28, 407–412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Holm A & Rune A Urine sampling techniques in symptomatic primary-care patients: a diagnostic accuracy review. BMC Fam Pract 17, 72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Łusiak-Szelachowska M, Weber-Dąbrowska B, Jończyk-Matysiak E, Wojciechowska R & Górski A Bacteriophages in the gastrointestinal tract and their implications. Gut Pathogens 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barr JJ et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A 110, 10771–10776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shan J et al. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci Rep 8, 5091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dabrowska K et al. Antitumor activity of bacteriophages in murine experimental cancer models caused possibly by inhibition of beta3 integrin signaling pathway. Acta Virol 48, 241–248 (2004). [PubMed] [Google Scholar]
- 133.Duerkop BA & Hooper LV Resident viruses and their interactions with the immune system. Nat Immunol 14, 654–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nieth A, Verseux C & Römer W A Question of attire: Dressing up bacteriophage therapy for the battle against antibiotic-resistant intracellular bacteria. Springer Sci Rev 3, 1–11 (2015). [Google Scholar]
- 135.Górski A et al. Phages and immunomodulation. Future Microbiol 12, 905–914 (2017). [DOI] [PubMed] [Google Scholar]
- 136.Dabrowska K et al. Immunogenicity studies of proteins forming the T4 phage head surface. J Virol 88, 12551–12557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Duerkop BA, Clements CV, Rollins D, Rodrigues JLM & Hooper LV A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A 109, 17621–17626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gama JA et al. Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS ONE 8, e59043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin DM, Koskella B & Lin HC Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 8, 162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Caldwell JA Bacteriophagy in urinary infections following the administration of the bacteriophage therapeutically. Arch Intern Med 41, 189 (1928). [Google Scholar]
- 141.Gill G & Young RF Therapeutic applications of phage biology: history, practice and recommendations In Emerging Trends in Antibacterial Discovery (eds Miller AA & Miller PF) 367–407 (Caister Academic Press, 2011). [Google Scholar]
- 142.Sybesma W et al. Bacteriophages as potential treatment for urinary tract infections. Front Microbiol 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khawaldeh A et al. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J Med Microbiol 60, 1697–1700 (2011). [DOI] [PubMed] [Google Scholar]
- 144.Leitner L et al. Bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: a randomized, placebo-controlled, double-blind clinical trial. BMC Urology 17, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Siddiq DM & Darouiche RO New strategies to prevent catheter-associated urinary tract infections. Nat Rev Urol 9, 305–314 (2012). [DOI] [PubMed] [Google Scholar]
- 146.Letkiewicz S et al. The perspectives of the application of phage therapy in chronic bacterial prostatitis. FEMS Immunol Med Microbiol 60, 99–112 (2010). [DOI] [PubMed] [Google Scholar]
- 147.Abedon ST, Kuhl SJ, Blasdel BG & Kutter EM Phage treatment of human infections. Bacteriophage 1, 66–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chan BK et al. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018, 60–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


