Abstract
Dogs are commonly used in human and veterinary pharmaceutical development. Physiologically based pharmacokinetic modeling using recombinant cytochrome P450 (CYP) enzymes requires accurate estimates of CYP abundance, particularly in liver. However, such estimates are currently available for only seven CYPs, which were determined in a limited number of livers from one dog breed (beagle). In this study, we used a label-free shotgun proteomics method to quantitate 11 CYPs (including four CYPs not previously measured), cytochrome P450 oxidoreductase, and cytochrome b5 in liver microsomes from 59 dogs representing four different breeds and mixed-breed dogs. Validation included showing correlation with CYP marker activities, immunoquantified protein, as well as CYP1A2 and CYP2C41 null allele genotypes. Abundance values largely agreed with those previously published. Average CYP abundance was highest (>120 pmol/mg protein) for CYP2D15 and CYP3A12; intermediate (40–89 pmol/mg) for CYP1A2, CYP2B11, CYP2E1, and CYP2C21; and lowest (<12 pmol/mg) for CYP2A13, CYP2A25, CYP2C41, CYP3A26, and CYP1A1. The CYP2C41 gene was detected in 12 of 58 (21%) livers. CYP2C41 protein abundance averaged 8.2 pmol/mg in those livers, and was highest (19 pmol/mg) in the only liver with two CYP2C41 gene copies. CYP1A2 protein was not detected in the only liver homozygous for the CYP1A2 stop codon mutation. Large breed–associated differences were observed for CYP2B11 (P < 0.0001; ANOVA) but not for other CYPs. Research hounds and Beagles had the highest CYP2B11 abundance; mixed-breed dogs and Chihuahua were intermediate; whereas greyhounds had the lowest abundance. These results provide the most comprehensive estimates to date of CYP abundance and variability in canine liver.
SIGNIFICANCE STATEMENT
This work provides the most comprehensive quantitative analysis to date of the drug-metabolizing cytochrome P450 proteome in dogs that will serve as a valuable reference for physiologically based scaling and modeling used in drug development and research. This study also revealed high interindividual variation and dog breed–associated differences in drug–metabolizing cytochrome P450 expression that may be important for predicting drug disposition variability among a genetically diverse canine population.
Introduction
Dogs are the most commonly used nonrodent species in pharmaceutical development (Tibbitts, 2003; Martignoni et al., 2006; Choi et al., 2011). The modern domestic dog has been divided through selective breeding into hundreds of isolated populations called breeds that were developed for desired phenotypic traits (Parker et al., 2017). Purpose-bred research Beagles are by far the most common breed of dog used in biomedical research because of their size, temperament, and ease of handling (Choi et al., 2011). Purpose-bred research hound and mongrel (mixed-breed) dogs are also used in biomedical research. A growing number of pharmaceuticals are being developed specifically for use in dogs. Although approval is often based on testing in homogenous populations of purpose-bred research dogs and a limited number of owned dogs, these drugs are intended for use in the genetically diverse general canine population.
Recombinant cytochrome P450 (CYP) enzymes are commonly used in human drug development to predict the contribution of different CYPs to drug metabolism (reaction phenotyping). This information is used to aid in the prediction of pharmacokinetics, drug interaction potential, safety, and efficacy of new chemical entities (Zientek and Youdim, 2015). Extrapolation of metabolic activities from recombinant CYPs to tissues requires an accurate estimate of the abundance of each CYP isoform in that tissue. Liver and intestines are the most important tissues involved in drug metabolism. Abundance values are also important for scaling beyond a single organ to whole-body clearance using physiologically based pharmacokinetic modeling leading to an estimation of population variability (Zientek and Youdim, 2015). Good CYP abundance estimates are available for human tissues (Achour et al., 2014), but only recently have accurate tissue estimates been published for dog CYPs. Liver and intestinal CYP abundance values are currently available for seven canine drug-metabolizing CYP isoforms, including CYP1A2, CYP2B11, CYP2C21, CYP2D15, CYP2E1, CYP3A12, and CYP3A26, which were obtained using tissues from purpose-bred research beagle dogs (Heikkinen et al., 2012, 2015).
There is some (albeit limited) evidence for breed-associated differences in CYP-mediated drug metabolism in dogs. Specifically, we have shown differences in the CYP2B11-dependent oxidation of propofol and benzyloxyresorufin by liver microsomes from beagle, greyhound, and mixed-breed dogs (Court et al., 1999; Hay Kraus et al., 2000). Beagles had significantly higher liver activities for both substrates compared with greyhounds, whereas mixed-breed dogs showed intermediate activities.
Previous studies evaluating CYP abundance in canine tissues did not include several CYPs of potential importance for drug metabolism, including CYP1A1, CYP2A13, CYP2A25, and CYP2C41. Recombinant CYP1A1 and CYP2C41 are commercially available, and several studies have reported catalytic activities for these isoforms (Shou et al., 2003; Locuson et al., 2009; Perez Jimenez et al., 2016, 2018). Although not yet commercially available, CYP2A13 and CYP2A25 expressed in a baculovirus–insect cell system were demonstrated in another study to have catalytic activities toward phenacetin and several other xenobiotic substrates (Zhou et al., 2010). Currently, it is not possible to extrapolate from these recombinant enzyme data to tissues to evaluate the contribution of these CYPs to drug metabolism in dogs.
The primary objective of this study was to provide the most comprehensive quantitative analysis to date of the drug-metabolizing CYP proteome in a moderately large (N = 59) and genetically diverse bank of canine liver microsomes. We employed an increasingly popular label-free shotgun proteomics method coupled with the total protein approach (TPA) that allows reliable absolute quantification of thousands of proteins without the need for labeled peptide standards (Cox and Mann, 2011). In addition to providing the first estimates of the abundance of CYP1A1, CYP2A13, CYP2A25, and CYP2C41 in canine liver, we also quantitated two important accessory proteins essential for CYP function, namely P450 oxidoreductase (POR) and cytochrome b5 (CYB5A). Finally, because our liver bank included samples from multiple dog breeds, including research Beagles, research hounds, greyhounds, Chihuahuas, and mixed-breed dogs, we evaluated whether the previously observed breed-related difference in CYP hepatic abundance was primarily restricted to one CYP isoform (i.e., CYP2B11) or affected other major CYP isoforms.
Materials and Methods
Chemicals and Reagents
Cytochrome c isolated from porcine heart was from Gold Biotechnology (St. Louis, MO). NADP+, NADPH, isocitrate dehydrogenase, DL-isocitrate, dextromethorphan, dextrorphan, thymol, trifluoroacetic acid, formic acid, and acetonitrile were from Sigma-Aldrich (St. Louis, MO). Urea and dithiothreitol were from Fisher Scientific (Pittsburgh, PA). Iodoacetamide and ammonium bicarbonate were the products of Acros Organics (Morris Plains, NJ). N-tosyl-L-phenylalanine chloromethyl ketone-treated trypsin was obtained from Worthington Biochemical (Freehold, NJ). Tramadol hydrochloride, O-desmethyltramadol hydrocholoride, O-desmethyltramadol-D6, and 4-hydroxypropofol were from Toronto Research Chemical (Toronto, ON, Canada). GW340416A, a chemical analog of bupropion, was provided by GlaxoSmithKline (Research Triangle Park, NC). Propofol was from Zeneca Pharmaceuticals (Wilmington, DE). Omeprazole was purchased from BeanTown Chemical (Hudson, NH), and omeprazole sulfone was purchased from Cayman Chemical (Ann Arbor, MI). Sodium hydroxide, potassium phosphate monobasic, potassium phosphate dibasic, and EDTA were from J. T. Baker (Center Valley, PA). Lysyl endopeptidase was from Wako Chemicals (Richmond, VA). Water Oasis HLB columns were from Waters (Milford, MA). Ultrapure water was from a Milli-Q Q-POD Millipore System (EMD Millipore, Burlington, MA).
Liver Microsomes
Liver tissue samples were from a bank of frozen dog livers maintained at Washington State University (Pullman, WA). Livers (DL-1 to DL-59) were from 59 healthy adult dogs that included 25 purpose-bred research Beagles (19 males and six females), 12 purpose-bred research hounds (all female), 13 mixed-breed dogs (seven males and six females), five greyhounds (three males and two females), and four Chihuahuas (three males and one female). Frozen liver samples from 14 research Beagles (11 males and three females) were purchased from Sekisui Xenotech (Kansas City, KS). The supplier indicated the dogs were from various sources, including Covance (Cumberland, VA), Xenometrics (Stilwell, KS), and Marshall BioResources (North Rose, TX). All remaining liver samples were obtained after euthanasia of dogs that were untreated control animals from various unrelated research studies. Although all dogs originated from the United States, the exact source of these latter animals was not available. Dogs were excluded if they had been treated with any pharmaceutical compound within two weeks before euthanasia. Other environmental exposures such as diet were unknown. Because all tissues were obtained from commercial sources, or were tissues that would have normally been discarded, use of these tissues was determined exempt from review by the Institutional Animal Care and Use Committee at Washington State University. Liver microsomes were prepared from the liver tissue samples, as previously described (Court et al., 1997). Microsomal protein concentrations were determined using the bicinchoninic acid assay (Thermo Fisher Scientific, Waltham, MA). Microsomes were stored at −80°C until use.
CYP1A2 and CYP2B11 Immunoblotting
Relative CYP1A2 and CYP2B11 protein contents were determined in dog liver microsomes DL-1 to DL-20 by adapting an immunoblotting method described previously (Court et al., 2003). Briefly, 5 µg microsomal protein was separated on 4%–15% gradient SDS acrylamide gels and electrophoretically transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, CA). Membranes were blocked for 1 hour at room temperature using 5% (w/v) nonfat dry milk in Tris-buffered saline–Tween. Rabbit anti-dog CYP2B11 serum (ɑ-PBD-2) was provided by J. Halpert (School of Pharmacy, University of Connecticut, Storrs, CT) (Duignan et al., 1987). Rabbit polyclonal anti-rat CYP1A2 serum (AB1255) was purchased from Chemicon Millipore (Temecula, CA). Blots were incubated with CYP2B11 or CYP1A2 antisera at 1:6000 or 1:500, respectively, overnight at 4°C. Blots were washed and then incubated with secondary horseradish peroxidase–conjugated goat anti-rabbit antisera (NEF812; PerkinElmer, Waltham, MA) at 1:10,000 dilution for 1 hour at room temperature, and chemiluminescent substrate (SuperSignal; Thermo Fisher Scientific) was applied. Blots were imaged by charge-coupled device camera (Kodak Image Station 440CF; Eastman Kodak, Rochester, NY), and bands were quantified using Kodak 1D Image Analysis Software (Eastman Kodak). Consistent protein loading and blot transfer were verified by Ponceau-S staining of blots. Standard curves for each CYP assayed were generated by loading serial dilutions of microsomes from those livers with the highest CYP2B11 and CYP1A2 content (based on preliminary experiments). Values were expressed relative to the liver with the lowest CYP content for the CYP being assayed. The reported results represent the average of three independent experiments.
CYP and Accessory Enzyme Activities
Tramadol O-Demethylation and N-Demethylation.
Tramadol O-demethylation and N-demethylation were measured in liver microsomes as marker reactions for CYP2D15 and CYP2B11 activities, respectively. We have previously shown that these reactions are primarily catalyzed by these CYP enzymes, and the metabolism assays were carried out as previously described (Perez Jimenez et al., 2016) using a final tramadol concentration of 5 µM and 0.2 mg/ml microsomal protein. Preliminary experiments confirmed the linearity of metabolite formation with respect to microsomal protein concentration and incubation times. All experiments were carried out in duplicate and repeated at least two indepndent times, and results were averaged.
Propofol Hydroxylation.
We have also previously shown that propofol hydroxylation is primarily catalyzed by canine CYP2B11 (Court et al., 1999; Hay Kraus et al., 2000). Measurement of propofol hydroxylation by liver microsomes was carried out as we previously described with slight modifications, as follows. A final concentration of 5 µM propofol and 0.05 mg/ml microsomal protein was used in the reaction with a 10-minute incubation time. Preliminary experiments confirmed the linearity of metabolite formation with respect to microsomal protein concentration and incubation times. All experiments were carried out in duplicate and repeated at least three independent times, and results were averaged. The high-pressure liquid chromatography column used was a 4 µM 150 × 2 mm Phenomenex Synergi Fusion-RP 80 Å (Torrance, CA). A gradient mobile phase (total flow of 0.4 ml/min) was used consisting of mobile phases A (100% acetonitrile) and B (80% 20 mM phosphate buffer and 20% acetonitrile, v/v). The gradient was as follows: linear gradient from 10% to 20% A over 10 minutes, 20%–46% A over 10 minutes, 46%–100% A over 2 minutes, and 100%–10% A over 1 minute.
Dextromethorphan O-Demethylation.
Dextromethorphan O-demethylation by the liver microsomes was used as a marker for CYP2D15 activity. This reaction has been previously reported to be selective for CYP2D15 (Shou et al., 2003). Dextromethorphan in methanol was added to microcentrifuge tubes to yield a final reaction concentration of 5 µM. Tubes were evaporated to dryness using a Savant SpeedVac Concentrator (Thermo Fisher Scientific). To reaction tubes, microsomal protein was added to achieve a final protein concentration of 0.1 mg/ml in 50 mM potassium phosphate buffer (pH 7.4). Reaction tubes were placed in a 37°C water bath for 3 minutes, and the reaction was started by adding 50 µl NADPH–regeneration system (0.5 mM NADP+, 3.75 mM DL-isocitrate, 1 U/ml isocitrate dehydrogenase, 5 mM MgCl2 in 50 mM potassium phosphate buffer, pH 7.4, final reaction concentration). The final reaction volume was 100 µl. The reaction was allowed to proceed for 10 minutes at 37°C and was stopped by the addition of 20 µl 1 N HCL. Ten microliters of 10 µM GW3404164 in methanol was added as the internal standard. Samples were vortexed and centrifuged for 10 minutes at 15,000g. Supernatant (10 µl) was analyzed using liquid chromatography (LC) tandem mass spectrometry (LC-MS/MS). Quantification of dextrorphan was based on calibration curves constructed using the peak area ratio of dextrorphan to the internal standard against dextrorphan concentrations using unweighted least squares linear regression. The rate of dextrorphan formation was calculated by dividing the amount of dextrorphan in the sample by the incubation time and the protein concentration present in the reaction. Preliminary experiments confirmed the linearity of metabolite formation with respect to microsomal protein concentration and incubation times. All experiments were replicated three independent times, and results were averaged.
The LC-MS/MS system consisted of an AB Sciex AP1 4000 (AB Sciex, Framingham, MA) connected to the liquid chromatography portion, a HP Agilent 1100 Series G1311A quaternary pump, G13221 degasser, and G1316A column compartment, as well as a HTC PAL autosampler (CTC Analytics AG, Zwingen, Switzerland). Data collection and integration were achieved using AB Sciex Analyst software (version 1.6). The LC-MS/MS system was configured with electrospray ionization in the positive mode, and data were acquired using multiple reaction monitoring of mass transition pairs at m/z 258/2/133.1 for dextrorphan and m/z 238/182 for the internal standard. The analytical column was the same as described for the propofol hydroxylation method. A gradient mobile phase was used consisting of mobile phases A (0.1% formic acid) and B (acetonitrile). The gradient was as follows: linear gradient from 80% to 5% A over 2 minutes, held at 5% A for 0.5 minute, followed by a step up to 80% A over 0.11 minutes. The column equilibrated for 3.4 minutes at 80% A prior to the next injection. Separation was carried out at 30°C with a flow rate of 0.4 ml/min.
Omeprazole Sulfonation.
Omeprazole sulfonation was used as a CYP3A marker activity based on reported human CYP3A selectivity (Andersson et al., 1993; Yamazaki et al., 1997). Omeprazole and microsomal protein, both in 50 mM phosphate buffer (pH 7.4), were added to microcentrifuge tubes to yield final reaction concentrations of 5 µM omeprazole and 0.2 mg/ml protein. Reaction tubes were placed in a 37°C water bath for 3 minutes, and the reaction was started by adding 50 µl of the previously described NADPH–regeneration system. The final volume of the reaction was 100 µl. The reaction was allowed to proceed for 15 minutes at 37°C and was stopped by the addition of 100 µl internal standard (1 µM pantoprazole in acetonitrile). Samples were vortexed and centrifuged for 10 minutes at 15,000g. Supernatant (10 µl) was analyzed using LC-MS/MS. Quantification of omeprazole sulfone was based on calibration curves constructed using the peak area ratio of omeprazole sulfone to the internal standard against omeprazole sulfone concentrations using 1/x weighted least squares linear regression. The rate of omeprazole sulfone formation was calculated by dividing the amount of omeprazole sulfone in the sample by the incubation time and the protein concentration present in the reaction. Preliminary experiments confirmed the linearity of metabolite formation for microsomal protein concentrations and incubation time. All experiments were replicated a minimum of three independent times, and results were averaged.
The LC-MS/MS system and configuration were the same as described for dextromethorphan O-demethylation. Data were acquired using multiple reaction monitoring of mass transition pairs at m/z 362/150.1 for omeprazole sulfone and m/z 384.2/200.1 for the internal standard. The analytical column was an Agilent (Santa Clara, CA) ZORBAX Exclipe XDB-C18 (5 µm, 2.1 × 50 mm). A gradient mobile phase was used consisting of mobile phases A (0.1% formic acid), B (methanol), and C (acetonitrile). The gradient was as follows: linear gradient from 90:9:1 (A:B:C, v/v) to 80:10:10 (A:B:C, v/v) over 1 minute, linear gradient to 55:22.5:22.5 (A:B:C, v/v) over 1.75 minutes, followed by a step to 90:9:1 (A:B:C, v/v/v) over 0.25 minute. The column equilibrated for 2 minutes at starting conditions prior to the next injection. Separation was carried out at 40°C with a flow rate of 1 ml/min.
Cytochrome c Reduction.
P450 oxidoreductase activity in the liver microsomes was measured by the reduction of cytochrome c. The assay was carried out as described by Guengerich et al. (2009) but modified to fit a microplate format using the SpectraMax i3 plate reader with Softmax Pro 6.3 software (Molecular Devices, San Jose, CA). Briefly, 5 µg total microsomal protein in 10 μL was used for each reaction. To each reaction, 190 µl cytochrome c dissolved in 300 mM potassium phosphate buffer, pH 7.8, containing 0.1 mM EDTA was added, resulting in a final reaction concentration of 30 µM cytochrome c. The reaction was started with 20 µl NADPH dissolved in water (final reaction concentration of 100 µM NADPH) and monitored at A550 by the plate reader in kinetic mode for 8 minutes at 22°C. The reaction rates were determined from the linear range of the kinetic trace and molar extinction coefficient of 21.1 mM−1 cm−1 for reduced cytochrome c. The experiment was replicated three independent times, and results were averaged.
Genotyping of CYP1A2 and CYP2C41 Polymorphisms.
Genomic DNA was isolated from 58 dog livers using DNAzol reagent following the manufacturer’s protocol (Invitrogen, Carlsbad, CA). One liver (DL-11) had insufficient tissue for DNA extraction. DNA samples were genotyped for two previously reported null CYP polymorphisms: the CYP1A2 premature stop codon mutation (c.1117C>T; Arg373Stop) (Tenmizu et al., 2004) and the CYP2C41 gene deletion polymorphism (Blaisdell et al., 1998). Assays were performed using a CFX96 Real-Time System with C1000 Touch Thermal Cycler with CFX Manager v. 3.0 software (Bio-Rad Laboratories). A custom-designed TaqMan Single nucleotide polymorphism genotyping assay (Applied Biosystems, Forest City, CA) for the CYP1A2 Arg373Stop variation was employed. Primer sequences were 5′-CCT TAA TGG AGG CCT TCA TCC-3′ and 5′-CAG AAG AAG CAG GCC TTA CCT-3′. Reporter sequences were 5′-VIC-ATC TTC CGA CAC ACC T-NFQ-3′ and 5′-FAM-ATC TTC TGA CAC ACC T-NFQ-3′. The assay was performed with TaqMan Genotyping Master Mix (Applied Biosystems) using 10 ng DNA in a 20 µl reaction. The cycling conditions were as follows: 95°C for 10 minutes for enzyme activation and initial denaturation, followed by 70 cycles of 95°C for 15 seconds and 60°C for 1.5 minutes. Allele calls were made using the CFX Manager software. Results were validated by direct sequencing of representative samples.
Predesigned TaqMan gene expression assays (Applied Biosystems) were used to determine CYP2C41 gene copy number with canine UGT1A as the reference gene. The CYP2C41 assay (Cf03054282_u1 ) amplified a 103-bp region within exon 8 of the CYP2C41 gene (Genbank IDs: mRNA-NM_001003334; gene-HF677515). The reference gene assay (Cf03987062_sH ) amplified a 98-bp region within the highly conserved exon 5 of the UGT1A gene [Genbank IDs: mRNA-NM_001003078; gene-NC_006607.3 (45081655..45161584)]. Paired (CYP2C41 and reference gene) assays were performed with TaqMan Gene Expression Master Mix (Applied Biosystems) using 10 ng DNA per 20 µl reaction. The cycling conditions were the same as described above for CYP1A2 genotyping. The amount of CYP2C41 DNA normalized to reference gene DNA content was calculated using the 2^(−ΔΔCt) method. Results were expressed relative to the liver sample with the highest CYP2C41 DNA content (DL-44), which was anticipated to contain two CYP2C41 gene copies. The reproducibility of results was verified by analysis of representative DNA samples with zero, one, and two CYP2C41 gene copies in three independent experiments. Observed CYP1A2 and CYP2C41 genotype frequencies were evaluated for consistency with the expected genotype frequencies (Hardy–Weinberg equilibrium distribution) by χ2 test with one degree of freedom.
Mass Spectrometry Quantitation of CYP and Accessory Enzymes
LC-MS/MS Proteomic Analysis.
Aliquots of 80 µg microsomal protein were used for proteomics sample preparation based on a previously reported Lys-C/trypsin combinatorial digest protocol (Glatter et al., 2012) with some modifications, as described by Shi et al. (2018a). LC-MS/MS analysis was performed on a TripleTOF 5600+ mass spectrometer (AB Sciex) coupled with an Eksigent 2D plus LC system (Eksigent Technologies, Dublin, CA). A trap-elute configuration was used for LC separation, which includes a trapping column (ChromXP C18-CL, 120 Å, 10 × 0.3 mm, 5 μm cartridge; Eksigent Technologies) and an analytical column (ChromXP C18-CL, 120 Å, 150 × 0.3 mm, 5 μm; Eksigent Technologies). Five micrograms of protein was injected for analysis from each sample, followed by a blank injection to prevent sample carryover. The LC method and ion source settings were identical to our previous report (Shi et al., 2018b).
Data-dependent acquisition was performed with a 250-ms time-of-flight mass spectrometry scan from 400 to 1250 Da, followed by tandem mass spectrometry scans in high sensitivity mode from 100 to 1500 Da for the top 30 precursor ions from the time-of-flight mass spectrometry scan (50-ms accumulation time, 10 ppm mass tolerance, charge state from +2 to +5, rolling collision energy, and dynamic accumulation). Former target ions were excluded from tandem mass spectrometry scan for 15 seconds.
Proteomics Data Analysis.
The proteomics data were analyzed using the MaxQuant software (version 1.6.1.0; Max Planck Institute of Biochemistry) with default settings. The dog proteome FASTA file with 25,491 entries (Proteome ID UP000002254, downloaded from UniProtKB on 1/31/2018) was used as reference.
Absolute protein quantification was based on the TPA method. It assumes that the total mass spectrometry (total MS) signal from all identified proteins in the dataset reflects the total protein amount, and the mass spectrometry (MS) signal from an individual protein corresponds to its abundance in the sample (Wiśniewski, 2017). Thus, the concentration of a protein (moles per gram total protein) was calculated by the following equation:
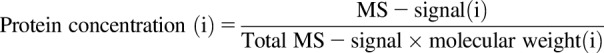 |
Statistical Analyses.
Shapiro–Wilk test for normality was used to determine the distribution shape of activity and protein abundance data. Most activity and protein abundance data sets were not normally distributed, warranting the use of nonparametric methods for statistical analyses. Relationships between protein abundance and enzyme activities were evaluated by calculation of the Spearman correlation coefficient. ANOVA on rank-transformed data was carried out to determine breed significance on protein expression. Mann–Whitney U tests for two groups and Dunn’s multiple comparison for three or more groups were used to compare enzyme expression differences between gender and breed, respectively. Statistical analyses were performed using Sigma Plot 12 software (Systat Software, San Jose, CA). For most statistical tests, a P value of less than 0.05 was considered statistically significant. For correlation analyses, a Rs value of less than 0.05 was considered indicative of a meaningful (moderate to strong) relationship between the variables being evaluated.
Results
Expression Profiles of 11 CYPs, POR, and CYB5A in Canine Liver Microsomes
A total of 59 canine liver specimens from four different dog breeds and mixed-breed dogs were assayed to examine interindividual and breed-related differences in absolute expression of 11 drug-metabolizing CYP enzymes (CYP1A1, CYP1A2, CYP2A13, CYP2A25, CYP2B11, CYP2C21, CYP2C41, CYP2D15, CYP2E1, CYP3A12, and CYP3A26) and two accessory proteins (POR and CYB5A) using a quantitative LC-MS/MS method. Table 1 summarizes the mean (CV%) microsomal abundance values for these data for all dog livers (N = 59), as well as a subset containing only research beagle dog livers (N = 25). Results for microsomal abundance values for each of these CYP isoforms and accessory proteins in individual dog livers are given in Supplemental Table 1.
TABLE 1.
Abundance of CYP enzymes and accessory proteins (P450 oxidoreductase, POR; cytochrome b5A, CYB5A) in a bank of liver microsomes measured by LC-MS/MS
Shown are the mean and CV% abundance values for the entire liver bank (N = 59) representing four different breeds and mixed-breed dogs, as well as for a subset of beagle dogs only (N = 25). Shown for comparison are abundance values reported by other investigators for the same CYPs in 18 beagle dogs by Heikkinen et al. (2015) using LC-MS/MS and in 12 beagle dogs by Eguchi et al. (1996) using ELISA.
| Protein | Mean (CV%) Liver Microsome Abundance (pmol/mg Protein) | |||
|---|---|---|---|---|
| This Study | Heikkinen et al. (2015) | Eguchi et al. (1996) | ||
| All Dogs | Research Beagles | Research Beagles | Research Beagles | |
| (N = 59) | (N = 25) | (N = 18) | (N = 12) | |
| CYP1A1 | 3.1 (276) | 3.3 (247) | — | — |
| CYP1A2 | 89 (105) | 123 (104) | 37 (60) | 16 (57)a |
| CYP2A13 | 11 (60) | 10 (66) | — | — |
| CYP2A25 | 8.1 (47) | 8.7 (39) | — | — |
| CYP2B11 | 79 (52) | 86 (47) | 35 (34) | 51 (14) |
| CYP2C21 | 54 (36) | 64 (32) | 70 (23) | 160 (21) |
| CYP2C41b | 8.2 (60) | 10 (48) | — | — |
| CYP2D15 | 143 (27) | 139 (29) | 56 (39) | — |
| CYP2E1 | 73 (28) | 73 (26) | 36 (41) | — |
| CYP3A12 | 124 (38) | 121 (28) | 93 (33) | 69 (29) |
| CYP3A26 | 3.7 (49) | 3.3 (46) | 2.7 (34) | — |
| POR | 40 (28) | 42 (27) | — | — |
| CYB5A | 431 (16) | 456 (17) | — | — |
Assumed to be primarily CYP1A2, but the ELISA method used did not distinguish between CYP1A1 and CYP1A2. “-” indicates that abundance of those proteins was not determined.
Note that the results given for CYP2C41 are for dogs that express at least one copy of the CYP2C41 gene (12 dogs total, including four Beagles).
Six of 11 CYP isoforms and both of the accessory proteins could be quantified in all 59 dog liver microsome samples. CYP1A2 and CYP2A13 were quantifiable in 58 samples, CYP3A26 in 55 samples, CYP1A1 in 23 samples, and CYP2C41 in 12 of 59 liver samples. Comparing average protein abundance of the 11 CYP isoforms and two accessory proteins in all liver samples, three proteins were highly expressed: CYB5A > CYP2D15 > CYP3A12 (more than 120 pmol/mg microsomal protein). Five proteins were intermediately expressed: CYP1A2 = CYP2B11 = CYP2E1 > CYP2C21 > POR (40–89 pmol/mg microsomal protein). The remaining five proteins had low expression: CYP2A13 = CYP2A25 = CYP2C41 > CYP3A26 = CYP1A1 (less than 12 pmol/mg microsomal protein). As indicated by CV% values, interindividual variability in protein abundance was high (105%–276%) for two CYPs: CYP1A1 > CYP1A2; intermediate (36%–67%) for seven CYPs: CYP2C41 = CYP2A13 > CYP2B11 = CYP3A26 = CYP2A25 > CYP3A12 = CYP2C21; and low (16%–28%) for two CYPs and the two accessory proteins: POR = CYP2E1 = CYP2D15 > CYB5A.
Correlation with Immunoblot Estimates, Marker Activities, and Null Alleles
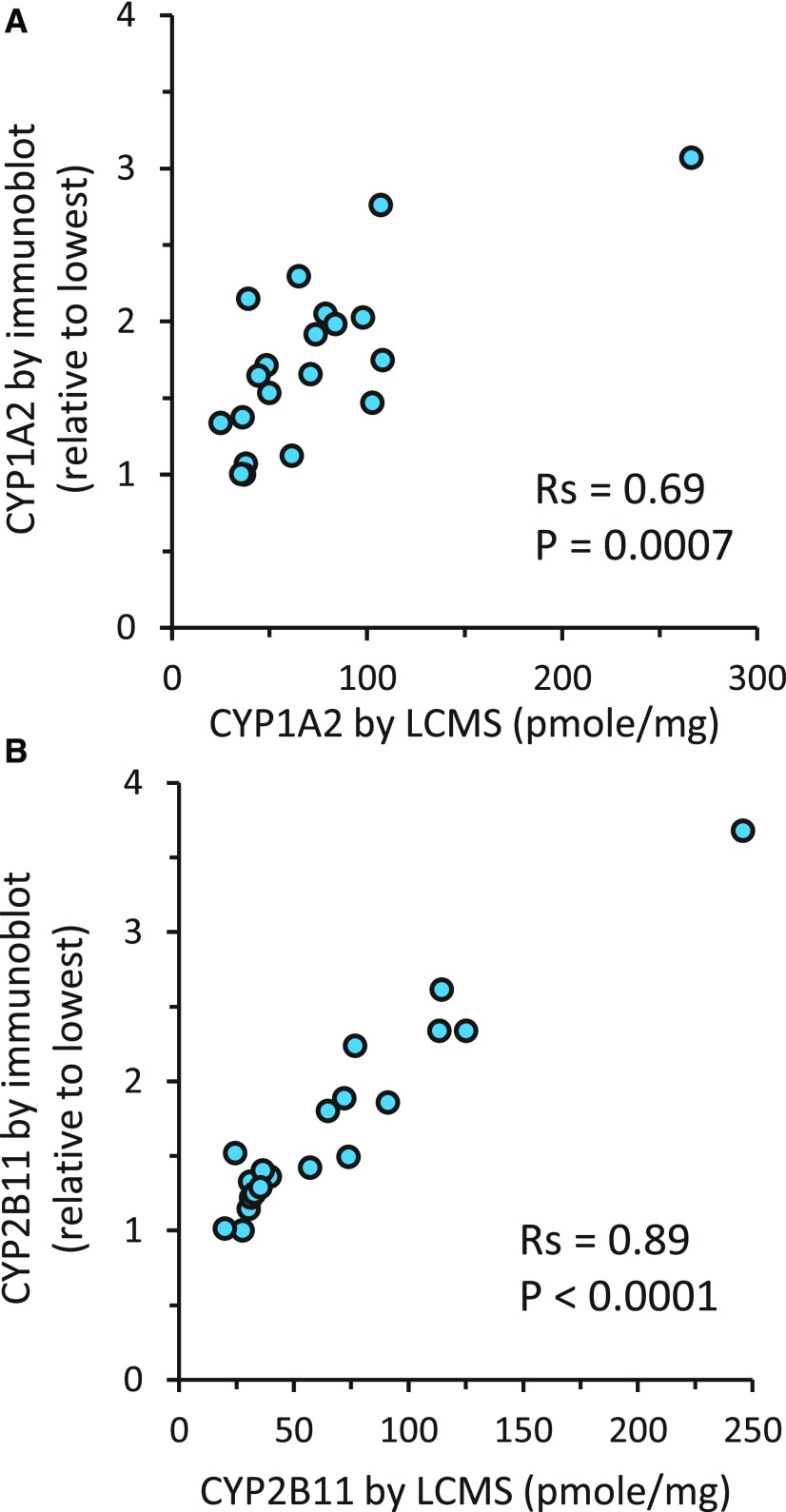
Additional validation of the current LC-MS/MS–derived abundance values was performed by correlation with immunoblot-quantified expression levels, marker enzyme activities, and null genotypes for selected CYPs determined in the same liver samples. Results of Spearman correlation analyses between immunoquantified (CYP1A2 and CYP2B11) and LC-MS/MS quantified protein concentrations are given in Table 2. Immunoquantified CYP1A2 abundance correlated most strongly with LC-MS/MS–quantified CYP1A2 (Rs = 0.69, P < 0.001; Fig. 1A), but poorly with concentrations of all other CYP and accessory proteins (Rs < 0.5). Similarly, immunoquantified CYP2B11 abundance correlated very strongly with LC-MS/MS–quantified CYP2B11 (Rs = 0.89, P < 0.001; Fig. 1B), but not with concentrations of any other CYP or accessory proteins (Rs < 0.4).
TABLE 2.
Correlation of the abundance of CYPs and accessory proteins (P450 oxidoreductase, POR; cytochrome b5A, CYB5A) measured by quantitative mass spectrometry with the abundance of CYP1A2 and CYP2B11 measured by semiquantitative immunoblot in the same set of dog liver microsomes (N = 20)
Shown are the Spearman correlation coefficients (Rs) and the associated P values for each correlation.
| Spearman Correlation Coefficient (P Value) | ||
|---|---|---|
| Immunoblot CYP1A2 | Immunoblot CYP2B11 | |
| CYP1A1 | 0.25 (0.29) | −0.08 (0.72) |
| CYP1A2 | 0.69 (0.0007) | 0.26 (0.27) |
| CYP2A13 | −0.24 (0.31) | −0.28 (0.22) |
| CYP2A25 | 0.23 (0.33) | 0.09 (0.71) |
| CYP2B11 | 0.33 (0.15) | 0.89 (<0.0001) |
| CYP2C21 | −0.04 (0.87) | 0.2 (0.4) |
| CYP2D15 | −0.24 (0.31) | −0.28 (0.23) |
| CYP2E1 | −0.28 (0.23) | −0.64 (0.003) |
| CYP3A12 | 0.32 (0.17) | 0.23 (0.32) |
| CYP3A26 | −0.06 (0.79) | 0.27 (0.26) |
| POR | 0.44 (0.05) | 0.13 (0.57) |
| CYB5A | 0.12 (0.4) | 0.22 (0.34) |
Fig. 1.
Correlation between protein abundance estimates of CYP1A2 (A) and CYP2B11 (B) measured by quantitative mass spectrometry and by semiquantitative immunoblot in the same set of dog liver microsomes (N = 20). Also shown are the Spearman correlation coefficients (Rs) and the associated P values for each correlation.
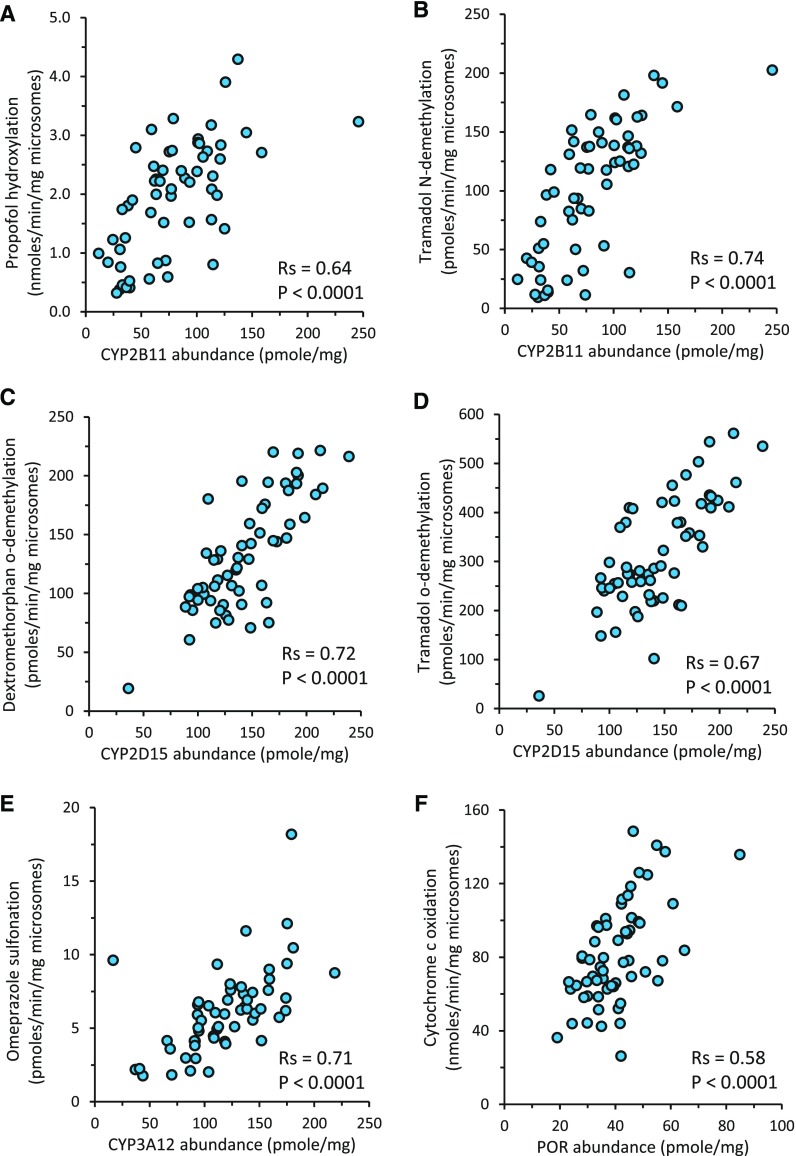
Correlation of marker activities for CYP2B11, CYP2D15, CYP3A12, and POR with LC-MS/MS abundance values measured in the same livers was assessed (Table 3). Both CYP2B11 markers, propofol hydroxylation (Rs = 0.64, P < 0.001; Fig. 2A) and tramadol N-demethylation (Rs = 0.74, P < 0.001; Fig. 2B), best correlated with CYP2B11 abundance. Moderate correlation was also found between propofol hydroxylation and CYB5A abundance (Rs = 0.58, P < 0.001), whereas weaker or no correlation was observed between both of the CYP2B11 marker activities and abundance estimates of other CYPs or accessory proteins (Rs < 0.5). Similarly, both CYP2D15 markers, including dextromethorphan O-demethylation (Rs = 0.72, P < 0.001; Fig. 2C) and tramadol O-demethylation (Rs = 0.67, P < 0.001; Fig. 2D), best correlated with CYP2D15 abundance. Much weaker or no correlation was observed between both of the CYP2D15 marker activities and abundance estimates of other CYPs or accessory proteins (Rs < 0.5). Omeprazole sulfonation, a CYP3A maker activity, best correlated with CYP3A12 abundance (Rs = 0.71, P < 0.0001; Fig. 2E), whereas weaker or no correlation was observed between omeprazole sulfonation and abundance estimates of other CYPs or accessory proteins (Rs < 0.5). Lastly, cytochrome c reduction activity (Rs = 0.58, P < 0.001; Fig. 2F) best correlated with POR abundance, but not with CYB5A or CYP abundance estimates (Rs < 0.5).
TABLE 3.
Correlation of CYP and accessory protein (P450 oxidoreductase, POR; cytochrome b5A, CYB5A) concentrations with marker enzyme activities measured in the same set of dog liver microsomes (N = 59)
Propofol hydroxylation and tramadol n-demethylation were used as markers of CYP2B11 activity; dextromethorphan O-demethylation and tramadol O-demethylation were used as markers of CYP2D15 activity; omeprazole sulfonation was used as a marker of CYP3A activity; whereas cytochrome c oxidation was used as a marker of POR activity. Activity data are presented as an average of multiple independent replicates (number of replicates for each assay described in Materials and Methods). The average variation between replicate activity assays was less than 20%. Shown are the Spearman correlation coefficients (Rs) and the associated P values for each correlation.
| Spearman Correlation Coefficient (P Value) | ||||||
|---|---|---|---|---|---|---|
| Propofol Hydroxylation | Tramadol N-Demethylation | Dextromethorphan O-Demethylation | Tramadol O-Demethylation | Omeprazole Sulfonation | Cytochrome c Oxidation | |
| CYP1A1 | −0.01 (0.9) | −0.14 (0.3) | −0.28 (0.03) | −0.19 (0.2) | 0.17 (0.2) | 0.28 (0.03) |
| CYP1A2 | 0.18 (0.2) | 0.05 (0.7) | −0.17 (0.2) | −0.16 (0.2) | 0.26 (0.05) | 0.38 (0.003) |
| CYP2A13 | 0.16 (0.2) | 0.16 (0.2) | 0.43 (0.0008) | 0.38 (0.003) | 0.15 (0.3) | −0.23 (0.09) |
| CYP2A25 | 0.09 (0.5) | −0.08 (0.5) | 0.15 (0.3) | 0.1 (0.5) | 0.17 (0.2) | 0.26 (0.05) |
| CYP2B11 | 0.64 (<0.0001) | 0.74 (<0.0001) | 0.22 (0.1) | 0.13 (0.3) | 0.43 (0.0009) | 0.07 (0.6) |
| CYP2C21 | 0.36 (0.005) | 0.18 (0.2) | 0.26 (0.05) | 0.31 (0.02) | 0.01 (0.9) | 0.43 (0.0008) |
| CYP2C41a | 0.36 (0.005) | 0.39 (0.003) | 0.24 (0.06) | 0.22 (0.1) | 0.08 (0.5) | −0.02 (0.9) |
| CYP2D15 | 0.32 (0.01) | 0.32 (0.01) | 0.72 (<0.0001) | 0.67 (<0.0001) | 0.23 (0.08) | −0.1 (0.4) |
| CYP2E1 | −0.06 (0.7) | −0.13 (0.3) | 0.15 (0.3) | 0.19 (0.2) | −0.13 (0.3) | 0.06 (0.6) |
| CYP3A12 | 0.49 (0.0002) | 0.42 (0.001) | 0.21 (0.1) | 0.07 (0.6) | 0.71 (<0.0001) | 0.19 (0.2) |
| CYP3A26 | 0.49 (0.0002) | 0.42 (0.0009) | 0.34 (0.009) | 0.24 (0.07) | 0.5 (<0.0001) | −0.07 (0.6) |
| POR | 0.4 (0.002) | 0.2 (0.1) | 0.01 (0.9) | 0.05 (0.7) | 0.47 (0.0002) | 0.58 (<0.0001) |
| CYB5A | 0.58 (<0.0001) | 0.39 (0.003) | 0.38 (0.003) | 0.29 (0.02) | 0.48 (0.0002) | 0.42 (0.001) |
Note that the results given for CYP2C41 are for the 12 dogs that express at least one copy of the CYP2C41 gene (four males and eight females).
Fig. 2.
Correlation between protein abundance estimates of CYP2B11 (A and B), CYP2D15 (C and D), CYP3A12 (E), and POR (F) with their respective enzyme marker activities propofol hydroxylation (A), tramadol N-demethylation (B), dextromethorphan O-demethylation (C), tramadol O-demethylation (D), omeprazole sulfonation (E), and cytochrome c oxidation (F) measured in the same set of dog liver microsomes (N = 59). Activity data are presented as an average of multiple independent replicates (number of replicates for each assay described in Materials and Methods). The average variation between replicate activity assays was less than 20%. Also shown are the Spearman correlation coefficients (Rs) and the associated P values for each correlation.
Dog livers were genotyped for two known CYP polymorphisms, the CYP1A2 premature stop codon mutation (Arg373Stop) and the CYP2C41 gene deletion polymorphism, each predicted to result in reduced (when heterozygous) or absent (when homozygous) amounts of the respective protein. One dog liver (DL-11) could not be genotyped because a DNA sample was unavailable. Observed CYP1A2 and CYP2C41 genotype frequencies did not deviate from the expected Hardy–Weinberg equilibrium distribution of genotype frequencies (P > 0.05; χ2 test with one degree of freedom).
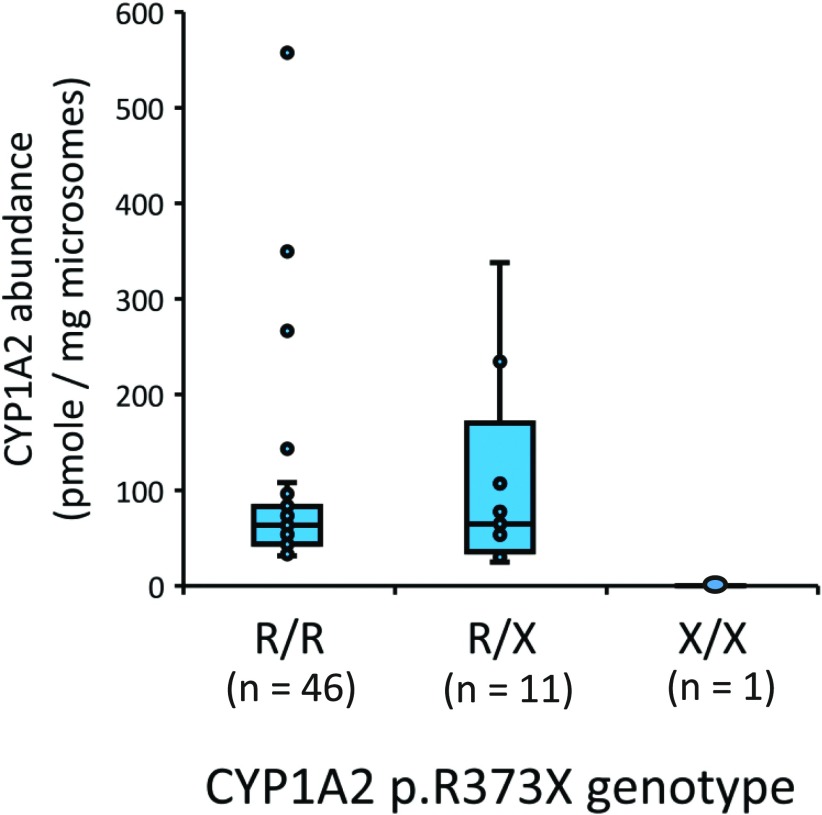
As shown in Fig. 3, one liver (DL-55) of 58 dog livers was determined to be homozygous for the CYP1A2 stop codon variant. Liver DL-55 was also the only liver sample that did not have detectable CYP1A2 protein. Genotyping of the other liver samples identified 11 heterozygous and 46 reference CYP1A2 genotype livers that had similar median CYP1A2 abundance estimates of 65 and 64 pmol/mg microsomal protein, respectively (P = 0.99; Mann–Whitney U test).
Fig. 3.
Effect of the CYP1A2 stop codon mutation (c.1117C>T; p.R373X) on the protein abundance of CYP1A2 measured by LC-MS/MS in a bank of dog liver microsomes (N = 59). Shown are box and whisker plots representing the median, 25th percentile, 75th percentile, range, and outlier points for each genotype. Below each genotype are the numbers of dogs with that particular genotype.
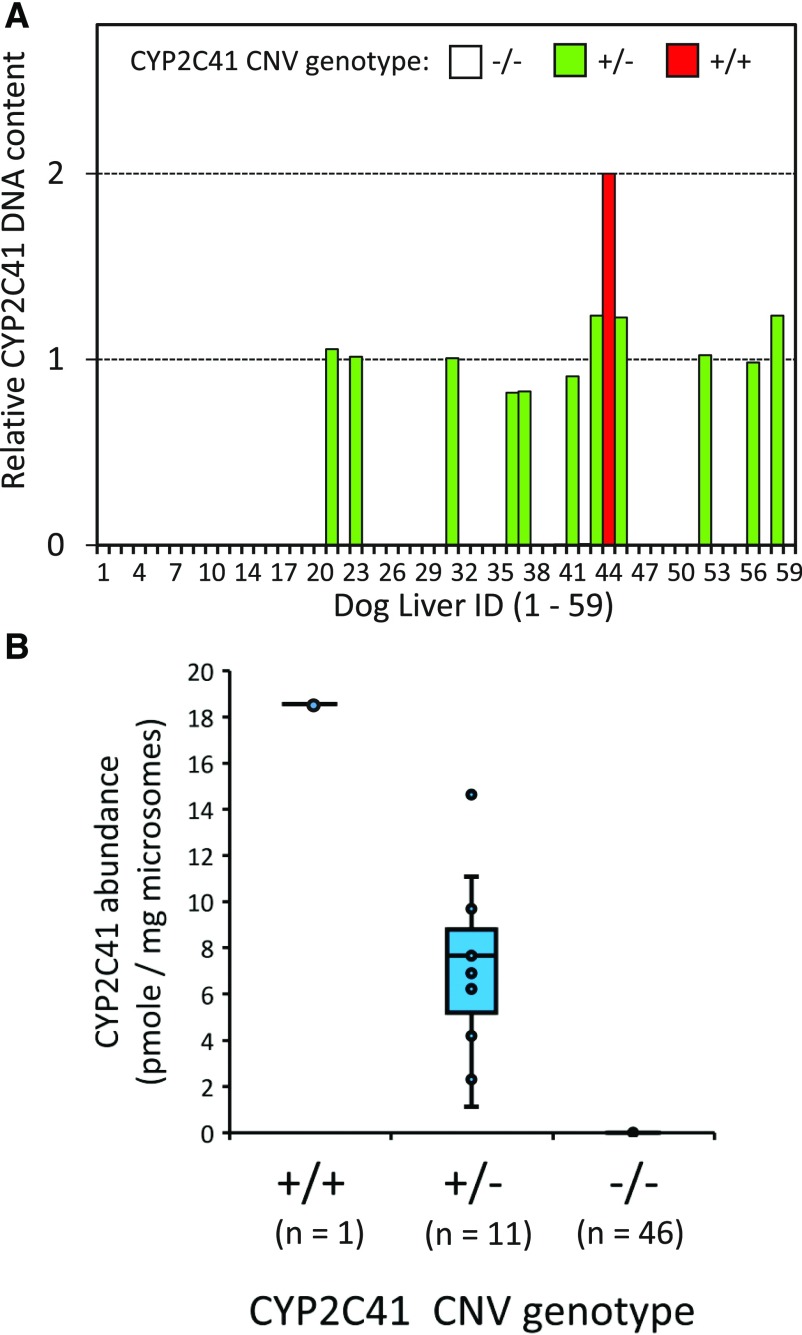
CYP2C41 genotyping showed that the majority of livers (46 of 58) lacked detectable CYP2C41 DNA, and were assigned the CYP2C41 null genotype (−/−) (Fig. 4A). Of the remaining 12 livers with detectable CYP2C41 DNA, one liver from a female purpose-bred research hound (DL-44) showed exactly twice the amount of CYP2C41 DNA when compared with the other 11 livers. Consequently, liver DL-44 was presumed to contain two CYP2C41 gene copies and was assigned the +/+ genotype. The remaining 11 livers were assigned the heterozygous +/− genotype. Figure 4B shows the distribution of CYP2C41 protein abundance among livers with each genotype. None of the 46 livers assigned the −/− genotype contained detectable amounts of CYP2C41 protein. The liver assigned the +/+ genotype (DL-44) contained the highest amount of CYP2C41 protein (18.6 pmol/mg microsomal protein), which was approximately twice the median abundance for livers assigned the +/− genotype (7.7 pmol/mg microsomal protein).
Fig. 4.
CYP2C41 copy number variation (CNV) and CYP2C41 protein abundance measured by LC-MS/MS in a bank of dog liver microsomes (N = 59). CYP2C41 CNV genotypes were determined using a real-time polymerase chain reaction assay that measured the amount of CYP2C41 gene (relative to a control gene) in DNA extracted from each liver sample (see Materials and Methods for details). As shown in (A), 46 livers were assigned the −/− genotype (CYP2C41 gene absent), 11 livers the +/− genotype (one CYP2C41 gene copy present), and one liver (44) was assigned the +/+ genotype (two CYP2C41 gene copies present). Shown in (B) are box and whisker plots of CYP2C41 protein abundance measured in the same set of livers. Plots represent the median, 25th percentile, 75th percentile, range, and outlier points for each genotype. Below each genotype are the numbers of dogs with that particular genotype.
Sex and Breed Differences in CYP, POR, and CYB5A Expression
Statistical approaches were used to evaluate whether there were differences in liver microsome CYP or accessory protein (POR and CYB5A) abundance related to the sex or breed of the donor dogs. As shown in Table 4, slightly higher median CYP2A13 (by 35%; P = 0.036, Mann–Whitney U test) and CYP2C21 (by 18%, P = 0.042, Mann–Whitney U test) protein concentrations were found in female compared with male dogs. Slightly lower median CYP2A25 (by 22%, P = 0.029, Mann–Whitney U test) protein concentration was also found in female compared with male dogs. No other sex-related differences in protein abundance were observed for any of the remaining CYPs, POR, or CYB5A (P > 0.05, Mann–Whitney U test).
TABLE 4.
Comparison of CYP and accessory protein (P450 oxidoreductase, POR; cytochrome b5A, CYB5A) abundance measured in liver microsomes from 32 male and 27 female dogs
Shown are the median and interquartile range (25%–75%) for each sex and the results (P value) of Mann–Whitney U test comparing sexes.
| Liver Microsome Abundance | |||
|---|---|---|---|
| Median (25%–75%) pmol/mg Protein | |||
| Protein | Males (N = 32) | Females (N = 27) | P Value |
| CYP1A1 | 0.1 (0.0–2.4) | 0.9 (0.0–3.2) | 0.11 |
| CYP1A2 | 64 (39–84) | 61 (43–90) | 0.86 |
| CYP2A13 | 8.3 (4.8–13.6) | 11 (7–17) | 0.036 |
| CYP2A25 | 8.1 (6.5–10.8) | 5.8 (4.9–9.0) | 0.029 |
| CYP2B11 | 73 (44–101) | 79 (48–116) | 0.37 |
| CYP2C21 | 51 (47–65) | 46 (36–57) | 0.042 |
| CYP2C41a | 4.3 (2.0–8.3) | 7.8 (7.5–10.0) | 0.21 |
| CYP2D15 | 136 (116–172) | 140 (117–167) | 0.92 |
| CYP2E1 | 72 (61–88) | 72 (61–79) | 0.55 |
| CYP3A12 | 115 (91–142) | 133 (100–152) | 0.23 |
| CYP3A26 | 3.1 (2.2–4.7) | 4.0 (3.0–5.2) | 0.10 |
| POR | 39 (30–45) | 40 (35–47) | 0.16 |
| CYB5A | 436 (382–499) | 429 (401–461) | 0.59 |
Note that the results given for CYP2C41 are for the 12 dogs that express at least one copy of the CYP2C41 gene (four males and eight females).
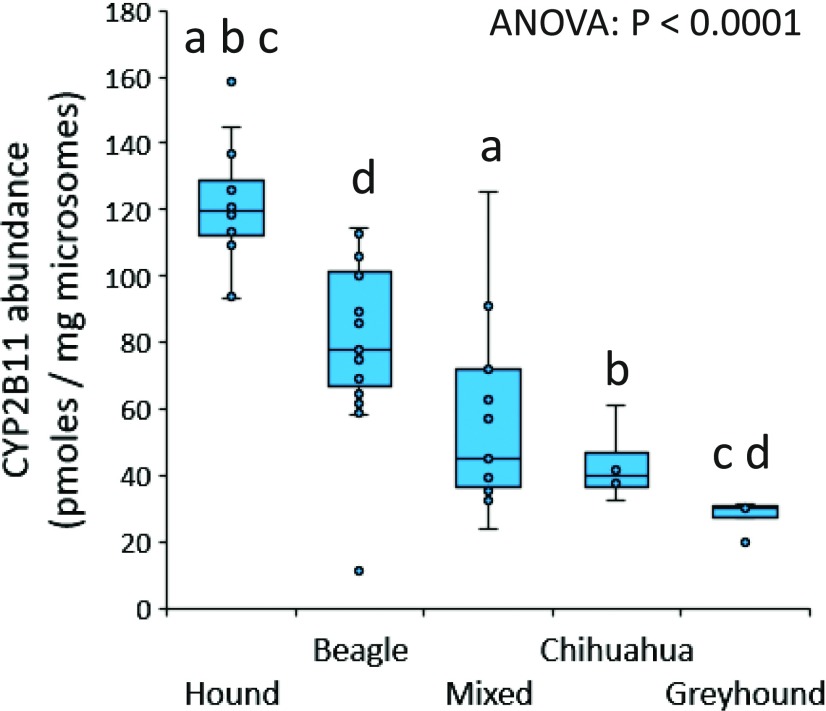
Dog breed–associated variability in protein abundance of each of the 11 CYPs and two accessory proteins is shown in Supplemental Fig. 1, A–M. Results of comparisons between dog breeds using Dunn’s multiple comparisons testing (conducted if initial ANOVA P value <0.05) are given in Table 5. As shown in Fig. 5, substantial differences in CYP2B11 abundance were found between multiple breeds (P < 0.0001). Compared with purpose-bred research hounds, median CYP2B11 abundance was lower in mixed-breed dogs (by 53%, P < 0.001), Chihuahuas (by 59%, P = 0.003), and greyhounds (by 77%, P < 0.001). Greyhounds also showed lower median CYP2B11 abundance (by 68%, P = 0.008) compared with purpose-bred research beagle dogs. Higher median CYP3A12 abundance (by 51%, P = 0.02) was observed in purpose-bred research hounds compared with mixed-breed dogs (Supplemental Fig. 1J; Table 5). Higher median CYP3A26 abundance (by 51%, P = 0.004) was also observed in purpose-bred research hounds compared with greyhounds (Supplemental Fig. 1K; Table 5). Conversely, lower CYP2C21 abundance (by 35%, P = 0.003) was observed in purpose-bred research hounds compared with purpose-bred research beagle dogs (Supplemental Fig. 1F; Table 5). No differences in protein abundance between dog breeds were observed for CYP1A1, CYP1A2, CYP2A13, CYP2A25, CYP2C41, CYP2D15, CYP2E1, POR, or CYB5A (Supplemental Fig. 1; Table 5).
TABLE 5.
Comparison of CYP and accessory protein (P450 oxidoreductase, POR; cytochrome B5A, CYB5A) abundance in liver microsomes from four different dog breeds and 13 mixed-breed dogs
Shown are the median and interquartile range (25%–75%) for each breed and mixed-breed dogs and the results of ANOVA on rank-transformed data. In instances where ANOVA results were significant (P < 0.05), Dunn’s test was used to isolate breed groups that were significantly different from each other in the abundance of that protein.
| Liver Microsome Abundance [Median (25%–75%) pmol/mg Protein] | |||||||
|---|---|---|---|---|---|---|---|
| Protein | Research Hounds (N = 12) | Research Beagles (N = 25) | Mixed-Breed Dogs (N = 13) | Chihuahuas (N = 4) | Greyhounds (N = 5) | ANOVA (P Value) | Dunn’s Test (P < 0.05) |
| CYP1A1 | 0.2 (0.0–1.0) | 0.0 (0.0–3.9) | 1.6 (0.2–4.9) | 1.6 (1.0–2.4) | 0.8 (0.2–1.5) | 0.39 | — |
| CYP1A2 | 53 (42–66) | 72 (54–107) | 61 (37–98) | 65 (43–85) | 48 (44–74) | 0.35 | — |
| CYP2A13 | 13 (11–16) | 8.6 (5.6–13.5) | 6.5 (4.3–9.3) | 9.3 (3.4–16.1) | 16 (15–17) | 0.043 | — |
| CYP2A25 | 5.4 (4.1–6.0) | 8.2 (6.5–11.2) | 9.8 (6.8–10.8) | 11.6 (7.9–14.5) | 6.8 (4.8–7.6) | 0.008 | — |
| CYP2B11 | 120 (112–129) | 78 (67–101) | 45 (37–72) | 40 (37–47) | 30 (28–31) | <0.0001 | a,b,c,d |
| CYP2C21 | 40 (36–44) | 58 (50–77) | 50 (47–56) | 48 (43–54) | 47 (39–48) | 0.006 | e |
| CYP2C41f | 7.8 (7.1–10.3) | 5.0 (2.0–9.4) | 9.7 | 6.2 | — | 0.37 | — |
| CYP2D15 | 167 (140–186) | 127 (100–181) | 138 (112–148) | 136 (115–167) | 131 (118–135) | 0.16 | — |
| CYP2E1 | 68 (59–76) | 72 (59–84) | 67 (62–80) | 72 (53–88) | 97 (81–99) | 0.35 | — |
| CYP3A12 | 145 (130–174) | 118 (108–139) | 93 (70–127) | 117 (107–132) | 91 (87–144) | 0.031 | a |
| CYP3A26 | 5.2 (4.5–6.2) | 3.0 (2.2–4.3) | 3.2 (2.6–5.2) | 3.1 (2.2–3.9) | 2.1 (0.6–3.0) | 0.002 | c |
| POR | 40 (36–42) | 42 (33–49) | 41 (28–51) | 39 (36–43) | 32 (30–35) | 0.44 | — |
| CYB5A | 436 (393–458) | 464 (425–506) | 402 (381–429) | 427 (380–442) | 382 (328–459) | 0.09 | — |
Research hounds vs. mixed-breed dogs.
Research hounds vs. Chihuahuas.
Research hounds vs. greyhounds.
Research Beagles vs. greyhounds.
Research hounds vs. research Beagles.
Note that the results given for CYP2C41 are for the 12 dogs that express at least one copy of the CYP2C41 gene (six research hounds, four Beagles, one mixed-breed, and one Chihuahua).
Fig. 5.
Dog breed–associated differences in liver microsome CYP2B11 abundance (picomoles per milligram of microsomal protein). Shown are box and whisker plots representing the median, 25th percentile, 75th percentile, range, and outlier points for each breed, including 12 purpose-bred research hounds, 25 purpose-bred research Beagles, 13 mixed-breed dogs, four Chihuahuas, and five greyhounds. Differences between breeds were analyzed by ANOVA on rank-transformed data with Dunn’s multiple comparisons test. Lowercase letters (a, b, c, or d) indicate breed groups that were significantly different from each other by Dunn’s test (P < 0.05 for groups with the same letter). Definitions of the letters are given in Table 5.
Discussion
This is the first study to use quantitative proteomics to characterize interindividual, sex, and breed variability in CYP drug-metabolizing enzyme content in a moderately large and diverse bank of canine liver microsomes. Furthermore, we provide the first estimates of the absolute abundance of CYP1A1, CYP2A13, CYP2A25, CYP2C41, POR, and CYB5A in canine liver microsomes, making this the most complete assessment of hepatic microsomal drug-metabolizing CYP and accessory proteins in the dog to date. The main findings were high interindividual variation (>100% CV) in expression of two CYPs (CYP1A1 and CYP1A2), minimal effect of the sex of the dog, and marked breed-associated variability for one of the major drug-metabolizing CYPs in dog liver (CYP2B11).
Average CYP abundance estimates in the current study were similar to values reported in the two previous studies (Eguchi et al., 1996; Heikkinen et al., 2015), generally differing by less than 2.5-fold (Table 1). Similarly, a study comparing human CYP abundances for hepatocyte and liver tissues measured by TPA with abundance values from two meta-analyses using other quantitative and semiquantitative techniques found that the median absolute fold difference was 2.2 with no trend of either over- or underestimating abundances (Vildhede et al., 2015). Explanations for discrepancies in CYP abundance between studies may include experimental technique differences associated with the preparation of the liver microsomes, the protein quantitation method (ELISA vs. surrogate peptide vs. TPA), biologic differences related to their differing genetic backgrounds, or nongenetic factors such as diet or other environmental exposures. As discussed below, it is noteworthy that the discrepant CYP in this study (CYP1A2) also displays substantial genetic polymorphisms and is highly inducible.
The Heikkinen study primarily used Beagles from a closed colony in New Jersey (United States), and the Eguchi study used Beagles from Japan, whereas our study used Beagles from multiple sources in the United States. We have previously observed (Court, 2013; Mealey et al., 2019) that the frequencies of the CYP1A2 stop codon mutation is higher in beagles from Japan (37%–39%), than Beagles from Germany (15%) or the United States (13%) (Mise et al., 2004; Tenmizu et al., 2004; Whiterock et al., 2007; Aretz and Geyer, 2011). Beagles in this study had a CYP1A2 stop codon frequency of 22%. Consequently, the lower CYP1A2 abundance reported by Eguchi may be a consequence of a higher prevalence of the stop codon mutation in Japanese beagles.
Our results confirm that CYP3A26 is a minor isoform in dog liver, as suggested by Heikkinen et al. (2015). Furthermore, CYP1A1 expression was undetectable (N = 36) or was quite low (N = 21) in most livers, averaging only about 3% to 4% of CYP1A2 expression, indicating that CYP1A1 is also a minor hepatic isoform in most dogs. This is consistent with results from a prior study that showed undetectable CYP1A1 mRNA and relatively low CYP1A2 mRNA by Northern blot in untreated dog livers (Uchida et al., 1990). However, two livers in this study, DL-05 (female mixed-breed) and DL-29 (female Beagle), had unusually high concentrations of CYP1A1 (52 and 39 pmol/mg, respectively; Supplemental Fig. 1A) as well as CYP1A2 (266 and 349 pmol/mg, respectively; Supplemental Fig. 1B). The prior study (Uchida et al., 1990) also showed significant induction of CYP1A1 and CYP1A2 mRNA levels in livers following treatment of dogs with a prototypical aryl hydrocarbon receptor. Although we excluded livers from dogs with a known recent history of pharmaceutical exposure, it is possible that high CYP1A1 and CYP1A2 variability between dogs resulted from differences in exposure to aryl hydrocarbons present in the diet or other environmental sources.
Both CYP2A13 and CYP2A25 showed relatively low levels of expression in all livers. The role of the CYP2A enzymes in drug metabolism in dogs is currently unclear. In the only study that characterized the function of these enzymes, recombinant CYP2A13 showed phenacetin O-deethylation activity (intrinsic clearance of 0.92 µl/min per picomole P450) that was comparable to CYP1A2 (1.1 µl/min per picomole P450) (Zhou et al., 2010). Based on this finding, it was suggested that metabolism by CYP2A13 could limit the utility of phenacetin O-deethylation as a specific marker activity for CYP1A2 in dog liver. However, extrapolation of these intrinsic clearances to liver microsomes using the average CYP2A13 and CYP1A2 abundances from this study (11 and 89 pmol/mg, respectively) results in values of 10 and 98 µl/min per milligram microsomal protein, respectively. This indicates that the contribution of CYP2A13 to phenacetin O-deethylation in liver microsomes would be only about 10% that of CYP1A2. Further studies of phenacetin O-deethylation using a full set of recombinant enzymes and other approaches are needed to resolve this issue.
In addition to showing significant correlations among marker activities, immunoblot estimates, and quantitative protein abundance of CYP2B11, CYP2D15, CYP3A12, and POR, we also validated our data through genotyping liver samples for the CYP1A2 stop codon and CYP2C41 gene deletion polymorphisms. As expected, no CYP1A2 or CYP2C41 protein was measured in livers that were homozygous variant genotype for the respective polymorphism. An assay for determining CYP2C41 copy number has not yet been reported, and so the protein abundance data also served to validate this genotyping assay.
Several reaction phenotyping studies using recombinant canine CYP enzymes identified diclofenac, S-mephenytoin, tramadol, and tramadol metabolites as CYP2C41 substrates (Shou et al., 2003; Locuson et al., 2009; Perez Jimenez et al., 2016, 2018). However, the other closely related CYP2C isoform in dogs (CYPC21) also metabolized these substrates to varying extents. Until this report, the liver content of CYP2C41 in dogs that possessed at least one copy of the gene was unknown, and thus the clinical importance of CYP2C41 (relative to CYP2C21 and other CYPs) for drug metabolism in dogs was unclear. Based on the CYP2C protein content determined in this study (Supplemental Table 1), the abundance of CYP2C41 relative to CYP2C21 in the 12 dogs expressing CYP2C41 averaged 20%, but ranged from 2% (in DL-56) to as high as 49% (in DL-44). The latter dog (DL-44) showed the highest CYP2C41 abundance (19 pmol/mg) and was the only dog that we identified with two CYP2C41 gene copies. Consequently, CYP2C41 could substantially contribute to drug metabolism in dogs expressing this gene, especially when reaction phenotyping studies for a drug show higher activity for recombinant CYP2C41 relative to other CYPs after correction for hepatic abundance.
This is the first report of the absolute abundance of POR and CYB5A in canine hepatic microsomes. POR content in dog liver microsomes (40 ± 11 pmol/mg) was approximately midway between reported estimates for human liver microsomes from a primarily white American population (mean 25 pmol/mg; Shirasaka et al., 2016) and those from a Chinese population (median 68 pmol/mg; Zhang et al., 2016). CYB5A content in dog liver microsomes (mean 431 ± 69 pmol/mg) was somewhat higher than the published estimate for liver microsomes from a Chinese population (median 270 pmol/mg; Zhang et al., 2015). Like human liver microsomes (Zhang et al., 2015, 2016; Shirasaka et al., 2016), interindividual variability for both accessory proteins in dog liver was relatively low. POR is the obligate electron donor to all microsomal CYP enzymes, whereas CYB5A enhances metabolism of multiple CYPs (Waskell and Kim, 2015). Low interindividual variability may be a reflection of their importance in the metabolism of xenobiotics and endogenous compounds.
Sex substantially influences hepatic expression of CYPs in some species, particularly rodents. Higher hepatic CYP3A4 expression has been reported for female versus male humans, whereas other CYPs are not consistently different (Zanger and Schwab, 2013). Previous studies of beagle livers found no differences between sexes in the protein abundance of CYP1A2, CYP2B11, CYP2C21, CYP2D15, CYP2E1, CYP3A12, or CYP3A26 (Eguchi et al., 1996; Heikkinen et al., 2015). Our findings largely agree with those findings. Small, albeit statistically significant differences were observed for CYP2A13 (higher in females), CYP2A25 (higher in males), and CYP2C21 (higher in females). However, given the small size of these differences (less than 30%), it is unlikely there would be any clinically meaningful effect.
To date, CYP content has been reported only for purpose-bred beagle dogs. Yet, there are more than 350 distinct breeds of dog worldwide (Parker et al., 2017). Hepatic CYP expression varies between humans with differing racial, ethnic, and geographic origins that are largely explained by genetic variation (Zanger and Schwab, 2013). Consequently, it is not surprising that breed-associated differences in hepatic CYP expression were found in this study. Large differences in CYP2B11 abundance were found between dog breeds, with research hounds and Beagles showing the highest abundance, mixed-breed dogs and Chihuahua exhibiting intermediate abundance, and greyhounds demonstrating the lowest abundance. Furthermore, our results suggest that breed differences may be minimal for the other drug-metabolizing CYPs that we assayed. However, our dog liver microsome bank only contained four different breeds plus mixed-breed dogs. Consequently, additional breed-related differences in abundance of drug-metabolizing CYPs may exist in other breeds not represented in our bank.
Breed-related differences in CYP2B11 could be explained by genetic polymorphism and/or differential exposure to enzyme inducers. Recent work in our laboratory (Martinez et al., 2018) indicates that low CYP2B11 protein abundance in greyhounds is caused, in part, by a variant located in the CYP2B11 3′-untranslated region that is associated with reduced translational efficiency. Other CYP2B11 polymorphisms could also explain higher CYP2B11 abundance in research Beagles and hounds, although this has not yet been studied. Interestingly, in addition to showing the highest CYP2B11 abundance (Fig. 5), research hounds also showed the highest CYP3A12 (Supplemental Fig. 1J) and CYP3A26 (Supplemental Fig. 1K) abundance. CYP2B and CYP3A enzymes share similar induction mechanisms involving the constitutive androstane receptor (CAR) and pregnane X receptor (PXR) (Graham et al., 2002). Consequently, higher CYP2B and CYP3A expression in research hounds could be a consequence of exposure to a CAR and/or PXR inducer. However, research hounds also had the lowest abundance of CYP2C21 (Supplemental Fig. 1F), which would argue against enzyme induction because CYP2C isoforms are also regulated by PXR and CAR. Dogs were excluded if they had a history of drug exposure within 2 weeks of euthanasia. However, the diet of each dog was unknown, and so we cannot exclude a role for diet in CYP2B11 abundance differences between breed groups.
In conclusion, the results of this study provide the most comprehensive estimates to date of the abundance and interindividual variability of the drug-metabolizing CYPs in canine liver. This information should serve as a useful reference resource for use in drug metabolism research as well as human and veterinary pharmaceutical development.
Abbreviations
- CAR
constitutive androstane receptor
- CYP
cytochrome P450
- LC
liquid chromatography
- LC-MS/MS
LC tandem mass spectrometry
- POR
P450 oxidoreductase
- PXR
pregnane X receptor
- TPA
total protein approach
Authorship Contributions
Participated in research design: Court, Martinez, H.-J. Zhu.
Conducted experiments: Martinez, Shi, Perez Jimenez, Z. Zhu.
Contributed new reagents or analytic tools: Shi, H.-J. Zhu.
Performed data analysis: Court, Martinez, Shi, H.-J. Zhu, Perez Jimenez.
Wrote or contributed to the writing of the manuscript: Court, Martinez, H.-J. Zhu.
Footnotes
S.E.M. and M.H.C. were supported by American Kennel Club Canine Health Foundation [Grants 2242 and 2529]. M.H.C. was also supported by the National Institutes of Health [Grant R01-GM-102130] and the William R. Jones endowment to Washington State University College of Veterinary Medicine. H.-J.Z. was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01-HL-126969].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Achour B, Barber J, Rostami-Hodjegan A. (2014) Expression of hepatic drug-metabolizing cytochrome p450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos 42:1349–1356. [DOI] [PubMed] [Google Scholar]
- Andersson T, Miners JO, Veronese ME, Tassaneeyakul W, Tassaneeyakul W, Meyer UA, Birkett DJ. (1993) Identification of human liver cytochrome P450 isoforms mediating omeprazole metabolism. Br J Clin Pharmacol 36:521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aretz JS, Geyer J. (2011) Detection of the CYP1A2 1117C>T polymorphism in 14 dog breeds. J Vet Pharmacol Ther 34:98–100. [DOI] [PubMed] [Google Scholar]
- Blaisdell J, Goldstein JA, Bai SA. (1998) Isolation of a new canine cytochrome P450 CDNA from the cytochrome P450 2C subfamily (CYP2C41) and evidence for polymorphic differences in its expression. Drug Metab Dispos 26:278–283. [PubMed] [Google Scholar]
- Choi SY, Hwang JS, Kim IH, Hwang DY, Kang HG. (2011) Basic data on the hematology, serum biochemistry, urology, and organ weights of beagle dogs. Lab Anim Res 27:283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH. (2013) Canine cytochrome P-450 pharmacogenetics. Vet Clin North Am Small Anim Pract 43:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Hay-Kraus BL, Hill DW, Kind AJ, Greenblatt DJ. (1999) Propofol hydroxylation by dog liver microsomes: assay development and dog breed differences. Drug Metab Dispos 27:1293–1299. [PubMed] [Google Scholar]
- Court MH, Krishnaswamy S, Hao Q, Duan SX, Patten CJ, Von Moltke LL, Greenblatt DJ. (2003) Evaluation of 3′-azido-3′-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism. Drug Metab Dispos 31:1125–1133. [DOI] [PubMed] [Google Scholar]
- Court MH, Von Moltke LL, Shader RI, Greenblatt DJ. (1997) Biotransformation of chlorzoxazone by hepatic microsomes from humans and ten other mammalian species. Biopharm Drug Dispos 18:213–226. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem 80:273–299. [DOI] [PubMed] [Google Scholar]
- Duignan DD, Sipes IG, Leonard TB, Halpert JR. (1987) Purification and characterization of the dog hepatic cytochrome P-450 isozyme responsible for the metabolism of 2,2′4,4′5,5′-hexachlorobiphenyl. Arch Biochem Biophys 255:290–303. [DOI] [PubMed] [Google Scholar]
- Eguchi K, Nishibe Y, Baba T, Ohno K. (1996) Quantitation of cytochrome P450 enzymes (CYP1A1/2, 2B11, 2C21 and 3A12) in dog liver microsomes by enzyme-linked immunosorbent assay. Xenobiotica 26:755–763. [DOI] [PubMed] [Google Scholar]
- Glatter T, Ludwig C, Ahrné E, Aebersold R, Heck AJR, Schmidt A. (2012) Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J Proteome Res 11:5145–5156. [DOI] [PubMed] [Google Scholar]
- Graham RA, Downey A, Mudra D, Krueger L, Carroll K, Chengelis C, Madan A, Parkinson A. (2002) In vivo and in vitro induction of cytochrome P450 enzymes in beagle dogs. Drug Metab Dispos 30:1206–1213. [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Martin MV, Sohl CD, Cheng Q. (2009) Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc 4:1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay Kraus BL, Greenblatt DJ, Venkatakrishnan K, Court MH. (2000) Evidence for propofol hydroxylation by cytochrome P4502B11 in canine liver microsomes: breed and gender differences. Xenobiotica 30:575–588. [DOI] [PubMed] [Google Scholar]
- Heikkinen AT, Friedlein A, Lamerz J, Jakob P, Cutler P, Fowler S, Williamson T, Tolando R, Lave T, Parrott N. (2012) Mass spectrometry-based quantification of CYP enzymes to establish in vitro/in vivo scaling factors for intestinal and hepatic metabolism in beagle dog. Pharm Res 29:1832–1842. [DOI] [PubMed] [Google Scholar]
- Heikkinen AT, Friedlein A, Matondo M, Hatley OJ, Petsalo A, Juvonen R, Galetin A, Rostami-Hodjegan A, Aebersold R, Lamerz J, et al. (2015) Quantitative ADME proteomics - CYP and UGT enzymes in the beagle dog liver and intestine. Pharm Res 32:74–90. [DOI] [PubMed] [Google Scholar]
- Locuson CW, Ethell BT, Voice M, Lee D, Feenstra KL. (2009) Evaluation of Escherichia coli membrane preparations of canine CYP1A1, 2B11, 2C21, 2C41, 2D15, 3A12, and 3A26 with coexpressed canine cytochrome P450 reductase. Drug Metab Dispos 37:457–461. [DOI] [PubMed] [Google Scholar]
- Martignoni M, Groothuis GM, de Kanter R. (2006) Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol 2:875–894. [DOI] [PubMed] [Google Scholar]
- Martinez MN, Court MH, Fink-Gremmels J, Mealey KL. (2018) Population variability in animal health: influence on dose-exposure-response relationships: Part I: Drug metabolism and transporter systems. J Vet Pharmacol Ther 41:E57–E67. [DOI] [PubMed] [Google Scholar]
- Mealey KL, Martinez SE, Villarino NF, Court MH. (2019) Personalized medicine: going to the dogs? Hum Genet 138:467–481. [DOI] [PubMed] [Google Scholar]
- Mise M, Hashizume T, Matsumoto S, Terauchi Y, Fujii T. (2004) Identification of non-functional allelic variant of CYP1A2 in dogs. Pharmacogenetics 14:769–773. [DOI] [PubMed] [Google Scholar]
- Parker HG, Dreger DL, Rimbault M, Davis BW, Mullen AB, Carpintero-Ramirez G, Ostrander EA. (2017) Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep 19:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Jimenez TE, Mealey KL, Grubb TL, Greene SA, Court MH. (2016) Tramadol metabolism to O-desmethyl tramadol (M1) and N-desmethyl tramadol (M2) by dog liver microsomes: species comparison and identification of responsible canine cytochrome P-450s (CYPs). Drug Metab Dispos 44:1963–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Jimenez TE, Mealey KL, Schnider D, Grubb TL, Greene SA, Court MH. (2018) Identification of canine cytochrome P-450s (CYPs) metabolizing the tramadol (+)-M1 and (+)-M2 metabolites to the tramadol (+)-M5 metabolite in dog liver microsomes. J Vet Pharmacol Ther 41:815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang X, Lyu L, Jiang H, Zhu HJ. (2018a) Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: focusing on drug-metabolizing enzymes. Drug Metab Pharmacokinet 33:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang X, Zhu H, Jiang H, Wang D, Nesvizhskii A, Zhu HJ. (2018b) Determining allele-specific protein expression (ASPE) using a novel quantitative concatamer based proteomics method. J Proteome Res 17:3606–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, Fohner A, Thornton TA, Isoherranen N, Unadkat JD, et al. (2016) Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J 16:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou M, Norcross R, Sandig G, Lu P, Li Y, Lin Y, Mei Q, Rodrigues AD, Rushmore TH. (2003) Substrate specificity and kinetic properties of seven heterologously expressed dog cytochromes p450. Drug Metab Dispos 31:1161–1169. [DOI] [PubMed] [Google Scholar]
- Tenmizu D, Endo Y, Noguchi K, Kamimura H. (2004) Identification of the novel canine CYP1A2 1117 C > T SNP causing protein deletion. Xenobiotica 34:835–846. [DOI] [PubMed] [Google Scholar]
- Tibbitts J. (2003) Issues related to the use of canines in toxicologic pathology--issues with pharmacokinetics and metabolism. Toxicol Pathol 31 (Suppl):17–24. [DOI] [PubMed] [Google Scholar]
- Uchida T, Komori M, Kitada M, Kamataki T. (1990) Isolation of cDNAs coding for three different forms of liver microsomal cytochrome P-450 from polychlorinated biphenyl-treated beagle dogs. Mol Pharmacol 38:644–651. [PubMed] [Google Scholar]
- Vildhede A, Wiśniewski JR, Norén A, Karlgren M, Artursson P. (2015) Comparative proteomic analysis of human liver tissue and isolated hepatocytes with a focus on proteins determining drug exposure. J Proteome Res 14:3305–3314. [DOI] [PubMed] [Google Scholar]
- Waskell L, Kim J. (2015) Electron transfer partners of cytochrome, in Cytochrome (Ortiz de Montellano P ed) p 450, Springer, Cham, Switzerland. [Google Scholar]
- Whiterock VJ, Delmonte TA, Hui LE, Orcutt TL, Sinz MW. (2007) Frequency of CYP1A2 polymorphism in beagle dogs. Drug Metab Lett 1:163–165. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR. (2017) Label-free and standard-free absolute quantitative proteomics using the “total protein” and “proteomic ruler” approaches. Methods Enzymol 585:49–60. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Inoue K, Shaw PM, Checovich WJ, Guengerich FP, Shimada T. (1997) Different contributions of cytochrome P450 2C19 and 3A4 in the oxidation of omeprazole by human liver microsomes: effects of contents of these two forms in individual human samples. J Pharmacol Exp Ther 283:434–442. [PubMed] [Google Scholar]
- Zanger UM, Schwab M. (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao N, Liu T, Fang Y, Qi B, Wen Q, Zhou J, Jia L, Qiao H. (2015) Effect of cytochrome b5 content on the activity of polymorphic CYP1A2, 2B6, and 2E1 in human liver microsomes. PLoS One 10:e0128547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Li ZH, Liu JY, Liu TT, Wang P, Fang Y, Zhou J, Cui MZ, Gao N, Tian X, et al. (2016) Correlation of cytochrome P450 oxidoreductase expression with the expression of 10 isoforms of cytochrome P450 in human liver. Drug Metab Dispos 44:1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Linnenbach AJ, Liu R, Luzietti RA, Harris JJ, Booth-Genthe CL, Grimm SW. (2010) Expression and characterization of dog cytochrome P450 2A13 and 2A25 in baculovirus-infected insect cells. Drug Metab Dispos 38:1015–1018. [DOI] [PubMed] [Google Scholar]
- Zientek MA, Youdim K. (2015) Reaction phenotyping: advances in the experimental strategies used to characterize the contribution of drug-metabolizing enzymes. Drug Metab Dispos 43:163–181. [DOI] [PubMed] [Google Scholar]