Significance
Echolocation is a prime example of convergent evolution, the independent gain of similar features in species of different lineages. Is phenotypic convergence driven by underlying molecular convergence? If so, could molecular convergence include contributions from highly constrained, often-pleotropic, coding regions? We develop a generalizable test that offers a resounding “yes” to both extensively debated questions. Our test highlights molecular convergence in genes regulating the cochlear ganglion of echolocating bats and whales, the skin of aquatic mammals, and the lung of high-altitude mammals. Importantly, the approach correctly dismisses confounding convergence-like patterns, such as those from sequence decay of vision genes in blind subterranean species, and is readily applicable to the thousands of genomes sequenced across the tree of life.
Keywords: convergent evolution, genome-wide functional enrichment tests, coding, echolocation, aquatic
Abstract
Distantly related species entering similar biological niches often adapt by evolving similar morphological and physiological characters. How much genomic molecular convergence (particularly of highly constrained coding sequence) contributes to convergent phenotypic evolution, such as echolocation in bats and whales, is a long-standing fundamental question. Like others, we find that convergent amino acid substitutions are not more abundant in echolocating mammals compared to their outgroups. However, we also ask a more informative question about the genomic distribution of convergent substitutions by devising a test to determine which, if any, of more than 4,000 tissue-affecting gene sets is most statistically enriched with convergent substitutions. We find that the gene set most overrepresented (q-value = 2.2e-3) with convergent substitutions in echolocators, affecting 18 genes, regulates development of the cochlear ganglion, a structure with empirically supported relevance to echolocation. Conversely, when comparing to nonecholocating outgroups, no significant gene set enrichment exists. For aquatic and high-altitude mammals, our analysis highlights 15 and 16 genes from the gene sets most affected by molecular convergence which regulate skin and lung physiology, respectively. Importantly, our test requires that the most convergence-enriched set cannot also be enriched for divergent substitutions, such as in the pattern produced by inactivated vision genes in subterranean mammals. Showing a clear role for adaptive protein-coding molecular convergence, we discover nearly 2,600 convergent positions, highlight 77 of them in 3 organs, and provide code to investigate other clades across the tree of life.
The evolutionary gain of similar traits in distantly related species has been proposed to be encoded, in some cases, by identical sequence substitution paths in their respective genomes (1, 2). While convergent phenotypes are seldom, if ever, the result of only (parallel or strictly) convergent molecular adaptations, several cases of identical amino acid changes underlying phenotypic convergence in distant species have been documented in recent years (3). For example, parallel evolution of prestin (SLC26A5) (4–7) and 4 other auditory genes (8–10) were identified in echolocating bats and toothed whales. However, these examples are few in number, limited in biological scope, and identified using candidate gene-based approaches.
Our recent ability to apply whole-genome sequencing to explore the genetic basis of natural complex adaptations (e.g., Fig. 1A) calls for the development of more systematic approaches for identifying adaptive molecular convergence patterns across genomes (11–14). Recent genome-wide screens illuminated sensory genes with sequence changes that segregate with echolocating mammals (11). Others highlighted positively selected genes that contain convergent amino acid substitutions (12) or identified parallel shifts in evolutionary rates in independent lineages of aquatic mammals (15). However, later analyses demonstrated that the genome-wide frequency of molecular convergence in echolocating mammals is similar to the frequency in nonecholocating control outgroups, suggesting that, in fact, there is no genome-wide protein sequence convergence in echolocation (16, 17).
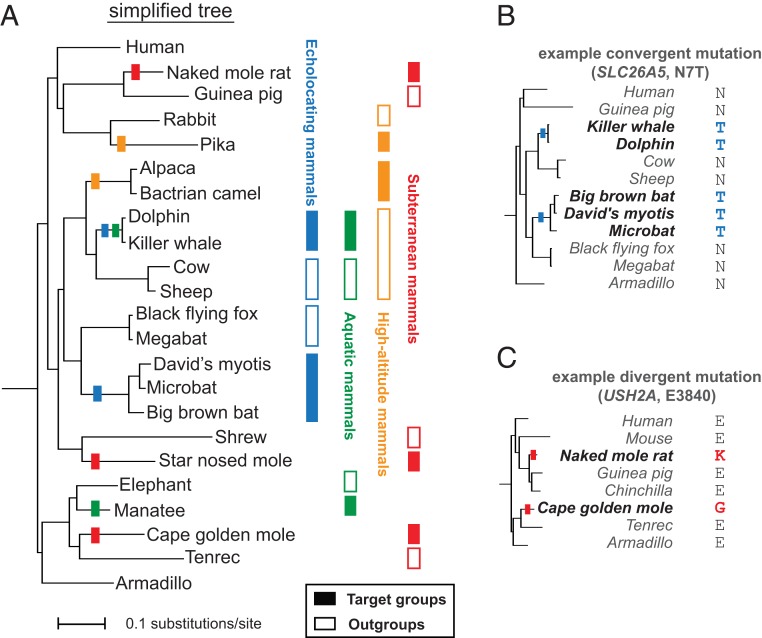
Fig. 1.
Screening for molecular convergence and divergence in the mammalian lineage. (A, Left) Simplified placental mammals phylogenetic tree. See SI Appendix, Fig. S1 for all 57 species used in the study. Colored rectangles highlight branches with independent phenotypic evolution of echolocation, aquatic, high-altitude, and subterranean lifestyles. (A, Right) Filled and empty rectangles represent target and outgroup species, respectively. We screened for (parallel or strictly) convergent and divergent amino acid substitutions along the branches leading from the last common ancestor of each target group and its outgroup to the target group itself. For example: (B) An N7T (asparagine to threonine in position 7) parallel convergent substitution in the hearing gene Prestin (SLC26A5) in echolocating mammals. (C) An E3840 divergent substitution in the vision/hearing gene Usherin (USH2A) in a pair of distantly related subterranean mammals.
Another key challenge in determining the molecular basis of phenotypic convergence is distinguishing adaptive convergent substitutions from molecular convergence-like patterns that do not contribute to the phenotypic convergence, but instead accumulate for other reasons (18). In particular, relaxation of evolutionary constraints (19, 20) or complete loss of a biological function (21) may lead to an accumulation of convergence-like (but in fact nonadaptive) substitutions through sequence relaxation in functionally related groups of genes.
Here, we demonstrate an unbiased approach that 1) identifies—in a genome-wide, function-agnostic manner—all convergent amino acid substitutions in target lineages relative to outgroups, 2) determines if these substitutions are statistically overrepresented in any (or none) of more than 4,000 tissues, and 3) checks for known functional congruence between the most-enriched tissues and the convergent phenotype.
In developing the approach, we considered molecular convergence (Fig. 1B) to include both parallel substitutions (i.e., identical substitutions in the 2 target lineages derived from the same ancestral amino acid) and strictly convergent substitutions (identical target substitutions derived from different ancestral amino acids). Following convention (22), we herein collectively refer to these both as convergent amino acids. Furthermore, we required that the exact same amino acid (the convergent amino acid) be present in every species from both target groups, and that no other amino acid identities be present in target group species. In target or outgroup clades containing 2 or more species (e.g., in the 3 echolocating bats), we allowed a missing amino acid alignment in some species, as long as the convergent amino acid was represented at least once and no other amino acids were present in the target clade.
Then, we devised a functional enrichment-based analysis that, rather than asking which individual genes contain (parallel or strictly) convergent amino acid substitutions, asked which (collection of genes, all affecting a) specific tissue or organ, if any, is most significantly affected by coding convergence. We emphasize that this approach is functionally agnostic (not candidate-based), as it considers convergent substitutions from all genes and tests for functional enrichment across thousands of tissues and organs (Methods and Fig. 2A). Furthermore, by screening for concurrent accumulation of divergent substitutions (Fig. 1C) in these same gene sets, we ensure that the test is specifically robust to otherwise-confounding sequence constraint relaxation.
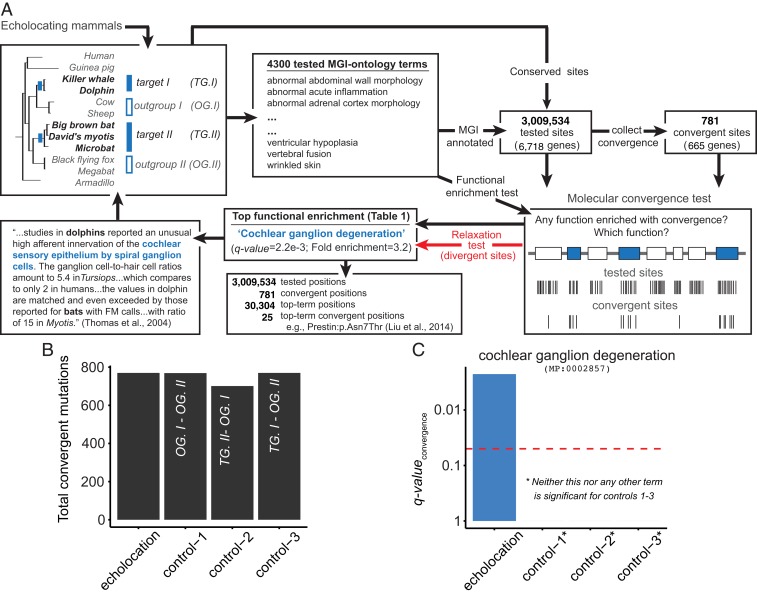
Fig. 2.
A molecular convergence test. Example application to echolocating mammals: (A, Top Left) We picked 2 target groups (TG) of species with a phenotypic convergence (echolocation here), and 2 outgroups (OG). (Top Middle) Our algorithm identified all cross-species conserved (and thus functionally important) amino acid positions in genes annotated for any of 4,300 specific MGI phenotype functions. (Top/Bottom Right) We then identified the subset of positions showing (parallel or strictly) convergent substitutions between our target groups and performed a hypergeometric test over positions to find the single most statistically enriched MGI function (if any) for amino acid convergence. We also tested for divergent substitutions between our target groups (red arrow). (Bottom Middle/Left) If the most-enriched convergent function is not also enriched for divergent substitutions, we declared an adaptive molecular convergence event and linked it back to the convergent species phenotypes. In this example, we discovered 25 convergence events in 18 genes regulating cochlear ganglion function in bats and whales. The cochlear ganglion prediction is particularly striking considering that 4,300 different functions were evaluated to arrive at a poster-child organ for phenotypic convergence between the 2 echolocating groups. (A and B) Comparable total number of convergent substitutions were observed in echolocating mammals (TG.I and TG.II in A, Top Left) as in 3 control sets formed by shuffling either or both target groups with their respective outgroups. (C) However, only the echolocating set of B showed a statistically significant enrichment for molecular convergence in the ontology term “cochlear ganglion degeneration.” In fact, none of the control experiments yielded any statistical enrichment across all 4,300 tested terms.
We applied our method to 3 different convergent trait gains (in echolocating, aquatic, and high-altitude mammals) and to 3 relaxation-based scenarios (vision loss in blind subterranean moles; Fig. 1A) and discovered coding molecular convergence affecting organs with clear functional convergence. We also provide the code underlying our approach so that others can use it to investigate the ever-increasing number of species now being sequenced.
Results
Convergent Amino Acid Substitutions Shared by Echolocating Bats and Cetaceans Are Enriched in Genes Regulating Cochlear Ganglion Function.
Toothed whales (Odontoceti) and certain bat lineages have independently evolved a sophisticated sonar system for 3-dimensional (3D) orientation and prey localization in conditions where vision is ineffective, such as at night or in turbid water (23). Starting with 57 placental mammal whole genomes available from University of California Santa Cruz (UCSC) (listed in SI Appendix, Table S1; complete phylogeny is given in SI Appendix, Fig. S1), we first selected 2 independent echolocating target groups (target 1: dolphin and killer whale; target 2: David’s myotis, microbat, and big brown bat). Our choice dictated 2 respective immediate outgroups (cloven-hoofed mammals and megabats; simplified phylogeny in Figs. 1A and 2A). We then looked for all human protein-coding genes that have an ortholog in many of the 56 other mammals, have at least 1 amino acid highly conserved across the orthologs, and are annotated in the MGI (Mouse Genome Informatics) phenotype ontology (24) for disrupting specific function when mutated in mouse models.
Following this procedure, we identified 3,009,534 conserved (and thus likely functionally important) amino acids across 6,718 genes annotated with 4,300 different MGI tissue-associated phenotypes (Fig. 2A and Methods). Scanning all conserved amino acids for (parallel or strictly) convergent substitutions in echolocating bats and whales that occurred exactly on the branch separating them from their outgroups (Methods and Fig. 1B) revealed 781 such events across 665 genes (summary statistics and complete list of discovered events in SI Appendix, Tables S2 and Dataset S1, respectively). We also analyzed 3 control scenarios, where one or both outgroups were swapped with their respective target group. The 3 control scenarios tested a very similar number of positions (2,986,063 to 2,987,561) and yielded a very similar number of convergent amino acid events (701 to 770 convergent substitutions in 593 to 658 genes), confirming previous observations that no excess genome-wide convergence exists between echolocating cetaceans and bats relative to their outgroups (16, 17) (Fig. 2B).
We then asked whether each convergent set of amino acids was statistically overrepresented in any particular MGI phenotype term by computing its q-value, a multiple-hypothesis-corrected P value (Methods). Strikingly, after testing and correcting for 4,300 MGI terms, the single most-enriched function for convergent substitutions in echolocating mammals is (the set of genes that, when mutated in mouse models, cause) cochlear ganglion degeneration (q-value = 2.2e-3, hypergeometric test; Table 1), with 25 convergent substitutions in 18 genes (listed in Table 2; also see Fig. 2A and Dataset S1). The cochlear ganglion is the group of nerve cells that facilitate the sense of hearing by sending a representation of sound from the cochlea to the brain. Strikingly, the cochlear ganglion is an exemplary organ of phenotypic convergence, as both dolphins and microbats display a significant increase in the ganglion-to-hair cell ratio in their cochlea compared to nonecholocating mammals (25, 26).
Table 1.
Finding molecular convergence in convergent phenotypes
| Convergent adaptation | Target species I* and II† | Top convergent term | Total convergent sites (genes) in top term | Top-term convergence enrichment (q-value, fold enrichment) | Total divergent sites (genes) in top term | Top-term divergence enrichment (q-value, fold enrichment) | Convergence (✓) or relaxation (×) |
| Echolocation | Bottlenose dolphin* | Cochlear ganglion degeneration | 25 (18) | q = 2.2e-03 fold = 3.2 | 15 (11) | q = 1 fold = 1.3 | ✓ |
| Killer whale* | |||||||
| Big brown bat† | |||||||
| Microbat† | |||||||
| David’s myotis bat† | |||||||
| Aquatic | Bottlenose dolphin* | Scaly skin | 27 (15) | q = 3.9e-06 fold = 4.1 | 12 (10) | q = 1 fold = 1.6 | ✓ |
| Killer whale* | |||||||
| Manatee† | |||||||
| High altitude | Alpaca* | Abnormal lung weight | 25 (16) | q = 1.9e-02 fold = 2.7 | 10 (5) | q = 1 fold = 0.8 | ✓ |
| Bactrian camel* | |||||||
| Pika† | |||||||
| Subterranean | Cape golden mole* | Short photoreceptor inner segment | 15 (6) | q = 3.9e-02 fold = 3.7 | 42 (8) | q = 3.9e-17 fold = 6.2 | × |
| Star-nosed mole† | |||||||
| Subterranean | Naked mole rat* | Abnormal eye electrophysiology | 73 (46) | q = 6.0e-07 fold = 2.3 | 103 (49) | q = 4.2e-08 fold = 2.1 | × |
| Star-nosed mole† | |||||||
| Subterranean | Cape golden mole* | Retinal photoreceptor degeneration | 30 (22) | q = 2.9e-02 fold = 2.4 | 57 (23) | q = 1.1e-09 fold = 3 | × |
| Naked mole rat† |
Each row describes the test for a different phenotypic convergence. To test positive for molecular convergence, the top enriched MGI phenotype term (of more than 4,000 tested) for convergent substitutions must not also be enriched for divergent substitutions. We discovered mostly novel convergent positions in cochlear, skin, and lung genes in echolocating, aquatic, and high-altitude mammals, respectively—while correctly identifying convergence accumulation in subterranean mammals’ vision genes as resulting from molecular relaxation (see main text).
Table 2.
Top-term convergent sites for echolocating, aquatic, and high-altitude mammal convergent enrichment tests
| Echolocating mammals (cochlear ganglion degeneration, q = 2.2e-3) | Aquatic mammals (scaly skin, q = 3.9e-6) | High-altitude mammals (abnormal lung weight, q = 0.019) | ||||||
| Gene | Amino acid substitution | Codon start position (GRC h38/hg38) | Gene | Amino acid substitution | Codon start position (GRC h38/hg38) | Gene | Amino acid substitution | Codon start position (GRC h38/hg38) |
| ADGRV1 | H2381R | chr5:90693897 | ABCA12 | H1013N | chr2:215000845 | ABCA3 | V3A | chr16:2326458 |
| ADGRV1 | E3384A | chr5:90725645 | ABCA12 | V1079I | chr2:214997752 | ACE2 | V748I | chrX:15564089 |
| CDH23 | R817Q | chr10:71811406 | ABCA12 | V1203I | chr2:214990717 | AP3B1 | S489N | chr5:78156264 |
| CDH23 | D820E | chr10:71811415 | ABCA12 | I1335T | chr2:214986700 | ARNTL | E332D | chr11:13372336 |
| CDH23 | N924S | chr10:71812589 | ABCA12 | G1396E | chr2:214983841 | ATR | A1828T | chr3:142498671 |
| GJB2 | F115Y | chr13:20189237 | ABCA12 | N1690S | chr2:214978374 | ESR1 | T483I | chr6:152094462 |
| KIT | A964T | chr4:54738516 | ABCA12 | I1729V | chr2:214975979 | FGF9 | T171S | chr13:21701319 |
| LOXHD1 | D212N | chr18:46538282 | ABCA12 | I2584V | chr2:214932670 | FOXP2 | Q206P | chr7:114629949 |
| LOXHD1 | R1094H | chr18:46477679 | CDSN | P45H | chr6:31117480 | GLI2 | F771Y | chr2:120988225 |
| MARVELD2 | V298I | chr5:69420277 | CDSN | G266R | chr6:31116817 | GLI2 | S1271N | chr2:120989725 |
| MCOLN3 | D542E | chr1:85019159 | CST6 | A52T | chr11:66012198 | GRB10 | L3V | chr7:50732314 |
| MPV17 | P106S | chr2:27312551 | KEAP1 | P3L | chr19:10500025 | GRB10 | K180R | chr7:50626943 |
| MYO15A | M3109L | chr17:18159956 | LDLR | L422V | chr19:11113355 | GRB10 | D549E | chr7:50593090 |
| NFKB1 | C447Y | chr4:102596176 | LRIG1 | E912A | chr3:66381513 | KIFAP3 | Q30H | chr1:170055379 |
| SLC17A8 | V109I | chr12:100380924 | MEFV | L590V | chr16:3243882 | LOX | Q44H | chr5:122077854 |
| SLC17A8 | R309K | chr12:100402617 | NFKBIZ | N219S | chr3:101853181 | NKX2-1 | A138T | chr14:36519034 |
| SLC26A5 | N7T | chr7:103421494 | NOTCH3 | P1026S | chr19:15180745 | NPC1 | F356L | chr18:23556501 |
| SLC26A5 | N308S | chr7:103397979 | NOTCH3 | P2309S | chr19:15160701 | NPC1 | I663M | chr18:23544485 |
| SLC4A7 | S181G | chr3:27436407 | RAG1 | N229S | chr11:36573989 | PRKDC | H738Y | chr8:47927816 |
| SRRM4 | G310E | chr12:119145537 | SLC39A2 | A14S | chr14:20999486 | PRKDC | K1407R | chr8:47889073 |
| SYNJ2 | D557E | chr6:158066587 | SPINK5 | S471F | chr5:148101889 | PRKDC | S1506G | chr8:47887601 |
| TMC1 | L192M | chr9:72751888 | TMEM79 | V201M | chr1:156285827 | PRKDC | K2762R | chr8:47830716 |
| TMPRSS3 | H186Q | chr21:42385423 | TRPV3 | R148Q | chr17:3543496 | PRKDC | S3546A | chr8:47794322 |
| TMPRSS3 | L385V | chr21:42376580 | TRPV3 | H160R | chr17:3542685 | ZNF521 | M237L | chr18:25227207 |
| USH1C | P387S | chr11:17520919 | TRPV3 | E207K | chr17:3542544 | ZNF521 | V1061I | chr18:25224735 |
| TRPV3 | R462H | chr17:3528852 | ||||||
| XPA | V234L | chr9:97675559 | ||||||
Every convergent amino acid site is shown with its gene name, amino acid substitution (ancestral position to derived), and UCSC “+” strand codon start position in genomic coordinates (GRCh38/hg38). See Datasets S1–S6 for a complete list of all convergent and divergent substitutions considered in each test and the respective Ensembl identifiers.
Indeed, 2 of the 25 convergent substitutions identified by our method (and, by design, all 18 genes we highlight) have already been experimentally shown to modulate cochlear function (discussed below). Moreover, despite great interest in the topic, most of the 25 positions we highlight have not been noted before in the context of echolocation. In contrast to the strong convergent enrichment detected in echolocators and despite considering a very similar total number of convergent events (Fig. 2B), the 3 ingroup-outgroup–switched control experiments do not exhibit statistically significant functional enrichment for molecular convergence in cochlear ganglion degeneration (Fig. 2C) or, in fact, in any tested ontology term. Our test is thus the first non-candidate-gene–based, unbiased approach to show a clear signature of molecular convergence in independent echolocating mammalian lineages.
Making the Convergent Enrichment Test Robust to Independent Relaxation.
We hypothesized that independent loss of phenotypic function (vestigialization) can produce misleading molecular convergence-like patterns, as genes related to the inactivated phenotype are freed from constraint and may (d)evolve more rapidly, producing more convergent events by chance alone. We tested this hypothesis using subterranean mammals whose visual system and associated genes are thought to have undergone regressive evolution (27, 28). We selected 3 independently evolved subterranean mammals, star-nosed mole, naked mole rat, and cape golden mole, along with 3 respective outgroups (Fig. 1A and SI Appendix, Fig. S1). Repeating the analysis described above (and in Fig. 2A), we performed 3 pairwise convergence tests, 1 for each pair of subterranean mammals (Methods). The 3 pairs yielded very similar results: 1,075 to 1,333 parallel or strictly convergent substitutions in 883 to 1,044 MGI-annotated genes (SI Appendix, Table S2 and Datasets S2–S4) with a top enriched term (q-value < 0.05 in all cases) directly related to vision: short photoreceptor inner segment, abnormal eye electrophysiology, and retinal photoreceptor degeneration (Table 1).
Next, we hypothesized that when the enrichment signal comes from a relaxation event (and not from a convergence event), the same set of genes will also experience a significant number of divergent substitutions of conserved amino acid positions (where both target groups underwent substitution to nonidentical amino acids; Fig. 1C). We thus collected all amino acid divergence events from the 3 pairs of moles (Methods and Fig. 1C). A total of 1,646 to 2,259 divergent amino acids were found in 1,197 to 1,437 MGI-annotated genes (SI Appendix, Table S2 and Datasets S2–S4). When repeating the amino acid enrichment test (Fig. 2A), this time for divergent positions, all 3 top convergent vision terms were also found to be highly significantly enriched for divergence (q-value < 1e-7 in all cases). In contrast, performing the divergence test on the echolocating targets and outgroups, which resulted in 1,140 divergent amino acids in 913 genes (SI Appendix, Table S2 and Dataset S1), suggested no enriched accumulation (q-value = 1) for the top convergent term, cochlear ganglion degeneration. Based on these heuristics, we deemed the echolocation signal as resulting from convergent evolution and the subterranean vision signals as likely stemming from independent relaxation (Table 1). To flag relaxation-derived patterns such as those observed in moles, we incorporated the divergent enrichment assay as an integral part of our test for the remainder of the study (Fig. 2A).
Molecular Convergence Test Flags Skin-Related Genes in Aquatic Mammals.
We employed the combined convergence and relaxation test to explore the molecular basis for convergent adaptations in aquatic mammals. We scanned for (parallel or strictly) convergent and divergent substitutions (831 and 954, respectively, summarized in SI Appendix, Table S2 and fully listed in Dataset S5) between toothed whales (bottlenose dolphin and killer whale) and manatee (Fig. 1A and SI Appendix, Fig. S1). The function most enriched for convergence is (the set of genes that, when mutated in mouse models, cause) scaly skin (q-value = 3.9e-6, 27 amino acids in 15 genes; Table 1 and listed in Table 2), which—importantly—is not statistically overrepresented with divergent substitutions (q-value = 1). Notably, of 27 total convergent substitutions annotated by the “scaly skin” term, 8 are located in ABCA12 (Table 2 and Fig. 3A), a skin keratinization gene implicated in ichthyosis (29, 30) but never previously studied for convergent evolution. Indeed, fully aquatic mammals have adapted to hydrodynamic movement by reducing pelage hair and, in some cases, dramatically increasing the turnover rate of the outermost epidermal cells of the skin (31). As in the echolocation test (Fig. 2C), for all ingroup-outgroup–switched control analyses, neither the scaly skin gene set nor any other tested terms were enriched for convergent substitutions (q-value = 1).
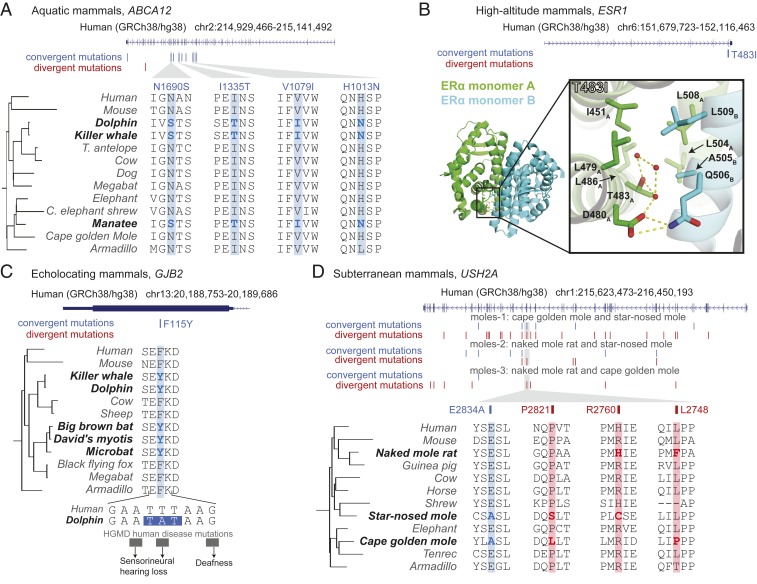
Fig. 3.
Example convergent and divergent substitutions identified. (A) A skin development gene ABCA12 exhibits 8 convergent substitutions and only a single divergent substitution in aquatic mammals. (B) The estrogen receptor ERα encoded by ESR1 contains the convergent substitution T483I in high-altitude mammals. An ERα structural model (Protein Data Bank ID code 3OS8) highlights the convergent substitution at the homodimeric interface (between monomers A and B, green and blue, respectively), suggesting an important functional role in regulating dimer stability. Thr483 interacts with surface polar residues D480A and Q506B through a polar contacts network, antagonizing the adjacent hydrophobic interaction that also occurs at the homodimeric interface. Thus, by substituting residue 483 to hydrophobic isoleucine, the adjacent hydrophobic interaction (involving nonpolar amino acids I451A, L479A, L486A, L504A, L508A, A505B, and L509B) would likely be strengthened, increasing dimer stability and promoting binding to estrogen-responsive elements. (C) A convergent substitution F115Y in GJB2 observed in echolocating mammals is central to 3 codons containing human hearing loss disease mutations, suggesting the residue’s importance in modulating hearing. (D) The vision gene USH2A contains multiple divergent and convergent substitutions in 3 tested pairs of moles. These changes likely accumulated as a result of relaxed purifying selection.
Molecular Convergence Test Flags Lung-Weight Genes in High-Altitude Mammals.
Finally, we employed the combined test (considering enrichment of both convergent and divergent substitution) to high-altitude adaptation in mammals. We defined the alpaca and Bactrian camel as one target group and pika as the second target (Fig. 1A and SI Appendix, Fig. S1). All 3 target species are well-adapted to altitudes higher than 3,000 feet above sea level (32–35). Our screen revealed 977 (parallel or strictly) convergent and 1,306 divergent substitutions (summarized and fully listed in SI Appendix, Tables S2 and Dataset S6, respectively). The function most enriched with convergent substitutions is (the set of genes that, when mutated in mouse models, cause) abnormal lung weight (q-value = 0.019, 25 amino acids in 16 genes; Table 1 and listed in Table 2), which is not enriched with divergent substitutions (q-value = 1; Table 1). All 3 ingroup-outgroup–switched control experiments did not yield enrichment for convergent substitutions in abnormal lung weight (q-value = 1) or in any other tested terms.
These results suggest adaptive molecular convergence of respiratory physiology in the target species. In support of this idea, we find a convergent event (Q44H) in LOX, a gene that is differentially expressed in high-altitude chickens (36) and a direct target of EPAS1, which encodes hypoxia-inducible factor 2-alpha and is thought to underlie high-altitude adaptation of Tibetans (37). Similarly, another convergent substitution we find (V748I) occurs in angiotensin converting enzyme-2 (ACE2), which is differentially expressed in cattle experiencing chronic hypoxia at high altitude compared to their lowland counterparts (38). Interestingly, humans and rodents born at low-oxygen conditions or high altitudes develop overall larger lungs and increased lung capacities (39, 40).
Direct Empirical Evidence for the Functional Importance of the Top-Term Convergent Substitutions.
Our analysis highlights convergent amino acid substitutions that appear to drive complex polygenic adaptations in independent echolocating, aquatic, and high-altitude lineages (Table 1). By intentional experimental design, these substitutions affect codons highly conserved across mammals and, in some cases, across all vertebrates (e.g., SI Appendix, Fig. S2). We, therefore, anticipate great functional significance for the substitutions we found and, indeed, several experimental studies, protein structural models, and human disease-causing variants directly involve our candidate positions in support of this hypothesis.
Direct experimental interrogation of convergent substitutions.
The motor protein prestin (SLC26A5) generates voltage-dependent changes to outer hair cell length and stiffness in response to mechanical stimuli from sound waves transmitted through the cochlea. These mechanical changes to outer hair cells provide energy to locally amplify sound waves in the cochlea, increasing the overall sensitivity and frequency selectivity of hearing (41). Patch-clamp experiments demonstrated that prestin orthologs from echolocating bats and whales function more similarly to each other than to orthologs from phylogenetically closer relatives (4). We highlight 2 echolocation-associated convergence events in prestin: N7T and N308S (Table 2). Prestin orthologs from non-echolocating bats and whales engineered to have the single convergent substitution N7T showed significant functional differences, all in the same direction of effect, and indeed became more functionally similar to the wild-type protein found in their echolocating relatives (4). Analogous interrogation of N308S yielded similar results (7).
We highlight 1 aquatic convergence event in XPA: V234L (Table 2). The DNA-binding protein XPA plays a critical role in the DNA-damage response by acting as a scaffold that organizes various domains of the nucleotide excision repair machinery. In fact, human mutations in XPA are associated with the most severe cases of xeroderma pigmentosum – a congenital disorder marked by extreme sensitivity to sunlight and dramatically increased rates of skin cancer. The convergent substitution V234L we identified in XPA of fully aquatic mammals is also observed as an extremely rare polymorphism in phenotypically normal human populations (42). In vitro experiments comparing the function of wild-type XPA protein to an otherwise identical protein except for a single V234L substitution demonstrated that the V234L variant shows improved repair of DNA adducts caused by a genotoxic compound (43). Since UV radiation also causes DNA adducts and since the open sea is largely devoid of sun protection, V234L may well improve repair of UV-induced DNA damage as a land-to-water adaptation in the skin of aquatic mammals.
Structural models highlighting convergent substitutions at functionally key residues.
Our analysis flagged the convergent substitution T483I in the ERα estrogen receptor encoded by ESR1 of high-altitude mammals. Because high-altitude-induced pulmonary hypertension disproportionately affects men more than women, it has been proposed that estrogen signaling is protective against hypoxia (44). Crystal structure-based modeling of ERα (45) suggests that the residue T483 is involved in a polar contact network at the interface between 2 ERα molecules forming the dimer conformation required for binding to estrogen-responsive elements (Fig. 3B) (46). Substituting position 483 to hydrophobic isoleucine would likely cause this residue to, instead, interact with and further stabilize an adjacent hydrophobic network also at the dimer interface, increasing overall dimer stability and up-regulating estrogen receptor signaling (Fig. 3B).
We highlight the aquatic convergent substitution A52T in the protease inhibitor Cystatin E/M (encoded by CST6), which is thought to promote skin barrier formation and protect against inflammatory skin conditions such as psoriasis and atopic dermatitis (47). A crystal structure of Cystatin E/M (48) suggests that the nonpolar side chain of alanine at position 52 is involved in hydrophobic interactions with nearby residues in the wild-type protein (SI Appendix, Fig. S4A). The convergent substitution of position 52 to a more polar threonine residue might thus antagonize the hydrophobic interaction and destabilize the protein. Given empirical evidence that destabilization of Cystatin E/M is required for its dimerization via a domain-swapping mechanism (49), we suggest that the convergent substitution A52T would ultimately promote Cystatin E/M dimerization and enhance its function as a protease inhibitor and regulator of cutaneous integrity.
In high-altitude lineages, we discover convergent substitutions in the genes ATR and PRKDC that encode kinases critical for the response to DNA damage (50), including adducts caused by oxidative stress which can result from hypoxia (51). Based on a cryo-electron microscopy structure of ATR in complex with ATR-interacting protein (ATRIP) (52), we propose that the convergent substitution at residue 1828 from hydrophobic alanine to polar threonine (A1828T) would strengthen interactions with nearby hydrophilic residues to stabilize conformation of the N-HEAT and FAT domains of ATR (SI Appendix, Fig. S4B). Such stabilization would likely increase ATR-mediated repair of reactive oxygen species-induced DNA adducts related to hypoxia. Similarly, in a crystal structure of DNA-PKcs (the kinase product of gene PRKDC) bound to Ku80 (53), we observe that the convergent substitution at residue 738 from a positively charged histidine to a nonpolar tyrosine occurs within the hydrophobic core of the protein. As such, we propose that H738Y would stabilize DNA-PKcs conformation and increase its DNA-repair activity (SI Appendix, Fig. S4C), improving cellular fitness in hypoxic conditions.
Human disease-causing variants at or adjacent to convergent substitutions.
We found evidence for exceptional functional importance for a number of additional amino acid residues in each of our 3 sets (echolocating, aquatic, and high-altitude mammals) where mutation at the convergent position, or at an immediately adjacent residue, is sufficient to trigger a severe monogenic disease in humans (SI Appendix, Table S3). For example, we observed the convergent substitution F115Y in GJB2 of echolocators. Mutations in both this codon, as well as both codons immediately before and after it (Fig. 3C), have individually been implicated in causing deafness or sensorineural hearing loss in humans (54). The aquatic mammal substitution L422V in LDLR is the exact residue altered by a human disease variant associated with hypercholesterolemia (SI Appendix, Table S3), which also manifests with skin lesions (55). Likewise, we identified multiple convergent, aquatic substitutions in TRPV3, a skin gene with roles in thermoregulation (56). One such convergent substitution in TRPV3 (R148Q) is the exact codon of a human disease variant underlying palmoplantar keratoderma (SI Appendix, Table S3).
Robustness of Enrichments to Individual Gene Omissions.
We tested the robustness of our top-term enrichment to omission of individual genes contributing to the term (Methods). For echolocation, the top term “cochlear ganglion degeneration” remains significantly enriched (q-value < 0.05) in 17 of 18 (>94%) gene-removal trials (with the 1 exception being the omission of CDH23, which results in a q-value of 0.053 while retaining a large fold enrichment of 2.9 for convergent substitutions). The “scaly skin” top term remains significantly enriched for aquatic mammals in all 15 of 15 (100%) gene-removal trials, including the removal of ABCA12 which contributes 8 convergent sites to the term (q-value = 0.00873). For high-altitude mammals, the top term “abnormal lung weight” remains significant in 14 of 16 (87.5%) trials. Similarly, our results are robust to perturbations in the requirement of ≥40 aligned species for a gene to be tested, as the top terms are unchanged for the same analysis using a more lenient requirement of either ≥30 or ≥35 aligned species.
Proximity of Adjacent Convergent Sites.
From analyzing the echolocating, aquatic, and high-altitude mammals, we find 437 total intragenic adjacent pairs of convergent amino acids. These pairs appear in proteins that vary widely in length (SI Appendix, Fig. S5A), and their frequency decreases with distance in codon space, when normalized to overall protein length (SI Appendix, Fig. S5B).
Discussion
Convergent phenotypic evolution is a striking and often-observed phenomenon. The extent to which phenotypic convergence is driven by molecular convergence is a fascinating question with implications on our understanding of the constraints and predictability of species evolution (13). This question is complicated by the fact that convergent lineages do not necessarily show an overall excess of convergent genomic events (Fig. 2B). Another highly debated issue relevant to this work concerns the contribution, if any, of often-pleiotropic protein-coding changes to convergent phenotypes (39, 57, 58). To address both questions, we devised a test to analyze distantly related pairs of echolocating, aquatic, and high-altitude mammals. The approach reveals excess accumulation of convergent coding substitutions in (gene sets regulating) organs with well-established roles in the context of the investigated phenotypic adaptation.
As is the case in many evolutionary scenarios, when a surprising number of constrained amino acids rapidly change, one must carefully weigh a switch from purifying selection to either positive selection or neutral drift. The subterranean mammals provided an excellent case study to fortify our test against the latter scenario. Whereas all 3 combinations of distantly related moles yielded a statistically significant accumulation of convergent substitutions in vision genes, all 3 sets also yielded a much stronger statistical signature for accumulated divergent substitutions in these same gene sets. For instance, divergent substitutions outnumber convergent changes in USH2A (Fig. 3D), which—along with other genes enriched for both convergence and divergence (including CRB1, ABCA3, GUCY2F, and PDE6C)—has previously been flagged for molecular relaxation in these species (19, 28). Indeed, this observation of ours could potentially explain why echolocating lineages do not share more convergent amino acids with each other than they do with their outgroups (Fig. 2B). Varying degrees of constraint relaxation will allow amino acid substitutions to different extents in different directions, including in seemingly convergent patterns, such that a fraction of what is technically defined as convergent (Fig. 1B) is not actually the result of adaptive pressure. A key to our approach is the ability to distinguish molecular convergence driving adaptation versus superficially similar phenomena from constraint relaxation.
Our results cannot, and should not, exclude additional noncoding, regulatory contribution to these same traits (59). Indeed, when a different high-altitude mammal, the Tibetan antelope, is analyzed as one target group with either Bactrian camel and alpaca (329 convergent coding substitutions) or pika (403 convergent coding substitutions) as the second target group, we do not observe any functional coding enrichment. As such, it is possible that the genomic basis of high-altitude adaptation in Tibetan antelope is mainly noncoding (60), or coding but not strictly convergent (61).
By nature, life-style adaptations like the ones we identify here are complex, multitissue, polygenic affairs. In this first derivation of our test, we examine only the single functional term (where one exists) most enriched for convergent substitutions that also does not exhibit excess accumulation of divergent changes. The rationale for doing so is severalfold: Consideration of just the top term focuses attention on specific (and, possibly, the most pronounced) aspects of convergent physiological adaptations—providing discernment that is not possible when one simply collects all convergent events and considers the multiple, often-pleiotropic functions of each affected gene, or when taking a candidate-based approach. We also find plenty of compelling candidates that can motivate extensive experimental inquiry into their significance beyond the promising functional studies we identified.
Recent advances in CRISPR-Cas–mediated DNA and RNA editing along with the development of in vitro organ models makes experimental evaluation of the top-term candidate substitutions easier and more powerful than ever before. Using just the 3 case studies presented here as starting examples, one could engineer and test—in a robust, isogenic manner—various combinations of our 77 candidate substitutions in human induced pluripotent stem cells (IPSCs). The modified IPSCs could be used to generate inner ear organoids containing functional hair cells (62) and determine whether their electrophysiological response to various stimuli is shifted to be more similar to that of echolocating mammals. IPSC-derived skin structures (63) from unmodified cells versus those engineered with convergent substitutions could test for changes in (ultra)structure, proliferation, or UV-induced DNA repair that more closely match the integument of fully aquatic mammals. Likewise, the mass, proliferation, and hypoxia tolerance (including DNA damage response) of 3D lung models (64) could be assayed to determine if high-altitude convergent substitutions affect cardiorespiratory characteristics beneficial for fitness at low-oxygen conditions.
Like other biological tests of significance, where the true underlying statistical distribution is beyond reach, our test could be extended in numerous ways (65), such as attempting to consider additional convergence-enriched functions that rank below the top term in statistical significance. It is not trivial, however, to implement such an extension, as the enrichment of other functionally related terms would not occur in a statistically independent manner (SI Appendix, Table S4). We, therefore, leave these considerations to future work and share our codebase for this purpose.
By analyzing a study that scored thousands of physiological and morphological traits for presence/absence in mammals (66), we estimate (Methods) that over a third (773/2,215, >34%) of the scored phenotypes are convergent, independently gained traits like echolocation (SI Appendix, Fig. S3B) and would be amenable to interrogation by our test. To deploy our test, one simply needs genome assemblies from a set of related species, a functionally annotated gene set for just 1 of the assemblies, and knowledge of phenotypic convergences among the species. Thousands of genomes across the tree of life are already publicly available at the National Center for Biotechnology Information alone (SI Appendix, Fig. S3A), and ongoing sequencing efforts at scales spanning large consortia to individual benchtops will generate many more. These genomes, combined with the increased ability to integrate phenotypic data into computational analyses (66–68), are ripe for exploration using screens like ours to identify sequence changes underlying a myriad of convergent phenotypes across the tree of life.
Methods
Code and Data Availability.
The code developed for the project is provided, along with required input files and detailed usage documentation, at https://bitbucket.org/bejerano/convergentevolution.
Species Set and Gene Set.
In this study we used genome assemblies of 57 species (listed in SI Appendix, Table S1) and their substitutions per site-weighted phylogenetic tree (SI Appendix, Fig. S1) from UCSC (genome.ucsc.edu). We used human genome assembly GRCh38/hg38 and Ensembl (ensembl.org) release 86 human protein-coding gene set as reference.
Finding Conserved Amino Acid Positions in Functionally Annotated Genes.
We started by picking a pair of independent clades (with 1 or multiple species) to serve as target groups for our convergent evolution test, and their associated outgroups. The 6 sets used in this paper are shown in SI Appendix, Fig. S1. We mapped all human genes, except a small fraction overlapping segmental duplication regions (from UCSC), to each of the other 56 species using UCSC liftOver chains (69). We then excluded genes that were not mapped to at least 1 species from each of the selected target and outgroups, as well as genes lacking any functional annotations in the MGI Phenotype Ontology (more details below). In addition, to focus our analyses on pan-mammalian genes, we required that a gene is aligned in at least 40 species. To evaluate the robustness of our results, we later repeated the test with ≥30 or ≥35 species thresholds.
We then used the alignments to determine the orthologous amino acid in each available species. Because cross-species exon boundaries sometimes shift for evolutionary or for alignment reasons, we excluded the first and last 2 amino acid positions in each exon from all downstream analyses. We then derived a set of testable amino acids from all remaining genes, where each amino acid is aligned in at least 1 species from each of the target and outgroups and is also conserved across all aligned species with Bayesian branch length score (BBLS) >0.9 (discussed below). We mapped only these genes back to the MGI ontology, removed ontology terms annotated by too few or too many genes (discussed below), and then excluded all genes and amino acids lacking functional annotations in the remaining set of ontology terms. The final number of ontology terms, genes, and amino acids used for testing each of the 6 species sets in the paper is described in SI Appendix, Table S2.
Cross-Species Amino Acid Conservation Score.
Using cross-species conservation as a hallmark of functional importance, we required that each amino acid position be aligned in at least 40 placental mammals. We then computed the total substitution per site branch length (BL) over which the dominant amino acid is conserved. This we converted to a BBLS which further takes into account the phylogenetic relatedness between species. We tested only amino acid positions conserved at BBLS above 0.9. BBLS was previously demonstrated to outperform BL scores (70) and is extensively discussed in Xie et al. (71).
Mouse Phenotype Ontology.
The MGI Phenotype Ontology catalogs spontaneous, induced, and genetically engineered mouse mutations and their associated phenotypes (24). Ontology data (containing 8,949 phenotypic terms; format version 1.2) was lifted over from mouse to human, resulting in 609,253 (canonicalized) gene–phenotype associations. To increase statistical power (by reducing the required multiple testing correction factor in our tests below) we ignored general ontology terms (those annotating more than 500 genes with at least 1 conserved position) or too-specific ontology terms (those annotating fewer than 10 such genes).
Calling Convergent and Divergent Substitutions.
Starting from an amino acid alignment at any tested position (derived above) we used PAML (V4.8) (72) to infer the most likely amino acid identity in the internal nodes of our 57 species eutherian phylogenetic tree (SI Appendix, Fig. S1). We scanned only for substitutions occurring along the branches leading from the last common ancestor of each target group and its outgroup to the target group itself.
The procedure for calling convergent substitution is described in the introduction. For divergent substitutions (Fig. 1C) we used similar logic: A pair of substitutions resulting in different amino acids in the 2 target groups was called a divergent substitution. In groups of more than 1 species, we allowed missing alignments (in either target or outgroup) but also allowed different amino acids within target groups, as long as the criteria of amino acid substitution along the branch from the common ancestor with the outgroup to the target itself is satisfied.
Enrichment Test over Amino Acids.
Different genes contribute to our test a different number of conserved amino acids, depending on gene length and its cross-species conservation. Therefore, rather than test ontology term enrichment over genes, we tested over the conserved amino acid positions, associating each gene’s ontology annotations to all of its amino acid positions we intended to test for convergence/divergence events. This is equivalent to a gene-based test where each gene is weighted by the number of positions tested in it.
For a given pair of target lineages, we started by determining the background set of conserved and MGI annotated amino acids as above (SI Appendix, Table S2). We used the hypergeometric test to calculate functional enrichment over this set. More specifically, the hypergeometric test was executed separately for each ontology term (π) with 4 parameters: 1) N, the total number of conserved and annotated amino acid positions (background); 2) Kπ, the total number of positions annotated by an ontology term (a subset of the background); 3) n, the total number of positions with convergent (or divergent) substitutions identified from the background; and 4) kπ, the intersection between 2 and 3. We computed a standard hypergeometric P value of the observed enrichment for term π as the fraction of ways to choose n (converged or diverged) amino acid positions without replacement from the entire group of N positions such that at least kπ of the n have ontology annotation π, using the formula below:
| [1] |
We corrected all P values for multiple testing by setting a Benjamini–Hochberg false discovery rate of 0.05 for significance. We also computed the fold enrichment for convergent (or divergent) substitutions labeled by a given term by dividing the observed fraction of convergent (or divergent) substitutions intersecting the tested term by the expected fraction of substitutions: (kπ/n)/(Kπ/N). We kept only those terms meeting all of the following criteria: q-value convergence < 0.05; q-value divergence ≥ 0.05; fold enrichment convergence > 2; fold enrichment divergence ≤ 2. Finally, we ranked all remaining terms, if any, by convergent q-value and then by convergent fold enrichment.
Enrichment Test Robustness to Top-Term Gene Omission.
To assess the robustness of the convergent enrichment for the top term, we recomputed the statistical significance and magnitude of enrichment for the top term when omitting, one by one, each gene contributing to that term. Gene omission entailed ignoring all of its amino acid positions for all steps of the calculation (as if the gene did not exist in the genome). The statistical thresholds (q-value convergence < 0.05; q-value divergence ≥ 0.05; fold enrichment convergence > 2; fold enrichment divergence ≤ 2) remained identical for this analysis.
Estimating the Abundance of Convergent Traits.
To estimate the abundance of phenotypic convergence, we obtained phenotypic data (66) from a large scale mammalian study in MorphoBank (project 773; https://morphobank.org/index.php/Projects/ProjectOverview/project_id/773) that scores 2,215 morphological, physiological, and behavioral presence/absence traits (labeled as “1” and “0,” respectively) across 86 extant and extinct mammals with an existing phylogenetic tree. We labeled as “convergent” the subset of traits that 1) switch from 0 → 1 (i.e., gained) independently in the tree at least twice and 2) after reconstructing the presence/absence of all of the nodes in the tree, the trait is inferred as “absent” in the ancestral head node of the phylogeny (thus reflecting a phenotypic innovation in species with “1”) with probability larger than 0.9, using a maximum likelihood tool available in R (package ape, using SYMreconstruction for ancestral state reconstruction, https://cran.r-project.org/web/packages/ape/index.html).
Protein Structural Models.
Protein structure figures were created using PyMol (https://pymol.org/2).
Supplementary Material
Acknowledgments
We thank the members of the G.B. laboratory, particularly A. M. Tseng for his help, and M. J. Berger, W. Heavner, H. M. Moots, J. H. Notwell, J. Birgmeier, K. A. Jagadeesh, B. Yoo, H. Guturu, and A.M. Wenger for technical advice and helpful discussions. We thank H. Clawson (UCSC) for assistance with mammalian genome alignments and the community at large for the availability of all the different genomes. We thank D. Cooper and P. Stenson from Human Gene Mutation Database for disease variant data. This work was funded in part by the National Science Foundation Graduate Research Fellowship Grant 1656518 (to H.I.C.) and NIH Grants U01MH105949 and R01HG008742, a Packard Foundation Fellowship, and a Microsoft Faculty Fellowship (to G.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The code developed for the project is provided, along with required input files and detailed usage documentation, at https://bitbucket.org/bejerano/convergentevolution.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818532116/-/DCSupplemental.
References
- 1.Stern D. L., The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Weinreich D. M., Delaney N. F., Depristo M. A., Hartl D. L., Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Storz J. F., Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet. 17, 239–250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z., Qi F.-Y., Zhou X., Ren H.-Q., Shi P., Parallel sites implicate functional convergence of the hearing gene prestin among echolocating mammals. Mol. Biol. Evol. 31, 2415–2424 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., et al. , Convergent sequence evolution between echolocating bats and dolphins. Curr. Biol. 20, R53–R54 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Li Y., Liu Z., Shi P., Zhang J., The hearing gene Prestin unites echolocating bats and whales. Curr. Biol. 20, R55–R56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y.-Y., Liu Z., Qi F.-Y., Zhou X., Shi P., Functional effects of a retained ancestral polymorphism in prestin. Mol. Biol. Evol. 34, 88–92 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Shen Y.-Y., Liang L., Li G.-S., Murphy R. W., Zhang Y.-P., Parallel evolution of auditory genes for echolocation in bats and toothed whales. PLoS Genet. 8, e1002788 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z., et al. , Parallel evolution of KCNQ4 in echolocating bats. PLoS One 6, e26618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., et al. , The voltage-gated potassium channel subfamily KQT member 4 (KCNQ4) displays parallel evolution in echolocating bats. Mol. Biol. Evol. 29, 1441–1450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker J., et al. , Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502, 228–231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foote A. D., et al. , Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sackton T. B., Clark N., Convergent evolution in the genomics era: New insights and directions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rey C., et al. , Detecting adaptive convergent amino acid evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chikina M., Robinson J. D., Clark N. L., Hundreds of genes experienced convergent shifts in selective pressure in marine mammals. Mol. Biol. Evol. 33, 2182–2192 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou Z., Zhang J., No genome-wide protein sequence convergence for echolocation. Mol. Biol. Evol. 32, 1237–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas G. W. C., Hahn M. W., Determining the null model for detecting adaptive convergence from genomic data: A case study using echolocating mammals. Mol. Biol. Evol. 32, 1232–1236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castoe T. A., et al. , Evidence for an ancient adaptive episode of convergent molecular evolution. Proc. Natl. Acad. Sci. U.S.A. 106, 8986–8991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prudent X., Parra G., Schwede P., Roscito J. G., Hiller M., Controlling for phylogenetic relatedness and evolutionary rates improves the discovery of associations between species’ phenotypic and genomic differences. Mol. Biol. Evol. 33, 2135–2150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcovitz A., Jia R., Bejerano G., “Reverse genomics” predicts function of human conserved noncoding elements. Mol. Biol. Evol. 33, 1358–1369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiller M., et al. , A “forward genomics” approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2, 817–823 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGhee G. R., Convergent Evolution: Limited Forms Most Beautiful (MIT Press, 2011). [Google Scholar]
- 23.Jones G., Echolocation. Curr. Biol. 15, R484–R488 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Eppig J. T., Blake J. A., Bult C. J., Kadin J. A., Richardson J. E.; Mouse Genome Database Group , The mouse genome database (MGD): Facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 43, D726–D736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas J. A., Moss C. F., Vater M., Echolocation in Bats and Dolphins (University of Chicago Press, 2004). [Google Scholar]
- 26.Wever E. G., McCormick J. G., Palin J., Ridgway S. H., The cochlea of the dolphin, Tursiops truncatus: Hair cells and ganglion cells. Proc. Natl. Acad. Sci. U.S.A. 68, 2908–2912 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim E. B., et al. , Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emerling C. A., Springer M. S., Eyes underground: Regression of visual protein networks in subterranean mammals. Mol. Phylogenet. Evol. 78, 260–270 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Walsh D. M., et al. , A novel ABCA12 mutation in two families with congenital ichthyosis. Scientifica (Cairo) 2012, 649090 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyama M., et al. , Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J. Clin. Invest. 115, 1777–1784 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons E. C. M., An Introduction to Marine Mammal Biology and Conservation (Jones & Bartlett Learning, Burlington, MA, ed. 1, 2012). [Google Scholar]
- 32.Reading R. P., Mix H., Lhagvasuren B., Blumer E. S., Status of wild Bactrian camels and other large ungulates in south-western Mongolia. Oryx 33, 247–255 (1999). [Google Scholar]
- 33.Vyas S., et al. , Reproductive status of Camelus bactrianus during early breeding season in India. Asian Pac. J. Reprod. 4, 61–64 (2015). [Google Scholar]
- 34.Bagchi S., Namgail T., Ritchie M. E., Small mammalian herbivores as mediators of plant community dynamics in the high-altitude arid rangelands of Trans-Himalaya. Biol. Conserv. 127, 438–442 (2006). [Google Scholar]
- 35.Kleinschmidt T., März J., Jürgens K. D., Braunitzer G., Interaction of allosteric effectors with α-globin chains and high altitude respiration of mammals. The primary structure of two tylopoda hemoglobins with high oxygen affinity: Vicuna (Lama vicugna) and alpaca (Lama pacos). Biol. Chem. Hoppe Seyler 367, 153–160 (1986). [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q., et al. , Genome resequencing identifies unique adaptations of Tibetan chickens to hypoxia and high-dose ultraviolet radiation in high-altitude environments. Genome Biol. Evol. 8, 765–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X.-H., et al. , Two functional loci in the promoter of EPAS1 gene involved in high-altitude adaptation of Tibetans. Sci. Rep. 4, 7465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F. Y., et al. , Effect of altitude chronic hypoxia on liver enzymes and its correlation with ACE/ACE2 in yak and migrated cattle [in Chinese]. Zhongguo Ying Yong Sheng Li Xue Za Zhi 31, 272–275 (2015). [PubMed] [Google Scholar]
- 39.Cooper K. L., Decoding the evolution of species. Science 356, 904–905 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Withers P. C., Cooper C. E., Maloney S. K., Bozinovic F., Neto A. P. C., Ecological and Environmental Physiology of Mammals (Oxford University Press, Oxford, ed. 1, 2016). [Google Scholar]
- 41.Zheng J., et al. , Prestin is the motor protein of cochlear outer hair cells. Nature 405, 149–155 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Mellon I., Hock T., Reid R., Porter P. C., States J. C., Polymorphisms in the human xeroderma pigmentosum group A gene and their impact on cell survival and nucleotide excision repair. DNA Repair (Amst.) 1, 531–546 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Porter P. C., Mellon I., States J. C., XP-A cells complemented with Arg228Gln and Val234Leu polymorphic XPA alleles repair BPDE-induced DNA damage better than cells complemented with the wild type allele. DNA Repair (Amst.) 4, 341–349 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Oshima K., Oka M., “Sex hormones” in Diagnosis and Treatment of Pulmonary Hypertension, Fukumoto Y., Ed. (Springer, 2017), pp. 55–65. [Google Scholar]
- 45.Bruning J. B., et al. , Coupling of receptor conformation and ligand orientation determine graded activity. Nat. Chem. Biol. 6, 837–843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar V., Chambon P., The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell 55, 145–156 (1988). [DOI] [PubMed] [Google Scholar]
- 47.Cheng T., et al. , The cystatin M/E-controlled pathway of skin barrier formation: Expression of its key components in psoriasis and atopic dermatitis. Br. J. Dermatol. 161, 253–264 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Dall E., Fegg J. C., Briza P., Brandstetter H., Structure and mechanism of an aspartimide-dependent peptide ligase in human legumain. Angew. Chem. Int. Ed. Engl. 54, 2917–2921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dall E., et al. , Structural and functional analysis of cystatin E reveals enzymologically relevant dimer and amyloid fibril states. J. Biol. Chem. 293, 13151–13165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maréchal A., Zou L., DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5, a012716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen B. P. C., Li M., Asaithamby A., New insights into the roles of ATM and DNA-PKcs in the cellular response to oxidative stress. Cancer Lett. 327, 103–110 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Rao Q., et al. , Cryo-EM structure of human ATR-ATRIP complex. Cell Res. 28, 143–156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sibanda B. L., Chirgadze D. Y., Ascher D. B., Blundell T. L., DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 355, 520–524 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Wattanasirichaigoon D., et al. , High prevalence of V37I genetic variant in the connexin-26 (GJB2) gene among non-syndromic hearing-impaired and control Thai individuals. Clin. Genet. 66, 452–460 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Pietroleonardo L., Ruzicka T., Skin manifestations in familial heterozygous hypercholesterolemia. Acta Dermatovenerol. Alp. Panonica Adriat. 18, 183–187 (2009). [PubMed] [Google Scholar]
- 56.Masamoto Y., Kawabata F., Fushiki T., Intragastric administration of TRPV1, TRPV3, TRPM8, and TRPA1 agonists modulates autonomic thermoregulation in different manners in mice. Biosci. Biotechnol. Biochem. 73, 1021–1027 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Hoekstra H. E., Coyne J. A., The locus of evolution: Evo devo and the genetics of adaptation. Evolution 61, 995–1016 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Berger M. J., Bejerano G., Comment on “A genetic signature of the evolution of loss of flight in the Galapagos cormorant.” bioRxiv:10.1101/181826 (8 September 2017).
- 60.Sackton T. B., et al. , Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364, 74–78 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Gonçalves G. L., et al. , Divergent genetic mechanism leads to spiny hair in rodents. PLoS One 13, e0202219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koehler K. R., et al. , Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 35, 583–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim Y., et al. , Establishment of a complex skin structure via layered co-culture of keratinocytes and fibroblasts derived from induced pluripotent stem cells. Stem Cell Res. Ther. 9, 217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y.-W., et al. , A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goeman J. J., Bühlmann P., Analyzing gene expression data in terms of gene sets: Methodological issues. Bioinformatics 23, 980–987 (2007). [DOI] [PubMed] [Google Scholar]
- 66.O’Leary M. A., et al. , The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339, 662–667 (2013). [DOI] [PubMed] [Google Scholar]
- 67.O’Leary M. A., Kaufman S., MorphoBank: Phylophenomics in the “cloud.” Cladistics 27, 529–537 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Lamichhaney S., et al. , Integrating natural history collections and comparative genomics to study the genetic architecture of convergent evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kent W. J., Baertsch R., Hinrichs A., Miller W., Haussler D., Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. U.S.A. 100, 11484–11489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wenger A. M., et al. , PRISM offers a comprehensive genomic approach to transcription factor function prediction. Genome Res. 23, 889–904 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie X., Rigor P., Baldi P., MotifMap: A human genome-wide map of candidate regulatory motif sites. Bioinformatics 25, 167–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.