Significance
The clustered regularly interspaced short palindromic repeat (CRISPR)-associated proteins (Cas) have been widely used for genome engineering. However, their off-target activities limit broad application. The small Cas9 ortholog from Staphylococcus aureus (SaCas9) can be packaged in the payload-limited adeno-associated viral (AAV) vector that is commonly used for in vivo gene editing. Nevertheless, there is still a lack of SaCas9 variants conferring high genome-wide specificity. Here, we report a rationally engineered SaCas9 variant (SaCas9-HF) with highly specific genome-wide activity in human cells without compromising on-target efficiency. SaCas9-HF can be delivered by AAV and show higher genome-wide specificity than wild-type SaCas9. Our finding provides an alternative to SaCas9 genome-editing applications requiring exceptional genome-wide precision.
Keywords: CRISPR-Cas9, SaCas9, off-target
Abstract
RNA-guided CRISPR-Cas9 proteins have been widely used for genome editing, but their off-target activities limit broad application. The minimal Cas9 ortholog from Staphylococcus aureus (SaCas9) is commonly used for in vivo genome editing; however, no variant conferring high genome-wide specificity is available. Here, we report rationally engineered SaCas9 variants with highly specific genome-wide activity in human cells without compromising on-target efficiency. One engineered variant, referred to as SaCas9-HF, dramatically improved genome-wide targeting accuracy based on the genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq) method and targeted deep sequencing analyses. Among 15 tested human endogenous sites with the canonical NNGRRT protospacer adjacent motif (PAM), SaCas9-HF rendered no detectable off-target activities at 9 sites, minimal off-target activities at 6 sites, and comparable on-target efficiencies to those of wild-type SaCas9. Furthermore, among 4 known promiscuous targeting sites, SaCas9-HF profoundly reduced off-target activities compared with wild type. When delivered by an adeno-associated virus vector, SaCas9-HF also showed reduced off-target effects when targeting VEGFA in a human retinal pigmented epithelium cell line compared with wild type. Then, we further altered a previously described variant named KKH-SaCas9 that has a wider PAM recognition range. Similarly, the resulting KKH-HF remarkably reduced off-target activities and increased on- to off-target editing ratios. Our finding provides an alternative to wild-type SaCas9 for genome editing applications requiring exceptional genome-wide precision.
Genome engineering technologies have enabled systematic interrogation of genome function and hold great potential for gene therapy (1–4). The clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein (Cas) system allows for efficient DNA modification when guided by a cRNA and in the presence of a protospacer adjacent motif (PAM). However, imperfect guide RNA–target DNA matching may also induce nuclease activity of Cas proteins, resulting in modifications at genomic loci other than the intended locus (5). This off-target activity could confound research results and constrain clinical utility. Two widely used Cas9 orthologs from Streptococcus pyogenes (SpCas9) and Staphylococcus aureus (SaCas9) have different levels of off-target activity (5–8). SaCas9 is compact and can be packaged in the payload-limited adeno-associated viral (AAV) vector that is commonly used for in vivo gene editing (6). While there are a handful of high-fidelity SpCas9 variants (9–14), no SaCas9 variant with high genome-wide specificity is available.
Two main strategies have been exploited to generate Cas9 variants with improved specificity. One is structure-guided protein engineering to modify amino acid residues in close contact with the target DNA strand or those interacting with the nontarget DNA strand (9–11). The other is through random mutagenesis followed by end-point selection or directed evolution (12–14). Studies employing these strategies mainly focused on SpCas9, except 1 on SaCas9, which generated eSaCas9 variants by modifying amino acid residues interacting with the nontarget DNA strand, leading to reduced activity at 3 predefined off-target sites, but these have unknown genome-wide specificity (10).
Cas9 recognition and binding of its target DNA sequence is a dynamic process that involves sequential conformational changes in functional domains between inactive and active states prior to concerted cleavage of both DNA strands (11, 15–17). Single-molecule Förster resonance energy transfer experiments on SpCas9 showed that the number of mismatched bases in the guide RNA–target DNA heteroduplex in the PAM-distal region was inversely correlated with the proportion of SpCas9 in the activated state (17). Wild-type SpCas9 amino acid residues proximal to the guide-RNA–target DNA interface could lower the threshold for activating the nuclease domain (11). Modification of these residues can raise the activation threshold and lead to a better discrimination between on- and off-target activity (enhanced proofreading) and, thus, improve specificity (11).
SaCas9 is much smaller than SpCas9 (1,053 vs. 1,368 a.a.), yet it still possesses robust nuclease activity (6). Despite sharing only 17% sequence identity with SpCas9, SaCas9 recognizes the PAM-distal region of the guide RNA–target DNA in a similar manner to SpCas9 (18). Based on the enhanced proofreading mechanism of SpCas9 and the molecular dynamic similarity of SpCas9 and SaCas9, we sought to improve the targeting accuracy of SaCas9 by modifying residues in close polar contact with the backbone of the target DNA strand in the PAM-distal region. Using genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq) (7) and targeted deep sequencing, we showed that 1 engineered variant dramatically reduced off-target cleavages without compromising on-target activity.
Results
Structure-Guided Protein Engineering for High-Fidelity SaCas9.
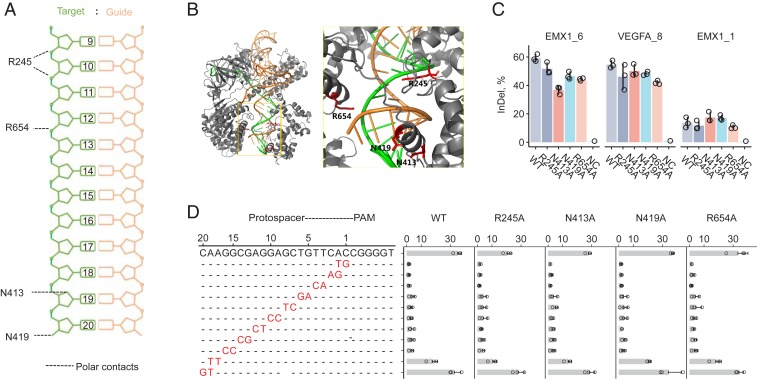
We studied the crystal structure of the SaCas9/sgRNA–target DNA complex and identified 4 amino acid residues (R245, N413, N419, and R654) forming polar contacts within a 3.0-Å distance from the target DNA strand (Fig. 1 A and B). Three of the residues were located in the recognition lobe, and the other 1 (R654) was in the RuvC-III domain. We first constructed 4 single amino acid mutants by alanine substitution and tested whether these mutants showed comparable on-target activities to those of wild-type SaCas9 of 3 human endogenous sites EMX1 site 6 (EMX1_6), VEGFA site 8 (VEGFA_8), and EMX1 site 1 (EMX1_1) using targeted deep sequencing (Fig. 1C). We chose these 3 targets because we wished to test the variants with both the canonical NNGRRT PAM [EMX1_6 and VEGFA_8 both were edited at high efficiencies (6, 8)] and a potentially targetable noncanonical NNARRT PAM (EMX1_1).
Fig. 1.
Identification and characterization of SaCas9 variants possessing single amino acid substitution at residues forming polar contacts with target DNA. (A) Schematic depicting SaCas9 residues in contact with the target DNA–guide RNA heteroduplex, labeled with protospacer positions (20 being most proximal to PAM). (B) Crystal structure of WT-SaCas9 interacting with the guide RNA–target DNA heteroduplex; close up of the active site showing those residues (red) forming polar contacts within 3.0-Å distance from the target DNA strand (green). (C) Percentage of InDel reads among all amplicon reads of targeted deep sequencing 3 human endogenous sites in HEK293T cells using WT, single mutant variants, and a no-Cas9 negative control (NC). (D) Fluorescence reduction of EGFP cells after gene editing by different SaCas9s using protospacer matched or mismatched sgRNAs (mean ± SD; n = 3).
Using targeted deep sequencing, we found that all 4 SaCas9 single mutants retained comparable on-target activities at an average 89% cleavage efficiency (range 69–122%) of WT-SaCas9 across the 3 human endogenous sites (Fig. 1C), except that the N413A variant showed moderate activity (62% of WT) in 1 target site, but still yielded a 38% insertion and/or deletion (InDel) outcome. At the noncanonical PAM NNARRT endogenous site EMX1_1, all 5 SaCas9s achieved a fair level of InDel editing (average 17%, range 12–22%), which was about 34% (range 28–39%) that of their counterparts targeting canonical PAM sites. InDel profiles introduced by the 4 single mutants were similar to that of WT-SaCas9, regarding frequencies of InDels along the spacer position (SI Appendix, Fig. S1).
We further used an enhanced green fluorescent protein (EGFP)-disruption assay to evaluate SaCas9 cleavage efficiency on expressed EGFP with fully matched and tiling 2-base mismatched guide sequences (Fig. 1D). The N413A, N419A, and R654A mutants possessed similar cleavage efficacy to that of WT-SaCas9 (range 78–105%), whereas R245A yielded 56% of the editing efficiency of WT-SaCas9. All SaCas9s tested were highly sensitive to mismatches between guide RNA and the target at PAM-proximal positions 1 through 6, relatively less sensitive at positions 7 through 18, and insensitive at positions 19 through 21. We observed no noticeable cleavage difference between WT and the 4 single-mutant SaCas9s using the mismatched guides.
Genome-Wide Targeting Specificity of Single Mutants.
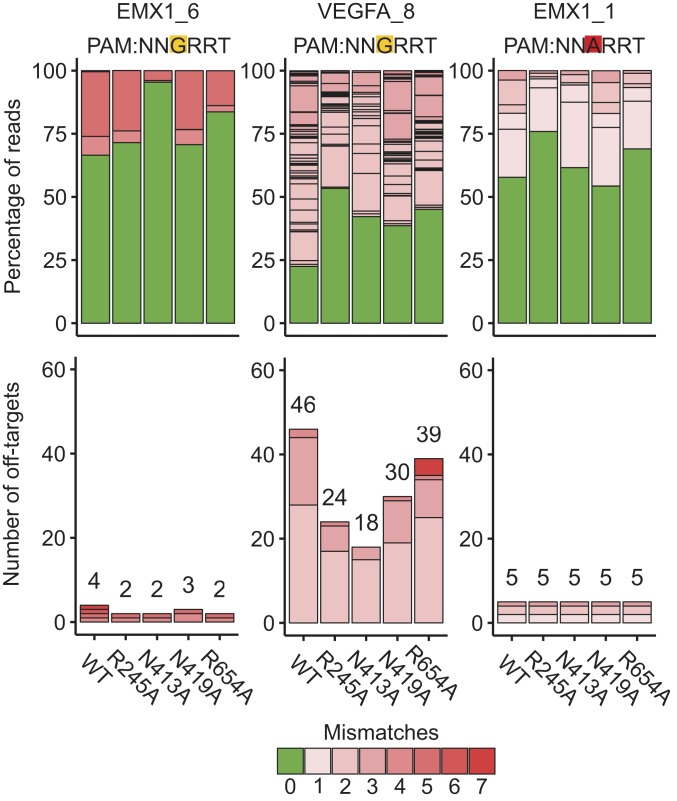
GUIDE-seq analysis showed that the 4 single mutants had improved specificity of varied levels at a canonical PAM site (EMX1_6), a known promiscuous site (VEGFA_8) and a noncanonical PAM site (EMX1_1) (Fig. 2 and SI Appendix, Fig. S2). The R245A mutant nearly halved the number of off-target sites at both of the canonical PAM sites, improved the on- to off-target read ratio and retained a comparable number of on-target GUIDE-seq reads (70%, 98%, and 84%, respectively, at the 3 sites) compared to WT-SaCas9. The other 3 single mutants exhibited improved on- to off-target ratios across the 3 sites, with varied numbers of on-target GUIDE-seq reads relative to WT-SaCas9 (17.1–216.0%). Notably, the R654A mutant appeared to possess several off-target sites containing noncanonical PAMs when targeting VEGFA_8 (SI Appendix, Fig. S2).
Fig. 2.
Genome-wide editing specificity of WT and single mutant SaCas9 variants at selected targets using GUIDE-seq. Percentage of edited reads detected by GUIDE-seq at the on-target site (green) and off-target sites (ordered by number of mismatches) among total edited reads by each SaCas9 (Top) and numbers of genome-wide off-target sites (Bottom).
Epistasis Effect of SaCas9 Residues on Targeting Specificity.
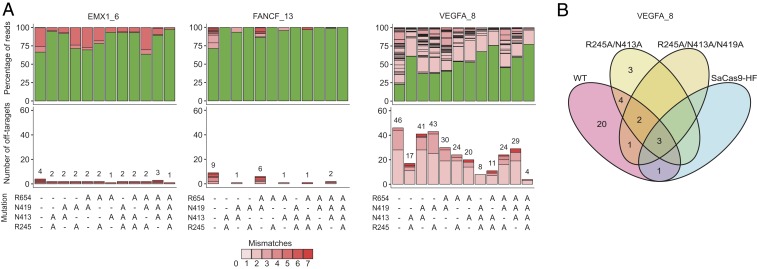
To comprehensively test for any combined effect in improving targeting specificity, we constructed all combinations of double, triple, and quadruple mutants from the 4 mutations. We performed GUIDE-seq on these multiple mutants targeting the above 2 canonical PAM sites and 1 additional site (FANCF_13) that showed the greatest off-target effects among the 5 canonical PAM sites observed previously (8). We found that mutants harboring R245A and N413A generally had lower numbers of off-target sites and higher on- to off-target ratios compared to those harboring N419A and R654A (Fig. 3A and SI Appendix, Fig. S3). The lowest off-target activity was observed for R245A/N413A among double mutants and R245A/N413A/N419 among triple mutants. Focusing on VEGFA_8, we observed that the best performing double (R245A/N413A), triple (R245A/N413A/N419A), and quadruple (hereafter referred to as SaCas9-HF) mutants shared 9, 6, and 4 off-target sites with WT-SaCas9, respectively (Fig. 3B and SI Appendix, Fig. S3). The 2 common VEGFA_8 off-target sites among all SaCas9 mutants were also among the top 5 most frequently cleaved sites by WT-SaCas9.
Fig. 3.
Genome-wide editing specificity of WT and combinatorial mutant SaCas9 variants at selected targets using GUIDE-seq. (A) Percentage of edited reads detected by GUIDE-seq at the on-target site (green) and off-target sites (ordered by number of mismatches) among total edited reads by each SaCas9 (Top), and numbers of genome-wide off-target sites (Bottom). (B) Venn diagram comparing off-target sites for WT, the best-performing double (R245A/N413A) and triple (R245A/N413A/N419A) mutant, and SaCas9-HF at the target VEGFA_8.
Genome-Wide Targeting Specificity of SaCas9-HF at Expanded Endogenous Sites.
To rigorously evaluate the genome-wide targeting specificity of SaCas9-HF, we first performed GUIDE-seq analyses on all 11 endogenous sites (6 canonical and 5 noncanonical PAMs) that were subjected to GUIDE-seq previously (8). Because the R245A mutant retained high on-target cleavage efficiency consistently across the sites tested, we also included R245A in this expanded evaluation.
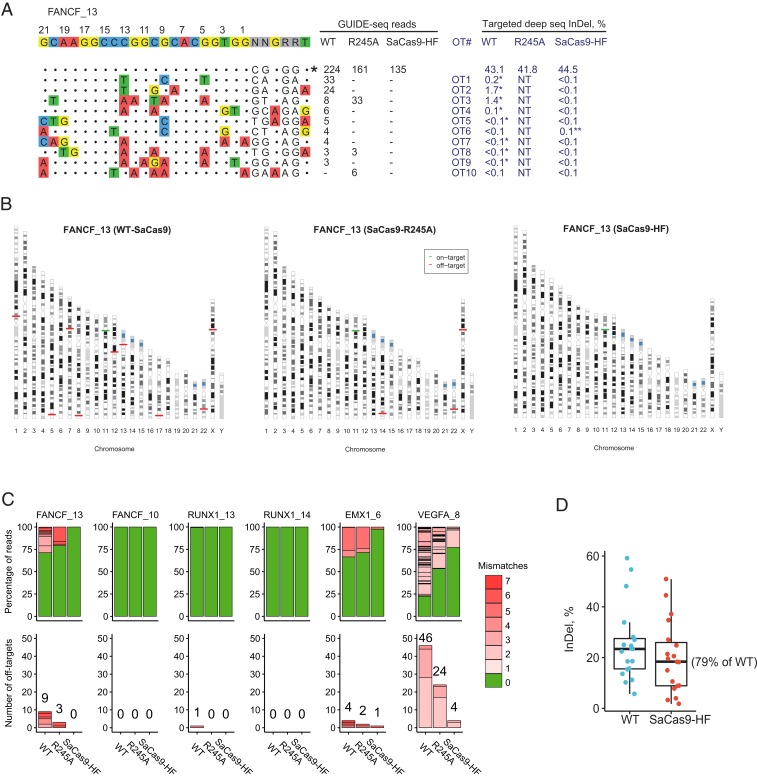
For a canonical PAM site FANCF_13, GUIDE-seq detected 9, 3, and 0 off-target sites in the WT, R245A, and SaCas9-HF-treated samples, respectively (Fig. 4 A–C). Targeted deep sequencing showed comparable on-target editing efficiencies for the 3 SaCas9s (WT 43.1%, R245A 41.8%, and SaCas9-HF 44.5%) (Fig. 4A). To validate the 10 off-target sites detected by GUIDE-seq, we performed targeted deep sequencing on WT-SaCas9 and SaCas9-HF treated samples. The top 4 off-target sites showed 0.1–1.7% InDels introduced by WT-SaCas9 and no detectable (<0.1%) InDels caused by SaCas9-HF (Fig. 4A). For the remaining 6 off-target sites, neither of the SaCas9s resulted in detectable InDels at the 0.1% cutoff level, which is the typical level of noise for next-generation sequencing (19). We thus performed manual inspection of the deep sequencing alignments using the Integrative Genomics Viewer (IGV) software (20). To facilitate visual identification, we ranked and sorted the alignments by the bases at the third and fourth positions of the protospacer, where most SaCas9-induced double strand breaks occur. Strikingly, the IGV analyses showed clear evidence of typical Cas9-edited InDels in all GUIDE-seq identified off-target sites in the WT-SaCas9 samples, including 4 InDels at 0.1–1.7% (OT1 to OT4) and 4 other InDels at <0.1% (OT5, OT7 through OT9), except 1 (OT6) at <0.1% (Fig. 4A). In contrast, for the SaCas9-HF treated samples, none of the sites had InDels, except 1 (OT6) showing a single-base G insertion in the context of GGGGG in IGV, which was likely due to the homopolymer sequencing issue of next-generation sequencing (21). Consistent with the GUIDE-seq results, the off-target site (OT10) identified only in the R245A sample showed no evidence of editing in WT-SaCas9 or SaCas9-HF samples. Among the sites FANCF_10, RUNX1_13, and RUNX1_14, almost no off-target activity was detected for the 3 SaCas9s, except that RUNX1_13 showed 1 off-target site with only 3 GUIDE-seq reads in the WT sample. This low level of editing was further confirmed by targeted deep sequencing, which showed in IGV an insertion of the 34-base dsODN introduced during GUIDE-seq.
Fig. 4.
Genome-wide editing on- and off-target activity of wild-type SaCas9, R245A, and SaCas9-HF at 6 canonical PAM sites using GUIDE-seq and targeted deep sequencing. (A) On- and off-target activity of the 3 SaCas9s at the target site FANCF_13. On-target site is indicated with “*” at the Right. Mismatched bases in off-target sites with the on-target site are colored. GUIDE-seq read counts for each SaCas9 are listed on the Right. InDel% detected in targeted deep sequencing for the on-target and off-target sites (OT1 through 10) (in dark blue) are listed. “NT” indicates off-target sites not tested using targeted deep sequencing; InDel% marked with “*” indicates edited reads confirmed by IGV visualization; “**” indicates an off-target site prone to false positives (homopolymer G) in targeted deep sequencing and thus its percentage was not calculated. (B) Ideogram presentation of on-target and off-target cleavages detected by GUIDE-seq at FANCF_13. (C) Percentage of edited reads detected by GUIDE-seq at on-target site (green) among total edited reads (Top) and the numbers of off-target sites (ordered by number of mismatches) (Bottom) for each SaCas9. (D) Percentage of InDel reads among all targeted deep sequencing reads at each of the 19 on-target sites, including 6 from C and 13 additional sites (from SI Appendix, Fig. S5), in HEK293T cells caused by WT-SaCas9 or SaCas9-HF editing.
Similarly, for the EMX1_6 and at the known promiscuous site VEGFA_8, the R245A variant rendered a moderate reduction and SaCas9-HF a dramatic reduction in off-target activities and increased on- to off-target read ratios when compared with WT-SaCas9 (Fig. 4C and SI Appendix, Fig. S4). Targeted deep sequencing confirmed 3 (OT1, OT2, and OT4) out of 4 off-target sites for EMX1_6 in the WT-SaCas9 sample and a single off-target site in the SaCas9-HF sample. Because VEGFA_8 had many off-target sites, to validate these results by targeted deep sequencing using a limited amount of materials (10 ng of DNA per off-target PCR), we selected 5 of the top 10 off-target sites randomly and another 5 with the lowest numbers of GUIDE-seq reads. One site (OT10) failed primer design and was thus not assessed. Targeted deep sequencing revealed results consistent with those of GUIDE-seq in 7 off-target sites tested (SI Appendix, Fig. S4), and the remaining 2 (OT7 and OT9) had highly repetitive sequences and showed InDels even in the control samples (from the AAV experiment described below), yet they still showed different InDel patterns among WT-SaCas9, SaCas9-HF, and the control (the latter 2 were similar).
Targeted deep sequencing evaluation for on-target efficiency at these 6 canonical PAM sites (Fig. 4A and SI Appendix, Fig. S4) showed that, relative to WT-SaCas9, the variants retained comparable on-target efficiencies (R245A: mean 97.6%, range 75–128%; SaCas9-HF: mean 80.0%, range 31–128%).
Next, to more comprehensively evaluate the genome-wide off-target activity of SaCas9-HF, we tested an additional panel of 13 targets examined in previous studies (6, 22). Seven sites (AAVS1_2, AAVS1_3, AAVS1_5, CCR5_1, EMX1_sg1, EMX1_sg5, EMX1_sg6) showed no or minimal off-target activities (1 or 2 off-target sites with a few GUIDE-seq reads) for both WT-SaCas9 and SaCas9-HF (SI Appendix, Fig. S5). However, in the remaining 6 sites with more off-target activities in WT-SaCas9 (mean off-target site number 9.8, range 3 to 18), SaCas9-HF significantly improved the off-target activities (mean off-target site number 3.0, range 1 to 5; 1-sided Mann–Whitney U test P value = 0.039). Further, targeted deep sequencing on all of the tested canonical sites (6 earlier and 13 additional) showed that SaCas9-HF had an average of 79% on-target activity compared to WT-SaCas9 (Fig. 4D).
For the 5 noncanonical PAM sites that were hypothesized to have low susceptibility to WT-SaCas9 editing, 1 to 4 off-target sites were still detected for WT-SaCas9, and this number was reduced to 0 to 2 for SaCas9-HF, further demonstrating the high fidelity of SaCas9-HF (SI Appendix, Fig. S6). Notably, all SaCas9s tested had some level of activity on EMX1_1, which contains an NNARRT PAM and a much lower level of activity on NNYRRT PAM sites.
Improved Specificity of KKH-SaCas9.
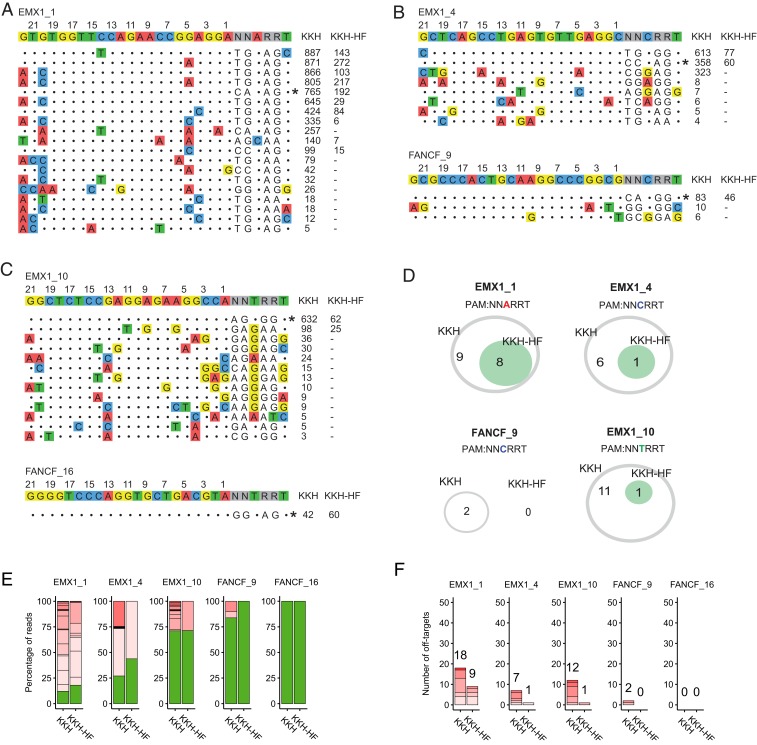
Next, we sought to test whether the SaCas9-HF mutations would bring enhanced targeting specificity to KKH-SaCas9 (8), which has broader targeting range than WT-SaCas9. We therefore constructed KKH-HF and compared its specificity with that of KKH-SaCas9 at all of the 11 endogenous target sites. We found that KKH-HF outperformed KKH-SaCas9 by profoundly reducing the number of off-target sites and increasing the on-target cleavage frequency at 4 of the 6 canonical PAM sites (SI Appendix, Fig. S7). At the 5 noncanonical PAM sites, KKH-HF dramatically reduced the number of off-target sites compared to KKH-SaCas9 (Fig. 5). While KKH-HF retained comparable on-target efficiency than KKH-SaCas9 for 2 sites (FANCF_9 and FANCF_16), it showed lower on-target activity at 3 other sites (over 50% reduction).
Fig. 5.
Genome-wide editing specificity of KKH-SaCas9 and KKH-HF at 5 noncanonical PAM sites. (A–C) GUIDE-seq detected on- and off-target sites by KKH-SaCas9 and KKH-HF when targeting 5 sites with noncanonical NNNRRT PAM. Read counts listed at Right represent number of GUIDE-seq reads. On-target site is indicated with “*”. Mismatched bases in off-target sites with the on-target site are colored. (D) Venn diagram shows the number of shared off-target sites by the 2 SaCas9s at each site. (E) Percentage of edited reads detected by GUIDE-seq at the on-target site (green) and off-target sites (ordered by number of mismatches) among total edited reads by KKH or KKH-HF. (F) The numbers of genome-wide off-target sites in E.
Effect of AAV Delivery on SaCas9-HF Activity.
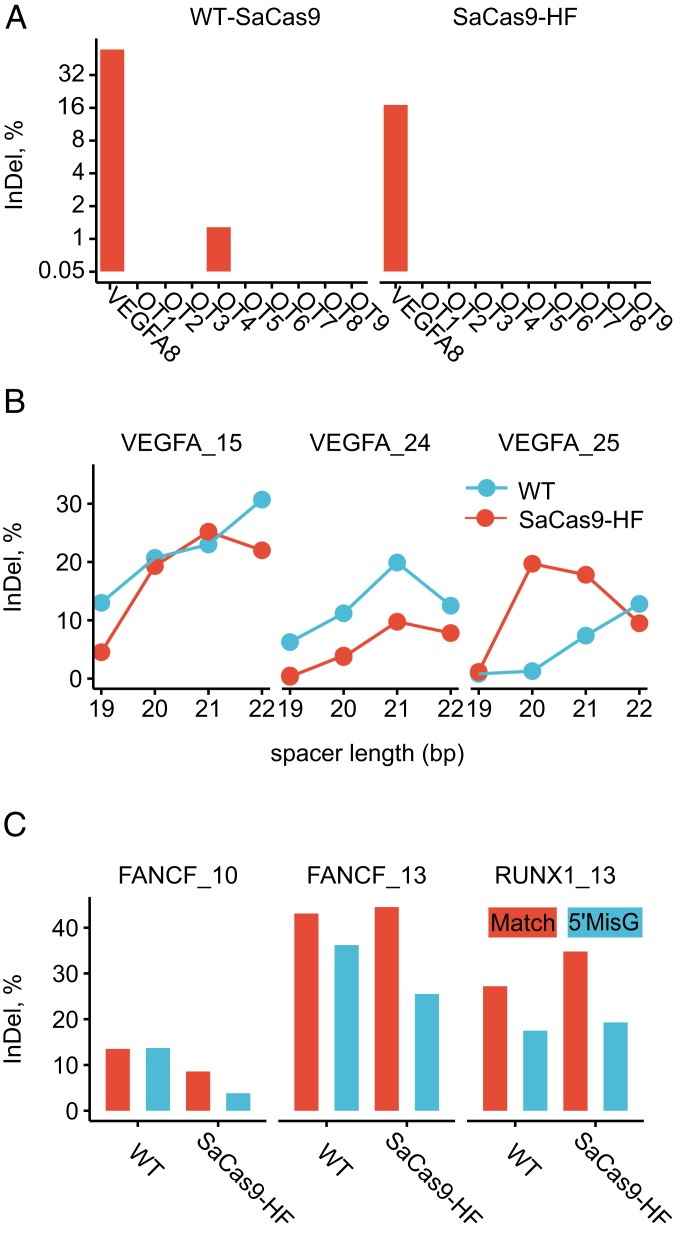
As one of the important advantages of SaCas9 is its ease of packaging into AAV for in vivo gene editing, we tested the on- and off-target activities of SaCas9 delivered by the AAV8 vector. We transduced AAV8-expressing SaCas9 and VEGFA_8-targeting sgRNAs in a human retinal pigmented epithelium cell line (ARPE19), because suppression of choroidal neovascularization by AAV-based antiangiogenic gene therapy was shown to be effective in a mouse model (23). Targeted deep sequencing showed an on-target efficiency of 50.9% in WT-SaCas9 and 18.4% (36.1% relative to WT) in SaCas9-HF (Fig. 6A). This relative on-target activity fell within the range (31–128%) of the earlier experiment. Because there are no well-examined off-target profiles for SaCas9 in ARPE cells, we assessed potential off-target activities at all 9 off-target sites tested in the previous HEK293T cells using targeted deep sequencing. Only 1 off-target site (OT4) was evident in WT-SaCas9, and none was found in SaCas9-HF, as confirmed by IGV (WT-SaCas9 showed insertions and dozens of reads with a single-base insertion at the third/fourth positions of the spacer), demonstrating improved targeting accuracy of AAV-delivered SaCas9-HF in ARPE cells.
Fig. 6.
Effect of AAV delivery, spacer length, and sgRNA design on SaCas9 performance. (A) InDel% measured by targeted deep sequencing in ARPE19 cells via AAV8 delivery at the target site VEGFA_8 and its 9 off-target sites (OT1 through 9) detected by GUIDE-seq earlier in HEK293T cells. (B) Relationship between InDel% (measured by targeted deep sequencing) and sgRNA spacer length (19–22 bp). (C) InDel% (measured by targeted deep sequencing) when perfectly matched and 5′ mismatched G sgRNAs were used.
Effect of Spacer Length and 5′ Starting Mismatched G on SaCas9-HF.
We designed sgRNAs with varying spacer length of 19 to 22 bases. The SaCas9s showed low on-target activities on all sites tested when sgRNAs with 19-base spacer were used (WT: mean 6.7%, SaCas9-HF mean 2.1%) (Fig. 6B); but comparable activities when 20 to 22 base spacers were used (WT means 11.3%, 16.7%, and 18.7%, respectively; SaCas9-HF means 14.3%, 17.6%, and 13.1%, respectively), consistent with a previous observation that SaCas9 works most efficiently with a spacer of 20 to 24 bases (22). On the other hand, the presence of a 5′ starting mismatched G in the sgRNA resulted in moderately reduced on-target activities for both WT-SaCas9 (relative to no mismatched G counterpart: mean 83.3%, range 64.3–101.5%) and SaCas9-HF (mean 52.5%, range 44.8–57.3%) (Fig. 6C) at the 3 testing sites.
Comparison with Other High-Fidelity Sa- and Sp-Cas9 Variants.
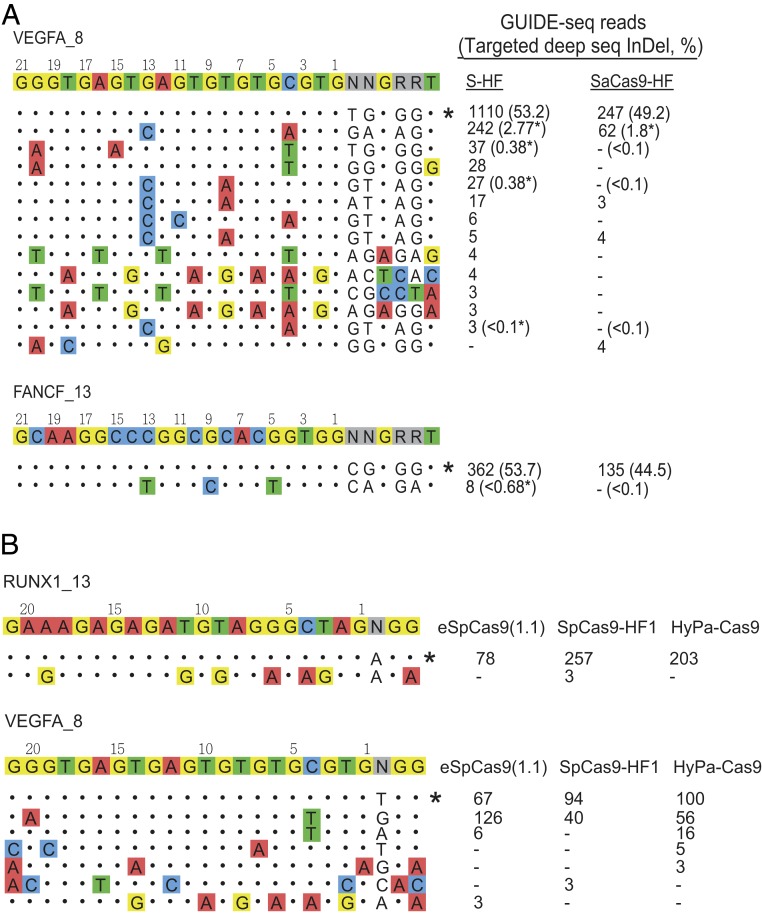
Slaymaker et al. identified a high-fidelity SaCas9 variant previously (R499A/Q500A/R654A/G655A, here referred to as S-HF) (10), but its genome-wide activities have not been reported. We thus compared S-HF with SaCas9-HF on 2 sites (VEGFA_8 and FANCF_13) having the highest off-target activities in WT-SaCas9 as characterized by both the previous (8) and current studies. For VEGFA_8, we detected 12 and 4 off-target sites in S-HF and SaCas9-HF, respectively (Fig. 7A). Interestingly, 4 of the S-HF off-target sites bear noncanonical PAMs, similar to our observation in the R654A single mutant (SI Appendix, Fig. S2). For FANCF_13, S-HF showed 1 off-target site, whereas SaCas9-HF showed no detectable off-target reads. We performed targeted deep sequencing on these S-HF off-target sites (the only site in FANCF_13 and selected 4 off-target sites that range from the top to the very bottom of the 12 off-target sites ordered by GUIDE-seq read count in VEGFA_8). Consistent with GUIDE-seq, the targeted deep sequencing and IGV analysis showed clear evidence of editing on all of the 5 sites tested (Fig. 7A). Further, targeted deep sequencing revealed comparable on-target activities of S-HF and SaCas9-HF at these 2 sites (S-HF mean 53.5%; SaCas9-HF mean 46.9%).
Fig. 7.
Comparisons of high fidelity Sa- and Sp-Cas9 variants. (A) On- and off-target sites detected by GUIDE-seq when targeting VEGFA_8 and FANCF_13 by S-HF and SaCas9-HF. On-target site is indicated with “*”. Mismatched bases in off-target sites with the on-target site are colored. GUIDE-seq read counts (percentage of InDel reads measured by targeted deep sequencing) for each SaCas9 are listed on the Right. InDel% for the on-target and off-target sites were measured by targeted deep sequencing. InDel% marked with “*” indicates edited reads confirmed by IGV visualization. (B) On- and off-target sites detected by GUIDE-seq when targeting RUNX1_13 and VEGFA_8 by eSpCas9(1.1), SpCas9-HF1, and HyPa-Cas9.
We further compared the performance of the high-fidelity variants of the 2 Cas9 orthologs (Sp- and Sa-Cas9) on mutually permissive PAM sites. When targeting RUNX1_13, none of the Cas9 variants showed detectable off-targets, except that SpCas9-HF1 showed minimal activity on 1 off-target site (Fig. 7B). For the VEGFA_8 target, high-fidelity SpCas9 variants consistently showed the same off-target site and 1 to 3 other low off-target activity sites, whereas SaCas9-HF showed a different off-target site and 3 other minimal off-target activity sites, suggesting comparable specificities among the 4 high-fidelity variants. The on- to off-target read ratios were highest for SaCas9-HF, followed by the 3 SpCas9 variants, and lowest in WT-SaCas9 (SI Appendix, Fig. S8).
Discussion
We have engineered a CRISPR Cas9 variant from Staphylococcus aureus (SaCas9-HF) that shows high genome-wide targeting accuracy without compromising on-target efficiency, as validated with rigorous evaluation of its on- and off-target activities across 24 endogenous sites.
The results of targeted deep sequencing combined with IGV inspection of InDels in a number of sites down to and below 0.1% provide compelling evidence that the off-target sites we identified using GUIDE-seq are bona fide target sites of WT-SaCas9. Theoretically, every 10 ng of DNA contains only 3 copies of mutant fragments when the mutation rate is 0.1%. Failure to confirm 1 out of 9 sites in FANCF_13 and 1 out of 4 sites in EMX1_6 by targeted deep sequencing might be due to undersampling of DNA fragments when the absolute copy of InDel fragments in the input DNA approaches zero. When InDel% is at the boundary of detection limit of a detection method, some of these off-target sites may not be detected in all experimental replicates. Since GUIDE-seq has a detection limit around 0.1% (7), a generally more sensitive method that can detect a large number of InDels below 0.1% such as CIRCLE-seq (24) would be helpful for ultrasensitive detection of Cas9 off-targets.
Because Cas9 activity is cell-type-specific owing in part to genomic locus accessibility and the integrity of double-stranded break repair pathways in particular cell type, the results from the ARPE cells lend support to the hypothesis that SaCas9-HF is highly precise in different cell lines. However, future studies on additional target sites and in additional cell types are needed to confirm comparable on-target efficiency and reduced off-target activity via AAV delivery and in more cell types.
SaCas9-HF shares the same mutation R654A with the enhanced specificity S-HF (10). S-HF contains 4 engineered residues that could weaken nonspecific SaCas9-DNA interaction and have shown dramatic activity reduction at 3 off-target sites known a priori. As in the scenario for SpCas9 shown previously, a simple combination of SpCas9-HF and eSpCas9 resulted in greatly impaired SpCas9 activity (11); thus we did not combine those S-HF mutations in our SaCas9-HF. Nonetheless, the R654 residue initially reported by S-HF is located in the RuvC-III domain. Interestingly, we found that both the R654A single mutant and S-HF led to some off-target activities on sites containing noncanonical PAMs. However, those off-target sites were not observed in SaCas9-HF, which shares the R654A mutation, and might be due to improved specificity imposed by 3 other mutations specific to SaCas9-HF. Similarly as Hypa-Cas9 (11), the best-performing triple mutant (R245A/N413A/N417A) has all engineered sites located in the recognition lobe domain of the Cas9 protein.
Reporter assays based on fluorescent protein gene disruption revealed that the WT-SaCas9 recognizes a NNGRRT PAM, with the third PAM position nucleotide showing low-level targetable T (6) or A (nearly 20% of G) (8) nucleotides, whereas another reporter assay showed a strict requirement for G at the third position and a complete absence of SaCas9 activity on non-G nucleotides (25). In contrast, in the human endogenous NNARRT PAM site we tested, SaCas9 can induce a fair level of cleavage (12–22% InDel).
The improved specificity of SaCas9-HF variants pertains to engineered KKH-HF over KKH-SaCas9, which has a broader PAM recognition range (NNNRRT) (8). However, a simple combination of high-fidelity mutations with PAM-broadening mutations might lead to “overengineering” as we observed occasionally reduced on-target activity of KKH-HF. Our results indicate that SaCas9-HF has the same tolerance for spacer length and similar restrictiveness on a 5′ starting mismatched G sgRNA as WT-SaCas9. Future studies employing combinatorial approaches to screen for a large number of protein mutations en masse, such as the CombiSEAL (26), would facilitate the development of SaCas9 variants with desired features.
Materials and Methods
Genome-wide off-targets of Cas9 editing were identified using GUIDE-seq (7) with minor modifications, including a redesign of the original half-functional adaptors (27) and placed sample index (index 2) at the head of read 1, following unique molecular index (SI Appendix, Methods). ARPE-19 cells expressing WT-SaCas9 or SaCas9-HF and VEGFA_8 sgRNA were transduced with AAV8 vectors.
Data.
Sequencing data are deposited under the European Nucleotide Archive (PRJEB31487).
Supplementary Material
Acknowledgments
We thank financial supports from Ming Wai Lau Centre of Reparative Medicine of Karolinska Institutet (Lau grant), City University of Hong Kong (internal grant), the National Natural Science Foundation of China (grant 81672098 to Z.Z. and 81770099 to J.S.), The Swedish Research Council (2016-02830 to Z.Z.), the Innovation and Technology Fund of Hong Kong Government (9440153 to Z.Z.), the Hong Kong Health and Medical Research Fund (05160296 to J.S.), the Hong Kong Research Grants Council (21101218 to J.S.), Shenzhen Science and Technology Innovation Fund (JCYJ20170413115637100 and JCYJ20170412152916724 to J.S.), and Sanming Project of Medicine in Shenzhen (SZSM201811092 to J.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Sequencing data reported in this article have been deposited in the European Nucleotide Archive (accession no. PRJEB31487).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906843116/-/DCSupplemental.
References
- 1.Jinek M., et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu P. D., Lander E. S., Zhang F., Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna J. A., Charpentier E., Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Maeder M. L., et al. , Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 25, 229–233 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Fu Y., et al. , High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran F. A., et al. , In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai S. Q., et al. , GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinstiver B. P., et al. , Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinstiver B. P., et al. , High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slaymaker I. M., et al. , Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J. S., et al. , Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casini A., et al. , A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 36, 265–271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J. K., et al. , Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 9, 3048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J. H., et al. , Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternberg S. H., LaFrance B., Kaplan M., Doudna J. A., Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palermo G., Miao Y., Walker R. C., Jinek M., McCammon J. A., CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc. Natl. Acad. Sci. U.S.A. 114, 7260–7265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dagdas Y. S., Chen J. S., Sternberg S. H., Doudna J. A., Yildiz A., A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 3, eaao0027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimasu H., et al. , Crystal structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross M. G., et al. , Characterizing and measuring bias in sequence data. Genome Biol. 14, R51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson J. T., et al. , Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minoche A. E., Dohm J. C., Himmelbauer H., Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 12, R112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedland A. E., et al. , Characterization of Staphylococcus aureus Cas9: A smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 16, 257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Askou A. L., et al. , Suppression of choroidal neovascularization by AAV-based dual-acting antiangiogenic gene therapy. Mol. Ther. Nucleic Acids 16, 38–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai S. Q., et al. , CIRCLE-seq: A highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie H., et al. , SaCas9 requires 5′-NNGRRT-3′ PAM for sufficient cleavage and possesses higher cleavage activity than SpCas9 or FnCpf1 in human cells. Biotechnol. J. 13, 1700561 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Choi G. C. G., et al. , Combinatorial mutagenesis en masse optimizes the genome editing activities of SpCas9. Nat. Methods 16, 722–730 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Zheng Z., et al. , Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 20, 1479–1484 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.