Significance
Toxic RNA molecules containing hundreds to thousands of CUG repeats cause myotonic dystrophy type 1 (DM1). Selectively blocking expression of this toxic CUG RNA has the potential to alleviate all downstream pathogenic consequences to the cell. To identify new hits and cellular targets that selectively regulate toxic CUG RNA levels, we developed a cell-based screening assay that permits the ratio-metric measurement of toxic CUG RNA abundance relative to an analogous control RNA. A pilot screen identified multiple microtubule inhibitors as hits that selectively regulate toxic CUG RNA levels in multiple DM1 models. These findings open avenues of research and therapy in DM1 and provide proof of concept for repeat-selective screening to identify unanticipated hits in other microsatellite expansion diseases.
Keywords: alternative splicing, myotonic dystrophy, muscleblind, transcription
Abstract
A CTG repeat expansion in the DMPK gene is the causative mutation of myotonic dystrophy type 1 (DM1). Transcription of the expanded CTG repeat produces toxic gain-of-function CUG RNA, leading to disease symptoms. A screening platform that targets production or stability of the toxic CUG RNA in a selective manner has the potential to provide new biological and therapeutic insights. A DM1 HeLa cell model was generated that stably expresses a toxic r(CUG)480 and an analogous r(CUG)0 control from DMPK and was used to measure the ratio-metric level of r(CUG)480 versus r(CUG)0. This DM1 HeLa model recapitulates pathogenic hallmarks of DM1, including CUG ribonuclear foci and missplicing of pre-mRNA targets of the muscleblind (MBNL) alternative splicing factors. Repeat-selective screening using this cell line led to the unexpected identification of multiple microtubule inhibitors as hits that selectively reduce r(CUG)480 levels and partially rescue MBNL-dependent missplicing. These results were validated by using the Food and Drug Administration-approved clinical microtubule inhibitor colchicine in DM1 mouse and primary patient cell models. The mechanism of action was found to involve selective reduced transcription of the CTG expansion that we hypothesize to involve the LINC (linker of nucleoskeleton and cytoskeleton) complex. The unanticipated identification of microtubule inhibitors as selective modulators of toxic CUG RNA opens research directions for this form of muscular dystrophy and may shed light on the biology of CTG repeat expansion and inform therapeutic avenues. This approach has the potential to identify modulators of expanded repeat-containing gene expression for over 30 microsatellite expansion disorders.
Myotonic dystrophy type 1 (DM1) is the most common form of adult-onset muscular dystrophy and is caused by the expansion of a CTG repeat tract within the 3′ untranslated region (3′ UTR) of the dystrophia myotonica protein kinase gene (DMPK) (1–3). Whereas unaffected individuals generally have fewer than 35 CTG repeats, DM1 patients have up to several thousand repeats, particularly in primarily postmitotic tissue such as skeletal muscle (4). Transcription of the mutant DMPK allele containing the CTG repeat expansion results in the synthesis of toxic CUG expansion RNA, r(CUG)EXP, that can fold into distinct hairpin structures and are bound by the muscleblind (MBNL) family of alternative splicing factors (5, 6). Nuclear sequestration of the MBNL family of proteins into ribonuclear foci by r(CUG)EXP results in the global missplicing of MBNL target pre-mRNAs directly causing symptoms of DM1. For example, missplicing of insulin receptor and chloride channel pre-mRNA leads to insulin resistance and myotonia, respectively (6–9). Studies have identified panels of commonly misspliced targets that correlate with disease severity in DM1 (10, 11). Reversal of missplicing of these targets is a strong predictor of therapeutic potential. For example, targeting the chloride channel exon 7a splice site by using antisense oligonucleotides to repress inclusion of this alternative exon restored muscle chloride channel protein levels and eliminated myotonia in DM1 cells and mouse models (12). Hence, missplicing observed in DM1 is an important biomarker for disease severity as well as for evaluating the efficacy of therapeutic interventions.
Currently, there are no Food and Drug Administration (FDA)-approved disease-targeting treatments for myotonic dystrophy. Many preclinical therapeutic studies focus on targeting nuclear r(CUG)EXP:MBNL aggregates using small molecules, antisense oligonucleotides, RNA interference, and proteins such as RNA-targeting Cas9 to free MBNL proteins and rescue DM1-associated missplicing (13–23). Several studies have targeted transcription of the expanded CTG repeat tract using small molecules and deactivated Cas9, providing proof of principle for blocking r(CUG)EXP production as a viable therapeutic strategy in DM1 (24–26). Using the CRISPR/Cas9 system, several studies have also demonstrated the potential for contraction or complete removal of the expanded CTG repeat tract (27–30).
In addition to hypothesis-based and targeted approaches to identify compounds and cellular targets to modulate toxic r(CUG)EXP levels, unbiased screens expedite the discovery of new compounds and can identify unanticipated cellular targets. Small-molecule and genetic screens have been employed in the search for therapeutics as well as to inform interesting new biology associated with the CTG repeat expansion (13, 31–36). However, a CTG repeat-selective screen targeting r(CUG)EXP production and/or stability in cells has not been reported. Selectivity for the expanded CTG repeat is important for the development of effective therapy, but can also reveal new biology of CTG and other repeat expansions. To address this gap in the field, we generated a DM1 HeLa cell model permitting the sensitive ratio-metric evaluation of r(CUG)480 levels relative to an r(CUG)0 control, enabling the identification of small molecules or cellular targets that selectively modulate r(CUG)EXP levels. We applied this cell line to a pilot screen of the LOPAC1280 (Sigma-Aldrich) chemical library targeting diverse cellular pathways, leading to the identification of multiple microtubule inhibitors as regulators of toxic r(CUG)EXP levels. We validated microtubule inhibition using the FDA-approved natural compound colchicine in the HSALR DM1 mouse model and in DM1 patient-derived myotubes showing a selective reduction of toxic r(CUG)EXP levels and partially correcting several DM1-associated missplicing events. This assay is a repeat-selective screen targeting r(CUG)EXP production/stability in cells and has yielded FDA-approved compounds, providing biological and therapeutic insight into DM1.
Results
Generation of a HeLa DM1 Cell Line for Repeat-Selective Screening.
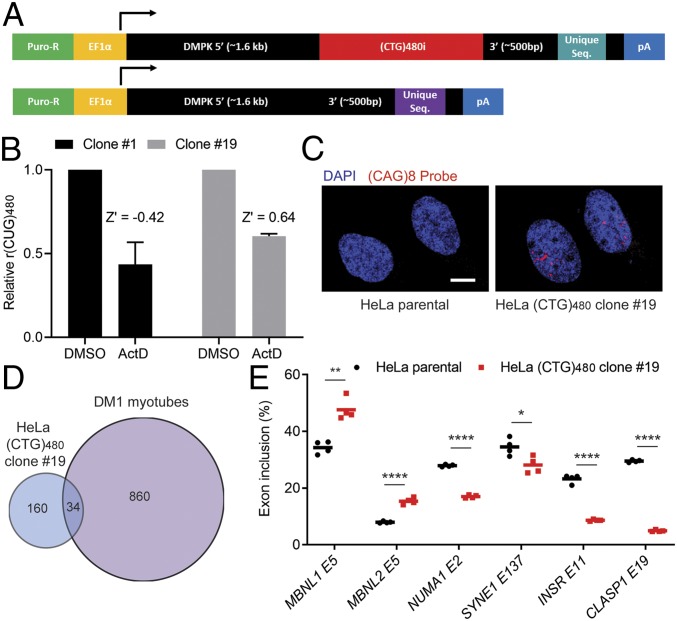
To screen for novel compounds and cellular targets that reduce expanded CUG RNA levels in a selective manner, we generated stable dual-construct HeLa cell lines. These cell lines contain an interrupted (CTG)480 repeat tract consisting of 24 modules of [(CTG)20(CTCGA)] and a (CTG)0 control tract; both contain exons 11–15 of human DMPK (37), each with a unique qPCR probe binding sequence 3′ of the repeat tract to distinguish the 2 transcripts produced (Fig. 1A). These constructs permit ratio-metric evaluation of repeat selectivity following treatments by measuring the abundance of r(CUG)480 relative to r(CUG)0. Multiplex qRT-PCR was performed where the same primer was used to reverse transcribe both RNA species, and the same primer pair was used in the qPCR reaction but with different fluorescent probes against the unique sequences to distinguish r(CUG)480 from r(CUG)0 in the same reaction (Fig. 1A and SI Appendix, Fig. S1). Both constructs were randomly integrated into the HeLa genome by using the PiggyBAC Transposon system. Several puromycin-resistant clones were expanded following single cell sorting, and the expression of r(CUG)480 and r(CUG)0 was measured in these lines (SI Appendix, Fig. S1). The HeLa (CTG)480 clonal lines robustly expressed both r(CUG)0 and r(CUG)480, while only marginal signal was detected in the HeLa parental cell line (SI Appendix, Figs. S1 and S2). All of the clones exhibited reduced cell viability to varying degrees relative to the HeLa parental line, which may be attributed to a combination of PiggyBAC cloning, single cell sorting, and transgene expression (SI Appendix, Fig. S3).
Fig. 1.
Establishing a DM1 HeLa cell line for repeat-selective screening. (A) Schematic of constructs used to generate a stable DM1 HeLa cell model for repeat-selective screening. Constructs are identical in sequence except for the presence of the (CTG)480i, an interrupted repeat tract consisting of 24 modules of [(CTG)20(CTCGA)] and unique sequence (21-bp qPCR probe target and 8-bp barcode). (B) ActD treatment (20 nM) overnight followed by multiplex RT-qPCR [r(CUG)480 relative to r(CUG)0] to determine CTG-selective response in HeLa (CTG)480 cell clones 1 and 19. Data are normalized to DMSO control treatments. Bars represent mean ± SD of 3 biological replicates. Z′ was calculated based on the differential response by C(t) values of (CTG)0 versus (CTG)480 to ActD following the method of Zhang et al. (41). (C) Fluorescence in situ hybridization for rCUGEXP foci using a Cy3-(CAG)8 probe and counterstained with DAPI for nuclei. (Scale bar, 10 µm.) (D) Overlap in alternative cassette exon missplicing events identified by using RNA-seq between HeLa clone 19 and DM1 myoblasts differentiated into myotubes (P < 0.0005, FDR < 0.1, |ΔPSI| > 0.15). (E) RT-PCR isoform analysis of the indicated alternative cassette exon events comparing HeLa (CTG)480 clone 19 to the parental HeLa cell line (mean, n = 4 biological replicates). *P < 0.05; **P < 0.01; ****P < 0.0001.
To test the potential use of the developed HeLa (CTG)480 clones in CTG repeat-selective screens, each clone was treated with actinomycin D (ActD), a natural compound that was previously shown to selectively bind to and reduce transcription from expanded CTG repeats in the low nanomolar range (25, 38–40) (SI Appendix, Fig. S4). Treatment with ActD resulted in a selective reduction of r(CUG)480 levels as expected in each of the clonal lines (SI Appendix, Fig. S4). Further testing of clones 1 and 19, which expressed the highest levels of r(CUG)480, and comparable expression of r(CUG)0 confirmed a highly reproducible response to ActD in clone 19 [Fig. 1B; Z′ = 0.64 from the method of Zhang et al. (41)], whereas clone 1 displayed greater variability, yielding a poor Z′ value (Fig. 1B). Based on the combination of robust expression of r(CUG)0 and r(CUG)480 at comparable levels, a highly reproducible response to ActD and favorable cell viability, clone 19 was a strong candidate for use in screening. Further characterization of clone 19 confirmed the presence of important cellular and molecular hallmarks of DM1, including ribonuclear foci and MBNL-dependent missplicing (Fig. 1 C–E). We also evaluated clones 1 and 11 and confirmed the presence of ribonuclear foci and MBNL-dependent missplicing in these additional HeLa DM1 model cell lines (SI Appendix, Figs. S5 and S6). RNA-sequencing (RNA-seq) analysis comparing HeLa (CTG)480 clone 19 to its parental line identified many alternative splicing changes of cassette exons (Fig. 1D and Dataset S1). We then compared these alternative splicing changes to those identified from RNA-seq in a DM1-patient-derived myoblast line that was differentiated into myotubes versus control myotubes, revealing overlapping alternative splicing changes in the 2 DM1 model cell lines (Fig. 1D and Dataset S2). Several known MBNL-dependent cassette exon alternative splicing events pertinent to DM1 (10, 11, 42, 43) in HeLa (CTG)480 clone 19 were validated by using RT-PCR (Fig. 1E). Taken together, these data show that the HeLa DM1 cell model recapitulates important molecular hallmarks of DM1 and, more importantly, permits sensitive measurement of CTG repeat selectivity.
Small-Molecule Screen for Selective Reduction of Expanded CUG RNA Identifies Multiple Microtubule Inhibitors.
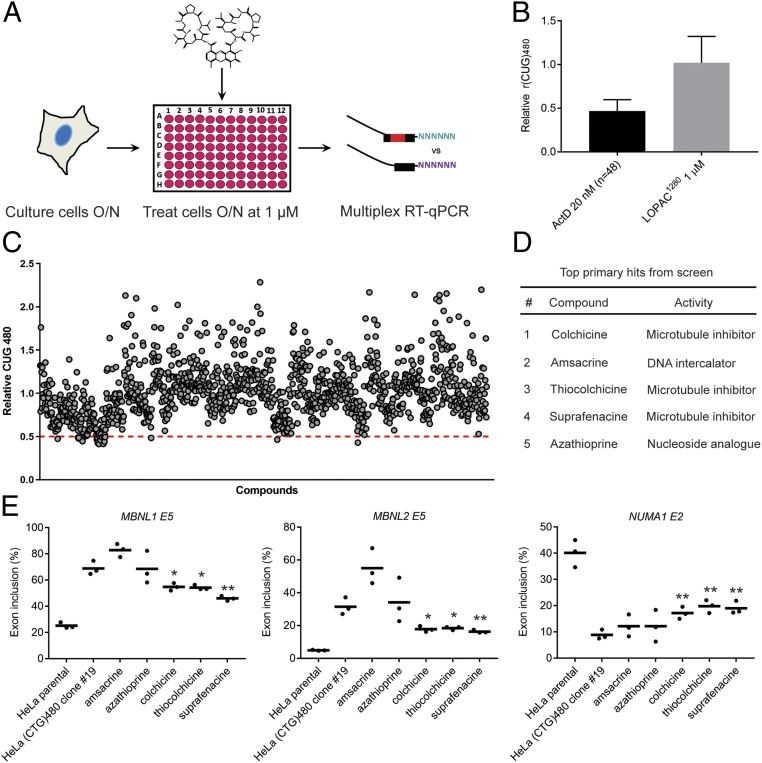
We carried out a pilot screen to identify compounds that selectively reduce r(CUG)480 levels in HeLa (CTG)480 clone 19 using the LOPAC1280 library (Sigma-Aldrich) that targets diverse cellular processes (Fig. 2A). Three dimethyl sulfoxide (DMSO) treatments and 3 ActD treatments (at 20 nM, where optimal CTG repeat selectivity was observed) were included on each plate of the LOPAC1280 (16 plates) as controls. The mean of the relative r(CUG)480 levels from the entire LOPAC1280 library treatments was ∼1.0, while treatment with ActD across the screen (n = 48) revealed that r(CUG)480 abundance was decreased ∼2-fold relative to r(CUG)0 following treatment (Fig. 2B). Based on ActD activity, a threshold of <0.5 r(CUG)480 relative to r(CUG)0 was set. Application of this cutoff revealed 20 primary hits that were rescreened and ranked relative to ActD (Fig. 2C and SI Appendix, Fig. S7). The top 5 ranked hits included 3 microtubule inhibitors (colchicine, thiocolchicine, and suprafenacine), a DNA intercalator (amsacrine), and a purine nucleoside analog (azathioprine) (Fig. 2D). These candidates were taken forward for further characterization.
Fig. 2.
Chemical screen identifies multiple microtubule inhibitors that selectively reduce r(CUG)480 levels. (A) Schematic of the CTG repeat-selective chemical screen. Following drug treatment (structure of ActD is shown as an example), cells are permeabilized, and RNA is directly taken forward from 96-well plates into RT-qPCR steps without the need for an RNA-extraction step. (B) Summary of LOPAC1280 screen on relative r(CUG)480 levels [normalized to r(CUG)0]. Mean of ActD treatments (20 nM, n = 48, ±SD) and mean of entire LOPAC1280 treatments (1 µM, n = 1,280 ± SD). (C) LOPAC1280 screen results of each individual drug treatment (1 µM) on relative r(CUG)480 levels [normalized to r(CUG)0]. The dashed red line indicates the 0.5 cutoff to identify primary hits falling below threshold. (D) Ranked list of top 5 primary hits from the LOPAC1280 screen. (E) RT-PCR isoform analysis of the indicated alternative cassette exon events in HeLa (CTG)480 clone 19 following treatment with each of the indicated compounds (1 µM for 48 h) (mean, n = 3 biological replicates). *P < 0.05; **P < 0.01.
We evaluated rescue of MBNL-dependent missplicing in clone 19 following treatment with the top 5 hits at the screening dose of 1 µM (Fig. 2E). Treatment with all 3 microtubule inhibitors revealed a partial rescue of missplicing in 3 MBNL-dependent events, while amsacrine and azathioprine treatments did not lead to any significant reversal (Fig. 2E). Furthermore, rescreening all 3 microtubule inhibitors in additional HeLa DM1 (CTG)480 clones produced a comparable selective reduction in r(CUG)480 levels (SI Appendix, Fig. S8).
Microtubule inhibitors are categorized as either destabilizing or stabilizing agents, depending on their binding site of tubulin and subsequent mode of action in deregulating microtubule polymers composed of αβ-tubulin heterodimers (44). Microtubule polymers are dynamic, and their activity is coordinated through both active polymerization and depolymerization of αβ-tubulin heterodimers. For example, it was shown that the involvement of microtubule dynamics in coordinating chromosome positioning during DNA repair is sensitive to both microtubule-stabilizing and -destabilizing drugs (45). All 3 microtubule inhibitors identified from the primary screen (colchicine, thiocolchicine, and suprafenacine) destabilize microtubules via the colchicine binding site of β-tubulin (44, 46). To test if both classes of microtubule drugs had a similar effect, we treated HeLa (CTG)480 clone 19 with paclitaxel (Taxol) and epothilone D, microtubule-stabilizing agents that function through binding to the taxane site of β-tubulin (44, 47) (SI Appendix, Fig. S9). There was a dose-dependent reduction in relative r(CUG)480 levels upon treatment with paclitaxel and epothilone D, supporting the role of both microtubule-stabilizing and -destabilizing agents in selectively reducing r(CUG)480 levels (SI Appendix, Fig. S9).
By developing and implementing a screen for compounds that selectively reduce abundance of transcripts containing expanded CUG repeats, we identified microtubule inhibitors as compounds that selectively reduce r(CUG)480 to levels sufficient to partially reverse several DM1-relevant missplicing events. Because a role for microtubule inhibitors in the selective regulation of r(CUG)EXP levels had not been described, we further evaluated microtubule inhibition in multiple DM1 models. We primarily focused on colchicine because it is an inexpensive, FDA-approved, natural therapeutic that is generally well-tolerated and is currently used in the clinic to treat gout and familial Mediterranean fever (FMF) (44, 48).
Validation of Colchicine In Vivo Using the HSALR DM1 Transgenic Mouse Model.
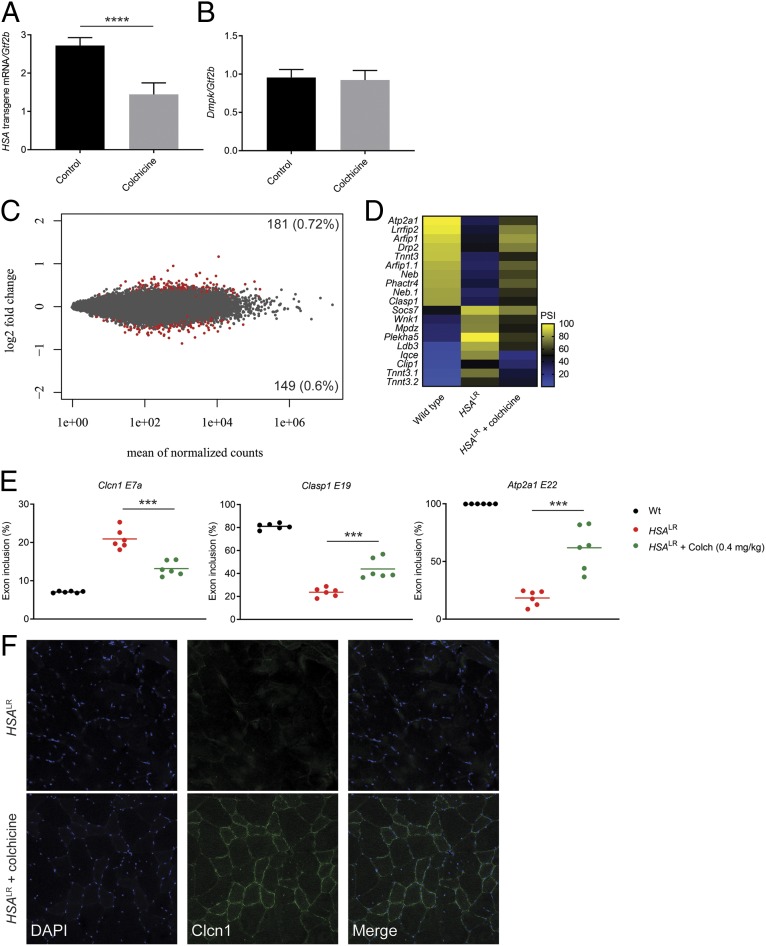
To determine whether microtubule perturbations have a similar effect on expanded CUG RNA levels in vivo, we tested the effects of colchicine treatment in a transgenic DM1 mouse model. The HSALR mouse model expresses ∼220 CUG repeats under the human skeletal actin promoter in the absence of DMPK sequence context (49). HSALR mice were treated with 0.4 mg/kg colchicine or phosphate-buffered saline (PBS) (control) daily for 14 d through i.p. injection, followed by RNA extraction from quadriceps muscle for analyses of gene expression and alternative splicing (Fig. 3). Treatment with colchicine resulted in a significant reduction in the relative abundance of HSA transgene mRNA containing the CUG expansion without affecting the levels of endogenous Dmpk transcript (Fig. 3 A and B). Importantly, RNA-seq analysis revealed very little global transcriptome change following colchicine treatment (Fig. 3C and Dataset S3). Approximately 1.3% of the transcriptome was altered, with only 1 transcript showing a greater than 2-fold change (Fig. 3C). In total, there were 181 genes up-regulated (0.72%) and 149 genes down-regulated (0.6%) (Fig. 3C). This number is an overestimate because the toxic CUG repeats cause transcriptional dysregulation (50). Comparing the transcriptional changes in the treated mice to mice without repeats reduced the total off-target events to 218 and showed that 113 transcripts reverted to expression levels comparable to mice without CUG repeats (SI Appendix, Fig. S10). Collectively, these data demonstrate that microtubule inhibition in vivo leads to a selective reduction in expanded CUG RNA levels without broadly affecting the transcriptome.
Fig. 3.
Colchicine treatment selectively reduces HSA transgene mRNA levels and partially rescues missplicing in the HSALR DM1 mouse model. (A and B) Effect of colchicine treatment (0.4 mg/kg) or PBS (control) for 14 d by i.p. injection on HSA transgene mRNA levels (A) or endogenous Dmpk levels (normalized to Gtf2b) (B) (n = 6 age- and gender-matched littermates in each treatment group, mean ± SD). ****P < 0.0001. (C) MA plot displaying gene-expression differences comparing colchicine-treated to PBS control-treated mice. Gene-expression change (log2 fold) is plotted against the mean of normalized counts. Red points represent changes with adjusted P < 0.1. (D) Heat map displaying the rescue of alternative cassette exon inclusion by mean normalized PSI identified from the top 100 missplicing events (top 50 cassette inclusion and top 50 exclusion events) in HSALR mice treated with colchicine compared to HSALR mice treated with PBS (control) and wild-type mice (P < 0.0005, FDR < 0.05). (E) RT-PCR isoform analysis of the indicated alternative cassette exon events following treatment with PBS-control or colchicine (mean, n = 6). ***P < 0.001. (F) Immunofluorescence against Clcn1 protein in colchicine-treated and PBS control-treated HSALR mouse quadricep muscle sections. DAPI staining for nuclei is shown in blue.
A selective reduction in HSA transgene mRNA containing the CUG expansion is expected to reverse MBNL-dependent missplicing pertinent to DM1. To evaluate potential rescue of missplicing from colchicine treatment, we analyzed RNA-seq data from wild-type and HSALR mice to identify a panel of the top 100 cassette exon missplicing events (top 50 cassette exon inclusion and top 50 exclusion events showing the greatest change in normalized exon percent spliced in [PSI] between wild-type and HSALR mice) (Dataset S4). Of these, we identified multiple missplicing events showing varying levels of rescue from 14 d of colchicine treatment (Fig. 3D). The rescue profile of these targets shows a partial reversal toward wild type when compared to control HSALR mice (Fig. 3D). Several of these include well-characterized targets in DM1 patients directly contributing to muscle phenotypes, such as Atp2a1, Tnnt3, and Clasp1 (10, 11) (Fig. 3D). We then validated rescue of MBNL-dependent missplicing using RT-PCR for Clasp1 and Atp2a1 events and the Clcn1 event known to cause myotonia through reduced muscle chloride channel protein levels when misspliced in DM (9, 12) (Fig. 3E). Staining for Clcn1 protein in muscle sections of the colchicine-treated HSALR mice confirmed increased Clcn1 protein in muscle membranes compared to control HSALR mice (Fig. 3F). Thus, colchicine treatment selectively reduces toxic expanded CUG RNA levels in vivo, leading to a partial correction of missplicing in a DM1 transgenic mouse model.
Validation of Colchicine in Primary DM1 Patient Cells.
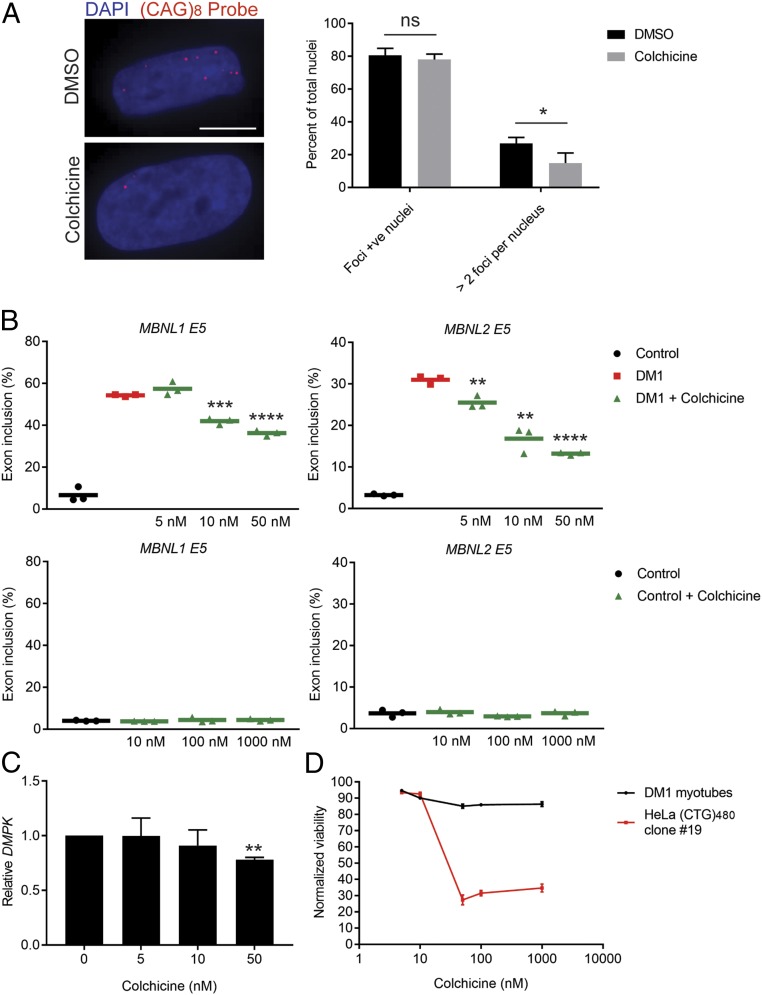
Having validated the effect of colchicine in 2 independent transgenic systems in vitro and in vivo, we next sought to test its effect in primary patient cells containing the repeat expansion within the endogenous genomic context. We used myoblasts derived from a DM1 patient containing an expansion of ∼1,900–3,000 CTG repeats in the 3′ UTR of DMPK (51). These primary myoblasts were differentiated into postmitotic myotubes, then treated with colchicine, and the effects on r(CUG)EXP foci and rescue of 2 established MBNL-dependent missplicing events (MBNL1 and MBNL2 exon 5) were measured (Fig. 4 A and B). There was a partial but significant reduction in both foci and missplicing following treatment (Fig. 4 A and B). The percentage of nuclei with more than 2 foci was significantly reduced following colchicine treatment (Fig. 4A), whereas the percentage of nuclei containing any foci was not significantly reduced. Measuring MBNL1 and MBNL2 exon 5 missplicing events revealed a partial but significant dose-dependent rescue (Fig. 4B). Increasing the colchicine treatment dose beyond 50 nM only marginally improved the rescue in missplicing, suggesting saturation of either the colchicine–tubulin interaction or of the downstream effector (SI Appendix, Fig. S11). Treatment of control myotubes that were cultured and differentiated in parallel did not affect splicing of either MBNL1 or MBNL2 exon 5, indicating an effect mediated through the CUG repeat expansion rather than directly acting on MBNL levels or its target pre-mRNA (Fig. 4B). Interestingly, treating fibroblasts derived from the same patient as the myoblasts (51) did not reverse missplicing of the INSR E11 event observed in this cell line (SI Appendix, Fig. S12). We observed a significant reduction in DMPK levels with 50 nM colchicine treatment in the patient-derived myotubes (Fig. 4C); however, in this assay, we were not able to differentiate between the mutant (CTG expansion) and nonmutant alleles. Taken together, we have demonstrated an effect of colchicine treatment on reducing r(CUG)EXP toxicity in primary DM1 patient cells.
Fig. 4.
Validation of colchicine treatment in DM1 patient cells. (A) Fluorescence in situ hybridization for r(CUG)EXP foci using a (CAG)8-Cy3 probe and counterstained with DAPI for nuclei. (Scale bar, 10 µm.) (A, Left) Representative images of DM1 myoblasts that were differentiated into myotubes and then treated with DMSO (control) or colchicine (50 nM) for 72 h. (A, Right) Quantification of the percentage of foci-positive nuclei and the percentage of nuclei containing more than 2 foci (mean ± SD n = 3 biological replicates, 231 DMSO-treated nuclei, 182 colchicine-treated nuclei). ns, not significant. *P < 0.05. (B) RT-PCR isoform analysis of the indicated alternative cassette exon events following treatment of DM1 myotubes (Upper) or control myotubes (Lower) with DMSO or colchicine at the indicated dose (mean, n = 3 biological replicates). **P < 0.01; ***P < 0.001; ****P < 0.0001. (C) Effect of colchicine treatment on relative DMPK levels (normalized to GAPDH) presented as a fold change from DMSO treatments (0 lane) (mean ± SD n = 3 biological replicates). **P < 0.01. (D) Cell-viability assay conducted on DM1 myotubes and HeLa (CTG)480 clone 19 treated with colchicine for 72 or 48 h, respectively, normalized to DMSO treatment (mean ± SD n = 3 biological replicates).
Clinical use of colchicine at low doses is generally well-tolerated and often used chronically to treat gout and as a life-long treatment for FMF, despite toxicity in some cases (44, 48, 52). To relate the effect of colchicine on rescuing CUG RNA toxicity to its effect on overall cell toxicity, we measured cell viability in both HeLa (CTG)480 clone 19 and differentiated DM1 myotubes in the dose range, where we observed significant rescue of missplicing (Fig. 4D). Consistent with the established sensitivity of cancer cells to microtubule inhibition (44), there was a clear induction of toxicity in HeLa (CTG)480 clone 19 with colchicine treatment from a concentration of 50 nM and beyond (Fig. 4D). In contrast, treatment of differentiated, postmitotic DM1 myotubes had only a marginal effect on viability across the dose range (Fig. 4D). These results support the effect of microtubule inhibition on r(CUG)EXP to be independent of the proliferation status of the cell or of toxicity to the cell.
Repeat Selectivity Mediated by Colchicine Treatment Occurs through a Mechanism Impacting Transcription.
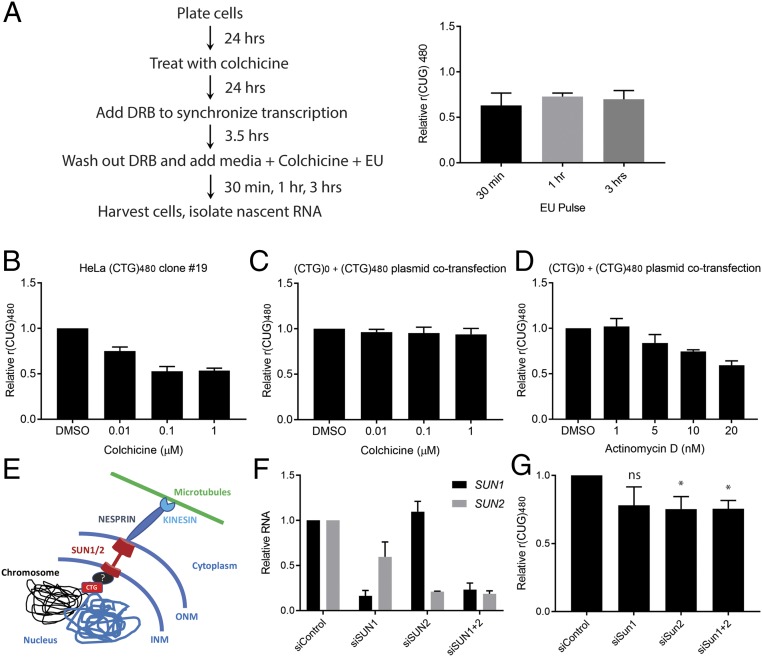
To address the mechanism through which microtubule inhibition was conferring a selective reduction in r(CUG)EXP transcripts, we first sought to separate an effect of microtubule inhibition on transcription from RNA stability. We carried out 5-ethynyl uridine (EU) pulse labeling and nascent RNA isolation in HeLa (CTG)480 clone 19 treated with colchicine. EU labeling was carried out with varying pulse times to differentiate transcription from stability/turnover of the nascent r(CUG)480 molecules (Fig. 5A). There was a comparable selective reduction in r(CUG)480 levels with colchicine treatment independent of EU pulse times. These results are consistent with colchicine treatment selectively inhibiting production of r(CUG)480 transcripts rather than selectively enhancing their rate of turnover or decay.
Fig. 5.
Selective effect of colchicine treatment on r(CUG)480 levels occurs through transcription potentially involving the LINC complex. (A) Schematic of HeLa (CTG)480 clone 19 colchicine treatment and nascent RNA isolation. DRB, 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside. Multiplex RT-qPCR was performed to determine relative levels of nascent r(CUG)480 [normalized to r(CUG)0 and compared to DMSO treatment] (mean ± SD; n = 3 biological replicates). (B and C) Effect of colchicine treatment at the indicated concentrations on relative r(CUG)480 levels [normalized to r(CUG)0] in HeLa (CTG)480 clone 19 (B) or HeLa parental line cotransfected with (CTG)0 and (CTG)480 constructs in expression plasmids (C). (D) Same experiment as in B, but treatment with ActD was at the indicated concentrations (mean ± SD n = 3 biological replicates). (E) Schematic of hypothetical LINC complex involvement in regulating the CTG expansion through an unknown factor (black oval with “?”). (F) HeLa (CTG)480 clone 19 was treated with control siRNA (siControl) or siRNA directed against SUN1/2 either individually (siSun1 or siSun2) or in combination (siSun1+2) for 96 h (n = 4 biological replicates). After treatment, mRNA levels of SUN1 and SUN2 were determined by qPCR. (G) Multiplex RT-qPCR was performed following siRNA treatments to determine effects on relative r(CUG)480 levels [normalized to r(CUG)0] and compared to siControl levels (mean ± SD; n = 4 biological replicates). *P < 0.05 (as assessed by one-way ANOVA followed by Tukey’s post hoc analysis). ns, not significant.
Notably, cotransfecting the parental HeLa cell line that was used to create HeLa (CTG)480 clone 19 with plasmids for ectopic expression of r(CUG)0 and r(CUG)480 did not recapitulate the repeat-selective effect (Fig. 5 B and C). This is in contrast to treatment with colchicine of HeLa (CTG)480 clone 19, where the repeat tract is integrated into the nuclear genome and a dose-dependent reduction in relative r(CUG)480 was evident (Fig. 5B). Cotransfection of the same plasmids into the parental HeLa line, followed by treatment with ActD, which selectively binds to CTG repeats and blocks transcription independent of nuclear context (25, 39, 40), retained the repeat-selective reduction in r(CUG)480 levels (Fig. 5D). These results highlight the benefit of screening at physiologically relevant conditions compared to transient overexpression systems.
Chromosomes are spatially organized within the nucleus and exhibit dynamic relocalization of regions undergoing transcription (53, 54). Microtubules can interact with and regulate chromosome dynamics via the LINC (linker of nucleoskeleton and cytoskeleton) complex to regulate nuclear functions such as transcription and DNA repair, highlighting an important cross-talk between the cytoskeleton and nuclear genome mediated by microtubules (45, 55–57). We hypothesized that microtubule dynamics facilitated by the LINC complex may be involved in selectively modifying transcription of CTG repeat expansions. The LINC complex is composed of NESPRINS in the cytoplasm that form physical contacts with SUN proteins present at the nuclear envelope, forming a cellular relay system to transmit cytoplasmic stimuli to the nucleus (56). Notably, we observed a significant increase in the inclusion of SYNE1 (encoding NESPRIN1) exon 137 in control myotubes treated with colchicine (SI Appendix, Fig. S13).
To directly test if LINC complex activity played a role in the selective reduction of r(CUG)480 levels resulting from microtubule perturbations, which could be through direct interaction with the CTG expansion, for example, at the nuclear lamina or potentially through an unknown factor (Fig. 5E), we treated HeLa (CTG)480 clone 19 with small interfering RNAs (siRNAs) against the LINC complex core components SUN1/2 either alone or in combination (Fig. 5F and SI Appendix, Fig. S14). There was a modest but significant reduction in relative r(CUG)480 levels upon SUN1/2 depletion (Fig. 5G). We assessed a panel of misspliced pre-mRNA targets to determine any potential rescue from this reduction in r(CUG)480 levels (SI Appendix, Fig. S15). Knockdown of SUN1/2 in combination in the HeLa parental cells resulted in alterations to splicing independent of the (CTG)480 tract and, hence, confounded our analysis of any potential rescue in missplicing in HeLa (CTG)480 clone 19 (SI Appendix, Fig. S15).
Discussion
We have established a DM1 HeLa cell line that enables the sensitive measurement of r(CUG)480 levels relative to an analogous r(CUG)0 control. This cell line can be used to screen compounds or cellular factors that have a selective effect on the r(CUG)480. We applied this cell line to a pilot screen of the LOPAC1280 chemical library, leading to the identification of multiple microtubule inhibitors as selective modulators of r(CUG)480 levels. We then validated the use of a microtubule inhibitor in the HSALR DM1 mouse model and in DM1 patient cells with colchicine, an FDA-approved, natural microtubule inhibitor currently used in the clinic. Our results provide proof of principle for the identification of compounds and cellular targets that selectively modulate r(CUG)EXP levels in DM1 using cell-based screening.
Our observations of a partial rescue in DM1-relevant missplicing in multiple models (Figs. 2E, 3 D and E, and 4B) warrants further evaluation of colchicine. This study is not sufficient to address the therapeutic efficacy of colchicine or of general microtubule inhibition for the treatment of DM1. It is important to determine if there is a positive trade-off between therapeutic efficacy in reducing DM1 symptoms in relation to the known toxicity from microtubule inhibition. As an example, and although very rare, myopathy has been reported in some individuals with compromised renal function who had been treated chronically with colchicine for gout (58, 59). Future long-term treatments in DM1 animal models, such as HSALR, at clinical doses to evaluate the reversal of DM1 phenotypes are a prerequisite to determine if any clinical studies are warranted.
Independent of therapeutic potential, the discovery of microtubule involvement in the selective reduction of r(CUG)EXP levels is interesting. By taking an approach that selectively measures toxic CUG RNA levels, we were able to identify a potential role of microtubule dynamics, which we postulate may be at least in part driven by interactions with the LINC complex. Compared to previous screens which have not reported these targets, our approach may provide an advantage by selectively measuring the toxic CUG RNA levels directly, rather than downstream consequences, such as ribonuclear foci or missplicing. Several recent studies have highlighted the importance of microtubules in regulating chromosome functions such as transcription, DNA replication, and repair (45, 57, 60–62). Many of these studies implicate an interaction between cytoplasmic microtubules and chromosomes via the LINC complex. Furthermore, telomere repeats are specifically targeted to the nuclear envelope through interactions with SUN1 protein to promote homolog pairing during meiosis in normal gametogenesis (63). Thus, one hypothesis is that microtubule perturbations may directly interfere with LINC complex-mediated transcriptional positioning of the expanded CTG repeat tract within the nucleus. Consistent with this hypothesis, we observed an effect of knocking down LINC complex components SUN1/SUN2 (Fig. 5 E–G). It was recently demonstrated that disease-associated tandem repeats, including CTG repeats, associate with boundaries of chromatin domains, with repeat expansions disrupting this nuclear organization (64). It is not known if perturbing this organization potentially through a LINC complex–microtubule axis may impact transcription of these repeat sequences. However, we cannot at this stage rule out the effect to be secondary. For example, de Lange and colleagues (45) identified a 53BP1, LINC complex, and microtubule dynamics-mediated activity facilitating chromatin mobility in double-strand-break repair of DNA, including telomere repeats. Intriguingly, another recent study found evidence for microtubule filaments present in the nucleus that function in DNA double-strand-break repair (62). Although the exact nature by which microtubules may interact with chromatin is not yet clear, in light of these results, one plausible hypothesis is that expanded CTG repeats undergo a similar DNA-repair process relying on microtubule dynamics. We may then observe reduced r(CUG)EXP levels as a consequence of microtubule perturbations, leading to transcriptional interference during failed repair of the expanded CTG repeat tract. However, microtubules are important for many functions within the cell, and, thus, to separate direct and secondary effects, future studies are necessary.
Although the precise mechanism through which microtubule inhibition selectively reduces r(CUG)EXP levels remains cryptic, the identification of hits that function in a CTG repeat-selective manner from a modest library of 1,280 compounds validates our approach and is encouraging for future studies involving larger compound libraries. Given the recent success of CRISPR/Cas9 knockout screens (65), we have established an amenable platform for such screens to probe genome-wide targets that selectively reduce r(CUG)EXP levels. Finally, as there are over 30 neurological and neuromuscular disorders involving microsatellite expansions, our approach to repeat-selective screening may be applicable to a wide variety of repeat expansion disorders.
Methods
Generation of Stable DM1 HeLa (CTG)480 Cell Lines.
Stable barcoded HeLa cell lines expressing r(CUG)0 and r(CUG)480 were generated as described (26). Briefly, (CTG)0 and (CTG)480 DMPK (exons 11–15) expression cassettes (37) containing a 22-bp unique sequence plus an additional 8-bp unique barcode for pooled RNA-seq were inserted into pAC156 (gift from Albert Cheng, The Jackson Laboratory, Farmington, CT), a plasmid with PiggyBac transposon terminal repeats as well as a puromycin selection cassette. Both plasmids were transiently transfected together with the PiggyBac mPB transposase into HeLa cells (ATTC) and selected by puromycin. Single cells were isolated by flow cytometry, and colonies were cultured in 96-well plates. Colonies were expanded and subjected to measurement of barcoded transcripts by amplicon sequencing to screen for integration and expression from both plasmids as described (26).
LOPAC1280 Screen.
Approximately 1 × 104 cells per well were plated and cultured overnight in 200 µL of medium containing Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin under standard conditions of 37 °C and 5% CO2. The next day, medium was removed and replaced with fresh medium (100 µL) containing drug at a final concentration of 1 µM or DMSO as a control. Following ∼24 h of treatment, cells were washed with PBS, and the plates were stored at −80 °C. The following day, plates were thawed, and each well was incubated with 20 µL of 0.25% Igepal buffer containing 10 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl on an orbital shaker for 5 min at room temperature. Two microliters of “lysate” was then directly taken forward to cDNA generation by using HT_RTprimer (IDT) (SI Appendix, Table S1) and SuperScript IV RT (Thermo Fisher).
qPCR was carried out on a Bio-Rad C1000 Touch Thermal Cycler by using Hot Start Taq 2× Master Mix (NEB) with HT_Forward and HT_Reverse primers (IDT) (SI Appendix, Table S1) and custom HT_Probe1 and HT_Probe2 fluorescent probes (IDT) (SI Appendix, Table S1). Data were analyzed by using the comparative (ΔΔCT) method. The levels of r(CUG)480 from drug treatments were normalized to r(CUG)0 and presented as relative mRNA levels by comparing to DMSO control treatments.
Cell Culture and Treatment of DM1 Myotubes.
Primary patient and control myoblast cell lines were derived from muscle biopsies under a University of Florida-approved Institutional Review Board (IRB) protocol with informed consent from all subjects. For cell culture of myoblasts, ∼1 × 105 cells were plated per well in 12-well plates in SkGM-2 BulletKit growth medium (Lonza). Cells were allowed to reach >90% confluency and then differentiated for 7 d to myotubes in DMEM/F-12 50/50 medium (Corning) supplemented with 2% (vol/vol) donor equine serum (HyClone). After 7 d, the differentiation medium was replaced with SkGM-2 growth medium, and the indicated concentrations of drug were added. Myotubes were harvested at 72 h of drug treatment. Quantitative real-time PCR was performed by using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) according to the package insert using primers (IDT) listed in SI Appendix, Table S2. Samples were run on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) and analyzed by using the comparative [(ΔΔ CT)] method.
Cell Culture and Treatment of DM1 Fibroblasts.
Primary patient and control fibroblast cell lines were derived from skin biopsies under a University of Florida-approved IRB protocol with informed consent from all subjects. Approximately 3.5 × 104 cells were plated per well in 24-well plates and cultured to confluence in DMEM supplemented with 15% FBS and 1% penicillin and streptomycin under standard conditions of 37 °C and 5% CO2. Once at confluence, medium was replaced with DMEM supplemented with 0.15% FBS and 1% penicillin and streptomycin with colchicine dissolved in DMSO added to the indicated concentrations, and cells were cultured under standard conditions of 37 °C and 5% CO2 for ∼48 h.
Colchicine Treatment in Mice.
Mouse handling and experimental procedures were conducted in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care. All of the procedures for animal experiments were approved by the institutional animal care and use committee of Osaka University. HSALR transgenic mice in line 20b (FVB inbred background) were described (49). Gender-matched homogeneous HSALR mice of 6–7 wk of age were treated with colchicine at a concentration of 0.4 mg/kg as a PBS solution by daily i.p. injection for 14 d. The control group received PBS. Mice were killed 1 d after the final injection, and vastus lateralis muscles (quadriceps) were obtained for splicing analysis. RNA extraction and cDNA preparation were carried out as described (13). Quantitative real-time PCR of human skeletal actin (HSA) transgene RNA and mouse Gtf2b and Dmpk RNA was performed by using the TaqMan Gene Expression (Thermo Fisher) assay on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) and analyzed by using the comparative (ΔΔ CT) method.
Fluorescence in Situ Hybridization Microscopy.
Cells were cultured in 8-chamber slides with or without drug treatments as indicated. Cells were fixed with 4% paraformaldehyde (PFA), permeabilized by using 70% ethanol, and prehybridized in the presence of tRNA (Thermo Fisher catalog no. AM7119) for 30 min at 37 °C. Cells were probed for 4 h at 50 °C with a Cy3-(CAG)8 probe (IDT). Slides were washed with 42 °C prewarmed 40% (vol/vol) formamide in 2× saline sodium citrate and mounted by using ProLong Diamond Antifade mountant with DAPI (Life Technologies). Nuclei were imaged on a Zeiss LSM 840 confocal scanning microscope with a 40× oil objective.
Immunofluorescence Microscopy.
The protocol was carried out as described with minor modifications (66). Frozen vastus lateralis (quadriceps) muscle from HSALR mice was sectioned into 8-µm slices onto slides and fixed with 4% PFA for 15 min at room temperature (RT), permeabilized by using 1:1 methanol:acetone (prechilled) for 5 min at −20 °C, and blocked by using Background Sniper for at least 30 min at RT (Biocare Medical). Slides were incubated with 1:100 rabbit anti-rat Clcn1 (Alpha Diagnostic International, catalog no. CLC11-A) overnight at 4 °C in 0.1× Background Sniper in Dulbecco’s PBS (DPBS) and washed 3× in DPBS for 5 min each at RT, and then samples were incubated with 1:1,000 goat anti-rabbit Alexa Fluor 488 (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at RT and mounted by using ProLong Diamond Antifade mountant with DAPI (Life Technologies). Samples were imaged on a Zeiss LSM 710 confocal scanning microscope with a 20× objective.
RT-PCR Splicing Analysis.
The protocol was carried out as described with minor modifications (66). RNA was isolated from cells by using an Aurum Total RNA mini kit (Bio-Rad) according to the package insert with on-column DNase1 treatment. RNA from mouse samples was TRIZol-extracted from quadriceps muscle of wild-type FVB mice, HSALR mice treated with either PBS (control) or 0.4 mg/kg colchicine as a PBS solution. RNA concentrations were determined by using a NanoDrop (Thermo) and reverse transcribed with SuperScript VI using random hexamer primers (IDT). The cDNA was then subjected to PCR for 32 cycles using the primer sets listed in SI Appendix, Table S1. Resulting PCR products were run via capillary electrophoresis on a Fragment Analyzer using the 1- to 500-bp DNF-905 kit (Advanced Analytical). Quantification was done by using the integration values of the electropherogram peaks corresponding to inclusion and exclusion products from the Prosize 2.0 software (Advanced Analytical). To determine the percent rescue of a given exon-skip (ES) event, Eq. 1 was used, where PSItreated = PSI of DM1 treated samples, PSIuntreated = PSI of DM1 untreated samples, and PSInon-DM = PSI of non-DM1 control samples.
| [1] |
Toxicity Analysis in Cell Culture.
The protocol was carried out as described with minor modifications (66). For myoblasts, ∼1 × 104 myoblasts were plated per well in 96-well plates in SkGM-2 BulletKit growth medium (Lonza). Cells were allowed to reach >90% confluency and then differentiated for 7 d to myotubes in DMEM/F-12 50/50 medium (Corning) supplemented with 2% vol/vol donor equine serum (HyClone). After 7 d of differentiation and 72 h of drug/DMSO treatment, medium was replaced with fresh SkGM-2 growth medium, and PrestoBlue cell viability reagent (Thermo Fisher) was added to the cells according to the package insert and incubated at 37 °C and 5% CO2, protected from light for 3 h. Absorbance at 570 and 600 nm was read on a BioTek Cytation 3 plate reader. The 570-nm/600-nm absorbance ratios were calculated for all samples with a background subtraction of the average 570-nm/600-nm values of no-cell plus drug/DMSO control wells.
For HeLa, ∼5 × 104 cells were plated per well in 96-well plates in DMEM with 10% FBS and 1% penicillin/streptomycin overnight. The next day, medium was replaced with fresh medium containing drug or DMSO (control) and incubated for 48 h, following which the medium with drug/DMSO was replaced with fresh medium and PrestoBlue cell viability reagent (Thermo Fisher) was added according to the package insert, and the cells were subsequently treated as described above.
RNA-Seq Library Preparation.
The protocol was carried out as described with minor modifications (66). RNA was isolated from cells by using an Aurum Total RNA mini kit (Bio-Rad) according to the package insert with on-column DNase1 treatment. For mouse experiments, RNA was TRIZol-extracted from quadriceps muscle of wild-type FVB mice and HSALR mice treated with either PBS only (control) or 0.4 mg/kg colchicine as a PBS solution. RNA quality was checked via capillary electrophoresis on Fragment Analyzer using the RNA Analysis DNF-471 kit, assuring RIN values greater that 8 (Advanced Analytical). The NEBNext Ultra II Directional RNA Library Prep Kit for Illumina with NEBNext rRNA Depletion Kit was used to prepare RNA-seq libraries, with a total of 500 ng of input RNA from each sample. The manufacturer’s protocols were followed, with the following exceptions: 40× adaptor dilutions were used; all bead incubations were done at RT; we used 4× lower concentrations of index primers; and 10 cycles of library amplification were performed. The resulting libraries were pooled in equimolar amounts, quantified by using the KAPA Library Quant Kit for Illumina, quality checked via capillary electrophoresis on a Fragment Analyzer using the NGS Analysis DNF-474 kit (Advanced Analytical), and sequenced using paired-end, 75-base-pair sequencing on the Illumina NextSeq 500 massively parallel sequencer at the University of Florida Center for NeuroGenetics.
Transcriptome Analysis from RNA-Seq Data.
The protocol was carried out as described with minor modifications (66). Raw reads were checked for quality and aligned to GRCm38.p5 mouse genome by using STAR (Version 2.5.1b) (67) and a .gtf file generated from Version M16 GENECODE gene models. Uniquely aligning paired sequences were input to Stringtie (Version 1.3.4d), and the prepDE.py script (offered with Stringtie package) was used to generate gene counts. Differential expression analysis was performed with DESeq2 (Version 1.16.1) (68). Differential expression was considered significant with P < 0.1. Of those events, a percent rescue of ≥10% was considered “Rescues” with colchicine treatment; “Off-target” gene expression events were those not in the WT versus HSALR events. To determine the rescue of a given differentially expressed gene, Eq. 2 was used, where WT_EXP = difference in expression of WT mice versus untreated HSALR mice, and drug_EXP = difference in expression of untreated HSALR mice versus HSALR mice treated with colchicine.
| [2] |
Splicing Analysis from RNA-Seq Data.
The protocol was carried out as described with minor modifications (66). Raw reads were checked for quality and aligned to GRCm38.p5 mouse genome by using STAR (Version 2.5.1b). After reads were aligned, rMATS (Version 3.2.5) (69) was used to analyze isoform abundances and compared to 3 wild-type samples (70). ES events were considered significant with a false discovery rate (FDR) < 0.1 and P < 0.01. Events were considered misspliced in the wild-type vs. HSALR datasets if the PSI change was ≥15% for a given ES event. To determine the percent rescue of a given ES event, Eq. 3 was used, where HSA_PSI = PSI of PBS control-treated HSALR mice, WT_PSI = PSI of wild-type mice, and drug_PSI = PSI of HSALR mice treated with colchicine.
| [3] |
EU Cell Treatments and Nascent RNA Isolation.
HeLa (CTG)480 clone 19 cells were seeded at a density of 1 × 105 cells per well of a 6-well plate. The next day, cells were treated with colchicine at a final concentration of 100 nM for ∼24 h. The following day, cells were treated with 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (Sigma-Aldrich) at a final concentration of 100 µM and incubated for ∼3.5 h. Cells were then washed in PBS two times and incubated in fresh DMEM supplemented with 10% FBS and 1% penicillin and streptomycin containing 100 nM colchicine, and EU (from the Click-iT Nascent RNA Capture Kit, Thermo Fisher) was added for 30 min, 1 h, or 3 h, and total RNA was isolated by using TRIzol reagent (Thermo Fisher) according to the manufacturer’s instructions. Nascent RNA was isolated by using the Click-iT Nascent RNA Capture Kit (Thermo Fisher) according to manufacturer’s protocols.
siRNA Cell Treatments.
HeLa (CTG)480 clone 19 cells were seeded at a density of 7.5 × 104 cells per well of a 12-well plate. The following day, siRNA SMARTpools (Dharmacon) were transfected by using Polyplus INTERFERin Transfection reagent according to the manufacturer’s protocols at a final concentration of 20 nM in Opti-MEM reduced-serum medium. The next day, medium was replaced with DMEM supplemented with 10% FBS and 1% penicillin and streptomycin; cells were cultured for an additional 72 h, and RNA was isolated from cells by using an Aurum Total RNA mini kit (Bio-Rad) according to manufacturer’s protocols with on-column DNase1 treatment.
Western Blots.
Protein was harvested by vortexing for 15 min at 4 °C in radioimmunoprecipitation assay buffer supplemented with 1 mM phenylmethylsulfonyl fluoride and 1× SigmaFast protease inhibitor (Sigma-Aldrich). After centrifugation at 12,000 rpm for 15 min at 4 °C, the supernatant was used to determine protein concentration with the Pierce BCA Protein Assay kit (Thermo). A total of 10 μg of protein was denatured for 5 min at 98 °C and run on a precast 10% sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis mini gel (Bio-Rad) at 200 V for ∼40 min in 1× running buffer (25 mM Tris base, pH 8.3, 192 mM glycine, and 0.1% [wt/vol] SDS). Gel was transferred onto low-fluorescence polyvinylidene fluoride membrane (Bio-Rad) for 1 h at 25 V in 1× transfer buffer (25 mM Tris base, pH 8.3, 192 mM glycine, and 20% [vol/vol] methanol). Membrane was blocked for 1 h by using SeaBlock (Thermo) and then incubated overnight with primary antibodies (1:1,000 SUN1 [Abcam catalog no. ab124770], 1:1,000 SUN2 [Abcam catalog no. ab124916], and 1:1,000 GAPDH [Abcam catalog no. ab14247]). Blots were incubated at RT for 1 h with secondary antibodies (1:7,500 goat anti-rabbit IRDye at 680 [Li-Cor] and 1:10,000 donkey anti-chicken IRDye at 800 [Li-Cor]), washed in Tris-buffered saline and Tween 20, and imaged on an Odyssey CLx imager (Li-Cor). Blots were analyzed by using ImageStudio Lite (Li-Cor).
RNA Sequencing.
All sequencing data detailed in this paper have been deposited in the Sequence Read Archive (SRA) database (accession no. SRP158284) (71).
Supplementary Material
Acknowledgments
We thank T. Reid and S. Shaughnessy for technical assistance in this study; M. Swanson for helpful comments on the manuscript; P. August and colleagues at Icagen for advice on screen design; G. Xia for myoblast and fibroblast cell lines; and A. Cheng for the pAC156 vector. This work was supported by a research grant from the Wyck Foundation and managed by the Myotonic Dystrophy Foundation (Grant 49 to J.A.B., E.T.W., Kausiki Datta, and Paul August); University of Florida institutional start-up funds (J.A.B. and E.T.W.); National Center of Neurology and Psychiatry Intramural Research Grant 29-4 (to M.N.); National Science Foundation Predoctoral Fellowship 2016204076 (to J.R.J.); and a Myotonic Dystrophy Foundation and Wyck Foundation postdoctoral fellowship (K.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.S. is a guest editor invited by the Editorial Board.
Data deposition: The RNA-seq data have been deposited in the Sequence Read Archive (SRA) database, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP158284).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901893116/-/DCSupplemental.
References
- 1.Brook J. D., et al. , Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 69, 385 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Harley H. G., et al. , Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature 355, 545–546 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Mahadevan M., et al. , Myotonic dystrophy mutation: An unstable CTG repeat in the 3′ untranslated region of the gene. Science 255, 1253–1255 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Thornton C. A., Johnson K., Moxley R. T. 3rd, Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann. Neurol. 35, 104–107 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Mooers B. H., Logue J. S., Berglund J. A., The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl. Acad. Sci. U.S.A. 102, 16626–16631 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller J. W., et al. , Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 19, 4439–4448 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho T. H., et al. , Muscleblind proteins regulate alternative splicing. EMBO J. 23, 3103–3112 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savkur R. S., Philips A. V., Cooper T. A., Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29, 40–47 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Mankodi A., et al. , Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 10, 35–44 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Nakamori M., et al. , Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol. 74, 862–872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner S. D., et al. , Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 12, e1006316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler T. M., Lueck J. D., Swanson M. S., Dirksen R. T., Thornton C. A., Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 117, 3952–3957 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warf M. B., Nakamori M., Matthys C. M., Thornton C. A., Berglund J. A., Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 106, 18551–18556 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arambula J. F., Ramisetty S. R., Baranger A. M., Zimmerman S. C., A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl. Acad. Sci. U.S.A. 106, 16068–16073 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rzuczek S. G., et al. , Precise small-molecule recognition of a toxic CUG RNA repeat expansion. Nat. Chem. Biol. 13, 188–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler T. M., et al. , Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 325, 336–339 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulders S. A., et al. , Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 106, 13915–13920 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojtkowiak-Szlachcic A., et al. , Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res. 43, 3318–3331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J. E., Bennett C. F., Cooper T. A., RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc. Natl. Acad. Sci. U.S.A. 109, 4221–4226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langlois M. A., et al. , Cytoplasmic and nuclear retained DMPK mRNAs are targets for RNA interference in myotonic dystrophy cells. J. Biol. Chem. 280, 16949–16954 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Sobczak K., Wheeler T. M., Wang W., Thornton C. A., RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy. Mol. Ther. 21, 380–387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bisset D. R., et al. , Therapeutic impact of systemic AAV-mediated RNA interference in a mouse model of myotonic dystrophy. Hum. Mol. Genet. 24, 4971–4983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batra R., et al. , Elimination of toxic microsatellite repeat expansion RNA by RNA-targeting Cas9. Cell 170, 899–912.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coonrod L. A., et al. , Reducing levels of toxic RNA with small molecules. ACS Chem. Biol. 8, 2528–2537 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siboni R. B., et al. , Actinomycin D specifically reduces expanded CUG repeat RNA in myotonic dystrophy models. Cell Rep. 13, 2386–2394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto B. S., et al. , Impeding transcription of expanded microsatellite repeats by deactivated Cas9. Mol Cell 68, 479–490.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cinesi C., Aeschbach L., Yang B., Dion V., Contracting CAG/CTG repeats using the CRISPR-Cas9 nickase. Nat. Commun. 7, 13272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Agtmaal E. L., et al. , CRISPR/Cas9-Induced (CTG⋅CAG)n repeat instability in the myotonic dystrophy type 1 locus: Implications for therapeutic genome editing. Mol. Ther. 25, 24–43 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Provenzano C., et al. , CRISPR/Cas9-Mediated deletion of CTG expansions recovers normal phenotype in myogenic cells derived from myotonic dystrophy 1 patients. Mol. Ther. Nucleic Acids 9, 337–348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dastidar S., et al. , Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells. Nucleic Acids Res. 46, 8275–8298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-López A., Llamusí B., Orzáez M., Pérez-Payá E., Artero R. D., In vivo discovery of a peptide that prevents CUG-RNA hairpin formation and reverses RNA toxicity in myotonic dystrophy models. Proc. Natl. Acad. Sci. U.S.A. 108, 11866–11871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A., et al. , Chemical correction of pre-mRNA splicing defects associated with sequestration of muscleblind-like 1 protein by expanded r(CAG)-containing transcripts. ACS Chem. Biol. 7, 496–505 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ketley A., et al. , High-content screening identifies small molecules that remove nuclear foci, affect MBNL distribution and CELF1 protein levels via a PKC-independent pathway in myotonic dystrophy cell lines. Hum. Mol. Genet. 23, 1551–1562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoskins J. W., et al. , Lomofungin and dilomofungin: Inhibitors of MBNL1-CUG RNA binding with distinct cellular effects. Nucleic Acids Res. 42, 6591–6602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia S. M., Tabach Y., Lourenço G. F., Armakola M., Ruvkun G., Identification of genes in toxicity pathways of trinucleotide-repeat RNA in C. elegans. Nat. Struct. Mol. Biol. 21, 712–720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F., et al. , A flow cytometry-based screen identifies MBNL1 modulators that rescue splicing defects in myotonic dystrophy type I. Hum. Mol. Genet. 26, 3056–3068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philips A. V., Timchenko L. T., Cooper T. A., Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280, 737–741 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Chastain P. D., 2nd, et al. , Anomalous rapid electrophoretic mobility of DNA containing triplet repeats associated with human disease genes. Biochemistry 34, 16125–16131 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Chen F. M., Binding of actinomycin D to DNA oligomers of CXG trinucleotide repeats. Biochemistry 37, 3955–3964 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Hou M. H., Robinson H., Gao Y. G., Wang A. H., Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 30, 4910–4917 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J. H., Chung T. D., Oldenburg K. R., A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Gates D. P., Coonrod L. A., Berglund J. A., Autoregulated splicing of muscleblind-like 1 (MBNL1) Pre-mRNA. J. Biol. Chem. 286, 34224–34233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sen S., et al. , Muscleblind-like 1 (Mbnl1) promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J. Biol. Chem. 285, 25426–25437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinmetz M. O., Prota A. E., Microtubule-targeting agents: Strategies to hijack the cytoskeleton. Trends Cell Biol. 28, 776–792 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Lottersberger F., Karssemeijer R. A., Dimitrova N., de Lange T., 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 163, 880–893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi B. H., et al. , Suprafenacine, an indazole-hydrazide agent, targets cancer cells through microtubule destabilization. PLoS One 9, e110955 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng H., Huang G., Synthesis and activity of epothilone D. Curr. Drug Targets 19, 1866–1870 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Kallinich T., et al. , Colchicine use in children and adolescents with familial Mediterranean fever: Literature review and consensus statement. Pediatrics 119, e474–e483 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Mankodi A., et al. , Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 289, 1769–1773 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Osborne R. J., et al. , Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 18, 1471–1481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia G., et al. , Generation of neural cells from DM1 induced pluripotent stem cells as cellular model for the study of central nervous system neuropathogenesis. Cell. Reprogram. 15, 166–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lachmann H. J., Periodic fever syndromes. Best Pract. Res. Clin. Rheumatol. 31, 596–609 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Shah S., et al. , Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rinn J., Guttman M., RNA function. RNA and dynamic nuclear organization. Science 345, 1240–1241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y. L., Burke B., LINC complexes and nuclear positioning. Semin. Cell Dev. Biol. 82, 67–76 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Meinke P., Schirmer E. C., LINC’ing form and function at the nuclear envelope. FEBS Lett. 589, 2514–2521 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Alam S. G., et al. , The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci. Rep. 6, 38063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dawson T. M., Starkebaum G., Colchicine induced rhabdomyolysis. J. Rheumatol. 24, 2045–2046 (1997). [PubMed] [Google Scholar]
- 59.Fernandez C., Figarella-Branger D., Alla P., Harlé J. R., Pellissier J. F., Colchicine myopathy: A vacuolar myopathy with selective type I muscle fiber involvement. An immunohistochemical and electron microscopic study of two cases. Acta Neuropathol. 103, 100–106 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Wang S., et al. , Mechanotransduction via the LINC complex regulates DNA replication in myonuclei. J. Cell Biol. 217, 2005–2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawrimore J., et al. , Microtubule dynamics drive enhanced chromatin motion and mobilize telomeres in response to DNA damage. Mol. Biol. Cell 28, 1701–1711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oshidari R., et al. , Nuclear microtubule filaments mediate non-linear directional motion of chromatin and promote DNA repair. Nat. Commun. 9, 2567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding X., et al. , SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863–872 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Sun J. H., et al. , Disease-associated short tandem repeats Co-localize with chromatin domain boundaries. Cell 175, 224–238.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kramer N. J., et al. , CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet. 50, 603–612 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jenquin J. R. Y., Yang H., Huigens R. W. 3rd, Nakamori M., Berglund J. A., Combination treatment of erythromycin and furamidine provides additive and synergistic rescue of mis-splicing in myotonic dystrophy type 1 models. ACS Pharmacol Transl Sci 2, 247–263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu T. D., Nacu S., Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen S., et al. , rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zygmunt D. A., et al. , Deletion of Pofut1 in mouse skeletal myofibers induces muscle aging-related phenotypes in cis and in trans. Mol. Cell. Biol. 37, e00426-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reddy K., et al. , Repeat-selective screening identifies microtubule inhibitors that reduce toxic CUG RNA. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/SRP158284. Deposited 16 August 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.