Significance
The Calvin–Benson (CB) cycle in plants, algae, and cyanobacteria fixes most of the carbon in most of the biomass on Earth. The CB cycle is regulated by the redox state, which enables it to be turned off in the dark. One part of this regulatory system is the small protein CP12, which binds to 2 essential CB-cycle enzymes in the dark, inactivating them. We have solved the structure of the complex between CP12 and the enzymes, explaining the mechanism of deactivation. Now that this is understood, this structure can be used as the starting point for modulating the redox regulation, which may have applications in improving crop productivity.
Keywords: redox regulation, carbon fixation, photosynthesis, Calvin–Benson cycle
Abstract
Plants, algae, and cyanobacteria fix carbon dioxide to organic carbon with the Calvin–Benson (CB) cycle. Phosphoribulokinase (PRK) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) are essential CB-cycle enzymes that control substrate availability for the carboxylation enzyme Rubisco. PRK consumes ATP to produce the Rubisco substrate ribulose bisphosphate (RuBP). GAPDH catalyzes the reduction step of the CB cycle with NADPH to produce the sugar glyceraldehyde 3-phosphate (GAP), which is used for regeneration of RuBP and is the main exit point of the cycle. GAPDH and PRK are coregulated by the redox state of a conditionally disordered protein CP12, which forms a ternary complex with both enzymes. However, the structural basis of CB-cycle regulation by CP12 is unknown. Here, we show how CP12 modulates the activity of both GAPDH and PRK. Using thermophilic cyanobacterial homologs, we solve crystal structures of GAPDH with different cofactors and CP12 bound, and the ternary GAPDH-CP12-PRK complex by electron cryo-microscopy, we reveal that formation of the N-terminal disulfide preorders CP12 prior to binding the PRK active site, which is resolved in complex with CP12. We find that CP12 binding to GAPDH influences substrate accessibility of all GAPDH active sites in the binary and ternary inhibited complexes. Our structural and biochemical data explain how CP12 integrates responses from both redox state and nicotinamide dinucleotide availability to regulate carbon fixation.
Plants regulate their carbon fixation and other reactions by redox state using it as a proxy for light (1). The product of the photosynthetic electron transport chain is reduced ferredoxin, which is used to produce NADPH in the cyanobacterial cytoplasm or chloroplast stroma. In the light, the chloroplast stroma is reducing, and in the dark, or when the light reactions are otherwise inactive, the stroma becomes oxidizing. Thioredoxin (2) proteins are reduced by ferredoxin and can then exchange a disulfide with a target. Example targets are the sedoheptulose (3) and fructose bisphosphatases (4), which have pairs of cysteines that oxidize to form disulfide bonds that inactivate the enzyme.
The carbon-fixation reaction is catalyzed by Rubisco, which carboxylates ribulose bisphosphate (RuBP). RuBP is produced by phosphoribulokinase (PRK) in the final regeneration reaction, in which a phosphate group is transferred from ATP to ribulose 5-phosphate (5). Plant-type PRK is dimeric and has 2 conserved disulfide bonds, one at the N terminus and the other near the C terminus close to the dimer interface (6, 7). The N-terminal disulfide bond inactivates most of the activity when formed (8–10), but may be reduced in plants by thioredoxin f (11).
The reduction step of the Calvin–Benson (CB) cycle is catalyzed by glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which uses NADPH to reduce bisphosphoglycerate (BPG) to glyceraldehyde 3-phosphate (GAP), the main product exit point of the cycle (12). In cyanobacteria, GAPDH has no disulfide bonds, instead redox regulation of GAPDH is driven by 2 disulfide bonds in the small (∼8 kDa) regulatory inhibitor protein CP12 (13, 14), which is disordered under reducing conditions (15). Under oxidizing conditions, 2 disulfide bonds form in CP12, ordering the C-terminal domain (16), enabling it to bind GAPDH, which is then inhibited.
The GAPDH-CP12 complex can then bind PRK, in an obligate sequential reaction (17). Previous structural studies of the GAPDH-CP12 complex resolved only a C-terminal fragment of CP12, so it remained unclear how CP12 regulated both GAPDH and PRK. A recent structure of a cyanobacterial CP12-CBS 2 domain protein resolved an N-terminal CP12-like region; however, the protein does not form a ternary complex with GAPDH and PRK (8). Calvin cycle GAPDH uses NADPH physiologically to reduce BPG, although NADH may also be a substrate.
CP12 is conserved in oxygenic phototrophs from cyanobacteria to plants (18). Plants often have more than 1 CP12 isoform, with tissue-specific patterns of expression (19). Plants also have 2 photosynthetic GAPDH isoforms GAP-A and GAP-B. GAP-A is similar to the cyanobacterial enzyme while GAP-B has a C-terminal extension homologous to the C-terminal GAPDH binding domain of CP12 (20). GAP-A and GAP-B form an A2B2 tetramer, which further aggregates to A8B8 on oxidation of the GAP-B CP12-like region (20). Information from cyanobacteria is still applicable to plants, as the cyanobacterial CP12 system is a subset of the plant regulatory system. The lack of structural information for full-length CP12 and CP12-bound regulatory complexes has prevented understanding of a key mechanism of CB-cycle redox regulation. To understand, at a molecular level, how the CB cycle is redox regulated in response to light, we solved crystal structures of a thermophilic cyanobacterial GAPDH with full-length CP12 and built an atomic model of the entire cyanobacterial GAPDH-CP12-PRK ternary complex using electron cryo-microscopy (cryoEM).
Results and Discussion
Structures of GAPDH with Different Nucleotides Bound and CP12 C-Terminal Regions.
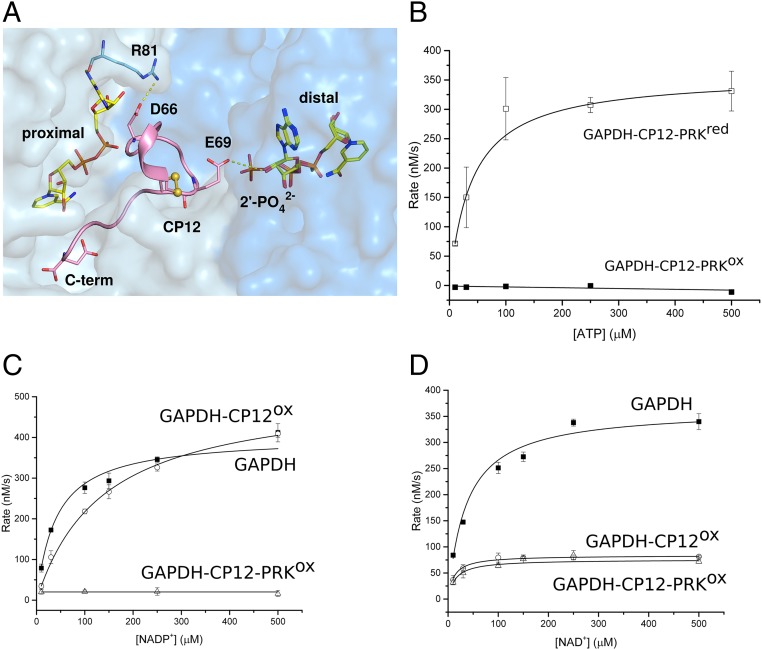
A previous structure of cyanobacterial GAPDH bound to CP12 had 4 CP12 per GAPDH tetramer (21), but the stoichiometry for eukaryotic complexes suggested by structure (22) and biochemistry (23) was 2. Working with recombinant proteins from the thermophilic cyanobacterium Thermosynechococcus elongatus, we found the cyanobacterial GAPDH4-CP122 complex was stable to gel-filtration and crystallized with this stoichiometry. We call the 2 CP12-occupied active sites proximal and the 2 unoccupied sites distal. In all our structures, and those published, the 2 proximal sites are in equivalent positions in the GAPDH tetramer. We obtained a GAPDH4-CP124 structure when GAPDH was incubated with 10-fold excess CP12 (SI Appendix, Fig. S1), so the physiological GAPDH-CP12 complex was unclear (21, 22). Only the C terminus of CP12 was visible in these structures. Our GAPDH had NAD+ bound from the Escherichia coli expression system, but this could be exchanged for NADP+ by turning over the enzyme. NADP(H) is apparently incompatible with CP12 binding, as the conserved CP12-Glu69 (SI Appendix, Figs. S2 and S3) (21, 22) clashes with the position of the 2′ phosphate group in the distal GAPDH active site (Fig. 1A), rendering NADP(H) and CP12 binding to GAPDH mutually exclusive. CP12-Glu69 is not only important for dinucleotide selection, but is also essential for ternary complex formation in Chlamydomonas (24).
Fig. 1.
Kinetics and coenzyme binding of GAPDH complexes. (A) View of GAPDH (NAD+)-CP12 with adjacent active sites. The CP12-bound (proximal) site makes extensive contacts with both NAD+ and GAPDH. CP12-Glu69 prevents clashes with NADPH binding to the distal site, as it would clash with the 2′-phosphate. (B) PRK activity for the ternary complex treated with either oxidized (GAPDH-CP12-PRKox; ■) or reduced DTT (GAPDH-CP12-PRKred; □). All reactions were measured in triplicate and fitted using Michaelis-Menten kinetics. Rate of NADP+ reduction (C) and NAD+ reduction (D) were measured for 275 µM GAPDH (■), 275 µM GAPDH-CP12 (○), and 63 µM GAPDH-CP12-PRK (△) complexes at increasing concentrations of nucleotide substrate. The maximal velocity (Vmax) with NADP+ is not inhibited in GAPDH-CP12, but is fully inhibited in GAPDH-CP12-PRK. In contrast, the Vmax with NAD+ is equally inhibited by GAPDH-CP12 and GAPDH-CP12-PRK.
Structure of Full-Length CP12 Bound to GAPDH.
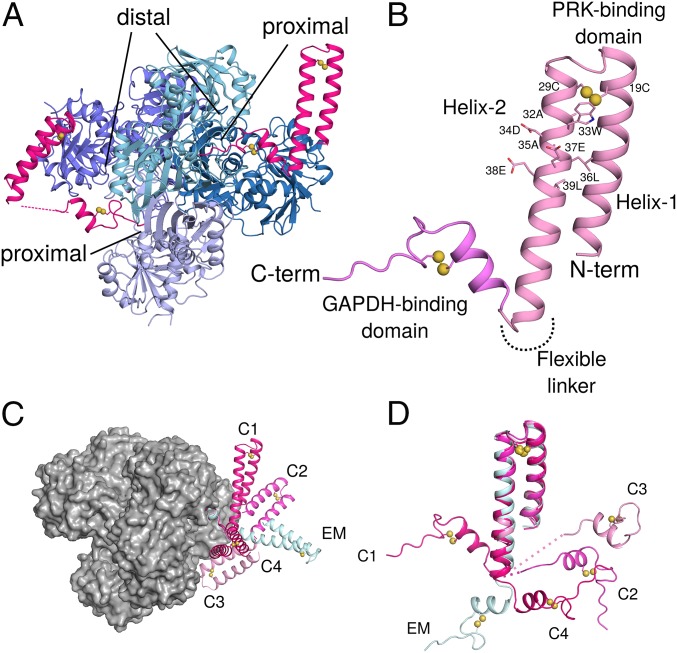
In 3 conditions, GAPDH-CP12 crystallized with one or more full-length CP12 visible in the electron density, at a best resolution of 2.1 Å. We obtained 4 crystallographically independent full-length oxidized CP12 molecules bound to GAPDH. CP12 comprises an N-terminal PRK binding domain (residues 1–52) and a C-terminal GAPDH binding domain (residues 55–75) connected by a fully flexible linker (Fig. 2). If the C terminus of CP12 is bound to GAPDH, the N-terminal domain can adopt any relative angle. Although the relative orientations of the N- and C-terminal domains of CP12 vary in our structures, the structures of both domains are conserved across 4 crystallographically independent CP12 molecules (Fig. 2 C and D).
Fig. 2.
Crystal structures of GAPDH-CP12 complex with full-length CP12. (A) GAPDH tetramer (blue) with 2 active sites bound by CP12 (pink). Residues 55–75 of CP12 are inserted in the active site of GAPDH, while 1–55 form a 2-helix PRK binding domain. (B) CP12 is formed of 2 domains connected by a flexible linker. The N-terminal PRK binding domain is formed of 2 anti-parallel helices connected by a disulfide bridge at the apex of the helices. Residues of the conserved CP12 AWDA(V/L)EEL motif are shown as sticks; disulfide bonds are indicated by yellow spheres. The C-terminal GAPDH binding domain is formed of a 2-turn helical region followed by the remaining C-terminal residues that insert into the GAPDH active site. This fold is also stabilized by a disulfide bridge. (C) The different conformations of CP12 observed in crystals (C1–C4) and cryoEM (EM) superposed on the C-terminal region, bound to a GAPDH tetramer (gray surface). (D) CP12 conformations superposed on the N-terminal PRK binding domain.
The CP12 N-terminal PRK binding domain is a 2-helix bundle, stabilized by a disulfide bridge between Cys-19 and 29 near the helical turn. The 2 helices bury a small hydrophobic core, including a short leucine zipper. Helix-2 (27-52) contains the conserved CP12 characteristic motif, AWDA(V/L)EEL (Figs. 2B and 3E) (25), which forms an acidic patch on the surface.
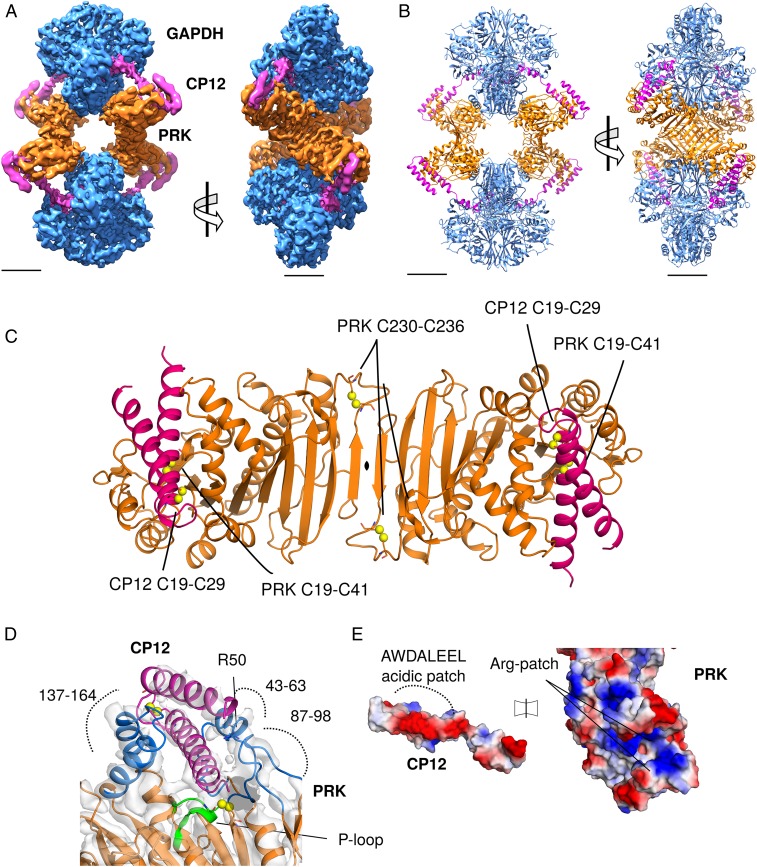
Fig. 3.
CryoEM reconstruction of GAPDH-CP12-PRK complex. (A) CryoEM maps of the complex with GAPDH (blue), PRK (orange), and CP12 (pink), (Scale bar, 30 Å.) (B) Cartoon view of the complex with 2 GAPDH tetramers and 2 PRK dimers forming an elongated diamond-shaped complex tethered by 4 CP12 chains. (C) Cartoon view of PRK dimer, with CP12 bound in the 2 active sites; disulfides in both proteins labeled. (D) CP12 (pink) sterically blocks PRK (orange) by binding in the active site (blue). The PRK active site cleft is formed of regions 147–164, 43–63, and 87–98. The PRK ATP-binding P-loop region (green) and Arg50 are indicated. (E) Electrostatic surface potential of CP12 and PRK interface regions, showing charge complementary. The CP12 conserved AWDA(V/L)EEL motif is highlighted.
Structure of GAPDH-CP12-PRK Complex.
We solved the structure of the full ternary GAPDH-CP12-PRK complex by cryoEM. The ternary complex was assembled by reconstituting recombinant GAPDH-CP12 with native PRK, partially purified from T. elongatus. PRK was purified from cell lysate and incubated with recombinant GAPDH-CP12 to form the ternary complex, which was isolated by size exclusion chromatography (SI Appendix, Figs. S4–S6). Frozen-hydrated samples were imaged in the electron microscope and used to generate a single-particle asymmetric C1 reconstruction with pseudo-D2 symmetry to 4.0 Å (FSC), with local resolution ranging from 3.6 Å to 6.2 Å (Fig. 3A and SI Appendix, Fig. S7). We used 3D classification to enrich for fully occupied CP12-PRK interfaces, then expanded the data according to the D2 symmetry and subjected these particles to 3D refinement with local search parameters to obtain our optimally featured density map (SI Appendix, Fig. S8). Our GAPDH-CP12 crystal structure together with models of PRK from an archaean (26) and cyanobacterium (7) were used as a basis for model building of the entire complex in Coot (27), and model refinement with Phenix (28) real space refine (Fig. 3B and SI Appendix, Table S2).
The inhibited GAPDH-CP12-PRK ternary complex has a hollow diamond-shaped architecture. GAPDH tetramers comprise 2 vertices separated by 200 Å, while the other 2 opposite vertices are formed by PRK dimers. Similar to our crystal structures of the binary complex, each GAPDH tetramer binds 2 copies of CP12. CP12 bridges the active sites of GAPDH and PRK, locking the complex in an inhibited conformation. The 4 CP12-PRK interaction interfaces were the least well-resolved regions of the reconstruction, having a worst resolution of 6.2 Å (SI Appendix, Fig. S7). Nevertheless, an atomic model of the entire complex could be fitted to the density. The GAPDH in the complex was similar to the crystal structure with NAD and 2 CP12 bound (RMSD 0.8 Å).
The CP12 N-terminal helical hairpin forms the interface with PRK. It has an RMSD of 0.9 Å (Fig. 2D) to the conformation present in the GAPDH-CP12 crystal structures and is in a conformation relative to GAPDH that we do not observe crystallographically (Fig. 2C). CP12 N-terminal domain is similar to that observed in the cyanobacterial CP12-CBS domain protein structure (8). Our full-length CP12 structures show that the oxidized N-terminal region of cyanobacterial CP12 is ordered as a helical pair prior to binding PRK, and oxidation via disulfide bridge formation primes this interaction. Given the similarities of all observed CP12 conformations together with high sequence conservation (SI Appendix, Fig. S2), we propose that this regulatory mechanism is evolutionarily conserved across cyanobacteria, algae, and plants.
Phosphoribulokinase Structure.
In the GAPDH-CP12-PRK complex, T. elongatus PRK is dimeric, as in solution (SI Appendix, Fig. S5E), and like other plant-type PRKs (6, 7). PRK has an alpha-beta-alpha sandwich fold where the central 9-strand beta-sheet is continuous across the dimer interface (Fig. 3C). The active site cleft lies between 3 loops (residues 137–164, 43–63, and 87–98). The N-terminal helical bundle of CP12 plugs this cleft and sterically blocks the active site (Fig. 3D). The charged patch created by the CP12 motif binds to complementary positively charged regions in the PRK active site (Fig. 3E), which have also been proposed to be important for negatively charged sugar phosphate substrate binding (6). Variants in the CP12 of Chlamydomonas reinhardtii, equivalent to Trp33, Glu37, and Glu38 in the conserved CP12 motif resulted in loss of complex formation (24). The conserved CP12-Trp33 is on the surface and packs against PRK, contradicting a prediction that it is buried (29). CP12-Glu33 makes a salt bridge with PRK-Arg164. In the PRK of C. reinhardtii, Arg64 is required for both ternary complex formation and full activity (30, 31). In our structure the equivalent residue, PRK-Arg50, is adjacent to the active site and contacts the outer face of the CP12 helical hairpin via the CP12 motif (Fig. 3D). Other interactions are hydrogen bonds between CP12-Asp34 to PRK-Ala60 backbone N, CP12-Glu14 to PRK-Lys54, and CP12-Gln40 to PRK-Asp146. The long flexible loop between residues 137–164 contains several active site residues, including Lys142, Asp146, Arg164. In a recent cyanobacterial PRK structure, it is only visible in one of the noncrystallographic dimers (7), and in our CP12-bound conformation, the loop is displaced outward relative to the crystal structure to accommodate the CP12 N-terminal domain.
Plant-type PRK has 2 conserved pairs of cysteine residues that form another tier of redox regulation to CP12 (32). Cys19 is in the middle of the ATP-binding Walker A motif (P-loop, residues 15–24, SI Appendix, Fig. S9) and forms a disulfide with Cys41. The loop containing Cys19 is flipped to bind Cys41 in our structure and the oxidized cyanobacterial PRK structure (7), but not in the reduced PRK structures from eukaryotes (6), or the equivalent loop in the nonredox regulated archaeal PRK (26) or homologous uridine kinases (33). When this disulfide bond is oxidized in free PRK, most activity is lost (8–10). When Cys19 is mutated to serine, although activity is retained, redox regulation is lost (32). We found no measurable PRK activity in the intact complex (Fig. 1B). Apart from the active site disulfide, PRK-Cys230 and Cys236 form a disulfide bond in the loop at the end of the sheet at the PRK dimer interface (Fig. 3C). This cysteine pair is nearly absolutely conserved in plant-type PRKs but is far from the active site and is not required for activity (34). Instead this disulfide is required for ternary complex formation in C. reinhardtii PRK (32). We predict that the formation of the PRK C-terminal disulfide locks the PRK dimer in a conformation that is competent to form the ternary complex. The dimer is flexible, as it is only linked by a strand–strand interaction. In our structure, the second monomer is rotated 8° relative to the cyanobacterial crystal structure. The final 10 C-terminal residues of PRK are not visible in the cryoEM model but form an extension that folds over the protein in the crystal structure. The displacement in the complex is presumably caused by the binding of GAPDH, which would otherwise clash with this region.
The sequential assembly of the inhibited ternary complex is driven by the relative strengths of interaction interfaces within the complex. In the ternary complex, PRK and GAPDH share a relatively small, nonconserved interface of only 330 Å2 buried surface area per GAPDH-PRK pair, with a predicted binding energy of −7 kcal/mol as calculated using the EBI-PISA server (35). We conclude that the ternary complex formation is dominated by the extensive contacts of GAPDH and CP12 (predicted binding energy −9.4 kcal/mol), and then between CP12 and PRK (predicted binding energy −7.4 kcal/mol). The GAPDH-PRK interactions may contribute to the stability of the complex, but are not sufficient for complex formation by themselves as PRK and CP12 also do not form a stable binary complex (17, 23, 36). We propose the sum of these relatively weak interactions has an avidity effect that makes the complex as a whole stable.
Enzyme Kinetics.
We used Michaelis–Menten kinetics to model the activities of GAPDH and PRK in their active and inhibited states (Fig. 1). PRK activity was assessed by measuring ADP production from ATP and ribulose 5-phosphate (Ru5P) in a coupled assay with ADP-hexokinase. PRK was completely inhibited in the ternary complex, where all of the active sites are blocked by CP12, and activity was restored after reduction with dithiothreitol (DTT) (Fig. 1B and SI Appendix, Table S3).
Although it remains possible that the difference in PRK activity observed between reduced and oxidized complexes may be due to the reduction of disulfide bonds present in CP12, PRK, or both, previous studies have shown that oxidized PRK alone retains ∼10% activity (8, 32)
GAPDH is a reversible enzyme, catalyzing the reduction of NAD(P)+ or oxidation of NAD(P)H. The physiological CB-cycle reaction is reduction of bisphosphoglycerate (BPG) by NADPH; however, the short-lived nature of BPG made it challenging to measure this activity accurately. Therefore, we measured GAPDH kinetics by an in situ reaction following NAD+ or NADP+ reduction by glyceraldehyde 3-phosphate GAP at 340 nm in a reverse of the physiological reaction.
We found that GAPDH activity with NADP+ was uninhibited in the GAPDH-CP12 complex, but was undetectable in the GAPDH-CP12-PRK complex. These data imply that NADPH can displace CP12 from the binary complex, but not the ternary complex, and that there is an important avidity stabilizing the GAPDH-CP12-PRK complex over GAPDH—CP12 effect from the complex formation. We found the apparent Km of GAPDH for NAD+ and NADP+ were similar, as were the apparent kcat values (Fig. 1 C and D and SI Appendix, Table S4). There is a small structural shift, mainly of Arg81 (Fig. 1A and SI Appendix, Fig. S10), in GAPDH depending on whether NAD+ or NADP+ is bound; however, this did not affect activity with either substrate. When CP12 was bound to GAPDH, activity decreased, and the enzyme was more specific for NAD+. The GAPDH-CP12 complex showed a much higher apparent Km for NADP+ than NAD+, which we attribute to CP12 being a competitive inhibitor for NADP+. The GAPDH-CP12 complex retained a high affinity for NAD+, although the rate of turnover was slower. These data can be explained by CP12 blocking the proximal GAPDH active sites and the steric hindrance of CP12 slowing NAD(H) binding to the remaining distal active sites. In the ternary complex, GAPDH activity with NAD+ was similar to the binary complex, implying that NAD+ can still access the open CP12 distal sites as in the GAPDH-CP12 binary complex.
These data suggest that cooperative avidity effects of ternary complex formation prevent NADP+ from dissociating CP12, which is bound more tightly in the ternary complex than the GAPDH-CP12 binary complex. For complex formation, NADP(H) must be replaced with NAD(H) (9, 10), suggesting that the formation of the GAPDH-CP12-PRK complex is integrating both redox and dinucleotide availability signals. The differential behavior of the complex to NADPH and NADH, where activity with the physiological substrate NADPH is more inhibited, shows that the complex function integrates responses from both redox state and nicotinamide dinucleotide availability.
GAPDH-CP12-PRK Complex Dissociation.
We tested the stability of the ternary complex using native gel electrophoresis and mass spectrometry after overnight incubation with combinations of NAD+, NADPH, ATP, and ADP (SI Appendix, Fig. S11). NADPH did not dissociate the cyanobacterial complex, which was consistent with our GAPDH kinetics data, where NADP+ reduction was inhibited in the ternary complex, and in contrast to reports on other species (9, 37, 38). In vitro, only reduction of disulfide bonds of CP12 and PRK with DTT reduced the disulfide bonds of CP12 and PRK, and dissociated the ternary complex (SI Appendix, Fig. S11). ATP and ADP did not apparently dissociate CP12 from PRK. In the homologous plant system, the complex is dissociated by a reduced thioredoxin, generated from ferredoxin by the light reactions (37).
Concluding Remarks.
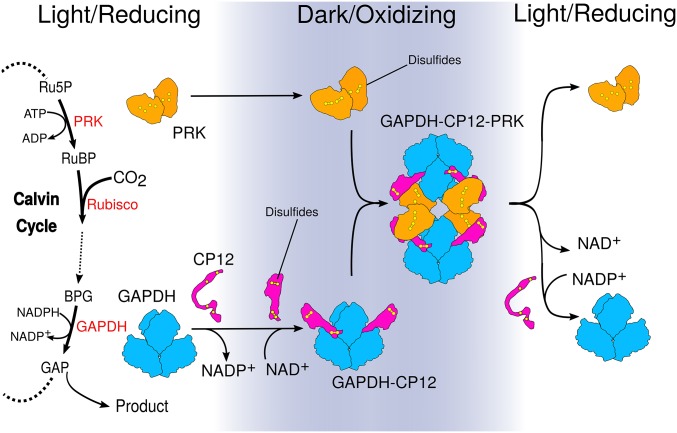
We investigated how structural changes in CP12 regulate substrate availability for carbon fixation in the CB cycle. We combined X-ray crystallography and single-particle cryoEM to determine the structures of GAPDH-CP12 and GAPDH-CP12-PRK. Our data provide a mechanism for how GAPDH and PRK activities are redox-regulated in response to light (Fig. 4). We show the structural basis for the conditional disorder of CP12 (16), where a redox-induced change causes a functional and structural switch. When bound to GAPDH, CP12 forms a disulfide-locked helical hairpin, which is ordered before interaction with PRK. GAPDH-CP12 has exchanged NADPH for NADH, and then captures PRK dimers to form a ternary complex that restricts production of RuBP, the substrate for Rubisco. The ternary complex prevents reduction of carboxylic acids to sugar by GAPDH with NADPH. Disulfide bonds within PRK and CP12 remain oxidized in the dark and maintain the inhibited complex. In the light, the complex is reactivated by reduction of CP12 and PRK by electrons from the photosynthetic electron transport chain. Our observations provide a redox-sensitive molecular mechanism, which also integrates signals of dinucleotide availability, that controls the “off switch” for how plants make biomass.
Fig. 4.
Model for how structural changes in redox-dependent complexes regulate carbon fixation. GAPDH and PRK are energy consuming enzymes of the Calvin–Benson cycle. The PRK catalyzed step directly precedes Rubisco carbon fixation. GAPDH (blue) activity is at the branch point between regeneration of RuBP or central metabolism. In the dark, the oxidizing environment causes intramolecular disulfide bridges to form within CP12 (pink) and PRK (orange). In parallel, NADP+ bound to GAPDH is exchanged for NAD+. CP12 binds to GAPDH, reducing its activity. Preordered CP12s subsequently recruit PRK, blocking PRK active sites to substrate and GAPDH active sites to NADP(H). When returning to the light, disulfide reduction dissociates the complex, releasing GAPDH and PRK.
Plants have evolved for effective reproduction under natural conditions, which is different to the maximum yield optimum desired in crops. Yield is closely correlated with photosynthetic carbon assimilation. A typical approach to augment yield is to improve partition into the harvested part of the plant. So far, photosynthetic efficiency has not been significantly improved in crop production, although reports have shown better growth with for example, CB-cycle enzyme overexpression (39), improved photorespiration (40) or improved photoprotection (41). In cyanobacteria, a CP12 knockout mutation improved flux through the CB cycle and productivity (42) but Arabidopsis and tobacco with anti-sense knockdown of CP12 were substantially impaired. CP12 overexpression was associated with chilling tolerance in a legume (43).
Driven by structural knowledge, it is possible that modulation of the GAPDH-CP12-PRK complex, and associated regulation in plants, will enable more subtle modifications that will increase carbon fixation in crops.
Materials and Methods
GAPDH and CP12 Expression and Purification.
GAPDH and CP12 were produced recombinantly in E. coli and purified by chromatography, then crystallized by vapor diffusion. Crystal structures were solved for GAPDH in different combinations with CP12, NAD+, and NADP+ (SI Appendix, Table S1). GAPDH activity was measured by following the reduction of NAD(P)+ in a linked enzyme assay.
Purification and Structure Solution of GAPDH-CP12-PRK Complex.
PRK was purified from T. elongatus cells by following the activity through chromatographic steps. PRK activity was measured using a coupled enzyme assay. The partially purified PRK was mixed with GAPDH-CP12 complex to yield the GAPDH-CP12-PRK complex, which was further purified by size-exclusion chromatography. The complex was adsorbed onto a holey-carbon grid, overlaid with a thin layer of amorphous carbon, then plunge-frozen in liquid ethane with a Vitrobot III robot. Final cryoEM data were collected at 300 keV on a Titan Krios microscope. The complex was reconstructed at 4.0 Å overall resolution in RELION (SI Appendix, Table S2), and an atomic model was built into the map and refined in real space.
Data Availability.
The atomic coordinates and structure factors for GAPDH-NAD+, GAPDH-NADP+, GAPDH-CP12-conf1-conf2, GAPDH-CP12-conf3, GAPDH-CP12-conf4, GAPDH-CP122 and GAPDH-CP124 have been deposited in the Protein Data Bank (PDB) under accession codes 6GFR, 6GFP, 6GFO, 6GHR, 6GHL, 6GFQ, and 6GG7, respectively. The GAPDH-CP12-PRK cryoEM map has been deposited in the Electron Microscopy Databank under EMDB-0071, and the atomic model has been deposited in the PDB, accession code 6GVE.
Supplementary Material
Acknowledgments
We thank Diamond for access and support of the CryoEM facilities at the UK national electron bio-imaging center (eBIC), (proposals EM19432 and EM18659), funded by the Wellcome Trust, Medical Research Council, and Biotechnology and Biological Sciences Research Council (BBSRC). We thank Diamond Light Source for X-ray beam time (proposal mx12579), and the staff of beam lines I02, I03, I24, and I04 for assistance with crystal testing and data collection. This work was supported by a BBSRC Doctoral Training Programme grant (BB/J014575/1 to C.R.M.). We thank project students Nishat Miah and Laura Briggs.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the PDB and EMDB repositories: PDB IDs 6GFP, 6GFQ, 6GFR, 6GFO, 6GG7, 6GHR, 6GHL, and 6GVE; EMDB ID EMD0071.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906722116/-/DCSupplemental.
References
- 1.Michelet L., et al. , Redox regulation of the Calvin-Benson cycle: Something old, something new. Front. Plant Sci. 4, 470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schürmann P., Buchanan B. B., The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10, 1235–1274 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Gütle D. D., et al. , Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc. Natl. Acad. Sci. U.S.A. 113, 6779–6784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiadmi M., Navaza A., Miginiac-Maslow M., Jacquot J.-P., Cherfils J., Redox signalling in the chloroplast: Structure of oxidized pea fructose-1,6-bisphosphate phosphatase. EMBO J. 18, 6809–6815 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miziorko H. M., Phosphoribulokinase: Current perspectives on the structure/function basis for regulation and catalysis. Adv. Enzymol. Relat. Areas Mol. Biol. 74, 95–127 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Gurrieri L., et al. , Arabidopsis and Chlamydomonas phosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle. Proc. Natl. Acad. Sci. U.S.A. 116, 8048–8053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson R. H., Hayer-Hartl M., Bracher A., Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301. Acta Crystallogr. F Struct. Biol. Commun. 75, 278–289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackenberg C., et al. , Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 7141–7146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamoi M., Miyazaki T., Fukamizo T., Shigeoka S., The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 42, 504–513 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Marri L., Trost P., Pupillo P., Sparla F., Reconstitution and properties of the recombinant glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase supramolecular complex of Arabidopsis. Plant Physiol. 139, 1433–1443 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandes H. K., Larimer F. W., Hartman F. C., The molecular pathway for the regulation of phosphoribulokinase by thioredoxin f. J. Biol. Chem. 271, 3333–3335 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Raines C. A., The Calvin cycle revisited. Photosynth. Res. 75, 1–10 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Pohlmeyer K., Paap B. K., Soll J., Wedel N., CP12: A small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol. Biol. 32, 969–978 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Wedel N., Soll J., Paap B. K., CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc. Natl. Acad. Sci. U.S.A. 94, 10479–10484 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gontero B., Maberly S. C., An intrinsically disordered protein, CP12: Jack of all trades and master of the Calvin cycle. Biochem. Soc. Trans. 40, 995–999 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Reichmann D., Jakob U., The roles of conditional disorder in redox proteins. Curr. Opin. Struct. Biol. 23, 436–442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graciet E., et al. , The small protein CP12: A protein linker for supramolecular complex assembly. Biochemistry 42, 8163–8170 (2003). [DOI] [PubMed] [Google Scholar]
- 18.López-Calcagno P. E., Howard T. P., Raines C. A., The CP12 protein family: A thioredoxin-mediated metabolic switch? Front. Plant Sci. 5, 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh P., Kaloudas D., Raines C. A., Expression analysis of the Arabidopsis CP12 gene family suggests novel roles for these proteins in roots and floral tissues. J. Exp. Bot. 59, 3975–3985 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fermani S., et al. , Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 104, 11109–11114 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumura H., et al. , Structure basis for the regulation of glyceraldehyde-3-phosphate dehydrogenase activity via the intrinsically disordered protein CP12. Structure 19, 1846–1854 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Fermani S., et al. , Conformational selection and folding-upon-binding of intrinsically disordered protein CP12 regulate photosynthetic enzymes assembly. J. Biol. Chem. 287, 21372–21383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marri L., et al. , Spontaneous assembly of photosynthetic supramolecular complexes as mediated by the intrinsically unstructured protein CP12. J. Biol. Chem. 283, 1831–1838 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Avilan L., et al. , CP12 residues involved in the formation and regulation of the glyceraldehyde-3-phosphate dehydrogenase-CP12-phosphoribulokinase complex in Chlamydomonas reinhardtii. Mol. Biosyst. 8, 2994–3002 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Stanley D. N., Raines C. A., Kerfeld C. A., Comparative analysis of 126 cyanobacterial genomes reveals evidence of functional diversity among homologs of the redox-regulated CP12 protein. Plant Physiol. 161, 824–835 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kono T., et al. , A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nat. Commun. 8, 14007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardebien F., Thangudu R. R., Gontero B., Offmann B., Construction of a 3D model of CP12, a protein linker. J. Mol. Graph. Model. 25, 186–195 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Avilan L., Gontero B., Lebreton S., Ricard J., Information transfer in multienzyme complexes–2. The role of Arg64 of Chlamydomonas reinhardtii phosphoribulokinase in the information transfer between glyceraldehyde-3-phosphate dehydrogenase and phosphoribulokinase. Eur. J. Biochem. 250, 296–302 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Roesler K. R., Marcotte B. L., Ogren W. L., Functional importance of arginine 64 in Chlamydomonas reinhardtii phosphoribulokinase. Plant Physiol. 98, 1285–1289 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thieulin-Pardo G., Remy T., Lignon S., Lebrun R., Gontero B., Phosphoribulokinase from Chlamydomonas reinhardtii: A Benson-Calvin cycle enzyme enslaved to its cysteine residues. Mol. Biosyst. 11, 1134–1145 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Tomoike F., Nakagawa N., Kuramitsu S., Masui R., Structural and biochemical studies on the reaction mechanism of uridine-cytidine kinase. Protein J. 34, 411–420 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Brandes H. K., Hartman F. C., Lu T.-Y., Larimer F. W., Efficient expression of the gene for spinach phosphoribulokinase in Pichia pastoris and utilization of the recombinant enzyme to explore the role of regulatory cysteinyl residues by site-directed mutagenesis. J. Biol. Chem. 271, 6490–6496 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Moparthi S. B., et al. , FRET analysis of CP12 structural interplay by GAPDH and PRK. Biochem. Biophys. Res. Commun. 458, 488–493 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Howard T. P., Metodiev M., Lloyd J. C., Raines C. A., Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc. Natl. Acad. Sci. U.S.A. 105, 4056–4061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wedel N., Soll J., Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate dehydrogenase complex dissociation. Proc. Natl. Acad. Sci. U.S.A. 95, 9699–9704 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Driever S., et al. , Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philos. Trans. R Soc. Lond. B Biol. Sci. 372, 20160384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.South P. F., Cavanagh A. P., Liu H. W., Ort D. R., Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kromdijk J., et al. , Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354, 857–861 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Kanno M., Carroll A. L., Atsumi S., Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria. Nat. Commun. 8, 14724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li K., et al. , Chloroplast protein 12 expression alters growth and chilling tolerance in tropical forage Stylosanthes guianensis (aublet) Sw. Front. Plant Sci. 9, 1319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for GAPDH-NAD+, GAPDH-NADP+, GAPDH-CP12-conf1-conf2, GAPDH-CP12-conf3, GAPDH-CP12-conf4, GAPDH-CP122 and GAPDH-CP124 have been deposited in the Protein Data Bank (PDB) under accession codes 6GFR, 6GFP, 6GFO, 6GHR, 6GHL, 6GFQ, and 6GG7, respectively. The GAPDH-CP12-PRK cryoEM map has been deposited in the Electron Microscopy Databank under EMDB-0071, and the atomic model has been deposited in the PDB, accession code 6GVE.