Significance
Pesticides and insecticides are crucial for agricultural productivity, global food security, and control of disease vectors. However, resistance to insecticides is a widespread and urgent problem, which leads to increased insecticide usage with dire consequences to the environment. A common resistance mechanism against 2 of the most widely used insecticide classes, organophosphates and carbamates, involves carboxylesterases. We applied covalent virtual screening to discover potent inhibitors for a common, resistance-mediating carboxylesterase. The compounds reversed insecticide resistance in 2 different pest species, representing a sustainable strategy that allows significant reduction of insecticide use without compromising their efficacy. Such synergists could have major economic and environmental benefits, and the approach demonstrated in this work should be applicable to additional resistance mechanisms.
Keywords: covalent docking, insecticide resistance, organophosphates, carboxylesterase, Lucilia cuprina
Abstract
Insecticides allow control of agricultural pests and disease vectors and are vital for global food security and health. The evolution of resistance to insecticides, such as organophosphates (OPs), is a serious and growing concern. OP resistance often involves sequestration or hydrolysis of OPs by carboxylesterases. Inhibiting carboxylesterases could, therefore, restore the effectiveness of OPs for which resistance has evolved. Here, we use covalent virtual screening to produce nano-/picomolar boronic acid inhibitors of the carboxylesterase αE7 from the agricultural pest Lucilia cuprina as well as a common Gly137Asp αE7 mutant that confers OP resistance. These inhibitors, with high selectivity against human acetylcholinesterase and low to no toxicity in human cells and in mice, act synergistically with the OPs diazinon and malathion to reduce the amount of OP required to kill L. cuprina by up to 16-fold and abolish resistance. The compounds exhibit broad utility in significantly potentiating another OP, chlorpyrifos, against the common pest, the peach–potato aphid (Myzus persicae). These compounds represent a solution to OP resistance as well as to environmental concerns regarding overuse of OPs, allowing significant reduction of use without compromising efficacy.
As the world population increases, agricultural productivity is essential for sustaining food security. This has been facilitated through the use of pesticides and insecticides for protecting crops and livestock (1). In terms of global health, insecticides are the first line of defense against many infectious diseases; they are especially important in developing countries, where insect vectors are responsible for nearly 20% of all infectious diseases (2). Insecticide-infused nets and residual spraying of dwellings are among the most effective means to control the spread of these diseases (3). However, the widespread use of insecticides has effectively selected for insects that are resistant to the toxic effects (4). Insecticide resistance is widespread and is an urgent global problem. Since the 1940s, the number of insect species with reported insecticide resistance has been rapidly increasing and recently passed 580 species (5). Resistance renders insecticides ineffective and leads to increased usage, with significant consequences to nontarget species and harm to agricultural workers (5, 6).
Organophosphates (OPs) and carbamates are two of the most widely used classes of insecticides (7). They inhibit acetylcholinesterase (AChE) at cholinergic neuromuscular junctions by phosphorylating/carbamylating the active site serine nucleophile (8). This leads to interminable nerve signal transduction and death (9). Resistance to OPs and carbamates has been documented in many insect species (4), with the most common mechanism of resistance involving carboxylesterases (CEs) (10). Resistance-associated CEs are either overexpressed to sequester insecticides (11) or mutated to gain a new hydrolase function (12, 13); both mechanisms allow CEs to intercept insecticides before they reach their target, AChE. The sheep blowfly Lucilia cuprina has become a model system for the study of insecticide resistance: resistance was first documented in 1966 (14), which was found to result from a Gly137Asp mutation in the gene encoding the αE7 CE (15). This resistance allele now dominates blowfly populations (16), and the equivalent mutation has been observed in a range of other OP-resistant fly species (13, 17). Recent work has shown that the wild-type (WT) αE7 protein also has some protective effect against OPs through its ability to sequester the pesticides (18).
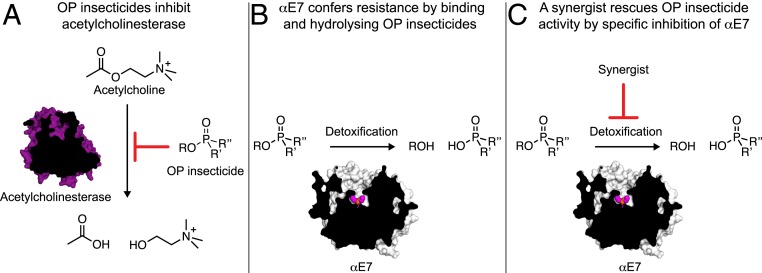
Recent attempts to overcome insecticide resistance have focused on the development of new insecticides with novel modes of action (19, 20). Although many of these new targets show promise, there are finite numbers of biochemical targets, and new targets are not immune to the problems of target site insensitivity and metabolic resistance. Synergists have been used in the past to enhance the efficacy of insecticides by inhibition of enzymes involved in insecticide detoxification; a prominent example of one of the few established insecticide synergists is piperonyl butoxide, a nonspecific inhibitor of cytochrome P450s, which is used to enhance the activity of carbamates and pyrethroids (21, 22). The idea of synergists can be taken further to specifically target enzymes that have evolved to confer metabolic resistance, thereby restoring the efficacy of the insecticide to preresistance levels. Thus, CEs such as αE7, being a relatively well-understood detoxification system, are ideal targets for the design of inhibitors to abolish insecticide resistance (Fig. 1).
Fig. 1.
Overview of synergists for OP insecticides. (A) OP insecticides inhibit AChE and prevent the hydrolysis of acetylcholine. (B) CEs, like αE7, rescue AChE activity by binding and hydrolyzing OP insecticides. (C) An inhibitor that outcompetes OPs for binding to CEs could act as a synergist to restore insecticide activity.
Here, we report the computational design of potent and selective covalent inhibitors of αE7 using DOCKovalent. DOCKovalent is a general method for screening large virtual libraries for the discovery of specific covalent inhibitors (23, 24). An initial screen of ∼23,000 boronic acids against the crystal structure of αE7 (25) identified picomolar to nanomolar inhibitors of WT αE7. Improving our understanding of the structure–activity relationships underlying the interaction enabled inhibitor optimization, resulting in potent inhibitors of both WT and the resistance-associated Gly137Asp enzymes. Bioassays of blowfly survival confirmed that the optimized inhibitors synergized with OP insecticides and abolished resistance. Their potential for broad-spectrum use was demonstrated by the ability of the inhibitors to synergize OPs against the peach–potato aphid Myzus persicae. The compounds were highly selective against human AChE and did not appear toxic when administered to mice. They can overcome resistance to cheap and available insecticides while lowering the overall amount of insecticide required by more than an order of magnitude. Such synergists could have major economic and environmental benefits, and the general approach demonstrated in this work should be applicable to additional CEs as a route to fight insecticide resistance.
Results
Virtual Screen of Boronic Acids Against LcαE7.
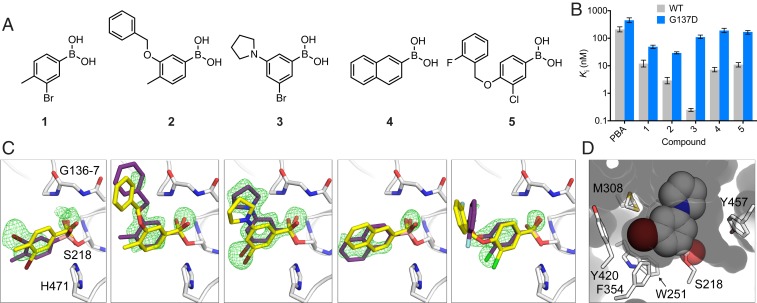
LcαE7 catalyzes the hydrolysis of fatty acid substrates via the canonical serine hydrolase mechanism (18, 25). Boronic acids are known to form reversible covalent adducts with the catalytic serine of serine hydrolases, which mimic the geometry of the transition state for carboxylester hydrolysis and therefore, bind with high affinity (26). We used DOCKovalent, an algorithm for screening covalent inhibitors, to screen a library of ∼23,000 boronic acids against the crystal structure of LcαE7 (Protein Data Bank [PDB] ID code 4FNG). Each boronic acid was modeled as a tetrahedral species covalently attached to the catalytic serine (Ser218) (SI Appendix, Fig. S1). After applying the covalent docking protocol, the top 2% of the ranked library was manually examined, and 5 compounds ranked between 8 and 478 were selected for testing (Fig. 2A).
Fig. 2.
Covalent docking predicts potent inhibitors of LcαE7. (A) Chemical structures of predicted LcαE7 inhibitors. Compounds 1 to 5 were ranked 8th, 169th, 202nd, 210th, and 478th in the DOCKovalent screen. (B) In vitro Ki inhibition constants for WT and Gly137Asp LcαE7 with PBA and compounds 1 to 5. Data are presented as ±95% confidence intervals for 3 repeat measurements of enzyme activity at each concentration of compound. (C) Compounds 1 to 5 (purple sticks) form covalent adducts with the catalytic serine of LcαE7 (Ser218). The omit mFO-DFC difference electron density is shown (green mesh contoured at 3 σ). The docking predictions (yellow sticks) are overlaid onto the corresponding cocrystal structures. Active site residues are shown as white sticks. (D) Surface view of LcαE7 active site, with compound 3 shown as spheres.
Potent Inhibitors of WT LcαE7.
The potency of the boronic acids was determined by enzymatic assays of recombinant LcαE7 with the model carboxylester substrate 4-nitrophenol butyrate (4-NPB). All 5 boronic acids exhibited Ki values lower than 12 nM (Fig. 2B and SI Appendix, Fig. S2), with the most potent compound (3) exhibiting a Ki value of 250 pM. While the 5 compounds are diverse, they all share a phenylboronic acid (PBA) substructure. PBA inhibits LcαE7 with a Ki value ∼2 to 3 orders of magnitude higher than the designed compounds (210 nM) (Fig. 2B).
Crystallography Validates Docking Pose Predictions.
We solved the cocrystal structures of compounds 1 to 5 with LcαE7 to assess the binding poses predicted by DOCKovalent (Fig. 2C and SI Appendix, Table S1). Difference electron density maps show the boronic acid compounds covalently bound to the catalytic serine (Fig. 2C). The fit of the boronic acids to the active site varies with substitution pattern; the 3,5-disubstitution of compound 3 is highly complementary, while the 3,4-disubstitution of the remaining compounds results in a suboptimal fit (Fig. 2D and SI Appendix, Fig. S3). The cocrystal structure of compound 3 reveals an unexpected trigonal planar adduct rather than the typical tetrahedral adduct (27) (SI Appendix, Fig. S4).
Comparison between the various cocrystal structures and the docked poses largely validates the DOCKovalent predictions (Fig. 2C). Compounds 3 and 5 were accurately predicted with rmsd values of only 1.11 and 1.61 Å, respectively. Docking predicted a flipped orientation of compound 1 with respect to the bromine substituent (rmsd 2.04 Å). For compound 2, the prediction of the benzyloxy substituent was not accurate (rmsd 3.01 Å). Finally, the naphthalene ring of 4 was flipped relative to the docking prediction (rmsd 2.02 Å), which required a change in conformation of Met308 (SI Appendix, Fig. S3).
Potent Inhibitors of Gly137Asp LcαE7.
The 2 most common CE-mediated insecticide resistance mechanisms involve increased protein expression or mutation to gain new catalytic (OP-hydrolase) functions. The Gly137Asp mutation is located in the oxyanion hole and positions a new general base to catalyze dephosphorylation of the catalytic serine (15, 28). Thus, compounds that inhibit WT LcαE7 as well as this common resistance-associated variant would increase the efficacy of OPs by targeting both detoxification routes. Encouraged by the activity of the boronic acids 1 to 5, we tested the compounds against the Gly137Asp variant of LcαE7 (Fig. 2B and SI Appendix, Fig. S5). The most potent compound was 2, exhibiting a Ki of 29 nM. The higher affinity of compound 2 compared with 3 (29 vs. 110 nM) may be due to increased steric interaction between the rigid pyrrolidinyl substituent of compound 3 compared with the flexible benzyloxy substituent of compound 2. Indeed, the decreased affinity of all compounds for Gly137Asp LcαE7 suggests that the Asp137 side chain impedes binding. This is consistent with the higher affinity of both OP and carboxylester substrates for WT LcαE7 relative to Gly137Asp LcαE7 (28).
Optimization of Gly137Asp LcαE7 Inhibition.
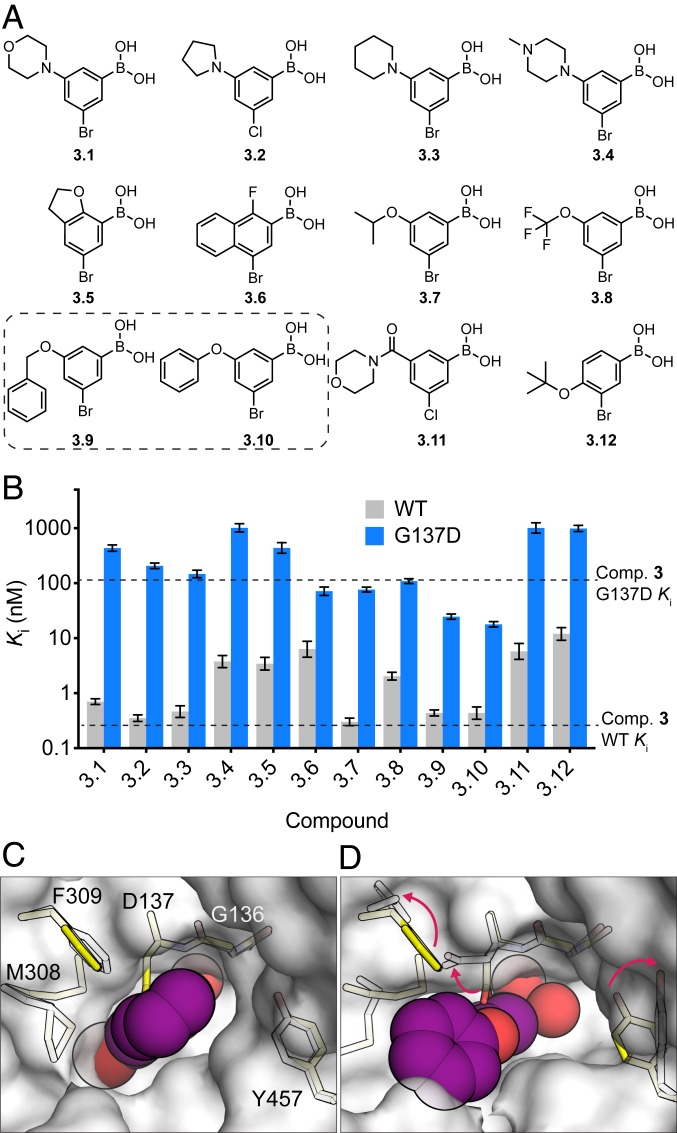
To improve Gly137Asp inhibition while maintaining good WT potency, we focused on elaborating compound 3, the most potent WT inhibitor. We purchased 12 commercially available analogs of 3-bromo-PBA (Fig. 3A). These compounds were chosen because they share the 3,5-disubsitution and include small and/or flexible substituents at the 5 position. We determined their Ki values for WT and Gly137Asp LcαE7, and while we did not find a more potent inhibitor of WT LcαE7, 6 of the 12 analogs exhibited picomolar Ki values (Fig. 3B and SI Appendix, Figs. S2 and S5). This establishes a stable structure–activity relationship between the 3,5-disubstituted PBA and high-affinity WT LcαE7 binding. Importantly, analogs 3.9 and 3.10, which possess the 3,5-disubstitution pattern and a benzyloxy or phenoxy substituent, exhibited 4.4- and 6.1-fold improvements, respectively, in inhibition of Gly137Asp LcαE7 compared with compound 3.
Fig. 3.
Second generation boronic acids are potent inhibitors of Gly137Asp LcαE7. (A) Chemical structures of compound 3 analogs. Compounds 3.9 and 3.10, which combine the disubstitution pattern of compound 3 and the relatively large flexible substituent of compound 2, are highlighted with a dashed box. (B) In vitro Ki inhibition constants for WT and Gly137Asp LcαE7 with compounds 3.1 to 3.12. Data are presented as ±95% confidence intervals for 3 repeat measurements of enzyme activity at each concentration of compound. (C) Surface view of the cocrystal structure of compound 3 with WT LcαE7 (white surface and sticks) overlaid with the apo Gly137Asp LcαE7 structure (yellow sticks). (D) The active site is rearranged in the cocrystal structure of compound 3.10 with Gly137Asp LcαE7 (white sticks and surface). The boronic acid compounds are shown as purple spheres.
To investigate the binding of the most potent inhibitor of Gly137Asp LcαE7, compound 3.10, we determined its cocrystal structure to 1.75 Å (SI Appendix, Fig. S6 and Table S1). The orientation of 3.10 is conserved compared with 3, with the 3-bromo substituent highly complementary to the active site and the 5-phenoxy substituent orientated toward the active site funnel (SI Appendix, Fig. S6). The active site has undergone a reorganization, with the Asp137, Phe309, and Tyr457 side chains adopting alternative conformations compared with the apo enzyme (Fig. 3C). The reorganization allowed the Asp137 side chain to avoid a steric clash with the phenoxy substituent. Surprisingly, compound 3.10 adopts a tetrahedral geometry rather than the trigonal planar geometry observed for compound 3 (SI Appendix, Fig. S6).
Selectivity Against Other L. cuprina Serine Hydrolases.
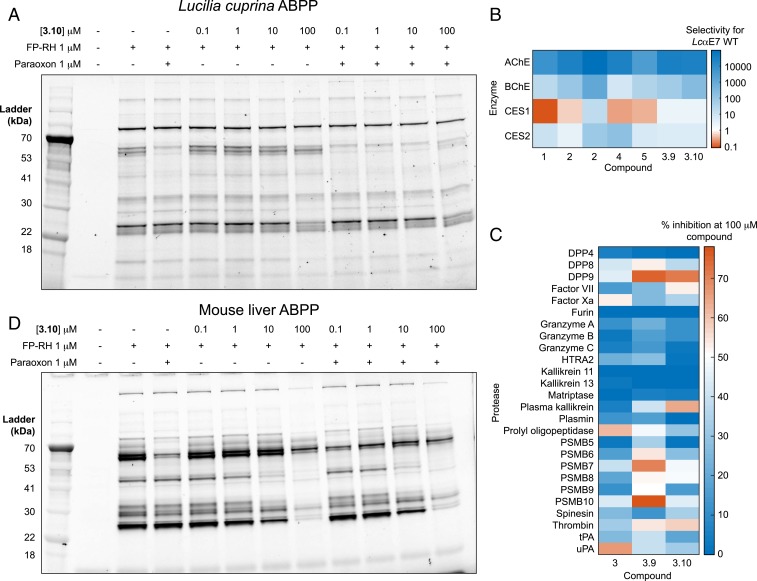
To explore the selectivity of the boronic acid compounds against other L. cuprina serine hydrolases, we performed an activity-based protein profiling (ABPP) experiment using a fluorophosphonate-rhodamine (FP-RH) probe (29). Lysate derived from larvae of the susceptible L. cuprina strain was pretreated with compound 3.10 in the presence or absence of paraoxon followed by incubation with the FP-RH probe and visualization by SDS-PAGE (Fig. 4A and SI Appendix, Fig. S7). Paraoxon was used in place of diazinon to eliminate the need for the OP to be activated by cytochrome P450s (30). In this ABPP experiment, a decrease in band intensity in a sample pretreated with paraoxon and/or boronic acid indicates that a serine hydrolase is being inhibited. In the sample pretreated with paraoxon, there was a substantial decrease in the intensity of several bands, including the elimination of a band between 60 and 70 kDa, which we tentatively assign to LcAChE (68 kDa [31]). In samples pretreated with compound 3.10, there was no significant serine hydrolase inhibition until the concentration of compound reached 100 μM. Although we failed to detect inhibition of a serine hydrolase with a molecular mass consistent with LcαE7 (66 kDa), this may be because the relatively low abundance of LcαE7 precludes detection (0.05% of total soluble protein of adult flies [32]). When lysate was pretreated with both compound 3.10 and paraoxon, the labeling profile was similar to the sample treated only with paraoxon (Fig. 4A). Overall, these results indicate that compound 3.10 is selective for LcαE7 over other L. cuprina serine hydrolases.
Fig. 4.
Boronic acid compounds are highly selective against L. cuprina, mouse, and human enzymes. (A) In-gel fluorescence ABPP of serine hydrolases in L. cuprina using an FP-RH probe. (B) Heat map showing compound selectivity against human AChE, BChE, CES1, and CES2. Selectivity was calculated as the ratio of Ki inhibition constants between WT LcαE7 and each of the human enzymes. (C) Heat map showing inhibition of 26 human serine and threonine proteases by compounds 3, 3.9, and 3.10. Percentage inhibition was determined at a single compound concentration (100 μM) in duplicate. SI Appendix, Table S3 shows assay conditions. (D) In-gel ABPP of serine hydrolases in mouse liver using an FP-RH probe.
Selectivity Against Human Hydrolases.
To characterize selectivity against human cholinesterases, compounds 1 to 5, 3.9, and 3.10 were assayed against human AChE and the related blood plasma enzyme butyrylcholinesterase (BChE) (Fig. 4B and SI Appendix, Fig. S8 and Table S2). The most promising compounds with respect to LcαE7 inhibition (3, 3.9, and 3.10) were at least 20,000-fold selective for WT LcαE7 over AChE. The compounds were less selective against BChE, with 1,000-, 60-, and 200-fold selectivity for compounds 3, 3.9, and 3.10, respectively. We also tested the compounds against human carboxylesterase 1 (CES1) and CES2 (Fig. 4B and SI Appendix, Fig. S9 and Table S2). These enzymes are responsible for the majority of hydrolase activity in the liver and small intestine, respectively, and their substrates include various xenobiotics and endogenous compounds (33). Compounds 3, 3.9, and 3.10 were potent inhibitors of CES1 with Ki values in the range of 1 to 4 nM (SI Appendix, Table S2). Moderate selectivity was observed against CES2, with Ki values in the range of 10 to 1,100 nM (Fig. 4B). Notably, the Ki values for compounds 3, 3.9, and 3.10 for CES1 and CES2 are an order of magnitude less than the published concentration of compound required to reduce enzyme activity to 50% (IC50) values for widely used and irreversible OPs (e.g., chlorpyrifos; 0.15 and 0.33 nM for CES1 and CES2, respectively) (34).
To explore the selectivity of the boronic acid compounds against other human serine hydrolases, we repeated the ABPP experiment using human embryonic kidney (HEK)HEK293 cell lysate (29) (SI Appendix, Fig. S10). We quantified the fluorescent labeling of 8 bands and found that none were diminished by more than 50% when the HEK293 cell lysate was pretreated with 100 nM of any of the 4 compounds tested (3, 3.7, 3.9, and 3.10). We also probed the selectivity of compounds 3, 3.9, and 3.10 against a panel of 26 human serine and threonine proteases (Fig. 4C and SI Appendix, Tables S3 and S4). At a 100 μM concentration of the compounds, 14 of 26 proteases retained at least 50% activity, and all proteases retained at least 20% activity. The higher concentration of compounds used in the ABPP experiment (400-fold) and protease panel (10,000-fold) relative to the Ki value of compound 3 for WT LcαE7 indicates that the compounds are highly selective for LcαE7 over these enzymes.
Mammalian Ex Vivo and In Vivo Toxicity Tests.
Although the compounds demonstrate high selectivity against human AChE and a protease panel (Fig. 4), the inhibition of CES1 and CES2 indicates that off-target interactions are possible. We extended the analysis of off-target toxicity by testing the toxicity of compounds 1 to 5, 3.9, and 3.10 against 9 human cell lines (SI Appendix, Figs. S11–S13). Toxicity was assessed by the concentration of compound required to reduce cell viability by 50% (EC50). The compounds were generally nontoxic except against the HeLa cell line, where EC50 values were between 2 and 38 μM. Compound 2 was most toxic, with EC50 values less than 50 μM for 5 of 9 cell lines tested.
We next examined the general toxicity of the boronic acid compounds using a model mammalian system of the C57BL/6 mouse. To assess mammalian toxicity, we evaluated the effects of the compounds on mice in an acute toxicity model, in which we administered the compounds at a very high dose of 300 mg/kg via oral gavage. Mice were monitored for 2 wk following administration, at which point surviving mice were killed and autopsied for assessment of internal organ damage. We tested compounds 3, 4, 3.7, 3.9, and 3.10 against 3 to 4 mice each. All mice survived and showed no clinical indication of toxicity. This demonstrates very good tolerance for the phenyl boronic acids as a general class. For context, the mouse LD50 (half maximal lethal concentration) of the OP chlorpyrifos is 62 mg/kg (35), and the oral LD50 for Donepezil, an AChE inhibitor used clinically, is 45 mg/kg (36). We repeated the ABPP experiment with mouse liver, kidney, and spleen (Fig. 4D and SI Appendix, Fig. S14). CEs are highly expressed in the liver and include a CE that is 78% identical to human CES1 (37). Two intense bands at ∼60 kDa were tentatively assigned as CEs. These hydrolases were inhibited when the mouse lysate was pretreated with 1 μM paraoxon; however, compound 3.10 only inhibited these hydrolases at 100 μM (Fig. 4D). Therefore, although compound 3.10 inhibits human CEs with low nanomolar affinity (SI Appendix, Table S2), high concentrations are required to block FP-RH labeling in mouse liver. This might be attributable to the difference between the enzymes or to the ABPP conditions. Overall, these results show that compounds from this class are relatively benign to mammals at high doses and are suitable for field use.
LcαE7 Inhibitors Synergistically Enhance Insecticides against Blowfly Larvae.
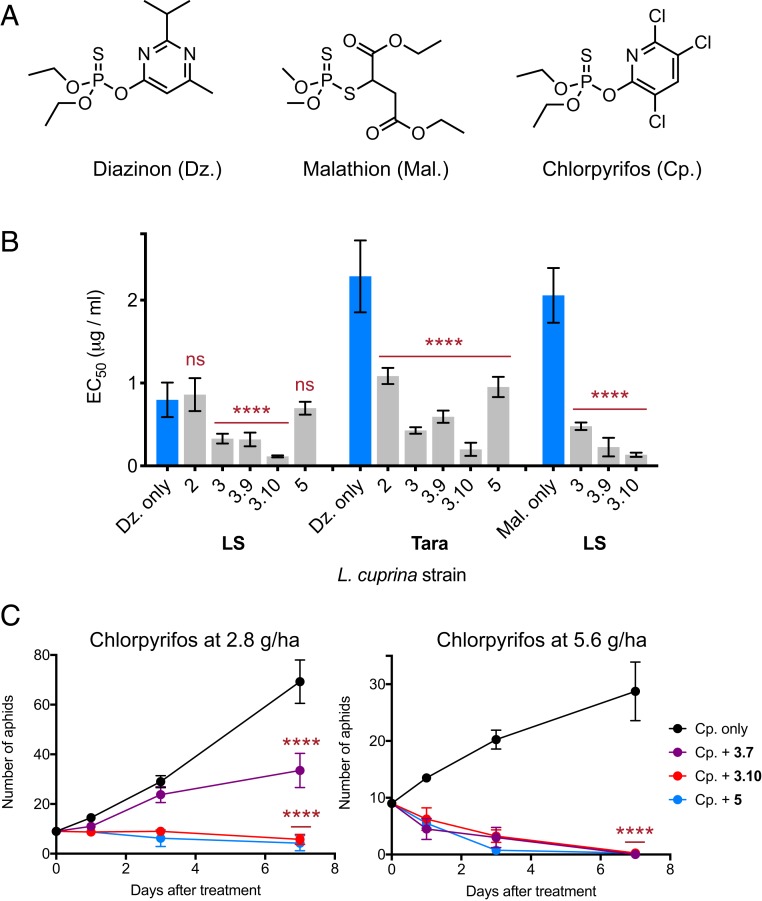
We then investigated whether the boronic acid compounds could act as synergists to restore the effectiveness of OP insecticides. We tested the compounds against 2 blowfly strains; a laboratory strain (LS), which is susceptible to the OP insecticide diazinon, and the field strain “Tara,” which is resistant to diazinon (Fig. 5A) (38). Compound efficacy was determined by treating blowfly larvae with diazinon over a range of concentrations in the presence or absence of the boronic acid compounds at constant concentration and comparing pupation (39). Compounds 2, 3, 3.9, 3.10, and 5 were selected for testing based on high potency against WT and/or Gly137Asp LcαE7 and their structural diversity. We initially tested the compounds by themselves and found that there was no significant difference between the fly pupation rates of either blowfly strain in the presence or absence of the boronic acids compounds (SI Appendix, Fig. S15). When the susceptible strain was treated with boronic acids combined with diazinon, synergism was observed for compounds 3, 3.9, and 3.10 (Fig. 5B and SI Appendix, Fig. S16). Compound 3.10 was the most effective, decreasing the amount of diazinon required to achieve a 50% reduction in pupation (EC50 value) 7.3-fold compared with a diazinon-only control (Fig. 5B). We observed similar synergism when the concentration of compound 3 was decreased from 1 to 0.25 or 0.06 mg/mL (SI Appendix, Fig. S17).
Fig. 5.
Boronic acid compounds synergize with OP insecticides. (A) Chemical structures of the active forms of the insecticides diazinon and malathion chlorpyrifos. (B) Compounds 3, 3.9, and 3.10 synergized diazinon and malathion against the susceptible LS blowfly strain, while all compounds tested synergized diazinon against the resistant Tara strain. EC50 values were calculated from 3 (diazinon) or 2 (malathion) repeat measurements of pupation rate, with 50 larvae at each diazinon/malathion concentration. EC50 values are presented as ±95% confidence intervals with the diazinon-only control compared with treatment with diazinon and the boronic acid compounds (1-way ANOVA followed by Dunnett’s multiple comparison test). ns, not significant. ****P < 0.0001. (C) Compounds 3.7, 3.10, and 5 synergized chlorpyrifos against the peach–potato aphid. After initial infestation with 9 adult aphids, the numbers of alive adults and new larvae were counted after 1, 3, and 7 d. Data are mean ± SEM for 4 replicate experiments with the chlorpyrifos-only control compared with treatment with chlorpyrifos and the boronic acid compounds (1-way ANOVA followed by Dunnett’s multiple comparison test). ****P < 0.0001.
Having demonstrated synergism between the boronic acids and an OP insecticide, we tested the compounds against the diazinon-resistant strain. Diazinon resistance is typically associated with the Gly137Asp mutation/resistance allele. We determined the genotype of the resistant strain and found that, although the susceptible strain carried only WT alleles (Gly137), the resistant strain carried both WT (Gly137) and mutant (Asp137) alleles (SI Appendix, Fig. S18). This is consistent with previous reports that duplications of the chromosomal region containing αE7 have occurred, meaning that resistant strains now carry copies of both WT and Gly137Asp LcαE7 (40). Resistance can be quantified by the increase in the insecticide EC50. The diazinon EC50 for the resistant strain was 2.9-fold higher compared with the susceptible strain (Fig. 5B and SI Appendix, Fig. S16). This decrease in OP efficacy is within the range previously reported for 8 different blowfly strains containing the Gly137Asp mutation (15), and it is sufficient to reduce OP protection in sheep from 16 wk to 4 to 6 wk (41) and lead to failure of OPs against third-instar larvae (42). When tested against the resistant strain, all of the boronic acids increased the efficacy of diazinon (Fig. 5B and SI Appendix, Figs. S16 and S17). The most effective compound (3.10) reduced the EC50 12-fold. Therefore, compared with controls treated only with diazinon, compound 3.10 abolished the 2.9-fold resistance in the resistance strain and rendered the resistant strain 4-fold more sensitive to diazinon compared with the susceptible strain.
Based on the in vitro enzyme inhibition profile (Fig. 2B), the higher level of diazinon synergism in the resistant strain with compound 3 vs. compound 2 is surprising. However, this discrepancy is consistent with our finding that the Tara strain carries both WT and Gly137Asp LcαE7 alleles (SI Appendix, Fig. S18); it has been shown that WT LcαE7 confers substantial OP protection to the blowfly (18). Specifically, a compound, such as 3.10, that effectively inhibits both LcαE7 variants would be the best synergist against this resistant strain. The synergism exhibited by compound 3.10 is, therefore, a function of the optimized Gly137Asp LcαE7 inhibition and retention of WT LcαE7 inhibition. This highlights the importance of both sequestration (via WT) and catalytic detoxification (via Gly137Asp) by LcαE7 in OP resistance.
We also tested the effects of compounds 3, 3.9, and 3.10 on the sensitivity of the LS to the OP insecticide malathion (Fig. 5B and SI Appendix, Fig. S16). Sensitivity to malathion and diazinon is qualitatively different. WT LcαE7 confers a low level of resistance to diazinon via high-affinity binding and slow hydrolysis; however, WT LcαE7 displays significant malathion hydrolase activity (13). This difference is evident in the similar EC50 values for the susceptible L. cuprina strain treated with malathion compared with the resistant strain treated with diazinon (Fig. 5B). Synergism with malathion was observed for all of the boronic acid compounds tested. Compound 3.10 was the most effective, reducing the EC50 16-fold compared with a malathion-only control (Fig. 5B).
LcαE7 Inhibitors Synergize with Chlorpyrifos against M. persicae.
To determine the potential for broad-spectrum use, we tested selected boronic acids in combination with chlorpyrifos against another important agricultural pest, the peach–potato aphid M. persicae. OP resistance in M. persicae is mediated by 2 overexpressed CE isozymes, E4 and FE4, which bind OPs with high affinity and act as insecticide “sponges” (43). Both E4 and FE4 are related to LcαE7 (50 and 53% amino acid similarity, respectively). We first tested the toxicity of the boronic acids in the absence of OP, which confirmed that the compounds have no significant toxicity by themselves (SI Appendix, Fig. S19). We then tested the efficacy of chlorpyrifos by treating adult aphids at 4 different concentrations equivalent to 0.7, 1.4, 2.8, and 5.6 g per hectare (ha). Significant killing only occurred at 5.6 g/ha (SI Appendix, Fig. S19). We then tested compounds 3.7, 3.10, and 5 (chosen on the basis of their structural diversity) alongside chlorpyrifos at 2.8 and 5.6 g/ha over a period of 7 d (Fig. 5C). These tests showed a significant increase in the efficacy of chlorpyrifos compared with the chlorpyrifos-only controls (Fig. 5C). While the 2.8-g/ha concentration of chlorpyrifos showed no significant killing efficacy by itself, addition of compounds 3.10 and 5 resulted in almost total extermination of the aphid population, and compound 3.7 resulted in ∼50% killing. For the 5.6-g/ha concentration, the addition of all 3 boronic acids results in almost full efficacy of the mixture. This significant increase in chlorpyrifos efficacy, with no optimization of the compounds for M. persicae, establishes the broad-spectrum potential for boronic acid compounds as a class of insecticide synergist that will allow significant reduction in the amount of OP that is required for the control of insect pests.
Discussion
The design of potent and selective enzyme inhibitors is a central challenge in chemical biology (44). Despite the prominence of high-throughput screening technologies, virtual compound screens provide an inexpensive and rapid alternative (45). Here, we implemented a virtual screen of 23,000 boronic acid compounds to identify inhibitors of LcαE7. The 100% hit rate from the initial screen, with all compounds exhibiting Ki values lower than 12 nM (Fig. 2B), is surprisingly high, even when compared with previous covalent virtual screens that exhibited high hit rates against diverse enzyme targets (23). The high hit rate can partly be attributed to the status of boronic acid compounds as potent inhibitors of serine hydrolases (46). Despite this general high-affinity binding, the specific modification to the PBA scaffold suggested by the virtual screen afforded up to 3 orders of magnitude improvement in LcαE7 inhibition. Indeed, although 74% of the screening library contained the PBA substructure, the docking still enriched it in the top hits (86% of the top 500). Therefore, although virtual screening methods will benefit from improvements to sampling and scoring reflected by the fact that there was no correlation between docking rank and inhibitor potency for the compounds from the initial screen (Fig. 2B), application of the DOCKovalent method allowed the rapid identification of potent inhibitors of LcαE7, with the initial testing of only 5 compounds. The initial screen also allowed us to optimize inhibitors for the Gly137Asp mutant by combining the features of compound 3, which conferred high-affinity WT binding, with the features of compound 2, which were tolerated by the Gly137Asp mutant. Additional improvements to the potency and selectivity of compounds 3.9 and 3.10 may be possible by modifying the benzyloxy/phenoxy substituents through medicinal chemistry. Alternatively, 3-bromo-PBA could be used as an “anchor” for the templated in situ synthesis of new inhibitors as has previously been described for AChE (47).
A major requirement for synergists to be practical is a benign safety and environmental profile. The main concern in terms of toxicity is selectivity against AChE. While the overall structures of LcαE7 and human AChE are similar (PDB ID code 4EY4 [48], 1.05 Å rmsd over 309 residues), structural differences in an active site loop mean that the bromo substituent of compound 3, which is highly complementary to the LcαE7 active site, is sterically occluded from the AChE active site by Phe295 (SI Appendix, Fig. S20). This results in between 20,000- and 100,000-fold selectivity for LcαE7 for compounds 3 and its derivatives. In practical terms, this means that the boronic acid compounds are at least 430-fold less potent inhibitors of AChE compared with the approved drug Donepezil, an AChE inhibitor used for the treatment of Alzheimer’s disease (49).
Having established that these compounds are unlikely to cause acute toxicity via inhibition of AChE, we also tested inhibition of BChE and CES1/2. The structural features that confer selective binding to AChE also explain the moderate selectivity against BChE. Although AChE and BChE both possess an active site loop that is not found in LcαE7, BChE has a smaller residue (Leu286) compared with AChE (Phe295), and this difference in size reduces the steric clash at this position and therefore, reduces selectivity (SI Appendix, Fig. S20). We also found low selectivity with the boronic acid compounds for LcαE7 over CES1/2 (Fig. 4B). The open binding pocket of CES1 (and most likely, CES2, although the structure is not known) reflects its broad substrate specificity (50) and allows the active site to accommodate the boronic acid compounds (SI Appendix, Fig. S20). Interestingly, despite the relatively open active site, CES1 possesses a similar active site loop to AChE, and this conserved feature might allow increased selectivity to be engineered. Despite the relatively low selectivity of the boronic acid compounds for CES1, OPs are even more potent inhibitors of CES1 and are irreversible inhibitors (e.g., chlorpyrifos has an IC50 value of 0.15 nM with CES1 [34]). Thus, the reversible inhibition of CES1/2 by the boronic acids is unlikely to cause toxicity in excess of that already observed for OPs. Furthermore, the main route of OP detoxification in humans, unlike insects, involves paraoxonases (e.g., PON1) (51, 52), meaning that substantial synergism of OP toxicity by the boronic acids is unlikely, as PON1 is not inhibited by boronic acids (53), and overall toxicity will be reduced, because an order of magnitude less OP would be required. Follow-up studies of potential synergism could involve dosing an animal model with both OP and the boronic acids simultaneously.
To investigate whether the boronic acids might have an unexpected toxic effect, we also tested the toxicity more broadly. We showed that the compounds were selective against human serine hydrolases using in-gel ABPP (SI Appendix, Fig. S10). We also demonstrated high selectivity against a panel of 26 serine and threonine proteases (Fig. 4C) and showed that the compounds have low toxicity against 9 human cell lines (SI Appendix, Fig. S11). The benign nature of the boronic acids was best demonstrated by their tolerance in mice; all tested compounds were fully tolerated at a high dose of 300 mg/kg. The risk of off-target effects of the boronic acids, including the possibility that they might synergize other esterified drugs, should be considered along with the potent, nonselective, and irreversible inhibition of serine hydrolases by OP insecticides. Because the boronic acid compounds would be used in combination with OPs, any toxicity associated with altered drug metabolism via esterase inhibition would be outweighed by the effect of cholinergic toxicity caused by the OP insecticides. Cholinergic toxicity is the main concern regarding OP insecticide use, and the synergists described in this work could allow an order of magnitude reduction in OP use without compromising efficacy. Moreover, there are already examples, such as the use of piperonyl butoxide as a synergist for pyrethroids, where compounds that inhibit human metabolic enzymes (cytochrome P450s in the case of piperonyl butoxide [22]) have been safely used for decades. Given that OP insecticides are used worldwide, with an estimated 9 million kg applied annually in the United States alone (7), the reduction in OP use enabled by the boronic acid synergists could have enormous health, environmental, and economic benefits.
Insecticides remain the primary measure for control of agricultural pests, such as the sheep blowfly and aphids, as well as disease vectors, such as mosquitoes. The constant evolution of pesticide resistance in almost all species makes the development of new approaches to prevent or abolish resistance of great importance. While there is hope for the development of new pesticides, there are a finite number of biochemical targets, and the use of synergists to knock out the resistance mechanisms and restore the effectiveness of OP insecticides is a viable alternative strategy. The compounds presented here increased the efficacy of diazinon by 12-fold against L. cuprina, which supports the utility of targeting CE-based resistance mechanisms. CEs have been associated with over 50 cases of pesticide resistance over the last 50 y (10). In addition to the high sequence conservation of metabolic insect CEs (10), the Gly > Asp mutation has been found in the equivalent position in 2 other insect pests (13, 17), indicating that there is the potential for boronic acid-based synergists to have broad-spectrum activity against a range of insect species. This potential was confirmed by the ability of the boronic acid compounds to synergize with OPs against the peach–potato aphid, M. persicae (Fig. 5C). An added benefit of boronic acid synergists is the potential protection from the evolution of resistance. Since boronic acids are transition-state analogs for the phosphorylation of the catalytic serine by OP insecticides, mutations that hinder boronic acid binding will also likely disrupt OP sequestration and/or hydrolysis. Despite this protection, other resistance mechanisms (although far less common than CE-mediated resistance) could still render OP insecticides ineffective, such as mutations to AChE that desensitize the enzyme to OPs (54).
The potent and selective CE inhibitors reported in this work represent a milestone in the use of virtual screening for inhibitor discovery in the context of combating pesticide resistance. We identified high-affinity boronic acid inhibitors of a key resistance enzyme and developed our understanding of the general structure–activity relationships that underlie the effectiveness of boronic acids with serine hydrolases, facilitating inhibitor optimization. The demonstration that the compounds effectively abolished OP insecticide resistance in L. cuprina and were also effective against M. persicae establishes the viability of this synergist-focused approach to combat pesticide resistance and restore the effectiveness of existing pesticide classes. The substantial increase in insecticide efficacy would allow more sustainable pesticide usage and reduce off-target environmental and health-related pesticide effects.
Materials and Methods
Covalent Virtual Screen.
DOCKovalent is a covalent adaptation of DOCK3.6. Given a pregenerated set of ligand conformations and a covalent attachment point, it exhaustively samples ligand poses around the covalent bond and selects the lowest-energy pose using a physics-based energy function that evaluates van der Waals and electrostatics interactions as well as penalizing for ligand desolvation. For the docking performed in this work, a boronic acids library of ∼23,000 commercially available compounds was used (available for docking via http://covalent.docking.org/).
Receptor preparation.
PDB ID code 4FNG was used for the docking. Ser218 was deprotonated to accommodate the covalent adduct, and the Oγ partial charge was adjusted to represent a bonded form. His471 was represented in its doubly protonated form.
Sampling parameters.
The covalent bond length was set to 1.5 ± 0.1 Å, and the 2 newly formed bond angles were set to Cβ-Oγ-B = 116.0 ± 5° and Oγ-B-Ligatom = 109.5 ± 5°; the boron atom was replaced by a carbon, as boron is not parameterized for some of the ligand preparation tool chain. As the covalent attachment atom is not scored during the docking, this replacement should not influence the results.
Candidate selection.
The docking results were reranked by calculated ligand efficiency (docking score divided by number of heavy atoms). The predicted docking poses for each of the top 500 results of this ranked list were then visually inspected. Based on this visual inspection, compounds were excluded based on considerations that are orthogonal to the docking scoring function, such as correct representation of the molecule: for instance, correct protonation state as validated by the Marvin suite (ChemAxon; https://chemaxon.com) and internal strain (ligand internal energy is not included in the scoring function). From the results that have passed the aforementioned filters, we further focused only on poses in which either of the boronic acid hydroxyls was predicted to occupy the oxyanion hole—a conserved trait or boronic acid inhibitors of serine hydrolses. This list was further clustered to select the most diverse candidates. Finally, intuition, price, and commercial availability guided the selection of the 5 reported candidates. Second generation compounds were selected based on the identified 3,5 phenyl-boronic acid substitution pattern from the CombiBlocks catalog.
Enzyme Expression and Purification.
His6-tagged proteins were expressed in BL21(DE3) Escherichia coli (Invitrogen) at 24 °C for 18 h. Cells were collected by centrifugation, resuspended in lysis buffer (300 mM NaCl, 10 mM imidazole, 50 mM Hepes (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.5), and lysed by sonication. Cell debris was pelleted by centrifugation, and the soluble fraction was loaded onto a HisTrap-HP Ni-Sepharose column (GE Healthcare). Bound protein was eluted with lysis buffer supplemented with 300 mM imidazole. Fractions containing the eluted protein were concentrated with a 30-kDa molecular mass cutoff centrifuge concentrator (Amicon) and loaded onto a HiLoad 26/60 Superdex-200 size exclusion column (GE Healthcare) preequilibrated with 150 mM NaCl and 20 mM Hepes, pH 7.5. Eluted fractions containing the monomeric protein were pooled for enzyme inhibition assays or crystallization. Protein concentration was determined by measuring the absorbance at 280 nm with an extinction coefficient calculated using the Protparam online server (55).
L. cuprina αE7 In Vitro Inhibition Assays.
Inhibition of WT LcαE7 and the Gly137Asp LcαE7 variant was determined by a competition assay between the native substrate analog 4-NPB (Sigma) and the boronic acid compounds. Initially, the Michaelis constant (KM) with 4-NPB was measured to determine an appropriate concentration of substrate for use in the competition assays. Reactions consisted of 178 μL assay buffer (100 mM NaCl, 20 mM Hepes, pH 7.5), 2 μL substrate in methanol (final concentrations ranged from 1,000 to 8 μM), and 20 μL enzyme (final concentrations were 2.5 nM for WT LcαE7 and 4 nM for Gly137Asp LcαE7). The enzymes were prepared in assay buffer supplemented with 4 mg/mL bovine serum albumin (BSA) (Sigma; catalog no. A8806) to maintain stability. Enzyme velocity was determined by measuring the formation of the 4-nitrophenolate product of hydrolysis (405 nm) for 4 min at room temperature using an Epoch microplate spectrophotometer (BioTek). Initial rates were corrected for nonenzymatic hydrolysis, and the Michaelis constant was determined by fitting the initial rates to the Michaelis–Menten equation using GraphPad Prism (SI Appendix, Fig. S21).
Enzyme inhibition with the boronic acid compounds was determined by assaying the initial rate of 4-NPB hydrolysis in the presence of either neat dimethyl sulfoxide (DMSO) or the boronic acid compounds serially diluted 1:3 in DMSO. Reactions consisted of 178 μL assay buffer supplemented with substrate (final concentrations were 15 μM for WT LcαE7 and 250 μM for Gly137Asp LcαE7), 2 μL neat DMSO or 2 μL compound (final concentrations ranged from 100 μM to 60 pM), and 20 μL enzyme (final concentrations were 0.5 nM for WT LcαE7 and 10 nM for Gly137Asp LcαE7). Product formation was monitored for 4 min, and the initial rates were corrected for nonenzymatic hydrolysis. To determine the concentration of boronic acid compounds required to inhibit 50% of esterase activity (IC50), a 4-parameter sigmoidal dose–response curve was fitted to percentage inhibition using GraphPad Prism (SI Appendix, Figs. S2 and S5). Ki values were determined using the Cheng–Prusoff equation assuming competitive inhibition (56).
Human AChE and Human BChE In Vitro Inhibition Assays.
Inhibition of human AChE (Sigma; catalog no. C0663) and BChE (Sigma; catalog no. B4186) by compounds 1 to 5, 3.9, and 3.10 was assayed using the Ellman method (57). Initially, the KM was determined for AChE with acetylthiocholine and BChE with butyrylthiocholine. Reactions consisted of 120 μL assay buffer supplemented with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB; Sigma), 40 μL assay buffer supplemented with substrate (final concentrations ranged from 1,000 to 8 μM), and 40 μL enzyme (final concentrations were 0.1 nM for AChE and 0.6 nM for BChE). The enzymes were prepared in assay buffer supplemented with 0.5 mg/mL BSA. Enzyme velocity was determined by measuring thiocholine formation (412 nm), and the Michaelis constant was determined as described previously (SI Appendix, Fig. S21). Human BChE shows substrate activation at high concentrations of butyrylthiocholine (58); hence, the highest 2 substrate concentrations (1 and 0.5 mM) were excluded for fitting the KM.
Enzyme inhibition was determined in a similar manner to LcαE7. Reactions consisted of 158 μL assay buffer supplemented with DTNB (final concentration of 300 μM) and substrate (final concentrations were 100 μM for both acetylthiocholine and butyrylthiocholine), 2 μL neat DMSO or 2 μL compound (final concentrations ranged from 2 mM to 400 pM), and 40 μL enzyme (final concentrations were 0.4 nM for AChE and 1.6 nM for BChE). Ki values were determined as described previously (SI Appendix, Fig. S8).
Human CES1 and CES2 In Vitro Inhibition Assays.
Inhibition of human CES1 (Sigma; catalog no. E0287) and CES2 (Sigma; catalog no. C4749) was determined for compounds 1 to 5, 3.9, and 3.10. The KM was determined with the substrate analog 4-nitrophenyl acetate (Sigma). Reactions consisted of 158 μL assay buffer, 2 μL substrate in methanol (final concentrations ranged from 1,000 to 8 μM), and 40 μL enzyme (final concentrations were 3 nM for CES1 and 0.1 nM for CES2). Enzymes were prepared in assay buffer supplemented with 0.5 mg/mL BSA. Enzyme velocity was determined by measuring 4-nitrophenolate formation (405 nm), and the Michaelis constant was determined as described previously (SI Appendix, Fig. S21).
Enzyme inhibition was determined in a similar manner to LcαE7. Reactions consisted of 158 μL assay buffer supplemented with substrate (final concentrations were 200 μM for CES1 and 100 μM for CES2), 2 μL neat DMSO or 2 μL compound (final concentrations ranged from 0.7 mM to 100 pM), and 40 μL enzyme (final concentrations were 3 nM for CES1 and 0.4 nM for CES2). Ki values were determined as described previously (SI Appendix, Fig. S9).
ABPP Assays.
L. cuprina lysate.
Samples were prepared by homogenizing third-instar larvae in 150 mM NaCl, 50 mM Tris, pH 8.0, and 1% Triton-X100 using a mortar and pestle. Samples were centrifuged (18,000 × g for 10 min at 4 °C), and the supernatant was stored at −80 °C.
Mouse lysate.
Mouse tissues (liver, kidney, and spleen) were Dounce homogenized in phosphate buffered saline (PBS) (pH 7.5) and centrifuged (1,400 × g for 5 min at 4 °C) to remove debris. The supernatant was then diluted with PBS and centrifuged (64,000 × g for 45 min) to obtain the soluble fraction. Samples was stored at −80 °C prior to use.
HEK cell lysate.
HEK293 cells were washed twice with media and then, washed twice with PBS. Cells were collected by centrifugation (200 × g for 5 min at 4 °C) and resuspended in radioimmunoprecipitation assay (RIPA) buffer (Sigma) supplemented with protease inhibitors (Pepstatin, Aprotinin, Leupeptin). The cells were incubated on ice for 5 min, and then, they were vortexed and centrifuged to collect cell debris (16,000 × g for 5 min at 4 °C). The supernatant was frozen at −80 °C.
Protein concentration.
Protein concentration was determined by bicinchoninic acid (BCA) (Thermo Fisher kit). Lysate (25 μL sample diluted 1:1, 1:10, or 1:100 in the appropriate buffer) was mixed with 200 μL BCA reagent (50:1 reagent A to reagent B). Standards consisted of an array of dilutions of BSA also in the appropriate buffer.
FP-RH ABPP assay.
Lysate samples at 1 mg/mL were preincubated with boronic acids (0, 0.1, 1, 10, 100 μM) in either the presence or absence of paraoxon (1 μM) for 10 min at 37 °C. The samples that included the FP-RH probe were than incubated with 1 μM probe for another 10 min at 37 °C after mixing by tapping. Each sample had a total volume of 25 μL. After the second incubation, 8.33 μL loading buffer was added to each sample to quench the labeling reaction. The samples were vortexed and incubated at 70 °C for 5 min; then, 10 μL each sample was run on an SDS/PAGE gel. Gels were imaged at 532 nm using a Typhoon fluorescence scanner.
Protease Selectivity Panel.
Compounds were tested for inhibition of a panel of 26 Ser/Thr proteases at a single-point concentration of 100 μM in duplicates by NanoSyn, Santa Clara. Test compounds were dissolved in 100% DMSO to make 10 mM stock. Final compound concentration in assay was 100 μM. Compounds were tested in duplicate wells at single concentration, and the final concentration of DMSO in all assays was kept at 1%. Five reference compounds (AEBSF, Carfilzomib, Granzyme B Inhibitor II, Dec-RVKR-CMK, and Teneligliptin hydrobromide) were tested in an identical manner with 8 concentration points at 5× dilutions. SI Appendix, Table S4 shows assay conditions.
Cellular Toxicity Assays.
A 7-point, 2-fold dose–response series with 100 μM as the upper limit and a DMSO-only control point was generated using an Echo 550 liquid handler (Labcyte Inc.) in 384-well plates. Subsequently, the human cell lines were seeded (1,000 cells per well) using a multidrop Combi (Thermo Fisher Scientific) on top of the compounds. Plates were then incubated at 37 °C and 5% CO2 for 48 h, on which cell viability was assessed by adding CellTiter-Glo (Promega) to the reaction. The luminescence signal was measured on a Pherastar FS multimode plate reader (BMG Labtech).
Mice Toxicity.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Weizmann Institute of Science. C57BL/6 7-wk-old female mice were purchased from Envigo and allowed to acclimatize to the animal facility environment for 2 wk before use for experimentation. Following acclimatization, mice were gavaged with 300 mg/kg body weight of the compound dissolved in sesame oil. After the substance had been administered, food was withheld for 2 h. Three animals were used for compounds 3.7, 3.10, and 4, and 4 animals were used for compounds 3 and 3.9. Animals were observed individually after dosing at least once during the first 30 min; periodically during the first 24 h, with special attention given during the first 4 h; and daily thereafter for a total of 14 d. Following the 2 wk, animals were killed and were subjected to histological examination; mice organs were fixed in 4% paraformaldehyde (BIO LAB; catalog no. 06450323), embedded in paraffin, sectioned (5-µm thick), and stained with hematoxylin and eosin.
Crystallization and Structure Determination.
Cocrystals of compounds 1 to 5 with the thermostable LcαE7 variant (25, 59) (LcαE7-4a; PDB ID codes 5TYP, 5TYO, 5TYN, 5TYL, and 5TLK) and compound 3.10 with Gly137Asp LcαE7-4a (PDB ID code 5TYJ) were grown using the hanging drop vapor-diffusion method. Reservoir solutions contained 100 mM sodium acetate (pH 4.6 to 5.1) and 15 to 26% polyethylene glycol (PEG) 2000 monomethyl ether (MME) or PEG 550 MME. Inhibitors prepared in DMSO were incubated with protein (7 mg/mL in 75 mM NaCl and 10 mM Hepes, pH 7.5) to achieve a 5:1 inhibitor to compound stoichiometric ratio. Hanging drops were set up with 2 μL reservoir and 1 μL protein, with crystals forming overnight at 19 °C. For cryoprotection, crystals were briefly immersed in a solution containing the hanging drop reservoir solution with the PEG concentration increased to 35% and then, vitrified at 100 K in a gaseous stream of nitrogen.
Diffraction data were collected at 100 K on either the MX1 or MX2 beam line at the Australian Synchrotron using a wavelength of 0.954 Å. Data were indexed, integrated, and scaled using XDS (60). High-resolution data were excluded when the correlation coefficient between random half datasets (61, 62) decreased below 0.3 in the highest-resolution shell. Phases were obtained by molecular replacement with the program Phaser (63) using the coordinates of apo-LcαE7-4a (PDB ID code 5CH3 [64]) as the search model. The initial model was improved by iterative model building with COOT (65) and refinement with phenix.refine (66). Inhibitor coordinates and restraints were generated with eLBOW (67). Crystallographic statistics are summarized in SI Appendix, Table S1.
To determine if the mutations present in the thermostable LcαE7-4a (25, 59) influenced the orientation or mode of inhibitor binding, the surface mutations present in LcαE7-4a were introduced into the WT background, and the protein was tested for crystallization. Two mutations (Lys530Glu and Asp83Ala) were sufficient to allow crystallization in the same conditions as described previously (PDB ID code 5TYM) (SI Appendix, Fig. S22).
L. cuprina Bioassays.
Two strains of L. cuprina were used: (i) a laboratory reference drug-susceptible strain, LS, derived from collections in the Australian Capital Territory over 40 y ago, with no history of exposure to insecticides; and (ii) a field-collected strain, Tara, resistant to diazinon and diflubenzuron (38). The LcαE7 gene was sequenced in each of the strains. Briefly, genomic DNA was prepared from 20 adult female flies from each strain using the DNeasy Blood and Tissue kit (Qiagen). PCR was performed with primers specific to the LcαE7 gene (68), and the product was cloned into a pGEM-T EASY vector (Promega). Eight clones of the susceptible strain and 10 clones of the resistant strain were sequenced using M13 forward and reverse primers.
The effect of compounds 2, 3, 5, 3.9, and 3.10 on the development of blowfly larvae in the presence of diazinon/malathion was assessed using a bioassay system in which larvae were allowed to develop from the first-instar stage until pupation on cotton wool impregnated with diazinon/malathion over a range of concentrations in the presence or absence of the compounds at constant concentrations (39). Each experiment utilized 50 larvae at each diazinon/malathion concentration. Experiments were replicated 3 times for diazinon and twice for malathion. The insecticidal effects were defined by measuring the pupation rate. The pupation rate dose–response data were analyzed by nonlinear regression (GraphPad Prism) to calculate EC50 values (with 95% confidence intervals) representing the concentration of diazinon/malathion (alone or in combination with compounds 2, 3, 5, 3.9, or 3.10) required to reduce the pupation rate to 50% of that measured in control assays. The effects of compounds 2, 3, 5, 3.9, and 3.10 were defined in 2 ways: (i) synergism ratio within each isolate = the EC50 for diazinon/malathion alone/EC50 for diazinon/malathion in combination with the compounds and (ii) resistance ratio = the EC50 for diazinon alone or in combination with the compounds for the Tara strain/EC50 for diazinon alone with the LS strain. Compounds were also tested without diazinon or malathion at 1 mg per assay.
M. persicae Bioassays.
Peach–potato aphids (M. persicae) were reared on small broad bean plants (Vicia faba, Aquadulce variety) under controlled conditions (15 °C/20 °C night/day, photoperiod: 14 h, 60% relative humidity). The bioassay was derived from the Insecticide Resistance Action Committee (IRAC) susceptibility test method number 019. Briefly, seeds of broad beans were sown in small pots and cultivated under controlled conditions (15 °C/20 °C night/day, photoperiod: 14 h) for 3 wk. Healthy, large, and flat leaves were selected from untreated plants and placed (abaxial face side up) on a petri dish lid for treatment. Leaves were treated by spraying with a Track sprayer equipped with flat fan nozzles TeeJet XR110015VS and calibrated to deliver 200 L/ha at 400 kPa and 4 km/h. Three leaves were used for each repetition. The leaves were then air dried for 1 h at 20 °C until completely dry. One leaf disk of 2.5 cm in diameter was cut in each treated leaf and placed, abaxial face side up, in a petri dish on a thick layer of sterile water agar medium. Each leaf disk was then infested with 3 adult aphids with a paint brush. Petri dishes were sealed with a perforated lid, with holes closed with nylon filter. One repetition was composed of a petri dish with 3 leaf disks and 9 aphids. Four repetitions were used for each condition. The petri dishes were incubated in the same controlled conditions as the breeding. Assessment of living aphids was made 1, 3, and 7 d after leaf infestation. Chlorpyrifos was tested at 0.7, 1.4, 2.8, and 5.6 g/ha alone or in combination with the boronic acid compounds at 0.2 mg/mL. Each boronic acid compound was also tested alone at 0.2 mg/mL, with the final concentration of DMSO at 1%. Water and 1% DMSO were used as negative controls.
Supplementary Material
Acknowledgments
We thank Paul Carr for assistance with X-ray crystallography and the Australian Synchrotron for beam time. We thank Benjamin Cravatt for providing us with the fluorescent serine hydrolase activity-based probe. This research was supported by an Australian Postgraduate Award (to G.J.C.), Australian Science and Industry Endowment Fund PF14-099 (to P.D.M. and C.J.J.), Australian Research Council Future Fellowship FT140101059 (to C.J.J.), Israel Science Foundation Grant 1097/16 (to N.L.), and German–Israeli Foundation Grant I-2483-302.5/2017 (to N.L.). Additionally, N.L. is the incumbent of the Alan and Laraine Fischer Career Development Chair.
Footnotes
Competing interest statement: G.J.C., C.J.J., and N.L., are inventors on a US patent application (62/443,825) for the described synergists.
This article is a PNAS Direct Submission.
Data deposition: All crystal structures reported here have been deposited in the Protein Data Bank (PDB; www.rcsb.org) under ID codes 5TYJ–5TYP. PDB validation reports are all available at www.rcsb.org.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909130116/-/DCSupplemental.
References
- 1.Cooper J., Dobson H., The benefits of pesticides to mankind and the environment. Crop Prot. 26, 1337–1348 (2007). [Google Scholar]
- 2.World Health Organization , Integrated Vector Management: Strategic Framework for the Eastern Mediterranean Region 2004–2010 (The WHO Regional Office for the Eastern Mediterranean, Cairo, Egypt, 2004). [Google Scholar]
- 3.World Health Organization , Pesticides and Their Application for the Control of Vectors and Pests of Public Health Importance (World Health Organization, Geneva, Switzerland, ed. 6, 2006). [Google Scholar]
- 4.Whalon M. E., Mota-Sanchez D., Hollingworth R. M., Global Pesticide Resistance in Arthropods, Whalon M. E., Mota-Sanchez D., Hollingworth R. M., Eds. (Cabi International, London, United Kingdom, 2008). [Google Scholar]
- 5.Sparks T. C., Nauen R., IRAC: Mode of action classification and insecticide resistance management. Pestic. Biochem. Physiol. 121, 122–128 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Eddleston M., et al. , Pesticide poisoning in the developing world–A minimum pesticides list. Lancet 360, 1163–1167 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Atwood D., Paisley-Jones C., Pesticides Industry Sales and Usage 2008–2012 Market Estimates (US Environmental Protection Agency, 2017). [Google Scholar]
- 8.Fukuto T. R., Mechanism of action of organophosphorus and carbamate insecticides. Environ. Health Perspect. 87, 245–254 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tafuri J., Roberts J., Organophosphate poisoning. Ann. Emerg. Med. 16, 193–202 (1987). [DOI] [PubMed] [Google Scholar]
- 10.Oakeshott J. G., Claudianos C., Campbell P. M., Newcomb R. D., Russell R. J., “Biochemical genetics and genomics of insect esterases” in Comprehensive Molecular Insect Science–Pharmacology, Gilbert L. I., Gill S. S., Eds. (Elsevier B.V., Oxford, United Kingdom, 2005), pp. 309–381. [Google Scholar]
- 11.Devonshire A. L., Field L. M., Gene amplification and insecticide resistance. Annu. Rev. Entomol. 36, 1–23 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Newcomb R. D., Gleeson D. M., Yong C. G., Russell R. J., Oakeshott J. G., Multiple mutations and gene duplications conferring organophosphorus insecticide resistance have been selected at the Rop-1 locus of the sheep blowfly, Lucilia cuprina. J. Mol. Evol. 60, 207–220 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Claudianos C., Russell R. J., Oakeshott J. G., The same amino acid substitution in orthologous esterases confers organophosphate resistance on the house fly and a blowfly. Insect Biochem. Mol. Biol. 29, 675–686 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Shanahan G. J., Development of a changed response in Lucilia cuprina (Wied.) to organophosphorus insecticides in New South Wales. Bull. Entomol. Res. 57, 93–100 (1966). [Google Scholar]
- 15.Newcomb R. D., et al. , A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc. Natl. Acad. Sci. U.S.A. 94, 7464–7468 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenzie J. A., Dieldrin and diazinon resistance in populations of the Australian sheep blowfly, Lucilia cuprina, from sheep-grazing areas and rubbish tips. Aust. J. Biol. Sci. 37, 367–374 (1984). [DOI] [PubMed] [Google Scholar]
- 17.de Carvalho R. A., Torres T. T., de Azeredo-Espin A. M. L., A survey of mutations in the Cochliomyia hominivorax (Diptera: Calliphoridae) esterase E3 gene associated with organophosphate resistance and the molecular identification of mutant alleles. Vet. Parasitol. 140, 344–351 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Birner-Gruenberger R., et al. , Functional fat body proteomics and gene targeting reveal in vivo functions of Drosophila melanogaster α-Esterase-7. Insect Biochem. Mol. Biol. 42, 220–229 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Mao Y.-B., et al. , Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Swale D. R., et al. , An insecticide resistance-breaking mosquitocide targeting inward rectifier potassium channels in vectors of Zika virus and malaria. Sci. Rep. 6, 36954 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews H. B., Casida J. E., Properties of housefly microsomal cytochromes in relation to sex, strain, substrate specificity, and apparent inhibition and induction by synergist and insecticide chemicals. Life Sci. I 9, 989–1001 (1970). [DOI] [PubMed] [Google Scholar]
- 22.Casida J. E., Mixed-function oxidase involvement in the biochemistry of insecticide synergists. J. Agric. Food Chem. 18, 753–772 (1970). [DOI] [PubMed] [Google Scholar]
- 23.London N., et al. , Covalent docking of large libraries for the discovery of chemical probes. Nat. Chem. Biol. 10, 1066–1072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.London N., et al. , Covalent docking predicts substrates for haloalkanoate dehalogenase superfamily phosphatases. Biochemistry 54, 528–537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson C. J., et al. , Structure and function of an insect α-carboxylesterase (αEsterase7) associated with insecticide resistance. Proc. Natl. Acad. Sci. U.S.A. 110, 10177–10182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koehler K. A., Lienhard G. E., 2-phenylethaneboronic acid, a possible transition-state analog for chymotrypsin. Biochemistry 10, 2477–2483 (1971). [DOI] [PubMed] [Google Scholar]
- 27.Diaz D. B., Yudin A. K., The versatility of boron in biological target engagement. Nat. Chem. 9, 731–742 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Mabbitt P. D., et al. , Conformational disorganization within the active site of a recently evolved organophosphate hydrolase limits its catalytic efficiency. Biochemistry 55, 1408–1417 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Leung D., Hardouin C., Boger D. L., Cravatt B. F., Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat. Biotechnol. 21, 687–691 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Kappers W. A., Edwards R. J., Murray S., Boobis A. R., Diazinon is activated by CYP2C19 in human liver. Toxicol. Appl. Pharmacol. 177, 68–76 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Chen Z., Newcomb R., Forbes E., McKenzie J., Batterham P., The acetylcholinesterase gene and organophosphorus resistance in the Australian sheep blowfly, Lucilia cuprina. Insect Biochem. Mol. Biol. 31, 805–816 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Whyard S., Russell R. J., Walker V. K., Insecticide resistance and malathion carboxylesterase in the sheep blowfly, Lucilia cuprina. Biochem. Genet. 32, 9–24 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Hatfield M. J., et al. , Carboxylesterases: General detoxifying enzymes. Chem. Biol. Interact. 259, 327–331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crow J. A., et al. , Inhibition of recombinant human carboxylesterase 1 and 2 and monoacylglycerol lipase by chlorpyrifos oxon, paraoxon and methyl paraoxon. Toxicol. Appl. Pharmacol. 258, 145–150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall W. K., Roberts J. R., “Ecotoxicology of chlorpyrifos” (Publication no. NRCC 16079, National Research Council of Canada, NRC Associate Committee on Scientific Criteria for Environmental Quality, Subcommittee on Pesticides and Related Compounds, Ottawa, ON, Canada, 1978).
- 36.Pfizer, “Material safety data sheet” (Version 4.0, Pfizer Pharmaceuticals Group, New York, NY, 2010), pp. 1–7.
- 37.Lian J., Nelson R., Lehner R., Carboxylesterases in lipid metabolism: From mouse to human. Protein Cell 9, 178–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levot G. W., Sales N., New high level resistance to diflubenzuron detected in the Australian sheep blowfly, Lucilia cuprina (Wiedemann) (Diptera: Calliphoridae). Gen. Appl. Entomol. 31, 43–46 (2002). [Google Scholar]
- 39.Kotze A. C., et al. , Histone deacetylase enzymes as drug targets for the control of the sheep blowfly, Lucilia cuprina. Int. J. Parasitol. Drugs Drug Resist. 5, 201–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth K. A., Boyce T. M., Russell R. J., Oakeshott J. G., MCE activities and malathion resistances in field populations of the australian sheep blowfly (Lucilia cuprina). Heredity 84, 63–72 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Levot G. W., Resistance and the control of sheep ectoparasites. Int. J. Parasitol. 25, 1355–1362 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Levot G. W., Sales N., Barchia I., In vitro larvicidal efficacy of flystrike dressings against the Australian sheep blowfly. Aust. J. Exp. Agric. 39, 457–464 (1999). [Google Scholar]
- 43.Devonshire A. L., Insecticide resistance in Myzus persicae: From field to gene and back again. Pestic. Sci. 26, 375–382 (1989). [Google Scholar]
- 44.Nature Chemical Biology, Probing questions. Nat. Chem. Biol. 11, 533 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Shoichet B. K., Virtual screening of chemical libraries. Nature 432, 862–865 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W., Gao X., Wang B., Boronic acid compounds as potential pharmaceutical agents. Med. Res. Rev. 23, 346–368 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Lewis W. G., et al. , Click chemistry in situ: Acetylcholinesterase as a reaction vessel for the selective assembly of a femtomolar inhibitor from an array of building blocks. Angew. Chem. Int. Ed. Engl. 41, 1053–1057 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Cheung J., et al. , Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 55, 10282–10286 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Tsurkan L. G., Hatfield M. J., Edwards C. C., Hyatt J. L., Potter P. M., Inhibition of human carboxylesterases hCE1 and hiCE by cholinesterase inhibitors. Chem. Biol. Interact. 203, 226–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bencharit S., et al. , Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1. J. Mol. Biol. 363, 201–214 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker C. H., Mackness M. I., “A” esterases and their role in regulating the toxicity of organophosphates. Arch. Toxicol. 60, 30–33 (1987). [DOI] [PubMed] [Google Scholar]
- 52.Furlong C. E., et al. , Role of paraoxonase (PON1) status in pesticide sensitivity: Genetic and temporal determinants. Neurotoxicology 26, 651–659 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Lorentz K., Wirtz W., Weiss T., Continuous monitoring of arylesterase in human serum. Clin. Chim. Acta 308, 69–78 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Russell R. J., et al. , Two major classes of target site insensitivity mutations confer resistance to organophosphate and carbamate insecticides. Pestic. Biochem. Physiol. 79, 84–93 (2004). [Google Scholar]
- 55.Gasteiger E., et al. , “Protein identification and analysis tools on the ExPASy server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Humana Press, Inc., Totowa, NJ, 2005), pp. 571–607. [Google Scholar]
- 56.Cheng Y., Prusoff W. H., Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973). [DOI] [PubMed] [Google Scholar]
- 57.Ellman G. L., Courtney K. D., Andres V. Jr, Feather-Stone R. M., A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 7, 88–95 (1961). [DOI] [PubMed] [Google Scholar]
- 58.Radić Z., Pickering N. A., Vellom D. C., Camp S., Taylor P., Three distinct domains in the cholinesterase molecule confer selectivity for acetyl- and butyrylcholinesterase inhibitors. Biochemistry 32, 12074–12084 (1993). [DOI] [PubMed] [Google Scholar]
- 59.Fraser N. J., et al. , Evolution of protein quaternary structure in response to selective pressure for increased thermostability. J. Mol. Biol. 428, 2359–2371 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karplus P. A., Diederichs K., Linking crystallographic model and data quality. Science 336, 1030–1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diederichs K., Karplus P. A., Better models by discarding data? Acta Crystallogr. D Biol. Crystallogr. 69, 1215–1222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Correy G. J., et al. , Mapping the accessible conformational landscape of an insect carboxylesterase using conformational ensemble analysis and kinetic crystallography. Structure 24, 977–987 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newcomb R. D., Campbell P. M., Russell R. J., Oakeshott J. G., cDNA cloning, baculovirus-expression and kinetic properties of the esterase, E3, involved in organophosphorus resistance in Lucilia cuprina. Insect Biochem. Mol. Biol. 27, 15–25 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.