Significance
The GluN1 subunit of NMDA receptors has multiple splice variants with distinct temporal and spatial expression patterns. The in vivo function of GluN1 splice variants is unknown. Here we provide evidence that N-terminal splicing of GluN1 regulates developmental remodeling of synaptic NMDA receptors and maturation of excitatory synapses. Using mice carrying a deletion of the exon 5 of Grin1, we show that the inclusion of the N-terminal cassette of GluN1 shortens the time course of NMDA receptor-mediated excitatory synaptic currents in the thalamus and cortex. Deletion of Grin1 exon 5 causes an overproduction of excitatory synapses in the cortex of young mice and an increase in seizure susceptibility in adult mice.
Keywords: NMDA receptor, alternative splicing, synaptic maturation, seizure, mouse model
Abstract
The majority of NMDA receptors (NMDARs) in the brain are composed of 2 GluN1 and 2 GluN2 subunits. The inclusion or exclusion of 1 N-terminal and 2 C-terminal domains of GluN1 results in 8 splicing variants that exhibit distinct temporal and spatial patterns of expression and functional properties. However, previous functional analyses of Grin1 variants have been done using heterologous expression and the in vivo function of Grin1 splicing is unknown. Here we show that N-terminal splicing of GluN1 has important functions in the maturation of excitatory synapses. The inclusion of exon 5 of Grin1 is up-regulated in several brain regions such as the thalamus and neocortex. We find that deletion of Grin1 exon 5 disrupts the developmental remodeling of NMDARs in thalamic neurons and the effect is distinct from that of Grin2a (GluN2A) deletion. Deletion of Grin2a or exon 5 of Grin1 alone partially attenuates the shortening of NMDAR-mediated excitatory postsynaptic currents (NMDAR-EPSCs) during early life, whereas deletion of both Grin2a and exon 5 of Grin1 completely abolishes the developmental change in NMDAR-EPSC decay time. Deletion of exon 5 of Grin1 leads to an overproduction of excitatory synapses in layer 5 pyramidal neurons in the cortex and increases seizure susceptibility in adult mice. Our findings demonstrate that N-terminal splicing of GluN1 has important functions in synaptic maturation and neuronal network excitability.
Activation of NMDA receptors (NMDARs) by glutamate at excitatory synapses opens cation-selective channels with high permeability to Ca2+ ions, and generates excitatory postsynaptic currents (EPSCs) with slow time course. NMDARs are heterotetramers assembled from the GluN1 subunit, 4 distinct GluN2 subunits (GluN2A, 2B, 2C, and 2D), and 2 GluN3 subunits (GluN3A and GluN3B) (1–3). The majority of NMDARs in the brain are tetramers composed of 2 GluN1 and 2 GluN2 subunits. The 4 GluN2 subunits show distinct spatial and temporal expression patterns (4, 5). Native NMDARs in neurons are highly diverse in functional properties, and much of this diversity has been attributed to the GluN2 subunit composition of the receptor (2).
Alternative splicing of Grin1 mRNA provides additional molecular diversity for NMDARs. The inclusion or exclusion of 1 N-terminal and 2 C-terminal domains of GluN1 results in 8 splicing variants that exhibit distinct temporal and spatial patterns of expression (6) and functional properties (7). The 2 C-terminal domains, both located intracellularly, have been implicated in the regulation of the retention by the endoplasmic reticulum and cell surface trafficking of NMDARs (8, 9). The N-terminal domain, encoded by the 63-bp exon 5 of Grin1, is located near the dimer interface of ligand-binding domains of NMDARs (10, 11). Studies using recombinant NMDARs show that the N-terminal splicing of Grin1 mRNA regulates agonist affinity, deactivation, and proton and zinc sensitivities of the receptor (11–16).
Synaptic NMDARs undergo dramatic changes throughout the brain during development. A common feature in the development of glutamatergic synapses is a reduction in time course of NMDAR-mediated EPSCs (NMDAR-EPSCs) during early life (17–19). Studies in the cortex and hippocampus have demonstrated that developmental change in EPSC kinetics is caused by a switch from GluN2B to GluN2A-containing NMDARs (20–24). GluN2A-containing NMDARs show faster deactivation kinetics than GluN2B-containing receptors (25–27). The switch from GluN2B to GluN2A causes important changes in NMDAR function such as altered intracellular signaling (28, 29).
It is unclear whether the switch from GluN2B to GluN2A is solely responsible for the developmental change in NMDAR kinetics. The inclusion of Grin1 exon 5 is up-regulated in the brain during early life and recombinant NMDARs composed of exon-5-containing GluN1 show lower affinity for glutamate (12, 13) and faster deactivation than those without exon 5 (15, 16). However, whether the N-terminal splicing of Grin1 is implicated in the maturation of synaptic NMDARs is unknown.
In this study, we investigate the roles of GluN2A and N-terminal splicing of Grin1 in developmental remodeling of NMDARs in the thalamus of the mouse. We find that both GluN2A and Grin1 exon 5 splicing are implicated in the maturation of NMDAR function at glutamatergic synapses. Grin1 exon 5 is predominantly involved in the early phase of receptor maturation, whereas GluN2A is responsible for the late phase. In addition, we show that Grin1 exon 5 splicing regulates the maturation of excitatory transmission in the cortex and seizure susceptibility of adult mice.
Results
Deletion of Grin2a Partially Blocked the Developmental Change in Decay Time of NMDAR-EPSCs.
Whether the switch from GluN2B to GluN2A is solely responsible for the developmental change in NMDAR kinetics remains unknown. We addressed this question at the lemniscal synapse in the thalamus of the mouse. This synapse, formed between an axon of the principal sensory trigeminal nucleus (Pr5) and a neuron in ventral posteromedial nucleus (VPm) of the thalamus, relays tactile information from large vibrissa to the layer 4 of the somatosensory cortex (S1). Previous studies have shown that this is an excitatory synapse with large NMDAR-EPSCs (30), and the decay time of NMDAR-EPSCs shortens markedly during the second week after birth (30).
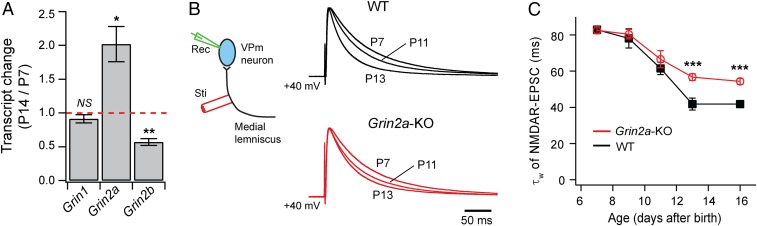
We performed RT-qPCR analysis of GluN2A, GluN2B, and GluN1 in the thalamus of wild-type C57BL/6J (B6) mice at P7 and P14 (Fig. 1A). GluN2A (Grin2a) and GluN2B (Grin2b) showed opposite changes, with the level of Grin2a transcript doubled between P7 and P14 and that of Grin2b reduced by 40% during the same period. In contrast, the level of total Grin1 transcript was not significantly different between P7 and P14. These results suggest that the expression of Grin2a is up-regulated in the thalamus during early life.
Fig. 1.
Up-regulation of GluN2A is responsible for the late phase of NMDAR maturation at the thalamic relay synapse. (A) Fold changes (mean ± SEM) in mRNA level (P14 over P7) detected by RT-qPCR. Data were obtained from 5 B6 mice each at P7 and P14. *P < 0.05; **P < 0.01; NS, P > 0.2, t test. (B) The diagram on the left shows the recording method. Traces on the right are normalized NMDAR-EPSCs recorded at +40 mV from WT and Grin2a-KO mice at P7, P11, and P13. Each trace is the averaged response from 10 to 15 cells. (C) Plots of weighted decay constant of NMDAR-EPSCs versus age for WT and Grin2a-KO. For each age group, data include 10–15 cells from 2 to 4 mice of either genotypes. ***P < 0.0001, Mann–Whitney U test.
Next, we examined NMDAR-EPSCs at the lemniscal synapse during early life in mice deficient of GluN2A and wild type (WT) control mice. Patch clamp recordings were obtained from VPm neurons in acute slices obtained from Grin2a-KO and WT littermates aged from P7 to P16, and EPSCs were evoked at lemniscal synapses by brief current pulses applied to the medial lemniscus (Fig. 1B). NMDAR-EPSCs were recorded at +40 mV. In WT mice, the decay time of NMDAR-EPSCs showed a marked decline between P7 and P13, and remained steady between P13 and P16 (Fig. 1 B and C). Deletion of Grin2a partially attenuated the reduction in NMDAR-EPSC decay time. In Grin2a-KO mice, the decay time constant of NMDAR-EPSCs was not different from that of WT neurons at P7, P9, and P11 (P > 0.05), but was much larger than that of WT neurons at P13 and P16 (Fig. 1 B and C). In other words, deletion of Grin2a had no effect on the decline of NMDAR-EPSC decay time that occurred between P7 and P11 but strongly attenuated the decline that occurred between P11 and P13. Quantitative RT-PCR analyses showed that deletion of Grin2a did not alter the expression of Grin2b or exon-5-containing Grin1 in the thalamus (SI Appendix, Fig. S1). These results suggest that at the lemniscal synapse, the shortening of NMDAR-EPSCs during early life involves multiple mechanisms, and GluN2A is selectively implicated in the late phase of NMDAR maturation.
N-Terminal Splicing of Grin1 mRNA Regulates the Decay Time of NMDAR-EPSCs.
Grin1 exon 5 is highly conserved across vertebrates. DNA sequences of Grin1 exon 5 are 100% identical between human, macaque, and mouse, and 95% identical between human and Xenopus (SI Appendix, Fig. S2), suggesting the importance of this exon in vertebrates. Previous studies have shown that in the rat, the inclusion of Grin1 exon 5 is up-regulated during early life in several brain regions including the thalamus (6, 31). To determine whether this up-regulation also occurs in the thalamus of the mouse, we performed RT-PCR and RT-qPCR using thalamic tissues of B6 mice. We found that the inclusion of this exon was significantly up-regulated in the thalamus during the second week after birth (SI Appendix, Fig. S3A). Between P6 and P14, transcripts of exon-5-containing Grin1 increased by 3-fold in the thalamus, while the total Grin1 transcript did not change (SI Appendix, Fig. S3B). These results show that the amount and proportion of GluN1 with exon 5 cassette to total GluN1 increases in the thalamus of the mouse during early life.
To analyze the in vivo function of Grin1 exon 5 splicing, we generated a mouse strain carrying a targeted deletion of Grin1 exon 5 (Grin1ΔEx5; Fig. 2A). Mice homozygous for the deletion were viable and did not show any overt phenotypes. RT-qPCR analyses of thalamic tissues showed that the deletion of Grin1 exon 5 does not alter the expression of Grin2a or Grin2b (Fig. 2B and SI Appendix, Fig. S4 A and B), up-regulation of Grin2a expression during the second week after birth (SI Appendix, Fig. S4C), or expression of Grin1 C-terminal variants (SI Appendix, Fig. S4 D and E).
Fig. 2.
Deletion of Grin1 exon 5 prolongs the time course of NMDAR-EPSCs at the thalamic relay synapse. (A) Generation of the mouse strain carrying targeted deletion of Grin1 exon 5 (Grin1ΔEx5). The Grin1 floxed Neo strain was generated by ES targeting in a C57BL/6J background. Exon 5 of Grin1 and the Pgk-Neo cassette was then excised using a germ line Cre driver. (B) RT-PCR for Grin1 exon 5, total Grin1, Grin2a, Grin2b, and the housekeeping gene Pgk using thalamic tissues from WT and Grin1ΔEx5/ΔEx5 mice. (C) Examples of NMDAR-EPSCs recorded at the lemniscal synapse from WT and Grin1ΔEx5/ΔEx5 mice at P7 and P16. EPSCs are normalized to the peak. (D) Weighted decay constants (τw) of NMDAR-EPSCs of WT (black), heterozygous (blue), and homozygous (red) mutant mice at various ages. Data are mean ± SEM from 12 to 28 cells in 3 or 4 mice. ***P < 0.0005, Mann–Whitney U or Kruskal-Wallis test.
We recorded NMDAR-EPSCs at the lemniscal synapse in homozygous Grin1ΔEx5/ΔEx5 and WT mice aged from P7 to P24. Deletion of Grin1 exon 5 significantly increased the decay time of NMDAR-EPSCs (Fig. 2 C and D). The effect of Grin1 exon-5 deletion is already present at P7, consistent with the expression data of previous studies (6). We also recorded from heterozygous Grin1ΔEx5/wt mice at P7, P11, and P16, and found that heterozygous deletion of Grin1 exon 5 caused an intermediate effect on the decay constant of NMDAR-EPSCs (Fig. 2D). Together, these results show that in addition to GluN2A, the developmental change in NMDAR-EPSC decay time is regulated by N-terminal splicing of Grin1 mRNA.
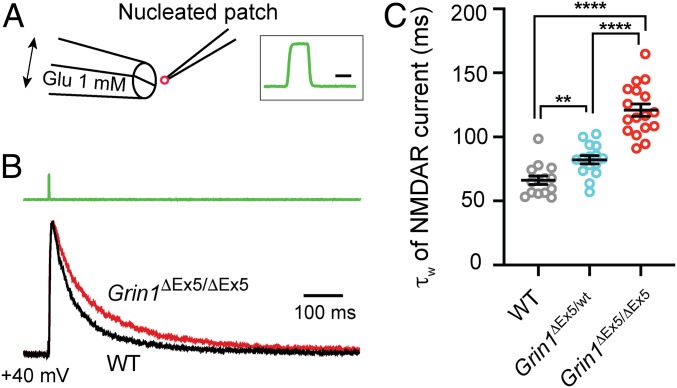
The decay time course of NMDAR-EPSCs is determined by the deactivation rate of postsynaptic NMDARs (32, 33) and the clearance of glutamate from extracellular space (34). To selectively examine the effect of Grin1-exon-5 deletion on NMDAR deactivation, we used a piezoelectric device to deliver short pulses of glutamate to nucleated outside-out patches from VPm neurons (Fig. 3A). In the constant presence of glycine and the AMPAR antagonist NBQX, a 2-ms pulse of glutamate evoked a robust NMDAR current in nucleated patches from VPm neurons (Fig. 3B). The deactivation phase of NMDAR-EPSCs was significantly slower in homozygous Grin1ΔEx5/ΔEx5 than WT neurons, whereas heterozygous neurons showed an intermediate effect (Fig. 3C). These results demonstrate that the deletion of Grin1 exon 5 leads to a much slower NMDAR-EPSC by reducing the deactivation rate of postsynaptic NMDARs.
Fig. 3.
Deletion of Grin1 exon 5 results in a slower NMDAR deactivation. (A) Schematic view of the experimental setup. Brief pulses (2 ms) of l-glutamate (Glu, 1 mM) were applied to nucleated patch using a piezo-electrical device. NBQX (10 μM) and glycine (100 μM) were present continuously. The insert on the right shows the speed of solution exchange when switching from ACSF to 30% ACSF. The 20–80% time was 117 μs for the onset and 108 μs for the offset. (Scale bar, 2 ms.) (B) Normalized NMDAR-mediated currents in response to 2-ms glutamate pulse recorded from nucleated patches obtained from a WT and a Grin1ΔEx5/ΔEx5 neuron at P14. The green trace on the top indicates the pulse application of glutamate. Recordings were performed at room temperature (21–23 °C). (C) Decay constants of NMDAR-mediated currents recorded from WT (14 cells from 3 mice), heterozygous (15 cells from 3 mice), and homozygous mutant neurons (17 cells from 4 mice) at P14-15. ****P < 0.0001, **P < 0.005, Mann–Whitney U test.
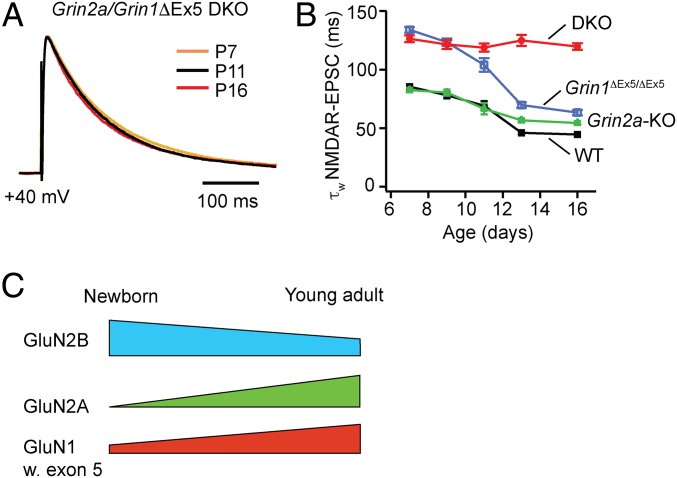
To determine whether GluN2A and Grin1 N-terminal splicing have distinct functions in the maturation of NMDARs at the synapse, we generated mice deficient of both GluN2A and exon-5-containing GluN1. Grin2a-KO/Grin1ΔEx5/ΔEx5 double mutant mice (DKO) were viable and did not show any overt phenotypes during early life. We recorded NMDAR-EPSCs at the lemniscal synapse in Grin2a/Grin1ΔEx5/ΔEx5 DKO mice aged from P7 to P16. Deletion of both Grin2a and Grin1 exon 5 completely abolished the developmental decline of NMDAR-EPSC decay time (Fig. 4 A and B). At P16, the decay constant of NMDAR-EPSCs at the lemniscal synapse of Grin2a/Grin1ΔEx5/ΔEx5 DKO mice was much larger than that of either Grin2a-KO or Grin1ΔEx5/ΔEx5 mice (Fig. 4B, P < 0.0001). These results show that GluN2A and N-terminal splicing of Grin1 have distinct functions in the regulation of the time course of NMDAR-mediated synaptic responses. Based on these results, we propose a model where both N-terminal splicing of GluN1 and up-regulation of GluN2A are implicated in NMDAR remodeling at the synapse (Fig. 4C).
Fig. 4.
Distinct roles of GluN2A and exon-5-containing GluN1 in developmental remodeling of NMDARs at thalamic relay synapses. (A) Normalized NMDAR-EPSCs recorded at VPm relay synapses from Grin1ΔEx5/Grin2a DKO mice at P7, P11, and P16. Each trace is the averaged responses from 13 to 22 cells. (B) Weighted decay constants (τw) of NMDAR-EPSCs of the 4 groups from P7 to P16. Data for Grin1ΔEx5/Grin2a DKO were from 13 to 22 cells from 3 to 4 mice for each time point. Data for Grin2a-KO and WT were the same as in Fig. 1C and data for Grin1ΔEx5 were the same as in Fig. 2D. (C) A model of developmental remodeling of NMDARs implicating both N-terminal splicing of Grin1 and up-regulation of Grin2a.
N-Terminal Splicing of Grin1 mRNA Regulates the Function and Number of Excitatory Synapses in the Neocortex.
NMDARs play important roles in the development of excitatory synapses in the brain. We hypothesize that deletion of Grin1 exon 5 would disrupt the maturation of excitatory synapses by altering NMDAR-mediated signaling. Thalamic relay synapses show a pronounced increase in the number of AMPA receptors (AMPARs) during early life (35) and this up-regulation of AMPARs is NMDAR dependent (36). We recorded AMPAR-EPSCs, NMDAR-EPSCs, AMPAR-mediated miniature EPSCs at the lemniscal synapse in WT, and Grin1ΔEx5/ΔEx5 mice at P15-16. There was no difference between the 2 groups in the amplitudes of AMPAR- or NMDAR-EPSCs, AMPAR/NMDAR ratio, or properties of mEPSCs (SI Appendix, Fig. S5). These results suggest that N-terminal splicing of Grin1 is not implicated in developmental up-regulation of AMPARs at lemniscal synapses.
Besides the thalamus, the inclusion of Grin1 exon 5 is up-regulated in several other regions in the brain. In the neocortex, exon-5-containing Grin1 transcripts are strongly expressed in layer 5 and 2/3 by 2 wk of age (6). We analyzed NMDAR-EPSCs in layer 5 pyramidal neurons in the somatosensory cortex. Neurons in Grin1ΔEx5/ΔEx5 mice showed a slower decay time of NMDAR-EPSCs (Fig. 5 A and B), suggesting that Grin1 exon 5 is implicated in developmental remodeling of synaptic NMDARs in these neurons. Next, we analyzed quantal synaptic transmission in layer 5 pyramidal neurons. Deletion of Grin1 exon 5 caused a 47% increase in the frequency of mEPSCs (Fig. 5 C and D) without any change in the amplitude (Fig. 5E), rise time, or decay time of mEPSCs. There was no change in the properties of miniature inhibitory postsynaptic currents (mIPSCs) in these neurons (SI Appendix, Fig. S6).
Fig. 5.
Deletion of Grin1 exon 5 leads to an increase in the number of excitatory synapses in layer 5 pyramidal neurons in the cortex. (A, Left) A schematic view of the recording method. (A, Right) Normalized NMDAR-EPSCs recorded from WT and Grin1ΔEx5/ΔEx5 mice at P17-18. (B) Weighted decay constants of NMDAR-EPSCs from WT (22 cells from 4 mice) and Grin1ΔEx5/ΔEx5 neurons (24 cells from 4 mice) at P17-18. ****P < 0.0001, Mann–Whitney U test. (C) Examples of mEPSCs recorded layer 5 pyramidal neurons in a WT and a Grin1ΔEx5/ΔEx5 mouse at P18. (Scale bars, 100 ms, 20 pA.) (D and E) Frequency and amplitude of mEPSCs from WT (33 cells from 4 mice) and mutant neurons (30 cells from 4 mice) at P17-18. ****P < 0.0001; NS, P > 0.05, Mann–Whitney U test. The mean rise time of mEPSCs was 0.48 ± 0.02 ms for WT and 0.48 ± 0.01 ms for mutant (P = 0.43). The mean decay time of mEPSC was 2.53 ± 0.05 ms for WT and 2.55 ± 0.05 ms for mutant (P = 0.81). (F) Examples of dendritic spines along apical dendrites of layer 5 pyramidal neurons from WT and Grin1ΔEx5/ΔEx5 at P18. (Scale bar, 4 μm.) (G and H) Spine density of apical and basal dendrites of WT (18 cells from 4 mice) and Grin1ΔEx5/ΔEx5 neurons (16 cells from 4 mice) at P17-18. ***P < 0.001, Mann–Whitney U test.
The increased mEPSC frequency in mutant neurons indicates that the number of excitatory synapses is increased. We labeled layer 5 pyramidal neurons with biocytin and analyzed dendritic spines (Fig. 5F). Spine density was significantly higher in mutant neurons and similar changes were found in apical and basal dendrites (Fig. 5 G and H). These results suggest that deletion of Grin1 exon 5 selectively enhances excitatory synaptic transmission in cortical neurons by increasing the number of excitatory synapses.
N-Terminal Splicing of Grin1 mRNA Regulates Seizure Susceptibility.
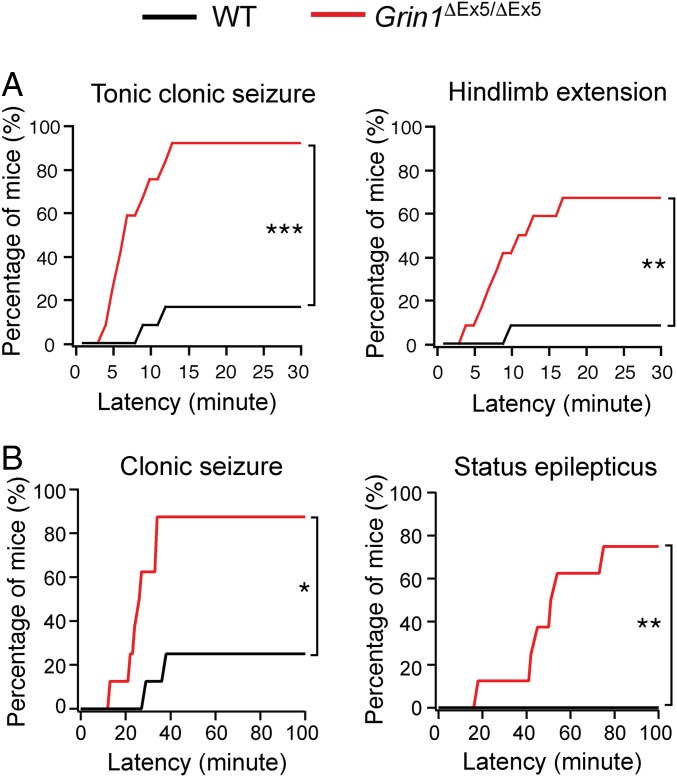
Abnormal regulation of excitatory transmission is implicated in epilepsy. The slower NMDAR-EPSC decay time and increased number of excitatory synapses in Grin1ΔEx5/ΔEx5 mice would result in hyperexcitation of neuronal circuits in these mice. On a B6 background Grin1ΔEx5/ΔEx5 mutant mice rarely showed spontaneous convulsive seizures. However, we reasoned that the enhanced NMDAR function and increased excitatory transmission in Grin1ΔEx5 mutant mice would make them more susceptible to seizures. To test this idea, we first examined seizure threshold of adult male Grin1ΔEx5/ΔEx5 and WT mice using pentylenetetrazol (PTZ). Grin1ΔEx5/ΔEx5 mice showed a much higher incidence of generalized tonic clonic seizures and hindlimb extension than WT mice (Fig. 6A). Similar results were obtained in female mice (SI Appendix, Fig. S7). Next, we examined seizure responses to kainic acid (KA). Seizure threshold was significantly lower in Grin1ΔEx5/ΔEx5 mutant mice than WT mice as measured by clonic seizures and status epilepticus (Fig. 6B). However, we did not find any cell death in the brains of mutant or WT mice injected with KA (SI Appendix, Fig. S8). Together, these results suggest that deletion of Grin1 exon 5 leads to increased seizure susceptibility in adult animals.
Fig. 6.
Deletion of Grin1 exon 5 decreases seizure threshold in adult mice. (A) Percentage of mice with seizure in response to a single dose of PTZ (50 mg/kg, i.p.). The Left shows the latency to the first episode of generalized tonic-clonic seizure; the Right shows the latency to seizure with hindlimb extension. Data were obtained from 12 male mice per genotype. **P < 0.01; ***P < 0.001, Fisher’s exact test. (B) Percentage of mice with seizure in response to a single dose of KA (15 mg/kg, i.p.). The Left shows the latency to the first episode of clonic seizure; the Right shows the latency to status epilepticus. Data were obtained from eight male mice per genotype. *P < 0.05; **P < 0.01, Fisher’s exact test.
Discussion
Using genetic manipulations in the mouse, we show that the deletion of Grin1 exon 5 disrupts the maturation of NMDARs at excitatory synapses. The effect of Grin1 exon-5 deletion on the time course of NMDAR-EPSCs is distinct from that of Grin2a deletion. The deletion of Grin1 exon 5 leads to an overproduction of excitatory synapses and decreased seizure threshold. Our findings suggest that N-terminal splicing of Grin1 has important functions in the maturation of excitatory synapses.
Previous studies showed that the inclusion of Grin1 exon 5 reduces affinities of glycine and glutamate binding (7, 16, 37), and sensitivities to proton (14) and zinc (13, 38, 39) of recombinant NMDARs. Consistent with the reduction in agonist binding affinity, NMDARs with Grin1 exon 5 have faster deactivation rates than those without the exon (15, 16). These findings suggest that N-terminal splicing of Grin1 may have important physiological functions. However, all these studies have been performed using heterologous expression and consequently, nothing is known about the function of Grin1 N-terminal splicing in vivo.
We have addressed this question by generating mice without Grin1 exon 5. We find that the deletion of Grin1 exon 5 leads to a much slower decay time of NMDAR-EPSCs at thalamic and cortical synapses and this effect is associated with a slower deactivation of NMDARs. Our results provide evidence that Grin1 N-terminal splicing has important functions in synaptic transmission.
Shortening of NMDAR-EPSC decay time is a common feature of glutamatergic synaptic maturation, and it has been attributed to the shift from GluN2B to GluN2A. Gray et al. have shown that the deletion of Grin2a completely abolishes the developmental shortening of NMDAR-EPSC decay time in CA1 pyramidal neurons (23), suggesting that the up-regulation of GluN2A is solely responsible for this change in these neurons. In contrast, we find that in VPm neurons, deletion of Grin2a or Grin1 exon 5 alone partially blocks the developmental shortening of NMDAR-EPSC decay time and deletion of both Grin2a and Grin1 exon 5 completely abolishes the change. These findings demonstrate that up-regulation of both GluN2A and GluN1 with exon 5 cassette contributes to the shortening of NMDAR-EPSC decay time in thalamic neurons. The difference between our results and those found in CA1 neurons may due to cell-type-specific regulation of Grin1 mRNA splicing. Unlike thalamic neurons, Grin1 exon 5 is expressed at very low levels in CA1 neurons (6). Previous studies have shown that while the expression of GluN1 lacking exon 5 cassette is high and ubiquitous in the brain during development, that of GluN1 with exon 5 cassette is selectively up-regulated in the thalamus, colliculus, cerebellum, and neocortex during early life (6, 31). N-terminal splicing of Grin1 mRNA may have important roles in the development and function of these brain regions.
Our results suggest that the acceleration in NMDAR-EPSC time course at the thalamic relay synapse is caused entirely by the up-regulation of exon-5-containing GluN1 and GluN2A. Deletion of Grin2a or Grin1 exon 5 does not affect the expression of each other. The effects of Grin2a and Grin1 exon 5 deletion are additive, indicating that Grin2a and Grin1 exon 5 have distinct roles. It is noted that unlike WT mice (Fig. 1A), mice with Grin1 exon-5 deletion did not show a significant reduction in Grin2b expression during the second week (SI Appendix, Fig. S4C). This indicates that the deletion of Grin1 exon 5 may reduce the ratio of GluN2A/GluN2B, thereby prolonging the time course of NMDAR-EPSCs. However, this possibility cannot explain the results from Grin2a-KO or Grin2a/Grin1ΔEx5/ΔEx5 DKO mice. From WT and Grin2a-KO data, we estimated that Grin1 exon 5 accounts for 71% of the change in NMDAR kinetics whereas Grin2a accounts for the remaining 29%. However, Grin1ΔEx5/ΔEx5 neurons showed a large acceleration in NMDAR kinetics between P7 and P16 (Fig. 2E), suggesting that GluN2A can partially compensate for the loss of Grin1 exon 5. In contrast to GluN2A, which shows ubiquitous expression in the brain, the expression of Grin1 exon 5 is more restricted (6). The role of exon-5-containing GluN1 in developmental acceleration of NMDAR kinetics is likely to be more selective.
The formation and maturation of excitatory synapses are strongly regulated by activity-dependent mechanisms. Deletion of GluN1 reduces spine density in layer 2/3 pyramidal neurons (40), whereas focal glutamate applications trigger de novo spine growth in these neurons and this effect requires activation of NMDARs (41). On the other hand, deletion of GluN1 leads to a 20% increase in spine density of layer 4 neurons in the cortex (42). These results suggest that the role of NMDARs in synapse development is cell-type specific. We find that deletion of Grin1 exon 5 has no effect on mEPSCs in VPm neurons, but significantly increases the frequency of mEPSCs and the density of dendritic spines in layer 5 pyramidal neurons. It should be mentioned that thalamic relay synapses are localized on the soma and dendritic shafts of VPm neurons (36, 43), which may be regulated by different mechanisms than synapses localized at dendritic spines. Dendritic spines are highly dynamic in the brain during postnatal development (44, 45). Up-regulation of exon-5-containing GluN1 during development may serve as a negative regulator of growth and maintenance of dendritic spines in selective brain regions.
Hyperfunction of NMDARs causes epileptic activities in brain tissues from rodents (46) and humans (47). Our results show that the deletion of Grin1 exon 5 enhances excitatory transmission by prolonging the time course of NMDAR-EPSCs and increasing the number of excitatory synapses. This may lead to hyperexcitation of the brain, which is consistent with our results on seizure susceptibility. Homozygous Grin1ΔEx5 mice showed lower seizure threshold than WT mice to both PTZ and KA, suggesting that Grin1 exon 5 splicing regulates network excitability in brain regions implicated in seizure induction.
The mammalian brain relies on alternative splicing to generate proteome complexity (48). A growing number of neurological diseases have been associated with abnormal splicing (49). Mutations in RBFOX1, a member of the RNA-binding Fox family of splicing factors, cause mental retardation, epilepsy, and autism spectrum disorder (50, 51). N-terminal splicing of Grin1 is a target of Rbfox1 and deletion of Rbfox1 in the mouse leads to increased seizure susceptibility and a reduction in the inclusion of Grin1 exon 5 (52). Our results suggest a role of Grin1 N-terminal splicing in brain disease. Several mutations of GRIN1 have been associated with neurodevelopmental disorders (53–55). However, none of these mutations are in exon 5 or affect exon 5 splicing. Patients carrying these mutations have severe epilepsy and profound developmental delay. In comparison, mice lacking Grin1 exon 5 appeared normal. Our findings suggest that human mutations that disrupt GRIN1 exon 5 or GRIN1 N-terminal splicing have mild phenotypes by themselves but may interact with other mutations or environmental insults to cause severe disease.
Materials and Methods
Animals, quantitative RT-PCR, electrophysiology, confocal imaging, seizure induction, and statistical analyses are described in SI Appendix. All experimental procedures were approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health Grant NS064013 (to Z-w.Z.). The Jackson Laboratory Cell Biology and Microinjection Services were subsidized by a National Cancer Institute Core Grant P30 CA034196. We thank Drs. Shigetada Nakanishi and Toshikuni Sasaoka for their permission to use the GluN2A knockout mice.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905721116/-/DCSupplemental.
References
- 1.Traynelis S. F., et al. , Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 62, 405–496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoletti P., Bellone C., Zhou Q., NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14, 383–400 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Sanz-Clemente A., Nicoll R. A., Roche K. W., Diversity in NMDA receptor composition: Many regulators, many consequences. Neuroscientist 19, 62–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monyer H., Burnashev N., Laurie D. J., Sakmann B., Seeburg P. H., Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron 12, 529–540 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Watanabe M., Mishina M., Inoue Y., Distinct spatiotemporal expressions of five NMDA receptor channel subunit mRNAs in the cerebellum. J. Comp. Neurol. 343, 513–519 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Laurie D. J., Seeburg P. H., Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J. Neurosci. 14, 3180–3194 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durand G. M., Bennett M. V., Zukin R. S., Splice variants of the N-methyl-D-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc. Natl. Acad. Sci. U.S.A. 90, 6731–6735 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mu Y., Otsuka T., Horton A. C., Scott D. B., Ehlers M. D., Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron 40, 581–594 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Horak M., Wenthold R. J., Different roles of C-terminal cassettes in the trafficking of full-length NR1 subunits to the cell surface. J. Biol. Chem. 284, 9683–9691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karakas E., Furukawa H., Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344, 992–997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regan M. C., et al. , Structural mechanism of functional modulation by gene splicing in NMDA receptors. Neuron 98, 521–529.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durand G. M., et al. , Cloning of an apparent splice variant of the rat N-methyl-D-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc. Natl. Acad. Sci. U.S.A. 89, 9359–9363 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollmann M., et al. , Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron 10, 943–954 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Traynelis S. F., Hartley M., Heinemann S. F., Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science 268, 873–876 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Rumbaugh G., Prybylowski K., Wang J. F., Vicini S., Exon 5 and spermine regulate deactivation of NMDA receptor subtypes. J. Neurophysiol. 83, 1300–1306 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Vance K. M., Hansen K. B., Traynelis S. F., GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J. Physiol. 590, 3857–3875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmignoto G., Vicini S., Activity-dependent decrease in NMDA receptor responses during development of the visual cortex. Science 258, 1007–1011 (1992). [DOI] [PubMed] [Google Scholar]
- 18.Hestrin S., Developmental regulation of NMDA receptor-mediated synaptic currents at a central synapse. Nature 357, 686–689 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Crair M. C., Malenka R. C., A critical period for long-term potentiation at thalamocortical synapses. Nature 375, 325–328 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Sheng M., Cummings J., Roldan L. A., Jan Y. N., Jan L. Y., Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368, 144–147 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Flint A. C., Maisch U. S., Weishaupt J. H., Kriegstein A. R., Monyer H., NR2A subunit expression shortens NMDA receptor synaptic currents in developing neocortex. J. Neurosci. 17, 2469–2476 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu H. C., Gonzalez E., Crair M. C., Barrel cortex critical period plasticity is independent of changes in NMDA receptor subunit composition. Neuron 32, 619–634 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Gray J. A., et al. , Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: Single-cell NMDA receptor subunit deletion in vivo. Neuron 71, 1085–1101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tovar K. R., Sprouffske K., Westbrook G. L., Fast NMDA receptor-mediated synaptic currents in neurons from mice lacking the epsilon2 (NR2B) subunit. J. Neurophysiol. 83, 616–620 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Vicini S., et al. , Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J. Neurophysiol. 79, 555–566 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Hansen K. B., Ogden K. K., Yuan H., Traynelis S. F., Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron 81, 1084–1096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun W., Hansen K. B., Jahr C. E., Allosteric interactions between NMDA receptor subunits shape the developmental shift in channel properties. Neuron 94, 58–64.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barria A., Malinow R., NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron 48, 289–301 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Kim M. J., Dunah A. W., Wang Y. T., Sheng M., Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron 46, 745–760 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Arsenault D., Zhang Z. W., Developmental remodelling of the lemniscal synapse in the ventral basal thalamus of the mouse. J. Physiol. 573, 121–132 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paupard M. C., Friedman L. K., Zukin R. S., Developmental regulation and cell-specific expression of N-methyl-D-aspartate receptor splice variants in rat hippocampus. Neuroscience 79, 399–409 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Lester R. A., Clements J. D., Westbrook G. L., Jahr C. E., Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature 346, 565–567 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Hestrin S., Sah P., Nicoll R. A., Mechanisms generating the time course of dual component excitatory synaptic currents recorded in hippocampal slices. Neuron 5, 247–253 (1990). [DOI] [PubMed] [Google Scholar]
- 34.Diamond J. S., Neuronal glutamate transporters limit activation of NMDA receptors by neurotransmitter spillover on CA1 pyramidal cells. J. Neurosci. 21, 8328–8338 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H., Liu H., Zhang Z. W., Elimination of redundant synaptic inputs in the absence of synaptic strengthening. J. Neurosci. 31, 16675–16684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z. W., Peterson M., Liu H., Essential role of postsynaptic NMDA receptors in developmental refinement of excitatory synapses. Proc. Natl. Acad. Sci. U.S.A. 110, 1095–1100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakanishi N., Axel R., Shneider N. A., Alternative splicing generates functionally distinct N-methyl-D-aspartate receptors. Proc. Natl. Acad. Sci. U.S.A. 89, 8552–8556 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paoletti P., Ascher P., Neyton J., High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J. Neurosci. 17, 5711–5725 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Traynelis S. F., Burgess M. F., Zheng F., Lyuboslavsky P., Powers J. L., Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J. Neurosci. 18, 6163–6175 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ultanir S. K., et al. , Regulation of spine morphology and spine density by NMDA receptor signaling in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 19553–19558 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon H. B., Sabatini B. L., Glutamate induces de novo growth of functional spines in developing cortex. Nature 474, 100–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Datwani A., Iwasato T., Itohara S., Erzurumlu R. S., NMDA receptor-dependent pattern transfer from afferents to postsynaptic cells and dendritic differentiation in the barrel cortex. Mol. Cell. Neurosci. 21, 477–492 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams M. N., Zahm D. S., Jacquin M. F., Differential foci and synaptic organization of the principal and spinal trigeminal projections to the thalamus in the rat. Eur. J. Neurosci. 6, 429–453 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Grutzendler J., Kasthuri N., Gan W. B., Long-term dendritic spine stability in the adult cortex. Nature 420, 812–816 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Holtmaat A. J., et al. , Transient and persistent dendritic spines in the neocortex in vivo. Neuron 45, 279–291 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Walther H., Lambert J. D., Jones R. S., Heinemann U., Hamon B., Epileptiform activity in combined slices of the hippocampus, subiculum and entorhinal cortex during perfusion with low magnesium medium. Neurosci. Lett. 69, 156–161 (1986). [DOI] [PubMed] [Google Scholar]
- 47.Avoli M., Louvel J., Pumain R., Olivier A., Seizure-like discharges induced by lowering [Mg2+]o in the human epileptogenic neocortex maintained in vitro. Brain Res. 417, 199–203 (1987). [DOI] [PubMed] [Google Scholar]
- 48.Li Q., Lee J. A., Black D. L., Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8, 819–831 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Scotti M. M., Swanson M. S., RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin C. L., et al. , Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 144B, 869–876 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Lal D., et al. , Extending the phenotypic spectrum of RBFOX1 deletions: Sporadic focal epilepsy. Epilepsia 56, e129–e133 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Gehman L. T., et al. , The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 43, 706–711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohba C., et al. , GRIN1 mutations cause encephalopathy with infantile-onset epilepsy, and hyperkinetic and stereotyped movement disorders. Epilepsia 56, 841–848 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Lemke J. R., et al. , Delineating the GRIN1 phenotypic spectrum: A distinct genetic NMDA receptor encephalopathy. Neurology 86, 2171–2178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zehavi Y., et al. , De novo GRIN1 mutations: An emerging cause of severe early infantile encephalopathy. Eur. J. Med. Genet. 60, 317–320 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.