Abstract
Cytochrome P450 (P450) 2S1 is one of the orphan P450s, known to be expressed but not having a defined function with an endogenous substrate or in drug oxidations. Although it has been clearly demonstrated to catalyze reductive reactions, its role in NADPH-dependent oxidations has been ambiguous. In our efforts to characterize orphan human P450 enzymes, we used an untargeted liquid chromatography-mass spectromterymetabolomic approach with recombinant human P450 2S1 and extracts of rat stomach and intestine, sites of P450 2S1 localization in humans and animals. The search yielded several candidates, including the product 19-hydroxyarachidonic acid. Subsequent 18O analysis and in vitro studies with commercial arachidonic acid and 19-hydroxyarachidonic acid were used to validate ω-1 hydroxylation of the former molecule as a NADPH- and O2-dependent reaction. Steady-state kinetic assays were done for ω-1 hydroxylation reactions of P450 2S1 with several other long-chain fatty acids, including arachidonic, linoleic, α-linolenic, eicosapentaenoic, and docosapentaenoic acids. Rates of hydroxylation were slow, but no detectable activity was seen with either medium-chain length or saturated fatty acids. P450 2S1 is known to be expressed, at least at the mRNA level, to the extent of some other non-3A subfamily P450s in the human gastrointestinal tract, and the activity may be relevant. We conclude that P450 2S1 is a fatty acid ω-1 hydroxylase, although the physiologic relevance of these oxidations remains to be established. The metabolomic approaches we employed in this study are feasible for orphan P450s and other enzymes, in regard to annotation of function, in mammals and other organisms.
SIGNIFICANCE STATEMENT
An untargeted mass spectrometry approach was utilized to identify ω-1 hydroxylation of arachidonic acid as an oxidative reaction catalyzed by human cytochrome P450 2S1. The enzyme also catalyzes the relatively slow ω-1 hydroxylation of several other unsaturated long-chain fatty acids.
Introduction
Enzymes play a vital role in almost all cellular metabolic activity and transduction systems. Cytochrome P450 enzymes (P450s) are a large family of hemoprotein enzymes (Ortiz de Montellano, 2015). They mainly catalyze mono-oxygenase reactions; however, some P450 members also show additional activities (Guengerich, 2001). P450s can act on a wide variety of endogenous and exogenous compounds and are responsible for the reported oxidations of ∼95% of all chemicals, including many drugs, steroids, fat-soluble vitamins, polyunsaturated fatty acids (PUFAs), and chemical carcinogens (Rendic and Guengerich, 2015). The Human Genome Project identified 57 human P450 genes; about five of these still do not have any known substrates or any clear physiologic function, that is, they are orphans (Guengerich, 2015).
Several approaches can be adopted to identify orphan P450 substrates and products. Predicting substrates from P450 crystal structures are difficult due to changes in P450 conformation upon ligand binding (Johnson and Stout, 2013). Trial-and-error methods, using small-molecule libraries, are a biased approach that can be incomprehensive as well as time consuming. The latter approach was successful in finding substrates for P450 2W1, but not for P450 2S1 (Wu et al., 2006). Sequence similarity to related P450s with known reactions is another approach that has been used with some limited success (Wu et al., 2006; Guengerich et al., 2011; Enright et al., 2015; Kramlinger et al., 2016). Global metabolite profiling (metabolomics) coupled with isotopic labeling has shown potential in the elucidation of P450 function and identifying endogenous substrates in an unbiased manner, at least in some cases (Tang et al., 2009; Xiao and Guengerich, 2012).
P450 2S1 is one of the human orphan P450s and is expressed predominantly in extrahepatic tissues. Expression of P450 2S1 was reported to be mainly in the epithelium of portal entry organs, for example, respiratory and gastrointestinal tract (Saarikoski et al., 2005b). Its expression can be regulated by endogenous and exogenous chemicals, at least in cell culture. Retinoic acid significantly elevated its expression at both the mRNA and protein level in several types of human epithelial cells (Smith et al., 2003). Culturing cells under hypoxic conditions resulted in elevation of P450 2S1 mRNA in mouse hepatoma Hepa-1 cells (Rivera et al., 2007), but not in human monocytes (Frömel et al., 2013). Expression was also elevated in response to agonists of the aryl hydrocarbon receptor, for example, 2,3,7,8-tetrachlorodibenzo-p-dioxin (Rivera et al., 2002) and components of cigarette smoke (Thum et al., 2006).
P450 2S1 is a controversial orphan P450 due to the inconsistency of some of the data in the literature. Depletion of P450 2S1 in a human bronchial epithelial cell line enhanced cell proliferation and migration in BEAs-2B cells (Madanayake et al., 2012). However, when nine P450 genes, including Cyp2s1, were knocked out in mice, no obvious phenotypic effects were observed (Wei et al., 2013). The only consistently documented NADPH-dependent reactions catalyzed by P450 2S1 have all been reductions (Nishida et al., 2010; Wang and Guengerich, 2012, 2013).
The aim of this study was to identify endogenous substrates for human P450 2S1. Toward this end, purified recombinant human P450 2S1 was incubated with rat stomach and intestine extracts (sites of P450 2S1 expression in rat), followed by untargeted metabolomic analysis, validated using isotopic labeling methodology (Sanchez-Ponce and Guengerich 2007; Tang et al., 2009). P450 2S1 catalyzed the oxidation of arachidonic acid into 19-hydroxy-5Z,8Z,11Z,14Z-eicosatetraenoic acid (19-HETE), the first substantiated report of a mono-oxygenation reaction catalyzed by P450 2S1 and dependent on its biologic redox partner NADPH-P450 reductase. P450 2S1 was also active with several other unsaturated fatty acids, and only ω-1 hydroxy PUFAs were formed.
Materials and Methods
Chemicals.
Lauric, myristic, palmitic, stearic, linoleic, α-linolenic, arachidonic, 19-HETE, eicosapentaenoic, docosapentaenoic, and docosahexaenoic acids and all-trans retinoic acid were purchased from Cayman Chemical (Ann Arbor, MI). Solvents, buffer components, and other reagents used for reactions or protein preparation were obtained from Millipore, Sigma-Aldrich, or Fisher Scientific (Hampton, NH), unless otherwise noted.
Tissue Samples and Enzymes.
Adult female rat stomach and intestine tissues (snap-frozen after dissection by supplier) were obtained from Cell Biologics (Chicago, IL). Recombinant human P450 2S1 (Wu et al., 2006), human cytochrome b5 (Guengerich, 2005), and rat NADPH-P450 reductase (Hanna et al., 1998) were expressed in Escherichia coli and purified, as reported previously. Recombinant mouse P450 2s1, used in a few experiments, was tailored in the N terminus in a similar construct as the human version (Wu et al., 2006) and was expressed and purified in the same way.
Tissue Extract Preparation.
Rat stomach and intestine tissue samples (320 mg) were thawed and rinsed with cold phosphate-buffered saline solution at 4°C. Homogenization was accomplished using a motor-driven Potter-Elvejehm device (Glas-Col, Terre Haute, IN) in 10 ml of a C2H5OH/CH3OH mixture (1:1, v/v) with repeated up-down passages for 5 minutes on ice, and the homogenates were then filtered through paper. The resulting extract was centrifuged for 10 minutes at 5 × 103 g. The organic supernatant was collected, evaporated under a stream of nitrogen, and immediately purged with argon and stored at −70°C. On the day of use, samples were reconstituted in 1 ml of C2H5OH (320 mg tissue equivalent/ml) and sonicated 3 × 30 seconds at a maximum power setting.
In Vitro Incubations of P450 2S1 with Tissue Extracts.
Incubation mixtures (1.0 ml) contained human P450 2S1 (1.0 μM), NADPH-P450 reductase (2.0 μM), cytochrome b5 (2.0 μM), L-α-dilauroyl-sn-glycero-3-phosphocholine (160 μM), a NADPH-generating system (10 mM glucose 6-phosphate, 0.5 mM NADP+, and 10 μg/ml yeast glucose-6-phosphate dehydrogenase), potassium phosphate buffer (100 mM, pH 7.4), thymol-free catalase (100 U per reaction mixture added to prevent artifacts resulting from H2O2) (Guengerich, 1978), and 40 μl ethanolic solution of a tissue extract (equivalent to 13 mg tissue). Reaction mixtures were incubated at 37°C for 1 hour and then terminated by adding 2 ml cold ethyl acetate and CHCl3 (1:1, v/v), followed by mixing for 1 minute with a vortex device and centrifugation at 3 × 103 g for 5 minutes. The organic layer was carefully removed and evaporated under a stream of nitrogen. Samples were reconstituted in 50 μl CH3CN containing 0.1% HCO2H and then adding 50 μl Milli-Q water containing 0.1% HCO2H (v/v), transferred into vials, and subjected to LC-MS analysis. Negative control runs did not contain either a NADPH-generating system or P450 2S1.
Anaerobic Incubations.
Anaerobic experiments were performed in Thunberg tubes, as previously described (Wang and Guengerich, 2012). When the desired level of anaerobicity was achieved (after five cycles of vacuum and Ar), a NADPH-generating system was added to the incubation mixture from the neck of the tube. The reaction mixtures were incubated at 37°C for 1 hour, and the reactions were terminated, as previously described (Wang and Guengerich, 2012).
Isotopic Labeling Experiments.
A method for 16O2/18O2 isotopic experiments was adapted from previous studies (Sanchez-Ponce and Guengerich, 2007; Tang et al., 2009). Following 10 alternating cycles of vacuum and argon, Thunberg tubes containing reaction mixtures were charged separately with 100% 16O2 and 97% 18O2, and the two samples were mixed together before extraction, as previously described. Reaction mixtures incubated under a 16O2 atmosphere served as control samples.
LC–High Resolution Mass Spectrometry Analysis of Tissue Extracts.
Samples (10 μl, equivalent to 1.3 mg tissue) were injected onto an Agilent Poroshell 120 SB-C18 column (2.1 mm × 100 mm, 2.7 μm; Agilent, Santa Clara, CA) at 40°C with a flow rate of 0.3 ml min−1. Chromatographic separation was achieved using a system with solvent A: H2O containing 0.1% HCO2H (v/v) and solvent B: CH3CN containing 0.1% HCO2H (v/v). Gradient conditions were set up as follows: 5% B (0 to 1 minute), linear gradient of B increased to 95% (1–12 minutes), held at 95% B (12–15.5 minutes), returned to the initial condition (5% B) for column equilibration (16–20 minutes). The ultra-perfomance liquid chromatography (UPLC) system was coupled to a Q Exactive HF hybrid quadrupole–orbitrap mass spectrometer with heated electrospray ionization (Thermo Fisher, San Jose, CA). Analysis was performed in both negative and positive ion modes. The full MS scan range was m/z 100–1000 Da at 30,000 resolution, 1e6 automatic gain control (AGC) target, and 250-ms ion injection time (time of accumulating ions per scan event) (IT) value. Tandem mass spectrometry (MS/MS) data-dependent fragmentation was triggered based on each full MS scan for the top five most intense ions at 15,000 resolution, 1e5 AGC target, and 50-ms IT value. The tune file source parameters were set as follows: N2 sheath gas flow 40 arbitrary units; auxiliary gas flow 10 arbitrary units; spray voltages at 3.5 kV in the positive ionization mode, 3.0 kV in the negative ionization mode; capillary temperature 320°C; S-lens RF value 60; probe heater temperature 200°C. All MS data were acquired within 1 week after an external mass calibration using both Pierce LTQ Velos positive and negative ion calibration solutions (Thermo Fisher Scientific). Data were then processed with the Qual browser module of Xcalibur 3.0 (Thermo Fisher).
Metabolite Data Processing and Analysis.
UPLC-MS/MS raw data were processed using Progenesis QI v.2.1 (Non-linear Dynamics, Newcastle, UK). Each raw data file was imported, followed by retention time (tR) alignment. Peak picking was performed on individual aligned runs by matching peaks in an aggregate data set that was created from all aligned runs. Following peak picking, the features (tR and m/z pairs) were reduced using both adduct and isotope deconvolution. Data were normalized to all observed features as an abundance ratio between the run being normalized. Putative annotations were determined within Progenesis using accurate mass measurements (<5 ppm error), isotope distribution similarity, and assessment of fragmentation spectra matching with Metlin (Smith et al., 2005) and Human Metabolome Database (Wishart et al., 2018).
Screening P450 2S1 Activity with Different Fatty Acids.
All PUFAs were purified prior to each of the P450 reactions, as follows: PUFAs (250 μg) were dissoslved in 40 μl CH3CN:water (1:1, v/v) containing 0.1% HCO2H (v/v). Samples were injected onto a preparative Phenomenex Luna C18 column (10 mm × 150 mm, 5 μm) (Phenomenex, Torrance, CA) at room temperature, with a flow rate of 4 ml min−1. Chromatographic separation was achieved using isocratic elution of 95% CH3CN in water containing 0.1% HCO2H (v/v). UV signals at 205 nm were recorded using an Agilent 1100 series diode array detector and used to collect purified PUFAs. UV signals at 235 nm were used to monitor PUFA hydroperoxy products (Graff et al., 1990). P450 reactions (50 μl) were prepared as described below, except that P450 2S1 (1 μM) was incubated with 40 μM of each of the following purified fatty acids individually: lauric, myristic, palmitic, stearic, linoleic, α-linolenic, arachidonic, eicosapentaenoic, docosapentaenoic, and docosahexaenoic acids (or all-trans retinoic acid). Reactions were done at 37°C for 30 minutes, terminated by adding 400 μl cold ethyl acetate containing 0.1% HCO2H (v/v), and then treated for analysis, as described above.
LC–High Resolution Mass Spectrometry Analysis of Fatty Acids.
Samples (20 μl) were injected onto a Hypersil Gold Vanquish C18 column (2.1 mm × 150 mm, 1.9 μm; Thermo Fisher) operating at 40°C with a flow rate of 0.3 ml min−1. Chromatographic separation was achieved using a system composed of solvent A: 10 mM ammonium acetate (pH 6.8) and solvent B: CH3CN. Gradient conditions were set up as follows: 45% B (0–2 minutes), linear gradient of B increasing to 75% (2–15 minutes), linear gradient of B increasing to 95% (15–16 minutes), held at 95% B (16–18 minutes), return to initial condition (5% B) for column equilibration (18–20 minutes). The UPLC was coupled to the same mass spectrometer with a heated electrospray ionization electrospray ionization source (Thermo Fisher). Analysis was performed in the negative ionization mode only using the following: 1) full MS scan range m/z 100–600 Da and 2) parallel reaction monitoring scanning at the exact mass of each of the parent molecules and its corresponding mono-oxygenation product with 30 eV collision energy and a m/z 1.6 isolation window. Both scans used the same 30,000 resolution, 2e5 AGC target, and 200-ms IT value. The tune file source parameters were the same as used in tissue extract analyses.
Kinetic Analysis of P450 Reactions and Derivatization Procedures.
Substrate concentrations ranging from 5 to 100 μM were used for steady-state kinetic studies. Reactions were incubated at 37°C for 30 minutes, and oxidation products were extracted, as described above. Pentafluorobenzyl (PFB) esters were prepared by dissolving dried (ethyl acetate) P450 reaction extract or 10 nmol PUFA in 20 μl CH3CN, 20 μl di-isopropylethylamine (in CH3CN, 1:9, v/v), and 2 μl PFB bromide. Each solution was incubated at 60°C for 30 minutes and then evaporated to dryness under a nitrogen stream. PFB esters of linoleic, α-linolenic, eicosapentaenoic, and docosapentaenoic acids were used to prepare calibration curves for the kinetic analysis of each of these oxidized PUFAs, respectively. PFB esters were reconstituted in 25 μl CH3CN and then 25 μl Milli-Q water, and then they were transferred into vials for LC-MS analysis.
Hyperbolic fits were done to solve for kcat and ksp (kcat/Km), which was analyzed directly instead of Km (Johnson, 2019) (see the reference for the advantages of this approach); nonlinear regression of hyperbolic fits was done using Prism software (GraphPad, San Diego, CA), using the input formula Y = [(ksp × X)/(1 + (ksp × X/kcat))].
LC–High Resolution Mass Spectrometry Analyses of PFB Ester Derivatives.
Samples (20 μl) were injected onto a Hypersil Gold Vanquish C18 column (2.1 mm × 150 mm, 1.9 μm) at 40°C, operating with a flow rate of 0.3 ml min−1. Chromatographic separation was achieved using solvent A, CH3CN:water (7:3, v/v) and solvent B, isopropanol:CH3CN (9:1, v/v). Gradient conditions were set up as follows: 5% B (0–2 minutes), linear gradient of B increasing to 95% (2–16 minutes), hold at 95% B (16–18 minutes), return to initial condition (5% B) for column equilibration (18–20 minutes). PFB ester derivatives were detected with the same mass spectrometer using electron capture atmospheric pressure chemical ionization (Lee et al., 2003). Analysis was performed in the negative ionization mode only, using both full MS and parallel reaction monitoring scan events with the same parameters used in screening P450 2S1 activity for different fatty acid oxidations. Atmospheric pressure chemical ionization tune file source parameters were set as follows: N2 sheath gas flow 60 arbitrary units; auxiliary gas flow 30 arbitrary units; 5.0 kV in the negative ionization mode; capillary temperature 320°C; S-lens RF value 100; probe heater temperature 300°C.
Results
Untargeted Search for P450 2S1-Derived Metabolites in Rat Stomach and Intestine Extracts.
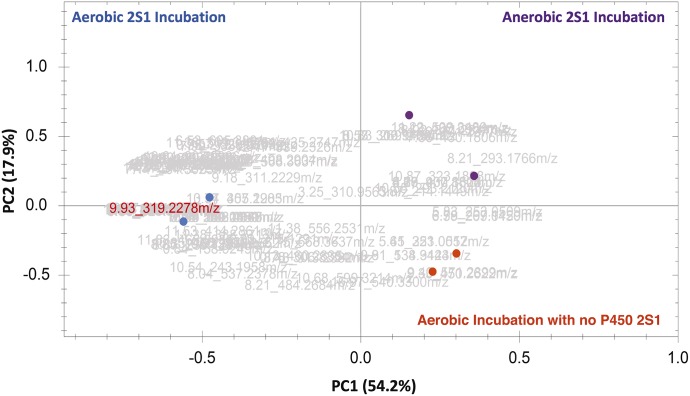
In the search for endogenous substrates for P450 2S1, the recombinant human enzyme was incubated with a tissue extract of rat stomach and intestine because of higher expression in these organs (Wang et al., 2011). Because P450 2S1 has shown reductive biotransformation activity (Wang and Guengerich, 2012, 2013), parallel incubations were done under anaerobic conditions. The reaction mixtures were analyzed in both negative and positive ion MS modes to profile as many metabolites as possible. The samples analyzed using negative ion modes demonstrated 1381 ion features. The number was filtered to 124 by selecting for a fold change ≥100 (calculated by Progenesis QI from combined abundance data) and excluding certain features, for example, lack of MS fragmentation and/or putative identification. Using the filtered features only, unsupervised principal component analysis was employed on the 124 putative metabolites to visualize clustering of the test runs away from both the peaks in the anaerobic incubation and negative control (Fig. 1). The number of peaks under consideration was reduced further, to eight, after eliminating putative identities related to any known xenobiotic or plant-derived metabolites. The samples analyzed using a positive ion mode did not show any separation among the groups studied.
Fig. 1.
Principal component analysis score plot showing separation between the three tested groups (n = 2). The loading plots showed 124 features in light gray (tR and m/z pairs) that were responsible for the clustering. The feature highlighted in red lettering is a m/z 319.2278 peak eluted at 9.93 minutes that was verified by isotopic labeling and identified later as 19-HETE.
Validation of Metabolomic Outcomes by Isotopic Labeling.
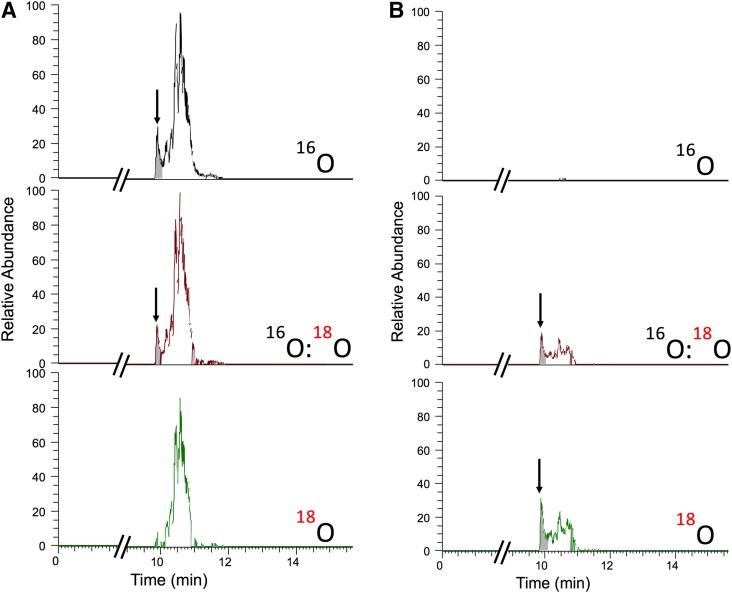
The remaining eight peaks (vide supra) were further investigated through doublet searches (M, M + 2) using 18O-based isotopic labeling experiments (Sanchez-Ponce and Guengerich, 2007; Tang et al., 2009). Incubation of rat stomach and intestinal extract with P450 2S1 with a 16O2/18O2 mixture resulted in observation of one doublet, m/z 319/321 (Fig. 2). The observed m/z 319 peak was not found under anaerobic incubations or in the negative control runs (Supplemental Fig. 1). A full m/z scan of the tR 9.93-minute peak showed the presence of both m/z 319.2286 and m/z 321.2326 peaks in samples incubated with a 16O2/18O2 mixture only (Supplemental Fig. 2). Samples analyzed using the positive ion mode did not demonstrate the characteristic isotopic signature using this isotopic labeling technique. High resolution mass spectrometry analysis of the observed doublet showed m/z 319.2284 in the incubation done under 16O2, corresponding to the tentative formula C20H31O3− (calculated mass of 319.2279, Δ ≤ 2 ppm). This doublet could be assigned to be an oxygenated product of arachidonic acid, based upon the observed high resolution mass fragmentation analysis of the products detected in the electrospray ionization (ESI) negative (Bylund et al., 1998) ion mode and a search of LIPIDMAPS database (http://www.lipidmaps.org). Incubation of P450 2S1 with arachidonic acid yielded a product eluted at the same tR and with the same fragmentation pattern as the product detected in the rat stomach/intestine extract incubation. PUFA autoxidation was minimized and better product sensitivity was obtained by applying the following procedures: 1) the arachidonic acid stock was purified prior to the P450 reaction, and 2) the chromatographic column and both the mobile phase nature and gradient were varied from that used in the untargeted incubation with rat tissue, to increase the separation. 19-HETE was verified as a product of incubation of P450 2S1 with arachidonic acid by comparison with a standard that eluted at the same tR and showed the same fragmentation pattern (Supplemental Fig. 3).
Fig. 2.
LC–high resolution mass spectrometry chromatograms of rat tissue extract incubated with human P450 2S1 enzyme and a NADPH-generating system. (A) Extracted ion chromatogram of m/z 319.2278 due to 16O2 incorporation; (B) extracted ion chromatogram of m/z 321.2320 due to 18O2 incorporation. The peak for 19-HETE at tR 9.9 is indicated with an arrow and shading. The scales of the chromatograms shown in (A and B) are identical (4.6 E6 full scale).
Fatty Acid Screening and Identification of ω-1 Hydroxy Products by LC-MS.
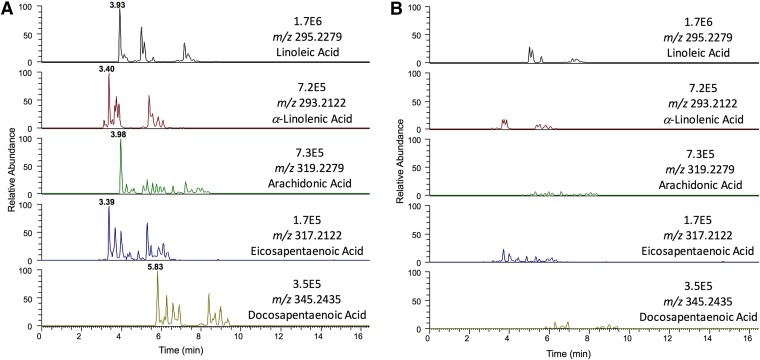
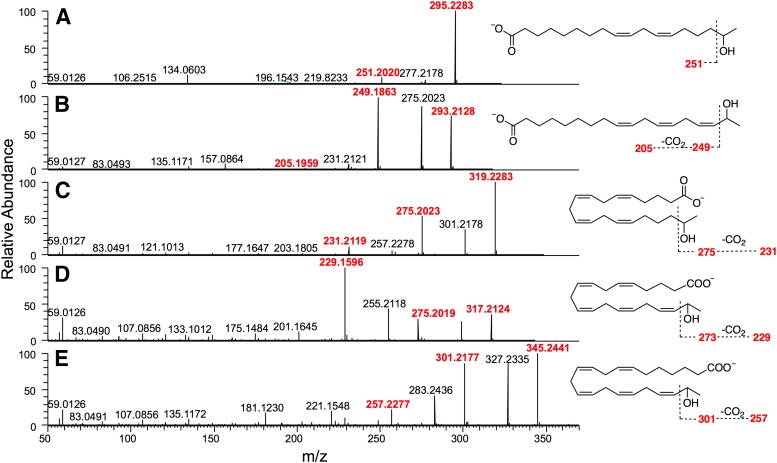
Validation of the metabolomic results showed that P450 2S1 catalyzed arachidonic acid mono-oxygenation, forming 19-HETE. Therefore, catalytic activity of P450 2S1 with other fatty acids (lauric, myristic, palmitic, stearic, linoleic, α-linolenic, arachidonic, eicosapentaenoic, docosapentaenoic, docosahexaenoic) and all-trans retinoic acid was investigated (Table 2). Only linoleic, α-linolenic, arachidonic, eicosapentaenoic, and docosapentaenoic acids formed a major product (Fig. 3). The rest of the fatty acids studied did not show any major differences from the corresponding negative controls. The observed molecular masses of the five main mono-oxygenated metabolites were observed at ≤2 ppm (Supplemental Table 1). Furthermore, the mass spectra of mono-oxygenated metabolites showed some noncharacteristic ion fragments corresponding to the neutral loss of H2O [M-1-18]− or the loss of both H2O and CO2 [M-1-62]−. Cleavage at the site of the hydroxyl group led to characteristic fragment ions that differed according to the position of the alcohol. The ω-1 hydroxylated derivatives, that is, secondary alcohols, exhibited a characteristic fragment [M-1-88]− resulting from the loss of two m/z 44 fragments corresponding to CH3-CHO and CO2 (Fer et al., 2008). All observed products (Fig. 4) and the 19-HETE standard (Supplemental Fig. 3) showed the distinctive fragment ion [M-1-88]− except the product derived from linoleic acid, which showed a loss of one m/z 44 fragment (Supplemental Figs. 4 and 5).
TABLE 2.
Steady-state kinetic analysis of ω-1 hydroxylated fatty acids by P450 2S1
| Fatty Acid | kcat (min−1)a | Km (μM)a | kcat/Km (min−1 M−1)a |
|---|---|---|---|
| Linoleic acid (C18:2) | 0.0017 ± 0.0001 | 30 ± 6 | 56 ± 7 |
| α-Linolenic acid (C18:3) | 0.00045 ± 0.00006 | 16 ± 7 | 28 ± 9 |
| Arachidonic acid (C20:4) | 0.0024 ± 0.0002 | 25 ± 6 | 97 ± 13 |
| Arachidonic acid, with mouse P450 2s1 | 0.0029 ± 0.0002 | 76 ± 13 | 38 ± 3 |
| Eicosapentaenoic acid (C20:5) | 0.0022 ± 0.0002 | 50 ± 11 | 44 ± 5 |
| Docosapentaenoic acid (C22:5) | 0.0020 ± 0.0004 | 91 ± 29 | 22 ± 4 |
±S.D.
Fig. 3.
Comparison of the extracted ion chromatogram of the exact mass of each mono-oxygenated PUFA incubated with human P450 2S1 (A) and without P450 2S1 (B). Each PUFA and its corresponding negative control are shown on the same scale. The ω-1 hydroxylated fatty acid peaks are indicated by tR values in (A); the minor peaks were also seen in the control experiments (B).
Fig. 4.
High resolution mass spectrometry (ESI−) mass fragmentation spectra of mono-oxygenated PUFAs formed in the presence of human P450 2S1. (A) Linoleic acid; (B) α-linolenic acid; (C) arachidonic acid; (D) eicosapentaenoic acid; (E) docosapentaenoic acid. The characteristic fragment ions are highlighted in red.
Kinetic Analysis of P450 2S1 Fatty Acid Oxidation Reactions.
Kinetic studies were performed for the five products (marked by tR in Fig. 3), with PUFA concentrations ranging from 5 to 100 μM and a reaction time of 30 minutes (Supplemental Fig. 6; Table 2). Although cytochrome b5 had been routinely included in the incubations in the possibility that it was important, subsequent assays showed that it did not stimulate the rate of ω-1 hydroxylation of arachidonic acid. Saturation of the steady-state rates was not achievable due to the tendency of PUFA to form micelles at high concentration in aqueous buffer (Serth et al., 1991). Hydroxylation (ω-1) of arachidonic acid was also observed with purified recombinant mouse P450 2s1, demonstrating that the mouse enzyme is similar in function.
Discussion
For more than 15 years since its discovery, a physiologic role for P450 2S1 has remained unknown. In this work, we used an untargeted metabolomic approach to identify candidate reactions, one of which is the ω-1 hydroxylation of arachidonic acid. This reaction was also seen with several other long-chain unsaturated fatty acids (Tables 1 and 2), and the chemical evidence for these reactions is unambiguous. No reaction was detected with medium-chain length or with saturated fatty acids. Nevertheless, the rates of these reactions are slow, and caveats must be associated with their significance in the absence of further evidence.
TABLE 1.
ESI LC-MS analysis of oxidation products of fatty acids produced by human P450 2S1
| Substrate | [M−] Product m/z | Major Fragment Ions m/z | Product |
|---|---|---|---|
| Arachidonic acid (C20:4) | 319.2274 | 301.2169, 275.2014, 231.2112 | ω-1 hydroxy |
| Linoleic acid (C18:2) | 295.2283 | 277.2178, 251.2020 | |
| α-Linolenic acid (C18:3) | 293.2128 | 275.2023, 249.1863, 205.1959 | |
| Eicosapentaenoic acid (C20:5) | 317.2125 | 299.2018, 273.1863, 229.1596 | |
| Docosapentaenoic acid (C22:5) | 345.2441 | 327.2335, 301.2177, 257.2277 | |
| Lauric acid (C12:0) | —a | —a | |
| Myristic acid (C14:0) | —a | —a | |
| Palmitic acid (C16:0) | —a | —a | |
| Stearic acid (C18:0) | —a | —a | |
| Docosahexaenoic acid (C22:6) | —a | —a |
Not detected.
In the Human Protein Atlas (https://www.proteinatlas.org/ENSG00000167600-CYP2S1/tissue), the highest levels of mRNA and protein expression of P450 2S1 are in the stomach, duodenum, and small intestine (Uhlén et al., 2015). Interestingly, skin does not show any expression of P450 2S1 at mRNA and protein levels in the Atlas, in contrast to previous reports (Smith et al., 2003; Saarikoski et al., 2005b). P450 2S1 mRNA levels in rat tissues follow similar patterns reported in mice and humans (Deb and Bandiera, 2009). Our working hypothesis was that a substrate might exist in the tissue in which the P450 2S1 is expressed. Toward this end, purified human recombinant P450 2S1 was incubated with adult rat stomach and small intestine extracts, followed by untargeted LC-MS metabolomic analysis, which was then verified by an isotopic labeling approach (Sanchez-Ponce and Guengerich, 2007; Tang et al., 2009) (Figs. 1 and 2). Linoleic, α-linolenic, arachidonic, eicosapentaenoic, and docosapentaenoic acids were identified as substrates for P450 2S1. The main oxidation products of the fatty acids formed by P450 2S1 were all characterized as ω-1 hydroxylated fatty acids (Figs. 3 and 4; Table 1). Mono-oxygenated arachidonic acid was the only fatty acid derivative observed in the metabolomic analysis and was confirmed using doublet peak analysis (M, M + 2) in an isotopic labeling experiment. The other PUFAs were not seen in the metabolomic analysis, presumably because they are mainly found in the diet. Linoleic acid and α-linolenic acids are essential fatty acids, abundant in the Western diet and green leafy vegetables. Eicosapentaenoic and docosapentaenoic acids are ω-3 fatty acids abundant in seafood. Steady-state kinetic analysis was done (Table 2), and the observed catalytic efficiencies (i.e., specificity constants, kcat/Km) imply that these are very slow P450-catalyzed mono-oxygenation reactions, but the five observed PUFAs were definitely oxidized by human P450s 2S1 and generated mainly ω-1 hydroxylated fatty acids.
The activity of recombinant human CYP2S1 has been studied in several laboratories, and the results from these studies have been conflicting. For instance, Bui et al. (2009, 2011) and Bui and Hankinson (2009) have reported that P450 2S1 is neither able to neither interact with NADPH-P450 reductase nor to accept electrons from the reductase. However, P450 2S1 was rapidly reduced by NADPH-P450 reductase (Xiao et al., 2011), and P450 2S1 is efficient in reductive biotransformation of the anticancer agent AQ4N (Nishida et al., 2010; Xiao et al., 2011), carcinogenic aromatic amines, and heterocyclic aromatic amines (Wang and Guengerich, 2013). All of the observed reductive reactions are NADPH-dependent.
P450 2S1 has been shown to catalyze NADPH-independent reactions involving cumene hydroperoxide (Mrízová et al., 2016) or lipid peroxide–supported oxidation of benzo[a]pyrene-7,8-dihydrodiol (Bui et al., 2009). Cumene hydroperoxide itself is not present endogenously and not physiologically relevant. Although lipid hydroperoxides are found endogenously and could potentially be relevant physiologically for CYP2S1, the concentrations used in vitro (50 µM) (Bui et al., 2009) are considerably greater than endogenous levels (3–500 nM) (Niki, 2009).
P450 2S1 has been previously reported to oxidize all-trans-retinoic acid (Smith et al., 2003) and naphthalene (Saarikoski et al., 2005a) in NADPH-dependent reactions, but others have failed to reproduce these results (Wu et al., 2006; Nishida et al., 2010). In this study, as before (Wu et al., 2006), we could not detect any products upon incubating P450 2S1 with all-trans-retinoic acid and NADPH. In our earlier studies, we did not observe any oxidative capability of P450 2S1 with arachidonic acid (Wu et al., 2006), but the limit of detection of the analysis method was only 0.15 nmol. In this study, the LC–high resolution mass spectrometry method had a limit of detection of ≤0.25 pmol 19-HETE. Frömel et al. (2013) reported that P450 2S1 was able to form bioactive epoxides from arachidonic, linoleic, and eicosapentaenoic acids in NADPH-dependent reactions, but did not provide any chromatographic or spectroscopic data to support the conclusion. In this study, we did not observe any epoxides upon incubating P450 2S1 with any of the screened PUFAs, and they clearly are separable in these systems (Sausville et al., 2018).
P450 2S1 is mainly expressed in extrahepatic epithelial cells (Rivera et al., 2002; Uhlén et al., 2015). Madanayake et al. (2012) reported that P450 2S1 plays a role in bronchial epithelial cell proliferation and migration by modulating prostaglandin E2 synthesis. Expression data have also been used to suggest that P450 2S1 may be associated with carcinogenesis: P450 2S1 immunoreactivity has been observed in breast (Murray et al., 2010), human epithelial colorectal (Kumarakulasingham et al., 2005), and squamous cell carcinomas (Saarikoski et al., 2005a). Additionally, P450 2S1 expression has been correlated with poor prognosis in ovarian, breast, and colorectal cancers (Downie et al., 2005; Kumarakulasingham et al., 2005; Murray et al., 2010).
The human gastrointestinal tract mainly expresses the following P450s: 3A4 > 2C9 > 2C19 ∼2D6 ∼2J2 (Paine et al., 2006). Based on the Human Protein Atlas database (https://www.proteinatlas.org/ENSG00000167600-CYP2S1/tissue), P450 2S1 RNA expression in the stomach is the highest among all aforementioned P450 (2S1 > > 2C9 > 2CJ2 > 2C19). P450 3A4 RNA expression was the highest in duodenum and small intestine (Supplemental Table 2). Duodenum showed high RNA expression of P450 3A4, followed by 2C9 > > 2S1 > 2C19 > 2J2 > 2D6. Small intestine exhibited high RNA expression of P450 3A4 as well, followed by 2J2 > 2C9 > 2S1 > 2C19 > 2D6. P450 2C19 forms 19-HETE and epoxides/diols as major metabolites, and P450 2C9 forms 19-HETE as a minor product (Bylund et al., 1998).
19-HETE is known to have an antagonist effect against the potent vasoconstrictor activity of 20-HETE (Quigley et al., 2000). ω-1 Hydroxylation of arachidonic acid has been previously reported for some human P450s, for example, 1A1 and 2U1. P450 1A1 is an arachidonic acid hydroxylase, forming primarily 19-HETE, and is also an eicosapentaenoic acid epoxygenase. P450 1A1 is expressed and is inducible by ligands of the aryl hydrocarbon receptor in the gastrointestinal tract, lung, placenta, brain, and smooth muscle cells (Schwarz et al., 2004). P450 2U1, most abundant in the human thymus and brain (Chuang et al., 2004; Siller et al., 2014), catalyzes the ω and ω-1 hydroxylations of arachidonic, docosahexaenoic, and docosahexaenoic acid but not fatty acids such as lauric acid or linoleic acid. P450 2C19 and P450 2E1 catalyze mainly ω-1 hydroxylation of fatty acids (Fer et al., 2008) when they are substrates.
In conclusion, our present work appears to be the first rigorously documented report of an in vitro mono-oxygenation reaction by P450 2S1 using its biologically relevant redox partner, mammalian NADPH-P450 reductase. Our study provides a successful application of the untargeted LC-MS metabolomics and the Progenesis QI program to identify endogenous substrates in tissue extracts for orphan human P450s such as P450 2S1, and five fatty acids were identified as substrates for human P450 2S1. The physiologic relevance of these reactions remains to be established. The current methodology can be adopted for untargeted metabolomic analysis of stomach, duodenum, and small intestine tissues from wild-type and P450 2S1 knockout mice to verify the presently observed results and to further investigate the physiologic relevance of these results regarding fatty acid oxidations in the gastrointestinal tract.
Acknowledgments
Thanks are extended to Prof. A. R. Brash for helpful suggestions, to Dr. M. W. Calcutt for technical assistance with some of the mass spectrometry work, and to K. Trisler for assistance in preparation of the manuscript.
Abbreviations
- 19-HETE
19-hydroxy-5Z,8Z,11Z,14Z-eicosatetraenoic acid
- AGC
automatic gain control
- IT
ion injection time (time of accumulating ions per scan event)
- P450 enzyme
cytochrome P450
- PFB
pentafluorobenzyl
- PUFA
polyunsaturated fatty acid
- tR
retention time
Authorship Contributions
Participated in research design: Fekry, Guengerich.
Conducted experiments: Fekry.
Contributed new reagents or analytic tools: Xiao, Berg.
Performed data analysis: Fekry, Guengerich.
Wrote or contributed to the writing of the manuscript: Fekry, Xiao, Berg, Guengerich.
Footnotes
This work was supported by National Institutes of Health [Grants R01 GM118122 (to F.P.G.) and T32 HL094296 (to J.Z.B.)].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Bui P, Imaizumi S, Beedanagari SR, Reddy ST, Hankinson O. (2011) Human CYP2S1 metabolizes cyclooxygenase- and lipoxygenase-derived eicosanoids. Drug Metab Dispos 39:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui PH, Hankinson O. (2009) Functional characterization of human cytochrome P450 2S1 using a synthetic gene-expressed protein in Escherichia coli. Mol Pharmacol 76:1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui PH, Hsu EL, Hankinson O. (2009) Fatty acid hydroperoxides support cytochrome P450 2S1-mediated bioactivation of benzo[a]pyrene-7,8-dihydrodiol. Mol Pharmacol 76:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund J, Ericsson J, Oliw EH. (1998) Analysis of cytochrome P450 metabolites of arachidonic and linoleic acids by liquid chromatography-mass spectrometry with ion trap MS. Anal Biochem 265 (1):55–68, doi: 10.1006/abio.1998.2897 . [DOI] [PubMed] [Google Scholar]
- Chuang SS, Helvig C, Taimi M, Ramshaw HA, Collop AH, Amad M, White JA, Petkovich M, Jones G, Korczak B. (2004) CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes omega- and (omega-1)-hydroxylation of fatty acids. J Biol Chem 279:6305–6314. [DOI] [PubMed] [Google Scholar]
- Deb S, Bandiera SM. (2009) Characterization and expression of extrahepatic CYP2S1. Expert Opin Drug Metab Toxicol 5:367–380. [DOI] [PubMed] [Google Scholar]
- Downie D, McFadyen MC, Rooney PH, Cruickshank ME, Parkin DE, Miller ID, Telfer C, Melvin WT, Murray GI. (2005) Profiling cytochrome P450 expression in ovarian cancer: identification of prognostic markers. Clin Cancer Res 11:7369–7375. [DOI] [PubMed] [Google Scholar]
- Enright JM, Toomey MB, Sato SY, Temple SE, Allen JR, Fujiwara R, Kramlinger VM, Nagy LD, Johnson KM, Xiao Y, et al. (2015) Cyp27c1 red-shifts the spectral sensitivity of photoreceptors by converting vitamin A1 into A2. Curr Biol 25:3048–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fer M, Corcos L, Dréano Y, Plée-Gautier E, Salaün J-P, Berthou F, Amet Y. (2008) Cytochromes P450 from family 4 are the main ω hydroxylating enzymes in humans: CYP4F3B is the prominent player in PUFA metabolism. J Lipid Res 49:2379–2389. [DOI] [PubMed] [Google Scholar]
- Frömel T, Kohlstedt K, Popp R, Yin X, Awwad K, Barbosa-Sicard E, Thomas AC, Lieberz R, Mayr M, Fleming I. (2013) Cytochrome P4502S1: a novel monocyte/macrophage fatty acid epoxygenase in human atherosclerotic plaques. Basic Res Cardiol 108:319. [DOI] [PubMed] [Google Scholar]
- Graff G, Anderson LA, Jaques LW. (1990) Preparation and purification of soybean lipoxygenase-derived unsaturated hydroperoxy and hydroxy fatty acids and determination of molar absorptivities of hydroxy fatty acids. Anal Biochem 188:38–47. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (1978) Destruction of heme and hemoproteins mediated by liver microsomal reduced nicotinamide adenine dinucleotide phosphate-cytochrome P-450 reductase. Biochemistry 17:3633–3639. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2001) Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol 14:611–650. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2005) Reduction of cytochrome b5 by NADPH-cytochrome P450 reductase. Arch Biochem Biophys 440:204–211. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2015) Human cytochrome P450 enzymes, in Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed (Ortiz de Montellano PR. ed) pp 523–785, Springer, New York. [Google Scholar]
- Guengerich FP, Tang Z, Cheng Q, Salamanca-Pinzón SG. (2011) Approaches to deorphanization of human and microbial cytochrome P450 enzymes. Biochim Biophys Acta 1814:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna IH, Teiber JF, Kokones KL, Hollenberg PF. (1998) Role of the alanine at position 363 of cytochrome P450 2B2 in influencing the NADPH- and hydroperoxide-supported activities. Arch Biochem Biophys 350:324–332. [DOI] [PubMed] [Google Scholar]
- Johnson EF, Stout CD. (2013) Structural diversity of eukaryotic membrane cytochrome p450s. J Biol Chem 288:17082–17090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KA. (2019) New standards for collecting and fitting steady state kinetic data. Beilstein J Org Chem 15:16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramlinger VM, Nagy LD, Fujiwara R, Johnson KM, Phan TT, Xiao Y, Enright JM, Toomey MB, Corbo JC, Guengerich FP. (2016) Human cytochrome P450 27C1 catalyzes 3,4-desaturation of retinoids. FEBS Lett 590:1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarakulasingham M, Rooney PH, Dundas SR, Telfer C, Melvin WT, Curran S, Murray GI. (2005) Cytochrome p450 profile of colorectal cancer: identification of markers of prognosis. Clin Cancer Res 11:3758–3765. [DOI] [PubMed] [Google Scholar]
- Lee SH, Williams MV, DuBois RN, Blair IA. (2003) Targeted lipidomics using electron capture atmospheric pressure chemical ionization mass spectrometry. Rapid Commun Mass Spectrom 17:2168–2176. [DOI] [PubMed] [Google Scholar]
- Madanayake TW, Fidler TP, Fresquez TM, Bajaj N, Rowland AM. (2012) Cytochrome P450 2S1 depletion enhances cell proliferation and migration in bronchial epithelial cells, in part, through modulation of prostaglandin E(2) synthesis. Drug Metab Dispos 40:2119–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrízová I, Moserová M, Milichovský J, Šulc M, Kizek R, Kubáčková K, Arlt VM, Stiborová M. (2016) Heterologous expression of human cytochrome P450 2S1 in Escherichia coli and investigation of its role in metabolism of benzo[a]pyrene and ellipticine. Monatsh Chem 147:881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD. (2010) Profiling the expression of cytochrome P450 in breast cancer. Histopathology 57:202–211. [DOI] [PubMed] [Google Scholar]
- Niki E. (2009) Lipid peroxidation: physiological levels and dual biological effects. Free Radic Biol Med 47:469–484. [DOI] [PubMed] [Google Scholar]
- Nishida CR, Lee M, de Montellano PR. (2010) Efficient hypoxic activation of the anticancer agent AQ4N by CYP2S1 and CYP2W1. Mol Pharmacol 78:497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz de Montellano PR, editor (2015) Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed, Springer, New York. [Google Scholar]
- Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC. (2006) The human intestinal cytochrome P450 “pie”. Drug Metab Dispos 34:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley R, Baum M, Reddy KM, Griener JC, Falck JR. (2000) Effects of 20-HETE and 19(S)-HETE on rabbit proximal straight tubule volume transport. Am J Physiol Renal Physiol 278:F949–F953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic S, Guengerich FP. (2015) Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol 28:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera SP, Saarikoski ST, Hankinson O. (2002) Identification of a novel dioxin-inducible cytochrome P450. Mol Pharmacol 61:255–259. [DOI] [PubMed] [Google Scholar]
- Rivera SP, Wang F, Saarikoski ST, Taylor RT, Chapman B, Zhang R, Hankinson O. (2007) A novel promoter element containing multiple overlapping xenobiotic and hypoxia response elements mediates induction of cytochrome P4502S1 by both dioxin and hypoxia. J Biol Chem 282:10881–10893. [DOI] [PubMed] [Google Scholar]
- Saarikoski ST, Rivera SP, Hankinson O, Husgafvel-Pursiainen K. (2005a) CYP2S1: a short review. Toxicol Appl Pharmacol 207 (2 Suppl):62–69. [DOI] [PubMed] [Google Scholar]
- Saarikoski ST, Wikman HA-L, Smith G, Wolff CH, Husgafvel-Pursiainen K. (2005b) Localization of cytochrome P450 CYP2S1 expression in human tissues by in situ hybridization and immunohistochemistry. J Histochem Cytochem 53:549–556. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ponce R, Guengerich FP. (2007) Untargeted analysis of mass spectrometry data for elucidation of metabolites and function of enzymes. Anal Chem 79:3355–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausville LN, Gangadhariah MH, Chiusa M, Mei S, Wei S, Zent R, Luther JM, Shuey MM, Capdevila JH, Falck JR, et al. (2018) The cytochrome P450 slow metabolizers CYP2C9*2 and CYP2C9*3 directly regulate tumorigenesis via reduced epoxyeicosatrienoic acid production. Cancer Res 78:4865–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Kisselev P, Ericksen SS, Szklarz GD, Chernogolov A, Honeck H, Schunck W-H, Roots I. (2004) Arachidonic and eicosapentaenoic acid metabolism by human CYP1A1: highly stereoselective formation of 17(R),18(S)-epoxyeicosatetraenoic acid. Biochem Pharmacol 67:1445–1457. [DOI] [PubMed] [Google Scholar]
- Serth J, Lautwein A, Frech M, Wittinghofer A, Pingoud A. (1991) The inhibition of the GTPase activating protein-Ha-ras interaction by acidic lipids is due to physical association of the C-terminal domain of the GTPase activating protein with micellar structures. EMBO J 10:1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller M, Goyal S, Yoshimoto FK, Xiao Y, Wei S, Guengerich FP. (2014) Oxidation of endogenous N-arachidonoylserotonin by human cytochrome P450 2U1. J Biol Chem 289:10476–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27:747–751. [DOI] [PubMed] [Google Scholar]
- Smith G, Wolf CR, Deeni YY, Dawe RS, Evans AT, Comrie MM, Ferguson J, Ibbotson SH. (2003) Cutaneous expression of cytochrome P450 CYP2S1: individuality in regulation by therapeutic agents for psoriasis and other skin diseases. Lancet 361:1336–1343. [DOI] [PubMed] [Google Scholar]
- Tang Z, Martin MV, Guengerich FP. (2009) Elucidation of functions of human cytochrome P450 enzymes: identification of endogenous substrates in tissue extracts using metabolomic and isotopic labeling approaches. Anal Chem 81:3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Erpenbeck VJ, Moeller J, Hohlfeld JM, Krug N, Borlak J. (2006) Expression of xenobiotic metabolizing enzymes in different lung compartments of smokers and nonsmokers. Environ Health Perspect 114:1655–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015) Proteomics: tissue-based map of the human proteome. Science 347:1260419. [DOI] [PubMed] [Google Scholar]
- Wang B, Robertson LW, Wang K, Ludewig G. (2011) Species difference in the regulation of cytochrome P450 2S1: lack of induction in rats by the aryl hydrocarbon receptor agonist PCB126. Xenobiotica 41:1031–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Guengerich FP. (2012) Bioactivation of fluorinated 2-aryl-benzothiazole antitumor molecules by human cytochrome P450s 1A1 and 2W1 and deactivation by cytochrome P450 2S1. Chem Res Toxicol 25:1740–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Guengerich FP. (2013) Reduction of aromatic and heterocyclic aromatic N-hydroxylamines by human cytochrome P450 2S1. Chem Res Toxicol 26:993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Li L, Zhou X, Zhang Q-Y, Dunbar A, Liu F, Kluetzman K, Yang W, Ding X. (2013) Generation and characterization of a novel Cyp2a(4/5)bgs-null mouse model. Drug Metab Dispos 41:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, et al. (2018) HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46:D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z-L, Sohl CD, Shimada T, Guengerich FP. (2006) Recombinant enzymes overexpressed in bacteria show broad catalytic specificity of human cytochrome P450 2W1 and limited activity of human cytochrome P450 2S1. Mol Pharmacol 69:2007–2014. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Guengerich FP. (2012) Metabolomic analysis and identification of a role for the orphan human cytochrome P450 2W1 in selective oxidation of lysophospholipids. J Lipid Res 53:1610–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Shinkyo R, Guengerich FP. (2011) Cytochrome P450 2S1 is reduced by NADPH-cytochrome P450 reductase. Drug Metab Dispos 39:944–946. [DOI] [PMC free article] [PubMed] [Google Scholar]