Significance
Proteins exist as ensembles of microstates governed by a free energy landscape, with multiple “excited” states coexisting with the minimum energy structure. These alternate folded and partially disordered states are continuously being accessed through protein dynamics and are key elements required for a comprehensive understanding of protein function and stability. Unfortunately, their low abundance makes these “invisible” states hard to characterize experimentally. A unique view of the hierarchy of unfolding states on the protein energy landscape was obtained here using pressure perturbation. Furthermore, pressure perturbation can directly identify empty protein cavities and determine the energetic penalty of filling these with water.
Keywords: protein stability, protein folding and cooperativity, unfolded state, high-pressure NMR
Abstract
Although many proteins possess a distinct folded structure lying at a minimum in a funneled free energy landscape, thermal energy causes any protein to continuously access lowly populated excited states. The existence of excited states is an integral part of biological function. Although transitions into the excited states may lead to protein misfolding and aggregation, little structural information is currently available for them. Here, we show how NMR spectroscopy, coupled with pressure perturbation, brings these elusive species to light. As pressure acts to favor states with lower partial molar volume, NMR follows the ensuing change in the equilibrium spectroscopically, with residue-specific resolution. For T4 lysozyme L99A, relaxation dispersion NMR was used to follow the increase in population of a previously identified “invisible” folded state with pressure, as this is driven by the reduction in cavity volume by the flipping-in of a surface aromatic group. Furthermore, multiple partly disordered excited states were detected at equilibrium using pressure-dependent H/D exchange NMR spectroscopy. Here, unfolding reduced partial molar volume by the removal of empty internal cavities and packing imperfections through subglobal and global unfolding. A close correspondence was found for the distinct pressure sensitivities of various parts of the protein and the amount of internal cavity volume that was lost in each unfolding event. The free energies and populations of excited states allowed us to determine the energetic penalty of empty internal protein cavities to be 36 cal⋅Å−3.
Protein stability and structural transitions are fundamental to all of biology, from the regulation of normal cellular activity to the onset of neurodegenerative diseases (1–4). The detailed investigation of the energetics and conformational dynamics is thus pivotal to detect labile states and bolster our understanding of protein stability and function in various contexts. However, the characterization of alternate states coexisting with the native state remains a challenge, as these are only marginally populated and transiently formed and therefore cannot be adequately studied by most experimental approaches (5). Propitiously, recent developments in NMR spectroscopy allow for the detection and structural investigation of “excited” and partially unfolded states of proteins at atomic resolution (5–12). In these so-called Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion NMR experiments, minor conformations are not directly observed spectroscopically, but can be inferred from the line broadening they cause on the signals of the observed ground state (5, 6, 13). By experimental variation of NMR pulse spacing, this exchange contribution shows a dispersion from which populations, exchange kinetics, and chemical shift information of the partaking states is extracted (5, 6, 13). In this way, populations and lifetimes of “invisible” states can be obtained, even if their occupancy is as low as 1%. Furthermore, alternative states may be elicited by pressure perturbation and studied by NMR spectroscopy (14, 15). Relative populations of excited states are shifted under pressure as a result of associated volume differences, which includes the loss of cavities or packing defects in the protein interior (16), cavity hydration (17–19), local and global unfolding (20–22), and structure relaxation from side-chain rearrangement (23). As internal cavities make a major contribution to the total volume change upon protein structural transition, hydrostatic pressure emerges as a key variable to the study of protein excited states (16). High-pressure (HP) NMR has already profoundly increased our understanding of protein stability, structure, dynamics, and function (24–32). This relative wealth of information has not, however, led to a consensus picture of the relation of protein thermodynamic stability and structural transitions and its origin, leading to apparently enigmatic descriptions (31–34). Here, we investigate a well-characterized variant of the protein lysozyme from phage T4 (T4L). A collection of invisible folded and partially unfolded states was identified by relaxation dispersion NMR spectroscopy and equilibrium hydrogen exchange (HX) measured at multiple pressures up to 2,500 bar. From the pressure-induced destabilization energies, the penalty of generating empty cavities or defects inside a tightly packed protein core could be determined. Our results are in excellent agreement with earlier reports of the energetic and structural consequences of cavity formation for the same protein (35). Our results shed a clear light on the energetics of cavities in protein stability and their consequences on structural transitions.
T4 Lysozyme: An Archetype of Protein Folding and Stability
A wealth of data exists on the stability, folding, dynamics, and structure of T4L and of multiple sequence variants in the scientific literature (36–45). Structurally, T4L has a well-defined 2-domain structure, with the N-terminal domain (NTD) (residues 13–65) packing against the C-terminal domain (CTD) (residues 1–12 and 66–164) (36). The domain boundary is somewhat ill defined but lies in the long helix C (comprising residues 63–80) that connects the 2 lobes (37). The 2 subdomains have distinct thermodynamic stabilities, giving rise to the possibility for T4L unfolding to initiate from at least 2 origins. Although T4L displayed apparent 2-state folding in bulk kinetic experiments (37, 38), an intermediate was identified when following folding kinetics in the presence of denaturant (43, 44). Intriguingly, Leu-to-Ala substitutions in T4L destabilize the protein to an extent that correlates with the size of the cavity introduced in the structure (35). Substitution of Leu for Ala at position 99 (L99A) in the sequence is highly destabilizing for T4L (35, 39), and the L99A variant contains 2 internal hydrophobic cavities of ∼150 Å3 (cavity 4) and 26 Å3 (cavity 3) in the CTD of the protein structure (Fig. 1A) (46).
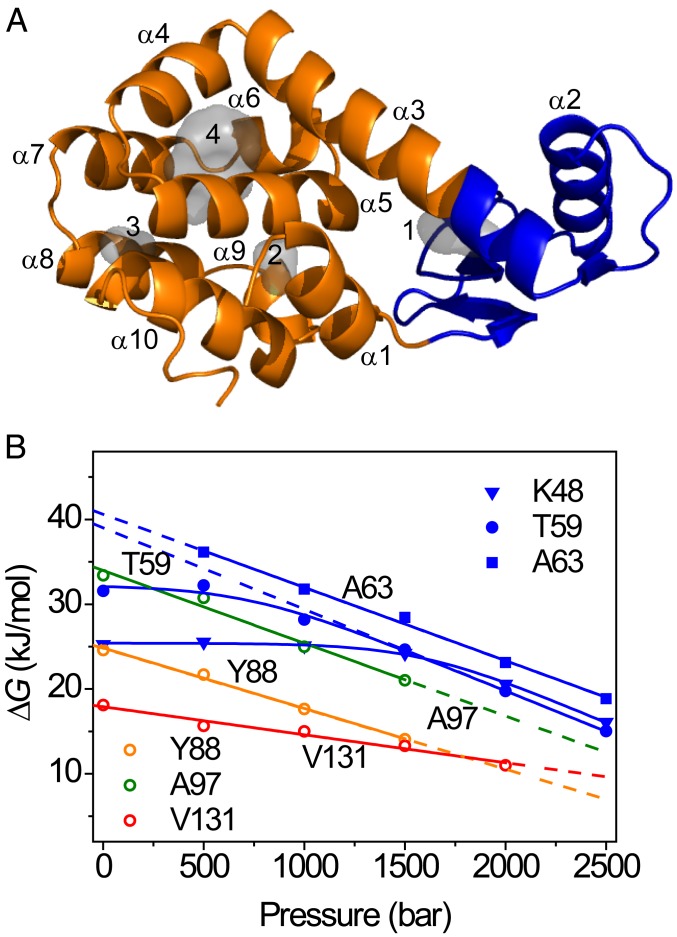
Fig. 1.
Pressure-dependent protein stability from NMR measurements. (A) Backbone ribbon representation of L99A T4L (PDB ID code 1L90). The gray space-filling shapes represent the hydrated, hydrophilic (1 and 2), and empty, hydrophobic cavities (3 and 4) in the structure. (B) Residue-specific stabilities determined as a function of pressure with native-state HX, shown for selected amide hydrogens (colored symbols). The lines represent fits to Eqs. 1 and 2 in the text.
Pressure-Driven Protein Destabilization and Unfolding Measured by HX
To investigate the role of cavities to protein stability, we used native-state HX to report on the thermodynamic stability to exchange with solvent for individual backbone amide hydrogens (7). The rate of exchange of amide protons with solvent deuterons occurs via rare events in which the peptide group becomes exchange competent as follows (47–50):
Here, kcl and kop are the closing and opening rates for the structural transition that leads to a competent conformation, and kint is the intrinsic rate for amide exchange with solvent (i.e., the rate expected for an unprotected, solvent-exposed peptide), estimated from empirical relations (51, 52). When exchange is rate-limited by structural opening (i.e., kcl >> kint, known as the EX2 condition) (49, 50), the observed amide exchange rates kex depend on the structural equilibrium that makes the exchangeable hydrogens solvent accessible, such that kex = Kopkint, where Kop = kop/kcl. Kop is thus determined via measurement of kex. In this manner, the free energy for the opening reaction, ΔGop = –RTlnKop, is accessible for each residue in the protein (49, 50). HX is ideally suited to detect states that have very low occurrence. For example, a residue with ΔGop = 40 kJ/mol has an equilibrium probability of unfolding equal to 10−7, which can be accurately determined from the strong retardation effect on amide exchange.
For L99A, we followed the decay of individual amide protons by 2D 1H–15N heteronuclear single-quantum coherence (HSQC) spectra, which were occasionally recorded in a period from hours to months exchange after a protein sample was exchanged from protonated to deuterated buffer. H/D exchange rates were measured in the range of 1 to 2,500 bar at 500-bar intervals, from exponential decay curves (SI Appendix, Fig. S1). In this range, pressure-induced structural changes, as measured by 1H–15N HSQC cross-peak position changes, were completely reversible, in agreement with previous high-pressure NMR (31, 32), fluorescence (40), electron paramagnetic resonance (EPR), and circular dichroism (CD) data for L99A (23). HX rates were subsequently converted to residue-specific stabilities, ΔGop, at each pressure. Because unfolding lowers the molar volumes of proteins, unfolded states are promoted by the application of hydrostatic pressure. Consequently, the dependence of the Gibbs free energy for protein unfolding (ΔG) follows Eq. 1:
| [1] |
where ∆G° is the free energy of unfolding at p0 = 1 bar. Eq. 1 is a good approximation if the isothermal compressibility difference between the folded and unfolded protein states is negligible over a moderate pressure range as applied here (53, 54). ∆G° and ∆V° can be obtained straightforwardly by fitting Eq. 1 to data of ∆G versus p. In this way, ∆G° values for L99A were determined from backbone amide HX rates recorded at multiple pressures, and examples are shown in Fig. 1B. A subset of residues shows a strong linear dependence of stability on pressure, indicative of an unfolding reaction that liberates significant volume. For example, the stability probed by Ala63 (α-helix 3) follows a straight line with pressure and can be fitted by a straight line with slope ∆V° = −86 mL/mol, which is equal to 144 Å3 when expressed per molecule. A strong pressure-dependent stability is also observed at Tyr88 (α-helix 4), with slope ∆V° = −71 mL/mol (118 Å3), again indicating a significant volume change associated with unfolding for regions in the C domain. The pressure-dependent stability for Val131 (α-helix 8) decreases much more gradually, suggesting that unfolding at this site is possible with less volume loss. SI Appendix, Fig. S2 shows that residues in the CTD show linear pressure responses, albeit with different slopes. The stability data fit into straight isotherms, indicative of a simple 2-state mechanism that exposes the amides to solvent. A range of values for ∆G° and ∆V° were obtained, and the 2 parameters showed to be very strongly correlated. This stratification demonstrates that the CTD consists of multiple segments that possess different stabilities and that liberate different amounts of volume in the process of domain unfolding. Convergently, residues in α-helix 1 show the same behavior (SI Appendix, Fig. S2A), in agreement with the documented structural and energetic integration of α-helix 1 in the CTD of T4L (37).
In contrast to the CTD, exchange for amide hydrogen atoms belonging to the NTD follow a completely different trend: As exemplified by Lys48 (α-helix 2) and Thr59 (turn before α-helix 3) in Fig. 1B, HX is pressure insensitive at low pressure but converts to become strongly pressure dependent at elevated pressure. This behavior can be rationalized as follows: Exchange can occur through multiple, simultaneous events where local, subglobal, and/or global unfolding reactions—with equilibrium constants Kop(local), Kop(subglobal), and Kop(global), respectively—all contribute, such that (7, 26):
| [2] |
Whereas local opening reactions may possess the largest Kop (smallest ΔGop) and dominate HX at ambient pressure, “latent” subglobal and global unfolding processes with large associated ∆V° may eventually come to prevail upon increasing the pressure (26). As Fig. 1B and SI Appendix, Figs. S3 and S4 show, such behavior is observed throughout the NTD. Local opening events remain poorly understood (37, 49). It has been suggested that local exchange may occur through “coughing” events in the native structure (55, 56), which transiently develop channels allowing temporary H2O association with the protein interior (57, 58), much like the diffusion of dioxygen through protein matrices (46, 59, 60). Once penetrated into the structure, exchange may occur through a relayed imidic exchange process (61). The observed lack of denaturant dependence for the NTD suggests that hydrophobic surface area is not exposed to solvent (SI Appendix, Figs. S3 and S4), in agreement with this interpretation (55, 56). Astonishingly, the large pressure-dependent destabilization of the CTD drives NTD unfolding at high pressure. This observation is in line with the known fact that the NTD is not stable in isolation (42). As cavities and packing defects are differentially distributed throughout protein structures, pressure provides a unique opportunity to delineate local and subglobal unfolding processes.
Domain Stability and Partially Unfolded States
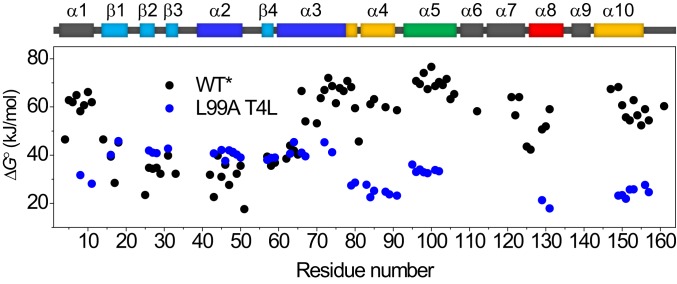
Fig. 2 shows a comparison of the extrapolated ∆G° as a function of sequence for L99A and wild-type (WT) T4L (all data recorded here is for the cysteine-free variant C54T/C97A, also known as WT*, and the L99A mutation is made in the WT* background) (37). Although fewer data points are available for L99A as a consequence of more residues exchanging in the dead time of buffer exchange compared to WT, the plot clearly shows that the cavity-creating mutation strongly and selectively affects the stability of the CTD. Whereas the CTD is more stable than the NTD for the WT protein (37), the Leu-to-Ala mutation inverts this situation.
Fig. 2.
Effect of mutating Leu99 to Ala on the local stability to unfolding ΔG° for T4L as determined by native-state HX. Black, WT* T4L (37); blue, L99A T4L.
The fitted values for ∆G° and ∆V° for the secondary-structure elements of L99A are given in Table 1 and SI Appendix, Tables S1 and S2. At ambient pressure, the most stable region of the protein encompasses the β-strands of the NTD, α-helix 2 and the N-terminal part of α-helix 3 (Table 1), with a communal stability of ∼41 kJ/mol. The remainder of the protein shows lower stabilities. The associated volume changes ∆V° for pressure-induced unfolding of this most stable core of L99A are similar and amount to approximately −100 mL/mol. As this is the largest value obtained for the protein, it is expected to reflect the consequence of global unfolding. Fitted volume changes and extrapolated free energies of stability for other regions of the protein are smaller, indicative of the fact that these involve only parts of the protein. The detection of partially unfolded forms (PUFs) by the pressure-dependent native-state HX data presented here is fully consistent with analogous observations made in denaturant-induced HX (7). Fig. 1 and SI Appendix, Figs. S2–S4 show a close correspondence with the data obtained by Englander for guanidinium-induced unfolding of cytochrome c (7). The important distinction being that in the present case protein unfolding is driven by the expulsion of void volume, rather than by more complex factors, such as differences in side-chain and backbone hydration that result from chaotropic agents (62) or the convoluted response from entropy and heat capacity differences that result from temperature variation. Furthermore, pressure and denaturant-induced unfolding of apocytochrome b562 has shown that the 3 regions of cooperative stability were the same, independent of the perturbation (26).
Table 1.
Calculated ΔVo and ΔGo for structural elements in L99A T4L
| Element | ΔGo, kJ/mol | ΔVo, mL/mol |
| α8 | 20 ± 2 | −35 ± 3 |
| α1, α4, and α10 | 26 ± 3 | −70 ± 9 |
| α5 | 34 ± 1 | −86 ± 10 |
| α2, α3, and β-strands | 41 ± 2 | −100 ± 8 |
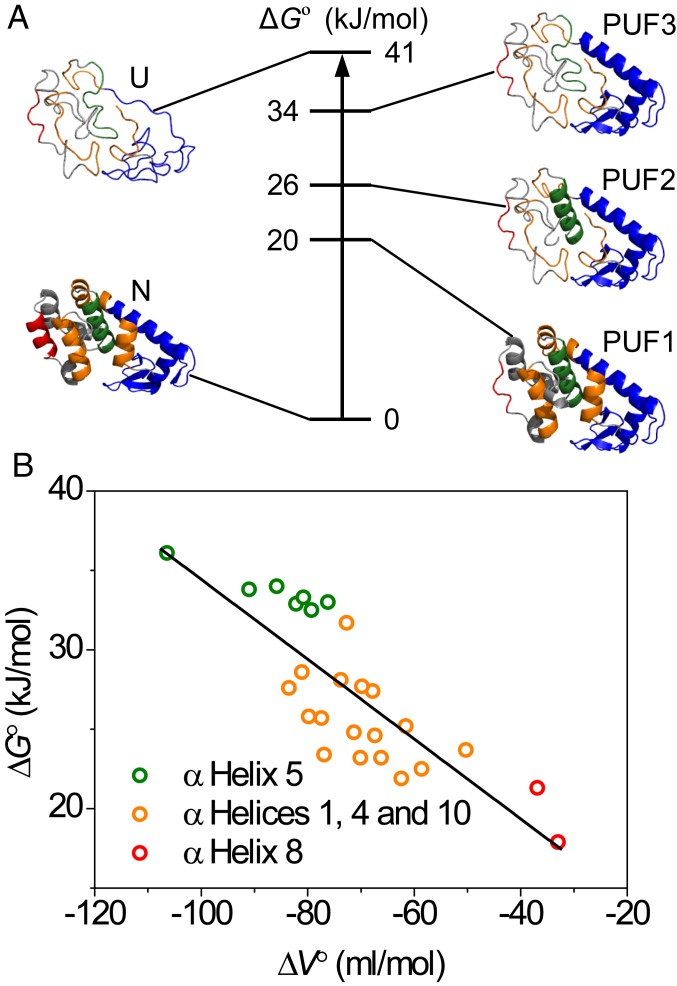
Based on the similar ∆G° and ∆V° values, secondary-structure elements that might belong to cooperative units of structure are grouped in Table 1. Using this further stratification, 3 PUFs were identified (Fig. 3A). Starting closest to the fully folded state, PUF1 shows an average stability of 20 kJ/mol and the smallest ∆V°, −35 mL/mol. This suggest that the unfolding probed by residues 129 and 131 (SI Appendix, Fig. S2E) involves the loss of α-helix 8. Based on the structure of L99A, the dissolution of α-helix 8 presumably leads to the concomitant exposure of cavity 3 to solvent. The observed volume difference, corresponding to 58 Å3, is larger than the size of cavity 3 (26 Å3). Reasons to explain this difference include neglect of the role of solvation volume differences and structural reorganization in PUF1, and the difficulty of accurately estimating small cavities and packing defects. Next, residues in α-helices 1, 4, and 10 show an average ∆G° of 26 kJ/mol and a larger ∆V° (−70 mL/mol), which results from partial CTD unfolding to produce PUF2. The volume change of 116 Å3 does not account for the loss of the total cavity volume in L99A, indicating that packing defects still remain in this PUF. Third, α-helix 5 constitutes the most stable region in the CTD and displays the largest observed ∆V° (−86 mL/mol), which is only very slightly lower than the volume change observed for residues in the NTD (Table 1). The unfolding of α-helix 5 produces PUF3, which is composed of a folded NTD and a partly folded CTD with a stability ∆G° of 34 kJ/mol (Fig. 3A). Finally, the unfolding of the NTD, comprising α-helix 2, the N-terminal part of α-helix 3 and the β-strands (Fig. 3A), produces the completely unfolded state U.
Fig. 3.
Residue-specific pressure-dependent stability data for L99A T4L. (A) Relative stabilities ΔG° for the native, partially and fully unfolded conformations of L99A T4L identified in this study. To the sides, protein structures are shown for a hierarchic model of protein unfolding. (B) Correlation of residue-specific ΔG° and ΔV° defines a broad range of stabilities for the CTD from which the cost of cavity formation was determined from a linear fit through the data.
It is important to note that none of the PUFs discussed here reach detectable concentrations under hydrostatic pressure. Notwithstanding their smaller free energy separation from the native state (Fig. 3A), pressure acts more strongly to increase the population of the fully unfolded state, such that it is predicted to be the dominant state to cross ∆G = 0 at elevated pressures (SI Appendix, Fig. S6), rendering the high-pressure unfolding transition effectively 2 state to bulk observations like CD and fluorescence spectroscopy. In addition, the apparent stability determined in bulk experiments for L99A is essentially that of the NTD. It is important to stress that the subregions detected by equilibrium native-state HX cannot define a folding pathway (63), but only identify the existence of states that are partially unfolded in equilibrium with the native state. However, as the PUFs demonstrate cooperative loss of structure and since proteins are hierarchic in their assembly (64), it is tempting to speculate that the PUFs identified in this study are synonymous with intermediate structures in a sequential loss of cooperative units and which potentially might be synonymous to foldons in the unfolding process (7, 65). At this point, this is speculation, but recent kinetic folding experiments have shown that equilibrium PUFs may indeed be congruent with the sequence of events in protein folding (7, 65, 66). Indeed, examples for the proteins Ubiquitin (67) and outer surface protein A (OspA) (21) have shown close identity of pressure-stabilized intermediates with kinetic intermediates in protein folding. The likelihood of hierarchic unfolding via PUFs identified in equilibrium experiments may be supported by the presence of a hierarchy of the 3D structure. In this view, the individual parts of a protein’s structure do not contain sufficient interactions to be stable in isolation, and the process of folding is driven by sequestration in the direction of lower total free energy (64).
The Energetic Cost of Protein Cavities
For L99A T4L, the NTD has a higher stability, requires at least part of the CTD to be folded, and contains structure that becomes exposed only upon global unfolding. The average volume change in the NTD (−100 mL/mol) is very similar to that determined for α-helix 5 of the CTD (−86 mL/mol) corroborating that the NTD does not contribute much to the volume difference, consistent with the fact that the hydrophilic cavities in the NTD are hydrated at ambient pressure (17). The value of −100 mL/mol translates to a loss of 166 Å3. This number is slightly lower than the volume of the hydrophobic cavities in the CTD in the X-ray structure (176 Å3). The large volume difference is, nonetheless, in agreement with at least the larger hydrophobic cavity being empty at ambient pressure, a result that is meanwhile supported by several lines of experimental evidence (17–19).
Fig. 3B and SI Appendix, Fig. S5 shows that ΔG° is strongly correlated with ΔV°, clearly attesting that unfilled (hydrophobic) cavities are net destabilizing. The stability loss for the CTD calculated from the slope in this plot is −0.25 kJ/mL, equivalent to a loss of 36 cal⋅Å−3. This value is at the high end of the 24–33 cal⋅Å−3 obtained from the pioneering study by Matthews and coworkers (35). However, as these authors note, estimating internal cavity volume from 3D crystal structures is highly nontrivial (35, 68), and many packing defects go undetected, such that 24 cal⋅Å−3 must be considered a lower limit. Our pressure-dependent protein stability data thus recapitulate the existence of empty internal cavities inside a protein core, and also establishes the energetic cost of creating such hydrophobic cavities in a folded protein.
Structure Relaxation
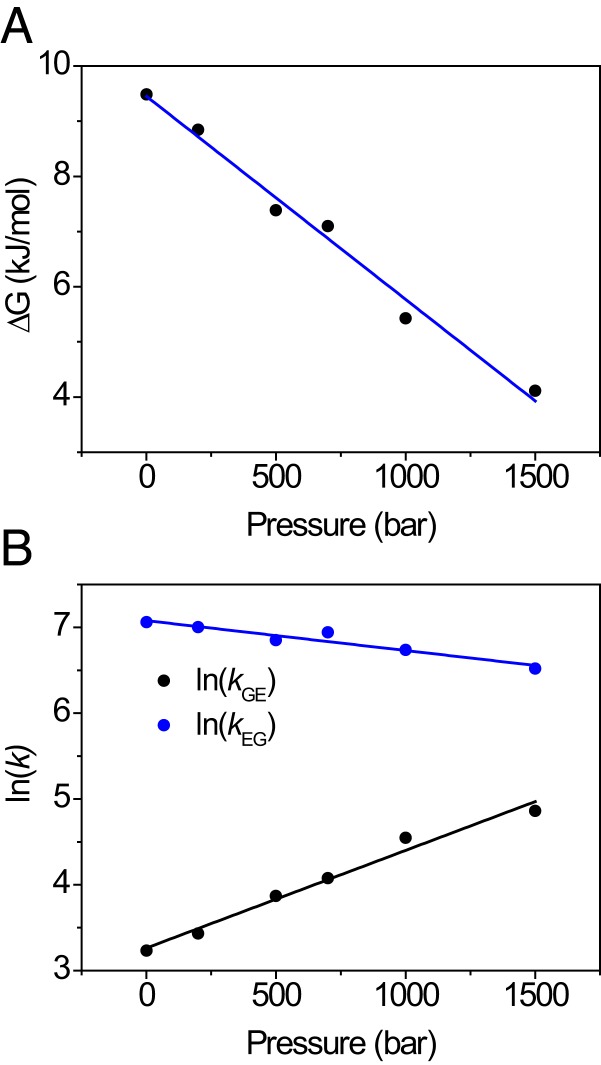
Finally, we turn to the question how pressure might promote alternate folded protein states through structure relaxation. L99A has previously been subject to intensive investigation, which has shown that the protein exists in 2 folded conformations that interconvert on the millisecond timescale (5, 6). At ambient pressure and temperature, the minor species is populated to ∼3% (i.e., 8.6 kJ/mol at 24 °C) and is coined an excited (E) state (5, 6). The structure of the E state has been determined, and shows that the ring of Phe114 is flipped inward, thereby partly filling the major cavity (5). It may thus be expected that structure relaxation will occur for L99A. Using pressure-dependent intensity changes in 1H–13C HSQC NMR, Maeno et al. (32) suggested that the excited state would indeed become increasingly populated under high pressure, whereas Nucci et al. (31) reached an alternative explanation from intensity changes in 1H–15N HSQC spectra, where the C domain would unfold. Addressing this controversy (33, 34), Hubbell and coworkers used CD spectroscopy to demonstrate that L99A does not undergo unfolding up to 2,500 bar, and deduced by electron–electron double-resonance EPR (DEER) of spin-labeled protein that the excited state of L99A becomes increasingly populated with pressure, with a partial molar volume difference equal to −36 mL/mol (23). We therefore measured relaxation dispersion NMR of backbone 15N nuclei (69, 70) to follow the ground (G)/excited (E) state equilibrium with pressure. The relaxation dispersion profiles were collectively fit to a single kinetic process, allowing the 15N chemical shift differences for each residue to be obtained. These agree well with published values (5), and similar values were obtained at all pressures (SI Appendix, Table S5). To ensure good fitting stability of the rate constants and populations, a single value for the chemical shift difference for each residue was subsequently used as a global parameter at all pressures. The pressure dependence of the free energy for the G↔E equilibrium is shown in Fig. 4A (SI Appendix, Table S3). The free energy separation follows a straight line with pressure, with a slope of –37 mL/mol, indistinguishable from the DEER result (23). Thus, there is collective evidence that increasing hydrostatic pressure causes a volume reduction of 60 Å3 through structure relaxation (23), and this process occurs in parallel with partial and complete unfolding, as detected by HX. This estimate of 60 Å3 is somewhat lower than the calculated van der Waals volume of a benzene ring (80 Å3) (71), but this is not unreasonable, as flipping the Phe114 side chain induces further local adjustments of the protein structure. Finally, it is worth mentioning that applying pressure results in a reduction in the rates for the G↔E conversion (Fig. 4B), as earlier proposed in the literature to explain the NMR signal loss with pressure observed in experiment (32).
Fig. 4.
Structure relaxation of L99A followed by relaxation dispersion NMR spectroscopy. (A) Decrease of ΔGGE with pressure indicates that E becomes increasingly populated; (B) forward (kGE) and reverse (kEG) rate constants with pressure show that the exchange rate kex = kGE + kEG decreases with pressure. Global fitting yields activation volumes ΔVGE0‡ = −28 ± 2 mL/mol, ΔVEG0‡ = 9 ± 1 mL/mol, and ΔVGE0 = −37 ± 2 mL/mol.
Conclusion
We show that pressure perturbation, coupled to CPMG relaxation dispersion and HX NMR spectroscopy presents a powerful approach to study the protein energy landscape. We are able to show that partial molar volume changes specifically drive structural transitions, which can involve alternatively folded as well as partly unfolded forms. Multiple PUFs were detected at all pressures, but these remain “hidden” states, refractory to direct observation due to their low occupancies. Complete unfolding was shown to involve a volume change of −100 mL/mol, coincident with the large volume of hydrophobic cavities in the CTD. These hydrophobic cavities are empty at ambient pressure, and, in solution, water does not occupy these spaces below 2.5-kbar pressure, attesting to the unfavorable character of hydrophobic hydration. The stabilities of the partially unfolded states show a continuum with pressure, from which the energetic penalty of hydrophobic cavities was computed to equal 36 cal⋅Å−3. As partial unfolding of labile protein regions is a major issue for industrial enzymes and biologicals as well as protein deposition diseases, the approach demonstrated here forms a powerful avenue to gain control to protein stability and unfolding at the atomic level.
Materials and Methods
Protein Sample Preparation.
The L99A T4L mutant was based on a cysteine-free background T4 lysozyme (C54T/C97A; WT*). The 15N-labeled L99A T4L samples were prepared at 0.4 mM in 50 mM phosphate buffer and 25 mM NaCl at pH 5.5.
Native-State HX.
The protein sample was buffer exchanged into 50 mM phosphate/D2O buffer, 25 mM NaCl (pH* 5.5, meter reading) using a 10-kDa MWCO Amicon centrifugal filter (Merck Millipore). The protein sample was measured at 24 °C on a 600-MHz spectrometer (Bruker BioSpin; AVANCE) under constant hydrostatic pressures of 1, 500, 1,000, 1,500, 2,000, and 2,500 bar.
15N Relaxation Dispersion Experiments.
15N-CPMG NMR relaxation dispersion data were measured on a 1 mM 15N-labeled L99A T4L sample at 25 °C under 6 different pressures (1, 200, 500, 700, 1,000, and 1,500 bar) using AVANCE 500- and 700-MHz spectrometers (Bruker BioSpin).
A detailed description of materials and methods is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Pramodh Vallurupalli and Prof. Lewis Kay (University of Toronto) for the kind gift of the T4 lysozyme L99A plasmid. This work was supported by Japan Society for the Promotion of Science KAKENHI Grant 25840025 (to R.K.); part of this work was performed using NMR spectrometers at the Danish Center for Ultra-High Field NMR Spectroscopy, supported by the Danish Ministry of Higher Education and Science (AU-2010-612-181). The acquisition of the HP system is supported by the Carlsberg Foundation (CF14-0636).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.E.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911181116/-/DCSupplemental.
References
- 1.Nicholls A., Sharp K. A., Honig B., Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Dobson C. M., Protein folding and misfolding. Nature 426, 884–890 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvignies G., et al. , Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature 477, 111–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulder F. A. A., Mittermaier A., Hon B., Dahlquist F. W., Kay L. E., Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 8, 932–935 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Bai Y., Sosnick T. R., Mayne L., Englander S. W., Protein folding intermediates: Native-state hydrogen exchange. Science 269, 192–197 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldwin A. J., Kay L. E., NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 5, 808–814 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Henzler-Wildman K. A., et al. , Intrinsic motions along an enzymatic reaction trajectory. Nature 450, 838–844 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Sugase K., Dyson H. J., Wright P. E., Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Tzeng S.-R., Kalodimos C. G., Dynamic activation of an allosteric regulatory protein. Nature 462, 368–372 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Vallurupalli P., Bouvignies G., Kay L. E., Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Mulder F. A. A., Hon B., Mittermaier A., Dahlquist F. W., Kay L. E., Slow internal dynamics in proteins: Application of NMR relaxation dispersion spectroscopy to methyl groups in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 124, 1443–1451 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Tugarinov V., Libich D. S., Meyer V., Roche J., Clore G. M., The energetics of a three-state protein folding system probed by high-pressure relaxation dispersion NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 54, 11157–11161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco R., Gil-Caballero S., Ayala I., Favier A., Brutscher B., Probing conformational exchange dynamics in a short-lived protein folding intermediate by real-time relaxation-dispersion NMR. J. Am. Chem. Soc. 139, 1065–1068 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Roche J., et al. , Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 6945–6950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L., Quillin M. L., Matthews B. W., Use of experimental crystallographic phases to examine the hydration of polar and nonpolar cavities in T4 lysozyme. Proc. Natl. Acad. Sci. U.S.A. 105, 14406–14411 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins M. D., Hummer G., Quillin M. L., Matthews B. W., Gruner S. M., Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc. Natl. Acad. Sci. U.S.A. 102, 16668–16671 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews B. W., Liu L., A review about nothing: Are apolar cavities in proteins really empty? Protein Sci. 18, 494–502 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamatari Y. O., Kitahara R., Yamada H., Yokoyama S., Akasaka K., High-pressure NMR spectroscopy for characterizing folding intermediates and denatured states of proteins. Methods 34, 133–143 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Kitahara R., et al. , A delicate interplay of structure, dynamics, and thermodynamics for function: A high pressure NMR study of outer surface protein A. Biophys. J. 102, 916–926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fossat M. J., et al. , High-resolution mapping of a repeat protein folding free energy landscape. Biophys. J. 111, 2368–2376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerch M. T., et al. , Structure-relaxation mechanism for the response of T4 lysozyme cavity mutants to hydrostatic pressure. Proc. Natl. Acad. Sci. U.S.A. 112, E2437–E2446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva J. L., Foguel D., Royer C. A., Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem. Sci. 26, 612–618 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Akasaka K., Probing conformational fluctuation of proteins by pressure perturbation. Chem. Rev. 106, 1814–1835 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Fuentes E. J., Wand A. J., Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry 37, 9877–9883 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Caro J. A., Wand A. J., Practical aspects of high-pressure NMR spectroscopy and its applications in protein biophysics and structural biology. Methods 148, 67–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nisius L., Grzesiek S., Key stabilizing elements of protein structure identified through pressure and temperature perturbation of its hydrogen bond network. Nat. Chem. 4, 711–717 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Williamson M. P., Kitahara R., Characterization of low-lying excited states of proteins by high-pressure NMR. Biochim. Biophys. Acta. Proteins Proteomics 1867, 350–358 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Charlier C., et al. , Study of protein folding under native conditions by rapidly switching the hydrostatic pressure inside an NMR sample cell. Proc. Natl. Acad. Sci. U.S.A. 115, E4169–E4178 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nucci N. V., Fuglestad B., Athanasoula E. A., Wand A. J., Role of cavities and hydration in the pressure unfolding of T4 lysozyme. Proc. Natl. Acad. Sci. U.S.A. 111, 13846–13851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeno A., et al. , Cavity as a source of conformational fluctuation and high-energy state: High-pressure NMR study of a cavity-enlarged mutant of T4 lysozyme. Biophys. J. 108, 133–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitahara R., Mulder F. A. A., Is pressure-induced signal loss in NMR spectra for the Leu99Ala cavity mutant of T4 lysozyme due to unfolding? Proc. Natl. Acad. Sci. U.S.A. 112, E923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wand A. J., Nucci N. V., Reply to Kitahara and Mulder: An ensemble view of protein stability best explains pressure effects in a T4 lysozyme cavity mutant. Proc. Natl. Acad. Sci. U.S.A. 112, E924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eriksson A. E., et al. , Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255, 178–183 (1992). [DOI] [PubMed] [Google Scholar]
- 36.Weaver L. H., Matthews B. W., Structure of bacteriophage T4 lysozyme refined at 1.7 Å resolution. J. Mol. Biol. 193, 189–199 (1987). [DOI] [PubMed] [Google Scholar]
- 37.Llinás M., Gillespie B., Dahlquist F. W., Marqusee S., The energetics of T4 lysozyme reveal a hierarchy of conformations. Nat. Struct. Biol. 6, 1072–1078 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Elwell M., Schellman J., Phage T4 lysozyme. Physical properties and reversible unfolding. Biochim. Biophys. Acta 386, 309–323 (1975). [DOI] [PubMed] [Google Scholar]
- 39.Baase W. A., Liu L., Tronrud D. E., Matthews B. W., Lessons from the lysozyme of phage T4. Protein Sci. 19, 631–641 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ando N., et al. , Structural and thermodynamic characterization of T4 lysozyme mutants and the contribution of internal cavities to pressure denaturation. Biochemistry 47, 11097–11109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shank E. A., Cecconi C., Dill J. W., Marqusee S., Bustamante C., The folding cooperativity of a protein is controlled by its chain topology. Nature 465, 637–640 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llinás M., Marqusee S., Subdomain interactions as a determinant in the folding and stability of T4 lysozyme. Protein Sci. 7, 96–104 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cellitti J., Bernstein R., Marqusee S., Exploring subdomain cooperativity in T4 lysozyme II: Uncovering the C-terminal subdomain as a hidden intermediate in the kinetic folding pathway. Protein Sci. 16, 852–862 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato H., Vu N. D., Feng H., Zhou Z., Bai Y., The folding pathway of T4 lysozyme: An on-pathway hidden folding intermediate. J. Mol. Biol. 365, 881–891 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson D. E., Lu J., McIntosh L., Dahlquist F. W., “The folding, stability and dynamics of T4 lysozyme: A perspective using nuclear magnetic resonance” in NMR of Proteins, Clore G., Gronenborn A., Eds. (MacMillan Press, 1993), pp. 258–304. [Google Scholar]
- 46.Kitahara R., Yoshimura Y., Xue M., Kameda T., Mulder F. A., Detecting O2 binding sites in protein cavities. Sci. Rep. 6, 20534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linderstrøm-Lang K., Deuterium exchange between peptides and water. Chem. Soc. Spec. Publ. 2, 1–20 (1955). [Google Scholar]
- 48.Hvidt A., Nielsen S. O., Hydrogen exchange in proteins. Adv. Protein Chem. 21, 287–386 (1966). [DOI] [PubMed] [Google Scholar]
- 49.Skinner J. J., Lim W. K., Bédard S., Black B. E., Englander S. W., Protein dynamics viewed by hydrogen exchange. Protein Sci. 21, 996–1005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krishna M. M. G., Hoang L., Lin Y., Englander S. W., Hydrogen exchange methods to study protein folding. Methods 34, 51–64 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Bai Y., Milne J. S., Mayne L., Englander S. W., Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molday R. S., Englander S. W., Kallen R. G., Primary structure effects on peptide group hydrogen exchange. Biochemistry 11, 150–158 (1972). [DOI] [PubMed] [Google Scholar]
- 53.Vidugiris G. J., Markley J. L., Royer C. A., Evidence for a molten globule-like transition state in protein folding from determination of activation volumes. Biochemistry 34, 4909–4912 (1995). [DOI] [PubMed] [Google Scholar]
- 54.Akasaka K., Matsuki H., High Pressure Bioscience: Basic Concepts, Applications and Frontiers (Springer, 2015). [Google Scholar]
- 55.Chamberlain A. K., Handel T. M., Marqusee S., Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat. Struct. Biol. 3, 782–787 (1996). [DOI] [PubMed] [Google Scholar]
- 56.Persson F., Halle B., How amide hydrogens exchange in native proteins. Proc. Natl. Acad. Sci. U.S.A. 112, 10383–10388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li R., Woodward C., The hydrogen exchange core and protein folding. Protein Sci. 8, 1571–1590 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woodward C., Simon I., Tüchsen E., Hydrogen exchange and the dynamic structure of proteins. Mol. Cell. Biochem. 48, 135–160 (1982). [DOI] [PubMed] [Google Scholar]
- 59.Kawamura T., et al. , Analysis of O2-binding sites in proteins using gas-pressure NMR spectroscopy: Outer surface protein A. Biophys. J. 112, 1820–1828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kitahara R., et al. , Nuclear magnetic resonance-based determination of dioxygen binding sites in protein cavities. Protein Sci. 27, 769–779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eriksson M. A. L., Härd T., Nilsson L., On the pH dependence of amide proton exchange rates in proteins. Biophys. J. 69, 329–339 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auton M., Holthauzen L. M. F., Bolen D. W., Anatomy of energetic changes accompanying urea-induced protein denaturation. Proc. Natl. Acad. Sci. U.S.A. 104, 15317–15322 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarke J., Itzhaki L. S., Fersht A. R., Hydrogen exchange at equilibrium: A short cut for analysing protein-folding pathways? Trends Biochem. Sci. 22, 284–287 (1997). [DOI] [PubMed] [Google Scholar]
- 64.Privalov P. L., Stability of proteins: Small globular proteins. Adv. Protein Chem. 33, 167–241 (1979). [DOI] [PubMed] [Google Scholar]
- 65.Maity H., Maity M., Krishna M. M. G., Mayne L., Englander S. W., Protein folding: The stepwise assembly of foldon units. Proc. Natl. Acad. Sci. U.S.A. 102, 4741–4746 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Englander S. W., Mayne L., The case for defined protein folding pathways. Proc. Natl. Acad. Sci. U.S.A. 114, 8253–8258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitahara R., Akasaka K., Close identity of a pressure-stabilized intermediate with a kinetic intermediate in protein folding. Proc. Natl. Acad. Sci. U.S.A. 100, 3167–3172 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Connolly M. L., Solvent-accessible surfaces of proteins and nucleic acids. Science 221, 709–713 (1983). [DOI] [PubMed] [Google Scholar]
- 69.Loria J. P., Rance M., Palmer A. G., A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999). [Google Scholar]
- 70.Tollinger M., Skrynnikov N. R., Mulder F. A. A., Forman-Kay J. D., Kay L. E., Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 123, 11341–11352 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Bondi A., van der Waals volumes and radii. J. Phys. Chem. 68, 441–451 (1964). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.