Significance
As a critical component of a healthy memory management system, forgetting has received increasing attention. Studies across multiple species support important roles of actin remodeling in forgetting. However, the underlying molecular mechanisms remain unclear. In Drosophila, Rac1 and Cdc42, 2 Rho GTPases that act as signaling hubs to coordinate actin remodeling, were reported to mediate the forgetting of labile and consolidated memories, respectively. Here, we showed that Rac1 and Cdc42 exert their effects on forgetting by acting through 2 different actin polymerization pathways, Rac1/SCAR/Dia and Cdc42/WASp/Arp2/3 complexes. These findings fill in the molecular landscape that link forgetting to actin remodeling at the cellular level and shed light on drug development that aims to tune forgetting to treat memory-related diseases.
Keywords: forgetting, memory, Drosophila, Rho GTPases, actin
Abstract
Different memory components are forgotten through distinct molecular mechanisms. In Drosophila, the activation of 2 Rho GTPases (Rac1 and Cdc42), respectively, underlies the forgetting of an early labile memory (anesthesia-sensitive memory, ASM) and a form of consolidated memory (anesthesia-resistant memory, ARM). Here, we dissected the molecular mechanisms that tie Rac1 and Cdc42 to the different types of memory forgetting. We found that 2 WASP family proteins, SCAR/WAVE and WASp, act downstream of Rac1 and Cdc42 separately to regulate ASM and ARM forgetting in mushroom body neurons. Arp2/3 complex, which organizes branched actin polymerization, is a canonical downstream effector of WASP family proteins. However, we found that Arp2/3 complex is required in Cdc42/WASp-mediated ARM forgetting but not in Rac1/SCAR-mediated ASM forgetting. Instead, we identified that Rac1/SCAR may function with formin Diaphanous (Dia), a nucleator that facilitates linear actin polymerization, in ASM forgetting. The present study, complementing the previously identified Rac1/cofilin pathway that regulates actin depolymerization, suggests that Rho GTPases regulate forgetting by recruiting both actin polymerization and depolymerization pathways. Moreover, Rac1 and Cdc42 may regulate different types of memory forgetting by tapping into different actin polymerization mechanisms.
Forgetting has been recently proposed to be a critical component of a healthy memory management system by providing flexibility and generalization ability (1, 2). Previous studies from invertebrates to vertebrates support that learning itself can activate signals which specifically accelerate the decay of a formed memory without affecting its acquisition (3–7). Such a process is termed active forgetting.
In Drosophila, 1-session training of olfactory aversive conditioning produces 2 memory forms that are distinguishable by their sensitivity to cold anesthesia (8). In wild-type flies, the cold-shock sensitive component, anesthesia-sensitive memory (ASM), lasts for up to 6 h; while the cold-shock resistant component, anesthesia-resistant memory (ARM), is more stable and lasts for over 1 d (9). Rac1, a Rho GTPase (10), regulates the rapid decay of ASM (3). Besides ASM, Rac1 also regulates a short-lived olfactory sensory memory in trace conditioning (11). In contrast, Cdc42, another Rho GTPase, regulates the forgetting of ARM. ARM is regarded as one of the consolidated memory forms in Drosophila (12), which gradually forms in the first hour after training, reaches a plateau at about 2 h, and undergoes slow decay thereafter (4). This later decay of ARM is modulated by repetitive training and is tied to the activity of Cdc42 (4).
Rac1 and Cdc42 are seated as signaling hubs that orchestrate actin rearrangement (13). In accordance with invertebrate studies, Rac1 was also reported to contribute to forgetting of multiple types of memories in mice (7, 14, 15) and rats (16). Still, how 2 similarly functioned Rho GTPases are involved in forgetting of different memory forms remains unknown. Rac1-mediated forgetting is partially explained by activating cofilin, an actin depolymerization factor, through a PAK/LIMK signaling cascade (3, 17). However, Rho GTPases are known to interact with numerous effectors (18, 19), and the downstream pathways that tie Rac1 and Cdc42 to actin remodeling and eventually forgetting are far from fully elucidated. Rho GTPases can also affect actin polymerization pathways. For example, 2 WASP family proteins, SCAR/WAVE and WASp, are known to transduce Rac1 and Cdc42 activity to the activation of Arp2/3 complex to promote actin polymerization (18, 20); but it is unclear how they may contribute to the forgetting functions of Rac1 and Cdc42.
Results
SCAR/WAVE Complex Regulates Labile Memory Forgetting in the MB Neurons.
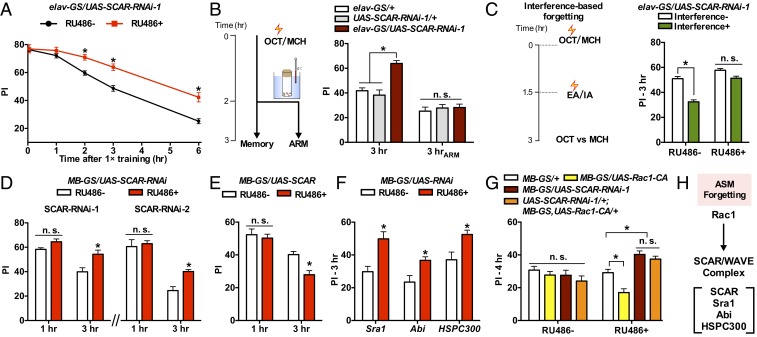
We first tested whether SCAR/WAVE complex affects ASM forgetting using RNAi in Drosophila. To avoid developmental defects, we restricted RNAi expression to the adult flies using elav-GS, a pan-neuronal conditional expression driver that depends on RU486 feeding (21). We examined memory retention curves after a 1-session olfactory aversive conditioning (9). SCAR-RNAi-expressing flies (RU486+) showed memory performance index (PI) similar to the uninduced controls without RU486 feeding (RU486−) shortly after training (3 min and 1 h, Fig. 1A), but the memory was significantly higher at later time points (2, 3, and 6 h, Fig. 1A). Additional experiments using attenuated training intensity (SI Appendix, Fig. S1A) confirmed that SCAR knockdown does not affect initial learning (assayed at 3 min after training). For more stringent controls, SCAR-RNAi-expressing flies showed higher memory retention when compared with their parental controls (elav-GS/+ and UAS-SCAR-RNAi-1/+, RU486+, Fig. 1B and SI Appendix, Fig. S1B). The phenotypes were again confirmed using an independent SCAR-RNAi line (SI Appendix, Fig. S1 C and D). The higher 3-h memory performance of SCAR-RNAi-expressing flies was blocked by cold-shock anesthesia (Fig. 1B and SI Appendix, Fig. S1E), suggesting that SCAR knockdown hampers the forgetting of the ASM component.
Fig. 1.
SCAR/WAVE regulates ASM forgetting in the MB neurons. (A) Memory retention curves. SCAR-RNAi-expressing flies (RU486+) showed slower memory decay compared with uninduced control. n = 8. (B) SCAR knockdown increased 3-h memory without affecting ARM component (3 hARM). Lightning symbol represents electric shock reinforcement. n = 8. (C) Interference-based forgetting was suppressed by RU486-induced expression of SCAR-RNAi. n = 8. (D and E) Memory performance in flies with SCAR knockdown (D) and overexpression (E) in the adult MB neurons. n = 8. (F) Effect of RNAi knockdown of different members in the SCAR complex on 3-h memory. n = 8. (G) RNAi knockdown of SCAR dominated the effect of constitutively active Rac1 (Rac1-CA) on 4 h memory. Rac1-CA-expressing flies (yellow bar, RU486+) had lower memory performance. Flies with coexpression of both Rac1-CA and SCAR-RNAi (orange bar, RU486+) showed memory performance higher than controls, and the performance was not different from flies expressing SCAR-RNAi alone. n = 8 to 12. (H) Data summary. Data are means ± SEM. *P < 0.05. n.s., nonsignificant.
We also tested the effect of SCAR knockdown on interference-based forgetting using a protocol as previously shown (3) (Fig. 1C). After an initial learning session, the trained flies were subjected to a second learning session with a novel pair of odors and the flies were evaluated for 3-h retention of the first learning. The introduction of a second learning session (interference+) lowered memory performance in control flies (RU486−), but had no significant effect in SCAR-RNAi-expressing flies (RU486+) (Fig. 1C).
We next found 3 lines of experimental evidence supporting the notion that SCAR is one of the downstream effectors of Rac1 in ASM forgetting. First, Rac1 and SCAR function in the same brain locus. Rac1-dependent forgetting has been mapped to the intrinsic mushroom body (MB) neurons (3), which play a critical role in olfactory learning and memory in Drosophila (22). We found that expressing SCAR-RNAi in the adult MB neurons using a MB-specific, inducible driver (MB-GS) (23) resulted in higher 3-h memory retention, but left 1-h memory intact (Fig. 1D). Conversely, overexpression of SCAR in the adult MB neurons did not affect 1-h memory but reduced 3-h memory retention (Fig. 1E), and the reduction was also blocked by cold anesthesia (SI Appendix, Fig. S1F). Second, RNAi knockdown of other proteins (Sra1, Abi, and HSPC300) in the SCAR/WAVE complex (20) confirmed the memory phenotype of SCAR at 3-h after training (Fig. 1F). Third, we performed a genetic epistasis experiment by combining the expression of a constitutively active mutant (Rac1-CA) and the SCAR-RNAi-1 in the MB neurons. Consistent with our previous report (3), Rac1-CA expression decreased 3-h memory (MB-GS/UAS-Rac1-CA, RU486+, Fig. 1G). The decrement was dominated by SCAR knockdown (UAS-SCAR-RNAi-1/+; MB-GS, UAS-Rac1-CA/+, RU486+, Fig. 1G). We therefore conclude that SCAR/WAVE complex functions downstream of Rac1-mediated ASM forgetting (Fig. 1H).
WASp Regulates ARM Forgetting in the MB Neurons.
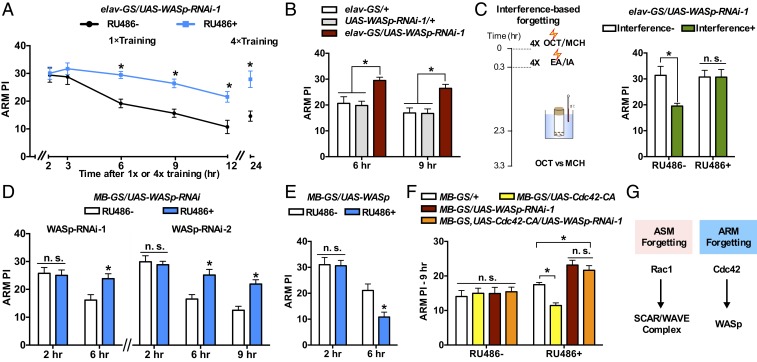
We also investigated the function of WASp in Cdc42-dependent ARM forgetting. To isolate ARM from ASM and to assay the retention of ARM (ARM PI) at different time points after training, we subjected flies to 2-min cold-shock treatment and then allowed flies to recover for 1 h at 25 °C before testing. We focus on the decay of ARM at 3 to 12 h after its formation reaches a plateau at about 2 h. Flies with conditional pan-neuronal WASp-RNAi expression (RU486+) showed slower ARM decay when compared with the RU486− controls. Their performance was higher when ARM was assayed at 6, 9, and 12 h after a 1-session training or at 24 h after a 4-session massed training (Fig. 2A). Note that the effect of WASp knockdown was specific to the later decay phase of ARM without affecting ARM retention at 2 and 3 h after a 1-session training (Fig. 2A) or at 2 h after attenuated training (SI Appendix, Fig. S2A), suggesting that WASp knockdown does not affect ARM formation. For more stringent controls, the higher ARM performance was confirmed by including the parental controls (Fig. 2B) and by using a second independent RNAi line (SI Appendix, Fig. S2C). ASM decay is likely not affected by WASp knockdown. WASp-RNAi-expressing flies had normal memory performance up to 3 h after a 1-session training (SI Appendix, Fig. S2B). Since ASM has considerable decay in the 3-h memory time window, the absence of effect of WASp knockdown on 3-h memory indicates that the formation and decay of ASM are independent of WASp. For better visualization, we subtracted ARM from the intact memory (without cold-shock treatment) to generate the ASM component. Despite a consistent increase of intact memory and ARM by WASp knockdown at 6 and 9 h after a 1-session training, an effect on the ASM component was not observed (Fig. 2B and SI Appendix, Fig. S2D).
Fig. 2.
WASp regulates ARM forgetting in the MB neurons. (A) ARM retention curves. WASp knockdown flies (RU486+) showed slower ARM decay. n = 8 to 11. (B) WASp-RNAi-expressing flies had higher ARM performance at 6 and 9 h after training compared with parental controls. n = 8. (C) Interference-based ARM forgetting was suppressed by RU486-induced expression of WASp-RNAi. Lightning symbol represents electric shock reinforcement. n = 8. (D and E) Memory performance in flies with WASp knockdown (D) and overexpression (E) in the adult MB neurons. n = 8. (F) RNAi knockdown of WASp dominated the effect of constitutively active Cdc42 (Cdc42-CA) on 9-h ARM. Cdc42-CA-expressing flies (yellow bar, RU486+) showed lower 9 hARM. Flies with coexpression of both Cdc42-CA and WASp-RNAi (orange bar, RU486+) showed memory performance higher than controls, and the performance was not different from flies expressing WASp-RNAi alone. n = 8 to 12. (G) Data summary. Data are means ± SEM. *P < 0.05. n.s., nonsignificant.
We also tested the requirement of WASp in interference-based forgetting of ARM using a retroactive interference paradigm used in our previous study (4). Flies received 4-session massed training, and immediately following the initial training, the trained flies were exposed to a second 4-session massed training with a novel odor pair. Testing of the ARM retention of the initial learning was performed at 3.3 h after the initial training. The interference learning reduced the performance in control flies (RU486−, Fig. 2C and SI Appendix, Fig. S2E), while such forgetting was suppressed in WASp-RNAi-expressing flies (RU486+, Fig. 2C and SI Appendix, Fig. S2E). Together, like Cdc42 (4), WASp is required for time-based and interference-based forgetting of ARM, and on the other hand, WASp is dispensable in ARM formation and ASM decay.
There are 2 additional lines of evidence supporting the idea that WASp functions downstream of Cdc42 in ARM forgetting. First, like Cdc42 (4), WASp-dependent forgetting also takes place in the MB neurons. Knockdown (Fig. 2D) and overexpression (Fig. 2E) of WASp in the adult MB neurons led to slower and accelerated ARM decay after a 1-session training. Second, we combined Cdc42 activation and WASp knockdown to test genetic interaction (Fig. 2F). Consistent with our previous finding (4), flies expressing constitutively active Cdc42 (MB-GS/UAS-Cdc42-CA, RU486+) had reduced ARM performance at 9 h after a 1-session training when compared with control flies (MB-GS/+, RU486+). The reduction was reversed by coexpression of WASp-RNAi (MB-GS, UAS-Cdc42-CA/UAS-WASp-RNAi-1, RU486+). Flies expressing both Cdc42-CA and WASp-RNAi-1 had a performance level similar to flies expressing WASp-RNAi alone, and both groups were higher than the MB-GS/+ control. The data suggest that WASp acts as a downstream effector of Cdc42 in ARM forgetting (Fig. 2G).
Arp2/3 Complex Is only Required in Forgetting of ARM but Not ASM.
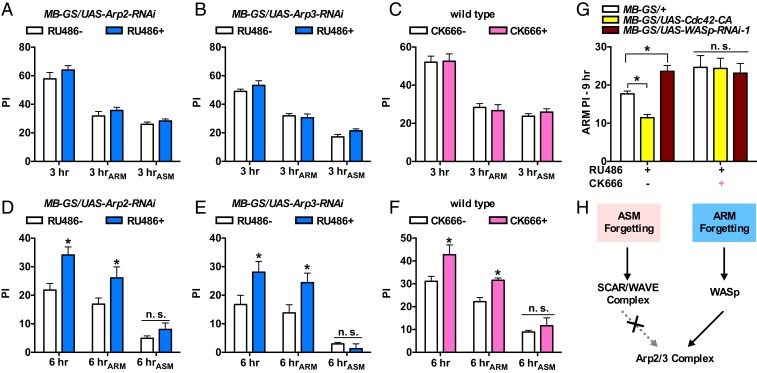
Arp2/3 complex is a known downstream effector that ties the Rac1/SCAR pathway and Cdc42/WASp pathway to actin polymerization (18, 20). And it has been reported to be important for forgetting in Caenorhabditis elegans (24). We next tested the role of Arp2/3 complex in forgetting by knocking down Arp2 and Arp3, 2 major members of this complex (25), and by feeding flies with 20 μM of CK666, a specific inhibitor of Arp2/3 complex that stabilizes the inactive state of the complex (26). The inhibition of Arp2/3 complex by both genetic and pharmacological methods led to higher 6-h memory (Fig. 3 D–F), but the 3-h memory was not affected (Fig. 3 A–C). The higher memory retention at 6 h is specific to ARM, while the ASM component is spared (Fig. 3 D–F). We additionally tested the dosage-dependent effect of CK666 feeding. Increased memory retention at 12 h was observed when flies were fed with CK666 higher than 5 μM (SI Appendix, Fig. S3B), while no effects were observed for memory retention at 3 h for all of the concentrations tested (up to 20 μM, SI Appendix, Fig. S3A). The specific effect on ARM forgetting indicates that Arp2/3 complex functions downstream of the Cdc42/WASp pathway. To test this, we combined the expression of constitutively active Cdc42-CA and WASp-RNAi with the pharmacological inhibition of Arp2/3 complex using CK666. ARM forgetting was accelerated by Cdc42-CA expression and slowed down by WASp-RNAi expression (Fig. 3G, CK666−). However, in the presence of CK666 feeding, there are no differences among control flies and flies expressing Cdc42-CA and WASp-RNAi, suggesting that the effect of Arp2/3 complex inhibition dominates those induced by Cdc42-CA and WASp-RNAi expression. These data support the idea that Arp2/3 complex is specifically required in Cdc42/WASp-mediated ARM forgetting but not in Rac1/SCAR-mediated ASM forgetting (Fig. 3H).
Fig. 3.
Arp2/3 complex functions in ARM forgetting but not ASM forgetting. (A and B) Knockdown of Arp2 (A) or Arp3 (B) in the adult MB neurons did not affect memory retention at 3 h. n = 8. (C) Pharmacological inhibition of the Arp2/3 complex (CK666+) did not affect 3-h retention of either ARM or ASM. n = 8. (D and E) Knockdown of Arp2 or Arp3 in the adult MB neurons resulted in higher ARM retention, but not ASM, at 6 h. n = 8. (F) The group with CK666 feeding showed higher 6-h retention of ARM but not ASM. n = 8. (G) Pharmacological inhibition of the Arp2/3 complex dominated Cdc42-CA- and WASp-RNAi-dependent effect in ARM forgetting. Without CK666 feeding (CK666−) and when compared with MB-GS/+ control group, Cdc42-CA-expressing flies showed lower 9 hARM, while WASp-RNAi-expressing files showed higher 9 hARM. Such effects were masked by CK666 feeding (CK666+). n = 8 to 10. (H) Data summary. Data are means ± SEM. *P < 0.05. n.s., nonsignificant.
Formin Dia Functions with Rac1/SCAR in ASM Forgetting.
To gain a better understanding of Rac1/SCAR-mediated ASM forgetting, we examined a number of interacting proteins of SCAR/WAVE complex (27–34) in a small-scale RNAi screen by knocking down these proteins in the MB neurons (SI Appendix, Fig. S4A). We found higher 3-h memory performance in flies expressing the RNAi of Diaphanous (dia), which encodes a formin family protein that induces linear actin polymerization (35).
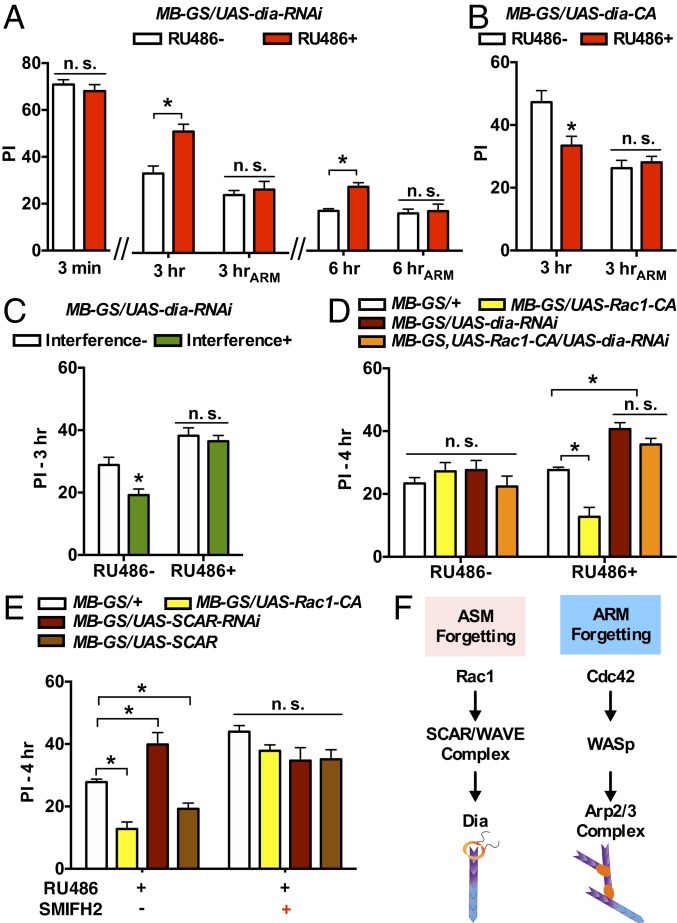
We further tested Dia’s role in ASM forgetting. Compared with the RU486− control, dia-RNAi-expressing flies (RU486+) showed normal memory performance at 3 min after a 1-session training, but memory decay at 3 and 6 h was slower (Fig. 4A). Such Dia-dependent slower memory decay was abolished by cold anesthesia (Fig. 4A), suggesting that Dia is required only for ASM forgetting. We also fed flies with 2.5 μM SMIFH2, a small molecule inhibitor of formin-dependent but not Arp2/3 complex-dependent actin polymerization (36). SMIFH2 also led to higher memory at 6 h after training and the phenotype is sensitive to cold anesthesia (SI Appendix, Fig. S4B), which is consistent with RNAi knockdown of Dia. Conversely, acute expression of a constitutively active form of Dia (UAS-dia-CA), which lacks the N-terminal regulatory sequence and the C-terminal autoinhibitory domain (37), reduced 3-h memory. The effect of Dia-CA was again blocked by cold anesthesia (Fig. 4B). Like SCAR, Dia is also required for interference-based forgetting (Fig. 4C). Thus, Dia bidirectionally regulates ASM forgetting in the MB neurons.
Fig. 4.
Identification of Dia as a downstream effector of Rac1/SCAR in ASM forgetting. (A) Knockdown of Dia in the adult MB neurons using RNAi (RU486+) did not affect immediate memory (3 min), but slowed down memory decay at 3 and 6 h. n = 8. (B) Expression of Dia-CA in the adult MB neurons (RU486+) reduced 3-h memory without affecting ARM. n = 8. (C) Interference-based forgetting of 3-h memory was suppressed in dia-RNAi-expressing flies (RU486+). n = 8. (D) Knockdown of Dia dominated Rac1-induced forgetting in 4-h memory. Flies with coexpression of both Rac1-CA and dia-RNAi (orange bar, RU486+) showed memory performance higher than controls, and the performance was not different from flies expressing dia-RNAi alone. n = 8 to 12. (E) Pharmacological inhibition of Dia dominated Rac1- and SCAR-induced forgetting. Pharmacological inhibition of Dia had no additive effect compared with knockdown of SCAR and brought decreased memory performance in Rac1-CA-expressing and SCAR-overexpression flies to a similar level. n = 8 to 12. (F) Working model. Data are means ± SEM. *P < 0.05. n.s., nonsignificant.
We next sought to determine the relationship between Dia- and Rac1/SCAR-mediated ASM forgetting using both genetic and pharmacological manipulations. The accelerated memory forgetting observed in Rac1-CA-expressing flies (yellow bar, RU486+) was dominated by the Dia effect (red bar, RU486+) in flies expressing both Rac1-CA and dia-RNAi (orange bar, RU486+) (Fig. 4D). Consistently, pharmacological inhibition of Dia also slowed down memory decay and masked the accelerated forgetting induced by Rac1-CA and SCAR overexpression (Fig. 4E). These data indicate that Dia functions downstream of Rac1/SCAR-mediated ASM forgetting in the MB neurons (Fig. 4F).
Rac1/SCAR/Dia-Dependent Forgetting Functions in the MB γ-Neurons.
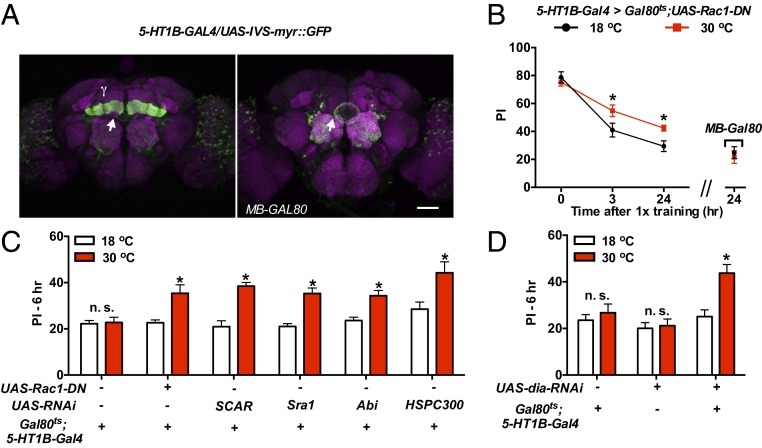
The MB neurons are divided into 3 major types: the γ-, α′/β′-, and α/β-neurons (SI Appendix, Fig. S5A), which have distinct roles in different phases and processes of olfactory memory (22). The MB-GS is a broad MB driver, which covers both the γ- and α/β-neurons (38) and is not suitable for differentiating different MB types. We hereby turned to the TARGET system (39) and used a temperature shift from 18 °C to 30 °C to inactivate the Gal80ts, a temperature-sensitive Gal4 inhibitor, and switched on the expression of a dominant-negative mutant (Rac1-DN). Limited by Gal4 lines, the initial Rac1 study narrowed down Rac1 forgetting the α/β- and γ-neurons (3). With the help of additional MB Gal4 lines (SI Appendix, Fig. S5B), we recently found that, besides the 2 pan-MB Gal4 drivers, OK107 and R13F02 (40), a γ-neuron driver, 5-HT1B-GAL4 (41), also led to slower memory decay that lasted up to 24 h (Fig. 5B and SI Appendix, Fig. S5C). In the MB, 5-HT1B-GAL4 drives expression exclusively in the γ-neurons; whereas weak expression can still be found elsewhere in the ellipsoid body and some scattered neurons in the antennal lobe and the optical lobe (Fig. 5A). However, the integration of a MB-Gal80 transgene (42) suppressed the expression specifically in the MB (Fig. 5A) and also blocked the memory increment at 24 h (Fig. 5B). We note that 5-HT1B-GAL4 has a higher expression level in the γ-neurons, which may explain the discrepancy as to why similar phenotypes were not observed in the initial study with 3 other γ-neuron drivers, 1471, NP1131, and 201Y. Consistent with the previous data, we did not detect a phenotype even when a previously used weak γ-neuron driver, 201Y, was combined with a α/β-neuron driver, C739 (SI Appendix, Fig. S5C). Besides memory decay, the inhibition of Rac1 in the γ-neurons with the strong driver 5-HT1B-GAL4 also inhibited forgetting in reversal learning and trace conditioning (SI Appendix, Fig. S5D), 2 paradigms that have previously been used to test Rac1’s roles in labile memory forgetting (3, 11).
Fig. 5.
Function of Rac1/SCAR/Dia forgetting pathway in the MB γ-neurons. (A) Expression patterns of 5-HT1B-GAL4. Besides the strong expression in the MB γ-neurons, the Gal4 also labels neurons outside the MB, including the ellipsoid body in the central complex (arrow). MB-Gal80 suppressed the expression of 5-HT1B-GAL4 in the MB neurons specifically. Green, mCD8-GFP reporter; magenta, nc82. (Scale bar, 50 μm.) (B) Memory retention curves. Conditional expression of Rac1-DN in the adult MB γ-neurons with the 5-HT1B-Gal4 driver was sufficient to slow down memory decay, and the effect was blocked by MB-Gal80 (24 h). n = 6 to 10. (C) Consistent with Rac1-DN, RNAi knockdown of SCAR/WAVE complex members in the MB γ-neurons led to higher memory retention at 6 h. n = 8 to 10. (D) Knockdown of Dia in the adult MB γ-neurons resulted in higher memory retention at 6 h. n = 6. Data are means ± SEM. *P < 0.05. n.s., nonsignificant.
RNAi knockdown of different members in the SCAR/WAVE complex in the γ-neurons with the 5-HT1B-Gal4 driver also suppressed forgetting (Fig. 5C). In addition, Dia knockdown in the γ-neurons (Gal80ts/+; 5-HT1B-Gal4/UAS-dia-RNAi, 30 °C) inhibited forgetting, compared with parental controls (Fig. 5D). These data further support the idea that ASM forgetting localizes in γ-neurons. We also explored WASp knockdown with the γ-neuron driver, 5-HT1B-Gal4, and the α/β-neuron driver, C739. However, we did not observe a phenotype in ARM at 6 h after a 1-session training (SI Appendix, Fig. S6). The MB neuron types required for ARM forgetting are yet to be identified.
Discussion
There are 3 major findings. First, 2 WASP family proteins, SCAR/WAVE and WASp, act as downstream effectors of Rac1-mediated ASM forgetting and Cdc42-mediated ARM forgetting, respectively. Second, although the Arp2/3 complex is a well-established effector that links activation of WASP family proteins to actin polymerization, it is only required in Cdc42/WASp-mediated ARM forgetting. Instead, formin Dia functions together with Rac1/SCAR in ASM forgetting. Third, feeding inhibitors of the Arp2/3 complex and Dia to fruit flies led to rather specific effects on ASM and ARM forgetting, raising the possibility of developing drugs on these molecular targets to treat memory-related diseases.
The effect of Rac1 on ASM forgetting has been tied to the activation of an actin depolymerization regulator cofilin presumably through a PAK/LIMK phosphorylation cascade (3, 17). However, actin dynamics is a balanced play that requires continuous turnover between polymerization and depolymerization (43). It is not known whether signaling pathways regulating actin polymerization also play a role. There are 3 families of proteins that nucleate and promote actin polymerization, Arp2/3 complex, WH2-domain proteins, and formin (18, 20). Our finding that Arp2/3 complex and formin Dia function in ARM and ASM forgetting suggests that both actin polymerization and depolymerization pathways contribute to forgetting. How Arp2/3 complex and Dia separately contribute to ARM and ASM forgetting remains an open question. It is yet to be determined whether these proteins have different expression or subcellular locations in the MB neurons. However, it is interesting that Arp2/3 complex and formins are specialized in different types of actin polymerization (18, 20, 44).
In our working model, Cdc42 activates Arp2/3 complex via a canonical pathway (Cdc42/WASp/Arp2/3 complex), while Rac1-mediated ASM forgetting depends on SCAR/WAVE complex. This complex, in addition to SCAR/WAVE, includes at least 4 other members: Sra-1, Abi, HSPC300, and Kette (20). These additional members are thought to hold SCAR/WAVE in the complex in an inactive state, until GTP-bound Rac1 binds to Sra-1 and relieves the inhibition (20). On the other hand, the intact complex is essential for the stability of the SCAR/WAVE protein as well (i.e., failure to keep the intact complex can lead to SCAR degradation) (45). This latter effect may explain our observation that RNAi knockdown of SCAR complex members has the same effect on inhibiting forgetting as the knockdown of SCAR. As a WASP family protein, SCAR/WAVE is able to associate with and activate Arp2/3 complex through its C-terminal region (46). However, RNAi knockdown of Arp2 and Arp3 and pharmacological inhibition of Arp2/3 complex specifically affects ARM forgetting, while no effects on ASM retention were observed. We therefore propose that Rac1/SCAR may function through Arp2/3 complex-independent mechanisms (47). SCAR/WAVE complex is reported to physically associates with Dia through one of its members, Abi, to regulate actin dynamics (48, 49). Our behavioral characterization of Dia knockdown and overexpression, as well as the genetic epistasis experiment, support the idea that Dia could be downstream of Rac1/SCAR in ASM forgetting. Details about the functional coordination between SCAR/WAVE and Dia therefore await further clarification.
Materials and Methods
Fly Strains.
Flies were reared at 25 °C and 60% relative humidity on a cornmeal medium under a 12/12 h light/dark cycle, except that flies in experiments using the TARGET system were reared at 18 °C. For details, see SI Appendix, SI Materials and Methods.
Behavioral Assays and Related Treatments.
Aversive olfactory conditioning was performed as previously described (3, 4). For details, see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jennifer A. Zallen, Dr. Ronald L. Davis, Dr. Ulrike Heberlein, Dr. Gerry Rubin, Dr. André Fiala, the Bloomington Stock Center, and the Tsinghua Fly Center for fly stocks; and the Developmental Studies Hybridoma Bank for reagents. We thank Dr. Guangshuo Ou, Dr. Xin Liang, Dr. Wantong Hu, and Dr. Yunchuan Zhang for helpful discussions. We are grateful to Bohan Zhao and Bo Lei for helpful comments on the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31700912, to Q.L.; 91632301, to Y.Z.), the Beijing Municipal Science and Technology Commission (Z161100002616010, to Y.Z.), and the Tsinghua-Peking Joint Center for Life Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.J.G. is a guest editor invited by the Editorial Board.
See Commentary on page 20807.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903763116/-/DCSupplemental.
References
- 1.Davis R. L., Zhong Y., The biology of forgetting-A perspective. Neuron 95, 490–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards B. A., Frankland P. W., The persistence and transience of memory. Neuron 94, 1071–1084 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Shuai Y., et al. , Forgetting is regulated through Rac activity in Drosophila. Cell 140, 579–589 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Zhang X., Li Q., Wang L., Liu Z. J., Zhong Y., Cdc42-Dependent forgetting regulates repetition effect in prolonging memory retention. Cell Rep. 16, 817–825 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Berry J. A., Cervantes-Sandoval I., Nicholas E. P., Davis R. L., Dopamine is required for learning and forgetting in Drosophila. Neuron 74, 530–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue A., et al. , Forgetting in C. elegans is accelerated by neuronal communication via the TIR-1/JNK-1 pathway. Cell Rep. 3, 808–819 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., et al. , Hippocampal activation of Rac1 regulates the forgetting of object recognition memory. Curr. Biol. 26, 2351–2357 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Quinn W. G., Dudai Y., Memory phases in Drosophila. Nature 262, 576–577 (1976). [DOI] [PubMed] [Google Scholar]
- 9.Tully T., Preat T., Boynton S. C., Del Vecchio M., Genetic dissection of consolidated memory in Drosophila. Cell 79, 35–47 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Luo L., Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173–180 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Shuai Y., Hu Y., Qin H., Campbell R. A., Zhong Y., Distinct molecular underpinnings of Drosophila olfactory trace conditioning. Proc. Natl. Acad. Sci. U.S.A. 108, 20201–20206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margulies C., Tully T., Dubnau J., Deconstructing memory in Drosophila. Curr. Biol. 15, R700–R713 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng J., Luo L., Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron 44, 779–793 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Lv L., Wang L., Zhong Y., Social isolation induces rac1-dependent forgetting of social memory. Cell Rep. 25, 288–295.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Dietz D. M., et al. , Rac1 is essential in cocaine-induced structural plasticity of nucleus accumbens neurons. Nat. Neurosci. 15, 891–896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang L., et al. , Inhibition of Rac1 activity in the hippocampus impairs the forgetting of contextual fear memory. Mol. Neurobiol. 53, 1247–1253 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Cervantes-Sandoval I., Chakraborty M., MacMullen C., Davis R. L., Scribble scaffolds a signalosome for active forgetting. Neuron 90, 1230–1242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo L., Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18, 601–635 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Ridley A. J., Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Takenawa T., Suetsugu S., The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37–48 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Osterwalder T., Yoon K. S., White B. H., Keshishian H., A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U.S.A. 98, 12596–12601 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keene A. C., Waddell S., Drosophila olfactory memory: Single genes to complex neural circuits. Nat. Rev. Neurosci. 8, 341–354 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Mao Z., Roman G., Zong L., Davis R. L., Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc. Natl. Acad. Sci. U.S.A. 101, 198–203 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadziselimovic N., et al. , Forgetting is regulated via Musashi-mediated translational control of the Arp2/3 complex. Cell 156, 1153–1166 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Rouiller I., et al. , The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hetrick B., Han M. S., Helgeson L. A., Nolen B. J., Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 20, 701–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricke R., et al. , Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr. Biol. 19, 1429–1437 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Chen X. J., et al. , Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev. Cell 30, 569–584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan R., et al. , The Abl pathway bifurcates to balance enabled and Rac signaling in axon patterning in Drosophila. Development 144, 487–498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin T. Y., et al. , Abi plays an opposing role to Abl in Drosophila axonogenesis and synaptogenesis. Development 136, 3099–3107 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Squarr A. J., et al. , Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J. Cell Biol. 212, 591–603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Z., et al. , Functional coordination of WAVE and WASP in C. elegans neuroblast migration. Dev. Cell 39, 224–238 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Koronakis V., et al. , WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. U.S.A. 108, 14449–14454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaipa B. R., et al. , Dock mediates Scar- and WASp-dependent actin polymerization through interaction with cell adhesion molecules in founder cells and fusion-competent myoblasts. J. Cell Sci. 126, 360–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castrillon D. H., Wasserman S. A., Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 120, 3367–3377 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Rizvi S. A., et al. , Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16, 1158–1168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somogyi K., Rørth P., Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Dev. Cell 7, 85–93 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Zhang X., Li Q., Wang L., Liu Z. J., Zhong Y., Active protection: Learning-activated Raf/MAPK activity protects labile memory from Rac1-independent forgetting. Neuron 98, 142–155.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 39.McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L., Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Jenett A., et al. , A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan Q., Lin F., Zheng X., Sehgal A., Serotonin modulates circadian entrainment in Drosophila. Neuron 47, 115–127 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Krashes M. J., Keene A. C., Leung B., Armstrong J. D., Waddell S., Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron 53, 103–115 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oda T., Iwasa M., Aihara T., Maéda Y., Narita A., The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Wallar B. J., Alberts A. S., The formins: Active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13, 435–446 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Kunda P., Craig G., Dominguez V., Baum B., Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13, 1867–1875 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Kurisu S., Takenawa T., The WASP and WAVE family proteins. Genome Biol. 10, 226 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sasaki N., Miki H., Takenawa T., Arp2/3 complex-independent actin regulatory function of WAVE. Biochem. Biophys. Res. Commun. 272, 386–390 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Ryu J. R., Echarri A., Li R., Pendergast A. M., Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol. Cell. Biol. 29, 1735–1748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng S., Bothe I., Baylies M., Diaphanous regulates SCAR complex localization during Drosophila myoblast fusion. Fly (Austin) 10, 178–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.