Abstract
Background
Long non-coding RNAs (lncRNAs) exert various functions in human cancers. However, the biological functions of lncRNAs in non-small cell lung cancer (NSCLC) are unknown. In the present study we investigated the tumor-suppressive role of lncRNA on chromosome 8p12 (TSLNC8) in the pathogenesis and progression of NSCLC.
Material/Methods
qRT-PCR was carried out to evaluate the expression of TSLNC8 in lung cancer cell lines. The effects of TSLNC8 on A549 cells proliferation, migration, and invasion were analyzed using CCK-8 assay, wound healing assay, Transwell assay, and Western blot analysis. We used flow cytometry to assess cell apoptosis, and cell autophagy was assessed by Western blot analysis and immunofluorescence staining. Levels of proteins in the IL-6/STAT3/HIF-1α pathway were measured by Western blot analysis.
Results
The results revealed that TSLNC8 was significantly downregulated in lung cancer cells compared to normal bronchial epithelial cells. Further experiments showed that overexpression of TSLNC8 in A549 cells significantly inhibited proliferation in a time-dependent manner and promoted cell apoptosis. We found that TSLNC8 overexpression suppressed cell migration and invasion, and upregulation of TSLNC8 regulated the protein levels of Beclin-1, p62, ATG14, and LC3-II and inhibited the IL-6/STAT3/HIF-1α signaling pathway.
Conclusions
lncRNA TSLNC8 remarkably inhibited the proliferation and migration and accelerated apoptosis of lung cancer cells by targeting the IL-6/STAT3/HIF-1α signaling pathway. TSLNC8 may be a potential therapeutic target for the diagnosis and treatment of NSCLC.
MeSH Keywords: Autophagy; Apoptosis; Carcinoma, Non-Small-Cell Lung; RNA, Long Noncoding
Background
Lung cancer is one of the most common and aggressive tumors worldwide, with extremely high morbidity and mortality rates [1]. Among diagnosed lung cancer cases, approximately 80–85% are non-small cell lung cancer (NSCLC) [2–4]. Although advances in diagnosis and treatment have been made in NSCLC, the 5-year survival rate and recurrence rate remain poor [5]. Previous studies have found that numerous genes are closely involved in the occurrence of lung cancer, but the underlying mechanisms have been unclear [6]. Therefore, it is of paramount importance to understand the molecular mechanisms involved in the pathogenesis of NSCLC to improve diagnosis and therapy for patients with lung cancer.
lncRNAs (long non-coding RNAs) are transcribed RNA molecules whose length is more than 200 nucleotides [7]. lncRNAs are involved in many biological activities in the body, such as angiogenesis, apoptosis, and migration [8,9]. Furthermore, mounting evidence shows that lncRNAs also act as regulatory factors in various cancers, including osteosarcoma [10], hepatocellular carcinoma [11], and glioblastoma [12]. In recent years, lncRNAs have been proved to exert important functions in lung cancer [13]. For instance, Yang et al. revealed that lncRNA PVT1 expression is significantly increased in NSCLC, and knockdown of PVT1 suppresses the proliferation, migration, and invasion of lung cancer cells [14]. In addition, Nie et al. showed that overexpression of lncRNA UCA1 improved cell growth, whereas UCA1 silencing attenuated the proliferation of NSCLC cells, and higher expression of UCA1 led to shorter survival time in NSCLC patients [15]. Shi et al. found that lncRNA GAS5 level is downregulated in cancerous tissues and functions as a tumor suppressor in lung cancer cells. Overexpression of GAS5 inhibits tumor cell growth and induces apoptosis, while knockdown of GAS5 accelerates cancer cell growth [16].
Tumor-suppressive lncRNA on chromosome 8p12 (TSLNC8), also named LINC00589, is one of various lncRNAs [17]. TSLNC8 was reported to be remarkably downregulated in human glioma tissues, suppressing proliferation and facilitating apoptosis in human glioma cells [18]. Furthermore, TSLNC8 exerts its tumor-suppressive function by inactivating the IL-6/STAT3 signaling pathway in liver cancer [17]. Therefore, TSLNC8 has been regarded as a tumor suppressor in some cancers; however, the biological role of TSLNC8 in human lung cancer remains unknown.
In this study, we assessed the expression of lncRNA TSLNC8 and its functional role in cell proliferation, migration, and apoptosis in NSCLC. Moreover, we also explored the inhibiting effects of lncRNA TSLNC8 on NSCLC cells through regulation of the IL-6/STAT3/HIF-1α signaling pathway.
Material and Methods
Cell culture
The human lung cancer cell lines A549 (CVCL_0023), H441 (CVCL_1561), and H1975 (CVCL_DG25) and normal human bronchial epithelial cell line HBE (CVCL_0287) were purchased from the American Type Culture Collection (ATCC, USA). All cells were cultured in RPMI-1640 medium (Gibco Co., USA) containing 10% fetal bovine serum (FBS; Invitrogen, USA), 100 μg/mL streptomycin, and 100 U/mL penicillin at 37°C with 5% CO2. To create hypoxic conditions and induce autophagy, cells were cultured in medium with 100 μmol/L CoCl2 for 24 h at 37°C with 5% CO2.
Cell transfection
For overexpression of TSLNC8, pcDNA-TSLNC8 and the empty vector (pcDNA) were constructed by Invitrogen. Then, the A549 cell lines were cultured in 6-well plates (1×106 cells/well) and transfected with pcDNA-TSLNC8 and pcDNA using Lipofectamine 2000 (Invitrogen-Life Technologies, USA) according to the manufacturer’s protocol. After transfection for 24 h, the cells were cultured in medium with 100 μmol/L CoCl2 for 24 h. Then the cells were processed for subsequent experimentation.
RNA extraction and qRT-PCR
Total RNAs were extracted from lung cancer cells using the Trizol reagent kit (Invitrogen). The concentration and quality of RNA were examined using NanoDrop 2000 (Thermo Scientific, Wilmington, DE). Total RNA was reverse-transcribed into cDNA using a Reverse Transcription System kit (Takara; Dalian, China). The TSLNC8 expression level was determined by RT-PCR using SYBR Premix ExTaq II kit (Takara) on an ABI PRISM 7900 system (Applied Biosystems, USA). GAPDH was used as the internal control. The PCR amplification program was the following: 95°C for 30 s, followed by 40 cycles of 95°C for 1 s and 60°C for 34 s. The sequences of primers were:
TSLNC8 forward, 5′-TGA TCC TCA TAG TAT AAT G-3′ and
reverse, 5′-AGT TCT TTA GCA GTA CATG-3′;
GAPDH forward, 5′-GTG GAC ATC CGC AAA GAC-3′ and
reverse, 5′-AAA GGG TGT AAC GCA ACTA-3′.
Cell Counting Kit-8 assay
Cell proliferation viability of A549 cells was evaluated by Cell Counting Kit-8 assay (CCK-8). Transfected cells were seeded at a density of 5×103 cells/well in 96-well plates and cultured at 37°C overnight. Then, we added 10 μL CCK-8 solution (Dojindo, Tokyo, Japan) to each well 0, 24, 48, and 72 h. After 2-h incubation, the absorbance was detected with a microplate reader (Bio-Rad) at a wavelength of 450 nm.
Cell apoptosis analysis
Cell apoptosis was detected using the FITC Annexin V Apoptosis Detection Kit I (Ribobio, Guangzhou, China) according to the manufacturer’s protocol. Briefly, cells were cultured in 6-well plates and were transfected for 48 h. Subsequently, cells were collected, washed with precooled phosphate-buffered saline (PBS), and re-suspended in binding buffer. Then, the cells were treated with Annexin V-FITC and propidium iodide for 20 min at room temperature in the dark. FlowJo software (Tree Star, Ashland, OR, USA) was used for apoptosis analysis.
Wound healing assay
To investigate cell migratory ability, transfected A549 cells were cultured in 6-well plates and grown until 80–90% confluence. An artificial scratch was created with a pipette tip, and wounded monolayer cells were washed 3 times with PBS. Following incubation for 24 h, images were captured and the numbers of migrated cells were counted using a light microscope (Nikon, Tokyo, Japan).
Cell invasion assays
The invasion capacity of A549 cells was investigated by Transwell assay. Matrigel (BD Biosciences) was added to the upper Transwell chambers (8.0 μm pore size, Costar, Shanghai, China) to form a matrix barrier. Then, the transfected cells were re-suspended in serum-free medium and plated into the upper chamber, while the bottom chamber was covered with the complete medium. After incubation at 37°C for 24 h, the non-invading cells above the membrane were wiped off. Afterwards, the invading cells were fixed with 100% methanol for 10 min and stained with 0.5% crystal violet for 10 min at room temperature. The numbers of invading cells in each well were counted from 5 visible fields using a microscope (Olympus Corp, Tokyo, Japan).
Western blot assay
Total proteins from transfected A549 cells were lysed with RIPA lysis buffer (Beyotime, Shanghai, China). The BCA Protein Assay Reagent Kit (Beyotime) was used to measure protein concentrations. Proteins were loaded onto 10% SDS-PAGE, then transferred onto PVDF membranes (Bio-Rad) and blocked with 5% skimmed milk for 2 h at room temperature. Subsequently, the membranes were incubated with primary antibodies (CDK2, 5 μg/ml, ab6433; cyclinE1, 1: 2000, ab71535; P21, 1: 3000, ab227443; MMP2, 1 μg/ml, ab37150; MMP9, 1: 1000, ab38898; Bcl-2, 1: 500, ab196495; Bax, 1: 500, ab53154; cleaved caspase3, 1: 500, ab49822; IL-6, 1: 500, ab6672; JAK2, 1: 1000, ab39636; p-STAT3, 1: 1000, ab30647; HIF-1α, 1: 1000, ab216842; STAT3, 1: 5000, ab119352; Beclin-1, 1 μg/ml, ab62557; p62, 1: 2000, ab155686; ATG14, 1 μg/ml, ab139727 and GAPDH, 1 μg/ml, ab37168, all from Abcam, overnight at 4°C. After washing 3 times with TBST, the membranes were incubated with goat anti-rabbit IgG (1: 10 000, ab175781, Abcam) or goat anti-mouse IgG (1: 10 000, ab175775, Abcam) secondary antibodies at room temperature for 2 h. ECL Western blotting kit (Bio-Rad lab, USA) was used to visualize the protein bands. GAPDH was used for an internal control.
Immunofluorescence staining
Transfected cells slides were fixed with 4% formaldehyde for 15 min and were permeabilized with 0.1% Trioton X-100 in PBS for 20 min on ice. After washing with PBS, the cells were incubated in a blocking solution composed of PBS containing 5% skimmed milk at room temperature for 1 h. Primary antibodies targeting LC3-II (Abcam) diluted 1: 200 in blocking solution were added to the slides at 4°C overnight. After washing 3 times with PBS, the cells were incubated with the blocking solution with Cy3-conjugated donkey anti-mouse immunoglobulin G antibody (Jackson Immuno Research Laboratories) at a dilution of 1: 600 for 2 h at room temperature. Finally, the slides were washed 3 times and photographed under a fluorescence microscopy (Olympus, Tokyo, Japan).
Statistical analysis
SPSS version 20.0 and GraphPad Prism 6 were used to perform statistical analysis. The two-tailed t test was used to analyze differences between 2 groups, while one-way analysis of variance (ANOVA) was used for multiple comparisons. Data are presented as the mean ±SD. A P value <0.05 was considered to indicate a statistically significant difference.
Results
TSLNC8 is significantly downregulated in lung cancer cell lines
To explore the role of TSLNC8 on lung cancer development, we first examined TSLNC8 RNA levels in lung cancer cells and normal human bronchial epithelial cells by quantitative real-time PCR. As presented in Figure 1, TSLNC8 was obviously downregulated in A549, H441, and H1975 cell lines, and the relative expression of TSLNC8 was reduced by 82.5%, 25.7%, and 66.4%, respectively, when compared with the normal cell lines. A549 cells showed the lowest expression level of TSLNC8 in the tested lung cancer cell lines. Therefore, A549 cells were selected for subsequent experiments.
Figure 1.
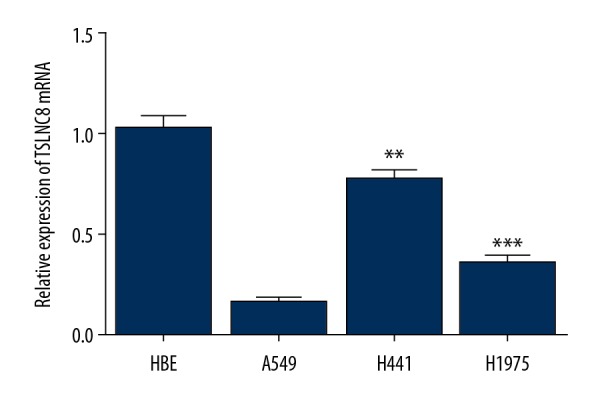
Expression of lncRNA TSLNC8 is decreased in lung cancer cell lines. Relative TSLNC8 levels in 3 lung cancer cell lines (A549, H441, and H1975) and normal human bronchial epithelial cells HBEs were detected by qRT-PCR. Each bar represents the mean ±SD calculated from 3 independent experiments. ** P<0.01, *** P<0.001 versus control.
Overexpression of TSLNC8 inhibits lung cancer cell proliferation
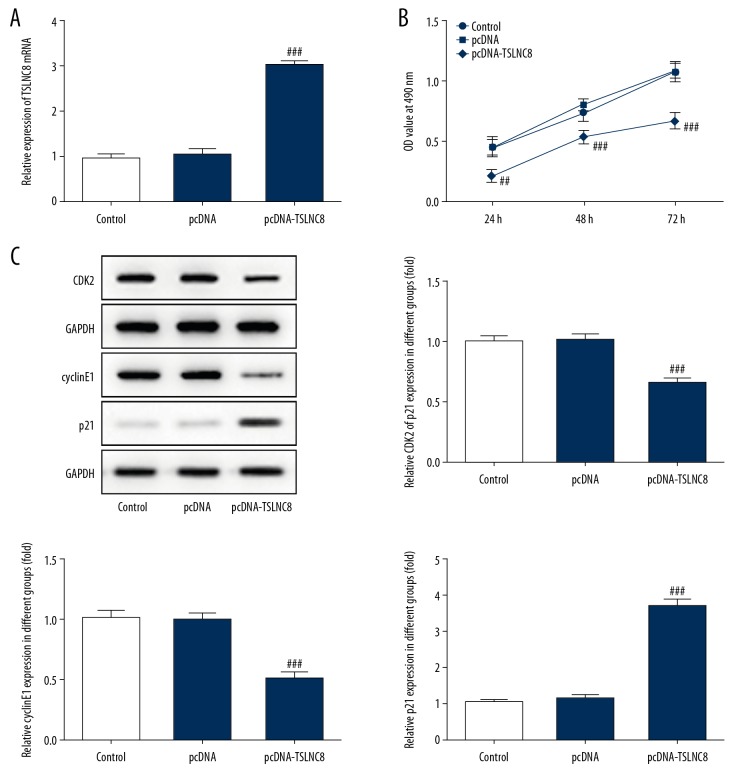
To investigate the influence of TSLNC8 on lung cancer cell proliferation, we overexpressed TSLNC8 in A549 cells (Figure 2A). The effect of TSLNC8 overexpression on proliferative ability in A549 cells was assessed by CCK-8 and Western blotting. The results from CCK-8 assay showed that TSLNC8 overexpression inhibited the growth of A549 cells at 24 h, 48 h, and 72 h, and the inhibitory rates were 54.2%, 34.1%, and 38.3%, respectively (Figure 2B). Consistent with the above results, the decreased levels of CDK2 and cyclinE1 and the increased p21 level in A549 cells were tested by Western blot assay. CDK2 and cyclinE1 activity was reduced by 55.2% and 50.9%, respectively, and p21 activity was increased by 267.9% (Figure 2C). These results indicate that TSLNC8 effectively suppressed lung cancer cell proliferation.
Figure 2.
Overexpression of TSLNC8 inhibits lung cancer cell proliferation. (A) lncRNA TSLNC8 expression levels were assessed by qRT-PCR. (B) Effect of pcDNA-TSLNC8 on proliferation of A549 cells was evaluated by CCK-8. (C) The expression of proteins involved in proliferation was estimated in A549 cells transfected with pcDNA-TSLNC8 or NC. Each bar represents the mean ±SD calculated from 3 independent experiments. ## P<0.01, ### P<0.001 versus the pcDNA group.
Overexpression of TSLNC8 inhibits lung cancer cell migration and invasion
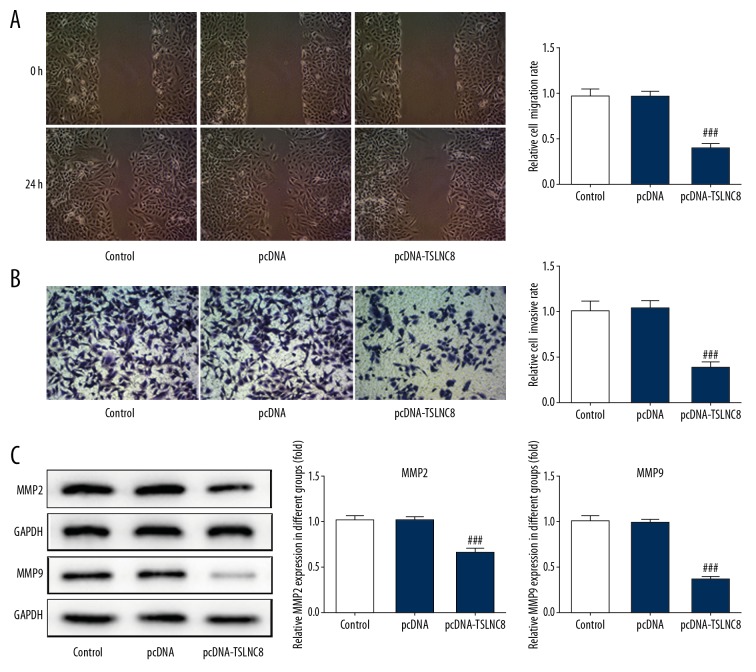
We studied the effects of TSLNC8 overexpression on lung cancer cell migration and invasion to identify the role of TSLNC8 in tumorigenesis. Wound healing assay revealed that upregulation of TSLNC8 markedly attenuated the cell migration capacity compared to the control, and cellular migration was inhibited by up to 60.5% (Figure 3A). Furthermore, Transwell assay indicated the number of invasive cells was significantly decreased after TSLNC8 was overexpressed, and the inhibitory rate was 62.4% (Figure 3B). Subsequently, the proteins levels of MMP2 and MMP9, which are involved in cell migration and invasion, respectively, were detected in A549 cells. According to the results of Western blot assay, the expressions of MMP2 and MMP9 were both greatly decreased and the activity of MMP2 and MMP-9 were reduced by 35.3% and 64.9% in TSLNC8-overexpressing A549 cells (Figure 3C). Thus, our results indicated that overexpression of TSLNC8 inhibited lung cancer cell migration and invasion.
Figure 3.
Overexpression of TSLNC8 suppresses lung cancer cell migration and invasion. (A) Wound healing assay was used to measure the effect of TSLNC8 overexpression on cell migration. Image magnification: 100×. (B) The invasive ability of TSLNC8 cells overexpressing A549 was analyzed via Transwell assays. Image magnification: 100×. (C) The levels of proteins involved in cell migration were assessed in pcDNA-TSLNC8-transfected A549 cells. Each bar represents the mean ±SD calculated from 3 independent experiments. ### P<0.001 versus the pcDNA group.
Upregulation of TSLNC8 facilitates lung cancer cell apoptosis
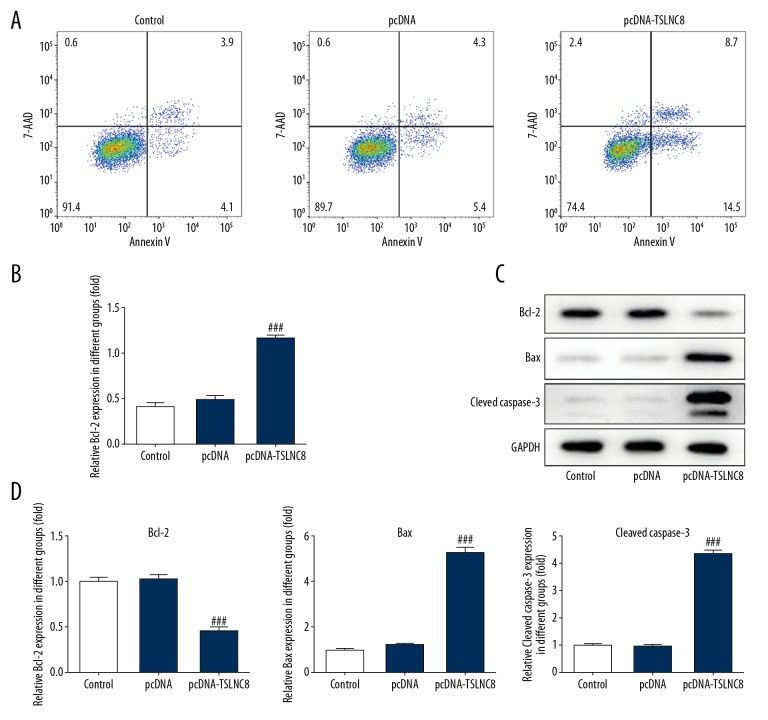
To further elucidate the role of TSLNC8 in tumorigenesis, cell apoptosis was detected by flow cytometry and Western blotting. As shown in Figure 4A and 4B, upregulation of TSLNC8 clearly promoted the apoptosis of A549 cells, and the apoptosis rate increased to 22.3% compared to the control and negative vector control. Moreover, decreased level of Bcl-2 and increased levels of Bax and cleaved caspase3 were observed in TSLNC8 overexpression, with 54.3% inhibition and 428.4% and 338.3% promotion, respectively. (Figure 4C, 4D). These results indicated the ability of TSLNC8 to accelerate apoptosis in A549 cells.
Figure 4.
(A–D) Upregulated TSLNC8 promotes lung cancer cell apoptosis. (A) Cell apoptosis was identified in TSLNC8 overexpressed A549 cells by flow cytometry. (B) TSLNC8 overexpression affected the levels of apoptosis-related proteins in A549 cells as determined by Western blot analysis. GAPDH was used as an internal reference. Each bar represents the mean ±SD calculated from 3 independent experiments. ### P<0.001 versus the pcDNA group.
Upregulated TSLNC8 expression suppresses autophagy in lung cancer cells
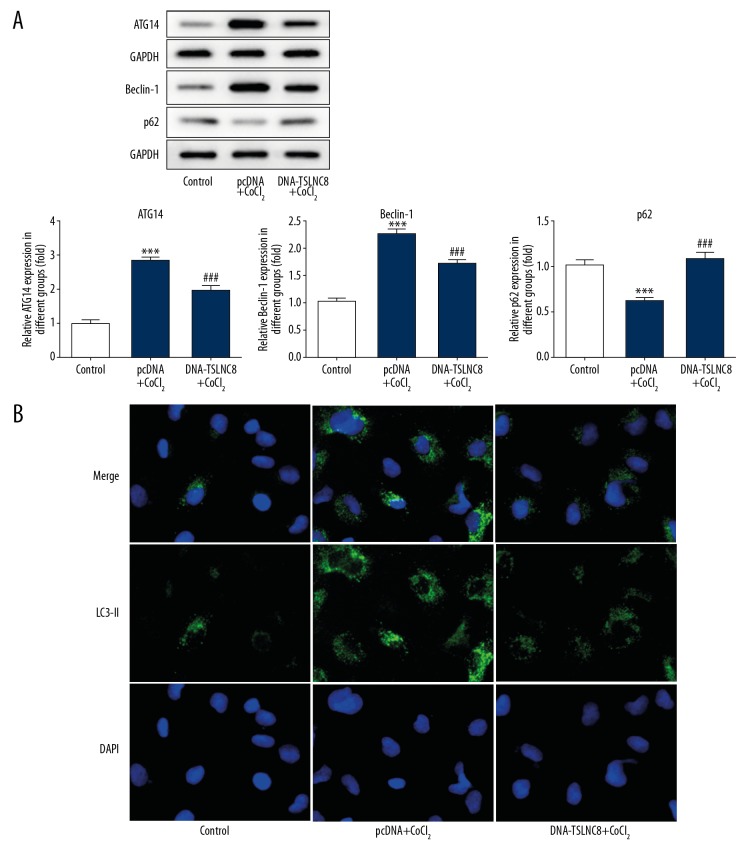
The above experiments revealed the relationship between TSLNC8 and cell apoptosis, which prompted us to explore whether TSLNC8 could affect autophagy in lung cancer cells. Cells were induced by CoCl2 for 24 h to establish a hypoxia model. The results demonstrated that the levels of ATG14 and Beclin-1 were significantly enhanced, whereas p62 was decreased in the negative vector control treated with CoCl2. The activity of ATG14 and Beclin-1 increased by 1.82-fold and 1.24-fold, while p62 was reduced by 55.5%. However, TSLNC8-overexpressing cells exhibited the opposite results and the activity of ATG14 and Beclin-1 were reduced by 31.4% and 23.2%, respectively, while p62 was increased by 58.7% compared with CoCl2 treatment (Figure 5A). In addition, immunofluorescence staining showed a significant increase in the expression of LC3-II in the control cells but there was a decrease in TSLNC8-overexpressing cells when treated by CoCl2 (Figure 5B). Taken together, these data suggest that TSLNC8 promotes cell apoptosis via inhibiting autophagy in lung cancer cells.
Figure 5.
TSLNC8 overexpression suppresses lung cancer cell autophagy. (A) Western blot assays were carried out to investigate the levels of ATG14, Beclin-1, and p62 in transfected A549 cells. (B) Immunofluorescence of LC3-II in A549 cells with pcDNA-TSLNC8 transfection. GAPDH was used as an internal reference. Each bar represents the mean ±SD calculated from 3 independent experiments. *** P<0.001 versus control; ### P<0.001 versus the pcDNA+CoCl2 group.
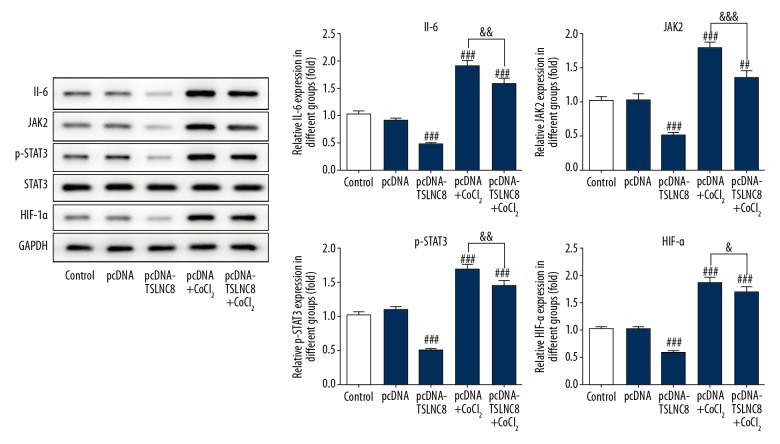
TSLNC8 overexpression suppresses autophagy by inhibiting the activation of IL-6/STAT3/HIF-1α pathway
We further explored the potential mechanism by which upregulation of TSLNC8 suppresses autophagy in lung cancer cells. According to the results of Western blotting, the protein levels of IL-6, JAK2, and HIF-1α and the phosphorylation of STAT3 in cells treated with CoCl2 were significantly higher than in the negative vector control, while the opposite results were observed in TSLNC8-overexpressing cells. Protein levels of IL-6, JAK2 p-STAT3, and HIF-1α in the pcDNA-TSLNC8+CoCl2 group were reduced by 17.4%, 24.3%, 14.4%, and 11.3% when compared with the pcDNA+CoCl2 group, but the total level of STAT3 did not change in each group (Figure 6). These results suggest that TSLNC8 overexpression inhibits A549 cell autophagy via regulation of the IL-6/STAT3/HIF-1α pathway.
Figure 6.
TSLNC8 inhibits autophagy by disrupting the IL-6/STAT3/HIF-1α pathway. The activities of IL-6, JAK2, HIF-1α, and p-STAT3 in A549 cells with or without CoCl2 were determined by Western blotting. GAPDH was used as an internal control. Each bar represents the mean ±SD calculated from 3 independent experiments. ## P<0.01, ### P<0.001 versus the pcDNA group. & P<0.05, && P<0.01, &&& P<0.001 versus the pcDNA+CoCl2 group.
Discussion
Accumulating evidence shows that several tumor-suppressor genes are located on chromosome 8, such as HTPAP (8p12) [19], SH2D4A (8p22) [20], and CSMD1 (8p23) [21]. Deletions in the short arm of chromosome 8 are closely associated with tumors [22–24]. In the present study we demonstrated that the lncRNA TSLNC8, located at 8p12, functions as a tumor suppressor in non-small cell lung cancer. The results suggest that TSLNC8 expression is significant downregulated in lung cancer cells and TSLNC8 overexpression suppresses cell proliferation and metastasis and promotes apoptosis resulting from autophagy. Importantly, TSLNC8 can inhibit autophagy via inactivation of the IL-6/STAT3/HIF-1α signaling pathway.
Cell apoptosis and autophagy play significant roles in homeostasis and in pathogenesis of diseases [25]. Autophagy has been shown to have a complex interplay with apoptosis [26]. Autophagy functions as a cell survival pathway to suppress apoptosis, but it also can lead to cell death in synergism with apoptosis or as a back-up mechanism for apoptosis [27]. In the present study we found that TSLNC8 suppresses cell proliferation, migration, and invasion, and promotes apoptosis in lung cancer cells. Considering the effect of TSLNC8 on cell apoptosis, we further investigated the relationship between autophagy and TSLNC8. Hypoxia can induce autophagy activation and quickly promote the processes of tumor growth [28]. We used CoCl2 to mimic hypoxia to induce cell autophagy, and the results showed that overexpression of TSLNC8 obviously inhibited A549 cells autophagy by regulating the levels of autophagy-related proteins Beclin-1, ATG14, and p62, as well as the level of LC3-II. Thus, TSLNC8 can promote cell apoptosis by inhibiting autophagy in lung cancer cells.
IL-6 is a pleiotropic inflammatory cytokine that is strongly associated with cancers [29,30]. IL-6 binds to its receptor and the complex activates Janus kinases (JAK), leading to the phosphorylation of STAT3 [31,32]. The IL-6/STAT3 signaling pathway has important biological roles in promoting tumor cell proliferation, invasion, and angiogenesis [33]. Mounting evidence shows that several subtypes of lung cancer can be suppressed by inhibiting the IL-6/STAT3 pathway. Jiang et al. found that IL-37 markedly inhibits cell invasion and decreases epithelial-mesenchymal transition in NSCLC by targeting the IL-6/STAT3 signaling pathway [34]. Yu et al. verified that large cell lung cancer cell lines were suppressed by plumbagin via suppressing the IL-6/STAT3 signaling pathway [35]. In the present study, the protein levels of IL-6 and JAK2 were significantly reduced and the phosphorylation of STAT3 was notably inhibited when TSLNC8 was overexpressed, while the total amount of STAT3 was unchanged. These results suggest that TSLNC8 acts as a tumor suppressor of NSCLC via regulating the expressions of IL-6 and its downstream genes, as well as the phosphorylation of STAT3.
HIF-1α is a transcription factor expressed in cells under hypoxia condition, and its activation is strongly linked to tumor growth and development [36], including lung cancers [37]. IL-6 induces the JAK2 pathway, which leads to increased activated p-STAT3. HIF-1α then interacts with activated STAT3 by blocking HIF-1α degradation and accelerating synthesis [29]. A recently discovered IL-6/STAT3/HIF1α autocrine loop has been found in multiple tumor types [10]. One study revealed that blocking Hsp90 disrupted phosphorylation of IGF-I and prevented IL-6 signaling cascades, which reduced the activation of STAT3 and HIF-1α in pancreatic cancer [38]. Another study showed that hypoxia-activated STAT3 regulates HIF-1α protein stability and enhances HIF-1-mediated expression of VEGF in renal carcinoma cells [39]. Consistent with the above, our results show that HIF-1α protein level is significantly inhibited by TSLNC8, along with the decreased expressions of p-STAT3 and IL-6. These data indicate that TSLNC8 exerts its tumor-suppressive activity through IL-6/STAT3/HIF-1α signaling pathways in NSCLC. However, the underlying mechanisms by which TSLNC8 suppresses IL-6 expression remains unclear, and we intend to explore this in further research.
Conclusions
In summary, our findings reveal for the first time that TSLNC8 works as a tumor suppressor through the IL-6/STAT3/HIF-1α signaling pathway in NSCLC by inhibiting cell metastasis and promoting apoptosis. As such, TSLNC8 may be an effective target for treating NSCLC.
Acknowledgements
We are grateful for the guidance of the hospital leader and the science and education teacher. We thank Jianxiang Wang, director of surgery, for strong support and help. We thank our department staff for their help. Thanks also to our spouses for support and encouragement.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Kaira K, Takahashi T, Murakami H, et al. The role of betaIII-tubulin in non-small cell lung cancer patients treated by taxane-based chemotherapy. Int J Clin Oncol. 2013;18(3):371–79. doi: 10.1007/s10147-012-0386-8. [DOI] [PubMed] [Google Scholar]
- 3.Wang P, Zhang L, Huang C, et al. Distinct prognostic values of alcohol dehydrogenase family members for non-small cell lung cancer. Med Sci Monit. 2018;24:3578–90. doi: 10.12659/MSM.910026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding K, Jiang J, Chen L, Xu X. Methylenetetrahydrofolate dehydrogenase 1 silencing expedites the apoptosis of non-small cell lung cancer cells via modulating DNA methylation. Med Sci Monit. 2018;24:7499–507. doi: 10.12659/MSM.910265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382(9893):720–31. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Wang QW, Zou ML, et al. Overexpressed lncRNA ZEB1-AS1 promotes cell invasion and angiogenesis through Wnt/β-catenin signaling in non-small cell lung cancer. Int J Clin Exp Pathol. 2017;10(3):3990–97. [Google Scholar]
- 7.Wang Y, Zhang Y, Yang T, et al. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144-3p in osteosarcoma cells. Oncotarget. 2017;8(35):59417–34. doi: 10.18632/oncotarget.19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Li C, Chen J, Zhang K, et al. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36(2):423–34. doi: 10.1159/000430109. [DOI] [PubMed] [Google Scholar]
- 10.Dong Z, Wang Y. LncRNA BLACAT1 accelerates the proliferation and migration of osteosarcoma cells through regulating STAT3. Pathol Res Pract. 2019;215(3):571–79. doi: 10.1016/j.prp.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Zhou Y, Huan L, et al. LncRNA MIR22HG inhibits growth, migration and invasion through regulating the miR-10a-5p/NCOR2 axis in hepatocellular carcinoma cells. Cancer Sci. 2019;110(3):973–84. doi: 10.1111/cas.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson CL, Dillon R, Devakumar A, et al. Quantitative phosphoproteomic analysis of the STAT3/IL-6/HIF1alpha signaling network: An initial study in GSC11 glioblastoma stem cells. J Proteome Res. 2010;9(1):430–43. doi: 10.1021/pr9007927. [DOI] [PubMed] [Google Scholar]
- 13.Sang H, Liu H, Xiong P, Zhu M. Long non-coding RNA functions in lung cancer. Tumour Biol. 2015;36(6):4027–37. doi: 10.1007/s13277-015-3449-4. [DOI] [PubMed] [Google Scholar]
- 14.Yang YR, Zang SZ, Zhong CL, et al. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7(10):6929–35. [PMC free article] [PubMed] [Google Scholar]
- 15.Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1–12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Li Z, Liu L, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67(1):171–87. doi: 10.1002/hep.29405. [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Yu X. Long noncoding RNA TSLNC8 suppresses cell proliferation and metastasis and promotes cell apoptosis in human glioma. Mol Med Rep. 2018;18(6):5536–44. doi: 10.3892/mmr.2018.9609. [DOI] [PubMed] [Google Scholar]
- 19.Ren N, Wu JC, Dong QZ, et al. Association of specific genotypes in metastatic suppressor HTPAP with tumor metastasis and clinical prognosis in hepatocellular carcinoma. Cancer Res. 2011;71(9):3278–86. doi: 10.1158/0008-5472.CAN-10-3100. [DOI] [PubMed] [Google Scholar]
- 20.Ploeger C, Waldburger N, Fraas A, et al. Chromosome 8p tumor suppressor genes SH2D4A and SORBS3 cooperate to inhibit interleukin-6 signaling in hepatocellular carcinoma. Hepatology. 2016;64(3):828–42. doi: 10.1002/hep.28684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Midorikawa Y, Yamamoto S, Tsuji S, et al. Allelic imbalances and homozygous deletion on 8p23.2 for stepwise progression of hepatocarcinogenesis. Hepatology. 2009;49(2):513–22. doi: 10.1002/hep.22698. [DOI] [PubMed] [Google Scholar]
- 22.Lai J, Flanagan J, Phillips WA, et al. Analysis of the candidate 8p21 tumour suppressor, BNIP3L, in breast and ovarian cancer. Br J Cancer. 2003;88(2):270–76. doi: 10.1038/sj.bjc.6600674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoehr R, Wissmann C, Suzuki H, et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84(4):465–78. doi: 10.1038/labinvest.3700068. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka K, Kikuchi-Yanoshita R, Muraoka M, et al. Suppression of tumorigenicity and invasiveness of colon carcinoma cells by introduction of normal chromosome 8p12-pter. Oncogene. 1996;12(2):405–10. [PubMed] [Google Scholar]
- 25.Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29(12):1717–19. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- 26.Kasprowska-Liskiewicz D. The cell on the edge of life and death: Crosstalk between autophagy and apoptosis. Postepy Hig Med Dosw (Online) 2017;71:825–41. doi: 10.5604/01.3001.0010.4672. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16(7):966–75. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 28.Jing X, Jiang T, Dai L, et al. Hypoxia-induced autophagy activation through NF-kappaB pathway regulates cell proliferation and migration to induce pulmonary vascular remodeling. Exp Cell Res. 2018;368(2):174–83. doi: 10.1016/j.yexcr.2018.04.026. [DOI] [PubMed] [Google Scholar]
- 29.Heikkila K, Ebrahim S, Lawlor DA. Systematic review of the association between circulating interleukin-6 (IL-6) and cancer. Eur J Cancer. 2008;44(7):937–45. doi: 10.1016/j.ejca.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 30.Skrinjar I, Brailo V, Vidovic-Juras D, et al. Evaluation of pretreatment serum interleukin-6 and tumour necrosis factor alpha as a potential biomarker for recurrence in patients with oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2015;20(4):e402–7. doi: 10.4317/medoral.20373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodge DR, Hurt EM, Farrar WL. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer. 2005;41(16):2502–12. doi: 10.1016/j.ejca.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Heinrich PC, Behrmann I, Muller-Newen G, et al. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334(Pt 2):297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu X, Luo F, Liu Y, et al. The IL-6/STAT3 pathway via miR-21 is involved in the neoplastic and metastatic properties of arsenite-transformed human keratinocytes. Toxicol Lett. 2015;237(3):191–99. doi: 10.1016/j.toxlet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Jiang M, Wang Y, Zhang H, et al. IL-37 inhibits invasion and metastasis in non-small cell lung cancer by suppressing the IL-6/STAT3 signaling pathway. Thorac Cancer. 2018;9(5):621–29. doi: 10.1111/1759-7714.12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu T, Xu YY, Zhang YY, et al. Plumbagin suppresses the human large cell lung cancer cell lines by inhibiting IL-6/STAT3 signaling in vitro. Int Immunopharmacol. 2018;55:290–96. doi: 10.1016/j.intimp.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 36.Kim HL, Cassone M, Otvos L, Jr, Vogiatzi P. HIF-1α and STAT3 client proteins interacting with the cancer chaperone Hsp90: Therapeutic considerations. Cancer Biol Ther. 2014;7(1):10–14. doi: 10.4161/cbt.7.1.5458. [DOI] [PubMed] [Google Scholar]
- 37.Kim IG, Lee JH, Kim SY, et al. Hypoxia-inducible transgelin 2 selects epithelial-to-mesenchymal transition and gamma-radiation-resistant subtypes by focal adhesion kinase-associated insulin-like growth factor 1 receptor activation in non-small-cell lung cancer cells. Cancer Sci. 2018;109(11):3519–31. doi: 10.1111/cas.13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang SA, Moser C, Gaumann A, et al. Targeting heat shock protein 90 in pancreatic cancer impairs insulin-like growth factor-I receptor signaling, disrupts an interleukin-6/signal-transducer and activator of transcription 3/hypoxia-inducible factor-1alpha autocrine loop, and reduces orthotopic tumor growth. Clin Cancer Res. 2007;13(21):6459–68. doi: 10.1158/1078-0432.CCR-07-1104. [DOI] [PubMed] [Google Scholar]
- 39.Jung JE, Lee HG, Cho IH, et al. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. FASEB J. 2005;19(10):1296–98. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]