Abstract
Multiprotein editosomes catalyze gRNA-specified insertion and deletion of uridines to create functional mitochondrial mRNAs in Trypanosoma brucei. Three functionally distinct editosomes are distinguished by their single KREN1, KREN2, or KREN3 RNase III endonuclease and, respectively, KREPB8, KREPB7, and KREPB6 partner proteins. These endonucleases perform the first catalytic step of editing, cleaving mRNA in diverse mRNA/gRNA heteroduplex substrates. We identified divergent and likely noncatalytic RNase III domains in KREPB4, KREPB5, KREPB6, KREPB7, KREPB8, KREPB9, and KREPB10 editosome proteins. Because known RNase III endonuclease functional domains are dimeric, the editing endonucleases may form heterodimers with one or more of these divergent RNase III proteins. We show here using conditional null cell lines that KREPB6, KREPB7, and KREPB8 are essential in both procyclic form (PF) and bloodstream (BF) cells. Loss of these proteins results in growth defects and loss of editing in vivo, as does mutation of their RNase III domain that is predicted to prevent dimerization. Loss of KREPB6, KREPB7, or KREPB8 also dramatically reduces cognate endonuclease abundance, as does the RNase III mutation, indicating that RNase III interactions with their partner proteins stabilize the endonucleases. The phenotypic consequences of repression are more severe in BF than in PF, indicating differences in endonuclease function between developmental stages that could impact regulation of editing. These results suggest that KREPB6, KREPB7, and KREPB8 form heterodimers with their respective endonucleases to perform mRNA cleavage. We also present a model wherein editosome proteins with divergent RNase III domains function in substrate selection via enzyme–pseudoenzyme interactions.
Keywords: RNA editing, trypanosomes, RNase III, mitochondria, endonuclease, developmental regulation
INTRODUCTION
Generating a functional mitochondrial transcriptome in Trypanosoma brucei requires extensive RNA editing, in which multiple guide RNAs (gRNAs) specify precise uridine (U) insertion and deletion at numerous editing sites (ESs) in 12 of the 18 mitochondrial (mt) encoded mRNAs (Stuart et al. 2005; Aphasizhev and Aphasizheva 2011; Read et al. 2016; Cruz-Reyes et al. 2018). Most of these edited mRNAs encode proteins of the oxidative phosphorylation (oxphos) system. The oxphos system is complete, necessary, and produces ATP in procyclic form (PF) parasites. However, oxphos composition and function are modified by developmentally regulated differential editing such that it hydrolyzes rather than synthesizes ATP in bloodstream form (BF) parasites which rely on glycolysis for ATP production (Vickerman 1985; Priest and Hajduk 1994; Hendriks et al. 2000; Gull 2001; Schneider 2001). The mechanism that developmentally regulates the editing of specific transcripts between the parasite's life cycle is unknown, but it reflects these stark differences in energy production between the BF and PF stages (Schnaufer et al. 2002). Editosomes, also called RNA Editing Core Complexes (RECCs), contain both catalytic and noncatalytic protein components that together perform numerous coordinated catalytic cycles of mRNA endonucleolytic cleavage, U insertion or deletion, and ligation at ESs. Each gRNA specifies the sites and numbers of inserted or deleted Us over a ∼35 nt region via Watson/Crick and G/U base-pairing, and most mRNAs require many gRNAs for their complete editing. Overall, the editing of the 12 mRNAs requires thousands of catalytic cycles and numerous gRNAs. The exacting precision by which the fully edited sequences are generated indicates highly specific recognition of the numerous distinct ESs, especially at the endonucleolytic cleavage step that initiates each catalytic cycle.
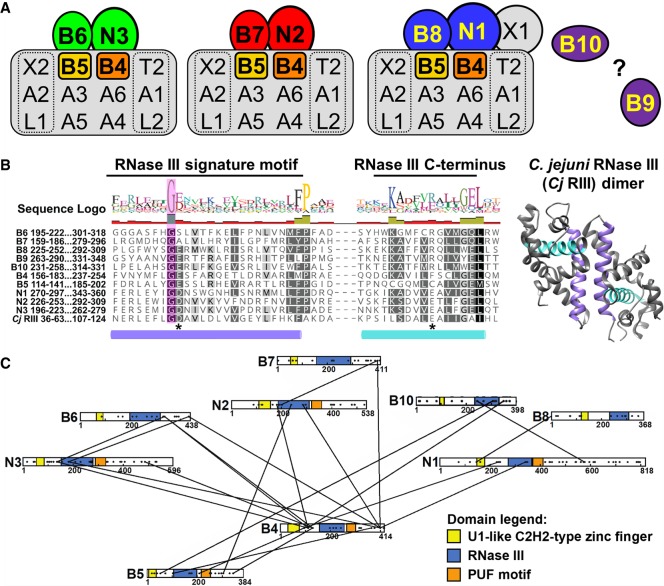
Three functionally and compositionally distinct editosomes are typified by mutually exclusive endonucleases KREN1, KREN2, and KREN3 (Fig. 1A; Panigrahi et al. 2006; Carnes et al. 2008, 2011, 2017; McDermott et al. 2016). KREN1 editosomes perform deletion editing, and uniquely possess the KREPB8 partner protein and catalytically active exoUase KREX1. KREN2 and KREN3 editosomes perform insertion editing with different substrate preferences and possess KREPB7 and KREPB6 partner proteins, respectively. All three endonucleases have a single catalytic RNase III domain, while their KREPB6, KREPB7, and KREPB8 partners have a single, highly divergent RNase III domain (McDermott et al. 2016) that lacks conserved amino acids required for catalysis, and is presumptively noncatalytic. The functions of KREPB6, KREPB7, and KREPB8 are unknown. Like the endonucleases, they also each contain a Zinc Finger (ZnF) motif in their amino-terminal region which has similarity to the U1C spliceosomal protein ZnF that facilitates both RNA and protein interactions (Heinrichs et al. 1990; Nelissen et al. 1994; Worthey et al. 2003; Carnes et al. 2011; Guo et al. 2012). RNAi knockdown studies in PF cells showed that KREPB7 and KREPB8 are essential for growth, while KREPB6 RNAi did not result in a growth defect. However, in all cases the RNAi knockdown may have been insufficient to generate a complete phenotype (Guo et al. 2012).
FIGURE 1.
RNase III domains have been identified in several editosome proteins. (A) Schematics of KREN1, KREN2, and KREN3 editosomes, with proteins containing an RNase III domain highlighted in color. Nonessential KREPB9 and KREPB10 are shown apart as they variably associate with editosomes. Editosomes contain 12 common core proteins, while KREN1, KREN2, and KREN3 and their partner proteins are mutually exclusive. (B) Alignment of RNase III domain sequences from KREPB4-B10, KREN1-N3, and C. jejuni (accession Q9PM40). A total of 299 sequences from 35 kinetoplastid species and strains were aligned using MUSCLE. Only T. brucei sequences and the RNase III signature motif/C-terminus are shown here for clarity. Colored cylinders denote the α-helices shown colored in the C. jejuni RNase III dimer structure (PDB ID 3O2R). A universally conserved glycine residue in the RNase III signature motif is shown shaded in magenta. Asterisks denote residues that are universally conserved in catalytic RNase IIIs, equivalent to D44 and E116 in C. jejuni. (C) Schematic showing location of crosslinks between RNase III domain containing editosome proteins identified in CXMS (McDermott et al. 2016). Domains are highlighted as indicated based on bioinformatics predictions (Worthey et al. 2003; Carnes et al. 2012b; McDermott et al. 2016). Black dots distributed across proteins indicate the positions of lysine residues available for CXMS crosslinking (McDermott et al. 2016).
Four other editosome proteins also have a single divergent, noncatalytic RNase III domain and U1 ZnF motifs. These are KREPB4 and KREPB5, which are essential for editosome integrity (McDermott et al. 2015a,b; McDermott and Stuart 2017), and KREPB9 and KREPB10, which are not essential in the stages tested in vitro, and are infrequently detected in editosome preparations (Wang et al. 2003; Worthey et al. 2003; Carnes et al. 2012b, 2018; Lerch et al. 2012; McDermott et al. 2015a,b; McDermott and Stuart 2017). The precise roles of the KREPB4, KREPB5, KREPB9, KREPB10 noncatalytic RNase III proteins in editing are unclear, but they may be associated with substrate interaction and endonuclease function. Thus, almost half of the editosome proteins have an RNase III domain, and their common domain structure suggests that they are all paralogs. All characterized RNase III endonucleases have a dimeric RNase III catalytic structure formed by two domains that are either in a single polypeptide (e.g., Dicer) or by the interaction of two polypeptides. Because a single KREN1, KREN2, or KREN3 endonuclease is present per editosome (Carnes et al. 2011), we hypothesized that catalytically active dimeric RNase IIIs in editosomes result from the interaction between the endonucleases and noncatalytic RNase III proteins (Carnes et al. 2011). We further suggested that the pairing of the functional RNase III domain with a noncatalytic RNase III domain enabled specific cleavage of the mRNA in the mRNA/gRNA heteroduplex rather than a staggered cleavage of both RNA strands as occurs with other RNase III endonucleases (Conrad et al. 2002; MacRae et al. 2007; Meng and Nicholson 2008; Nicholson 2014). Cross linking Mass Spectrometry (CXMS) studies showed proximities between most of the RNase III proteins (Fig. 1C), including within the KREN1/KREPB8, KREN2/KREPB7, and KREN3/KREPB6 protein pairs. This suggests that dimerization of the pairs of RNase III domains could form an active mRNA strand specific catalytic site (McDermott et al. 2016). Furthermore, the presence of multiple potential RNase III heterodimers within the same complex may enable the recognition of diverse RNA substrates.
In addition to interactions with partner endonucleases and other RNase III domain proteins, KREPB6, KREPB7, and KREPB8 preferentially associate with the heterotrimeric deletion subcomplex (KREX2–KREPA2–KREL1) common to all editosomes. In contrast, the endonucleases associate with the heterotrimeric insertion subcomplex (KRET2–KREPA1–KREL2). Tandem Affinity Purification (TAP)-tagged KREPB6, KREPB7, or KREPB8 isolated complexes from conditional null (CN) BF cells in which the partner endonuclease is knocked down contain the deletion subcomplex and lose the insertion subcomplex (Carnes et al. 2011). Reciprocally, TAP-tagged KREN1, KREN2 and KREN3 isolated editosomes from RNAi PF cells in which the partner protein, KREPB8, KREPB7, or KREPB6, respectively, is knocked down contain insertion subcomplexes and lose deletion subcomplexes (Guo et al. 2012). The results of CXMS experiments were also consistent with these interactions (McDermott et al. 2016). Thus, while KREPB6, KREPB7, and KREPB8 associate with the mutually exclusive editing endonucleases, they also interact with other proteins common to all editosomes.
Overall editosome composition appears to be essentially the same in BF and PF cells (Carnes et al. 2011), but functional differences have been observed between these stages, implying that editosomes have features that facilitate developmental control of editing (McDermott et al. 2015a,b, 2016; McDermott and Stuart 2017). We have previously shown that point mutations in KREPB5, including in its RNase III domain, differentially affect cell growth, editosome integrity and RNA editing between BF and PF stages, revealing life-cycle differences in KREPB5 and RNase III function (McDermott et al. 2015a,b). Furthermore, loss of KREPB5 and KREPB4 differentially affect editosome integrity between BF and PF (McDermott et al. 2015b; McDermott and Stuart 2017). Therefore, editosome complexes and components, including the RNase III domain containing proteins are implicated in the mechanisms that control differential editing during the life cycle. The roles of KREPB6, KREPB7, and KREPB8 in BF are unstudied, but we hypothesize that their functions may differ between BF and PF.
To further address the roles of KREPB6, KREPB7, and KREPB8, we generated both BF and PF CN cell lines for each of these proteins and analyzed the consequences to growth and editing in vivo. The knockdowns almost completely eliminated expression of the target gene in both life cycle stages, resulting in growth inhibitions thus showing that KREPB6, KREPB7, and KREPB8 proteins are essential. The repression resulted in dramatic editing defects in vivo in all cell lines. Western analyses after repression of KREPB6, KREPB7, or KREPB8 revealed loss of partner endonuclease in both BF and PF, with the exception of KREN1 after KREPB8 repression in PF. Similarly, exclusive expression of KREPB6, KREPB7, or KREPB8 with mutations designed to prevent RNase III dimerization was also lethal and reduced partner endonuclease abundance, again with the exception of KREN1 in PF. This result is consistent with the hypothesis that RNase III domain dimerization is required for endonuclease stability and function. The different consequences of KREPB8 repression in BF versus PF add to a growing list of evidence that editosomes have distinct life cycle-dependent characteristics (McDermott et al. 2015a,b, 2016; McDermott and Stuart 2017). Together, the evidence presented here demonstrates the critical roles played by KREPB6, KREPB7, and KREPB8 in editing, and suggests a mechanism of RNase III domain dimerization with partner endonucleases to regulate cleavage activity at numerous distinct ESs in vivo.
RESULTS
KREPB6, KREPB7, and KREPB8 are essential and required for RNA editing in BF and PF T. brucei
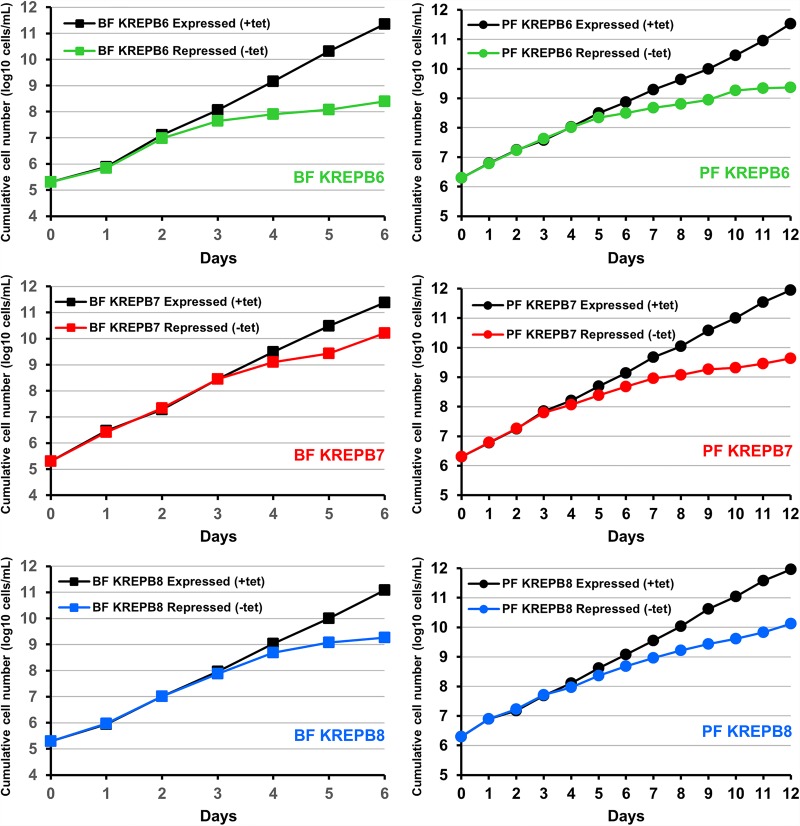
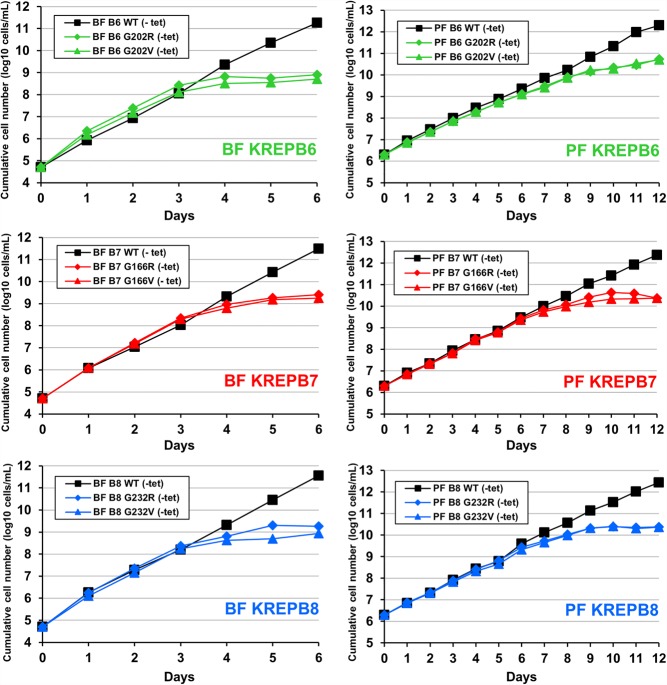
To examine the functions of the KREPB6, KREPB7, and KREPB8 proteins in both BF and PF life-cycle stages, we created BF and PF cell lines that are conditionally null (CN) for the expression of each of these proteins. We deleted both endogenous alleles in these cells and inserted a tetracycline (tet)-regulatable wild-type (WT) allele into the ribosomal DNA (rDNA) locus. Depletion of KREPB6, KREPB7, or KREPB8 by tet withdrawal resulted in severe growth inhibition in both BF and PF CN cells (Fig. 2). These results unambiguously show that KREPB6, KREPB7, and KREPB8 are essential for growth in both BF and PF cells.
FIGURE 2.
KREPB6, KREPB7, and KREPB8 are essential for in vitro growth in BF and PF cells. Cumulative growth of BF (left panels) or PF (right panels) CN cells in which tet-regulatable WT KREPB6, KREPB7, or KREPB8 is conditionally expressed or repressed. Data are representative of two independent experiments.
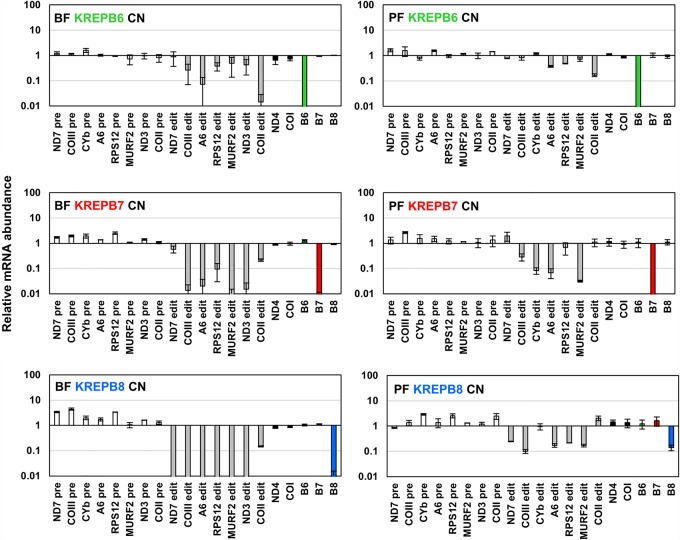
The abundances of edited, pre-edited, and never-edited RNAs were assessed by RT-qPCR in BF and PF CN cells in which KREPB6, KREPB7, or KREPB8 were repressed relative to cells in which they were expressed (Fig. 3). RT-qPCR analysis showed that mRNAs coding for KREPB6 and KREPB7 were reduced by >99% upon withdrawal of tet from BF for 48 h and PF CN cells for 96 h, that is, prior to growth arrest (Fig. 2). KREPB8 was reduced by >99% upon withdrawal of tet from BF, and by ∼85% in PF (Fig. 3). We were unable to detect edited CYb mRNA in BF or edited ND3 mRNA in PF cells, regardless of whether they were expressing or repressing KREPB6, KREPB7, or KREPB8. These results are consistent with what is known about differential editing of these transcripts between the different life cycle stages of T. brucei (Schnaufer et al. 2002). Upon KREPB6, KREPB7, or KREPB8 repression, levels of all assayed never-edited mRNAs remained essentially unchanged in both BF and PF, indicating that the observed defects do not affect all mitochondrial mRNAs. The levels of pre-edited mRNAs were either unchanged or slightly increased, as has been previously observed upon depletion of essential editosome components (Carnes et al. 2005, 2008, 2017; Trotter et al. 2005; Salavati et al. 2006; Guo et al. 2008, 2012; Ernst et al. 2009; McDermott et al. 2015b; McDermott and Stuart 2017).
FIGURE 3.
Real-time PCR analysis shows loss of KREPB6, KREPB7, or KREPB8 expression, and impact on RNA editing in both BF and PF CN cells. Relative RNA abundance is shown for KREPB6, KREPB7, KREPB8, and never-edited mRNAs COI and ND4 (black bars), pre-edited mRNAs (white bars), and edited mRNAs (gray bars). For each target amplicon, the relative change in RNA abundance was determined by using telomerase reverse transcriptase (TERT) mRNA as an internal control, with BF or PF CN cell lines that have either KREPB6, KREPB7, or KREPB8 repressed compared to the same cell line in which KREPB6, KREPB7, or KREPB8 was expressed. Data are shown as means ± SEM from two independent experiments.
Repression of KREPB6 in BF led to substantial reductions in edited COIII, A6, RPS12, MURF2, ND3, and COII mRNA levels (51%–99%), while levels of edited ND7 were slightly decreased. In PF, levels of edited ND7, COIII, CYb, and MURF2 were unaffected, while RPS12 and A6 were decreased by ∼50%–60%. Edited COII mRNA exhibited the greatest relative reduction in abundance in both BF and PF as it was decreased by 99% in BF and 84% in PF, which resembles results observed after repression of KREN3 (Carnes et al. 2008, 2017). Although the reduction in edited COII mRNA in PF appears less relative to BF, these differences may reflect the greater steady-state abundance of edited COII mRNA in PF due to developmental regulation, rather than differences in KREPB6 function. Indeed, this result is consistent with previous studies that have reported greater effects on editing in BF than in PF following editosome protein repression (McDermott et al. 2015a,b; McDermott and Stuart 2017), implying inherent differences in editing between life cycle stages.
KREPB7 depletion in BF resulted in reductions of >90% in all assayed edited mRNAs with the exceptions of COII and ND7 which were less severely affected. A similar result was observed in PF where edited COII and ND7 levels were essentially unchanged, while those of COIII, CYb, A6, and MURF2 were reduced by 71%–97%. A notable difference between BF and PF results after KREPB7 repression is that the abundance of edited RPS12, which was almost unaltered in PF is reduced ∼90% after KREPB7 depletion in BF. Aside from the previously observed general trend of editing being more sensitive to editosome perturbations in BF (McDermott et al. 2015a,b; McDermott and Stuart 2017) the significance of the drastic difference in edited RPS12 abundance between BF and PF is unclear.
Repression of KREPB8 resulted in large reductions in all assayed edited mRNAs in BF (>99%) and PF (75%–90%) with the exceptions of CYb and COII, correlating with a lack of deletion editing sites in CYb and COII. In PF, CYb levels were unchanged, while those of COII were increased approximately twofold. In BF, COII levels were decreased by ∼85%, despite lacking deletion sites that are cleaved by KREN1 (Benne et al. 1986). Previously published analyses of BF cell lines lacking KREN1 have shown increased supernumerary U insertion at both canonical and noncanonical editing sites, which could interfere with the generation of fully edited sequences in transcripts that lack canonical deletion sites (Carnes et al. 2017). Thus, KREN1 editosomes appear to remove extra Us at insertion sites thereby indicating proofreading during editing (Igo et al. 2002; Trotter et al. 2005; Ringpis et al. 2010; Carnes et al. 2012a; Cruz-Reyes et al. 2018). Differences between BF and PF editing after KREPB8 repression could be due to differences in the extent of KREPB8 knockdown, however this seems unlikely. Although knockdown of KREPB8 mRNA is incomplete in PF, the knockdown of editing of most transcripts is severe and the impact on growth is strikingly similar compared to KREPB6 and KREPB7 knockdowns (Fig. 2).
Thus, loss of KREPB6, KREPB7, or KREPB8 results in considerable defects in editing in vivo. The observed editing defects are largely consistent with those previously observed upon loss of KREN3, KREN2, and KREN1, respectively, suggesting that KREPB6, KREPB7, and KREPB8 are required for their partner endonuclease function (Carnes et al. 2005, 2008, 2017; Trotter et al. 2005; Guo et al. 2012).
KREPB6, KREPB7, and KREPB8 are critical for cognate endonuclease abundance
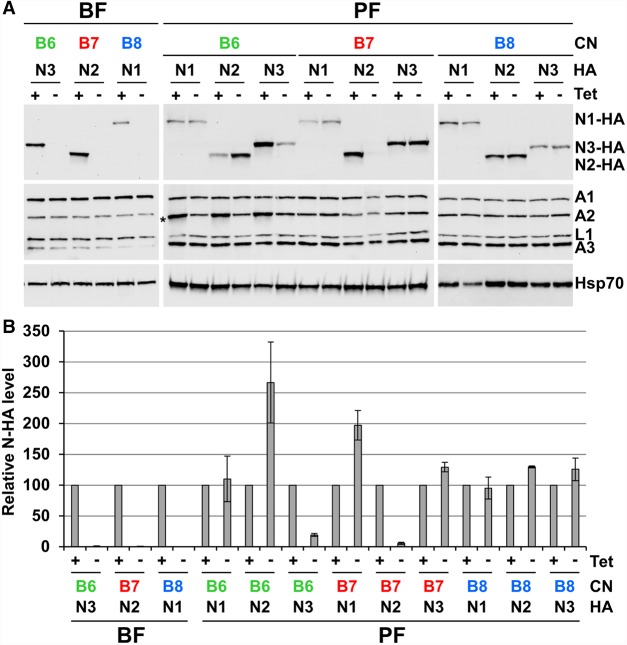
To assess the impact of KREPB6, KREPB7, or KREPB8 repression on editosome protein abundances and complex integrity, total and fractionated lysates from CN cell lines were analyzed by western blotting (Fig. 4; Supplemental Fig. 1). The KREPB6, KREPB7, and KREPB8 CN cell lines were modified for these experiments by introducing carboxy-terminal HA-epitope tags into the endogenous locus of each cognate endonuclease to permit their detection. Western analysis was performed on total lysates of cells that were grown in the presence or absence of tet for 48 h (BF) or 96 h (PF) (Fig. 4A). Blots were probed using antibodies to the HA-epitope tag for endonuclease detection, a mixture of monoclonal antibodies against editosome proteins KREPA1, KREPA2, KREL1, and KREPA3, or mitochondrial Hsp70 antibody as a control for loading and fractionation. Loss of KREPB6, KREPB7, or KREPB8 did not significantly affect the abundances of KREPA1, KREPA2, KREL1, and KREPA3 in BF or PF. However, repression of KREPB6, KREPB7, or KREPB8 resulted in complete loss of cognate endonuclease signal in BF lysates. Repression of KREPB6 or KREPB7 also resulted in large reductions in cognate endonuclease signal in PF lysates. However, repression of KREPB8 did not alter the amount of KREN1-HA detected in PF. RT-qPCR analysis of mRNAs encoding the endonucleases in PF CN cell lines confirmed that changes in cognate endonuclease abundances were occurring at the protein, but not the mRNA level (Supplemental Fig. 2). Abundances of HA-tagged noncognate endonucleases were also assessed in the PF CN cell backgrounds, revealing increases in abundances of noncognate endonucleases following partner B-protein repression e.g. KREN2-HA in KREPB6 CN, and KREN1-HA in KREPB7 CN (Fig. 4B).
FIGURE 4.
Western blot analyses show KREPB6, KREPB7, and KREPB8 are critical for endonuclease abundance in BF and PF. The KREPB6, KREPB7, and KREPB8 CN cell lines were modified by introducing HA-epitope tags into the endogenous loci of each endonuclease. (A) Western analysis of total lysates (equivalent of 1 × 107 BF cells/lane, and 5 × 106 PF cells/lane) from BF and PF CN cells in which the tet-regulatable WT KREPB6, KREPB7, or KREPB8 alleles were expressed or repressed for 2 and 4 d, respectively. The western blots were probed with monoclonal antibodies against the HA-epitope tag on the cognate partner endonucleases, the editosome proteins KREPA1, KREPA2, KREL1, and KREPA3, and quantified by densitometry using mitochondrial Hsp70 for normalization. Asterisk denotes position of the TAP-tagged regulatable KREPB6 protein in PF, which is also detected by our editosome protein monoclonal antibodies. (B) Western quantification of the relative change in abundance of each endonuclease was determined in BF or PF CN cell lines that have either KREPB6, KREPB7, or KREPB8 repressed compared to the same cell line in which KREPB6, KREPB7, or KREPB8 was expressed. Data are shown as means ± SEM from two independent experiments (four experiments for PF KREPB6/KREN2-HA, KREPB7/KREN2-HA, and KREPB8/KREN1-HA).
Glycerol gradient fractionation of lysates from CN cell lines revealed that the sedimentation of the common editosome proteins KREPA1, KREPA2, KREL1, and KREPA3 were unchanged following KREPB6, KREPB7, or KREPB8 repression in both BF and PF cells (Supplemental Fig. 1). Despite the considerable loss of the HA tagged cognate endonuclease following KREPB6 and KREPB7 knockdown in PF (see Fig. 4) the remaining tagged KREN3 and KREN2 sedimented in a peak ∼20S region along with the other editosome proteins (Supplemental Fig. 1). This was also the case for tagged KREN1 whose abundance was unchanged following loss of KREPB8, and also for the noncognate endonucleases (Supplemental Fig. 3). Thus, in all cases the endonucleases that were present were associated with editosomes.
Together, these results indicate that loss of KREPB6, KREPB7, and KREPB8 reduces the steady-state abundances of their cognate endonucleases in one or both life cycle stages, without significantly affecting complex integrity or endonuclease association with editosomes. Thus, the interactions between KREPB6–KREN3, KREPB7–KREN2, and KREPB8–KREN1 reinforce endonuclease stability. These results are also consistent with the abundances of the editing endonucleases being in stoichiometric balance, with competition between KREPB6–KREN3, KREPB7–KREN2, and KREPB8–KREN1 for stabilizing interactions with common editosome core proteins. For example, we speculate that the loss of KREN3 editosomes after KREPB6 repression results in stabilization of KREN2, as it no longer competes for limiting amounts of editosome common core proteins. The different consequences after KREPB8 repression in BF versus PF CN cells with respect to KREN1 abundance is also striking and adds to the list of editosome components implicated in the processes that control differential editing throughout the T. brucei life cycle.
Mutation of the RNase III domain prevents KREPB6, KREPB7, and KREPB8 function
To determine whether the RNase III domain is required for KREPB6, KREPB7, or KREPB8 function, we compared exclusive expression of mutant RNase III alleles to WT in the background of our CN cell lines. Because these domains lack conserved catalytic amino acids, the introduced mutations were designed to disrupt dimerization of RNase III domains by changing the universally conserved glycine (G) in the signature motif (Fig. 1) to a larger, charged amino acid, arginine (R). Steric constraints with another α-helix in the RNase III fold are predicted to prevent amino acids larger than glycine from maintaining the proper orientation of the dimerization interface. The same substitution in this conserved glycine residue was also previously shown to disrupt KREPB4 and KREPB5 function, as well as KREPB9 and KREPB10 association with editosomes (Carnes et al. 2012b, 2018; McDermott et al. 2015a,b; McDermott and Stuart 2017). Exclusive expression of G202R mutant KREPB6, G166R mutant KREPB7, and G232R mutant KREPB8 was lethal in both BF and PF, while control WT alleles permitted normal growth (Fig. 5). Replacing the conserved glycine with a smaller, uncharged valine was also lethal, likely reflecting the significant steric limitations of this position required for RNase III dimerization. The ability of these RNase III domains to form dimers is therefore critical for function. To assess whether the carboxy-terminal portion of the RNase III domain was similarly essential, we independently mutated R310, R288, and R301 residues to either alanine or glutamic acid in KREPB6, KREPB7, and KREPB8, respectively. The arginine at this position aligns with a glutamic acid essential for catalysis in all characterized RNase III endonucleases (Fig. 1). Exclusive expression of each of these mutant alleles was also lethal (Supplemental Fig. 4), indicating that the entire modeled RNase III fold is essential for KREPB6, KREPB7, and KREPB8 function.
FIGURE 5.
The highly conserved glycine in the RNase III domains in KREPB6, KREPB7, and KREPB8 is essential for in vitro growth in BF (left panels) and PF (right panels) cells. Cumulative growth of BF and PF CN cells that constitutively express V5-tagged WT, or RNase III Glycine (G) to Arginine (R) or Valine (V) mutant versions of KREPB6, KREPB7, or KREPB8 and in which the tet-regulatable WT alleles were repressed. Data are representative of two independent experiments.
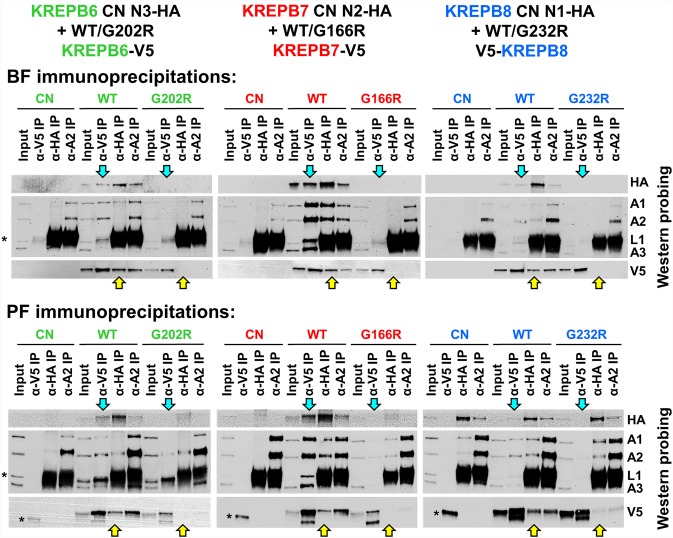
To further address the impact of RNase III mutation in KREPB6, KREPB7, and KREPB8, we analyzed immunoprecipitates (IPs) from BF and PF CN cell lines with HA-tagged cognate endonucleases that also exclusively express V5-tagged WT or RNase III G to R mutant KREPB6, KREPB7, or KREPB8 alleles (Fig. 6). Lysates were prepared from cells that were grown without tet for 2 d for BF lines, or for 4 d for PF lines to eliminate expression of the regulatable WT protein. For IPs we used (i) anti-KREPA2 antibody that is present in all three editosomes, (ii) anti-HA antibody that isolates proteins associated with tagged cognate endonuclease, or (iii) anti-V5 antibody that isolates proteins associated with exclusively expressed mutant or WT KREPB6, KREPB7, or KREPB8 (Fig. 6). KREPA1, KREPA2, and KREPA3 were detected in anti-KREPA2 IPs from BF and PF CN cell lysates, and from those that were exclusively expressing WT or RNase III G to R mutant alleles. This indicates that loss or RNase III domain mutation in KREPB6, KREPB7, and KREPB8 does not grossly disrupt editosome integrity in BF or PF, which is consistent with the editosome protein abundance and sedimentation studies (Fig. 4; Supplemental Fig. 1). However, we observed reduced or lack of coimmunoprecipitated V5-tagged RNase III mutant compared to V5-tagged WT KREPB6, KREPB7, and KREPB8 proteins with anti-KREPA2. This indicates that RNase III domain mutations in KREPB6, KREPB7, and KREPB8 affect their ability to associate with editosomes. This is further supported by reduced coimmunoprecipitation of KREPA1, KREPA2, KREL1, and KREPA3 editosome core proteins in anti-V5 IPs from V5-tagged RNase III mutant versus WT KREPB6, KREPB7, and KREPB8. The reductions in the steady state abundances of KREN3, KREN2, or KREN1 in input lysates due to the RNase III domain mutations were similar to those resulting from the loss of KREPB6, KREPB7, or KREPB8 in BF, and of KREN3 and KREN2 in PF (Fig. 4). Because HA-tagged KREN3, KREN2, or KREN1 is largely or completely absent in these cases where mutant KREPB6, KREPB7, or KREPB8 is respectively exclusively expressed, coimmunoprecipitation of these proteins could not be detected. This result indicates that the RNase III domains of KREPB6, KREPB7, and KREPB8 are important for the stabilization of their cognate endonucleases in one or both life-cycle stages, depending on the editosome type.
FIGURE 6.
Western blots of editosome IPs from BF and PF CN cells with V5-tagged WT or mutant KREPB6, KREPB7, and KREPB8 and HA-tagged KREN3, KREN2, or KREN1 endonucleases. The cells either had KREPB6, KREPB7, or KREPB8 expression eliminated (CN) or exclusively expressed mutant G-R or WT RNase III V5-tagged KREPB6, KREPB7, or KREPB8. The IPs and input lysates were probed with monoclonal antibodies against the HA-epitope tag on the endonucleases, the V5-tag on the WT or mutant KREPB proteins, and the KREPA1, KREPA2, KREL1, and KREPA3 editosome proteins. For comparisons of G-R mutant relative to WT KREPB protein, cyan arrows highlight loss of cognate endonuclease (HA probing) in KREPB protein IPs, while yellow arrows highlight loss of cognate KREPB protein (V5 probing) in endonuclease IPs. Asterisks indicate heavy chains of IP antibodies.
DISCUSSION
Results from conditional KREPB6, KREPB7, and KREPB8 null cell lines demonstrate that these proteins and their divergent RNase III domains are essential for growth and editing in vivo in both BF and PF cells. With the notable exception of KREPB8 repression in PF cells, our results show that cognate endonuclease abundance is linked to partner B protein expression, indicating stabilizing interactions between KREPB6/KREN3, KREPB7/KREN2, and KREPB8/KREN1. The resilience of KREN1 levels after KREPB8 repression in PF implies that the additional presence of the KREX1 exoUase in KREN1 editosomes contributes to the stabilization. Despite gross similarities in editosome composition in BF and PF parasites (Carnes et al. 2011), evidence for significant developmental differences in editosome function continues to mount (McDermott et al. 2015a,b; McDermott and Stuart 2017) and the differential stability of KREPB8/KREN1 editosomes between life cycle stages adds to this list. Unidentified inherent differences in editosomes between the stages provide a new paradigm for the long mysterious basis for the edited RNA transcriptome differences between BF and PF cells. Because RNase III cleavage is the first catalytic step in the editing process, it is a likely point of regulation. The noncatalytic RNase III domains in KREPB6, KREPB7, and KREPB8 are shown here to be critical for both in vivo function and abundance of partner endonucleases. In addition, editosomes contain eight proteins with RNase III domains (KREN1, KREN2, KREN3, KREPB4, KREPB5, KREPB6, KREPB7, and KREPB8) as well as two others (KREPB9 and KREPB10) that are infrequently associated with editosomes (Worthey et al. 2003; Carnes et al. 2005, 2008, 2012b, 2018; Trotter et al. 2005; Panigrahi et al. 2006; Lerch et al. 2012; McDermott et al. 2015b, 2016; McDermott and Stuart 2017). This highlights the importance of the endonuclease cleavage step in RNA editing, its potential as a point of regulation and the possible roles of these various RNase III proteins in the regulation of editing, including differentially between life cycle stages.
The effects on in vivo RNA editing upon the loss of KREPB6, KREPB7, or KREPB8 mirror the defects previously observed after repression of their partner endonucleases. This is likely due to the reductions in the steady-state abundances of their cognate endonucleases in one or both life cycle stages, which did not significantly affect the levels of other editosome proteins or complex integrity. Strong reductions in KREN1, KREN2, and KREN3 levels occurred respectively after KREPB8 repression in BF and KREPB7 and KREPB6 repression in BF and PF. These reductions occurred at the level of protein stability, as mRNA levels were unaltered (Supplemental Fig. 2). The KREPB6/KREN3, KREPB7/KREN2, and KREPB8/KREN1 interactions therefore appear to prevent endonuclease turnover. The lack of altered KREN1 abundance in PF upon repression of KREPB8 may reflect stabilizing interactions between KREN1 and KREX1, which is uniquely present in KREN1 editosomes. This possibility is supported by the identification of numerous proximal crosslinks between these proteins in CXMS experiments (McDermott et al. 2016). In addition, KREN1 can also crosslink to KREPB10, which has been isolated in KREN1 complexes that lack KREPB8 (Fig. 1; Guo et al. 2012; Lerch et al. 2012; McDermott et al. 2016). The similarity between KREPB8 and KREPB10 sequences suggest that there may be functional or associative overlap between these proteins in certain circumstances which may be related to the persistence of KREN1 in the absence of KREPB8 in PF. KREPB10 and KREPB8 have 32% sequence identity and are adjacent on T. brucei chromosome 8 suggesting a relatively recent gene duplication event (Lerch et al. 2012). In all cell lines, the reduction of endonuclease abundance was more severe in BF compared to PF, a developmental difference that underscores the recent appreciation of the differential editosome function between T. brucei life cycle stages (McDermott et al. 2015a,b, 2016; McDermott and Stuart 2017). While it is now clear that such functional differences exist, the mechanism by which they operate remains elusive. Because the noncatalytic RNase III protein KREPB5 has been shown to have developmentally distinct functionality (McDermott et al. 2015a,b), the same phenomenon may also be at work with KREPB6, KREPB7, and KREPB8.
Our results provide new insights into editosome interactions that stabilize the endonucleases. KREPB6 knockdown results in a large reduction in cognate KREN3, and a corresponding large increase in noncognate KREN2. Similarly, KREPB7 knockdown results in a large reduction of cognate KREN2 and increased abundances of noncognate KREN1 and KREN3. These results expand on previously observed increases in insertion (KREN2 or KREN3) or deletion (KREN1) cleavage activity in vitro following repression of KREPB6, KREPB7, or KREPB8 in PF RNAi cell lines (Guo et al. 2012). Furthermore, repression of KREN1, KREN2, or KREN3 in PF RNAi, BF CN, or BF mutant γ-ATP synthase cell lines also increases the activities of the remaining endonucleases in vitro and in vivo (Trotter et al. 2005; Carnes et al. 2008, 2017). In addition, editosome complexes from cells expressing only a single endonuclease show altered gradient sedimentation, while those from cells expressing two endonucleases sediment normally, apparently compensating for the loss of one endonuclease (Carnes et al. 2017). Together these results are consistent with the concept that endonuclease levels and activities are in stoichiometric balance as a result of competition for limiting amounts of editosome common core proteins. Our data demonstrate that this balance requires stabilizing interactions between KREPB6/KREN3, KREPB7/KREN2, and KREPB8/KREN1. Thus, endonuclease protein abundance is stabilized by at least two types of interactions: similar interactions with editosome common core proteins and specific interactions with cognate partner B protein (McDermott et al. 2016).
Critically, the mechanism by which KREPB6, KREPB7, and KREPB8 function requires the noncatalytic RNase III domain, implicating the dimerization with other editosome RNase III domains as an essential feature in RNA editing. Mutation of the universally conserved Gly in the RNase III domain was designed to disrupt the ability to form dimers and has previously been shown to prevent the function of editosome RNase III proteins (Carnes et al. 2012b, 2018; McDermott et al. 2015a,b; McDermott and Stuart 2017). Here, exclusive expression of mutant KREPB6, KREPB7, or KREPB8 is incapable of supporting normal growth in either BF or PF, while WT protein permits normal growth (Fig. 5; Supplemental Fig. 4). More importantly, this mutation in the RNase III domain of KREPB6, KREPB7, or KREPB8 dramatically reduces or eliminates cognate endonuclease abundance (Fig. 6), which mirrors endonuclease loss after repression (Fig. 4). Except for KREN1-HA in PF cells, the HA-tagged endonuclease is notably reduced or absent in cells that exclusively express the G to R RNase III mutation. As discussed above, the presence of KREX1 or KREPB10 may stabilize KREN1 in the absence of KREPB8 in PF. However, despite the persistence of KREN1-HA when KREPB8-G232R-V5 is exclusively expressed, co-isolation of these two proteins was dramatically reduced in IPs relative to WT control. Thus, the RNase III domain KREPB6, KREPB7, and KREPB8 plays a central role in facilitating interactions with cognate endonuclease. Furthermore, RNase III mutation prevents robust association of KREPB6, KREPB7, and KREPB8 with editosomes, indicating that RNase III dimerization is the mechanism by which these proteins interact with editosomes.
The demonstration that the noncatalytic RNase III domain in KREPB6, KREPB7, and KREPB8 is critical for the function of these proteins provides evidence for a potential mechanism for editing endonuclease activity at ESs. These data, in combination with prior results (McDermott et al. 2016; McDermott and Stuart 2017), suggest arrangements among RNase III editosome proteins that are analogous to characterized enzyme–pseudoenzyme interactions (Willert et al. 2007; Nguyen et al. 2013, 2015; Kafková et al. 2017; Murphy et al. 2017; Patel et al. 2018), where pseudoenzymes are catalytically inactive but interact with and regulate the activity of catalytic paralogs. Furthermore, we hypothesize that heterodimers of catalytically active endonucleases with proteins containing noncatalytic RNase III domains would cleave only the mRNA strand in a mRNA–gRNA duplex, leaving the gRNA intact. There is evidence that the editing process consumes gRNAs, as knockdown or inactivation of editosome proteins KREPA6 and KRET2 results in gRNA accumulation (Aphasizheva et al. 2014). However, the mechanism responsible for editing-mediated gRNA turnover is unknown, and it may be unrelated to the cleavage activities of editosomes. A clearer view on gRNA metabolism requires additional experimentation.
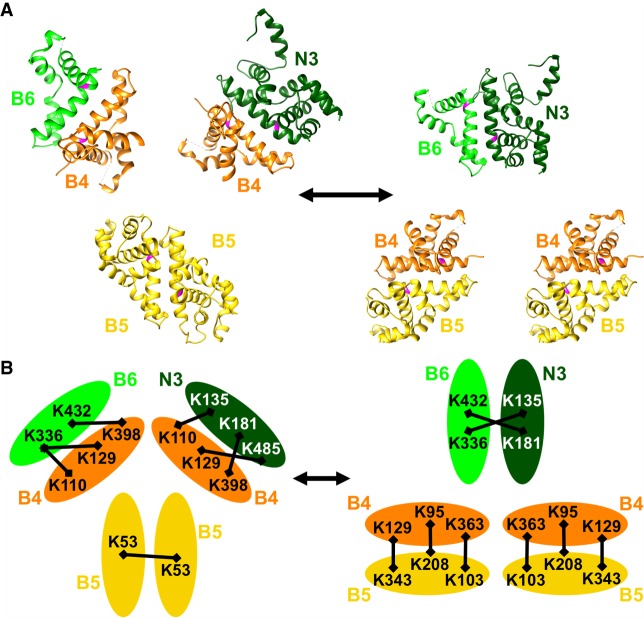
Given the number of potential RNase III enzyme–pseudoenzyme interactions within editosomes, we also extend our model to propose that pairing between RNase III domains shifts among different RNase III proteins (Fig. 7). The consequences of this shift would subsequently promote or repress endonuclease activity in the context of distinct substrates. Multiple lines of evidence are consistent with this model. Coexpression experiments have indicated that endonucleases KREN1, KREN2, or KREN3 are present as a single copy per editosome, while KREPB4 and KREPB5 are present as multiple copies per editosome (Carnes et al. 2011; McDermott and Stuart 2017) (McDermott S and Stuart K, unpubl.). The presence of multiple copies of KREPB5 was also indicated by our CXMS experiments (McDermott et al. 2016), where Lys53 in one peptide of KREPB5 was crosslinked to Lys53 in another peptide of KREPB5, indicating that two copies of KREPB5 appear to be in proximity. Additionally, many of the lysines that specifically crosslink KREPB6 to KREN3, KREPB7 to KREN2, and KREPB8 to KREN1 are the same lysines found to crosslink KREPB4 to either KREPB6, KREPB7, KREPB8, KREN1, KREN2, or KREN3, in a similar pattern (Fig. 7). For example, observed crosslinks between KREPB4 and KREPB6 were B4Lys110–B6Lys336 and B4Lys398–B6Lys432, and the same residues in KREPB4 also crosslink to KREN3, B4Lys110–N3Lys135 and B4Lys398–N3Lys181. Observed crosslinks between KREPB6 to KREN3 are B6Lys336–N3Lys135 and B6Lys432–N3Lys181, similar to the arrangement each protein has with KREPB4. One explanation for these data is that RNase III domain interactions between KREPB4 and either KREPB6, KREPB7, KREPB8, KREN1, KREN2, or KREN3 are similar to the RNase III domain interactions between KREPB6/KREN3, KREPB7/KREN2, and KREPB8/KREN1. In T. brucei, enzyme–pseudoenzyme regulation occurs in polyamine synthesis, deoxyhypusine synthesis, and protein arginine methylation (Willert et al. 2007; Nguyen et al. 2013, 2015; Kafková et al. 2017; Murphy et al. 2017; Patel et al. 2018). Considering the number of noncatalytic RNase III proteins in editosomes, pseudoenzyme regulation of endonuclease activity is a compelling hypothesis, and would also provide a mechanism to recognize a range of distinct substrates (McDermott and Stuart 2017). Furthermore, while the existence of the three distinct KREPB6/KREN3, KREPB7/KREN2, and KREPB8/KREN1 editosomes is apparent, their precise composition and detailed structural conformations may be quite dynamic as the very numerous distinct but similar editing sites are processed.
FIGURE 7.
Proposed model for RNase III interactions within editosomes. Top panel (A) shows RNase III domain structures modeled on C. jejuni 3O2R for KREPB4 (orange), KREPB5 (yellow), KREPB6 (light green), and on 1O0W for KREN3 (dark green) (McDermott et al. 2016) in two configurations: On the left, two KREPB5 domains dimerize while KREPB4 forms dimers with KREPB6 and KREN3; and on the right, two dimers of KREPB4 with KREPB5 form while KREPB6 and KREN3 dimerize. Universally conserved glycine residue critical for function shown in magenta in each domain. Interchange between these configurations could facilitate ES recognition and cleavage by KREN3 using enzyme–pseudoenzyme regulation of catalytic activity. Bottom panel (B) shows crosslinks identified by CXMS between KREPB4, KREPB5, KREPB6, and KREN3 (represented by ovals), all of which are consistent with configurations in the model shown in the top panel (McDermott et al. 2016). Crosslinked Lys residues (K) are indicated by their position in the protein, and crosslinks depicted by black bars. Similar configurations are proposed for KREPB7-KREN2 and KREPB8-KREN1 but are not shown.
MATERIALS AND METHODS
Growth of cells in vitro
BF cells were grown in HMI-9 (Hirumi and Hirumi 1989) with 10% FBS at 37°C, 5% CO2. PF cells were grown in SDM-79 (Brun and Schönenberger 1979) with 10% FBS at 27°C. For growth curve analysis, cell density was measured using a Coulter Counter. BF were reseeded at 2 × 105 cells/mL in 10 mL every day, while PF were reseeded at 2 × 106 cells/mL in 10 mL every 2 d. Transfections of BF cell lines with the Amaxa Nucleofector (Lonza), and of PF cell lines with the BTX transfection device (Harvard Apparatus, Inc.), were carried out as previously described (Merritt and Stuart 2013). Concentrations of drugs used for selection and tet-regulated expression of transgenes are as follows. For BF: 2.5 µg/mL G418, 5 µg/mL hygromycin, 2.5 µg/mL phleomycin, 0.5 µg/mL tet, 0.1 µg/mL puromycin. For PF: 15 µg/mL G418, 25 µg/mL hygromycin, 2.5 µg/mL phleomycin, 0.5 µg/mL tet, 1 µg/mL puromycin, 10 µg/mL blasticidin, 25 µg/mL ganciclovir, 100 µg/mL nourseothricin.
Generation of BF and PF conditional null cell lines
WT KREPB6, KREPB7, and KREPB8, each including the stop codon and flanked in frame with attB Gateway recombination sites, were PCR amplified using primers described in Supplemental Table 1. BP Clonase II (Thermo Fisher Scientific) were used to transfer the PCR products into the Gateway entry vector pDONR221. LR Clonase II (Thermo Fisher Scientific) was then used to transfer the sequences into the Gateway destination vector pLEW100v5(BLE)GW (Merritt and Stuart 2013), which allows for tet-regulatable expression from the rRNA locus. The resulting pLEW100v5(BLE)-KREPB6, B7, and B8 plasmids were linearized with NotI, transfected into PF 29.13, or BF SM427 cells, and transgenic lines were selected by phleomycin resistance. The KREPB8 tet-regulatable copy plasmid transfected into BF SM427 was made in a similar way, except that the open reading frame was cloned into pLEW79 (Wirtz et al. 1999) using the HindIII and BamHI restriction sites. Preparation of DNA constructs and transfections for endogenous allele knockouts using blasticidin and puromycin drug cassettes in the regulatable PF cell lines were generated and carried out as previously described for the generation of the PF KREPB5 CN cell line (McDermott et al. 2015b), using primers described in Supplemental Table S1. DNA constructs for allele knockouts in the regulatable BF cell line were generated by PCR amplification of hygromycin and puromycin drug cassettes from pyrFEKO-HYG (Addgene plasmid #24020; George Cross) or pSM09 (for BF KREPB8) (Merritt and Stuart 2013), and pSM07 (Merritt and Stuart 2013). Targeting sequences were also amplified using sequences described in Supplemental Table S1, and combined with drug cassettes in fusion PCR reactions (Merritt and Stuart 2013). The resulting constructs were transfected stepwise into the regulatable BF cell line, transgenic lines selected by puromycin and hygromycin resistance, and correct insertion of knockout cassettes assessed by PCR. The drug selection cassettes in the resulting CN cell lines were excised using transient expression of Cre recombinase and selection with ganciclovir (Invivogen) as previously described (McDermott et al. 2015b; Carnes et al. 2017).
Epitope tagging
PF and BF KREPB6, KREPB7, and KREPB8 CN cell lines carrying HA-tagged versions of KREN1, KREN2, or KREN3, were generated by in situ tagging with a 3× HA epitope tag. Constructs for transfection were generated by a single round of PCR as previously described (Oberholzer et al. 2006), using the pMOTag4H-crelox-PUR plasmid (Addgene plasmid #24028; George Cross), a forward primer containing sequence preceding the stop codon of the target endonuclease, and a reverse primer containing the reverse complement of the 3′UTR of the target endonuclease. Transgenic lines were selected by puromycin resistance. Primers are described in Supplemental Table S1. Transient expressions of pLEW100Cre_del_tetO (Addgene plasmid #24019; George Cross) and ganciclovir (Invivogen) selection were used as previously described (McDermott et al. 2015b) to excise and allow reuse of the puromycin drug resistance cassette in BF.
Generation of exclusive expression cell lines
WT KREPB6 and KREPB7, lacking the stop codon, and WT KREPB8 containing the stop codon, and all flanked in frame with attB Gateway recombination sites, were PCR amplified using primers described in Supplemental Table S1 and transferred into pDONR221. pENTR-KREPB6 and -B7 (-stop) were used in LR reactions with the destination vectors pHD1344tub(NAT or PAC)GW-Cterm3V5, which allow for constitutive expression of carboxy-terminal 3xV5 tagged proteins in the β-tubulin locus. pENTR-KREPB8 (+stop) was used in LR reactions with the destination vectors pHD1344tub(NAT or PAC)GW-Nterm3V5, which allow for constitutive expression of amino-terminal 3xV5 tagged proteins in the β-tubulin locus (Carnes et al. 2018). The resulting pHD1344tub(NAT or PAC)-KREPB6 or B7-Cterm3V5 plasmids, and pHD1344tub(NAT or PAC)-Nterm3V5-KREPB8 plasmids were used as templates for site-directed mutagenesis (QuikChange II kit; Agilent) using forward and reverse primers listed in Supplemental Table S1. NotI digested plasmids were transfected into the BF and PF KREPB6, B7, and B8 CN cell lines containing HA-tagged endonucleases. Transgenic BF lines were selected by puromycin resistance, while transgenic PF lines were selected by nourseothricin resistance, and constitutive expression of KREPB6, or B7-3xV5, and 3xV5-KREPB8 confirmed by western blotting.
Fractionation on glycerol gradients
Glycerol gradient fractionation was carried out on total cell lysates from 2 × 109 PF or BF cells in the presence or absence of 0.5 µg/mL tet. Following lysis in 650 µL lysis buffer (10 mM Tris-HCl pH 7.2, 10 mM MgCl2, 100 mM KCl, 1% Triton X-100) and centrifugation (12,000 rpm, 10 min, 4°C), cleared lysates were loaded onto 11 mL 10%–30% glycerol gradients and centrifuged at 38,000 rpm for 5 h at 4°C in a SW40Ti rotor (Beckman). Twenty-four fractions of 500 µL were collected from top to bottom, flash frozen in liquid nitrogen and stored at −80°C.
Immunoprecipitation
Cleared lysate was prepared by lysis of 2 × 108 cells in 1 mL IPP150 (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Nonidet P-40, Roche Complete protease inhibitors) with 1% Triton X-100, followed by centrifugation at 12,000 rpm at 4°C. For each anti-V5 immunoprecipitation, 0.5 mL cleared lysate (1 × 108 cells) was incubated overnight with 1 μL of rabbit antibody (Rockland Immunochemicals; Supplemental Table S2) specific for the V5 epitope tag. 12.5 μL of Protein G Mag Sepharose Xtra (GE Healthcare) were washed twice with 1 mL of 1× PBS, 0.1% BSA and once with 1 mL IPP150. Beads were then incubated for 4 h rotating at 4°C with cleared lysate/antibody. Anti-KREPA2 and anti-HA endonuclease immunoprecipitations were alternatively carried out by incubation of 0.5 mL cleared lysate (1 × 108 cells) with goat anti-mouse IgG dynabeads (ThermoFisher Scientific) that had been coated in mAb P1H3-D7 (Panigrahi et al. 2001a) (Supplemental Table S2) as previously described (Panigrahi et al. 2001a,b; Ernst et al. 2003) or in mAb 2-2.2.14 (ThermoFisher Scientific). After incubation, the supernatant was removed, and beads were washed four times with 1 mL IPP150. Complexes bound to beads were eluted with 100 μL of 2× SDS sample buffer, heated for 5 min at 95°C.
SDS-PAGE and western blotting
SDS-PAGE loading buffer was added to cleared whole cell lysates, or to samples containing purified protein complexes and resolved on 10% SDS-PAGE gels (Criterion Tris-HCl, Bio-Rad). For Western analysis, resolved proteins were transferred to Immobilon-P PVDF membranes (Millipore), and probed using antibodies described in Supplemental Table S2. Blots were developed with an enhanced chemiluminescence kit (Thermo Scientific) per the manufacturer's instructions and imaged using the FluorChem E system (ProteinSimple) or x-ray film (Kodak).
RNA isolation and RT-qPCR analysis
Total RNA was harvested using TRIzol and treated with TURBO DNase (Life Technologies) according to manufacturer's instructions. RNA integrity was confirmed using a RNA nanochip on a BioAnalyzer (Agilent Technologies). Two micrograms of total RNA were reverse-transcribed using TaqMan Reverse Transcription Reagents and MultiScribe Reverse Transcriptase (Life Technologies), preamplified in multiplex Specific Target Amplification (STA) reactions using TaqMan PreAmp Master Mix (Life Technologies) and treated with Exonuclease I (NEB). The abundance of reference, never-edited, pre-edited and edited transcript cDNAs were then analyzed by high-throughput real-time PCR on the BioMark HD system as previously described (McDermott et al. 2015b). Primers are described in Supplemental Table S1. Calculations of RNA levels in samples following tet withdrawal (for 48 h in BF or 96 h for PF), relative to the presence of tet, were done using the 2 [−ΔΔC(T)] method (Livak and Schmittgen 2001) using TERT as an internal reference. Technical duplicates of each cDNA sample were assayed for each target and internal reference per experiment and C(T) data averaged before performing the 2 [−ΔΔC(T)] calculation. Experiments were repeated using two or three biological replicates.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paige Haas, Sibani Das, and Amy Pelz for assistance with creating cell lines. This work was supported by National Institutes of Health grant R01AI014102 to K.S.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.071258.119.
REFERENCES
- Aphasizhev R, Aphasizheva I. 2011. Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer. Wiley Interdiscip Rev RNA 2: 669–685. 10.1002/wrna.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R. 2014. RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol 34: 4329–4342. 10.1128/MCB.01075-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826. 10.1016/0092-8674(86)90063-2 [DOI] [PubMed] [Google Scholar]
- Brun R, Schönenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop 36: 289–292. [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. 2005. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci 102: 16614–16619. 10.1073/pnas.0506133102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. 2008. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol 28: 122–130. 10.1128/MCB.01374-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Soares CZ, Wickham C, Stuart K. 2011. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J Biol Chem 286: 19320–19330. 10.1074/jbc.M111.228965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Lewis Ernst N, Wickham C, Panicucci B, Stuart K. 2012a. KREX2 is not essential for either procyclic or bloodstream form Trypanosoma brucei. PLoS ONE 7: e33405 10.1371/journal.pone.0033405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Schnaufer A, McDermott SM, Domingo G, Proff R, Steinberg AG, Kurtz I, Stuart K. 2012b. Mutational analysis of Trypanosoma brucei editosome proteins KREPB4 and KREPB5 reveals domains critical for function. RNA 18: 1897–1909. 10.1261/rna.035048.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, McDermott S, Anupama A, Oliver BG, Sather DN, Stuart K. 2017. In vivo cleavage specificity of Trypanosoma brucei editosome endonucleases. Nucleic Acids Res 45: 4667–4686. 10.1093/nar/gkx116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, McDermott SM, Stuart K. 2018. RNase III domain of KREPB9 and KREPB10 association with editosomes in Trypanosoma brucei. mSphere 3: e00585-17 10.1128/mSphereDirect.00585-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C, Schmitt JG, Evguenieva-Hackenberg E, Klug G. 2002. One functional subunit is sufficient for catalytic activity and substrate specificity of Escherichia coli endoribonuclease III artificial heterodimers. FEBS Lett 518: 93–96. 10.1016/S0014-5793(02)02653-4 [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J, Mooers BHM, Doharey PK, Meehan J, Gulati S. 2018. Dynamic RNA holo-editosomes with subcomplex variants: insights into the control of trypanosome editing. Wiley Interdiscip Rev RNA 9: e1502 10.1002/wrna.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst NL, Panicucci B, Igo RP Jr, Panigrahi AK, Salavati R, Stuart K. 2003. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol Cell 11: 1525–1536. 10.1016/S1097-2765(03)00185-0 [DOI] [PubMed] [Google Scholar]
- Ernst NL, Panicucci B, Carnes J, Stuart K. 2009. Differential functions of two editosome exoUases in Trypanosoma brucei. RNA 15: 947–957. 10.1261/rna.1373009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull K. 2001. The biology of kinetoplastid parasites: insights and challenges from genomics and post-genomics. Int J Parasitol 31: 443–452. 10.1016/S0020-7519(01)00154-0 [DOI] [PubMed] [Google Scholar]
- Guo X, Ernst NL, Stuart KD. 2008. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei. Mol Cell Biol 28: 6939–6953. 10.1128/MCB.01115-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Carnes J, Ernst NL, Winkler M, Stuart K. 2012. KREPB6, KREPB7, and KREPB8 are important for editing endonuclease function in Trypanosoma brucei. RNA 18: 308–320. 10.1261/rna.029314.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Bach M, Lührmann R. 1990. U1-specific protein C is required for efficient complex formation of U1 snRNP with a 5′ splice site. Mol Biol Rep 14: 165 10.1007/BF00360459 [DOI] [PubMed] [Google Scholar]
- Hendriks E, van Deursen FJ, Wilson J, Sarkar M, Timms M, Matthews KR. 2000. Life-cycle differentiation in Trypanosoma brucei: molecules and mutants. Biochem Soc Trans 28: 531–536. 10.1042/bst0280531 [DOI] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol 75: 985–989. 10.2307/3282883 [DOI] [PubMed] [Google Scholar]
- Igo RP Jr, Weston DS, Ernst NL, Panigrahi AK, Salavati R, Stuart K. 2002. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot Cell 1: 112–118. 10.1128/EC.1.1.112-118.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafková L, Debler EW, Fisk JC, Jain K, Clarke SG, Read LK. 2017. The major protein arginine methyltransferase in Trypanosoma brucei functions as an enzyme-prozyme complex. J Biol Chem 292: 2089–2100. 10.1074/jbc.M116.757112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch M, Carnes J, Acestor N, Guo X, Schnaufer A, Stuart K. 2012. Editosome accessory factors KREPB9 and KREPB10 in Trypanosoma brucei. Eukaryot Cell 11: 832–843. 10.1128/EC.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. 2007. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 14: 934–940. 10.1038/nsmb1293 [DOI] [PubMed] [Google Scholar]
- McDermott SM, Stuart K. 2017. The essential functions of KREPB4 are developmentally distinct and required for endonuclease association with editosomes. RNA 23: 1672–1684. 10.1261/rna.062786.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Carnes J, Stuart K. 2015a. Identification by random mutagenesis of functional domains in KREPB5 that differentially affect RNA editing between life cycle stages of Trypanosoma brucei. Mol Cell Biol 35: 3945–3961. 10.1128/MCB.00790-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Guo X, Carnes J, Stuart K. 2015b. Differential editosome protein function between life cycle stages of Trypanosoma brucei. J Biol Chem 290: 24914–24931. 10.1074/jbc.M115.669432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Luo J, Carnes J, Ranish JA, Stuart K. 2016. The architecture of Trypanosoma brucei editosomes. Proc Natl Acad Sci 113: E6476–E6485. 10.1073/pnas.1610177113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Nicholson AW. 2008. Heterodimer-based analysis of subunit and domain contributions to double-stranded RNA processing by Escherichia coli RNase III in vitro. Biochem J 410: 39–48. 10.1042/BJ20071047 [DOI] [PubMed] [Google Scholar]
- Merritt C, Stuart K. 2013. Identification of essential and non-essential protein kinases by a fusion PCR method for efficient production of transgenic Trypanosoma brucei. Mol Biochem Parasitol 190: 44–49. 10.1016/j.molbiopara.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Mace PD, Eyers PA. 2017. Live and let die: insights into pseudoenzyme mechanisms from structure. Curr Opin Struct Biol 47: 95–104. 10.1016/j.sbi.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Nelissen RL, Will CL, van Venrooij WJ, Lührmann R. 1994. The association of the U1-specific 70 K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J 13: 4113–4125. 10.1002/j.1460-2075.1994.tb06729.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S, Jones DC, Wyllie S, Fairlamb AH, Phillips MA. 2013. Allosteric activation of trypanosomatid deoxyhypusine synthase by a catalytically dead paralog. J Biol Chem 288: 15256–15267. 10.1074/jbc.M113.461137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S, Leija C, Kinch L, Regmi S, Li Q, Grishin NV, Phillips MA. 2015. Deoxyhypusine modification of eukaryotic translation initiation factor 5A (eIF5A) is essential for Trypanosoma brucei growth and for expression of polyprolyl-containing proteins. J Biol Chem 290: 19987–19998. 10.1074/jbc.M115.656785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson AW. 2014. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip Rev RNA 5: 31–48. 10.1002/wrna.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M, Morand S, Kunz S, Seebeck T. 2006. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol 145: 117–120. 10.1016/j.molbiopara.2005.09.002 [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Gygi SP, Ernst NL, Igo RP Jr, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, et al. 2001a. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol Cell Biol 21: 380–389. 10.1128/MCB.21.2.380-389.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Carmean N, Igo RP Jr, Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, et al. 2001b. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol Cell Biol 21: 6833–6840. 10.1128/MCB.21.20.6833-6840.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. 2006. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA 12: 1038–1049. 10.1261/rna.45506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MM, Volkov OA, Leija C, Lemoff A, Phillips MA. 2018. A dual regulatory circuit consisting of S-adenosylmethionine decarboxylase protein and its reaction product controls expression of the paralogous activator prozyme in Trypanosoma brucei. PLoS Pathog 14: e1007404 10.1371/journal.ppat.1007404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. 1994. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J Bioenerg Biomembr 26: 179–191. 10.1007/BF00763067 [DOI] [PubMed] [Google Scholar]
- Read LK, Lukeš J, Hashimi H. 2016. Trypanosome RNA editing: the complexity of getting U in and taking U out. Wiley Interdiscip Rev RNA 7: 33–51. 10.1002/wrna.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringpis GE, Aphasizheva I, Wang X, Huang L, Lathrop RH, Hatfield GW, Aphasizhev R. 2010. Mechanism of U insertion RNA editing in trypanosome mitochondria: the bimodal TUTase activity of the core complex. J Mol Biol 399: 680–695. 10.1016/j.jmb.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati R, Ernst NL, O'Rear J, Gilliam T, Tarun S Jr, Stuart K. 2006. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA 12: 819–831. 10.1261/rna.2244106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A, Domingo GJ, Stuart K. 2002. Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int J Parasitol 32: 1071–1084. 10.1016/S0020-7519(02)00020-6 [DOI] [PubMed] [Google Scholar]
- Schneider A. 2001. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int J Parasitol 31: 1403–1415. 10.1016/S0020-7519(01)00296-X [DOI] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem Sci 30: 97–105. 10.1016/j.tibs.2004.12.006 [DOI] [PubMed] [Google Scholar]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. 2005. A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell 20: 403–412. 10.1016/j.molcel.2005.09.016 [DOI] [PubMed] [Google Scholar]
- Vickerman K. 1985. Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull 41: 105–114. 10.1093/oxfordjournals.bmb.a072036 [DOI] [PubMed] [Google Scholar]
- Wang B, Ernst NL, Palazzo SS, Panigrahi AK, Salavati R, Stuart K. 2003. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot Cell 2: 578–587. 10.1128/EC.2.3.578-587.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert EK, Fitzpatrick R, Phillips MA. 2007. Allosteric regulation of an essential trypanosome polyamine biosynthetic enzyme by a catalytically dead homolog. Proc Natl Acad Sci 104: 8275–8280. 10.1073/pnas.0701111104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101. 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. 2003. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res 31: 6392–6408. 10.1093/nar/gkg870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.