Significance
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder of the gastrointestinal tract, and the worldwide prevalence of IBD rapidly continues to rise. It has been widely accepted that genetic predisposition remains an important risk factor of IBD in addition to microbiota, dietary environment, and immune response. In this study, we demonstrated that retinoic acid-related orphan receptor α (RORα) controls the inflammatory signaling network to maintain homeostasis in vivo in intestinal epithelium. Our study strongly proposes that RORα is a therapeutic target for the treatment of IBD.
Keywords: epigenetic regulation, inflammation, RORα, HDAC3, NF-kB signaling
Abstract
Retinoic acid-related orphan receptor α (RORα) functions as a transcription factor for various biological processes, including circadian rhythm, cancer, and metabolism. Here, we generate intestinal epithelial cell (IEC)-specific RORα-deficient (RORαΔIEC) mice and find that RORα is crucial for maintaining intestinal homeostasis by attenuating nuclear factor κB (NF-κB) transcriptional activity. RORαΔIEC mice exhibit excessive intestinal inflammation and highly activated inflammatory responses in the dextran sulfate sodium (DSS) mouse colitis model. Transcriptome analysis reveals that deletion of RORα leads to up-regulation of NF-κB target genes in IECs. Chromatin immunoprecipitation analysis reveals corecruitment of RORα and histone deacetylase 3 (HDAC3) on NF-κB target promoters and subsequent dismissal of CREB binding protein (CBP) and bromodomain-containing protein 4 (BRD4) for transcriptional repression. Together, we demonstrate that RORα/HDAC3-mediated attenuation of NF-κB signaling controls the balance of inflammatory responses, and therapeutic strategies targeting this epigenetic regulation could be beneficial to the treatment of chronic inflammatory diseases, including inflammatory bowel disease (IBD).
Intestinal epithelial cells (IECs) serve as physical barriers to separate huge microbial populations from immune cells and cross-talk with residual immune cells to secrete inflammatory cytokines for the efficient clearing of pathogens to maintain intestinal homeostasis (1, 2). Given that IECs undergo dynamic physiological stresses, including inflammation, control of balanced inflammatory responses is important in IECs to prevent tissue injury and microbial invasion (3, 4). Since excessive inflammation suppresses reconstitution and regeneration of the epithelial physical barrier, leading to severe tissue damage in the intestinal epithelium (5), the attenuation of inflammation is critical for epithelial reconstitution (6). However, the molecular mechanism of how the attenuation of inflammation is regulated at the transcriptional level in IECs still remains unclear.
The nuclear factor κB (NF-κB) family members consist of p65/Rel A, p50/NF-κB1, p52, Rel B, and c-Rel, and the p50-p65 heterodimer is the most abundant form (7–9). The p50-p65 heterodimer serves as a master regulator to maintain inflammatory response in IECs as well as immune cells (10–12). NF-κB transcriptional activity is tightly controlled by various posttranslational modifications in IECs, providing a binding platform for cofactors and regulating the activity and target specificity of NF-κB (13–15). Histone acetyltransferases (HATs), including CREB binding protein (CBP)/p300, introduce acetylation on the p50-p65 heterodimer as well as histones for transcriptional activation of NF-κB target genes (16). Bromodomain-containing protein (BRD) family members recognize acetylated substrate and promote the recruitment of HATs to increase the expression of inflammatory cytokine genes (17). Thus, inhibition of acetylation of the p50-p65 heterodimer has been widely considered as a potential therapeutic strategy for the treatment of many inflammatory diseases, such as inflammatory bowel disease (IBD) (18).
Members of the bromodomain and extraterminal domain (BET) family have been shown to bind acetylated chromatin for induction of gene expression via facilitating access to transcriptional coactivators (19, 20). BRD4, a member of the BET family, binds acetylated p65 and increases the transcriptional activity of p65 for the activation of inflammatory response (21, 22). Given that the blockade of epigenetic function of BRD4 is a promising therapeutic strategy to suppress inflammatory response, it is fundamental to understand epigenetic regulation of how BRD4 function is regulated to suppress NF-κB signaling.
Retinoic acid-related orphan receptor α (RORα), a member of the orphan nuclear receptor family, plays an important role in various physiological functions, such as embryonic development, cellular differentiation, metabolic pathway, and cancer (23–26). Previously, we have reported that RORα has a tumor-suppressive function by repressing Wnt/β-catenin signaling, leading to inhibition of colorectal cancer growth, and by increasing p53 stability, leading to apoptosis of cancer cells upon the DNA damage signal (27, 28). RORα activates the expression of many target genes for the regulation of circadian rhythm, whereas REV-ERBs repress them in a circadian manner (29). Also, RORα modulates the expression of pattern recognition receptors in response to the circadian rhythm (30). On the other hand, RORα functions as a negative feedback regulator of NF-κB signaling for the inflammatory responses (31), although the mechanisms of how RORα suppresses NF-κB signaling in IECs remains unrevealed.
Here, we report that RORα acts as a critical epigenetic regulator to maintain the balance of inflammation in IECs using IEC-specific RORα-deficient (RORαΔIEC) mice. RORαΔIEC mice exhibit excessive inflammation and severe tissue damage, and the reconstitution of damaged colonic tissue is remarkably impaired in RORαΔIEC mice compared with wild-type (WT; RORαf/f) mice during dextran sulfate sodium (DSS)-induced colitis. Genome-wide transcriptome analysis reveals that RORα functions as a transcriptional repressor of inflammatory genes by attenuating NF-κB signaling in IECs, indicating that RORα plays a key role in maintaining intestinal homeostasis to prevent intestinal inflammation. Our data strongly demonstrate that RORα is crucial for maintaining intestinal homeostasis via attenuation of NF-κB signaling for the regeneration of damaged lesions.
Results
RORα Attenuates Excessive Intestinal Inflammation to Maintain Intestinal Homeostasis.
To explore the function of RORα in IEC homeostasis and intestinal inflammation, we generated RORαΔIEC mice and compared RORα expression in IECs. The messenger RNA (mRNA) and protein levels of endogenous RORα were significantly depleted in IECs isolated from RORαΔIEC mice compared with their RORαf/f littermate controls (Fig. 1 A and B and SI Appendix, Fig. S1A). Fed with a regular chow diet, RORαΔIEC mice did not exhibit any significant phenotypic differences, including body weights (Fig. 1C).
Fig. 1.
RORα attenuates intestinal inflammation to maintain intestinal homeostasis during DSS-induced colitis. (A) mRNA levels of Rora in IECs from RORαf/f and RORαΔIEC mice. The mRNA levels were normalized to β-actin expression. Rel., relative. (B) Protein levels of RORα in IECs from RORαf/f and RORαΔIEC mice. β-Actin was used as a loading control. (C) Average body weight in RORαf/f and RORαΔIEC mice (8-wk-old females; n = 8∼10 per group). (D) RORαf/f and RORαΔIEC mice were given 2% DSS in their drinking water for 7 d and then given regular drinking water for an additional 7 d. Changes in body weight of RORαf/f and RORαΔIEC mice treated with DSS are shown. This experiment was repeated twice with similar results (n = 8∼10 per group). (E) Colon length was measured in RORαf/f and RORαΔIEC mice on day 0 (n = 3 per group) and day 14 (n = 7∼10 per group) after DSS treatment. (F) Genomic DNA from mesenteric lymph node was isolated, and bacterial content was measured by 16s rDNA using quantitative real-time PCR (qRT-PCR) (n = 6∼7 per group). The 16s rDNA levels were normalized to mouse genomic L32 expression. (G) RORαf/f and RORαΔIEC mice were given 2% DSS in their drinking water for 5 d (Day 5) followed by regular drinking water for 3 d (Day 8). Representative images of hematoxylin and eosin (H&E) staining of colon sections from RORαf/f and RORαΔIEC mice are shown after 0, 5, and 8 d of 2% DSS. (Scale bar, 100 μm.) (H) Immunohistochemistry was performed in colon sections from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS, and representative confocal images are shown. The proportion of Ki-67+ IECs was determined from image analysis and represented as optical density. DAPI, blue; Ki-67, red. (Scale bar, 100 μm.) (I) Immunohistochemistry was performed in colon sections from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS. Representative F4/80 (macrophage marker)-, Gr-1 (neutrophil marker)–, and CD4 (T cell marker)-stained sections (green) of colon are shown. DAPI, blue. (Scale bar, 100 μm.) (J) Gene expression profile involved in inflammation from the colon of RORαf/f and RORαΔIEC mice after 8 d of 2% DSS as determined by qRT-PCR. The mRNA levels were normalized to L32 expression (n = 5∼8 per group). (K) Representative bright-field images of enteroids cultured for 7 d. (Scale bar, 200 μm.) (L) Immunohistochemistry was performed in enteroids cultured for 14 d, and representative Ki-67 images are shown. DAPI, blue; Ki-67, green. (Scale bar, 100 μm.) (M) mRNA levels of intestinal stem cell marker genes in IECs from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS (n = 4 per group). *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis was performed using an unpaired t test. Data are from 3 independent experiments (A, D, E, F, H, J, and H; mean ± SEM) or are representative of 3 independent experiments with similar results (G, I, K, and L).
To evaluate the physiological roles of epithelial RORα during colitis pathogenesis, RORαΔIEC mice were subjected to a DSS-induced experimental colitis model that resembles pathologies of ulcerative colitis in human patients (32, 33). Mice were administered 2% DSS in drinking water for 7 d (injury phase), followed by drinking water without DSS for an additional 7 d (recovery phase) to allow regeneration of intestinal epithelium (Fig. 1D). After administration of DSS, the body weight of RORαΔIEC mice steadily decreased, and by day 10, RORαΔIEC mice had lost more than 20% of their initial body weight. In contrast, RORαf/f mice exhibited minimal body weight loss and recovered body weight quickly when fed with regular drinking water (Fig. 1D). While colonic lengths are comparable between genotypes under normal conditions (day 0), DSS-injured RORαΔIEC mice exhibited significant shortening of colon length (Fig. 1E). To evaluate epithelial barrier integrity, we examined intestinal permeability by quantitative real-time PCR to detect the bacterial 16S ribosomal DNA (rDNA) gene in mesenteric lymph node. A substantial increase of 16S rDNA of bacteria was observed in mesenteric lymph nodes from DSS-injured RORαΔIEC mice compared with RORαf/f mice (Fig. 1F), indicating that markedly increased gut permeability in RORαΔIEC mice is due to severe disruption of intestinal epithelium.
We next examined histological features of colonic tissues with hematoxylin and eosin staining. While no histological features were observed for the first 5 d of the DSS-injury phase, severe colonic ulceration and epithelium disruption were observed in DSS-injured RORαΔIEC mice on day 8 (recovery phase) compared with DSS-injured RORαf/f mice (Fig. 1G), suggesting that RORα plays a pivotal role in maintaining intestinal epithelial homeostasis under intestinal inflammation. Consistent with the results stated above, histological scores, disease activity indices, and survival rates exhibited that RORαΔIEC mice were more susceptible to DSS-induced intestinal inflammation (SI Appendix, Fig. S1 B–D). To determine the regeneration capacity of disrupted epithelium in DSS-injured RORαΔIEC mice, we examined cell proliferation using Ki-67+ staining. Although RORαf/f and RORαΔIEC mice exhibited comparable intensity of Ki-67+ fluorescence under normal conditions, DSS-injured RORαΔIEC mice exhibited significantly fewer Ki-67+ cells compared with RORαf/f mice, indicating that the regeneration capacity of the intestinal epithelium was substantially impaired in RORαΔIEC mice (Fig. 1H).
To confirm the severity of intestinal inflammation, we stained colon tissue sections to determine macrophage (F4/80), neutrophil (Gr-1), and T lymphocyte (CD4) infiltration. Compared with little or no change observed under normal conditions, DSS-injured RORαΔIEC mice displayed increased immune cell infiltration in colonic tissues (Fig. 1I), indicating that RORαΔIEC mice had sustained intestinal inflammation during the recovery phase in DSS-induced colitis. Accordingly, mRNA levels of proinflammatory cytokine genes, including Il-1b and Tnfa, dramatically increased in colon tissues from DSS-injured RORαΔIEC mice (Fig. 1J). These data indicate that RORα functions to attenuate intestinal inflammation for restoring intestinal epithelium during DSS-induced colitis.
Next, we performed an organoid culture experiment after isolating the distal colon crypts from RORαf/f and RORαΔIEC mice in order to evaluate the functions of RORα in ex vivo conditions. Despite severe defects in the IEC regenerative capacity of DSS-injured RORαΔIEC mice, RORα-depleted organoids did not show any significant difference in their microscopic appearance, proliferation, and organoid formation efficiency (Fig. 1 K and L and SI Appendix, Fig. S2A).
To evaluate the functions of RORα in intestinal stem cell activity, we examined the gene expression profiles involved in intestinal stem cell and differentiation markers. We analyzed stem cell marker gene expression of RORαf/f and RORαΔIEC IECs on day 0 and day 8 of DSS treatment. Expression of the intestinal stem cell marker genes Lgr5, Tert, and Lrig1 in IECs of RORαf/f and RORαΔIEC mice did not show any significant difference (Fig. 1M). Consistent with the gene expression pattern in IECs, the gene expression profiles of intestinal stem cell and differentiation markers were quite similar between RORαf/f and RORαΔIEC organoids (SI Appendix, Fig. S2 B and C), implying that deficiency of RORα in IECs appears to have little or no significant effect on the function of intestinal stem cells.
Genome-Wide Transcriptome Analysis of Genes Affected by RORα Depletion in IECs.
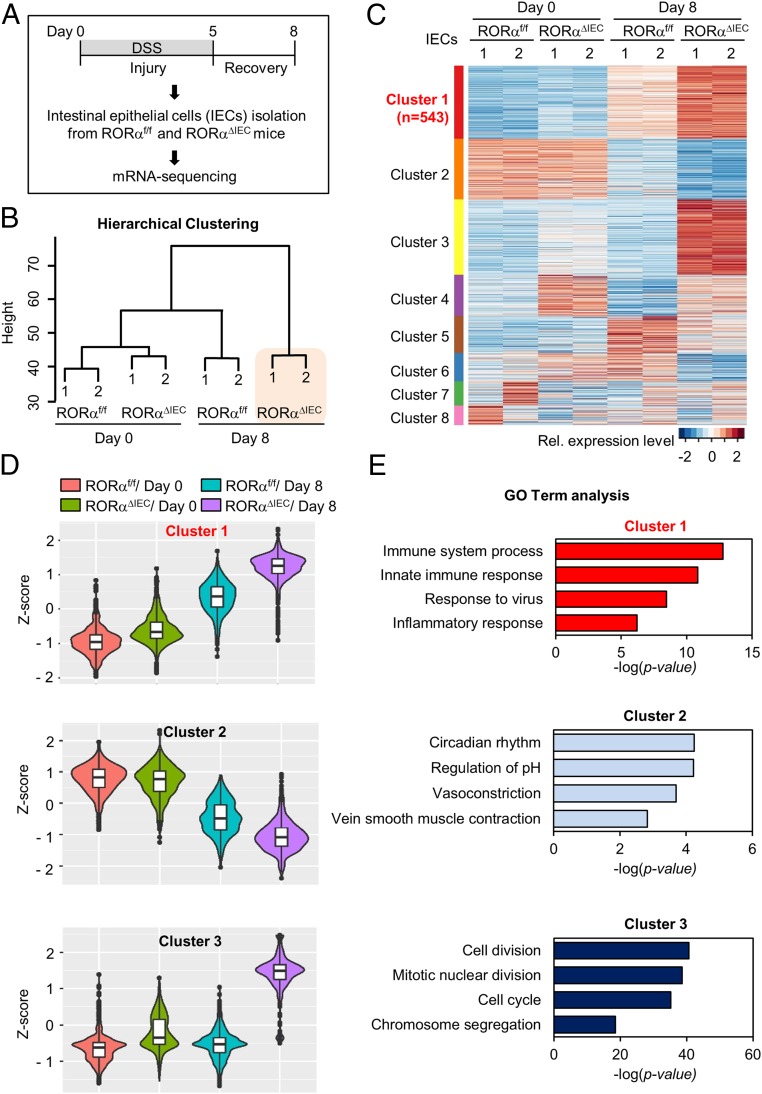
To elucidate the roles of RORα in transcriptional regulation in IECs, we analyzed gene expression profiles of IECs from RORαf/f and RORαΔIEC mice on day 3 of recovery after DSS treatment (Fig. 2A). Consistent with little or no phenotypic change in RORαΔIEC mice with a regular chow diet (Fig. 1 A–C), the number of differentially expressed genes (DEGs) in RORαΔIEC IECs compared with RORαf/f IECs under normal conditions was only 62 genes (|log2FC| ≥ 1, adjusted P ≤ 0.05), suggesting that RORα function is more important under perturbed conditions (SI Appendix, Fig. S3). Next, we performed unsupervised hierarchical clustering in order to see which samples had the most distinct or similar transcriptome. Interestingly, the gene expression profile of IECs from RORαΔIEC mice with DSS treatment was not closely clustered with any other profiles and showed a rather distinct transcriptome (Fig. 2B). These data indicate that the effect of RORα depletion on the transcriptional network is particularly significant in DSS-induced colitis.
Fig. 2.
Genome-wide transcriptome analysis of genes affected by RORα depletion in IECs. (A) Flow chart showing the strategy of RNA-sequencing analysis. (B) Unsupervised hierarchical clustering of the top 5% variably expressed genes (Euclidean distance metric). The dendrogram shows similarity between each transcriptome. (C) Heatmap of k-means clustering of the top 5% variably expressed genes (n = 2,668). Genes were grouped into 8 clusters on the basis of expression similarity. Rel., relative. (D) Violin box plots of clusters 1 to 3. Box plots show distribution of z-scores from each group in clusters 1 to 3. (E) Functional GO analysis of cluster 1. Immune and inflammatory responses are the most enriched terms in clusters 1 to 3. The immune system process and innate immune response genes are significantly enriched terms in cluster 1. Circadian rhythm genes are the most enriched in cluster 2. Cell division genes are significantly enriched in cluster 3.
To interrogate the transcriptional signatures affected by genotypes and/or conditions of the 4 profiles (RORαf/f/day 0, RORαf/f/day 8, RORαΔIEC/day 0, and RORαΔIEC/day 8), we applied unsupervised k-means clustering for genes. Clusters 1 and 3 showed prominent up-regulation of target genes in IECs from RORαΔIEC mice with DSS treatment (Fig. 2C). The box plots for the relative gene expression levels of the genes in cluster 1 exhibited a marginal increase in RORαf/f and RORαΔIEC IECs on day 0 but an amplified up-regulation on day 8 (Fig. 2D), which is in agreement with phenotypic observations such as changes in body weight and colon length. In contrast, the box plots for cluster 3 displayed solely up-regulation in IECs from RORαΔIEC mice with DSS treatment (Fig. 2D). Surprisingly, functional gene ontology (GO) analysis showed that immune and inflammatory responses are the most enriched terms in cluster 1, whereas cluster 3 has cell cycle-related terms as the top hits, thus revealing the key role of RORα as a transcriptional repressor of inflammatory genes upon DSS treatment (Fig. 2E).
RORα Functions as a Transcriptional Repressor of Inflammatory Genes in DSS-Induced Colitis.
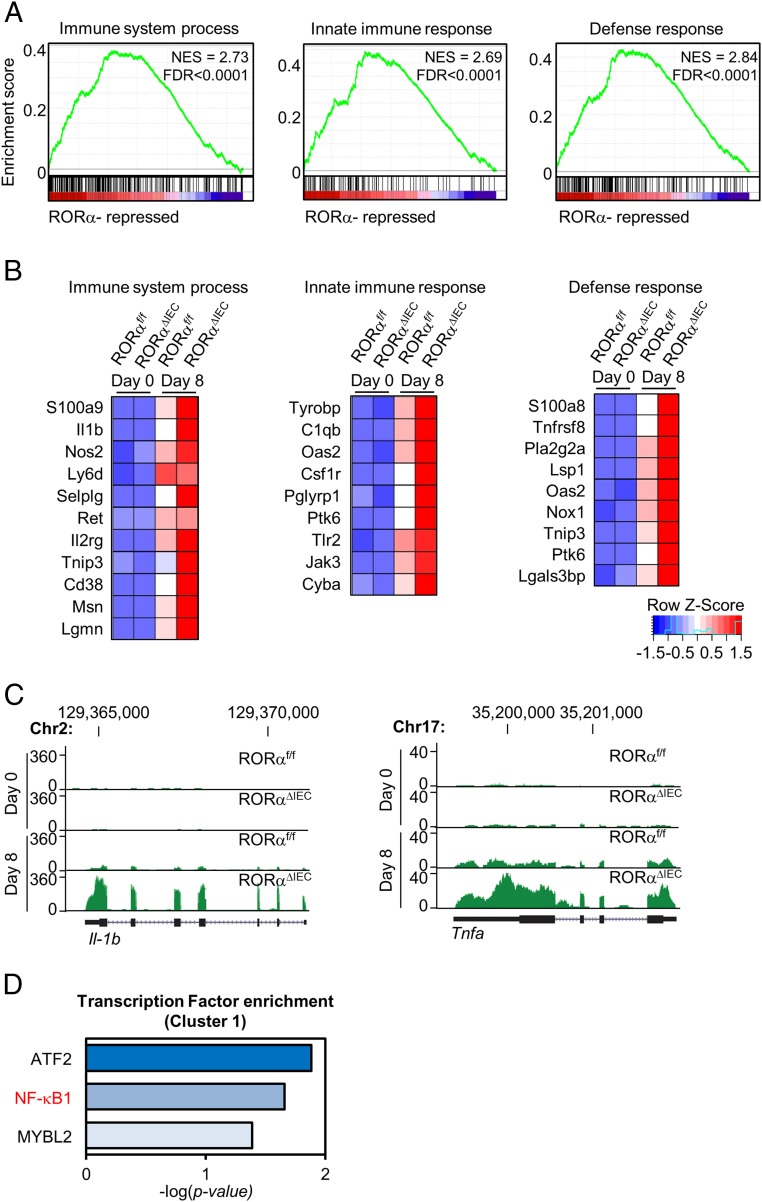
To validate the function of RORα as a transcriptional repressor and identify its direct or indirect target genes, we performed gene set enrichment analysis (GSEA) through ranking genes by Pearson correlation with the phenotypic labels based on cluster 1 (Fig. 3A). The results evidently showed that the gene sets involved in the immune response and cytokine production are significantly enriched in accordance with the more severe phenotype observed in RORαΔIEC mice with DSS treatment (SI Appendix, Fig. S4). Furthermore, the genes contributing to the enrichment are likely to be the direct and indirect targets regulated by RORα under the perturbed conditions, as supported by the expression pattern of target genes (Fig. 3B). Despite high expression of Il-1b and Tnfa in RORαΔIEC mice, Il-1b and Tnfa turned out to be the representative targets repressed by RORα in DSS-induced colitis (Fig. 3C). Consistent with transcriptome analysis, transcription factor enrichment analysis exhibited that p50/NF-κB1 is the major transcription factor of the genes in cluster 1, suggesting that RORα plays a key role in modulating NF-κB transcriptional activity (Fig. 3D). Taken together, analysis of phenotypic signatures within the gene expression profiles demonstrated that RORα represses the transcription of inflammatory genes in DSS-induced colitis.
Fig. 3.
RORα functions as a transcriptional repressor of inflammatory genes in DSS-induced colitis. (A) Representative GSEA results (ranking genes by the Pearson correlation, with the phenotypic labels based on cluster 1). FDR, false discovery rate; NES, normalized enrichment score. (B) Genes contributing to the enrichment for each term. (C) Screen shot from the UCSC Genome Browser (GB). The GB shows RNA-sequencing coverage plots and individual reads for Il-1b and Tnfa. (D) Transcription factor enrichment analysis in cluster 1. Hypergeometric P values were calculated. The bars represent the enrichment scores: −log10(P value).
Recruitment of RORα on the NF-κB Target Promoters Is Important for the Attenuated Transcription of Inflammatory Genes.
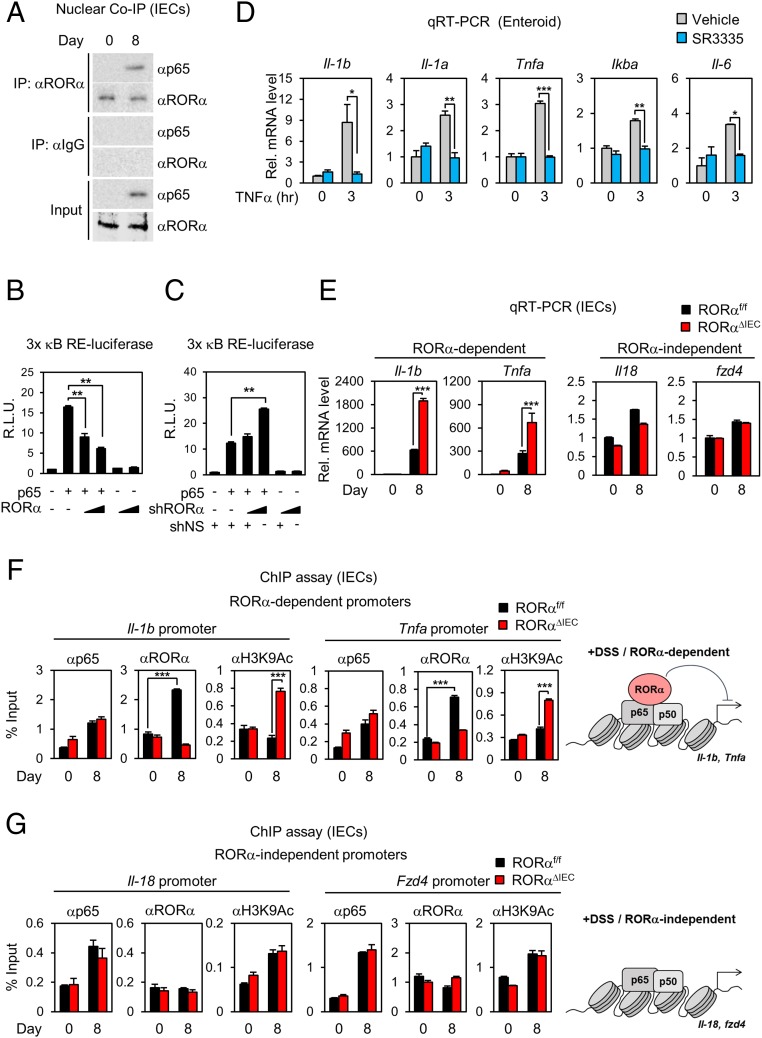
To explore how NF-κB transcriptional activity is dysregulated in RORαΔIEC mice, we performed immunoblot analysis to examine protein levels of p65 and inhibitor of κB (IκB) in IECs isolated from RORαf/f and RORαΔIEC mice with DSS injury for 5 d to induce intestinal inflammation. DSS-injured RORαf/f and RORαΔIEC mice were then administered regular drinking water for 3 d to allow tissue regeneration. The major subunit of NF-κB, p65, substantially localized in the nucleus of IECs, and IκB protein levels were markedly reduced in the cytoplasm of IECs from both DSS-injured RORαf/f and RORαΔIEC mice (SI Appendix, Fig. S5). However, protein levels of RORα in both the cytoplasm and nucleus of IECs from RORαf/f mice were unaffected by DSS treatment (SI Appendix, Fig. S5). Interestingly, RORα mutually interacted with p65 in IECs isolated from DSS-injured RORαf/f mice (Fig. 4A). Given that day 8 is the period of the recovery phase for tissue regeneration during DSS-induced colitis, these data clearly indicate that interaction of RORα with p65 is crucial for tissue regeneration during the period of recovery in DSS-induced colitis.
Fig. 4.
Recruitment of RORα on the NF-κB target promoters is crucial for the attenuation of intestinal inflammation in DSS-induced colitis. (A) Coimmunoprecipitation (Co-IP) assay of RORα with p65 was conducted in nuclear fractions of IECs from RORαf/f mice after 8 d of 2% DSS (n = 5 per group). The effect of overexpression of RORα (B) and knockdown of RORα (C) on 3× κB RE-luciferase reporter activity is shown. R.L.U., relative luminometer units. (D) Quantitative real-time PCR (qRT-PCR) analyses of NF-κB target genes in organoids from RORαf/f and RORαΔIEC mice treated with TNF-α and SR3335. Rel., relative. (E) qRT-PCR analyses of RORα-dependent and RORα-independent NF-κB target genes (up-regulated genes after 2% DSS) in IECs from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS (n = 4 per group). The mRNA levels were normalized to L32 expression. ChIP assays on RORα-dependent (F) or RORα-independent (G) target promoters in IECs from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS (n = 5 per group) are shown. Schematic model of RORα-dependent and -independent target promoters are shown. **P < 0.01, ***P < 0.001. Statistical analysis was performed using an unpaired t test. Data are from 3 independent experiments (C–G; mean ± SEM).
To evaluate the underlying molecular mechanism of RORα on NF-κB signaling, we performed a reporter assay using luciferase plasmid containing 3× NF-κB response element (RE) in the regulatory region. While overexpression of RORα substantially suppressed the luciferase activity (Fig. 4B), knockdown of RORα further enhanced the luciferase activity (Fig. 4C). Interestingly, treatment with tumor necrosis factor-α (TNF-α) markedly induced expression of genes involved in proinflammatory cytokines in RORα-deficient organoids compared with WT organoids (SI Appendix, Fig. S6A). Consistently, we have observed that RORα-deficient organoids were more susceptible to TNF-α–induced disruption of organoids (SI Appendix, Fig. S6B). Induction of NF-κB target genes, such as TNF-α and interleukin-6 (IL-6), were markedly elevated in the RORα-deficient colorectal cancer cell line DLD-1 (SI Appendix, Fig. S7A). To examine whether a synthetic inverse agonist of RORα is capable of suppressing NF-κB transcriptional activity, we tested SR3335 in TNF-α–treated murine organoids. While TNF-α treatment induced the expression of numerous NF-κB target genes, such as Il-1b, Il-1a, Tnfa, Ikba, and Il-6, SR3335 markedly reversed the induction of inflammatory genes activated by TNF-α or lipopolysaccharide (LPS) in intestinal murine organoids as well as human colorectal cancer cell lines, such as DLD1 and SW620 (Fig. 4D and SI Appendix, Fig. S7 B and C). To examine whether RORα-mediated transcriptional repression is crucial for the attenuation of NF-κB transcriptional activity, we examined NF-κB target gene expressions in clusters 1 and 5. While expression of NF-κB target genes, including Il-1b and Tnfa in cluster 1 (RORα-dependent genes), largely increased in DSS-injured RORαΔIEC mice, expression of NF-κB target genes in cluster 5, including Il-18 and Fzd4 (RORα-independent genes), failed to increase in both RORαf/f and RORαΔIEC mice (Fig. 4E). Therefore, we propose that RORα-mediated attenuation of NF-κB target gene expression in cluster 1 is fundamental to the restoration of intestinal epithelium during tissue injury.
To evaluate whether RORα is critical for attenuation of NF-κB signaling to promote tissue regeneration, we performed chromatin immunoprecipitation (ChIP) assays with anti-p65, RORα, and acetylated K9 of histone 3 (H3K9Ac) antibodies in IECs. ChIP assay revealed that recruitment of p65 was not changed on the NF-κB target promoters, including Il-1b and Tnfa, in IECs isolated from DSS-injured RORαf/f and RORαΔIEC mice (Fig. 4F). However, recruitment of RORα on the NF-κB target promoters was remarkably increased in DSS-injured RORαf/f mice, but not in RORαΔIEC mice, during the recovery phase (Fig. 4F), indicating that RORα-mediated transcriptional repression of NF-κB target genes is critical for tissue regeneration in the damaged tissues. In contrast, RORα deficiency led to increased H3K9Ac levels, but not p65 levels, on the promoters of Il-1b and Tnfa (RORα-dependent promoters) on day 8 (Fig. 4F). On the other hand, occupancy of p65 and H3K9Ac was not changed between genotypes on the promoters of Il-18 and Fzd4 from cluster 5 (RORα-independent promoters) (Fig. 4G). These data indicate that RORα-mediated attenuation of NF-κB target gene expression is critical for the restoration of intestinal epithelium under DSS-induced colitis.
RORα/Histone Deacetylase 3 Dismiss BRD4/CBP on the NF-κB Target Promoters for the Attenuation of Intestinal Inflammation.
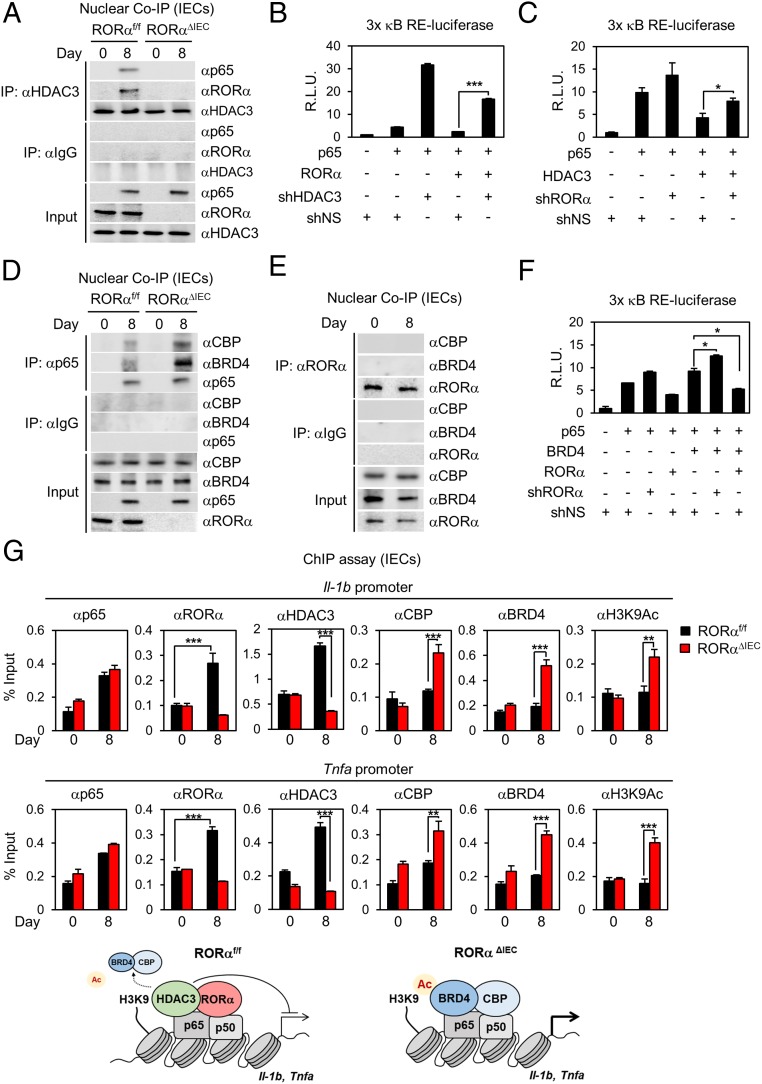
We have previously reported that RORα specifically interacts with histone deacetylase 3 (HDAC3) to suppress transcriptional activity of peroxisome proliferator-activated receptor-γ to control hepatic lipid homeostasis (34). Interestingly, RORα and HDAC3 composed the transcriptional corepressor complex with p65 in IECs from DSS-treated RORαf/f mice, which was largely dependent on the presence of RORα (Fig. 5A). To determine if HDAC3 is required for RORα-mediated repression of NF-κB activity, we measured 3× NF-κB RE-luciferase activity. Knockdown of HDAC3 substantially increased luciferase activity (Fig. 5B). Consistently, knockdown of RORα largely reversed HDAC3-mediated repression of NF-κB transcriptional activity (Fig. 5C). These data indicate that HDAC3 exerted a repressive function on NF-κB transcriptional activity in the presence of RORα.
Fig. 5.
RORα/HDAC3 dismiss BRD4/CBP on the NF-κB target promoters for the attenuation of intestinal inflammation. (A) Coimmunoprecipitation (Co-IP) assay of HDAC3 with p65 and RORα was conducted in nuclear fractions of IECs from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS (n = 5 per group). The effect of overexpression of RORα and knockdown of HDAC3 (B) or overexpression of HDAC3 and knockdown of RORα (C) on 3× κB RE-luciferase reporter activity with p65 is shown. R.L.U., relative luminometer units; sh, short hairpin. (D) Co-IP assay of p65 with CBP and BRD4 was conducted in nuclear fractions of IECs from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS (n = 5 per group). (E) Co-IP assay of RORα with CBP and BRD4 was conducted in nuclear fractions of IECs from RORαf/f mice after 8 d of 2% DSS (n = 5 per group). (F) Effect of RORα on 3× κB RE-luciferase reporter activity with p65 and BRD4. (G) ChIP assays were performed on the Il-1b and Tnfa promoters in IECs from RORαf/f and RORαΔIEC mice after 8 d of 2% DSS (n = 5 per group). Promoter occupancy by p65, RORα, HDAC3, CBP, BRD4, and H3K9Ac was analyzed. *P < 0.05, **P < 0.01, ***P < 0.001. Statistical analysis was performed using an unpaired t test. Data are from 3 independent experiments (B, C, F, and G; mean ± SEM).
Since RORα-mediated attenuation of NF-κB signaling requires HDAC3, we compared 4 DEG sets in our study with the DEG of WT versus HDAC3ΔIEC mice (35). We found further evidence that RORα and HDAC3 share a few target genes under inflammatory conditions (SI Appendix, Fig. S8). Since the platforms to measure the gene expression profile of HDAC3ΔIEC (35) and RORαΔIEC mice are different (microarray vs. next generation sequencing), it is unfair to apply the same P value cutoff to identify DEGs. Thus, DEGs in HDAC3ΔIEC mice were defined and fixed (n = 1,418), and our DEGs were then cumulatively taken from the most significant genes and compared with the DEGs in HDAC3ΔIEC mice. Although P values of the hypergeometric test for overlap tend to be exaggerated, RORα deletion, along with DSS treatment, increases overlaps with DEGs in HDAC3ΔIEC mice, indicating that RORα regulates shared target genes with HDAC3 upon DSS-induced injury. Interestingly, newly appeared common DEGs of RORαΔIEC and HDAC3ΔIEC mice upon DSS treatment have shown significant enrichment of GO terms related to immune response. This analysis clearly supports that RORα and HDAC3 cooperatively regulate the same target genes, such as inflammatory genes in DSS-induced colitis (SI Appendix, Fig. S9 A and B).
We next examined whether RORα impedes the assembly of the coactivator complex required for the induction of NF-κB target genes. BRD4 is a member of the BET family that recognizes acetylated lysine residues (20, 36–38). It has been reported that mutual interaction of BRD4/CBP and p65 is required to induce NF-κB target gene expression (21). Interestingly, assembly of BRD4/CBP and p65 was substantially increased in IECs isolated from DSS-injured RORαΔIEC mice compared with those of DSS-injured RORαf/f mice (Fig. 5D), indicating that NF-κB activity was enhanced in IECs from DSS-injured RORαΔIEC mice. Given that endogenous RORα failed to interact with BRD4 or CBP in IECs (Fig. 5E), RORα may compete with BRD4/CBP for the interaction with p65 to attenuate NF-κB target gene expression. A reporter assay with 3× NF-κB RE-luciferase reporter activity revealed that introduction of BRD4 enhanced p65-mediated luciferase activity (Fig. 5F). While RORα introduction suppressed p65/BRD4-mediated luciferase activity, knockdown of RORα enhanced p65/BRD4-mediated luciferase activity (Fig. 5F), indicating that RORα interferes with assembly of p65/BRD4/CBP coactivator complexes to suppress NF-κB transcriptional activity. These data indicate that RORα competes with BRD4/CBP for interaction with p65 and requires HDAC3 for the attenuation of NF-κB transcriptional activity.
To evaluate whether recruitment of RORα/HDAC3 is critical for dismissal of BRD4/CBP on the NF-κB target promoters, we performed a ChIP assay with anti-p65, RORα, HDAC3, CBP, BRD4, and H3K9Ac antibodies in IECs. Indeed, recruitment of p65 was not changed on the NF-κB target promoters, while RORα/HDAC3 occupancy was remarkably increased in IECs isolated from DSS-injured RORαf/f mice during the recovery phase (Fig. 5G). In contrast, the occupancy of BRD4/CBP coactivators was dramatically increased on the NF-κB target promoters in RORα-deficient IECs (Fig. 5G). Although DSS-injured RORαf/f mice attenuated inflammatory signaling to allow tissue regeneration in the intestinal tract, failure of RORα/HDAC3 recruitment further allowed BRD4/CBP occupancy on the NF-κB target promoters, resulting in highly activated inflammation during tissue regeneration in IECs from RORαΔIEC mice.
Discussion
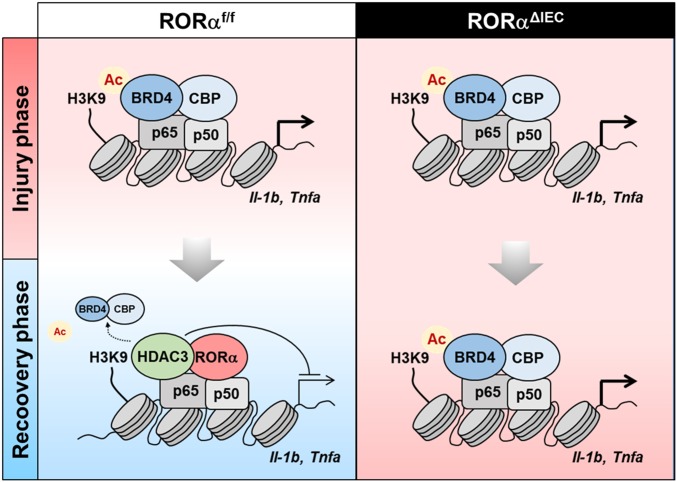
In this study, we identify RORα as a negative regulator of intestinal inflammation. Intestine-specific RORα-deficient mice with DSS-induced intestinal inflammation developed unusually stimulated inflammatory responses and exacerbated colitis. Tissue regeneration in the damaged colon was remarkably dysregulated in RORαΔIEC mice. RORα acts as a checkpoint specifically inhibiting p65 transcriptional activity in the inflammatory signaling pathway, which is essential for the regeneration of IECs in damaged tissues (Fig. 6). Our study provides a detailed mechanism at the transcriptional level of inflammatory responses that are crucial for controlling regeneration of the intestinal epithelium. The success or failure of tissue regeneration depends on the regulation of stem cell activation and the balance of complex inflammatory responses (39). Epithelial RORα appears to contribute to successful tissue regeneration by attenuation of NF-κB activation. Our results suggest distinct immune-regulatory roles of RORα in control of DSS-driven intestinal inflammation.
Fig. 6.
Schematic models shows RORα function as a transcriptional repressor of inflammatory genes in DSS-induced colitis. The schematic models illustrates the role of RORα in IECs after tissue injury. RORα attenuates NF-κB signaling via HDAC3 recruitment and dismisses BRD4/CBP on the NF-κB target promoters for attenuation of intestinal inflammation to restore the epithelial barrier (Left). Failure of RORα/HDAC3 recruitment further allowed BRD4/CBP occupancy on the NF-κB target promoters, leading to highly activated inflammation and impaired tissue regeneration in IECs (Right).
Previous studies have shown that RORα controls the immune system via lymphocyte development (40). Rag1−/− mice reconstituted with CD4+ splenocytes from RORα-deficient mice exhibited less susceptibility to experimental autoimmune encephalomyelitis than mice reconstituted with WT CD4+ splenocytes, suggesting that loss of RORα function greatly reduces the susceptibility to the development of autoimmune disease (41). Consistently, Sg mice having spontaneous RORα mutation have exhibited less susceptibility to ovalbumin-induced lung inflammation (42). Together, these observations suggest that RORα may provoke an allergic inflammatory response in different autoimmune disease animal models. Surprisingly, Sg mice have shown hypersusceptibility to LPS-induced inflammation (43). Moreover, recent studies have shown an antiinflammatory function of RORα in sepsis using Sg mice (44). However, the detailed molecular mechanisms of how RORα controls immune responses to maintain immune homeostasis are still controversial.
Genetic susceptibility is a well-known trigger of IBD pathogenesis in humans (45). Over the past few decades, remarkable achievement has widened our understanding of genetic contributions to IBD. Genome-wide association studies (GWAS) have identified novel single-nucleotide polymorphisms in genes such as Nod2, Il23r, and Atg16l1 as key susceptibility genes for the pathogenesis of IBD in humans (46, 47). GWAS has shown that RORα is a potential candidate genetic factor for the development of autoimmune diseases, including asthma (48). Thus, it is plausible that genetic variants of the Rora gene may confer genetic susceptibility to IBD pathogenesis in humans. Furthermore, a systemic analysis of circadian genes using a genome-wide complementary DNA microarray from human patients with Crohn’s disease and ulcerative colitis indicates that RORα is a novel therapeutic target for the treatment of IBD (49).
Analysis of specific-up-regulated DEGs in DSS-injured RORαΔIEC mice has revealed significant hits in cell division and cell cycle as well as inflammation, implying that the up-regulated gene expression profiles of cell division and cell cycle did not correlate with in vivo phenotypes of slow cell proliferation during the recovery phase in DSS-injured RORαΔIEC mice (Fig. 2 D and E). It would be plausible that severe and sustained inflammation in IECs from DSS-injured RORαΔIEC mice overwhelmed proliferative capability and disrupted the functional stepwise process of wound healing. Effective wound healing should follow sequential stages: inflammation, proliferation, and remodeling. Unlike RORαf/f mice, the gene signatures in the RORαΔIEC mice showed simultaneous inflammation and proliferation upon DSS treatment, which are abnormal conditions in the epithelium during the wound-healing process. Thus, the high expression of genes involved in cell cycle and division would not positively correlate with epithelial cell proliferation under the severe inflammation induced by DSS treatment.
We identify HDAC3 as an indispensable corepressor for RORα-mediated attenuation of NF-κB signaling in IECs. Consistent with our observation, recent studies have shown that intestine-specific HDAC3 deficient mice exhibited impaired gene regulation to maintain intestinal homeostasis in IECs, including decreased basal expression of genes associated with antimicrobial defense (35). Critically, deletion of HDAC3 in IECs led to loss of Paneth cells, impaired IEC function, and dysbiosis in the composition of commensal bacteria (35). Although the specific mechanisms through which IEC-intrinsic RORα expression regulates commensal bacteria-derived signals to maintain IEC responses remain to be determined, these data indicate that RORα may play a critical role to modulate the expression of genes, including antimicrobial peptides, and keep balance of symbiosis of commensal bacteria to maintain inflammatory signaling in IECs.
To delineate whether RORα is capable of directly regulating NF-κB target gene expression, we performed a ChIP assay using putative RORE motifs on IL-1β promoters. We observed that RORα failed to be recruited to the putative RORE on IL-1β promoters in both RORαf/f and RORαΔIEC mice (SI Appendix, Fig. S10), suggesting that RORα modulates NF-κB target genes in trans-mechanisms by directly binding to p65 for the suppression of p65-mediated transcriptional activities and recruiting numerous corepressor complexes, such as HDAC3.
In conclusion, our data indicate that RORα requires HDAC3 to attenuate NF-κB signaling to maintain intestinal homeostasis and tissue regeneration during DSS-induced colitis. Our findings provide a direct link between RORα and HDAC3 to maintain intestinal homeostasis under diverse environmental stresses. Therefore, therapeutic strategies designed to modulate RORα activity would be beneficial for the treatment of intestinal inflammation, such as ulcerative colitis.
Materials and Methods
Additional methods are provided in SI Appendix.
Mice.
Previously described RORαf/f mice were bred to mice expressing Cre-recombinase under the control of the villin promoter (The Jackson Laboratory) to generate RORαΔIEC (50). All mice used were 8 to 10 wk old. The sample sizes for all animal studies are indicated in each figure legend. Mice were housed in a specific pathogen-free Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility under controlled conditions of temperature (25 °C) and light (12 h light/12 h dark, lights switched on at 7:00 AM). Food and water were available ad libitum. All mice used in these experiments were backcrossed to C57BL/6 mice for at least 8 generations. Animals for each group of experiments were chosen randomly. The primers used in PCR analysis for genotyping floxed alleles are as follows: forward 5′-GCTTGTGGGTTTCTCCTACA-3′ and reverse 5′-GCAGCAAGTGTTGTGTCCCA-3′.
All animal studies and procedures were approved by the Institutional Animal Care and Use Committee of the National Cancer Center Research Institute.
Antibodies and Reagents.
The following commercially available antibodies were used: Anti-RORα (PA5-11224) antibody was purchased from Thermo Fisher Scientific, anti-HDAC3 (ab7030) and anti-H3K9Ac (ab4441) antibodies were purchased from Abcam, anti-p65 (no. 8242) antibody was purchased from Cell Signaling Technology, anti-lamin A/C (sc-6215) and anti-CBP (sc-583) antibodies were purchased from Santa Cruz Biotechnology, anti–β-actin (A1978) antibody was purchased from Sigma, antitubulin (LF-PA0146A) antibody was purchased from Abfrontier, and anti-BRD4 (A301-985A) antibody was purchased from Bethyl Laboratories.
Murine Colitis Model.
DSS (catalog no. 160110; MP Biomedicals; relative molecular mass of 36,000 to 50,000 Da) was added to drinking water at 2% (wt/vol) for either 5 or 7 d, and mice were then allowed to recover by drinking water for an additional 3 or 7 d.
RNA-Sequencing Analysis.
Total RNAs were extracted from RORαΔIEC and RORαf/f IECs on day 0 and day 8 using a Qiagen RNeasy Kit, and the mRNA-sequencing libraries were prepared with an Illumina TruSeq RNA Library. One hundred-base pair paired-end reads were produced using an Illumina HiSeq 2000 system. Adapter sequences were trimmed from the raw data using Trimmomatic v0.36 (51). The trimmed reads were aligned to the mm10 genome assembly and the transcriptome using STAR v2.5.3a (52), along with GENCODE M16 annotation. RSEM v1.3.0 (53) was used to calculate the expected read counts and transcripts per millon (TPM) for each sample from the corresponding BAM files. The read counts were processed using DESeq2 v1.18.1 (54) to identify DEGs, and GO analysis was performed using DAVID v6.8 (55). The TPM values were log2-transformed and used for following downstream analyses, such as hierarchical clustering, k-means clustering, and GSEA (version 2.0) (56, 57). The phenotype label for GSEA was set to RORαf/f on day 0: RORαf/f on day 8: RORαΔIEC on day 0: RORαΔIEC on day 8 = 1:2:1:3, and the Pearson correlation coefficient was then calculated per gene for ranking. Finally, C2 and C5 gene sets in MSigDB (molecular signature database) v6.2 were used for the enrichment score.
Statistical Analysis.
For animal studies, the sample size for experiments was determined empirically based on previous studies to ensure appropriate statistical power. Animals for each group of experiments were chosen randomly. No animals were excluded from statistical analysis, and the investigators were not blinded in the studies. Statistical analysis of different groups was performed using the Student’s unpaired t test. Bars in graphs in accompanying figures indicate mean ± SEM. GraphPad Prism 5 was used for all analyses.
Supplementary Material
Acknowledgments
We thank the National Cancer Center Animal Sciences Branch for excellent guidance and assistance with the performed mouse experiments. This work was supported by the Creative Research Initiatives Program (Research Center for Epigenetics Code and Diseases, Grant NRF-2017R1A3B1023387), the Bio & Medical Technology Development Program (Grant NRF-2018M3A9E2023523), and the Science Research Center Program (Grant NRF-2016R1A5A1010764) (all to S.H.B.); Korea Mouse Phenotyping Project Grant 2013M3A9D5072550 and Bio & Medical Technology Development Program Grant 2017M3A9F3046538 (to S.F.); Collaborative Genome Program Grant NRF-2017M3C9A5031595 (to D.P.); National Research Foundation Grant NRF-2019R1C1C1008181 (to D.P.) and Grant NRF-2019R1C1C1004022 (to D.K.); and National Cancer Center Grant NCC-1310100 (to H.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE121977).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907595116/-/DCSupplemental.
References
- 1.Abraham C., Medzhitov R., Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140, 1729–1737 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goto Y., Ivanov I. I., Intestinal epithelial cells as mediators of the commensal-host immune crosstalk. Immunol. Cell Biol. 91, 204–214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alverdy J., Zaborina O., Wu L., The impact of stress and nutrition on bacterial-host interactions at the intestinal epithelial surface. Curr. Opin. Clin. Nutr. Metab. Care 8, 205–209 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Chaplin D. D., Overview of the immune response. J. Allergy Clin. Immunol. 125 (suppl. 2), S3–S23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karin M., Clevers H., Reparative inflammation takes charge of tissue regeneration. Nature 529, 307–315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Sadi R., Boivin M., Ma T., Mechanism of cytokine modulation of epithelial tight junction barrier. Front. Biosci. 14, 2765–2778 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oeckinghaus A., Ghosh S., The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 1, a000034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vallabhapurapu S., Karin M., Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Caamaño J., Hunter C. A., NF-kappaB family of transcription factors: Central regulators of innate and adaptive immune functions. Clin. Microbiol. Rev. 15, 414–429 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wullaert A., Bonnet M. C., Pasparakis M., NF-κB in the regulation of epithelial homeostasis and inflammation. Cell Res. 21, 146–158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh S., Karin M., Missing pieces in the NF-kappaB puzzle. Cell 109 (suppl.), S81–S96 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Steinbrecher K. A., Harmel-Laws E., Sitcheran R., Baldwin A. S., Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J. Immunol. 180, 2588–2599 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Kim D., et al. , PKCα-LSD1-NF-κB-signaling cascade is crucial for epigenetic control of the inflammatory response. Mol. Cell 69, 398–411.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Huang B., Yang X. D., Lamb A., Chen L. F., Posttranslational modifications of NF-kappaB: Another layer of regulation for NF-kappaB signaling pathway. Cell. Signal. 22, 1282–1290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribet D., Cossart P., Pathogen-mediated posttranslational modifications: A re-emerging field. Cell 143, 694–702 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dekker F. J., van den Bosch T., Martin N. I., Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov. Today 19, 654–660 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Muller S., Filippakopoulos P., Knapp S., Bromodomains as therapeutic targets. Expert Rev. Mol. Med. 13, e29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S. C., Sundaram C., Reuter S., Aggarwal B. B., Inhibiting NF-κB activation by small molecules as a therapeutic strategy. Biochim. Biophys. Acta 1799, 775–787 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeRoy G., Rickards B., Flint S. J., The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell 30, 51–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filippakopoulos P., Knapp S., The bromodomain interaction module. FEBS Lett. 586, 2692–2704 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Huang B., Yang X. D., Zhou M. M., Ozato K., Chen L. F., Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 29, 1375–1387 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou Z., et al. , Brd4 maintains constitutively active NF-κB in cancer cells by binding to acetylated RelA. Oncogene 33, 2395–2404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J. M., et al. , EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell 48, 572–586 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Vogel M. W., Sinclair M., Qiu D., Fan H., Purkinje cell fate in staggerer mutants: Agenesis versus cell death. J. Neurobiol. 42, 323–337 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Doulazmi M., et al. , Cerebellar purkinje cell loss during life span of the heterozygous staggerer mouse (Rora(+)/Rora(sg)) is gender-related. J. Comp. Neurol. 411, 267–273 (1999). [PubMed] [Google Scholar]
- 26.Wong S. H., et al. , Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13, 229–236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J. M., et al. , RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol. Cell 37, 183–195 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Kim H., et al. , DNA damage-induced RORα is crucial for p53 stabilization and increased apoptosis. Mol. Cell 44, 797–810 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Duez H., Staels B., The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab. Vasc. Dis. Res. 5, 82–88 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Mukherji A., Kobiita A., Ye T., Chambon P., Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153, 812–827 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Delerive P., et al. , The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2, 42–48 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clapper M. L., Cooper H. S., Chang W. C., Dextran sulfate sodium-induced colitis-associated neoplasia: A promising model for the development of chemopreventive interventions. Acta Pharmacol. Sin. 28, 1450–1459 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Eichele D. D., Kharbanda K. K., Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 23, 6016–6029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K., et al. , RORα controls hepatic lipid homeostasis via negative regulation of PPARγ transcriptional network. Nat. Commun. 8, 162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alenghat T., et al. , Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature 504, 153–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong F., Chiu L. Y., Miller K. M., Acetylation reader proteins: Linking acetylation signaling to genome maintenance and cancer. PLoS Genet. 12, e1006272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung M., et al. , Affinity map of bromodomain protein 4 (BRD4) interactions with the histone H4 tail and the small molecule inhibitor JQ1. J. Biol. Chem. 289, 9304–9319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman S., et al. , The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 31, 2641–2652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aurora A. B., Olson E. N., Immune modulation of stem cells and regeneration. Cell Stem Cell 15, 14–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang H. S., et al. , Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol. Genomics 43, 818–828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jetten A. M., Retinoid-related orphan receptors (RORs): Critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl. Recept. Signal. 7, e003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaradat M., et al. , Modulatory role for retinoid-related orphan receptor alpha in allergen-induced lung inflammation. Am. J. Respir. Crit. Care Med. 174, 1299–1309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stapleton C. M., et al. , Enhanced susceptibility of staggerer (RORalphasg/sg) mice to lipopolysaccharide-induced lung inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 289, L144–L152 (2005). [DOI] [PubMed] [Google Scholar]
- 44.García J. A., et al. , Disruption of the NF-κB/NLRP3 connection by melatonin requires retinoid-related orphan receptor-α and blocks the septic response in mice. FASEB J. 29, 3863–3875 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Khor B., Gardet A., Xavier R. J., Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J. C., Parkes M., Genome-wide association studies and Crohn’s disease. Brief. Funct. Genomics 10, 71–76 (2011). [DOI] [PubMed] [Google Scholar]
- 47.de Lange K. M., Barrett J. C., Understanding inflammatory bowel disease via immunogenetics. J. Autoimmun. 64, 91–100 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Solt L. A., Griffin P. R., Burris T. P., Ligand regulation of retinoic acid receptor-related orphan receptors: Implications for development of novel therapeutics. Curr. Opin. Lipidol. 21, 204–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmieri O., et al. , Systematic analysis of circadian genes using genome-wide cDNA microarrays in the inflammatory bowel disease transcriptome. Chronobiol. Int. 32, 903–916 (2015). [DOI] [PubMed] [Google Scholar]
- 50.el Marjou F., et al. , Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.