Abstract
Circulating tumor cells (CTCs) have received a great deal of scientific and clinical attention as a biomarker for diagnosis and prognosis of many types of cancer. Given their potential significance in clinics, a variety of detection methods, utilizing the recent advances in nanotechnology and microfluidics, have been introduced in an effort of achieving clinically significant detection of CTCs. However, effective detection and isolation of CTCs still remain a tremendous challenge due to their extreme rarity and phenotypic heterogeneity. Among many approaches that are currently under development, this review paper focuses on a unique, promising approach that takes advantages of naturally occurring processes achievable through application of nanotechnology to realize significant improvement in sensitivity and specificity of CTC capture. We provide an overview of successful outcome of this biomimetic CTC capture system in detection of tumor cells from in vitro, in vivo, and clinical pilot studies. We also emphasize the clinical impact of CTCs as biomarkers in cancer diagnosis and predictive prognosis, which provides a cost-effective, minimally invasive method that potentially replaces or supplements existing methods such as imaging technologies and solid tissue biopsy. In addition, their potential prognostic values as treatment guidelines and that ultimately help to realize personalized therapy are discussed.
Keywords: circulating tumor cells (CTCs), nanotechnology, dendrimer, cell rolling, multivalent binding, liquid biopsy
Graphical abstract
1. Introduction
Currently available methods for cancer diagnosis and prognosis include medical imaging techniques, solid tissue biopsies, and liquid biopsies targeting biomarkers in patients’ blood [1–5]. Medical imaging techniques, including magnetic resonance imaging (MRI), computed tomography (CT), fludeoxyglucose-positron emission tomography (FDG-PET), and ultrasound tomography scans, are currently used as front-line standards to detect abnormal tissues as signs of primary cancer as well as recurrence of cancers [6–8]. However, relatively low sensitivity, required use of radioactive contrast agents, and high cost have hindered their frequent applications to patients for effective monitoring on disease progress and efficacy of applied therapeutics [9–11]. Following the discovery of abnormal tissues using the imaging techniques, solid tissue biopsies are typically performed to determine the pathology and the clinical stage of the disease. However, its invasive nature often results in discomfort/pain to patients and the risk of complications, such as bleeding, infection, and, rarely, tumor spreading along the track of the needle [12, 13]. In addition, information obtained from tissue collected from a single location at a given time provides only limited snap-shot of the tumor which does not reflect the heterogeneity and dynamicity of tumors [14–16]. Liquid biopsies based on detection of biomarkers in cancer patients’ blood have recently emerged as a potentially alternative way to overcome the limitations of the aforementioned methods, because they allow clinicians to frequently monitor therapeutic responsiveness and cancer recurrences with minimally invasive procedure and low cost [17, 18]. For instance, screening of prostate-specific antigen (PSA) and carcinoembryonic antigen (CEA) has shown clinical success in diagnosis of prostate and colorectal cancers, respectively [19–22]. However, these molecular biomarkers have clinical significance only for a few specific cancer types, and do not provide other prognostically valuable information, such as genetic mutation in cancers [23]. To address the limitations, novel biomarkers that provide valuable information for cancer diagnosis/prognosis and that enable liquid biopsy, various genetic analysis, and ultimately personalized medicine would obviously be desirable to fill the clinically unmet need.
Liquid biopsy via detection of circulating tumor cells (CTCs) from peripheral blood of patients is a promising alternative to detect primary and metastatic tumors since cancer metastasis is frequently mediated by CTCs that are shed from primary tumor sites [8, 24–31]. Furthermore, it has been reported that CTC counts well correlate with clinical stage, metastasis, and recurrence of cancer [24, 25, 32]. CTC detection from cancer patients’ blood also allows efficient monitoring of the biomarker as patient compliance to frequent blood drawing is typically high due to its minimally invasive and cost-effective nature. Additionally, detected CTCs have potential to provide genetic information of heterogeneous and dynamic tumors, which would be useful to develop a personalized therapy [33, 34]. Consequently, the CellSearch™ system was developed to detect CTCs from clinical specimens and is the first FDA-approved CTC detection system for metastatic breast (2004/2006), colorectal (2007), and prostate cancers (2008) [35]. CellSearch™ utilizes immunoaffinity-based detection and separation of CTCs. Briefly, a blood layer containing mononuclear cells, so-called buffy coat, is separated from 7.5 mL of peripheral blood of patients, followed by incubation with magnetic nanoparticles coated with anti-epithelial-cell-adhesion-molecule (aEpCAM) [24, 26]. This step results in magnetization of CTCs that express EpCAM (not expressed by normal hematologic cells), followed by separation of CTCs using a magnet. The captured CTCs are immunostained against epithelial cell-specific cytokeratin (CK), leukocyte-specific CD45, and nucleus (DAPI), and CK positive, CD45, negative, and DAPI positive cells are identified as CTCs [36]. This method has shown a degree of clinical success, i.e., having a threshold number of 5 out of 7.5 mL of blood as an indicator of patient survival [37]. However, CellSearch™ suffers from its limited sensitivity, often failing to provide actionable information to physicians, resulting in a very low prescription rate [38, 39]. Since then, various CTC detection methods have been introduced to improve the CTC detection sensitivity and specificity. Among those, capture systems, such as size-based trapping system (e.g., ISET) [40, 41], immunoaffinity-based fluidic system (e.g., NanoVelcro Chips and CTC-Chip) [30, 42–45], and immunostaining-based cytometry system (Epic HD-Chip) [46, 47], have shown promising results. However, an improved CTC detection method with clinically significant sensitivity and specificity toward CTCs are still in need to achieve early detection and reliable prognosis of cancer progress.
In response to the need of a CTC-detection technology enabling highly sensitive and specific capture, we have developed a platform using a biomimetic nanotechnology approach that integrates biomimicking cell rolling and multivalent binding engineered via dendrimer-based nanotechnology and microfluidics engineering [27, 28, 48]. Our capture system is unique in four aspects. First, it utilizes naturally occurring cell rolling that is often found in many of initial interactions between flowing cells in the blood and endothelial layers in events such as inflammation, stem cell homing, and CTC transmigration [49–51]. Cell rolling mediated via human recombinant E-selectin pulls the fast-flowing cells down to the capture surface, thereby increasing the chance of the cells to interact with the surface. Second, immobilization of capture agents that are linked to dendrimers significantly elevates capture efficiency and specificity through dendrimer-mediated multivalent binding effect. Third, the surface platform that combines cell rolling and multivalent binding also allows to employ virtually any antibodies that are specific to CTCs. Finally, the three unique aspects are engineered to be integrated into a single capture platform through applications of nano-scale dendrimers and fluidics engineering. The combination of all four aspects has been proven effective in capturing CTCs, as reported in our in vitro data and preliminary clinical data [27, 28, 48, 52]. This review paper highlights the concept and development of our biomimetic platform for improved CTC detection sensitivity and selectivity, covering from its CTC capture mechanisms to its application in monitoring therapeutic effects and clinical outcomes.
2. Biomimetic nanotechnology to improve CTC capture
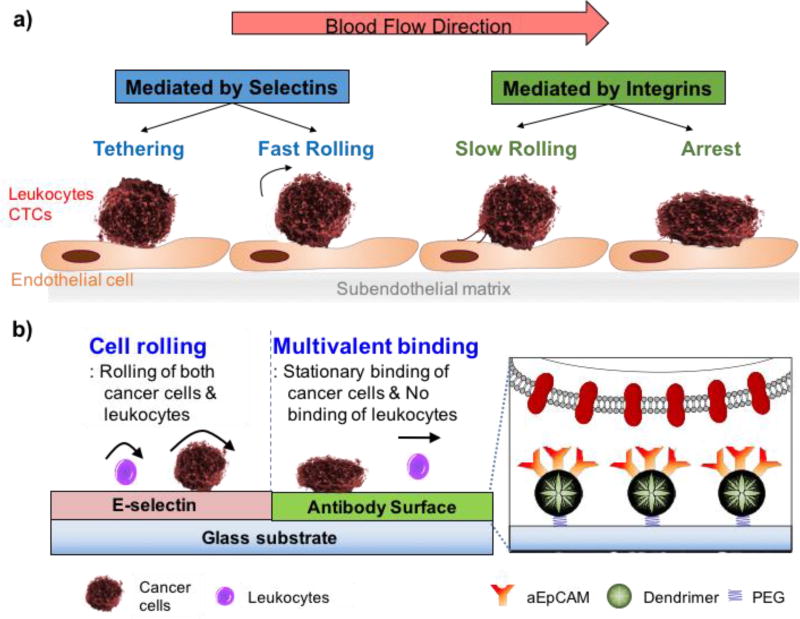
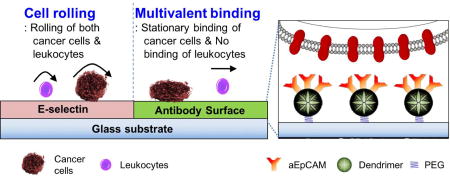
The adhesion of CTCs to endothelial cells is precisely regulated by the micromechanical and kinetic properties of molecular binding interactions with cell adhesion molecules, extracellular matrix components, and chemokines under the local circulatory hemodynamics in specialized microvasculature niches [53–55]. The physiological interactions between CTCs and an endothelium in the bloodstream, as illustrated in Figure 1a, could be classified into two stages: initial surface binding with fast association/dissociation kinetics, resulting in cell rolling; and stationary, tight adhesion steps [54, 56]. To detect and isolate target CTCs at a great sensitivity and specificity, we tried to mimic the concurrent rolling and firm adhesion in the physiological interactions on our biofunctionalized surface using E-selectin and aEpCAM – one of the most frequently used CTC capture agents – for a proof-of-concept study. Moreover, to substantially increase the firm adhesion kinetics, poly(amidoamine) (PAMAM) dendrimers were also used to immobilized aEpCAM to mediate strong multivalent binding effect [27, 57]. Using this configuration, we assessed the following three hypotheses: i) E-selectin-mediated cell rolling can efficiently recruit flowing cells from bulk flow to the capture surface with distinct biofunctions (selectin to induce rolling and aEpCAM to statically capture target cells); ii) the binding strength and stability between aEpCAM and CTCs can be substantially enhanced via dendrimer-mediated multivalent binding; and iii) the sequential binding events of cell rolling and multivalent binding can be micropatterned on a single platform surface to enhance overall capture efficiency of the surface (Figure 1b).
Figure 1.
Schematic diagrams of the biological interaction between CTCs and endothelium (a) and our biomimetic approach for CTC capturing on a micropatterned surface using iterative cell rolling and multivalent stationary adhesion (b). The inset diagram represents aEpCAM-immobilized dendrimers, flexible polymer nanolinkers, by which the multivalent binding effect can be achieved through locally concentrated aEpCAM.
2.1. Cell rolling for efficient cell recruitment from bulk flow
During the process of CTC extravasation, CTCs often bind to vascular endothelium in a manner analogous to leukocyte homing to sites of inflammation and homing of hematopoietic progenitor cells, which is initiated via a transient adhesion of flowing cells to the endothelium, known as cell rolling [58]. A family of selectins on the vascular endothelial cell surface, E-selectin (CD62E), L-selectin (CD62L), and P-selectin (CD62P), has been known to be mainly involved in the molecular mechanism mediating shear-resistant adhesive interactions with membrane ligands on the carcinoma cell surface [59–62]. The rapid turnover of selectin–ligand bonds, due to their fast on- and off-rates along with their remarkably high tensile strengths, enables them to mediate cell tethering and rolling in shear flow [28]. Among them, E-selectin, an inducible endothelial cell-surface glycoprotein [63], was chosen to induce CTC rolling on our biomimetic platform, since it is being involved in the adhesion and homing of various types of cancer cells such as prostate [58], breast [51, 64, 65], small cell lung cancer cells [66], and colon [61, 67, 68] carcinoma cells.
Recombinant human E-selectin Fc chimera proteins, a hybrid protein of human IgG1 constant domains (Fc) and E-selectin binding domains through genetic engineering of a fusion gene, were immobilized on an epoxy-functionalized glass surface. Under flow conditions, human breast cancer and leukemia cell lines, MCF-7 as a CTC model and HL-60 as a leukocyte model, respectively, were efficiently recruited from bulk flow to the capture surface, and rolled in a shear stress-dependent way [48, 69]. However, given that a large class of hematological cells, including leukocytes, platelets, neutrophils, mesenchymal and hematopoietic stem cells, and metastatic cancer cells all exhibit rolling on E-selectin, CTC detection that is solely based on cell rolling has limitations to achieve a capture device with sufficient specificity to CTCs.
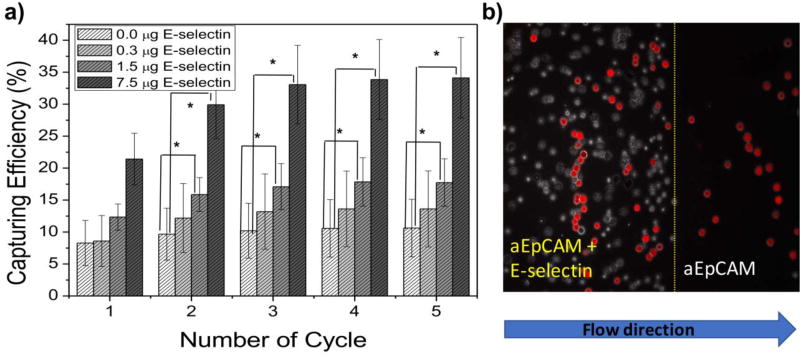
To provide sufficient specificity to the surface, the most commonly used CTC-specific antibody, aEpCAM, was co-immobilized to differentiate target tumor cells out of the rolling cell population. The surface immobilization of E-selectin and aEpCAM was confirmed by X-ray photoelectron scattering (XPS) and fluorescence microscopy using fluorophore-conjugated antibodies. XPS analysis showed an increase in carbon and nitrogen compositions and decreased silicon content from the functionalized glass surface upon the surface functionalization. Fluorescence-labeled proteins, such as fluorescein-anti-E-selectin (green fluorescence) and APC-anti-EpCAM (red fluorescence), were also used to confirm the surface functionalization with E-selectin using a fluorescence microscope, as previously shown in our earlier report [27]. The immobilized proteins maintained their characteristic biological adhesive functions, i.e., cell rolling by E-selectin and tumor cell-specific stationary binding by aEpCAM, as tested using in vitro cell lines under flow conditions (Figure 2 a). Note that the sole use of E-selectin would not provide specific capture of tumor cells as many hematological cells (e.g., leukocytes, neutrophils, and other inflammatory cells), in addition to various stem cells and CTCs, can all roll on the surface. The addition of E-selectin can induce the rolling of various cell types to be readily accessible by aEpCAM that recognizes/captures tumor cells, resulting in substantially enhanced capture efficiency of tumor cells by more than 3-fold enhancement, compared to the surface with aEpCAM alone (Figure 2 b). The E-selectin-induced tumor cell rolling most likely maximizes the chance of the tumor cells to interact with aEpCAM on the surface, resulting in effective stationary binding and improved detection sensitivity of CTCs. These results proved our first hypothesis, i.e., efficient recruiting of flowing cells to the capture surface via cell rolling, thereby enhancing capture efficiency of the surface.
Figure 2.
The improved capture efficiencies of the surfaces immobilized with the mixtures of anti-EpCAM and E-selectin (a) and a representative image of HL-60 and DsRED-transfected MCF-7 cells (red cells) on patterned E-selectin/anti-EpCAM coated surfaces (b). (a) Based on the numbers of DsRED-MCF-7 cells injected and recovered using a flow chamber, the capture efficiency was calculated at a shear stress of 0.16 dyn/cm2. As increasing E-selectin concentration, the capture efficiency of the surfaces was further enhanced by up to 3 folds, which was statistically higher than the surface functionalized with aEpCAM alone (one-factor ANOVA, Error bars: standard error, * p < 0.05). (b) From the mixture with HL-60 (a leukocyte model: white), DsRED-transfected MCF-7 cells (a CTC model: red) on the anti-EpCAM coated region of the patterned surface with E-selectin and anti-EpCAM were efficiently isolated. (Copyright WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Ref. 27)
2.1.1. Connection to the literature: the attenuated tumor growth and metastasis in selectin-deficient animals
Our concept to improve the CTC detection sensitivity through E-selectin-mediated cell rolling is highly supported by other literature regarding how E-selectin involves efficient cell recruitment in cancer metastasis. For instance, the metastasis of human colon cancers to lung was significantly reduced in E-selectin-deficient, severe combined immunodeficient (SCID) mice [61], and E-selectin expressing on bone marrow endothelial cells (BMECs) leads the predilection of prostate tumor metastasis to bone, compared to other tissue microvessels [58, 70, 71]. In addition, our approach to monitor the interaction between cancer cells and recombinant E-selectin in shear flow resulted in elucidating some of the molecular and biophysical mechanisms of CTC adhesion to endothelial cells via E-selectin. For example, MCF-7 cells express none of previously known ligands to E-selectin, such as E-selectin ligand-1 (ESL-1) [72], P-selectin glycoprotein ligand-1 (PSGL-1) [73, 74], sialyl Lewis X (sLex), sialyl Lewisa (sLea) [75, 76], CD43[77], hematopoietic cell E- and L-selectin ligand (HCELL; a specialized glycoform of CD44) [70], β2 integrins [78], and CD44v4[79]. By a series molecular and cellular experiments, we were able to identify CD24 overexpressed on MCF-7 cells as an E-selectin ligand for the first time to our knowledge [27]. Given the oncological significance of E-selectin, better understanding of E-selectin ligands on the cancer cells would help to potentially find a way to prohibit the spreading of primary cancers in patients and provide guidelines for developing anti-metastatic therapies by disrupting the E-selectin receptor–ligand bonds, which is well reviewed in a previous publication [70].
2.2 Dendrimer-mediated multivalent binding for strong, specific CTC capture
As mentioned above, the stationary binding between aEpCAM and tumor cells was observed, along with the cell rolling on E-selectin under flow. The aEpCAM-based tumor cell enrichment was critical to detection specificity, especially for extremely rare CTCs among numerous hematological cells. However, it is well known that the expression level of EpCAM on CTCs is not always maintained high, unlike some of the in vitro tumor cells, due to intrinsic heterogeneity of tumors and epithelial-mesenchymal-transition (EMT). By introducing multivalent binding effect through increasing localized density of surface-immobilized aEpCAM, we attempted to increase the binding strength and detection specificity of the rare, heterogeneous CTCs on our capture surfaces.
2.2.1. Multivalent binding effect
Multivalent binding is the simultaneous binding of multiple ligands to multiple receptors in biological systems, which is observed in many physiological and pathological processes, including the attachment of viral, parasitic, mycoplasmal, and bacterial pathogens to the surface of a host cell during the infection process [80–82]. These multiple interactions impart the substantial increase of collective binding strength (avidity) for the interaction of a relatively weakly bound ligand and its receptor without increasing the affinity of single, monovalent ligand-receptor interactions [81]. The following cooperative, localized bindings at adjacent sites significantly influence the equilibrium between association and dissociation of a ligand from the initial site on the receptor at the first binding [83]. The binding strength and stability of multivalent ligands to surface-bound receptors can be affected by the composition and distribution of ligands on the surface of multivalent binding mediators [84]. Thus, the physiochemical and biological properties of the mediators to induce the multivalent bindings are critical to achieve significantly strong binding events, which will ultimately increase the capture efficiency.
2.2.2. Uniqueness of PAMAM dendrimers
Given their high surface-to-volume ratios, various types of nanoparticles, such as inorganic nanoparticles, linear polymers, branched polymers, dendritic polymers, and supramolecular assemblies, could surface immobilize aEpCAM at high local density, and thus could serve as mediators for multivalent binding, as summarized in our previous review paper [85]. Among these nanoparticles, poly(amidoamine) (PAMAM) dendrimers have been reported as an excellent mediator for multivalent binding effect because preorganization/orientation of ligands, polymer backbone topology, and easy deformability of the macromolecules all contribute to strong multivalent binding to cell surfaces [86–88]. Nano-scale PAMAM dendrimers are hyperbranched, chemically well-defined, flexible, spherical macromolecules with a high number of peripheral functional groups, thereby allowing multifunctionalization through a variety of conjugation chemistries. There unique aspects of the dendrimers enable precise control over cellular interactions and molecular recognitions through multivalent binding [85, 89]. Such strong binding has been more commonly used to improve targeting efficiency of dendrimer-based drug delivery systems [57, 90, 91]. Similarly, one could imagine that the advantages of enhanced binding avidity through dendrimers could significantly improve the detection sensitivity and specificity particularly in capturing human disease-related rare cells in blood (e.g., <0.1% subpopulation), such as CTCs, which our group attempted to achieve for the first time to our knowledge.
2.2.3. Enhanced binding strength
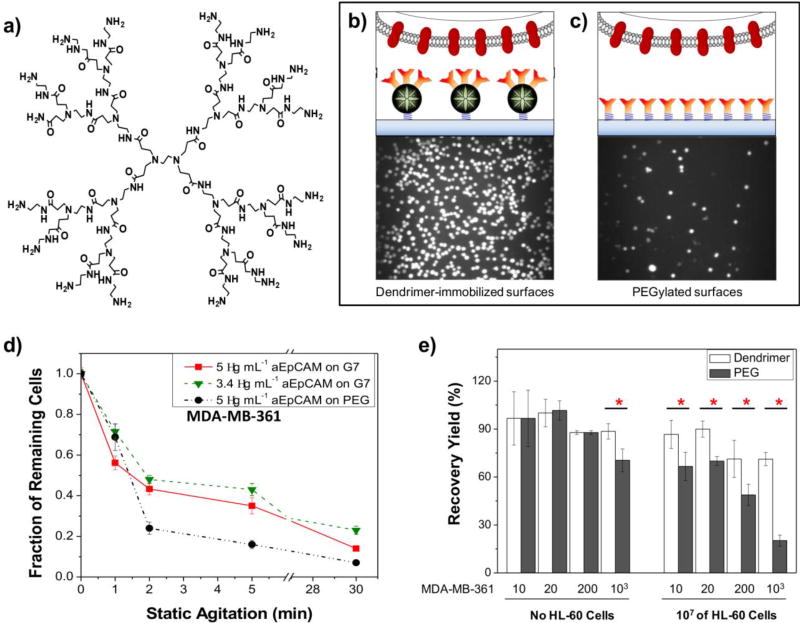
To create a highly sensitive surface via the multivalent binding effect, we employed generation 7 (G7) PAMAM dendrimers due to their adequate size (8 nm in diameter) and surface functional groups (512 theoretically) to accommodate multiple aEpCAM (around 5.5 nm in diameter of Fc region) per dendrimer. We first tested the multivalent interactions between aEpCAM-G7 PAMAM dendrimer conjugates and recombinant EpCAM-immobilized gold surfaces via a direct, quantitative analysis using surface plasmon resonance (SPR). Remarkably, compared to free aEpCAMs, the binding strength of the aEpCAMs conjugated on dendrimers with recombinant EpCAM were enhanced by up to ~106-fold (Figure 3a).
Figure 3.
Dendrimer-mediated multivalent binding for enhanced detection of CTCs. (a) The chemical structures of generation 2 (G2) of PAMAM dendrimers. Note that the chemical structure of G2 PAMAM dendrimer is given for structural simplicity instead of G7 used for our biomimetic platforms. (b–c) Experimental setup for comparison of the bound cell numbers on dendrimer (b)- and linear polymer, polyethylene glycol (PEG, c)-immobilized surfaces. Multivalent binding effect induced via multiple ligands-functionalized dendrimers can be used to enhance the detection sensitivity. (d–e) Compared to the surface with aEpCAM-conjugated linear polymers, the dendritic nanoparticle-immobilized platform significantly increased the binding stability with tumor cells (d) and improved detection specificity of tumor cells from a mixture with 107 HL-60 cells (e, Error bars: standard error (n=3), * p < 0.05). (Copyright WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim, Ref. 27)
Although the aEpCAM-G7 PAMAM dendrimer conjugates dramatically enhanced binding kinetics through multivalent binding, we could not directly immobilize the conjugates on the epoxy-functionalized glass slide due to the flexibility and peripheral multifunctional groups of PAMAM dendrimers and 3-dimentional orientation of aEpCAM. To induce the multivalent binding on our capture surface, a layer-by-layer approach was used to build the dendrimer-functionalized surface through a sequential immobilization of a linear polymer, NH2-PEG-COOH, and partially carboxylated G7 PAMAM dendrimers onto epoxy-functionalized glass slides [27, 28, 92]. The spacer PEG polymers and reduced amine groups of PAMAM dendrimers likely allowed to prevent spreading of PAMAM dendrimers on the epoxy-functionalized surface and keep dendrimers spherical after immobilized. The aEpCAM molecules were finally added on top of the dendrimer-functionalized surfaces, likely maximizing the surface availability of aEpCAM at a high local density, owing to the localized, multiple functional groups provided by the dendrimers.
The dendrimer-based configuration of the capture surfaces exhibited dramatically enhanced cell adhesion and binding stability of three breast cancer cell lines (MDA-MB-361, MCF-7, and MDA-MB-231) we tested, compared to the control surfaces with the linear PEG-aEpCAM conjugates (Figure 3b). Furthermore, a significantly higher number of bound cancer cells, particularly MDA-MB-231 cells which have lower EpCAM expression than the other two cell lines, remained attached on the dendrimer-coated surface after strong agitation (up to 15.2 fold), further confirming the multivalent cell capture.[27] The non-specific binding between HL-60 cells and a capture platform was relatively negligible, indicating that selectivity and specificity of the capture surfaces were also improved.[27] These results validated our 2nd hypothesis, i.e., dendrimer-mediated multivalent binding effect would significantly enhance the binding kinetics between EpCAM on tumor cells and aEpCAM used as a capture agent.
2.2.4. Connections to the literature: Multivalent binding effect to enhance the therapeutic and diagnostic efficiency
The enhanced targeting efficiency and binding strength/stability through dendrimer-mediated multivalent binding has been used to design a wide range of potential therapeutics in Pharmacology and targeted drug delivery systems [86, 93–95]. The biological multivalent inhibitors are responsible for modulation of potency and affinity and have yielded quantitative measurements of binding avidities with 1–9 orders of enhancement, compared to monovalent inhibitors [84, 96]. For instance in targeted drug delivery systems, G5 PAMAM dendrimers were multifunctionalized with therapeutic (methotrexate), targeting (folate), and imaging (radiolabel or fluorophore) agents and evaluated in a mouse tumor xenograft model using folate receptor (FR)-overexpressing KB cells [90]. The targeting efficiency of folate on the multifunctional dendrimer conjugates primarily to the tumor and liver was trackable using the attached imaging agents over the course of 4 days, resulting in the enhancement of therapeutic efficacy of methotrexate as measured by a reduction in tumor volume and decreased off-target toxicity. Another example is the 5-flurouracil (5-FU) delivery system using folate-targeted PEGylated PAMAM dendrimers. Compared to non-PEGylated dendrimers 5-FU (tmax=1–2.5 hrs), the 5-FU-encapsulated PEGylated dendrimers showed prolonged retention of 5-FU (tmax=2.5–5 hrs) and its anti-tumor efficacy, which was significantly safer and more effective at decreasing tumor volume [97]. Multivalent ligand-functionalized dendrimers have been applied successfully in vivo for anticancer drug delivery on numerous occasions and have been reviewed elsewhere [89, 98].
Multivalent binding has not only been utilized to enhance targeting efficacy as mentioned above, rather it has also been leveraged to develop highly sensitive devices to detect target molecules and sustain the trapped target molecules strongly and securely for diagnostic purposes [99]. These dendrimer-based, multivalent binding materials can be injected for analysis in vivo for direct detection, mixed with specimens in vitro, or immobilized on the surface of solid supports for in vitro analysis. As a contrast agents for in vivo diagnosis, dendrimers conjugated with targeting agents (e.g. antibodies, aptamers, nucleotides, proteins, and small molecules) were used to encapsulate or conjugate imaging agents (e.g. MRI contrast agents or fluorescent dyes) [100–102]. A dendrimer-immobilized substrate was used for the detection of DNA [103], toxins [104], antigens [105]. For instance, the dendrimer functionalization on substrates was able to enhance the capture of oligomers by 2-fold, as observed in nucleic acid hybridization experiments using fluorophore-labeled complementary oligonucleotide targets, compared to the untreated intact substrate [106].
2.3. A combination of the two biomimetic approaches
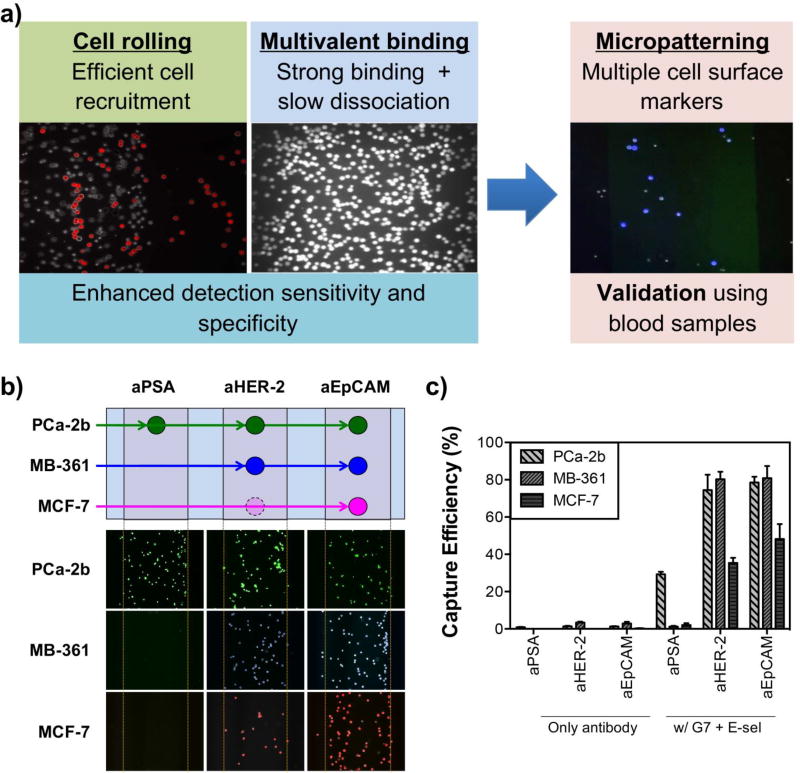
As described above, it is obvious that the two biomimetic approaches, cell rolling and multivalent binding, individually enhance the surface capture of tumor cells in vitro. We then reached a next question: what if we integrate the both approaches into a single substrate to more faithfully mimic physiological complexity to better capture CTCs? To answer this question, E-selectin-mediated cell rolling and dendrimer-mediated multivalent binding were engineered onto a single multifunctional platform via micropatterning (Figure 4a). The E-selectin-coated patterns would be effective in pulling CTCs along with leukocytes out of the bulk flow and induce the cell rolling behaviors on the surface. This step would substantially increase the chance of the rolling cells to be interacting with the next patterns functionalized with the CTC-specific antibody-dendrimer conjugates, resulting in a strong binding formation between the CTCs and the surface. Our hypothesis (hypothesis 3) was proven correct: unlike the surface marker-dependent stationary binding of tumor cells, leukocytes and HL-60 cells passed through the channel without stationary capture on the antibody pattern after rolling on the E-selectin patterns. The geometry of the patterns of E-selectin and aEpCAM was optimized in terms of angle of the E-selectin patterns and length between the E-selectin and capture antibody patterns, as previously reported [28, 107]. The engineered multifunctional surface exhibited a significant enhancement in capture efficiency by up to 7-fold, compared to the surfaces with CTC-specific antibodies only [27, 28, 48, 92].
Figure 4.
Micropatterning of E-selectin and aEpCAM-dendrimers for combining the effects of rolling and multivalent binding under flow. (a) Schematic illustration of the surface marker-dependent cell capture using aPSA, aHER-2, and aEpCAM. (b) The capture patterns of the three cell lines labeled with three different fluorescent colors: green for MDA-PCa-2b, blue for MDA-MB-361, and red for MCF-7. The lower capture efficiency of MCF-7 cells for aHER-2 due to low HER-2 expression of MCF-7 cells were presented as a dotted, faded circle in the schematic illustration (a). (c) A combination of dendrimers and E-selectin (a cell rolling inducing agent), along with multiple antibodies achieved highly sensitive differential detection of tumor cells (up to 82%, Error bars: standard error (n=4)). (Copyright American Chemical Society, Ref. 28)
One can ask questions about the cell mixture included on the rolling population, which may decrease the purity of CTCs out of the captured cells. Those rolling cells (HL-60 for in vitro studies and leukocytes/inflammatory cells in our clinical studies) on E-selectin could be easily removed using a simple washing step with PBS buffer-supplemented with Ca2+ chelating agent ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid (EGTA) because the interaction between E-selectin and cells is Ca2+-dependent. Importantly, the washing step did not induce any noticeable detachment of CTCs captured on the dendrimer-coated regions due to the ultralow dissociation constants achieved through the multivalent binding effect. This series of CTC isolation steps resulted in a highly sensitive, specific capture of tumor cells and CTCs from cell mixtures or blood samples.
We also wanted to introduce multiple antibodies into a single capture platform to effectively capture CTCs with high phenotypic heterogeneity [108] and biological plasticity frequently found during the metastatic process [109, 110]. In addition to aEpCAM, we successfully immobilized additional cancer cell-specific markers, such as human epidermal growth factor receptor-2 (HER-2) [111] and prostate specific antigen (PSA) [112]. To mimic the heterogeneity of CTC samples, three tumor cell lines, MDA-PCa-2b, MCF-7, and MDA-MB-361 cells were used to test the multifunctional surfaces. Depending on the surface expression of the ligands on each of the cell lines, differential detection of the cells was achieved, as appeared in patterns that were pre-determined with different capture antibodies [28]. Following the in vitro experiments, we used tumor cell-spiked human blood samples to demonstrate that our multifunctional surface is functional and effective capture tumor cells with the background of human blood cells. As expected, a significantly enhanced capture of tumor cells in human blood was achieved, up to 82% capture efficiency (~10-fold enhancement than a surface with the antibodies alone) and up to 90% purity (Figure 4b). Taken together, these results indicate that our approach taking advantage of cell rolling and multivalent binding significantly enhance sensitivity and specificity of CTC capture and is expandable to multiple antibodies, accommodating virtually any antibodies to be used as capture agents. Although our extensive data assure that the efficient cell capture by combination of cell rolling, multivalent binding, and multiple antibodies, the effect of multiscale patterning with more precise geometries, such as lengths and angles of the protein patterns, on CTC capture could potentially further improve the device functionality, which is the subject of further studies.
3. Clinical significance of CTC capture and its relation to clinical outcomes
3.1. Prognostic/therapeutic applications
Our biomimetic approach clearly enhances the detection sensitivity, selectivity, and purity of CTC capture, compared to the control surface using a conventional approach solely based on antibodies. The clinical applications of this device are to monitor the change of CTCs as a biomarker for therapeutic effect monitoring (section 3.1.1) and to evaluate novel therapeutic agents under development (section 3.1.2, Figure 5a). For clinical studies, three antibodies, aEpCAM, HER-2, and antibodies against epidermal growth factor receptor (aEGFR), were chosen as a cocktail to consistently generate the high capture efficiency as they are commonly expressed by many of cancer cells [113].
Figure 5.
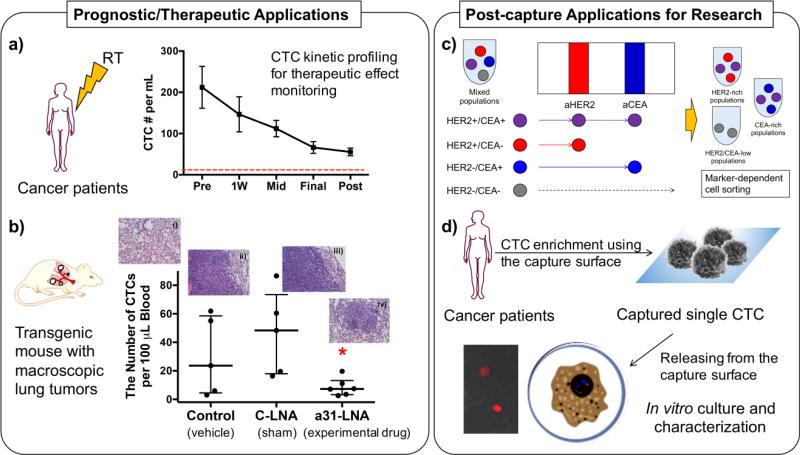
Various applications of our biomimetic platform with high capture sensitivity and specificity. (a) CTC capture in cancer patients undergoing RT. From 24 cancer patients in a pilot study, CTCs in their peripheral blood were able to detecte with high capture sensitivity and specificity on the biomimetic platform. The CTC kinetic profiles before and after RT clearly showed its potential for therapeutic effect monitoring. (Ref. 113) (b) CTC numbers detected from three groups including the transgenic mice treated with an experimental drug, anti-miR-31 LNA (a31-LNA). The number of CTCs in transgenic mice with macroscopic lung tumors after a31-LNA treatment significantly decreased compared to the other two control groups (n=5, mean ± S.E., *p<0.05). Representative histology images of lung tissues of the mice were taken at 20× magnification: i) healthy lung and ii) lung tumors formed in the vehicle-treated control; iii) lung tumors formed in the control LNA (C-LNA) group; and iv) shrunk lung tumor and close-to-the-normal lung tissue morphology found in the a31-LNA group. (Copyright American Chemical Society, Ref. 52) (c) A schematic illustration of the surface marker-dependent bindings of cancer cell models for cell sorting. The enhanced capture efficiency on the multifunctional surfaces can be developed as a diagnostic platform for rare CTC, which could provide the information about cell surface markers for personalized cancer therapy and be used for cell sorting approaches in vitro. (d) A schematic diagram of the procedure how to establish in vitro CTC-originated cancer cell line from cancer patients’ blood. The image in the inset was obtained from the ex vivo culture of MDA-MB-231 cells from xenograft mice, after recovering using the biomimetic platform.
3.1.1. Monitoring of Therapeutic effects
On the biomimetic device functionalized with a cocktail of aEpCAM, aHER-2, and aEGFR, the improved sensitivity and specificity of CTC capture enable to investigate the clinical significance of CTCs and their kinetic profiles in cancer patients before, during, and after treatments [114]. We have conducted a clinical pilot study by recruiting cancer patients with diverse cohorts who undergo radiotherapy (RT) treatment [114]. For CTC kinetic profiling, 24 patients diagnosed with rectal, cervical, prostate, or head and neck primary carcinoma were enrolled in our pilot study. The median age of the enrolled patients was 58 years old (range, 42–84), including 7 (29%) female and 17 (71%) male patients. Their peripheral blood was collected prior to the radiotherapy (RT) and at the each of radiotherapy (RT), including during the first week of RT, mid-way through RT (Mid-RT), and during the last week of RT (End-RT), and after completion of RT. This is in sharp contrast to previous reports using CellSearch™ where CTCs were not detected from ~1/3 of the patients with the comparable cohort [115–117]. The combined use of multivalent binding via PAMAM dendrimers and cell rolling dramatically improved CTC capture sensitivity and specificity (captured CTCs (CK+/CD45−/DAPI+) among all captured cells), respectively. The CTC kinetic profiles well correlated to the clinical outcomes. The number of CTCs gradually decreased throughout RT in 18 patients with complete clinical and/or radiographic response. In contrast, pathologic residual disease was found from 3 patients with elevated CTC numbers. These results indicate that the CTC counts using our biomimetic platform allow reliable monitoring on CTC changes during and after treatment, opening a potential avenue of using CTCs as a biomarker to monitor therapeutic efficacy.
3.1.2. Efficacy test for experimental drugs in vivo
One of the clear advantages of our approach is that it provides a modular, platform technology. The dendrimer-based CTC detection platform was adapted for lung cancer CTCs by immobilizing aEGFR, and used to measure murine CTCs from peripheral blood of cyclin-E overexpressing (CEO) transgenic mice. A lack of an effective detection method for lung CTCs presents a substantial challenge to elucidate the value of CTCs as a diagnostic or prognostic indicator in lung cancer, particularly in non-small cell lung cancer (NSCLC). Given that 85% of the tumor cells from NSCLC patients overexpress epidermal growth factor receptor (EGFR) [118], aEGFR was chosen as a capture agent for this study [52]. Following in vitro confirmation using the murine lung cancer cell lines (a wild-type cyclin E-driven lung cancer cell line (ED-1) and invasive ED1-SC harvested from tumor of FVB/N mice after subcutaneous injection of ED-1 cells), CEO transgenic mice were employed as an in vivo lung tumor model to assess specificity and sensitivity of the capture surface. Note that aberrant cyclin E expression is a negative prognostic indicator in the NSCLC patients, and the CEO mice have a high incidence and rapid onset of lung carcinogenesis [119, 120]. Our surfaces functionalized with the EGFR-dendrimer conjugates demonstrated that the numbers of CTCs in blood from the CEO mice were significantly higher than those from the healthy controls (on average 75.3 ± 14.9 vs. 4.4 ± 1.2 CTCs/100 µL of blood, p<0.005), indicating the high level of sensitivity of the modified capture system. We then investigated the function of the surfaces as a therapeutic effect monitoring tool to evaluate a new engineered antagonist (locked nucleic acid) against microRNA-31 (anti-miR-31). A significant decrease in the CTC numbers from the CEO mice upon a treatment using a novel anti-miR-31 locked nucleic acid (LNA) was observed, compared to a vehicle treatment and a control-LNA treatment (p<0.05) [52]. Our detection system also demonstrated its efficiency in monitoring therapeutic effect of a novel CDK2/9 inhibitor in lung cancer, as shown in our recent publication [121]. All in all, our results using the in vivo lung cancer models confirm that our new CTC detection technology has great potential to be used as a diagnostic and prognostic tool for lung cancer and offers a promising way to monitor cancer progress and responsiveness to therapeutic interventions.
3.2. Post-capture applications for research
Additional features of this device include the ability to collect CTCs from whole blood under continuous flow without labeling or damaging CTCs (section 3.2.1, Figure 5c). Therefore, the collected CTCs could be extracted and potentially be culture expanded to be the subject for further analysis, such as genetic profiling and tests for patient-specific responses to various therapeutic options (section 3.2.2, Figure 5d).
3.2.1. Surface marker-dependent cell sorting
As described above, one of the unique characteristic of our engineered platform is that the capture mechanism is solely based on the surface expression of adhesive proteins from CTCs. The binding strength between the CTC proteins and the antibodies immobilized on our surfaces could be significantly augmented via dendrimers and cell rolling, thus maximizing the overall capture efficiency [28]. This advantage would enable us to exploit virtually any antibodies to achieve differential detection of CTCs sorted based on their surface protein profiles. As a proof-of-concept study, various cell lines, e.g., prostate cancer (MDA-PCa-2b) and breast cancer (MDA-MB-361 and MCF-7) cells, were tested in mixture as well as after being spiked into human blood using a multipatterned surface with different markers. For example, MDA-PCa-2b cells, the only PSA-positive cell line among the three CTC models, bound primarily on the aPSA-coated region at 91–100% purity from the cell mixtures with PSA-negative cells. MDA-MB-231 were bound to the patterns coated with aHER-2 and aEpCAM, whereas MCF-7 cells were primarily adhered to the aEpCAM-coated region [28]. It is noteworthy that the two breast cancer cell lines, MCF-7 and MDA-MB-231, exhibited different binding behaviors on the aHER-2 pattern, which is not surprising as MCF-7 cells do not express a high level of HER-2 whereas MDA-MB-231 cells do [122, 123]. Interestingly, MCF-7 cells showed noticeably increased binding to aHER-2 after addition of E-selectin, which was not observed on the same surface without E-selectin. This is an indication that E-selectin-based cell rolling likely improves the capture efficiency of antibodies even though the cells express a low level of the corresponding surface receptors. Our results indicate that our CTC capture platform may achieve the surface marker-dependent tumor cell differentiation from cell mixtures and potentially from patients’ blood without cell labeling (Figure 5c) [28, 85, 92].
3.2.2. In vitro culture of CTCs
Post-capture analysis and culture expansion of the captured CTCs from blood specimens would be helpful to find CTC biomarkers and establish in vitro cell models for CTCs. Upon sensitive CTC capture, molecular analysis of the extracted CTCs would be particularly important, as it would provide genetic information to understand the invasive cancer in detail, which will ultimately help to develop an effective treatment, and even cure, for the debilitating metastatic cancers. To understand the molecular characteristics of CTCs, in vitro cell lines derived from clinical CTCs could be established upon the isolation of live CTCs from metastatic cancer patients [124, 125]. After mycobacteria-free confirmation and genome mapping (characterization), the captured CTCs could be expanded and established as cell lines for subsequent in vitro studies [124, 126]. Other groups have been successful in culture expanding CTCs. For example, CTCs isolated from breast cancer patients were expanded to ex vivo culture. The cultured CTCs were tested drug susceptibility in vitro and in mouse xenografts by directly inoculating the cultured CTCs to show potential therapeutic targets [34]. In our previous publication, the CTCs captured on our multifunctional platforms were still live as being stained with live cell tracking markers, which meant negligible damage to the cells from the capture procedure [28]. Moreover, CTCs were captured on our capture surfaces at high purity, which would allow more selective enrichment of CTC-containing cell population. The DsRED-transfected MDA-MB-231 cells recovered from the blood of xenografted mice using our biomimetic platform were alive and successfully cultured in tissue culture well plates in our pilot study (Figure 5d).
Subsequent cell culture and single cell analysis post efficient CTC capture would enable us to extract valuable clinical information from individual patients, ultimately allowing personalized medicine. The critical issue is how to isolate CTCs from the surfaces without damaging the cells. Although a simple treatment with trypsin exhibited partial success in our case, novel methods are required to efficiently collect the captured CTCs in their intact form. The methods of high potential that could be applicable to our devices include the approaches using stimuli-responsive polymers that cleave upon exposure to external stimuli, such as light, temperature, pH, and physical stress [127–129]. Additionally, enzyme-degradable materials, such as alginate hydrogel, DNA, or aptamer, could also be used to engineer the CTC capture surface to release the cells upon exposure to specific enzymes, such as alginate lyase, DNase, or endonuclease, respectively [130, 131]. Our next generation devices that are under investigation will incorporate such surface release mechanisms to effectively collect the captured CTCs in their intact form for a series of post-capture analyses.
4. Summary and Future Perspectives
Considering all the benefits that CTC-based liquid biopsy can potentially provide, a highly sensitive and selective device for CTC capture would obviously beneficial. Our recent studies in an effort of integrating biomimicry and nanotechnology on a chip have resulted in a highly reliable CTC-capture platform with excellent yield and selectivity. The series of investigations using in vitro cancer cell suspension, cancer cell-spiked blood samples, and cancer patient specimens, as highlighted herein, clearly demonstrate the uniqueness and advantages of our CTC capture technology exploiting multivalent binding and cell rolling. Although more extensive clinical studies are still required, highly sensitive and specific CTC detection achieved using our detection method has shown a great potential to provide valuable clinical insight into the progress of metastatic cancers in individual patients, to monitor responses of patients during currently available cancer therapy as well as novel experimental treatments. Besides accurate CTC enumeration, our capture system can also potentially provide additional features, such as label-free cell differentiation and isolation of live CTCs, which would lead to molecular profiling of the captured CTCs and ultimately to personalized medicine against debilitating metastatic cancers.
To successfully translate our CTC detection technology to a clinical setting, additional investigations are necessary. First, clinical studies need to be expanded for various types of cancers and cancer therapies to further validate our system. The expanded clinical studies will allow us to confirm our device in terms of the accuracy of kinetic profiling of CTC numbers and its correlation with clinical outcomes. The CTC identification step will also have to be validated for each of the cohorts. The current DAPI+/CD45−/CK+ standard has been reported to often provide false values, as there are some of non-specific immunostaining of cells [132, 133]. Second, although our surface functionalization is based on simple chemical reactions, the consistency in quality control and device fabrication should be warranted to achieve a high level of controllability, scalability, and reproducibility. Third, tailoring the design of the capture system to different cancer types would be required. It is well known that the phenotypes of CTCs significantly vary depending on their origins and status of their phenotypic changes upon transition such as EMT [134, 135]. A right choice of mixtures of capture agents would be required to make our capture system effective in capturing highly heterogeneous CTCs. Through these efforts for clinical translation, we expect our CTC device to be implemented for the routine use in point-of-care testing and ultimately play a key role in achieving personalized treatments for cancer patients. This biomimetic nanotechnology platform could be broadly applicable to a variety of liquid biopsies by efficiently capturing and isolating other biomarkers, such as other types of rare cells, exosomes, proteins, and DNA/RNA not only from peripheral blood but also from other human specimens.
Acknowledgments
This work was supported by National Cancer Institute (NCI)/National Institutes of Health (NIH) under grant # R01-CA182528 (SH) and National Science Foundation (NSF) under grant # DMR-1409161 (SH).
Disclosure statement
SH and AZW are co-founders of Capio Biosciences Inc., a biotech startup that is commercializing the biomimetic CTC detection technology (CapioCyte™).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dixon J, Anderson T, Lamb J, Nixon S, Forrest A. Fine needle aspiration cytology, in relationships to clinical examination and mammography in the diagnosis of a solid breast mass. British Journal of Surgery. 1984;71:593–596. doi: 10.1002/bjs.1800710809. [DOI] [PubMed] [Google Scholar]
- 2.Augsburger JJ, Shields JA, Folberg R, Lang W, O'Hara BJ, Claricci JD. Fine needle aspiration biopsy in the diagnosis of intraocular cancer: cytologic-histologic correlations. Ophthalmology. 1985;92:39–49. doi: 10.1016/s0161-6420(85)34068-x. [DOI] [PubMed] [Google Scholar]
- 3.Sidransky D. Nucleic acid-based methods for the detection of cancer. Science. 1997;278:1054–1058. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- 4.Trimboli P, Nasrollah N, Guidobaldi L, Taccogna S, Modica DDC, Amendola S, Romanelli F, Lenzi A, Nigri G, Centanni M. The use of core needle biopsy as first-line in diagnosis of thyroid nodules reduces false negative and inconclusive data reported by fine-needle aspiration. World journal of surgical oncology. 2014;12:61. doi: 10.1186/1477-7819-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredsøe J, Rasmussen AK, Thomsen AR, Mouritzen P, Høyer S, Borre M, Ørntoft TF, Sørensen KD. Diagnostic and Prognostic MicroRNA Biomarkers for Prostate Cancer in Cell-free Urine. European Urology Focus. 2017 doi: 10.1016/j.euf.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Lehman CD, Gatsonis C, Kuhl CK, Hendrick RE, Pisano ED, Hanna L, Peacock S, Smazal SF, Maki DD, Julian TB. MRI evaluation of the contralateral breast in women with recently diagnosed breast cancer. N Engl J Med. 2007;2007:1295–1303. doi: 10.1056/NEJMoa065447. [DOI] [PubMed] [Google Scholar]
- 7.Saslow D, Boetes C, Burke W, Harms S, Leach MO, Lehman CD, Morris E, Pisano E, Schnall M, Sener S. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography. CA: a cancer journal for clinicians. 2007;57:75–89. doi: 10.3322/canjclin.57.2.75. [DOI] [PubMed] [Google Scholar]
- 8.Bar-Shalom R, Yefremov N, Guralnik L, Gaitini D, Frenkel A, Kuten A, Altman H, Keidar Z, Israel O. Clinical performance of PET/CT in evaluation of cancer: additional value for diagnostic imaging and patient management. Journal of nuclear medicine. 2003;44:1200–1209. [PubMed] [Google Scholar]
- 9.de Rooij M, Hamoen EH, Witjes JA, Barentsz JO, Rovers MM. Accuracy of magnetic resonance imaging for local staging of prostate cancer: a diagnostic meta-analysis. European urology. 2016;70:233–245. doi: 10.1016/j.eururo.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 10.De Visschere P, Lumen N, Ost P, Decaestecker K, Pattyn E, Villeirs G. Dynamic contrast-enhanced imaging has limited added value over T2-weighted imaging and diffusion-weighted imaging when using PI-RADSv2 for diagnosis of clinically significant prostate cancer in patients with elevated PSA. Clinical radiology. 2017;72:23–32. doi: 10.1016/j.crad.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Head J N.R.-M.D. Cooperative. Dynamic contrast-enhanced MRI detects acute radiotherapy-induced alterations in mandibular microvasculature: prospective assessment of imaging biomarkers of normal tissue injury. Scientific reports. 2016;6 doi: 10.1038/srep29864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature biotechnology. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 13.Papanastassiou I, Gerochristou M, Zafeiris C, Patoulias I, Ntallas D, Isaakidis D, Papageorgiou G. Needle tract seeding after core biopsy in a knee tumor; should biopsy tract be excised? Journal of musculoskeletal & neuronal interactions. 2016;16:261. [PMC free article] [PubMed] [Google Scholar]
- 14.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer research. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England journal of medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature reviews Clinical oncology. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 18.Pantel K, Alix-Panabières C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer research. 2013;73:6384–6388. doi: 10.1158/0008-5472.CAN-13-2030. [DOI] [PubMed] [Google Scholar]
- 19.Catalona WJ, Richie JP, Ahmann FR, M’Liss AH, Scardino PT, Flanigan RC, Dekernion JB, Ratliff TL, Kavoussi LR, Dalkin BL. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: results of a multicenter clinical trial of 6,630 men. The Journal of urology. 1994;151:1283–1290. doi: 10.1016/s0022-5347(17)35233-3. [DOI] [PubMed] [Google Scholar]
- 20.Schröder FH, Carter HB, Wolters T, van den Bergh RC, Gosselaar C, Bangma CH, Roobol MJ. Early detection of prostate cancer in 2007: part 1: PSA and PSA kinetics. European urology. 2008;53:468–477. doi: 10.1016/j.eururo.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 21.Duffy M, van Dalen A, Haglund C, Hansson L, Klapdor R, Lamerz R, Nilsson O, Sturgeon C, Topolcan O. Clinical utility of biochemical markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines. European journal of cancer. 2003;39:718–727. doi: 10.1016/s0959-8049(02)00811-0. [DOI] [PubMed] [Google Scholar]
- 22.Mackay A, Patel S, Carter S, Stevens U, Laurence D, Cooper E, Neville A. Role of serial plasma CEA assays in detection of recurrent and metastatic colorectal carcinomas. Br Med J. 1974;4:382–385. doi: 10.1136/bmj.4.5941.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulmert D, Serio AM, O'Brien MF, Becker C, Eastham JA, Scardino PT, Bjork T, Berglund Gr, Vickers AJ, Lilja H. Long-term prediction of prostate cancer: prostate-specific antigen (PSA) velocity is predictive but does not improve the predictive accuracy of a single PSA measurement 15 years or more before cancer diagnosis in a large, representative, unscreened population. Journal of Clinical Oncology. 2008;26:835–841. doi: 10.1200/JCO.2007.13.1490. [DOI] [PubMed] [Google Scholar]
- 24.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;2004:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 25.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. Journal of clinical oncology. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 26.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clinical cancer research. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 27.Myung JH, Gajjar KA, Saric J, Eddington DT, Hong S. Dendrimer-Mediated Multivalent Binding for the Enhanced Capture of Tumor Cells. Angewandte Chemie International Edition. 2011;50:11769–11772. doi: 10.1002/anie.201105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myung JH, Gajjar KA, Chen J, Molokie RE, Hong S. Differential detection of tumor cells using a combination of cell rolling, multivalent binding, and multiple antibodies. Analytical chemistry. 2014;86:6088–6094. doi: 10.1021/ac501243a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warkiani ME, Khoo BL, Wu L, Tay AKP, Bhagat AAS, Han J, Lim CT. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nature protocols. 2016;11:134–148. doi: 10.1038/nprot.2016.003. [DOI] [PubMed] [Google Scholar]
- 30.Chen J-F, Zhu Y, Lu Y-T, Hodara E, Hou S, Agopian VG, Tomlinson JS, Posadas EM, Tseng H-R. Clinical applications of NanoVelcro rare-cell assays for detection and characterization of circulating tumor cells. Theranostics. 2016;6:1425. doi: 10.7150/thno.15359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen P-i, Morgan B, Trautwein J. Inertial focusing for tumor antigen–dependent and–independent sorting of rare circulating tumor cells. Science translational medicine. 2013;5:179ra147–179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, Matera J, Allard WJ, Doyle GV, Terstappen LW. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clinical Cancer Research. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 33.Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer research. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 34.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welinder C, Jansson B, Lindell G, Wenner J. Cytokeratin 20 improves the detection of circulating tumor cells in patients with colorectal cancer. Cancer letters. 2015;358:43–46. doi: 10.1016/j.canlet.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 36.Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clinical cancer research. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 37.Raimondi C, Gradilone A, Naso G, Cortesi E, Gazzaniga P. Clinical utility of circulating tumor cell counting through CellSearch®: the dilemma of a concept suspended in Limbo. OncoTargets and therapy. 2014;7:619. doi: 10.2147/OTT.S46200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rack B, Schindlbeck C, Andergassen U, Schneeweiss A, Zwingers T, Lichtenegger W, Beckmann M, Sommer H, Pantel K, Janni W. Use of circulating tumor cells (CTC) in peripheral blood of breast cancer patients before and after adjuvant chemotherapy to predict risk for relapse: The SUCCESS trial. Journal of Clinical Oncology. 2010;28:1003–1003. [Google Scholar]
- 39.Bidard F-C, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado R, Mavroudis D, Grisanti S, Generali D, Garcia-Saenz JA, Stebbing J. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. The Lancet Oncology. 2014;15:406–414. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 40.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K, Capron F, Franco D, Pazzagli M, Vekemans M. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. The American journal of pathology. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. British journal of cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei Ki, Sun J, Sherman DJ, Behrenbruch CP, Wu H. Three-Dimensional Nanostructured Substrates toward Efficient Capture of Circulating Tumor Cells. Angewandte Chemie. 2009;121:9132–9135. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Liu K, Liu J, Yu ZTF, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angewandte Chemie International Edition. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stott SL, Hsu C-H, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proceedings of the National Academy of Sciences. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werner SL, Graf RP, Landers M, Valenta DT, Schroeder M, Greene SB, Bales N, Dittamore R, Marrinucci D. Analytical validation and capabilities of the epic CTC platform: enrichment-free circulating tumour cell detection and characterization. Journal of Circulating Biomarkers. 2015;4:3. doi: 10.5772/60725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Physical biology. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Myung JH, Launiere CA, Eddington DT, Hong S. Enhanced tumor cell isolation by a biomimetic combination of E-selectin and anti-EpCAM: implications for the effective separation of circulating tumor cells (CTCs) Langmuir. 2010;26:8589–8596. doi: 10.1021/la904678p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrence MB, Springer TA. Leukocytes roll on a selectin at physiologic flow rates: distinction from and prerequisite for adhesion through integrins. Cell. 1991;65:859–873. doi: 10.1016/0092-8674(91)90393-d. [DOI] [PubMed] [Google Scholar]
- 50.Rüster B, Göttig S, Ludwig RJ, Bistrian R, Müller S, Seifried E, Gille J, Henschler R. Mesenchymal stem cells display coordinated rolling and adhesion behavior on endothelial cells. Blood. 2006;108:3938–3944. doi: 10.1182/blood-2006-05-025098. [DOI] [PubMed] [Google Scholar]
- 51.Giavazzi R, Foppolo M, Dossi R, Remuzzi A. Rolling and adhesion of human tumor cells on vascular endothelium under physiological flow conditions. Journal of Clinical Investigation. 1993;92:3038. doi: 10.1172/JCI116928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Myung JH, Roengvoraphoj M, Tam KA, Ma T, Memoli VA, Dmitrovsky E, Freemantle SJ, Hong S. Effective capture of circulating tumor cells from a transgenic mouse lung cancer model using dendrimer surfaces immobilized with anti-EGFR. Analytical chemistry. 2015;87:10096–10102. doi: 10.1021/acs.analchem.5b02766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gakhar G, Navarro VN, Jurish M, Lee GY, Tagawa ST, Akhtar NH, Seandel M, Geng Y, Liu H, Bander NH. Circulating tumor cells from prostate cancer patients interact with E-selectin under physiologic blood flow. PloS one. 2013;8:e85143. doi: 10.1371/journal.pone.0085143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nature Reviews Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim M-Y, Oskarsson T, Acharyya S, Nguyen DX, Zhang XHF, Norton L, Massagué J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nourshargh S, Hordijk PL, Sixt M. Breaching multiple barriers: leukocyte motility through venular walls and the interstitium. Nature reviews Molecular cell biology. 2010;11:366–378. doi: 10.1038/nrm2889. [DOI] [PubMed] [Google Scholar]
- 57.Hong S, Leroueil PR, Majoros IJ, Orr BG, Baker JR, Holl MMB. The binding avidity of a nanoparticle-based multivalent targeted drug delivery platform. Chemistry & biology. 2007;14:107–115. doi: 10.1016/j.chembiol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 58.Dimitroff CJ, Lechpammer M, Long-Woodward D, Kutok JL. Rolling of human bone-metastatic prostate tumor cells on human bone marrow endothelium under shear flow is mediated by E-selectin. Cancer research. 2004;64:5261–5269. doi: 10.1158/0008-5472.CAN-04-0691. [DOI] [PubMed] [Google Scholar]
- 59.McEver R. Selectins: novel receptors that mediate leukocyte adhesion during inflammation. Thrombosis and haemostasis. 1991;65:223–228. [PubMed] [Google Scholar]
- 60.Tedder T, Steeber D, Chen A, Engel P. The selectins: vascular adhesion molecules. The FASEB Journal. 1995;9:866–873. [PubMed] [Google Scholar]
- 61.Köhler S, Ullrich S, Richter U, Schumacher U. E-/P-selectins and colon carcinoma metastasis: first in vivo evidence for their crucial role in a clinically relevant model of spontaneous metastasis formation in the lung. British journal of cancer. 2010;102:602–609. doi: 10.1038/sj.bjc.6605492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miles A, Liaskou E, Eksteen B, Lalor PF, Adams DH. CCL25 and CCL28 promote α 4 β 7-integrin-dependent adhesion of lymphocytes to MAdCAM-1 under shear flow. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2008;294:G1257–G1267. doi: 10.1152/ajpgi.00266.2007. [DOI] [PubMed] [Google Scholar]
- 63.Kansas GS. Selectins and their ligands: current concepts and controversies. Blood. 1996;88:3259–3287. [PubMed] [Google Scholar]
- 64.Yuan K, Kucik D, Singh RK, Listinsky CM, Listinsky JJ, Siegal GP. Alterations in human breast cancer adhesion-motility in response to changes in cell surface glycoproteins displaying α-L-fucose moieties. International journal of oncology. 2008;32:797–807. [PMC free article] [PubMed] [Google Scholar]
- 65.Shaker OG, El-Deen MAA, El-Rahim MTA, Talaat RM. Gene expression of E-selectin in tissue and its protein level in serum of breast cancer patients. Tumori. 2006;92:524. doi: 10.1177/030089160609200610. [DOI] [PubMed] [Google Scholar]
- 66.Richter U, Schröder C, Wicklein D, Lange T, Geleff S, Dippel V, Schumacher U, Klutmann S. Adhesion of small cell lung cancer cells to E-and P-selectin under physiological flow conditions: implications for metastasis formation. Histochemistry and cell biology. 2011;135:499–512. doi: 10.1007/s00418-011-0804-4. [DOI] [PubMed] [Google Scholar]
- 67.Tremblay P-L, Huot J, Auger FA. Mechanisms by which E-selectin regulates diapedesis of colon cancer cells under flow conditions. Cancer research. 2008;68:5167–5176. doi: 10.1158/0008-5472.CAN-08-1229. [DOI] [PubMed] [Google Scholar]
- 68.Woodward J. Crossing the endothelium: E-selectin regulates tumor cell migration under flow conditions. Cell adhesion & migration. 2008;2:151–152. doi: 10.4161/cam.2.3.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong S, Lee D, Zhang H, Zhang JQ, Resvick JN, Khademhosseini A, King MR, Langer R, Karp JM. Covalent immobilization of p-selectin enhances cell rolling. Langmuir. 2007;23:12261–12268. doi: 10.1021/la7014397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barthel SR, Gavino JD, Descheny L, Dimitroff CJ. Targeting selectins and selectin ligands in inflammation and cancer. Expert opinion on therapeutic targets. 2007;11:1473–1491. doi: 10.1517/14728222.11.11.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tözeren A, Kleinman HK, Grant DS, Morales D, Mercurio AM, Byers SW. E-selectin-mediated dynamic interactions of breast-and colon-cancer cells with endothelial-cell monolayers. International journal of cancer. 1995;60:426–431. doi: 10.1002/ijc.2910600326. [DOI] [PubMed] [Google Scholar]
- 72.Steegmaier M, Levinovitz A, Isenmann S, Borges E. The E-selectin-ligand ESL-1 is a variant of a receptor for fibroblast growth factor. Nature. 1995;373:615. doi: 10.1038/373615a0. [DOI] [PubMed] [Google Scholar]
- 73.Paul AA, Geng JG, Darwin A, Raycroft L, Li M, Elhammer ÅP. Partial characterization of the N-linked oligosaccharide structures on P-selectin glycoprotein ligand-1 (PSGL-1) Cell research. 2001;11:28–36. doi: 10.1038/sj.cr.7290063. [DOI] [PubMed] [Google Scholar]
- 74.Zou X, Patil VRS, Dagia NM, Smith LA, Wargo MJ, Interliggi KA, Lloyd CM, Tees DF, Walcheck B, Lawrence MB. PSGL-1 derived from human neutrophils is a high-efficiency ligand for endothelium-expressed E-selectin under flow. American Journal of Physiology-Cell Physiology. 2005;289:C415–C424. doi: 10.1152/ajpcell.00289.2004. [DOI] [PubMed] [Google Scholar]
- 75.Stevenson JL, Varki A, Borsig L. Heparin attenuates metastasis mainly due to inhibition of P-and L-selectin, but non-anticoagulant heparins can have additional effects. Thrombosis Research. 2007;120:S107–S111. doi: 10.1016/S0049-3848(07)70138-X. [DOI] [PubMed] [Google Scholar]
- 76.Insug O, Otvos L, Kieber-Emmons T, Blaszczyk-Thurin M. Role of SA–Le a and E-selectin in metastasis assessed with peptide antagonist. Peptides. 2002;23:999–1010. doi: 10.1016/s0196-9781(02)00024-4. [DOI] [PubMed] [Google Scholar]
- 77.Richards RL, Moss J, Alving CR, Fishman PH, Brady RO. Choleragen (cholera toxin): a bacterial lectin. Proceedings of the National Academy of Sciences. 1979;76:1673–1676. doi: 10.1073/pnas.76.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moser M, Bauer M, Schmid S, Ruppert R, Schmidt S, Sixt M, Wang H-V, Sperandio M, Fässler R. Kindlin-3 is required for β2 integrin–mediated leukocyte adhesion to endothelial cells. Nature medicine. 2009;15:300–305. doi: 10.1038/nm.1921. [DOI] [PubMed] [Google Scholar]
- 79.Yarishkin OV, Ryu HW, Park JY, Yang MS, Hong SG, Park KH. Sulfonate chalcone as new class voltage-dependent K+ channel blocker. Bioorg Med Chem Lett. 2008;18:137–140. doi: 10.1016/j.bmcl.2007.10.114. [DOI] [PubMed] [Google Scholar]
- 80.Lee RT, Lee YC. Affinity enhancement by multivalent lectin–carbohydrate interaction. Glycoconjugate journal. 2000;17:543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]
- 81.Mammen M, Choi S-K, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angewandte Chemie International Edition. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 82.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Designing a polyvalent inhibitor of anthrax toxin. Nature biotechnology. 2001;19:958–961. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 83.Haes AJ, Van Duyne RP. A nanoscale optical biosensor: sensitivity and selectivity of an approach based on the localized surface plasmon resonance spectroscopy of triangular silver nanoparticles. Journal of the American Chemical Society. 2002;124:10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- 84.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. Influencing receptor− ligand binding mechanisms with multivalent ligand architecture. Journal of the American Chemical Society. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 85.Myung JH, Tam KA, Park SJ, Cha A, Hong S. Recent advances in nanotechnology-based detection and separation of circulating tumor cells. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2016;8:223–239. doi: 10.1002/wnan.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 87.Pagé D, Zanini D, Roy R. Macromolecular recognition: effect of multivalency in the inhibition of binding of yeast mannan to concanavalin A and pea lectins by mannosylated dendrimers. Bioorganic & medicinal chemistry. 1996;4:1949–1961. doi: 10.1016/s0968-0896(96)00177-0. [DOI] [PubMed] [Google Scholar]
- 88.Ashton PR, Anderson DW, Brown CL, Shipway AN, Stoddart JF, Tolley MS. The synthesis and characterization of a new family of polyamide dendrimers. Chemistry-A European Journal. 1998;4:781–795. [Google Scholar]
- 89.Sunoqrot S, Bae JW, Pearson RM, Shyu K, Liu Y, Kim D-H, Hong S. Temporal control over cellular targeting through hybridization of folate-targeted dendrimers and PEG-PLA nanoparticles. Biomacromolecules. 2012;13:1223–1230. doi: 10.1021/bm300316n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kukowska-Latallo JF, Candido KA, Cao Z, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR. Nanoparticle targeting of anticancer drug improves therapeutic response in animal model of human epithelial cancer. Cancer research. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 91.Makidon PE, Nigavekar SS, Bielinska AU, Mank N, Shetty AM, Suman J, Knowlton J, Myc A, Rook T, Baker JR., Jr Characterization of stability and nasal delivery systems for immunization with nanoemulsion-based vaccines. Journal of aerosol medicine and pulmonary drug delivery. 2010;23:77–89. doi: 10.1089/jamp.2009.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Launiere C, Gaskill M, Czaplewski G, Myung JH, Hong S, Eddington DT. Channel surface patterning of alternating biomimetic protein combinations for enhanced microfluidic tumor cell isolation. Analytical chemistry. 2012;84:4022–4028. doi: 10.1021/ac2033408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kiessling LL, Pohl NL. Strength in numbers: non-natural polyvalent carbohydrate derivatives. Chemistry & biology. 1996;3:71–77. doi: 10.1016/s1074-5521(96)90280-x. [DOI] [PubMed] [Google Scholar]
- 94.Quintana A, Raczka E, Piehler L, Lee I, Myc A, Majoros I, Patri AK, Thomas T, Mulé J, Baker JR. Design and function of a dendrimer-based therapeutic nanodevice targeted to tumor cells through the folate receptor. Pharmaceutical research. 2002;19:1310–1316. doi: 10.1023/a:1020398624602. [DOI] [PubMed] [Google Scholar]
- 95.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nature biotechnology. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 96.Kitov PI, Bundle DR. On the nature of the multivalency effect: a thermodynamic model. Journal of the American Chemical Society. 2003;125:16271–16284. doi: 10.1021/ja038223n. [DOI] [PubMed] [Google Scholar]
- 97.Singh P, Gupta U, Asthana A, Jain NK. Folate and folate− PEG− PAMAM Dendrimers: synthesis, characterization, and targeted anticancer drug delivery potential in tumor bearing mice. Bioconjugate chemistry. 2008;19:2239–2252. doi: 10.1021/bc800125u. [DOI] [PubMed] [Google Scholar]
- 98.Bae CW, Cho YH, Hong SH, Kim JH, Lee JK, Kim CJ. The anatomical location and course of the facial nerve in vestibular schwannomas : a study of 163 surgically treated cases. J Korean Neurosurg Soc. 2007;42:450–454. doi: 10.3340/jkns.2007.42.6.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Woller EK, Walter ED, Morgan JR, Singel DJ, Cloninger MJ. Altering the strength of lectin binding interactions and controlling the amount of lectin clustering using mannose/hydroxyl-functionalized dendrimers. Journal of the American Chemical Society. 2003;125:8820–8826. doi: 10.1021/ja0352496. [DOI] [PubMed] [Google Scholar]
- 100.Cheng Z, Thorek DL, Tsourkas A. Gadolinium-Conjugated Dendrimer Nanoclusters as a Tumor-Targeted T1 Magnetic Resonance Imaging Contrast Agent. Angewandte Chemie International Edition. 2010;49:346–350. doi: 10.1002/anie.200905133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Z, Huang P, Zhang X, Lin J, Yang S, Liu B, Gao F, Xi P, Ren Q, Cui D. RGD-conjugated dendrimer-modified gold nanorods for in vivo tumor targeting and photothermal therapy. Molecular pharmaceutics. 2009;7:94–104. doi: 10.1021/mp9001415. [DOI] [PubMed] [Google Scholar]
- 102.Li Z, Huang P, He R, Lin J, Yang S, Zhang X, Ren Q, Cui D. Aptamer-conjugated dendrimer-modified quantum dots for cancer cell targeting and imaging. Materials letters. 2010;64:375–378. doi: 10.1166/jnn.2010.2217. [DOI] [PubMed] [Google Scholar]
- 103.Stears RL, Getts RC, Gullans SR. A novel, sensitive detection system for high-density microarrays using dendrimer technology. Physiological Genomics. 2000;3:93–99. doi: 10.1152/physiolgenomics.2000.3.2.93. [DOI] [PubMed] [Google Scholar]
- 104.Tang J, Huang Y, Zhang C, Liu H, Tang D. Amplified impedimetric immunosensor based on instant catalyst for sensitive determination of ochratoxin A. Biosensors and Bioelectronics. 2016;86:386–392. doi: 10.1016/j.bios.2016.06.080. [DOI] [PubMed] [Google Scholar]
- 105.Ajikumar PK, Ng JK, Tang YC, Lee JY, Stephanopoulos G, Too H-P. Carboxyl-terminated dendrimer-coated bioactive interface for protein microarray: High-sensitivity detection of antigen in complex biological samples. Langmuir. 2007;23:5670–5677. doi: 10.1021/la063717u. [DOI] [PubMed] [Google Scholar]
- 106.Benters R, Niemeyer C, Wöhrle D. Dendrimer-activated solid supports for nucleic acid and protein microarrays. Chem Bio Chem. 2001;2:686–694. doi: 10.1002/1439-7633(20010903)2:9<686::AID-CBIC686>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 107.Karnik R, Hong S, Zhang H, Mei Y, Anderson DG, Karp JM, Langer R. Nanomechanical control of cell rolling in two dimensions through surface patterning of receptors. Nano letters. 2008;8:1153–1158. doi: 10.1021/nl073322a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Attard G, Jameson C, Moreira J, Flohr P, Parker C, Dearnaley D, Cooper CS, De Bono JS. Hormone-sensitive prostate cancer: a case of ETS gene fusion heterogeneity. Journal of clinical pathology. 2009;62:373–376. doi: 10.1136/jcp.2008.061515. [DOI] [PubMed] [Google Scholar]
- 109.Gorges TM, Tinhofer I, Drosch M, Röse L, Zollner TM, Krahn T, von Ahsen O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hsieh S-M, Look MP, Sieuwerts AM, Foekens JA, Hunter KW. Distinct inherited metastasis susceptibility exists for different breast cancer subtypes: a prognosis study. Breast Cancer Research. 2009;11:R75. doi: 10.1186/bcr2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matsumoto M, Shigeta A, Furukawa Y, Tanaka T, Miyasaka M, Hirata T. CD43 collaborates with P-selectin glycoprotein ligand-1 to mediate E-selectin-dependent T cell migration into inflamed skin. The Journal of Immunology. 2007;178:2499–2506. doi: 10.4049/jimmunol.178.4.2499. [DOI] [PubMed] [Google Scholar]
- 112.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Science translational medicine. 2012;4:127rv123–127rv123. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Subik K, Lee J-F, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung M-C, Bonfiglio T, Hicks DG. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast cancer: basic and clinical research. 2010;4:35. [PMC free article] [PubMed] [Google Scholar]
- 114.Myung JH, Caster JM, Eblan MJ, Wang K, Roche K, Park S, Tam KA, Miller SM, Chen RC, Zhang T, Tepper JE, Chera BS, Wang AZ, Hong S. Sensitive and specific capture of circulating tumor cells (CTCs) using a biomimetic chip. 2017 Submitted. [Google Scholar]
- 115.Grisanti S, Almici C, Consoli F, Buglione M, Verardi R, Bolzoni-Villaret A, Bianchetti A, Ciccarese C, Mangoni M, Ferrari L. Circulating tumor cells in patients with recurrent or metastatic head and neck carcinoma: prognostic and predictive significance. PloS one. 2014;9:e103918. doi: 10.1371/journal.pone.0103918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gröbe A, Blessmann M, Hanken H, Friedrich RE, Schön G, Wikner J, Effenberger KE, Kluwe L, Heiland M, Pantel K. Prognostic relevance of circulating tumor cells in blood and disseminated tumor cells in bone marrow of patients with squamous cell carcinoma of the oral cavity. Clinical Cancer Research. 2014;20:425–433. doi: 10.1158/1078-0432.CCR-13-1101. [DOI] [PubMed] [Google Scholar]
- 117.Nichols AC, Lowes LE, Szeto CC, Basmaji J, Dhaliwal S, Chapeskie C, Todorovic B, Read N, Venkatesan V, Hammond A. Detection of circulating tumor cells in advanced head and neck cancer using the CellSearch system. Head & neck. 2012;34:1440–1444. doi: 10.1002/hed.21941. [DOI] [PubMed] [Google Scholar]
- 118.Hirsch F, Varella-Garcia M, Cappuzzo F. Predictive value of EGFR and HER2 overexpression in advanced non-small-cell lung cancer. Oncogene. 2009;28:S32. doi: 10.1038/onc.2009.199. [DOI] [PubMed] [Google Scholar]
- 119.Freemantle SJ, Dmitrovsky E. Cyclin E transgenic mice: discovery tools for lung cancer biology, therapy, and prevention. Cancer prevention research. 2010;3:1513–1518. doi: 10.1158/1940-6207.CAPR-10-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Müller-Tidow C, Metzger R, Kügler K, Diederichs S, Idos G, Thomas M, Dockhorn-Dworniczak B, Schneider PM, Koeffler HP, Berdel WE. Cyclin E is the only cyclin-dependent kinase 2-associated cyclin that predicts metastasis and survival in early stage non-small cell lung cancer. Cancer research. 2001;61:647–653. [PubMed] [Google Scholar]
- 121.Kawakami M, Mustachio LM, Rodriguez-Canales J, Mino B, Roszik J, Tong P, Wang J, Lee JJ, Myung JH, Heymach JV. Next-Generation CDK2/9 Inhibitors and Anaphase Catastrophe in Lung Cancer. JNCI: Journal of the National Cancer Institute. 2017;109 doi: 10.1093/jnci/djw297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kourtidis A, Srinivasaiah R, Carkner RD, Brosnan MJ, Conklin DS. Peroxisome proliferator-activated receptor-γ protects ERBB2-positive breast cancer cells from palmitate toxicity. Breast cancer research. 2009;11:R16. doi: 10.1186/bcr2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Camirand A, Lu Y, Pollak M. Co-targeting HER2/ErbB2 and insulin-like growth factor-1 receptors causes synergistic inhibition of growth in HER2-overexpressing breast cancer cells. Medical Science Monitor. 2002;8:BR521–BR526. [PubMed] [Google Scholar]
- 124.Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, Goodman JC, Groves MD, Marchetti D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Science translational medicine. 2013;5:180ra148–180ra148. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cayrefourcq L, Mazard T, Joosse S, Solassol J, Ramos J, Assenat E, Schumacher U, Costes V, Maudelonde T, Pantel K. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer research. 2015;75:892–901. doi: 10.1158/0008-5472.CAN-14-2613. [DOI] [PubMed] [Google Scholar]
- 126.Zhang Z, Xu J, Hong B, Chen X. The effects of 3D channel geometry on CTC passing pressure– towards deformability-based cancer cell separation. Lab on a Chip. 2014;14:2576–2584. doi: 10.1039/c4lc00301b. [DOI] [PubMed] [Google Scholar]
- 127.Tehfe M-A, Dumur F, Graff B, Clément J-L, Gigmes D, Morlet-Savary F, Fouassier J-P, Lalevée J. New Cleavable Photoinitiator Architecture with Huge Molar Extinction Coefficients for Polymerization in the 340–450 nm Range. Macromolecules. 2013;46:736–746. [Google Scholar]
- 128.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T. Capture and stimulated release of circulating tumor cells on Polymer-Grafted silicon nanostructures. Advanced materials. 2013;25:1547–1551. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang H, He J, Zhang M, Tao Y, Li F, Tam KC, Ni P. Biocompatible and acid-cleavable poly (ε-caprolactone)-acetal-poly (ethylene glycol)-acetal-poly (ε-caprolactone) triblock copolymers: synthesis, characterization and pH-triggered doxorubicin delivery. Journal of Materials Chemistry B. 2013;1:6596–6607. doi: 10.1039/c3tb21170c. [DOI] [PubMed] [Google Scholar]
- 130.Hatch A, Hansmann G, Murthy SK. Engineered alginate hydrogels for effective microfluidic capture and release of endothelial progenitor cells from whole blood. Langmuir. 2011;27:4257–4264. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu H, Xu S, He Z, Deng A, Zhu J-J. Supersandwich cytosensor for selective and ultrasensitive detection of cancer cells using aptamer-DNA concatamer-quantum dots probes. Analytical chemistry. 2013;85:3385–3392. doi: 10.1021/ac303789x. [DOI] [PubMed] [Google Scholar]
- 132.Ahr A, Scharl A, Müller M, von Minckwitz G, Gätje R, Pantel K, Kaufmann M. Cross-reactive staining of normal bone-marrow cells by monoclonal antibody 2E11. International journal of cancer. 1999;84:502–505. doi: 10.1002/(sici)1097-0215(19991022)84:5<502::aid-ijc9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 133.Fritschy JM. Is my antibody-staining specific? How to deal with pitfalls of immunohistochemistry. European Journal of Neuroscience. 2008;28:2365–2370. doi: 10.1111/j.1460-9568.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- 134.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen CL, Mahalingam D, Osmulski P, Jadhav RR, Wang CM, Leach RJ, Chang TC, Weitman SD, Kumar AP, Sun L. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. The Prostate. 2013;73:813–826. doi: 10.1002/pros.22625. [DOI] [PMC free article] [PubMed] [Google Scholar]